-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Neocentromeres Come of Age
article has not abstract
Published in the journal: Neocentromeres Come of Age. PLoS Genet 5(3): e32767. doi:10.1371/journal.pgen.1000370
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1000370Summary
article has not abstract
Sixteen years ago, the discovery of a newly formed, ectopic centromere in a human [1],[2] was a turning point for centromere research. Whereas previously centromeres had been thought of as immovable and unchanging, embedded in vast tracts of tandemly repeated DNA, this new centromere—or neocentromere—lacked any characteristic centromeric DNA sequences and had formed in a gene-rich area of the genome. Essentially, a fully functional centromere had spontaneously arisen where no centromere had any right to be, complete with all the necessary centromere proteins and epigenetic marks required for the creation of a complex DNA/protein structure. Neocentromere formation remains one of the most astonishing examples of epigenetic change within the genome.
Since this discovery, neocentromeres (not to be confused with the “classical” facultative neocentromeres, which were originally described in maize (reviewed in [3]) have been shown to be a means of centromere repositioning during karyotype evolution and speciation in vertebrates, with evidence suggesting a similar role in plants (for review, see [4]). Clearly, there is an evolutionary advantage in being able to form new centromeres, and this process has been conserved. However, an understanding of the mechanisms of neocentromere formation remains elusive.
It was this question that Ketel et al., in this issue of PLoS Genetics, set out to answer [5]. The authors based their study on the pathogenic fungus Candida albicans, which has small, simple, regional centromeres flanked by inverted repeats, and extremely high rates of homologous recombination. Their approach was to specifically remove the centromeric DNA on Chromosome V by replacing it with URA3, a selectable marker gene, and observe the positioning and frequency of neocentromeres that resulted via chromatin immunoprecipitation for the fundamental centromere marker protein CENP-A (Figure 1A).
Fig. 1. Formation of neocentromeres in C. albicans. 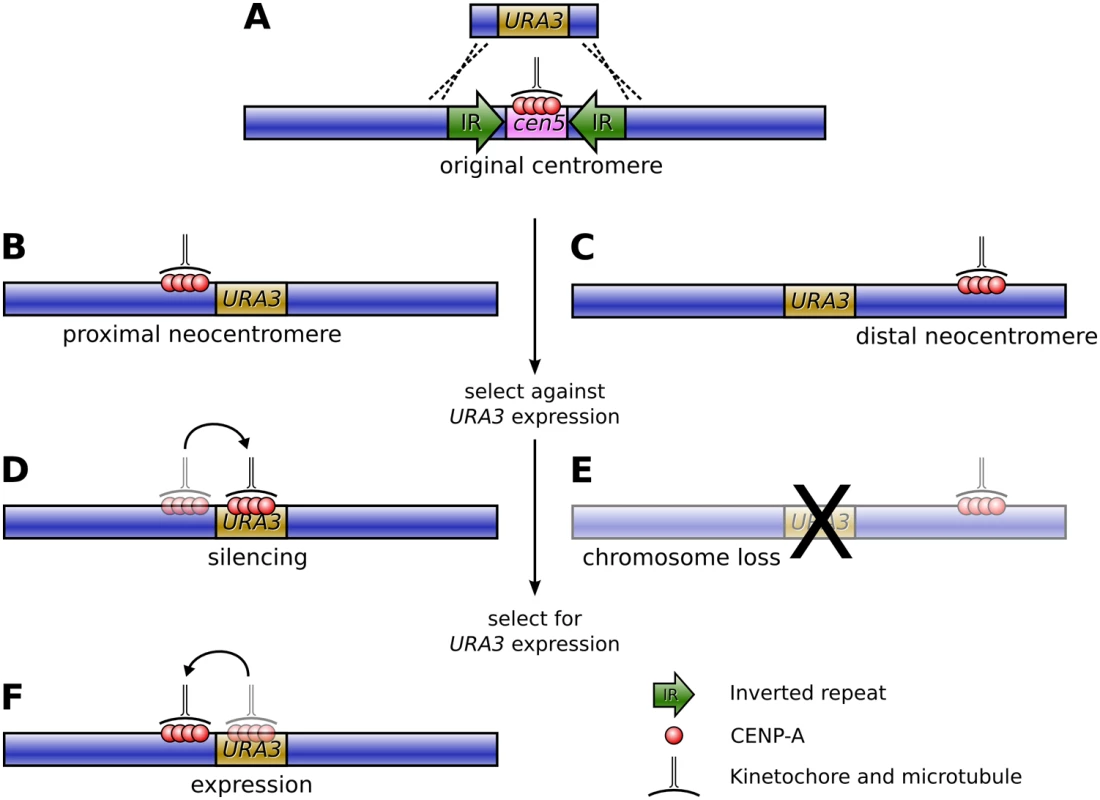
(A) The existing centromere on Chromosome V, together with the surrounding inverted repeats, is replaced with the URA3 gene via homologous recombination, resulting in neocentromere formation either proximal (B) or distal (C) to the original centromere. Selection against URA3 expression results in either chromosome loss (E) or silencing of URA3 through centromere shifting (D). If resistant colonies from the latter case are again grown on uridine-deficient media, a second shift in the position of the centromere restores URA3 expression (F). The results were striking: the authors found an extremely high frequency of neocentromere formation (with neocentromeres forming in all transformants) at multiple possible locations along Chromosome V. Essentially, these neocentromeres fell into two distinct classes: proximal neocentromeres, which formed close to the location of the original, excised centromere (Figure 1B); and distal neocentromeres, which formed at all other locations on the chromosome (Figure 1C). Although experimentally induced neocentromere formation has been previously investigated in flies [6],[7], plants [8], and other fungi [9], this is the first example, to our knowledge, where neocentromeres have been found to form at seemingly random chromosomal locations, similar to human neocentromeres (Figure 2).
