-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Short Day–Mediated Cessation of Growth Requires the Downregulation of AINTEGUMENTALIKE1 Transcription Factor in Hybrid Aspen
Day length is a key environmental cue regulating the timing of major developmental transitions in plants. For example, in perennial plants such as the long-lived trees of the boreal forest, exposure to short days (SD) leads to the termination of meristem activity and bud set (referred to as growth cessation). The mechanism underlying SD–mediated induction of growth cessation is poorly understood. Here we show that the AIL1-AIL4 (AINTEGUMENTALIKE) transcription factors of the AP2 family are the downstream targets of the SD signal in the regulation of growth cessation response in hybrid aspen trees. AIL1 is expressed in the shoot apical meristem and leaf primordia, and exposure to SD signal downregulates AIL1 expression. Downregulation of AIL gene expression by SDs is altered in transgenic hybrid aspen plants that are defective in SD perception and/or response, e.g. PHYA or FT overexpressors. Importantly, SD–mediated regulation of growth cessation response is also affected by overexpression or downregulation of AIL gene expression. AIL1 protein can interact with the promoter of the key cell cycle genes, e.g. CYCD3.2, and downregulation of the expression of D-type cyclins after SD treatment is prevented by AIL1 overexpression. These data reveal that execution of SD–mediated growth cessation response requires the downregulation of AIL gene expression. Thus, while early acting components like PHYA and the CO/FT regulon are conserved in day-length regulation of flowering time and growth cessation between annual and perennial plants, signaling pathways downstream of SD perception diverge, with AIL transcription factors being novel targets of the CO/FT regulon connecting the perception of SD signal to the regulation of meristem activity.
Published in the journal: Short Day–Mediated Cessation of Growth Requires the Downregulation of AINTEGUMENTALIKE1 Transcription Factor in Hybrid Aspen. PLoS Genet 7(11): e32767. doi:10.1371/journal.pgen.1002361
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002361Summary
Day length is a key environmental cue regulating the timing of major developmental transitions in plants. For example, in perennial plants such as the long-lived trees of the boreal forest, exposure to short days (SD) leads to the termination of meristem activity and bud set (referred to as growth cessation). The mechanism underlying SD–mediated induction of growth cessation is poorly understood. Here we show that the AIL1-AIL4 (AINTEGUMENTALIKE) transcription factors of the AP2 family are the downstream targets of the SD signal in the regulation of growth cessation response in hybrid aspen trees. AIL1 is expressed in the shoot apical meristem and leaf primordia, and exposure to SD signal downregulates AIL1 expression. Downregulation of AIL gene expression by SDs is altered in transgenic hybrid aspen plants that are defective in SD perception and/or response, e.g. PHYA or FT overexpressors. Importantly, SD–mediated regulation of growth cessation response is also affected by overexpression or downregulation of AIL gene expression. AIL1 protein can interact with the promoter of the key cell cycle genes, e.g. CYCD3.2, and downregulation of the expression of D-type cyclins after SD treatment is prevented by AIL1 overexpression. These data reveal that execution of SD–mediated growth cessation response requires the downregulation of AIL gene expression. Thus, while early acting components like PHYA and the CO/FT regulon are conserved in day-length regulation of flowering time and growth cessation between annual and perennial plants, signaling pathways downstream of SD perception diverge, with AIL transcription factors being novel targets of the CO/FT regulon connecting the perception of SD signal to the regulation of meristem activity.
Introduction
The ability to adapt to changes in the environment is crucial to the survival of both animals and plants. Plants, unlike animals, are sessile organisms and have therefore evolved highly sophisticated mechanisms to anticipate seasonal changes and modulate their patterns of growth and development. Day length is one of the key environmental cues utilised by plants to anticipate seasonal changes and regulates several key developmental transitions associated with plant adaptation and reproduction. One of the most fascinating examples of this is provided by perennial plants, e.g. the long-lived trees of the boreal forest, in which the day length signal regulates the developmental transition from active growth to a more resilient dormant state prior to the onset of winter [1]. These perennial plants anticipate the approach of winter by detecting the reduction in day length (i.e. the short day signal, or SD signal) in the autumn and when the day length falls below the critical day length required for the promotion of growth, cell division in the meristems ceases [2]. The most visible indicator of short day–induced growth cessation is the formation of a bud that encloses the apical meristem and leaf primordia [3]. The importance of day length sensing for the survival of perennial plants is illustrated by the increased mortality due to delayed growth cessation in transgenic hybrid aspen plants that are unable to sense reductions in day length [4].
Intriguingly, there are numerous similarities at the regulatory level between day length mediated control of growth cessation in perennial plants and one of the most well studied developmental transitions in plants - the transition from vegetative growth to floral development. For example, key flowering time regulators such as the CONSTANS (CO), FLOWERING LOCUS T (FT) and the group of photoreceptors known as PHYTOCHROMES (PHYs) that are involved in day length mediated regulation of flowering time regulation in Arabidopsis [5], [6], [7], [8], are all also involved in SD–induced growth cessation in trees [4], [9], [10], [11]. In poplar species two closely related orthologs of FT (FT1 and FT2) have been found and recent analysis in hybrid aspen clone 353 indicates that FT2 could be primarily involved in SD–mediated growth cessation whereas FT1 is primarily involved in flowering [11]. In hybrid aspen (clone T89, used in this study), it has been shown that short day mediated downregulation of FT gene expression (FT1 and FT2) triggers the induction of growth cessation whereas overexpression of FT1 eliminates the plants' ability to respond to the SD signal and thus prevents timely growth cessation [9]. Both CO and PHYTOCHROME A (PHYA) act upstream of the FT genes in SD-mediated induction of growth cessation in much the same manner as in the flowering transition in Arabidopsis [9]. Subjecting aspen trees to conditions in which the peak of CO expression occurs in the dark (e.g. under SD conditions) brings about rapid downregulation of FT2 expression leading to the induction of growth cessation response [9]. These findings indicate evolutionary conservation of the day length response pathway between annual plants such as Arabidopsis and perennial trees such as hybrid aspen.
