Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in
Despite the apparent complexity of ageing, animal lifespan can be extended. Activity of Forkhead Box O (FoxO) transcription factors can prolong survival of organisms ranging from the budding yeast to the fruit fly, and FoxO gene variants are linked to human longevity. FoxOs extend lifespan by driving complex, widespread changes in gene expression. Their primary targets include a second tier of transcriptional regulators, but it remains unclear how these secondary regulators are involved in the anti-ageing programmes orchestrated by FoxOs in vivo. To elucidate the role of this second tier, we identify a transcription factor called Anterior open (Aop) as directly regulated by the single Drosophila melanogaster FoxO protein (dFOXO) in the adult fly gut. Under certain circumstances, such as co-activation of the Pointed (PNT) transcription factor, dFOXO can be detrimental to lifespan. The role of Aop is to protect from this negative synergistic effect. Additionally, activation of AOP in the fly adipose tissue can robustly extend lifespan. Our study reveals a complex interplay between two evolutionarily conserved transcriptional regulators and dFOXO in lifespan. This significance of this interplay may extend to other physiological processes where these transcription factors play important roles.
Published in the journal:
Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in. PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004619
Category:
Research Article
doi:
https://doi.org/10.1371/journal.pgen.1004619
Summary
Despite the apparent complexity of ageing, animal lifespan can be extended. Activity of Forkhead Box O (FoxO) transcription factors can prolong survival of organisms ranging from the budding yeast to the fruit fly, and FoxO gene variants are linked to human longevity. FoxOs extend lifespan by driving complex, widespread changes in gene expression. Their primary targets include a second tier of transcriptional regulators, but it remains unclear how these secondary regulators are involved in the anti-ageing programmes orchestrated by FoxOs in vivo. To elucidate the role of this second tier, we identify a transcription factor called Anterior open (Aop) as directly regulated by the single Drosophila melanogaster FoxO protein (dFOXO) in the adult fly gut. Under certain circumstances, such as co-activation of the Pointed (PNT) transcription factor, dFOXO can be detrimental to lifespan. The role of Aop is to protect from this negative synergistic effect. Additionally, activation of AOP in the fly adipose tissue can robustly extend lifespan. Our study reveals a complex interplay between two evolutionarily conserved transcriptional regulators and dFOXO in lifespan. This significance of this interplay may extend to other physiological processes where these transcription factors play important roles.
Introduction
Forkhead Box O (FoxO) transcription factors (TFs) play a key, evolutionarily conserved role in ageing. Drosophila melanogaster has a single FoxO orthologue (dfoxo) and increasing its activity in certain tissues is sufficient to extend fly lifespan [1]–[4] Furthermore, both dfoxo and the Caenorhabditis elegans othologue, daf-16, are strictly required for lifespan extension upon reduction in insulin/IGF-like signalling (IIS) [5], [6]. This evolutionary conservation appears to extend all the way to yeast on one side, where forkhead-like factors can extend lifespan [7], and to humans on the other, where certain variants of the FoxO3A locus are robustly correlated with longevity [8]–[12].
FoxOs control a plethora of traits at both organismal and cellular levels, including control of cell cycle, cell death, growth and metabolism. In all cases, FoxOs can be viewed as acting to preserve homeostasis [13]. Indeed, numerous processes are remodelled by activation of FoxOs, through regulation of a large number of direct and indirect targets, all acting in concert to preserve homeostasis in old age and extend animal lifespan [14]–[19].
Several studies have examined the targets of FoxOs. A striking finding of these studies is that FoxOs control a range of other cellular regulators. These include secreted endocrine factors, components of intracellular signalling pathways and several TFs [14], [16]–[20]. Transcriptional feedback within the signalling pathway plays a role [21], but in most cases the functions of these other regulators remain unknown, both in isolated cells and, more importantly, in vivo.
The putative roles of TFs regulated by FoxOs are particularly intriguing. Numerous studies have shown that FoxOs interact with a number of unrelated TFs, in a number of ways, with important consequences for the output of both interacting partners [13], [22]. These TFs include Myc, p53, Smads, ß-catenin, and numerous nuclear hormone receptors [22]–[27]. Hence, there is a potential for the TFs regulated by FoxO to profoundly alter FoxO's functional output through interactions with FoxO itself. However, it remains unclear what the role of these interactions is in the whole animal, in vivo and, specifically, what is their role in lifespan?
In this study we set out to elucidate the role played in lifespan by a TF directly regulated by dFOXO. We identify an E-twenty six (ETS) - family transcriptional repressor, Anterior open (Aop), as regulated by dFOXO in the adult Drosophila gut. Aop is the functional orthologue of the human Etv6 gene and, in Drosophila, it is known to counteract the activity of an ETS activator, Pointed (Pnt). We show that Aop acts to prevent the detrimental effects of co-activation of dFOXO and PNT in adult Drosophila gut, and we present evidence that this interaction is mediated by binding to the same genomic locations as dFOXO. AOP activation on its own in the adult fat body can also robustly extend lifespan. Our study reveals a complex interplay between evolutionarily conserved ETS-family TFs and dFOXO in longevity. The significance of this interplay may extend to other physiological processes.
Results
dFOXO regulates distinct genes but similar functions in the adult gut and fat body
dFOXO, like its mammalian orthologues, controls gene expression in a tissue-specific manner [19], [28]–[30]. Hence, to investigate the functional interplay between dFOXO and one of its target TFs, we turned our attention to a tissue-specific, adult-inducible, lifespan-relevant system. Over-expression of dfoxo using the RU486-inducible, S1106 Geneswitch driver [31], robustly extends lifespan [1], [4], [32]–[34]. S1106 restricts dfoxo induction to two specific adult fly organs: the midgut and abdominal fat body (subsequently referred to as gut and fat body; Figure S1A) [31], the latter functionally equivalent to mammalian white adipose tissue and liver. Both have an evolutionarily conserved role in aging [35], [36], and it is currently unclear whether activation of dfoxo in either organ alone is sufficient to extend lifespan. For these reasons, we chose to identify the TFs regulated by dFOXO in both of these organs.
We micro-dissected mid-guts or carcass-associated thoracic/abdominal fat body of S1106>dfoxo females (+/− RU486) and determined their mRNA profiles using Affymetrix gene expression arrays (ArrayExpress accession number: E-MTAB-1020). In each case, we controlled for the changes associated with induction of the driver alone (S1106 +/− RU486). 447 genes were differentially expressed in the gut (p value cut-off of 0.00285 corresponding to FDR of 5%, Figure 1A). We detected fewer significant changes in the fat body, 87 differentially regulated genes (p value cut-off 0.0022, FDR 20%, Figure 1A), most-likely due to the difficulty of dissecting this loosely-associated tissue. The full list of genes regulated by dFOXO, as well as all other lists mentioned in the paper, are given in Dataset S1. The list included some well-known targets of dFOXO, such as initiation factor 4E binding protein (4ebp) [37], [38] and the Drosophila insulin receptor (dInR) [37], both activated in the gut, and the insulin-regulated kinase Akt [17], [39], induced in both the gut and fat body.
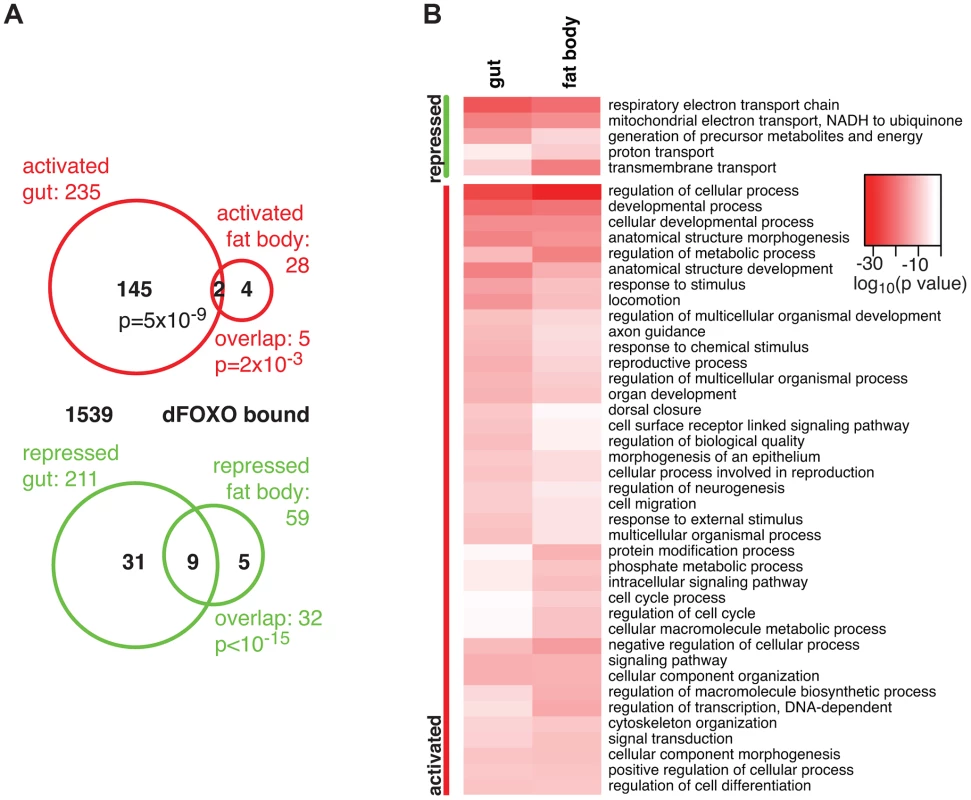
The overlap between genes regulated in the same direction in gut and fat body was significant (p = 0.002 for up-regulated, p<10−15 for down-regulated genes), but by no means complete (Figure 1A), indicating that dFOXO regulates both related and unrelated sets of genes in the two tissues. This did not appear to be caused by weaker signal obtained from the fat body, since changes unique to the fat body were also detected (e.g. Obp99b; see later). To further explore this, we examined the functions regulated by dFOXO in the two tissues using Catmap, an approach that can detect co-ordinated, subtle changes over many genes, rather than depending on an arbitrarily chosen, differential expression p-value cut-off [40]. Although some differences occurred, the biological process Gene Ontology (GO) categories differentially expressed in the two tissues were similar (Figure 1B), indicating that dFOXO regulates similar functions in the gut and in the fat body.