Fig. 2. Organisms in which neocentromere formation has been reported. 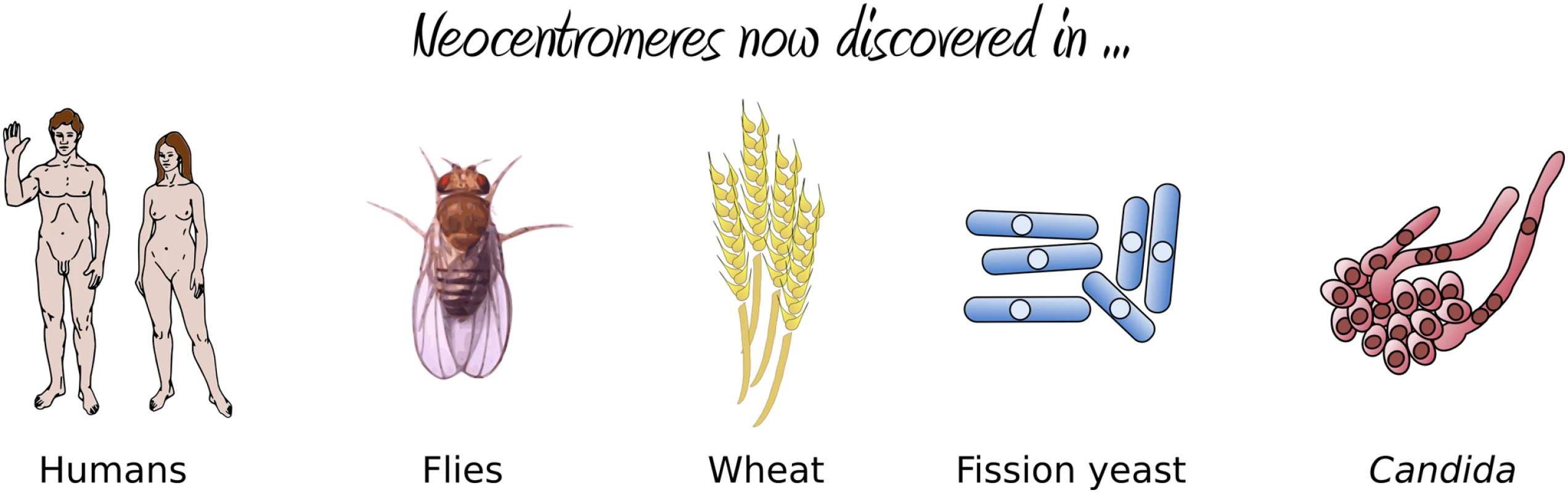
From left to right are: humans (reviewed in [4]), flies [6],[7], wheat [8], Schizosaccharomyces pombe [9], and C. albicans [5]. In most cases, the size of the neocentromeres was similar to a normal C. albicans centromere, albeit with reduced quantities of CENP-A. Would the resulting neocentromeres be less stable during mitosis? To find out, Ketel et al. used a standard assay to gauge chromosome stability, growing the transformant strains on 5-FOA media, which is toxic to Ura+ cells. Those transformants with distal neocentromeres became resistant through loss of the neocentric chromosome at a rate comparable to control strains, suggesting that Candida neocentromeres suffered no loss of mitotic stability (Figure 1E).
However, transformants with proximal neocentromeres (near the selectable marker gene) became FOA-resistant at a much higher rate. Astonishingly, though, this was not due to higher rates of chromosome loss. In these strains the neocentromere had shifted onto the URA3 gene, thereby silencing URA3 expression (Figure 1D). Furthermore, moving the resistant strains back onto media selective for uridine synthesis resulted in the neocentromere shifting away from the gene and URA3 expression being restored (Figure 1F).