Despite the evolutionary conservation of early acting components involved in day length regulated growth cessation and flowering time, considerable lacunae remain in our understanding of SD mediated regulation of growth cessation at the molecular level. Particularly, the factors targeted by SD signal downstream of the early acting components such as PHYA and the CO/FT regulon in regulating growth cessation responses remain unknown. The critical role of these hitherto unknown downstream targets of SD signal has become evident from the analysis of growth cessation response in hybrid poplar where they have been shown to be important for regulating the variation in timing of growth cessation responses [12]. Thus a key question that remains unanswered is; How does SD mediated downregulation of FT2 expression lead to the induction of growth cessation response? Answering this question would require the identification of targets of SD signal downstream of the CO/FT regulon in trees and elucidating their role in the regulation of growth cessation responses. The network of genes involved in day length regulation of the floral transition is well defined, and downstream targets of the CO/FT pathway like SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1) and floral meristem identity genes FRUITFUL (FUL) and APETALA1 (AP1) are known (reviewed in [13]). In contrast the targets of the SD signal downstream of the CO/FT module in the growth cessation response can not simply be deduced from extrapolation of knowledge of floral transition related genes given the difference between the growth cessation process and floral transition.
To identify the downstream targets of the SD signal in growth cessation we have previously analysed global transcriptional changes associated with this process [14], [15]. One of the genes whose transcript is strongly downregulated during growth cessation is a Populus homolog of the Arabidopsis gene AINTEGUMENTA (ANT)[16]; the Populus homolog is henceforth referred to as AINTEGUMENTALIKE1 (AIL1). The expression data along with a proposed role of ANT in the regulation of cell cycle [17], [18], [19] suggested that AIL1 (and most likely the other closely related members AIL2-AIL4 of this sub-family) is a potential downstream target of the SD signal transduced via the CO/FT module in the regulation of growth cessation response in perennial trees. We tested this hypothesis by investigating the regulation of AIL1 expression by SD signal in transgenic hybrid aspen plants that are perturbed in the SD response. In a complementary approach we investigated the short day mediated regulation of growth cessation response in transgenic hybrid aspen plants that either maintain high levels of AIL1 or AIL3 expression even after SD treatment or have reduced expression of AIL1. Finally we identified downstream targets of the AIL1 transcription factor in the apex. Taken together, these analyses show that AIL genes are targets of the SD signal downstream of the CO/FT module and their down-regulation is necessary for short day regulated growth cessation in hybrid aspen plants.
Results
Downregulation of the expression of the AINTEGUMENTALIKE (AIL) gene family coincides with the cessation of growth and bud set following SD treatment in Populus
The Populus genome contains 13 genes belonging to the ANT-subgroup of the AP2 transcription factor family [20]. Four of these genes are here designated as AIL1-AIL4 (AINTEGUMENTALIKE 1-4) as they belong to the same clade as the Arabidopsis ANT transcription factor (Figure S1). We investigated the expression of AIL1 as well as the expression of the related genes AIL2-AIL4 in the apex of hybrid aspen plants after SD treatment (Figure 1A and Figure S2). RT-PCR data indicates that AIL1 (Figure 1A) as well as AIL2-AIL4 expression (Figure S2) are downregulated along with that of cell cycle markers CYCD3 : 2 and CYCD6 : 1 after SD treatment (Figure 1B, panels B and C) and this downregulation coincides with the cessation of growth and bud set in the apex of hybrid aspen T89 trees [21].
Fig. 1. Expression of AIL1 and D-type cyclins in the apex during short day treatment. 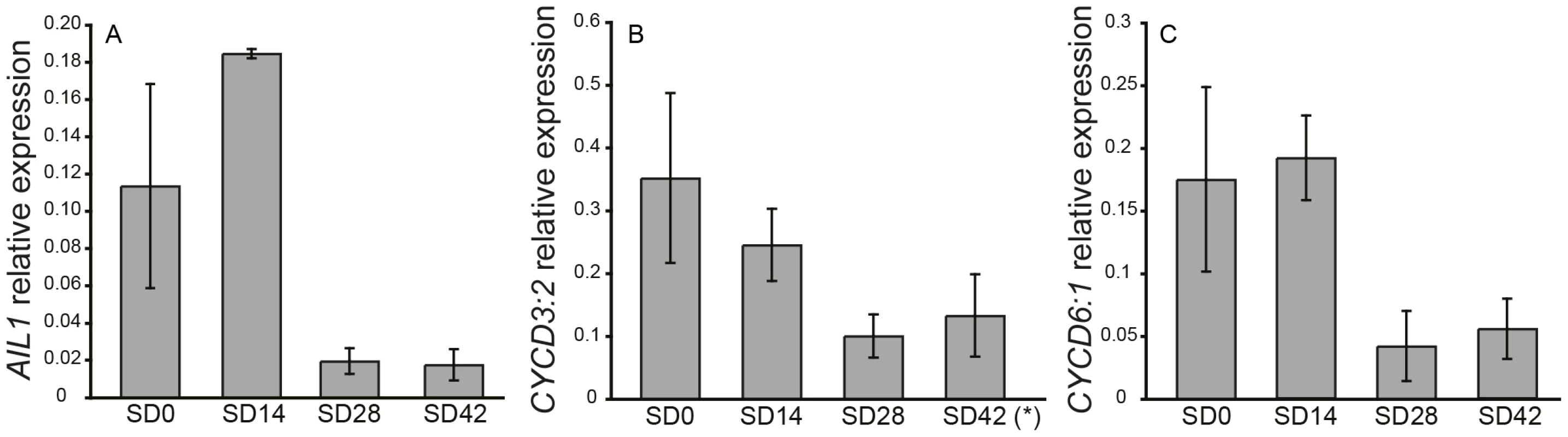
Expression of (A) AIL1, (B) CYCD3:2* and (C) CYCD6:1 in the apex of wild type hybrid aspen analyzed by real-time PCR after 0 (0SD), 14 (14SD), 28 (28SD) and 42 (42SD) days of short day treatment (8 h day). X-axis indicates time in short days and Y-axis indicates the ratio of gene expression relative to that of the reference gene (UBQ). (Error bars = Standard deviation of three biological replicates). *one sample excluded due to poor specificity of amplification based on melt curve analysis. Downregulation of AIL gene expression is perturbed in hybrid aspen plants with altered SD response
We then investigated the effect of perturbed SD perception or response on the regulation of AIL gene expression after SD treatment. For this we used transgenic hybrid aspen that are unable to respond to the SD signal due to overexpression of PHYA or FT1 cDNA as well as plants that are hypersensitive to the SD signal and undergo premature growth cessation due to the downregulation of FT expression (FTRNAi) [4], [9]. Since all 4 AIL genes are highly similar and displayed similar expression pattern after SD treatment, we chose to perform detailed analysis of AIL1 regulation. RT-PCR analysis indicated that the downregulation of AIL1 expression after SD treatment is severely attenuated in the apex of PHYA and FT1 overexpressors in contrast with the wild type (Figure 2A). In contrast the FTRNAi plants that respond more rapidly to SD treatment than wild type [9] display a stronger and earlier reduction in the expression of AIL1 (Figure 2B). These results strongly suggest that AIL1 expression is a potential downstream target of the SD signal transduced via the CO/FT module in cessation of growth and bud set in the apex of hybrid aspen.