Notably, dFOXO strongly repressed respiratory electron transport chain components in both the gut and fat body (p = 4×10−25 and p = 1×10−21, respectively) and, in particular, components of complex I, which transfers electrons from NADH to ubiquinone (p = 2×10−19 and p = 2×10−17). Indeed, part of the effect of dFOXO on lifespan could be mediated by its repression of the components of complex I, because reducing the electron flow through this complex, by bypassing it, can extend fly lifespan [41], [42].
5 TFs are directly regulated by dFOXO in the adult gut
We identified a total of 16 TFs regulated by induction of dfoxo in either the gut or the fat body, including p53 and the nuclear hormone receptor HR96 (see Dataset S1 for the full list). To further narrow down the set of interesting candidates, we isolated the TFs encoded by genes directly bound and regulated by dFOXO, thus identifying the immediate second tier of regulators. We determined the genomic regions bound by dFOXO using a GFP-dFOXO fusion protein that is functional in lifespan-extension [33]. We prepared chromatin from RU486-fed S1106>GFP-dfoxo female flies and pulled down the DNA associated with the fusion protein, and hence restricted to gut and fat body, using an anti-GFP antibody. As a control, we performed chromatin immunoprecipitation (ChIP) using the same antibody on chromatin prepared from females over-expressing dfoxo alone (S1106>dfoxo + RU486).
To confirm that we were detecting tissue-restricted binding, we compared the GFP-dFOXO bound sites with those that we previously identified in whole adults or S2 cells [17]. The dInR locus is transcribed from three promoters under tight spatio-temporal control [43]. In whole flies, dFOXO is detected as bound in the coding region of the gene and is absent from the P1 or P3 regions [17] (Figure 2A). The functional significance of this binding in the 3′ region in Drosophila is unclear but the mammalian FoxO proteins are able to act at great distances [20]. On the other hand, in serum-starved S2 cells dFOXO is bound to the P1 promoter but not the coding or P3 regions [17] (Figure 2A). In contrast, GFP-dFOXO expressed in the adult gut and fat body was bound to the P3 promoter of dInR (Figure 2A), revealing a different pattern of binding in these tissues.
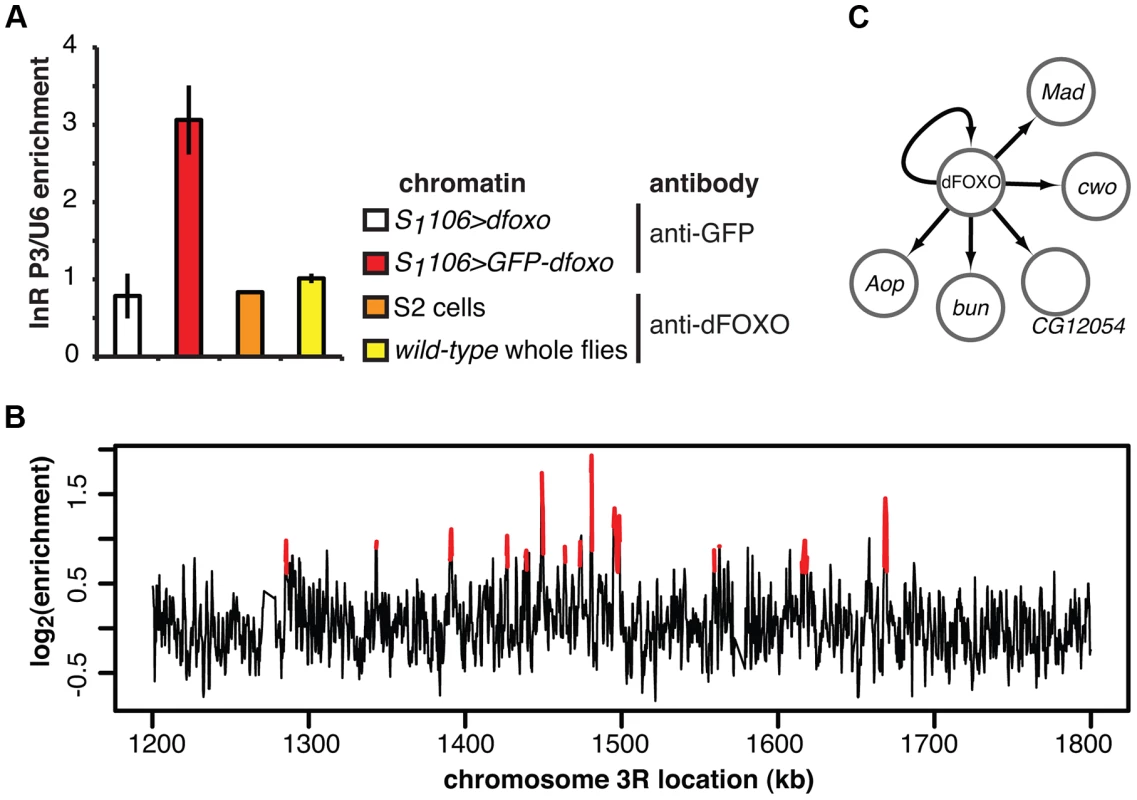
We hybridised three biological repeats of the experimental and control ChIP samples to tiling arrays. We identified ∼1400 genomic regions bound by GFP-dFOXO in the gut and/or the fat body (ArrayExpress accession number: E-MTAB-1021; for examples of peaks identified see Figure 2B; for a list of all bound locations see Dataset S1), with highly reproducible ChIP-chip signal across the three biological replicates (Figure S1B). The regions occupied by dFOXO in the gut and fat body were different from those previously identified in whole flies (Figure S1C), further confirming that we were detecting tissue-restricted binding. They were predominantly located in the 5′ end of genes (Figure S1D), indicating promoter-proximal binding, in contrast to enhancer binding observed with mammalian FoxO3 [20]. The regions bound by dFOXO were enriched for forkhead-like binding motifs (Figure S1E), confirming conservation of in vivo binding-sequence preference.
Finally, we identified the genes that are likely to be directly regulated by dFOXO based on their transcriptional responsiveness to induction of dfoxo and proximity (<1 kb) to a GFP-dFOXO bound site (Figure 1A). Genes up-regulated in the gut were specifically enriched for the GFP-dFOXO bound genes (p = 5×10−9). This is consistent with the predominant function of dFOXO as a transcriptional activator, conserved in its worm and mammalian orthologues [16], [19], [20]. We also observed GFP-dFOXO binding in the vicinity of the genes regulated by dfoxo in the fat body (Figure 1A), but this overlap was not significant. Note that numerous GFP-dFOXO-bound sites could not be associated with specific transcriptional events. This has been observed previously for FoxO factors in flies and other organisms [16], [17], [19], [20], [28], and it is currently unclear whether this is due to technical limitations in associating expression changes to binding events, whether FoxOs detected on these sites are poised for activation under a different set of conditions, or whether some of these sites are not functional.
The set of direct dFOXO targets included five sequence-specific TFs: clockwork orange (cwo), mothers against dpp (Mad), bunched (bun), anterior open (Aop) and CG12054 (summarised in Figure 2C). All of these were activated by dFOXO in the gut (Figure S1F, for Aop see also Figure 3). dFOXO also bound its own locus, and inducing the transgenic and intron-less dfoxo in S1106>dfoxo females resulted in an increase in unspliced dfoxo (Figure S1G), indicating that dFOXO self-activates (Figure 2C).
dFOXO induces an ETS transcriptional repressor in the adult gut
Among the TFs discovered as directly activated by dFOXO in the gut, we were especially interested in Aop (a.k.a. Yan). This ETS-family transcriptional repressor has a clear human orthologue in the Etv6 (a.k.a. Tel) gene [44]. Aop and Etv6 display clear conservation of function, with Aop involved in tracheal sprouting in flies while Etv6 is involved in the equivalent process of endothelial sprouting in mammals [45]. Interestingly, mammalian FoxOs and Etv6 act in similar physiological processes: both are tumour suppressors required for maintenance of adult haematopoietic stem cells [30], [46]; indicating that the previously-uncharacterised, functional interplay between the two factors may be evolutionarily conserved. Importantly, Drosophila presents a unique opportunity to examine the physiological functions of Aop/Etv6, because there is no known orthologue in yeast or worm.