Does this mean, then, that centromeres are incompatible with gene expression? Experiments such as the current work and recent reports in fission yeast [10]—where genes inserted within centromeric chromatin were similarly down-regulated—would suggest that this is the case. But these results are somewhat contradicted by results in human cells, where at both a neocentromere [11] and artificially generated chromosomes [12],[13] gene expression has been demonstrated despite the presence of CENP-A. Such observations may point to a different chromatin environment between humans and fungi at centromeres. Alternatively, it is possible that centromeric chromatin is merely impermissible to high levels of gene transcription—both experiments in fungi reported very low levels of reporter gene transcription still occurring. But such observations are intriguing considering recent reports of transcription at centromeres [14], and investigation of the precise relationship between centromeric chromatin and transcription is likely to become an important research focus in the future.
A key question regarding neocentromere formation has been whether there are any DNA sequence motifs required for a new centromere to arise. Using the three distal neocentromeres isolated in this study, Ketel et al. were unable to find any common sequence between the three regions. The only similarity, indeed, seemed to be that all neocentromeres formed within intergenic regions on the chromosome—not surprising, perhaps, considering the negative effect that centromeric chromatin appears to have on gene expression in Candida. It is unfortunate, though, that so few distal neocentromeres were analysed, making it impossible to tell if C. albicans has “hotspots” of neocentromere similar to those found on human chromosomes [4]. And what of the large number of proximal neocentromeres that arose? The high frequency of proximal neocentromere formation makes these neocentromeres difficult to explain through an occasional shifting or spreading of the centromeric signal. Perhaps there are other epigenetic marks conducive to centromere formation that lie outside of the excised cen5 region.
So what can we conclude from this research? Clearly, C. albicans provides an excellent model system for studying the process of neocentromere formation, and the current work throws up many new questions regarding both the process of centromere formation and its impact upon transcription. What this work undeniably demonstrates, though, is that the ability to form neocentromeres is common from fungi to humans and is clearly an integral part of the genome.
Zdroje
1. VoullaireLE
SlaterHR
PetrovicV
ChooKH
1993 A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet 52 1153 1163
2. du SartD
CancillaMR
EarleE
MaoJI
SafferyR
1997 A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet 16 144 153 doi:10.1038/ng0697-144
3. DaweRK
HiattEN
2004 Plant neocentromeres: fast, focused, and driven. Chromosome Res 12 655 669 doi:10.1023/B:CHRO.0000036607.74671.db
4. MarshallOJ
ChuehAC
WongLH
ChooKHA
2008 Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82 261 282 doi:10.1016/j.ajhg.2007.11.009
5. KetelC
WangHSW
McClellanM
BouchonvilleK
SelmeckiA
2009 Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5(3) e1000400 doi:10.1371/journal.pgen.1000400
6. WilliamsBC
MurphyTD
GoldbergML
KarpenGH
1998 Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet 18 30 37
7. MaggertKA
KarpenGH
2001 The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158 1615 1628
8. NasudaS
HudakovaS
SchubertI
HoubenA
EndoTR
2005 Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci U S A 102 9842 9847 doi:10.1073/pnas.0504235102
9. IshiiK
OgiyamaY
ChikashigeY
SoejimaS
MasudaF
2008 Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321 1088 1091 doi:10.1126/science.1158699
10. CastilloAG
MelloneBG
PartridgeJF
RichardsonW
HamiltonGL
2007 Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet 3 e121 doi:10.1371/journal.pgen.0030121
11. SafferyR
SumerH
HassanS
WongLH
CraigJM
2003 Transcription within a functional human centromere. Mol Cell 12 509 516
12. NakashimaH
NakanoM
OhnishiR
HiraokaY
KanedaY
2005 Assembly of additional heterochromatin distinct from centromere-kinetochore chromatin is required for de novo formation of human artificial chromosome. J Cell Sci 118 5885 5898 doi:10.1242/jcs.02702
13. LamAL
BoivinCD
BonneyCF
RuddMK
SullivanBA
2006 Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A 103 4186 4191 doi:10.1073/pnas.0507947103
14. Bouzinba-SegardH
GuaisA
FrancastelC
2006 Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A 103 8709 8714 doi:10.1073/pnas.0508006103
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2009 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Neocentromeres Come of Age
- Mitotic Recombination: Why? When? How? Where?
- Life, Death, Differentiation, and the Multicellularity of Bacteria
- Capturing the Spectrum of Interaction Effects in Genetic Association Studies by Simulated Evaporative Cooling Network Analysis
- Measures of Autozygosity in Decline: Globalization, Urbanization, and Its Implications for Medical Genetics
- Ciliary Beating Recovery in Deficient Human Airway Epithelial Cells after Lentivirus Gene Therapy
- Parallel Germline Infiltration of a Lentivirus in Two Malagasy Lemurs
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Neocentromeres Come of Age
- Capturing the Spectrum of Interaction Effects in Genetic Association Studies by Simulated Evaporative Cooling Network Analysis
- Mitotic Recombination: Why? When? How? Where?
- Life, Death, Differentiation, and the Multicellularity of Bacteria
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