Fig. 2. Expression of AIL1 in the apex of transgenic lines with aberrant SD response. 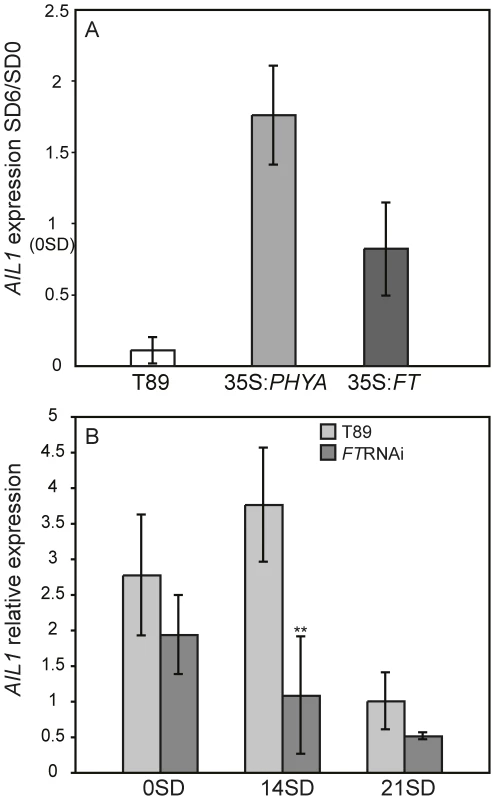
(A) AIL1 expression in the apex of wild type hybrid aspen (T89), transgenic hybrid aspen overexpressing PHYA (35S:PHYA) [4] or FT1 (35S:FT1) [9] after 6 weeks of SD treatment (8 h day). Expression levels (represented on Y-axis) for each genotype are normalised against their expression at SD0 (prior to the start of short day treatment). (Error bars = standard deviation of three separate experiments). (B) AIL1 expression in the apex of wild type (T89) and FTRNAi [9] during the first three weeks in SD (8 h day). After two weeks (SD14) AIL1 expression is significantly lower in FTRNAi compared to that in the wild type (T89) (p<0.05). X-axis indicates time in short days (SD) and Y-axis indicates the ratio of candidate gene expression relative to the average expression of two reference genes (UBQ and TIP41-like). (Error bars = Standard deviation of three biological replicates). AIL1 expression in hybrid aspen apex is confined to zones of actively dividing cells
We further investigated expression of AIL1 in different tissues and found that AIL1 is primarily expressed in the apical region of hybrid aspen (Figure 3A). Since the downregulation of AIL1 gene expression was closely associated with cessation of growth and bud set, we analysed the domain of AIL1 gene expression in the apex. For this analysis we generated transgenic hybrid aspen expressing a transcriptional fusion between 2.5 Kb upstream sequence of AIL1 gene from Populus trichocarpa and the uidA (b-glucoronidase/GUS) reporter gene. In the transgenic hybrid aspen, the reporter gene expression was mostly confined to the zone of dividing cells in the apex, the provascular tissues and the leaf primordia (Figure 3B). The expression pattern of pAIL1:UidA reporter construct correlates well with the previously described expression pattern of CYCA1 which serves as a marker of dividing cells in the apex of hybrid poplar plants [22] indicating that AIL1 expression is associated primarily with cell proliferation.
Fig. 3. Analysis of tissue-specific AIL1 expression pattern. 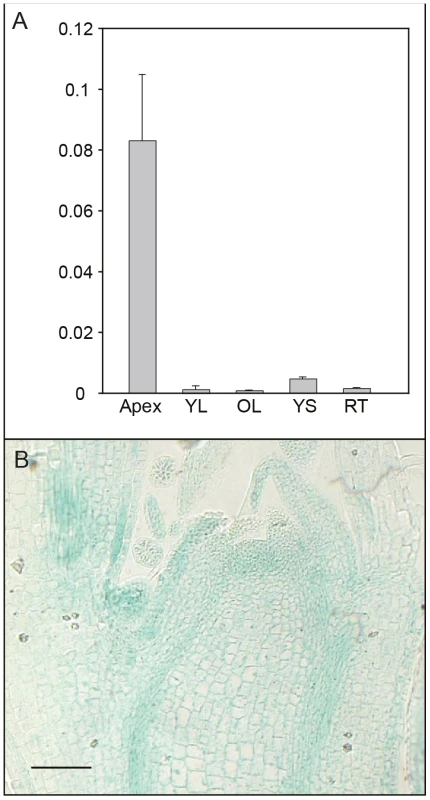
(A) AIL1 expression across the different tissues; apex, young leaf (YL), old leaf (OL), young stem (YS), old stem (OS) and root tip (RT) in tissue culture grown wild type hybrid aspen. All expression values (on Y-axis) are ratios of AIL1 expression relative to the reference (UBQ). (Error bars = Standard deviation of three biological replicates) (B) GUS activity in actively growing hybrid aspen apex expressing the uidA (GUS) gene under control of the AIL1 promoter. Perturbing AIL gene expression affects SD–mediated regulation of growth cessation response
Analysis of AIL1 gene expression suggested that its downregulation could be important for the SD mediated cessation of growth and bud set. We tested this hypothesis by generating transgenic hybrid aspen that would maintain high levels of AIL1 expression even after SD treatment in contrast with the wild type by expressing AIL1 cDNA under the control of the 35S promoter (these transgenic lines are henceforth referred to as AIL1oe lines). Several independent lines were obtained and tested for high expression of AIL1 (Figure S3A shows data for two chosen lines); two lines were chosen for detailed analysis of their response to SD treatment. Unlike the wild type plants that undergo growth cessation and form an apical bud after 6 week of SD treatment, the apices of AIL1oe fail to undergo proper growth cessation and bud set after 6 weeks of SD treatment (Figure 4A–4C). We also generated transgenic hybrid aspen overexpressing AIL3 (Figure S3) to investigate whether other members of the gene family share the same function (these transgenic lines are henceforth referred to as AIL3oe lines). AIL3oe display a similar phenotype to AIL1oe during SD treatment (Figure 4D–4F).