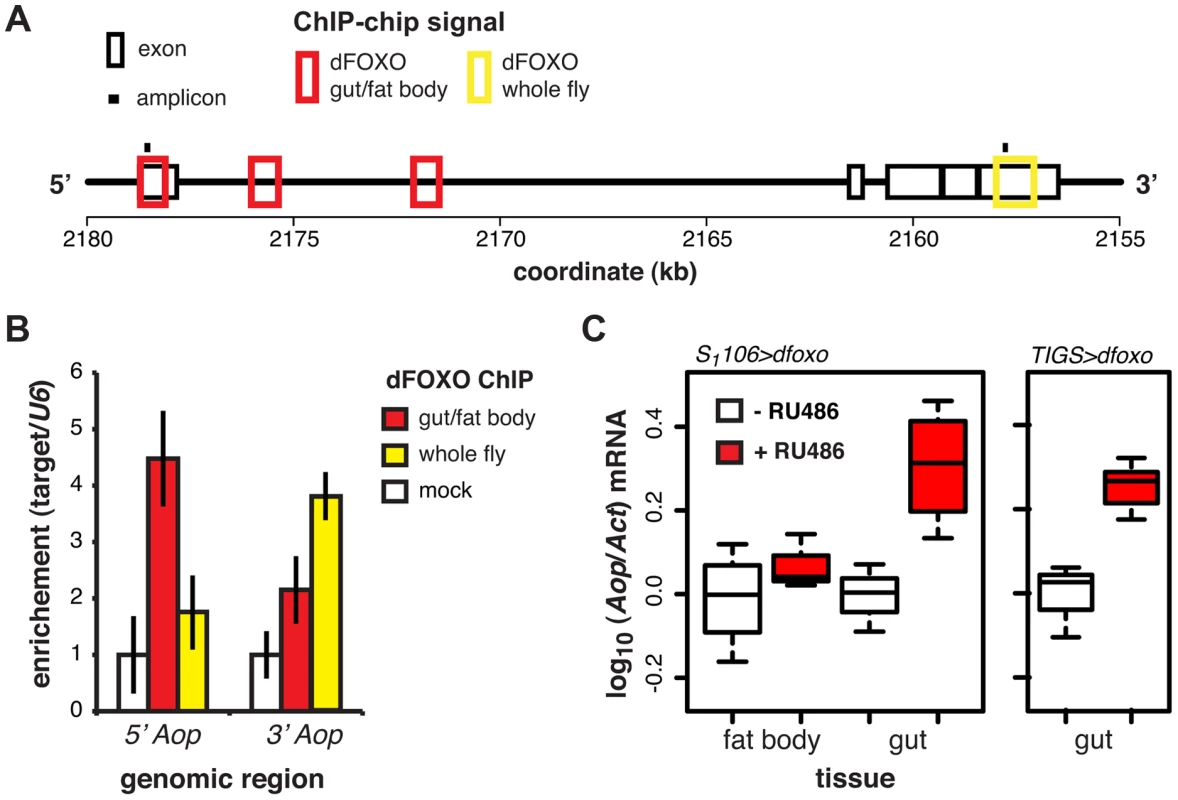
ChIP-chip revealed that GFP-dFOXO bound in the promoter-proximal, 5′ end of Aop, specifically in the 1st exon and 1st intron (Figure 3A). The binding to the 1st exon in the gut/fat body was confirmed with qPCR (Figure 3B). Similarly to dInR, we previously observed dFOXO bound to the coding region (3′ end) of this gene in whole flies (see [17] and Figure 3A and B). The functional significance of this 3′-end binding is unclear [17].
Induction of dfoxo in the gut and the fat body resulted in significant induction of Aop transcript only in the gut (p = 0.002, Figure 3C). To confirm the statistical significance of this tissue-restricted effect of dfoxo, we analysed the Aop expression data with a mixed-effects linear model and found a significant difference in the way the two tissues respond to RU486 (p<0.05). To confirm that these effects were cell autonomous, we induced dfoxo solely in the gut using an RU486-inducible, gut-specific driver, TIGS [31]. Feeding RU486 to TIGS>dfoxo females also resulted in induction of Aop transcript in the gut (Figure 3C). Hence, dFOXO induces Aop transcription in the gut, most likely through direct binding to the Aop promoter in gut cells.
Aop prevents the detrimental co-activation of dfoxo and Pointed
To understand the relationship between dfoxo and its target TF, Aop, we next examined the physiological role played by Aop in the context of tissue-restricted dfoxo induction and lifespan. Since Aop is an essential gene [47], [48], we chose to knock it down in the adult gut and fat body using a short-hairpin RNAi construct with no predicted off-targets [49]. Driving this construct with a ubiquitous, constitutive driver (daughtelessGAL4) resulted in the expected embryonic lethality. Inducing this construct in the adult gut and fat body, using the S1106 driver, reduced the levels of Aop mRNA by ∼70% (p = 0.04, Figure S2A) but had no major effect on lifespan (Figure 4A), revealing that, in a wild-type fly, Aop in these tissues is not limiting for survival.
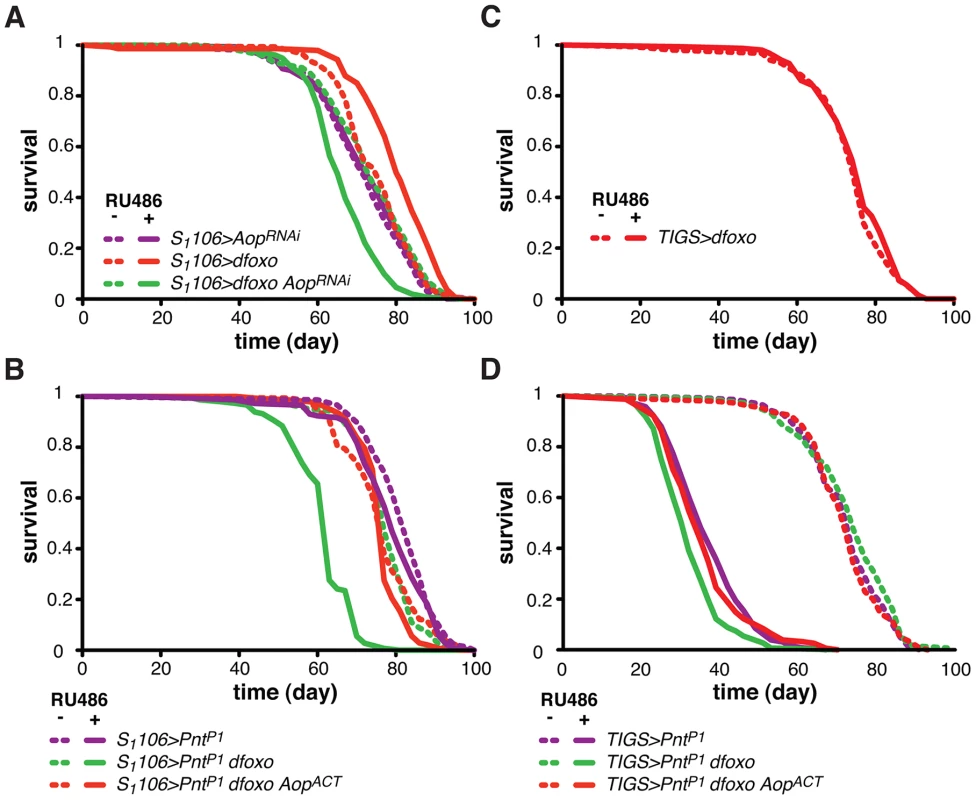
To test if Aop is required for the dfoxo-induced longevity, we simultaneously expressed dfoxo and knocked-down Aop. Surprisingly, while dfoxo alone extended lifespan, this combined treatment was detrimental to the fly (Figure 4A), indicating that Aop is required to prevent some toxic effect of dfoxo activation [39]. To determine the statistical significance of this synergistic effect, we used Cox Proportional Hazards (CPH) analysis [50]. This type of survival analysis can determine the significance of individual covariates as well as their interaction. We used “genotype” and “RU486” as individual covariates and found that the response to RU486 was significantly different in S1106>dfoxo AopRNAi genotype from the response in either S1106>dfoxo or S1106> AopRNAi genotypes (interaction of “genotype” and “RU486”, p = 5×10−15 and p = 1×10−7, respectively). This confirmed the synthetic interaction between the loss of Aop and induction of dfoxo. Importantly, expressing a control RNAi construct targeting GFP with S1106 driver did not prevent dfoxo from extending lifespan (Figure S2B).
We next examined the mechanism underlying this toxic effect of inducing dfoxo in the absence of Aop. Aop's activity is known to counteract that of Pointed (Pnt), an ETS-family transcriptional activator [51], [52]. These two TFs regulate the same genes through binding to the same regulatory elements but with opposing outcomes [51]–[53]. Hence, the synthetic toxicity of Aop loss- and dfoxo gain-of-function indicated that co-activation of Pnt and dfoxo could be highly detrimental. To test this, we induced the constitutively active form of Pnt (PntP1) [51] in the adult gut and fat body. This resulted in a marginal negative effect on lifespan (p = 0.06) and, similarly to the phenotype of Aop loss and dfoxo gain, the co-induction of dfoxo and PntP1 was highly detrimental (Figure 4B).
It was possible that dFOXO induced the transcription of Aop in order to avert co-activation of PNT and the resulting detrimental synergistic effect. Indeed, the detrimental effect of combined dFOXO and PNT activity was completely rescued by additional activation of AOP, achieved by expression of a constitutively active form of Aop [48], called AopACT, in addition to dfoxo and PntP1 (Figure 4B). The significance of this effect was confirmed by CPH analysis, which showed that the response to RU486 in S1106>PntP1 genotype was significantly different from that in S1106>PntP1 dfoxo genotype (p<10−15), but not from the response observed in S1106>PntP1 dfoxo AopACT (p = 0.5). Importantly, induction of AopACT did not interfere with the induction of dfoxo or PntP1 in the S1106>PntP1 dfoxo AopACT females compared to the S1106>PntP1 dfoxo females (Figure S2C).
dFOXO regulates Aop transcription specifically in the gut (Figure 3C). For this reason we next examined whether the genetic interactions we observed between dfoxo and Aop/Pnt could be recapitulated with the gut-specific TIGS driver. This driver has been successfully used by others to determine the contribution of the gut to S1106-triggered phenotypes [54]. The gut is a highly regionalised organ with several cell types [55], [56] and we confirmed that the two drivers activate transgene expression in similar regions (Figure S3A), both driving expression in the enterocytes (Figure S3B). Feeding RU486 to TIGS>dfoxo females had no significant effect on lifespan (Figure 4C), even though dfoxo was induced in the gut (Figure S1A) and activated the expression of Aop (Figure 3C). With the caveat that the levels of expression achieved with TIGS and S1106 in the gut are not identical (Figure S1A, S3A) and the two drivers may not drive in completely overlapping subsets of gut cells (Figure S3A and B), our data indicate that activation of dFOXO in the gut alone is insufficient for lifespan extension.