Fig. 4. Bud formation in wild type, AIL1 (AIL1oe), and AIL3 (AIL3oe) overexpressing transgenic hybrid aspen plants after 6 weeks in short days. 
Six weeks of SD-treatment (8 h light) leads to bud formation in wild type T89 (A), whereas no bud set is observed in AIL1oe line 2B or 3B (B and C). Similarly in SD treatment consisting of 14 hours day length, bud formation is observed after 6 weeks SD-treatment in apices of wild type T89 (D) whereas no bud set is observed in AIL3oe line 9 (E) or AIL3oe line 10 (F). Position of the apex is indicated by white arrows. We also investigated the effect of downregulation of AIL gene expression on SD mediated bud set. The functional redundancy between AIL genes suggested by similar regulation and effect on bud set in AIL1oe and AIL3oe plants lead us to generate transgenic hybrid aspen plants in which the expression of all 4 AIL genes was targeted for downregulation using artificial microRNA (amiRNA). Two amiRNA constructs (255 and 256) were expressed in hybrid aspen and two lines (255-6 and 256-23) with reduced expression of AIL1 gene expression (Figure S4) were selected for further analysis of growth cessation response. The transition from active growth to bud set after SD treatment in the wild type and lines 255-6 and 256-23 was investigated using a method separating different stages of bud development [23]. Compared to the wild type, the lines 255-6 and 256-23 displayed a more rapid transition from active growth to bud set and a majority of the plants in the two transgenic lines made the transition to intermediate and late stage of bud set at least a week (or more) earlier than the wild type plants after SD treatment (Figure S5). This data along with the perturbed growth cessation response in AIL1oe and AIL3oe plants indicate that downregulation of AIL gene expression is necessary for SD mediated growth cessation response.
AIL1 is the downstream target of SD signal in the activation of growth cessation responses
The altered growth cessation responses in AIL1oe plants could either be due to the failure of these plants to perceive the short day signal or to a failure in properly responding to it. To distinguish between these two possibilities we compared the response of FT2 expression to SD treatment in the leaves of wild type and AIL1oe. Downregulation of FT2 expression in the leaves is the earliest known marker for the detection of the SD signal and recent results have implicated its downregulation in SD mediated growth cessation [9], [11], [12]. Our data show that both the wild type (Figure 5A) and AIL1oe lines (Figure 5B) exhibit similar decreases in their levels of FT2 transcripts following SD treatment. This result indicates that unlike FT genes, AIL1 does not act early in SD response but is rather a downstream target of the SD signal.
Fig. 5. FT2 expression is rapidly downregulated in wild type (T89) and AIL1oe after transfer to short days. 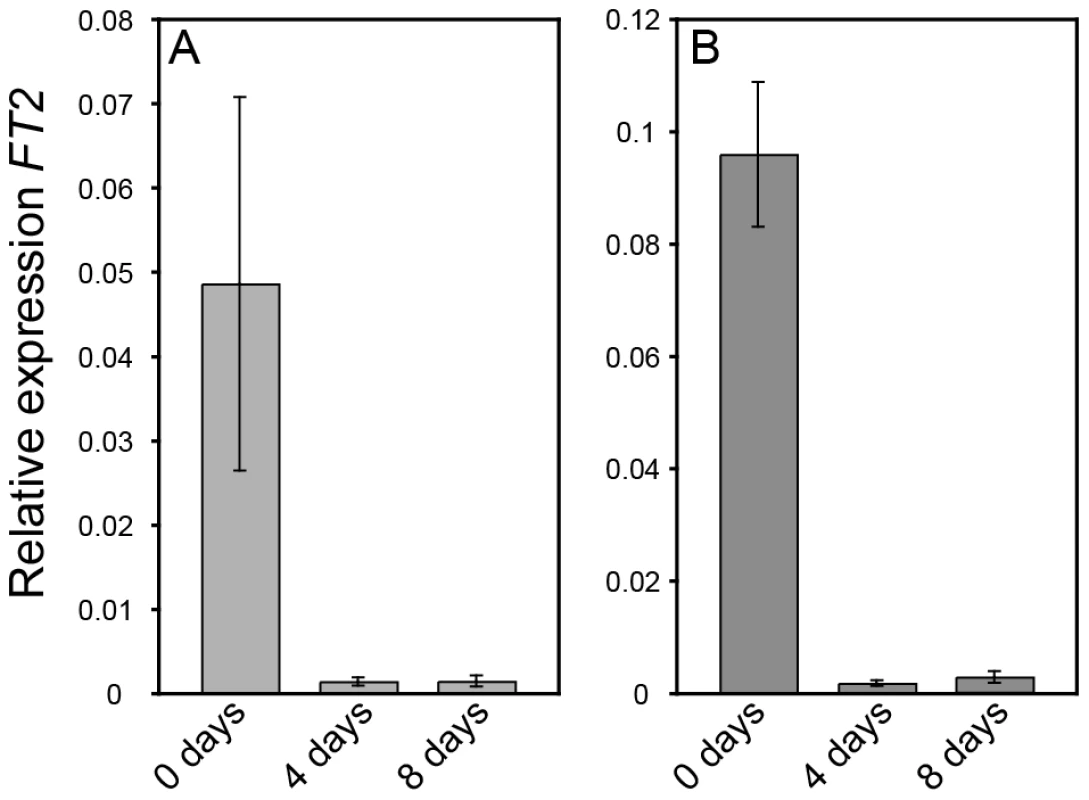
RT-PCR analysis of FT2 transcript level in the leaves of the wild type (A) and AIL1oe (B) after 0, 4 and 8 short days (8h day). Y-axis indicates the ratio of FT2 expression relative to that of the reference gene (UBQ) and time in short days is shown on the X-axis. (Error bars: Standard deviation of three biological replicates.) AIL1 is a potential regulator of D-type cyclin gene expression
The expression of several cell proliferation related genes, e. g. D-type cyclins, that are key cell cycle regulators [24], [25], [26], [27] is downregulated in a similar manner to AIL genes during SD mediated cessation of growth in hybrid aspen [15], Figure 1B, 1C). We therefore investigated whether the AIL1 transcription factor could be involved in the regulation of the D-type cyclin genes and if so whether their expression is perturbed in AIL1oe lines after SD treatment. Therefore we analysed the expression of two D-type cyclins, CYCD3 : 2 and CYCD6 : 1 after SD treatment in AIL1oe plants after 6 weeks of SD treatment. Our RT-PCR data (Figure 6) showed that while the expression of CYCD3 : 2 and CYCD6 : 1 is downregulated in the wild type after 6 weeks of SD treatment, this was not the case in the AIL1oe plants. This result indicates that AIL1 could be involved in the regulation of D-type cyclins and the failure to downregulate AIL1 expression after SD treatment leads to a corresponding failure to downregulate the expression of these key cell cycle regulators.