On the other hand, the induction of PntP1 solely in the adult gut was highly detrimental for the fly (Figure 4D), while RU486 had no effect on the lifespan of the TIGS-alone or UAS-PntP1-alone controls (Figure S3C and D). The interaction between dfoxo and Pnt revealed with the TIGS driver was similar to that observed with S1106: induction of dfoxo exacerbated the toxicity of PntP1 and this, in turn, could be remedied by further induction of AopACT (Figure 4D). While the magnitude of this synergistic effect was smaller with TIGS than S1106, CHP analysis confirmed that the response to RU486 in TIGS>PntP1 females was significantly different from that in TIGS>PntP1 dfoxo (p<10−4), but not in TIGS>PntP1 dfoxo AopACT females (p = 0.8). Hence, the dFOXO-PNT-AOP interaction can be mapped specifically to the gut. Note, however, that we cannot exclude the possibility of a similar interaction also occurring in the fat body.
Overall, our data are consistent with dFOXO up-regulating Aop transcription in the gut in order to counteract the activity of PNT. In this way, dFOXO prevents the detrimental effect that would result from dFOXO being active at the same time as PNT in that organ.
dFOXO and AOP share binding locations
Intuitively, the genetic interactions observed between dfoxo and Aop/Pnt were not consistent with a simple, linear cascade of dFOXO acting on AOP/PNT to influence lifespan (Figure 5A). To formally examine this, we used Boolean network modelling. We created network configurations with dFOXO, AOP, PNT and “lifespan” as nodes that can take values of 0 or 1 (inactive/short life or active/long life) and described the relationships between them using Boolean logic operators. We then perturbed the network by fixing either dFOXO or PNT or both as “active” and examined the probability that lifespan will take on the value of 1 (long life) after 1000 state transition using Markov chain simulations. This type of modelling formalised what was intuitively evident: the synergistic negative effect of dfoxo and Pnt co-activation could be recapitulated by a network circuit that includes a feed-forward loop between dfoxo and Pnt with a negative effect on lifespan (described by a NAND operator), but not by a linear cascade (Figure 5A).
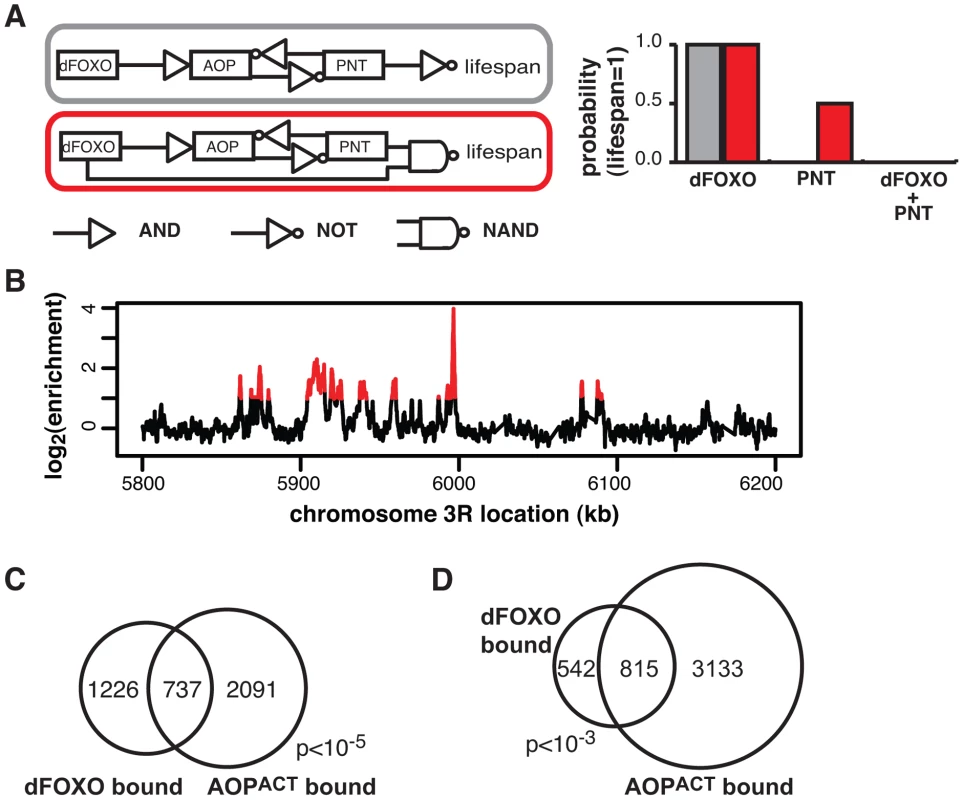
Feed-forward loops involve co-regulation of the same targets, and are often accompanied by extensive overlaps in genomic sites bound by the TFs [57]. To gather further evidence for the existence of this feed-forward loop, we determined the relationship between the sites bound by the AOP/PNT couple and dFOXO. We performed ChIP-chip on flies expressing FLAG-AOPACT, since both PNT and AOP are known to bind the same sequences, competing for the same sites [51]–[53].
To facilitate the comparisons with dFOXO, we used the S1106 driver and the RU486 inducer, with the control IP performed with an anti-FLAG antibody on chromatin from flies expressing untagged AOPACT (ArrayExpress accession number: E-MTAB-1306). We discovered ∼4000 genomic regions bound by FLAG-AOPACT neighbouring some 3000 genes, including the dfoxo locus, with good correlation between biological repeats (Figure S4A). An example of peaks is given in Figure 5B. The binding locations were associated with genes and tended to occur in the 5′ region (Figure S4B), consistent with a role in promoter-proximal regulation of transcription. Similarly to the recently published AOP binding locations observed in larvae [58], we found that AOP tended to bind long stretches of DNA, longer than dFOXO (Figure S4C). At the same time, over 80% of the regions bound in adult gut/fat body were distinct from those bound in larvae (Figure S4D).
We found a significant and substantial overlap in the genes bound by GFP-dFOXO and those bound by FLAG-AOPACT (p<10−5, Figure 5C), indicating that AOP/PNT and dFOXO may regulate numerous genes in common. Furthermore, dFOXO did not just bind in the vicinity of the same genes but actually to the same regions of the DNA: 60% of GFP-dFOXO-bound regions directly overlapped regions of FLAG-AOPACT binding (Figure 5D). This striking and significant (p<10−3) overlap in sites bound corroborates the existence of a feed-forward loop between AOP/PNT and dFOXO. Furthermore, the genetic interactions we observed between dfoxo and Pnt/Aop are likely to be mediated by functional interactions on the shared regulatory regions.
Pnt and dfoxo synergistically affect lipid metabolism
The strength of the negative synergistic effect observed between dfoxo and Pnt prompted us to seek out its physiological basis. To initiate this investigation, we looked at the GO categories that are over-represented in genes bound by FLAG-AOPACT. The most over-represented functional category was “lipid particle” (p = 3×10−17, Figure 6A), a GO category that includes lipid droplets, sites of cellular lipid storage. This was also the most over-represented category in the set of genes bound by both FLAG-AOPACT and GFP-dFOXO (p = 3×10−5, Figure 6A), indicating that PNT/AOP and dFOXO together regulate lipid metabolism.
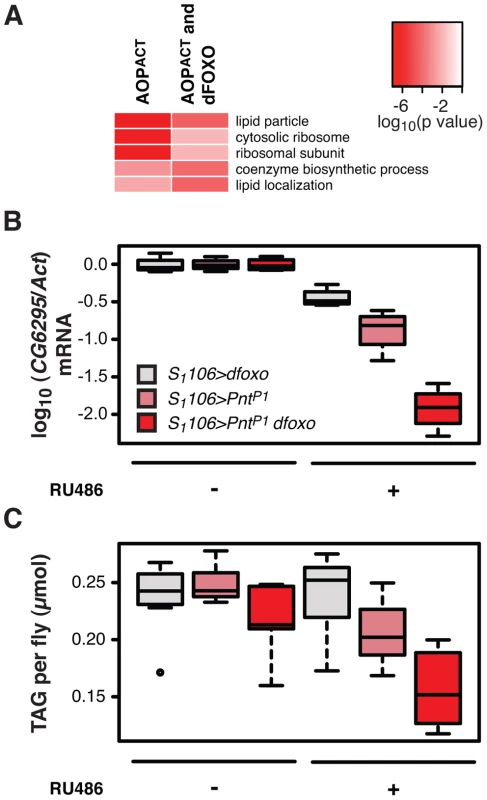
Activation of dFOXO in the aging gut represses two gastric lipases, encoded by lipA (a.k.a. magro) and CG6295 genes [39]. These two lipases are thought to facilitate assimilation of ingested lipids, and their repression by dFOXO results in a decrease in lipid stores and a shortened lifespan [39]. We observed the detrimental effects of dFOXO activation in the gut to be dependent on simultaneous activation of PNT (Figure 4B, C and D), prompting us to examine whether the expression of either lipA or CG6295 was synergistically regulated by dfoxo and Pnt. We used the S1106 driver, where the synergistic effect on lifespan is pronounced, and found that both dfoxo and PntP1 resulted in repression of CG6295, 3-fold and 8-fold, respectively (Figure 6B). When the two TF were co-expressed, the reduction in CG6295 was even greater, reaching 80-fold reduction after only 5 days of transgene induction (Figure 6B). Indeed, a linear model revealed a significant difference in the way the three genotypes responded to RU486 (p<10−4), and the amount of CG6295 transcript was significantly different between S1106>PntP1 + RU486 or S1106>dfoxo + RU486 and S1106>PntP1 dfoxo + RU486 conditions (p<10−4). Hence dfoxo and Pnt synergistically repress the CG6295 lipase. The effect of the two TFs on lipA was similar but smaller, and there was no significant difference between S1106>dfoxo + RU486 and S1106>PntP1 dfoxo + RU486 conditions (p>0.05, Figure S5A). Note that neither AOPACT nor dFOXO bind in the vicinity of CG6295 and hence the effects on the expression of this gene are likely to involve an intermediate factor.
To examine whether the synergistic effect on the expression of the CG6295 lipase had a significant physiological consequence, we determined the levels of triacylglycerol (TAG) stores in flies expressing dfoxo, Pnt or both, under the control of the S1106 driver. We found that the levels of TAG paralleled the levels of CG6295 transcript (Figure 6C) and, using a linear model, we confirmed that the effects of RU486 on the TAG levels in the 3 genotypes were significantly different (p = 0.01), with the TAG being significantly more depleted in S1106>PntP1 dfoxo flies than in the other two genotypes after only 5 days of RU486 feeding (p<0.02, Figure 6C).
The effect on CG6295 expression and TAG levels could be symptomatic of a more general loss of gut integrity. The function of the gut as a barrier is a good surrogate for overall gut integrity and is important for survival [59]. The effectiveness of this barrier can be assessed by feeding flies a food containing a dye (fluoresceine) and scoring the number of flies in which the gut is unable to exclude this dye from the whole body (“smurf” phenotype) [59]. We found that driving dfoxo and PntP1 for 3 weeks using S1106 did not increase the proportion of smurfs in the population (Figure S5B), while the effect on lipid metabolism was evident after only 5 days (Figure 6C). This confirmed that the effect on lipid metabolism was not due to a general loss of gut integrity. In addition, we also observed no significant changes in feeding behaviour of S1106>PntP1 dfoxo females after RU486 treatment, as assessed with the proboscis-extension assay (Figure S5C).
Based on these data, we propose that the detrimental synergy between dfoxo and Pnt arises from a mis-regulation of lipid metabolism resulting in a profound drop in TAG stores. Indeed, S1106>PntP1 dfoxo flies were starvation sensitive after 5 days of RU486 feeding, and this sensitivity could be reversed by co-induction of AopACT (Figure S5D).
Activation of AOP alone can extend lifespan
We next focused on other possible roles of Aop in Drosophila lifespan. Expression of the activated form of Aop in the flies co-expressing dfoxo and PntP1 with S1106 driver had a substantially beneficial effect on lifespan (Figure 4B). This was due, in part, to its role in preventing dFOXO and PNT co-activation, but we hypothesised that activation of AOP might additionally increase the lifespan of wild type flies. We induced expression of the activated form of AOP (AopACT) using the S1106 driver and the RU486 inducer in an otherwise wild-type adult female. This resulted in significant lifespan-extension (p = 2×10−10), increasing the median by 14% and maximal lifespan by 11% (Figure 7A), while RU486 feeding had no effect on the lifespans of the driver- or transgene-alone controls (Figure S6A and B). This effect was robust and observed in six independent experiments, performed in the course of three years, with average median lifespan extension of 12% (Figure 7B). Furthermore, independently generated, FLAG-tagged AopACT also extended lifespan (Figure S6C).
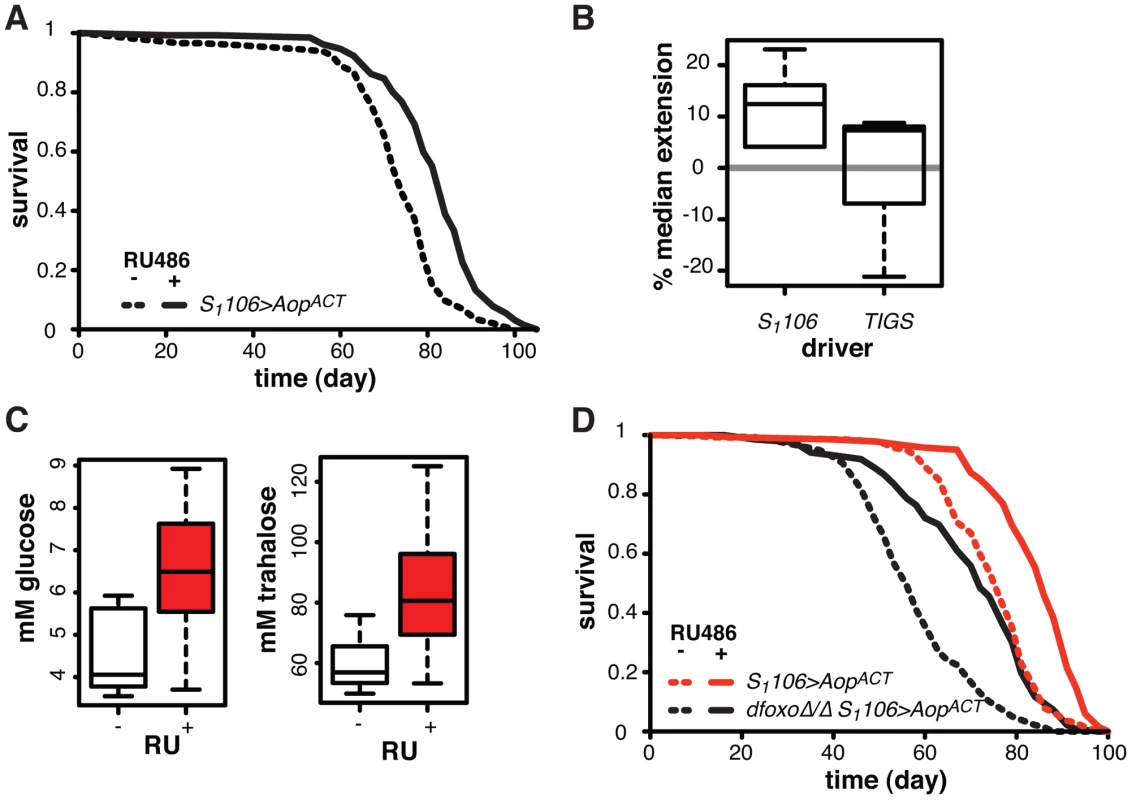
To examine whether this lifespan-extending effect could be localised to the same tissues where the interaction between dfoxo and Pnt/Aop occurs, we drove AopACT with the gut specific TIGS driver. Induction of AopACT using the TIGS driver did not extend lifespan (Figure 7B), despite transgene induction in the gut with both TIGS and S1106 drivers (Figure S6D). Although the induction levels were lower with TIGS (Figure S6D), they must still be physiologically relevant because, even with this driver, AopACT could partially remedy the toxicity of Pnt and dfoxo co-activation (Figure 4D).
Our data are consistent with two roles of Aop in lifespan modulation: (1) Aop can counteract the negative effect of dfoxo and Pnt co-activation in the gut, (2) its additional activation in the fat body extends wild-type lifespan. The lifespan extension by AopACT in flies co-expressing dfoxo and PntP1 under S1106 control most likely combines the two beneficial effects of Aop activation, since the effect appears greater in that context than in the otherwise wild-type female (median increased by 22% between S1106>dfoxo PntP1 + RU486 and S1106>dfoxo PntP1 AopACT + RU486, Figure 4B, versus 12% average extension in wild-type, Figure 7B).
We examined other phenotypes triggered in S1106>AopACT females by RU486 feeding, focusing on phenotypes often associated with lifespan extension. RU486 feeding did not cause any significant changes in starvation, hydrogen peroxide or DDT resistance; any changes to whole body trehalose, glycogen or lipid content; feeding or fecundity (Figure S7). However, we did find that induction of AopACT in the adult gut and fat body resulted in increased circulating sugars (Figure 7C), a diabetic phenotype potentially indicative of slightly reduced IIS. Pnt is known to promote IIS in larvae and its gain-of-function stimulates clearance of circulating sugars [60], consistent with the converse phenotype we observed in S1106>AopACT adults.
Since FoxOs are inhibited by IIS [61], Aop could act to reduce IIS and activate dFOXO, thereby increasing the lifespan of otherwise wild-type females. However, AopACT extended lifespan in complete absence of dfoxo (Figure 7D), and CPH analysis found no evidence for a difference in the response to RU486 in dfoxoΔ/Δ S1106>AopACT versus S1106>AopACT females (p = 0.2). Hence, Aop extends wild-type lifespan independently of dfoxo.
dFOXO and AOP share targets in the fat body
We further examined the function of Aop in the fat body, where its activity appears important for lifespan but where it is not regulated by dfoxo. We found that AOP transcript and protein levels were strongly induced in the fat body when S1106>AopACT females were fed RU486 (Figure S8A), but in only a portion (∼26±6%) of the fat body cells, suggesting heterogeneity of the tissue (Figure 8A, for quantification see Figure S8B). This mosaic expression was not due to the driver, because S1106 drove GFP expression in all the cells of the tissue (Figure S8C).