Fig. 6. Differential regulation of D-type cyclin expression in the AIL1oe lines compared to the wild type (T89) after SD-treatment. 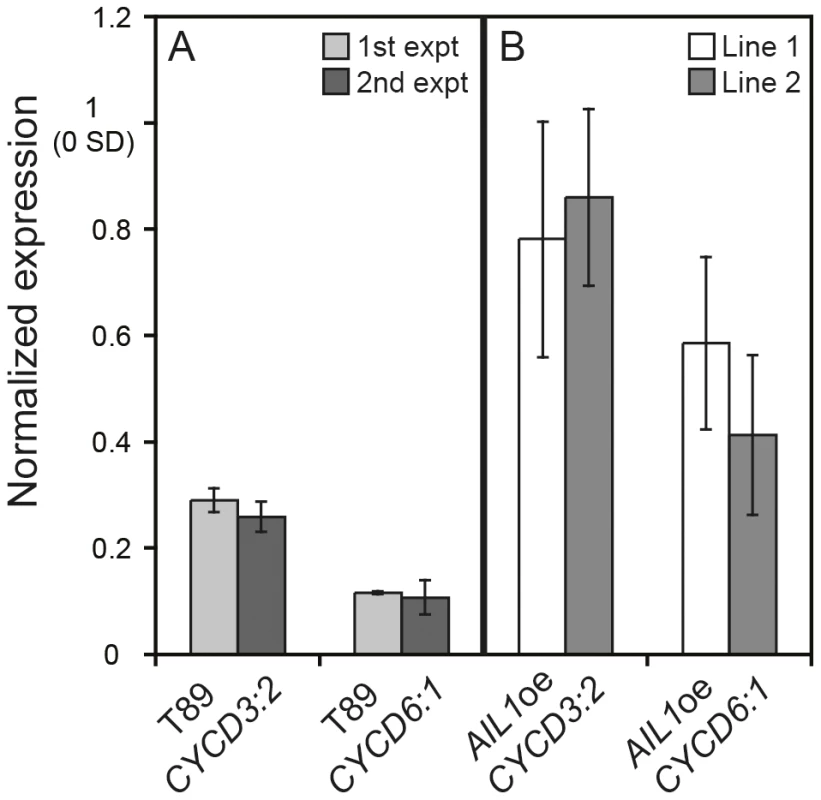
(A) Expression of CYCD3:2 and CYCD6:1 in T89 after 6 weeks SD (8 h day). Results from two separate experiments are shown. All expression values normalised to expression at 0 SD. (Error bars: SD of three technical replicates). (B) Expression of CYCD3:2 and CYCD6:1 in AIL1oe after 6 weeks SD. Results from two lines are shown. Expression values normalised to expression at 0 SD. (Error bars = Standard deviation of three biological replicates). AIL1 transcription factor can interact with D-type cyclin promoters
The observation that CYCD3 : 2 and CYCD6 : 1 expression after SD treatment is perturbed in AIL1oe plants prompted us to investigate whether the AIL1 transcription factor can interact with CYCD promoters from hybrid aspen using electrophoretic mobility shift assays (EMSA). We expressed HA-tagged AIL1 protein in Arabidopsis protoplasts and used the extracts in gel shift assays using 3 different different fragments from a hybrid aspen CYCD3 : 2 promoter (results from two fragments are shown here). Our data show that extracts containing AIL1 protein specifically display a gel shift with the promoter fragment consisting of 200 bp of sequence situated upstream of the start codon of CYCD3 : 2 (Figure 7). Together with the CYCD3 : 2 and CYCD6 : 1 gene expression data, the gel-shift analysis strongly suggests these cyclin genes might be potential downstream targets of the AIL1 transcription factor in hybrid aspen.
Fig. 7. Interaction of HA-tagged AIL1 with CYCD 3:2 promoter fragment by electrophoretic mobility shift assay (EMSA). 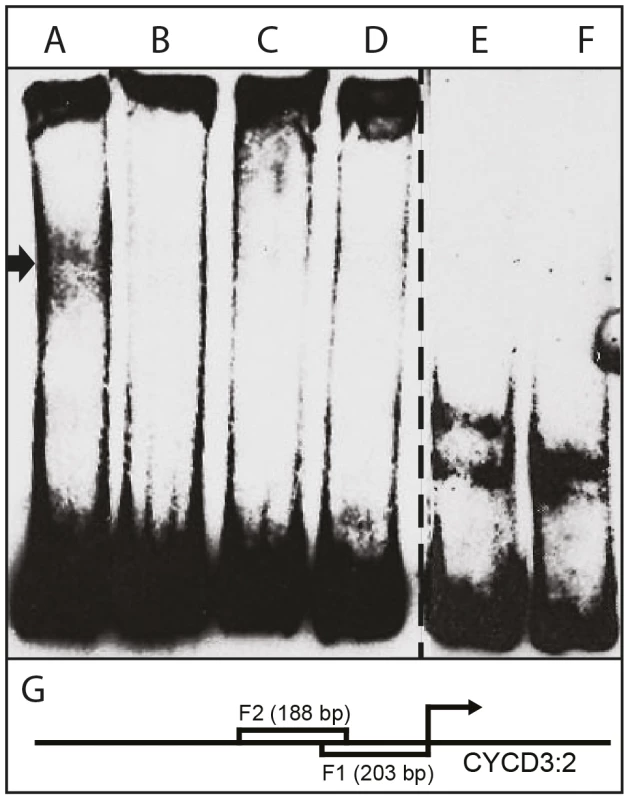
(A) AIL1 + 5 fmol biotin- labeled CYCD 3:2 promoter fragment F1, (B) AIL1 + biotin- labeled CYCD 3:2 promoter fragment F1+ 500 fmol unlabeled F1, (C) AIL1 + 5 fmol biotin- labeled CYCD 3:2 promoter fragment F2, (D) AIL1 + Biotin- labeled CYCD 3:2 promoter fragment F2+ 500 fmol unlabeled F2, (E) biotin-labeled F1, (F) biotin-labeled F2 and (G) F1 and F2 in relation to the start codon of CYCD 3:2. Lanes A-D have additional non-specific competitor added to the reaction mix. Cell extracts from protoplast expressing HA-tagged AIL1 were used for gel-shift analysis. Additional lanes between D and F on the gel are omitted from the picture. Discussion
Short day mediated cessation of growth and budset prior to the onset of winter is a key developmental transition that is critical to the survival of perennial plants in boreal forest [1]. In this work, we identify AIL genes belonging to the AP2 transcription factor family as downstream targets of the SD signal transduced via the CO/FT module and that downregulation of their expression is necessary for cessation of growth and bud set in hybrid aspen.
In poplar, there are 4 closely related AIL genes and our data indicates that all four AIL genes could have similar function at least in SD mediated growth cessation response as suggested by the similar phenotypes of AIL1 and AIL3 overexpressing plants as well as in plants in which these genes are targeted for downregulation. However we cannot exclude the possibility that there could be functional differences between the different AIL genes with respect to other biological processes as we have not been able to specifically downregulate individual genes of the AIL family and study the effect on growth and development so far. It is particularly important to note in this respect that even closely related genes can diverge both in expression profiles and at a functional level as suggested by the careful analysis of FT1 and FT2 in poplar species which indicate that FT1 could be primarily involved in reproductive growth whereas FT2 controls growth cessation [11].
Several observations suggest that the downregulation of AIL gene expression following SD treatment is necessary for the activation of growth cessation responses. AIL1 (and most likely other genes of this family as well) is primarily expressed in dividing and meristematic cells in hybrid aspen (Figure 3) and the downregulation of their expression coincides temporally with the SD-mediated induction of growth cessation responses in hybrid aspen (Figure 1 and Figure 4), including the termination of elongation growth, bud set and the downregulation of core cell cycle genes such as the D-type cyclins (Figure 1). Furthermore, AIL1 downregulation after SD treatment is attenuated in FT or PHYA overexpressors (Figure 2) that fail to respond properly to SD treatment [4], [9]. Importantly, growth cessation response is perturbed when AIL1 or AIL3 expression is maintained at high levels even after SD treatment and earlier bud set is observed in transgenic hybrid aspen with reduced expression of AIL1 (Figure 4 and Figure S5). All of these results are consistent with the AIL genes being downstream targets of the SD signal in the control of the growth cessation response.
Three hypotheses can be proposed to explain the role of AIL genes in SD mediated control of growth cessation response and why growth cessation response is perturbed when the AIL1 or AIL3 expression is maintained at high level even after SD treatment as in AIL1oe and AIL3oe lines. Firstly, the AIL genes could act upstream of FT2, in which case the increased expression of AIL1 as in AIL1oe could counteract the downregulation of FT2 by the SD signal. However, this hypothesis is incompatible with the observation that the downregulation of FT2 subsequent to SD treatment proceeds as normal in AIL1oe lines (Figure 5). Alternatively, the AIL genes could act independently of FT2 and their increased expression in AIL1oe and AIL3oe could prevent the downregulation of the targets of FT2 following SD treatment. Alternatively, the AIL genes are the targets of SD signal downstream of CO/FT regulon leading to their downregulation after SD treatment. Our results support the latter hypothesis because SD treatment results in the downregulation of the AIL gene expression and this downregulation of AIL gene expression is severely attenuated in plants that overexpress FT1. Further evidence for the connection between the CO/FT regulon and AIL1 expression was obtained by analysis of FTRNAi lines that respond more rapidly than the wild type to SDs and in these lines, AIL1 expression is significantly reduced compared to wild type after 2 weeks of SD treatment (Figure 2B). Finally the downregulation of the AIL1 expression leads to earlier transition from active growth to bud set strongly suggesting that the AIL genes are the downstream targets of the SD signal. Thus our results suggest a mechanism in which AIL genes act downstream of the CO/FT regulon and that downregulation of AIL gene expression culminates in growth cessation and bud set after SD treatment.
FT has been shown to act as a transcriptional co-regulator in Arabidopsis [28]. In poplar species, two FT genes are present of which FT2 is rapidly downregulated after SD treatment; thus FT2 could either directly regulate AIL at the transcriptional level in hybrid aspen or alternatively downstream targets of FT2 could regulate AIL gene expression. Our data supports the latter suggestion because the kinetics of downregulation of FT2 and AIL gene expression subsequent to SD treatment is not consistent with direct regulation of AIL gene expression by FT2. While FT2 is typically downregulated within 3–7 days in the leaves after the commencement of SD treatment ([9], [12], Figure 5), it takes 2–3 weeks until downregulation of the AIL genes becomes apparent in the apex (Figure 1). Moreover, induction of FT2 in the leaves has little effect on expression of most of AIL genes [11], which might again suggest an indirect regulation of AIL expression by FT2. Thus these results suggest that there may be one or more genes that are direct targets of FT2 and act upstream of the AIL genes regulating their expression in the apex. Determining the identity of these targets of FT2 and regulators of AIL expression in the apex is an important objective for future research in this area. The downstream targets of FT in daylength mediated regulation of flowering time such as SOC1 and the floral meristem identity genes FUL and AP1 [13] are well known. However these are unlikely to be the targets of the CO/FT regulon in the regulation of the AIL genes in the apex unless the tree homologs of these genes have acquired novel functions and have been recruited to regulate meristem activity by controlling AIL gene expression.