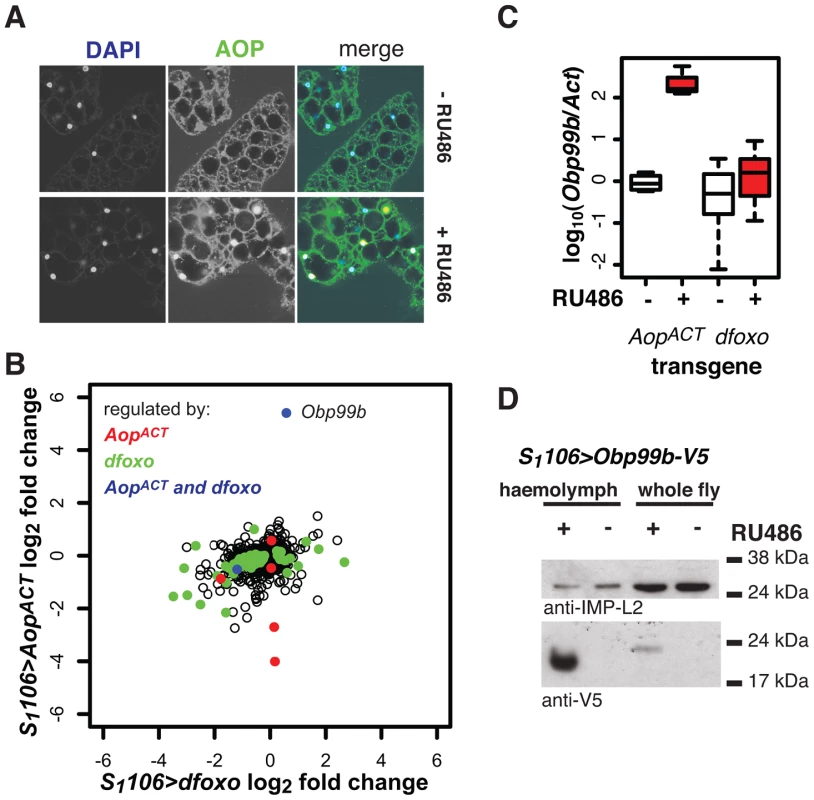
To elucidate the function of Aop in the fat body we used microarray analysis on dissected tissue to identify changes in gene expression upon RU486 feeding in S1106>AopACT females (ArrayExpress accession number: E-MTAB-927). The induction of AOPACT in only a few cells of the fat body precluded identification of many genes with significant changes upon RU486 feeding. We identified 8 genes (p value cut off of 0.00017, corresponding to 20% FDR, Figure 8B). Two of these were also regulated by dfoxo (Figure 8B), and this modest overlap was statistically significant (p = 2×10−3). The functions regulated by the two factors, as determined by Catmap analysis (Figure S9A) were also partially overlapping. For example, both dfoxo and AopACT had a negative effect on respiratory electron transport chain (p = 10−21 and p = 7×10−9 respectively), while AopACT alone had an effect on carbohydrate metabolism (p = 9×10−18 for AopACT, p>0.05 for dfoxo) (Figure S9A). Interestingly, we found that the lifespan benefits from the induction of AopACT, dfoxo or both were not significantly different (Figure S9B). This indicated that the effects of the two factors are not additive and, in turn, that the two are likely to affect lifespan via the same or related physiological processes, in an otherwise wild-type background.
Interestingly, the most strongly up-regulated gene in the fat body upon AopACT induction was also up-regulated by dFOXO in the same tissue, albeit less strongly (Figure 8B). These observations were confirmed by qPCR (Figure 8C). This gene encodes a signal peptide followed by an odorant binding protein (Obp) domain - Obp99b. Non-sensory Obps are thought to bind and carry lipophilic compounds, fulfilling similar functions to the mammalian lipocalin family proteins, rather than acting in odour perception [62], [63]. Obp99b could represent a humoral factor, induced by both dfoxo and Aop, mediating inter-tissue communication. Such inter-tissue communication is known to be important in the physiological effects of tissue-restricted dfoxo induction [4]. To establish that this Obp was actually secreted in vivo we expressed a transgene encoding a V5-tagged Obp99b with the S1106 driver. Feeding RU486 to these flies confirmed the predicted enrichment of Obp99b-V5 in the haemolymph (Figure 8D) and revealed that its transcriptional up-regulation is enough to release it in circulation. Hence, in the fat body, AopACT and dfoxo regulate a common humoral factor, whose function warrants further investigation.
Discussion
Our study clearly demonstrates the in-vivo complexity of interactions that occur between FoxOs and the TFs they regulate. It emphasises the need to untangle the tissue-specific transcriptional networks within which FoxOs act in order to understand the role of FoxO factors in lifespan.
We show that dFOXO activates the transcription of an ETS repressor, Aop, in adult Drosophila gut to antagonise an ETS activator, Pnt, and avert the detrimental effects of the co-activation of dFOXO and PNT. This interaction most likely takes place at the gene promoters/enhances where AOP may directly displace PNT from a dFOXO bound region. Whether this interaction is facilitated by a direct protein-protein interaction between dFOXO and AOP, or by juxtaposition of dFOXO and AOP binding sites, remains to be determined.
Others have shown that induction of dfoxo solely in the gut is detrimental for lifespan [39]. In our outbred, wild-caught background, this detrimental effect is conditional on co-activation of Pnt. Despite these differences, the deleterious physiological outcome is likely to be based on mis-regulation of lipid metabolism genes and a drop in TAG stores in both cases. In parallel, the activation of AOP in the gut/fat body of otherwise wild-type females has only two phenotypes: increase in lifespan and increase in circulating sugars. The increase in circulating sugars is a metabolic phenotype, generally not detrimental to fly lifespan as it can be observed in a number of long-lived IIS mutants [64], [65]. Together with the effects observed upon dfoxo/Pnt co-activation, it indicates that the Aop/Pnt couple have metabolic functions in the adult fly.
FoxOs are regulated by AKT [61], while ETS factors, such as AOP and PNT, are regulated by ERK [51]. In turn, both ERK and AKT are activated in response to activation of receptor tyrosine kinases (RTK) [66]. Viewed in this context, the interaction between dFOXO, inhibited by AKT, and AOP, inhibited by ERK, insures that the two branches down-stream of RTKs are coordinated. Our findings reveal the mis-coordination of the two branches to be highly detrimental, with the cross-talk between dfoxo and Aop set up to prevent it. This mechanism for coordination of the activities of the two branches, via interactions between FoxO and ETS factors, is potentially relevant in numerous contexts beyond Drosophila lifespan.
Dissecting the relationship between dFOXO and AOP led us to the discovery of a second beneficial role for AOP and the identification of Aop as a longevity determinant in its own right. Aop has an important and well-established role in Drosophila development. The gene was initially isolated as encoding a negative regulator of neural development, in the context of the Drosphila eye [47]. Subsequent studies revealed it to be a general inhibitor of differentiation [48], [67]. Its role in the adult has not been investigated and its effect on lifespan was completely unsuspected.
Several contingent observations lead us to speculate that the role of Aop as a fly longevity gene, as well as its interaction with dfoxo, will be conserved in its mammal orthologue Etv6: Aop and Etv6 display clear conservation of function in other physiological processes [45]; the common roles of dfoxo and Aop in fly lifespan are paralleled by shared roles of FoxO factors and Etv6 in mammals [30], [46]; the neuroendocrine axes controlling growth are important in mammalian lifespan [68] and Etv6 has been identified as associated with human height in genome-wide association studies [69], [70].
Materials and Methods
Fly lines, husbandry, lifespan and physiological assays
S1106 [1], [31], TIGS [31] UAS-dfoxo [1], UAS-GFP-dfoxo [32], UAS-AopACT [48], UAS-Obp99b-V5.5 [71], UAS-PntP1 [51], dfoxoΔ94 [6] and UAS-GFPRNAi [72] were backcrossed at least 6 times into the wild-type, outbred, Dahomey population carrying the w1118 mutation, which had been cured of Wolbachia infection over five years ago, and frequently outcrossed back into the same wild-type population. The UAS-AopRNAi construct was obtained from the TRiP collection (HMS01256) [49], and was tracked by PCR during backcrosses as above. The Dahomey stock was collected in 1970 in Dahomey (now Benin) and has been kept in population cages maintaining its lifespan and fecundity at levels similar to freshly caught stocks. Combinations of transgenes/mutants were created using standard fly genetic techniques while avoiding population bottlenecks. To create UAS-FLAG-AopACT, the AopACT open reading frame was amplified from genomic DNA of flies carrying UAS-AopACT (primers are given in Protocol S1) and cloned into pENTR-D-TOPO vector, confirmed by sequencing and transferred to pTFW P-element-based vector, integrated into the fly genome and the transgene backcrossed as above.
The lines were maintained, and all experiments performed, at 25°C with 60% humidity and 12 h∶12 h light∶dark cycle, on sugar-yeast-agar (1SYA) food [73]. Experimental flies developed at standardised densities and once-mated females we sorted on day two of adulthood onto food containing 200 µM RU486 (Sigma) or control food as required. Lifespans were performed as described [1]. Heamolymph extraction and measurements of circulating glucose and trahalose, and other phenotyping, were performed as described [4], [74], [75]. Examination of the “smurf” phenotype was carried out using fluorescein in 1SYA +/− RU486 as described [59], except the flies were kept on the food for 18 h.