AIL1 is expressed in dividing cells (Figure 2), can potentially interact with the promoters of D-type cyclins (Figure 7) and maintaining high level of AIL expression prevents the downregulation of D-type cyclin expression after SD treatment (Figure 6) suggesting that AIL1 has a role in regulation of key cell cycle regulators. Indeed, data from Arabidopsis also shows that the putative AIL1 ortholog ANT can positively regulate cell division as its overexpression leads to increased duration of cell division [17]. We therefore propose that the downregulation of AIL gene expression after SD treatment leads to the downregulation of a subset of D-type cyclins such as CYCD3 : 2 and CYCD6 : 1. The downregulation of the expression of core cell cycle regulators such as the abovementioned cyclins would then culminate in cessation of growth and bud set. However it is unlikely that the D-type cyclins are the only targets of AIL1 because the expression of several other cell cycle genes is also downregulated after SD treatment [15]. Additionally transcriptional network analysis indicates that several other cell cycle genes might be regulated by the AIL1 transcription factor [29]. Moreover, preliminary investigations suggest that altering CYCD3 : 2 expression alone is not sufficient to activate the growth cessation response.
Substantial progress has recently been made in understanding how SD signal is perceived, and downregulation of FT2 expression after SD treatment has been identified as a key early event in the induction of growth cessation response [9], [11]. However, the components targeted by SD signal downstream of the CO/FT regulon in the induction of growth cessation response have remained elusive, especially factors that would link the downregulation of FT2 expression to cessation of growth. Indeed analyses of hybrid poplar clones that differ in timing of bud set have suggested an important role for such factors in differential growth cessation [12]. Our finding that the AIL genes are the targets of the SD signal that is transduced via the CO/FT module in growth cessation response and bud set therefore represents an important step in elucidating the mechanism underlying this key developmental transition in perennial plants as this links the CO/FT module to the regulation of cell cycle through the AIL genes in SD mediated cessation of growth and bud set.
The CO/FT module is an important component of the molecular machinery that allows plants to respond to changes in day length, and its role in day length mediated control of flowering time is well established [30]. Therefore it was not surprising that the same CO/FT module is also involved in controlling the timing of SD-mediated growth cessation in perennial trees, as this is another key developmental transition that is regulated by the day length signal. However, given that flowering and growth cessation processes are distinct morphologically, it appears unlikely that the downstream targets of this module in the regulation of flowering would be the same as those involved in the growth cessation response. Our findings suggest that the AIL transcription factors, which have the potential to regulate the expression of cell cycle genes, were co-opted at some point in evolutionary history to serve as mediators of the day length signal. This co-option would have allowed the versatile CO/FT module to regulate a novel developmental transition. These results demonstrate that an evolutionary “mix and match” strategy involving combining different regulatory modules can allow a small number of regulatory modules to control a wide range of diverse biological processes. In conclusion, our data demonstrates the divergence of the regulatory pathway downstream of the conserved CO/FT module between day length controlled floral transition and growth cessation response and identifies AIL1 as a potential regulator of cell cycle related genes and a novel target of the short day signal downstream of the CO/FT module in regulation of growth cessation in perennial trees.
Materials and Methods
Plant material and growth conditions
Cuttings of hybrid aspen (Populus tremula x tremuloides) clone T89 (wild type) and the transgenic lines were grown in half-strength Murashige/Skoog medium (½ MS) under sterile conditions for approximately 4 weeks and then transferred to soil. After four weeks in greenhouse the plants were moved to growth chambers (18 hour light/6 hour night, 20°C). After one week the chamber settings were shifted to short day conditions (8 hour light/16 hour night, 20°C or 14 hour light/10 hour night, 20°C). Growth cessation was determined by measurement of elongation growth and/or bud set. Pictures of apices to assess bud formation were taken using Canon EOS digital camera. For tissue specific expression analysis of AIL genes, samples were taken from tissue culture grown plants 4 weeks after cuttings were transferred to new media.
Identification of AIL-genes and phylogenetic analysis
AIL-genes were identified by blasting the Arabidopsis AINTEGUMENTA gene (AT4G37750) against the Populus genome. Gene models (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) were manually chosen based on intron-exon structure (JGI protein ID for each model can be found in Figure S1). Sequences were aligned and a bootstrapped phylogenetic tree generated using ClustalX [31]. The phylogenetic tree was visualised using TreeView (http://darwin.zoology.gla.ac.uk/~rpage/treeviewx/).
Generation of AIL1oe and AIL3oe lines
The full length cDNA for AIL1 transcription factor was cloned into the donor vector pDONR201 (Invitrogen.com) before transfer into the destination vector pK2GW7 [32]. The resulting vectors were introduced into agrobacterium GV3101pmp90RK [33] followed by the transformation of hybrid aspen clone T89 [34]. The same strategy was used to generate AIL3oe lines with the exception of entry clone construction that in this case was performed using the pENTR/D-TOPO cloning kit (Invitrogen.com).
GUS promoter analysis
The AIL1 promoter was amplified using the primers: FW: CACCCGGGGAATGATAGGCTGACAA and RP:CCCAAAATCTTGCCTACTTCC and cloned into the pENTR/D-TOPO vector (Invitrogen.com). The fragment was transferred into the pK2GWFS7 binary vector [32]. The construct was transformed into hybrid aspen using Agrobacterium mediated transformation as described before [34]. Apices from transgenic lines expressing the reporter gene were collected from greenhouse grown trees approx. 5 weeks after potting. The apices were incubated approx. 3 h at 37°C in GUS-solution (1 mm X-gluc, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, 50 mm sodium phosphate buffer (pH 7.0), and 0.1% (v/v) Triton X-100). The samples were then rinsed with water, dehydrated to 50% (v/v) ethanol, fixed for 10 min in FAA (5% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol), and cleared in 100% (v/v) ethanol. Once cleared, the samples were embedded in LR-White/10% PEG 400 resin in polypropylene capsules (TAAB) The apices were then sectioned on a Microm HM350 microtome (Microm International GmbH, Germany) at approx 20 µm, floated on water, heat-fixed to glass slides, mounted in Entellan neu (Merck, Germany) Sections were visualized with Zeiss Axioplan light microscope and captured with a digital camera, AxioCam together with the Axiovision 4.5 software (Zeiss, Germany).
RNA isolation and real-time PCR analysis
Total RNA from poplar apices was extracted using the Aurum Total RNA kit (Bio-Rad). Care was taken to collect tissue samples for RNA isolation at the same time of the day (usually between 13–16 PM) for each experiment. 100–500 ng of RNA was DNase treated with RNase free DNaseI (Fermenta) and used for cDNA synthesis using iScript cDNA synthesis kit (BioRad) or qScript cDNA synthesis kit (Quanta BioSciences). Reference genes were validated using GeNorm Software [35]. The reference gene chosen was UBQ in all experiments except for the analysis of the overexpression of AIL1 and AIL3 in the AIL1oe and AIL3oe lines, where 18S rRNA was used as the reference gene. Analysis of expression in FTRNAi used two reference genes, UBQ and TIP-41 like. SYBR green (Bio-Rad or Quanta BioSciences) was used as non-specific probe in all reactions and relative expression values were calculated using the Δ-ct-method [9]. A complete list of primers used in RT-PCR analysis can be found in Table S1.