Chromatin immunoprecipitation
Biological triplicates were done for all fly chromatin preparations. For each experiment all the batching was done so that the treatments to be compared were carried out in parallel. Chromatin was prepared from 7-day old S1106>GFP-dfoxo or S1106>FLAG-AopACT (experimental) or S1106>dfoxo or S1106>AopACT (mock control) females fed RU486-containing food from day two of adulthood, as follows: 1000 females were crushed to a fine powder under liquid nitrogen and re-suspended in 6 ml of PBS supplemented with Protease Inhibitors Cocktail (10 µl per ml, Sigma). Cross-linking was performed with 0.5% formaldehyde for 10 min and quenched with addition of 1.5 ml of 2.5 M glycine. The cross-linked chromatin was recovered by centrifugation and washed twice with FA/SDS buffer (50 mM Hepes-KOH pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Na deoxycholate, 0.1% SDS, 1% Triton –X-100 and 1 mM PMSF) re-suspended in the same and incubated for 1 h at 4°C. Chromatin was again recovered by centrifugation and sheared to an average size of 400 bp by sonication, giving on average 6 ml of chromatin in FA/SDS.
For anti-GFP IPs, anti-GFP antibody (Abcam, 1 µl per IP) was bound to Protein-G Dynabeads (Invitrogen) and incubated for 2 h at room temperature with 450 µl of chromatin. Beads were washed once with FA/SDS, 3 times with FA/SDS containing 500 mM NaCl, once with 10 mM Tris-HCl pH 8, 250 mM LiCl, 1 mM EDTA, 1% NP40 and 0.5% Na deoxycholate, and once with TE. For array hybridisation, the entire IP after volume reduction, or 50 ng of total chromatin DNA, were amplified two times (Whole Genome Amplification kit, Sigma) as per manufacturer's instructions. The material from the IP was hybridised against the input material. The labelling and hybridisations were carried out by Nimblegen Systems, using custom Drosophila whole-genome tiling arrays with probes spaced approximately every 300 bp, as described [76]. ChIP-chip data were normalized using the LIMMA package [77] in Bioconductor [78], applying loess normalization within each array and quantile normalization between arrays. Replicate information was pooled by taking the median probe value for each set of arrays and was smoothed along each chromosome using a running median within a window of three probes. Experimental signal was adjusted by mock control (pre-immune serum) data by direct subtraction of median probe intensity values. Peaks were called using the Ringo package [79] in R, using a y0 threshold of 0.97 and a distance cut-off of 600 bp.
Anti-FLAG IPs were performed essentially as above, except anti-FLAG antibody (Sigma, 3 µl per IP) was used on 1350 µl of chromatin. Subsequent processing was the same except that 100 µM dUTP was spiked in during amplification, and chromatin and IP were separately hybridized to Affymetrix Drosophila Tiling 2.0R arrays. The IP to chromatin ratios were computationally determined after normalisation using the Starr package [80], and peaks identified as above, except that smoothing was over 5 probes.
Genes were associated to peaks if any gene feature occurred within 1 kb upstream or downstream of the outermost peak probes. The overlap between regions bound by GFP-dFOXO and FLAG-AOPACT was compared to the random distribution by bootstrap analysis (10000 iterations looking for marginal basepair overlap in block bootstrap) [81] and the Z-score and p value calculated.
Chromatin was also prepared from wild-type female flies and serum-starved S2 cells and IP performed with the affinity-purified anti-dFOXO antibody [32] as described [17]. Primers used for qPCR are given in Protocol S1.
Microarray and mRNA expression analysis
For microarray analysis of dfoxo regulated genes at least four and up to five biological repeats of 7-day old S1106 or S1106>dfoxo females that had been fed RU486-containing or control food, were dissected removing either the mid-gut (50 per sample), or the abdominal/thorasic fat body (25 per sample) as associated with the cuticle, and the RNA extracted with RNeasy (Qiagen). The fat body dissections were performed by isolating and opening the fly thorax and/or abdomen, removing the internal organs with minimal disturbance of the fat body and keeping the fat body associated with the cuticle. We estimate that such dissection of the fat body results in ∼80% purity. For microarray analysis of AopACT regulated genes four biological repeats of 7-day old S1106>AopACT females that had been fed RU486-containing or control food, were dissected removing the abdominal fat body (25 per sample) as associated with the cuticle and the RNA extracted with RNeasy (Qiagen). All dissections and subsequent processing were done in batches so that the samples to be compared were always processed in parallel. The RNA was further processed into cRNA using standard Affymetrix protocols and hybridized to the Affymetrix Drosophila Genome 2.0 Genechip at SHWFGF/Glasgow Polyomics (University of Glasgow). The data were analysed in R. They were summarised and normalised using RMA as implemented in the LIMMA package [82]–[84]. Differential expression was assessed using linear models and the empirical Bayes moderated t-statistic implemented in LIMMA. For dfoxo-regulated gene analysis the effects in the linear model were: “dissection batch”, “S1106+RU486”, “S1106>dfoxo” and “S1106>dfoxo+RU486”; for AopACT-regulated genes: “dissection batch” and “S1106>AopACT+RU486”. Present/Absent calls were performed with Mas5 and transcripts present on at least four arrays for fat body, or at least five for gut were kept for further analysis. FDR was controlled using the described procedure [85]. For Gene Ontology enrichment, Catmap [40], or DAVID EASE [86] analyses were performed.
For qPCR analysis RNA was isolated from five to 10 midguts, five fat bodies or five guts and fat bodies with TRIZOL, converted into cDNA and qPCR performed as described [17]. The primers are given in Protocol S1.
Boolean network modeling
The network was modelled using BoolNet in R [87].
Immunofluorescence and western blots
Abdominal fat bodies as associated with the cuticle were dissected and immunofluorescence was performed using a monoclonal mouse anti-AOP antibody [48] at 1∶100 dilution using the described protocol [74]. For GFP detection, dissected body parts were fixed for 10 min before visualisation. Images were captured on Zeiss LSM 700, and quantified using ImageJ. For western blots, proteins were extracted from whole flies, or from 10 fat bodies, with TCA, or obtained by denaturing adult haemplymph in Lemmini buffer [74], separated by SDS-PAGE, transferred to nitrocellulose and probed with anti-V5 antibody (1∶500, Sigma), anti-IMP-L2 (1∶2500) [74], anti-ACTIN (1∶1000, Abcam) or anti-AOP (1∶500).
Statistical analysis
Statistical analyses were performed in JMP (version 9) software (SAS Institute), R or Excel (Microsoft). qPCR data were log-transformed to fit a normal distribution and analysed with ANOVA, linear models or mixed effects linear models, followed by selected pair-wise comparisons using t-test. Survival analysis was performed with Log-rank, CPH or mixed-effects CPH methods in JMP, or in R using survival and coxme packages (Terry Therneau, http://CRAN.R-project.org/package=survival, http://CRAN.R-project.org/package=coxme). Analysis of smurf data was performed with a generalised linear model in R and quasibinomial distribution. Analysis of feeding was performed in JMP, using a generalised linear model, binomial distribution and over-dispersion adjustment. Significance of set overlaps was determined with Hypergeometric distribution. Details of tests performed are given in figure legends.
Array data
The unprocessed array data are available from ArrayExpress under accession numbers: E-MTAB-1020, E-MTAB-1021, E-MTAB-927, and E-MTAB-1306.
Supporting Information
Zdroje
1. GiannakouME, GossM, JungerMA, HafenE, LeeversSJ, et al. (2004) Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305: 361.
2. HwangboDS, GershmanB, TuMP, PalmerM, TatarM (2004) Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429: 562–566.
3. DemontisF, PerrimonN (2010) FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143: 813–825.
4. AlicN, TulletJ, NiccoliT, BroughtonS, HoddinottMP, et al. (2014) Cell-nonautonomous effects of dFOXO/DAF-16 in ageing. Cell Rep 6: 608–616.
5. KenyonC, ChangJ, GenschE, RudnerA, TabtiangR (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464.
6. SlackC, GiannakouME, FoleyA, GossM, PartridgeL (2011) dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell 10: 735–748.
7. PostnikoffSD, MaloME, WongB, HarknessTA (2012) The yeast forkhead transcription factors fkh1 and fkh2 regulate lifespan and stress response together with the anaphase-promoting complex. PLoS Genet 8: e1002583.
8. KuningasM, MagiR, WestendorpRG, SlagboomPE, RemmM, et al. (2007) Haplotypes in the human Foxo1a and Foxo3a genes; impact on disease and mortality at old age. Eur J Hum Genet 15: 294–301.
9. WillcoxBJ, DonlonTA, HeQ, ChenR, GroveJS, et al. (2008) FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A 105: 13987–13992.
10. FlachsbartF, CaliebeA, KleindorpR, BlancheH, von Eller-EbersteinH, et al. (2009) Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A 106: 2700–2705.
11. AnselmiCV, MaloviniA, RoncaratiR, NovelliV, VillaF, et al. (2009) Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res 12: 95–104.
12. PawlikowskaL, HuD, HuntsmanS, SungA, ChuC, et al. (2009) Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell 8: 460–472.
13. EijkelenboomA, BurgeringBM (2013) FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol 14: 83–97.
14. MurphyCT, McCarrollSA, BargmannCI, FraserA, KamathRS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283.
15. McElweeJJ, SchusterE, BlancE, PiperMD, ThomasJH, et al. (2007) Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol 8: R132.
16. SchusterE, McElweeJJ, TulletJM, DoonanR, MatthijssensF, et al. (2010) DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol 6: 399.
17. AlicN, AndrewsTD, GiannakouME, PapatheodorouI, SlackC, et al. (2011) Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol 7: 502.
18. BaiH, PingK, HernandezAM, TatarM (2013) Activin Signaling Targeted by Insulin/dFOXO Regulates Aging and Muscle Proteostasis in Drosophila. PLoS Genet 9 (11) e1003941.
19. WebbAE, PollinaEA, VierbuchenT, UrbanN, UcarD, et al. (2013) FOXO3 shares common targets with ASCL1 genome-wide and inhibits ASCL1-dependent neurogenesis. Cell Rep 4: 477–491.
20. EijkelenboomA, MokryM, de WitE, SmitsLM, PoldermanPE, et al. (2013) Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol Syst Biol 9: 638.
21. PuigO, TjianR (2005) Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev 19: 2435–2446.
22. van der VosKE, CofferPJ (2008) FOXO-binding partners: it takes two to tango. Oncogene 27: 2289–2299.
23. BouchardC, MarquardtJ, BrasA, MedemaRH, EilersM (2004) Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J 23: 2830–2840.
24. NemotoS, FergussonMM, FinkelT (2004) Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 306: 2105–2108.
25. OggS, ParadisS, GottliebS, PattersonGI, LeeL, et al. (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999.
26. SeoaneJ, LeHV, ShenL, AndersonSA, MassagueJ (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117: 211–223.
27. EssersMA, de Vries-SmitsLM, BarkerN, PoldermanPE, BurgeringBM, et al. (2005) Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science 308: 1181–1184.
28. TelemanAA, HietakangasV, SayadianAC, CohenSM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32.
29. PaikJH, KolliparaR, ChuG, JiH, XiaoY, et al. (2007) FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323.
30. TothovaZ, KolliparaR, HuntlyBJ, LeeBH, CastrillonDH, et al. (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339.
31. PoirierL, ShaneA, ZhengJ, SeroudeL (2008) Characterization of the Drosophila gene-switch system in aging studies: a cautionary tale. Aging Cell 7: 758–770.
32. GiannakouME, GossM, JacobsonJ, VintiG, LeeversSJ, et al. (2007) Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging Cell 6: 429–438.
33. GiannakouME, GossM, PartridgeL (2008) Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: not required, but its activity modulates the response. Aging Cell 7: 187–198.
34. MinKJ, YamamotoR, BuchS, PankratzM, TatarM (2008) Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell 7: 199–206.
35. LibinaN, BermanJR, KenyonC (2003) Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502.
36. BluherM, KahnBB, KahnCR (2003) Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299: 572–574.
37. PuigO, MarrMT, RuhfML, TjianR (2003) Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev 17: 2006–2020.
38. JungerMA, RintelenF, StockerH, WassermanJD, VeghM, et al. (2003) The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol 2: 20.
39. KarpacJ, BiteauB, JasperH (2013) Misregulation of an adaptive metabolic response contributes to the age-related disruption of lipid homeostasis in Drosophila. Cell Rep 4: 1250–1261.
40. BreslinT, EdenP, KroghM (2004) Comparing functional annotation analyses with Catmap. BMC Bioinformatics 5: 193.
41. BahadoraniS, ChoJ, LoT, ContrerasH, LawalHO, et al. (2010) Neuronal expression of a single-subunit yeast NADH-ubiquinone oxidoreductase (Ndi1) extends Drosophila lifespan. Aging Cell 9: 191–202.
42. SanzA, SoikkeliM, Portero-OtinM, WilsonA, KemppainenE, et al. (2010) Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc Natl Acad Sci U S A 107: 9105–9110.
43. Casas-TintoS, MarrMT2nd, AndreuP, PuigO (2007) Characterization of the Drosophila insulin receptor promoter. Biochim Biophys Acta 1769: 236–243.
44. RoukensMG, Alloul-RamdhaniM, MoghadasiS, Op den BrouwM, BakerDA (2008) Downregulation of vertebrate Tel (ETV6) and Drosophila Yan is facilitated by an evolutionarily conserved mechanism of F-box-mediated ubiquitination. Mol Cell Biol 28: 4394–4406.
45. RoukensMG, Alloul-RamdhaniM, BaanB, KobayashiK, Peterson-MaduroJ, et al. (2010) Control of endothelial sprouting by a Tel-CtBP complex. Nat Cell Biol 12: 933–942.
46. HockH, MeadeE, MedeirosS, SchindlerJW, ValkPJ, et al. (2004) Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev 18: 2336–2341.
47. LaiZC, RubinGM (1992) Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell 70: 609–620.
48. RebayI, RubinGM (1995) Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81: 857–866.
49. NiJQ, ZhouR, CzechB, LiuLP, HolderbaumL, et al. (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8: 405–407.
50. CoxDR (1972) Regression models and life-tables. J R Stat Soc Ser B Stat Methodol 34: 187–220.
51. O'NeillEM, RebayI, TjianR, RubinGM (1994) The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78: 137–147.
52. BrunnerD, DuckerK, OellersN, HafenE, ScholzH, et al. (1994) The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature 370: 386–389.
53. HalfonMS, CarmenaA, GisselbrechtS, SackersonCM, JimenezF, et al. (2000) Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103: 63–74.
54. ReraM, BahadoraniS, ChoJ, KoehlerCL, UlgheraitM, et al. (2011) Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab 14: 623–634.
55. BuchonN, OsmanD, DavidFP, FangHY, BoqueteJP, et al. (2013) Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep 3: 1725–1738.
56. MarianesA, SpradlingAC (2013) Physiological and stem cell compartmentalization within the Drosophila midgut. Elife 2: e00886.
57. NegreN, BrownCD, MaL, BristowCA, MillerSW, et al. (2011) A cis-regulatory map of the Drosophila genome. Nature 471: 527–531.
58. WebberJL, ZhangJ, CoteL, VivekanandP, NiX, et al. (2013) The relationship between long-range chromatin occupancy and polymerization of the Drosophila ETS family transcriptional repressor Yan. Genetics 193: 633–649.
59. ReraM, ClarkRI, WalkerDW (2012) Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A 109: 21528–21533.
60. ZhangW, ThompsonBJ, HietakangasV, CohenSM (2011) MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet 7: e1002429.
61. BrunetA, BonniA, ZigmondMJ, LinMZ, JuoP, et al. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868.
62. RothemundS, LiouYC, DaviesPL, KrauseE, SonnichsenFD (1999) A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands. Structure 7: 1325–1332.
63. GrahamLA, DaviesPL (2002) The odorant-binding proteins of Drosophila melanogaster: annotation and characterization of a divergent gene family. Gene 292: 43–55.
64. GronkeS, ClarkeD-F, BroughtonS, AndrewsTD, PartridgeL (2010) Molecular evolution and functional characterisation of Drosophila insulin-like peptides. PLoS Genet 6: e1000857.
65. BroughtonSJ, PiperMD, IkeyaT, BassTM, JacobsonJ, et al. (2005) Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A 102: 3105–3110.
66. LemmonMA, SchlessingerJ (2010) Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134.
67. RoggeR, GreenPJ, UranoJ, Horn-SabanS, MlodzikM, et al. (1995) The role of yan in mediating the choice between cell division and differentiation. Development 121: 3947–3958.
68. FontanaL, PartridgeL, LongoVD (2010) Extending healthy life span–from yeast to humans. Science 328: 321–326.
69. GudbjartssonDF, WaltersGB, ThorleifssonG, StefanssonH, HalldorssonBV, et al. (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40: 609–615.
70. Lango AllenH, EstradaK, LettreG, BerndtSI, WeedonMN, et al. (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838.
71. FujiiS, AmreinH (2002) Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J 21: 5353–5363.
72. RoignantJY, CarreC, MugatB, SzymczakD, LepesantJA, et al. (2003) Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9: 299–308.
73. BassTM, GrandisonRC, WongR, MartinezP, PartridgeL, et al. (2007) Optimization of dietary restriction protocols in Drosophila. J Gerontol A Biol Sci Med Sci 62: 1071–1081.
74. AlicN, HoddinottMP, VintiG, PartridgeL (2011) Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell 10: 137–147.
75. BroughtonS, AlicN, SlackC, BassT, IkeyaT, et al. (2008) Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS One 3: e3721.
76. ChoksiSP, SouthallTD, BossingT, EdoffK, de WitE, et al. (2006) Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell 11: 775–789.
77. SmythGK, SpeedT (2003) Normalization of cDNA microarray data. Methods 31: 265–273.
78. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
79. ToedlingJ, SkylarO, KruegerT, FischerJJ, SperlingS, et al. (2007) Ringo–an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8: 221.
80. ZacherB, KuanPF, TreschA (2010) Starr: Simple Tiling ARRay analysis of Affymetrix ChIP-chip data. BMC Bioinformatics 11: 194.
81. BickelP, BoleyN, BrownJ, HuangH, ZhangN (2010) Subsampling Methods for Genomic Inference. Ann Appl Stat 4: 1660–1697.
82. BolstadBM, IrizarryRA, AstrandM, SpeedTP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193.
83. IrizarryRA, BolstadBM, CollinF, CopeLM, HobbsB, et al. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15.
84. IrizarryRA, HobbsB, CollinF, Beazer-BarclayYD, AntonellisKJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264.
85. BenjaminiY, HochbergY (1995) Controlling the False Discovery Rate: A practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Stat Methodol 57: 289–300.
86. Huang daW, ShermanBT, LempickiRA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57.
87. MusselC, HopfensitzM, KestlerHA (2010) BoolNet–an R package for generation, reconstruction and analysis of Boolean networks. Bioinformatics 26: 1378–1380.
Štítky
Genetika Reprodukčná medicínaČlánok vyšiel v časopise
PLOS Genetics
2014 Číslo 9
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
Najčítanejšie v tomto čísle
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