Generation of transgenic hybrid aspen plants with reduced expression of AIL genes
To downregulate the expression of AIL genes, artificial microRNAs were designed using the online tools at http://wmd.weigelworld.org/cgi-bin/mirnatools. Briefly primers (Table S3) were used to generate artificial microRNAs directed against all the 4 AIL genes and cloned into the plant transformation vector pK2GW7 according to the cloning protocol at http://wmd3.weigelworld.org/. Two different miRNA contructs (named 255 and 256) were made and transformed into hybrid aspen clone T89 as described earlier. Following transformation several hybrid aspen lines with reduced expression of AIL1 were obtained and one line each for the two constructs 255 and 256 were selected for the analysis of bud set after SD treatment (lines 255-6 and 256-23).
Analysis of bud set in hybrid aspen plants with reduced expression of AIL genes
Bud set was scored using the method described by [23]. We used a score of 3 to indicate active growth (complete lack of bud set) and 0 to indicate a completely closed bud and score of 2 or 1 to indicate intermediate stages. For this analysis, bud set was scored every 7 days in a minimum of 5 or more plants for a period of 7 weeks.
Generation of HA-tagged AIL1 and overexpression in Arabidopsis protoplasts
AIL1 full length cDNA was amplified using the following primers: pttAIL1(EcoRI) FW - CATGGAATTCATGAAATCTACGGGTGATAA and pttAIL1(SalI) RP-CATGGTCGACTTCTCCTTTTCCTTGGTTCATGC. The resulting fragments were cloned into pRT104-3xHA [36]. Transfection into Arabidopsis protoplasts were performed as described [36], [37] using 8 µg of purified plasmid. Cells were lysed in a lysis buffer containing 25 mM Tris-HCL (pH 7.5), 50 mM KCl, 1 mM EDTA, 10% Glycerol 1 mM DTT, 0.1% Igepal and 1X PIC (Protease Inhibitor Cocktail). After centrifugation the supernatant was collected and immediately frozen in liquid nitrogen. The expression of the HA-tagged AIL1 protein was confirmed with western blot and resulting cell extracts were used for subsequent analysis.
Generation of labeled CycD3 : 2 promoter fragments
CycD3 : 2 promoter sequences were identified using the JGI populus genome database (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). Approx. 200 base pair fragments were amplified using primers specified in Table S2. The fragments were gel-purified using E.Z.N.A. Gel Purification Kit (Omega Bio-Tek) followed by phenol-chloroform extraction and ethanol precipitation prior to use in gel-shift assays. Five pmol of purified fragments were biotin labeled using the Biotin 3′ End DNA Labeling Kit (Pierce). Labeling and labeling efficiency determination was performed according to the manufacturers recommendation.
Electrophoretic mobility shift assay
The biotin-labelled promoter fragments were mixed with protoplast cell extracts containing AIL1-HA or control extracts from non-transfected protoplasts. For the binding reaction the following conditions were used: 10 µl protoplast cell extract, 0.5 µl biotin-labelled DNA (10 fmol/µl), 0.4 µl non-specific competitor (poly (dI:dC), 1 mg/ml), 0.5 µl BSA (20 mg/ml) and lysis buffer to a total of 20 µl. For specific competition, 500 fmol non-labelled fragment was added to the reaction. Binding was performed on ice for 10 min followed by 30 min in room temperature. The samples were run on a non-denaturing polyacrylamide gel (5%-0.5xTBE) and transferred to a Hybond N+ membrane (GE Healthcare, Sweden). Crosslinking and detection was performed using the LightShift Chemiluminescent EMSA kit (Pierce.com).
Supporting Information
Zdroje
1. RohdeABhaleraoRP 2007 Plant dormancy in the perennial context. Trends in Plant Science 12 217 223
2. NitschJP 1957 Growth responses of woody plants to photoperiodic stimuli. Proceedings of the American Society for Horticultural Science 70 512 525
3. RohdeA 2002 PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in Poplar. Plant Cell 14 1885 1901
4. OlsenJEJunttilaONilsenJErikssonMEMartinussenI 1997 Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant Journal 12 1339 1350
5. BagnallDJKingRWWhitelamGCBoylanMTWagnerD 1995 Flowering responses to altered expression of phytochrome in mutants and transgenic lines of Arabidopsis thaliana (L) Heynh. Plant Physiology 108 1495 1503
6. PutterillJRobsonFLeeKSimonRCouplandG 1995 The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell 80 847 857
7. KardailskyIShuklaVKAhnJHDagenaisNChristensenSK 1999 Activation tagging of the floral inducer FT. Science 286 1962 1965
8. KobayashiYKayaHGotoKIwabuchiMArakiT 1999 A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960 1962
9. BohleniusHHuangTCharbonnel-CampaaLBrunnerAMJanssonS 2006 CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040 1043
10. KozarewaIIbanezCJohanssonMOgrenEMozleyD 2010 Alteration of PHYA expression change circadian rhythms and timing of bud set in Populus. Plant Molecular Biology 73 143 156
11. HsuC-YAdamsJPKimHNoKMaC 2011 FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proceedings of the National Academy of Sciences of the United States of America 108 10756 10761
12. ResmanLHoweGJonsenDEnglundMDruartN 2010 Components acting downstream of short day perception regulate differential cessation of cambial activity and associated responses in early and late clones of hybrid poplar. Plant Physiology 154 1294 1303
13. TurckFFornaraFCouplandG 2008 Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology 59 573 594
14. RuttinkTArendMMorreelKStormeVRombautsS 2007 A molecular timetable for apical bud formation and dormancy induction in Poplar. Plant Cell 19 2370 2390
15. KarlbergAEnglundMPetterleAMolnarGSjodinA 2010 Analysis of global changes in gene expression during activity-dormancy cycle in hybrid aspen apex. Plant Biotechnology 27 1 16
16. ElliottRCBetznerASHuttnerEOakesMPTuckerWQJ 1996 AINTEGUMENTA, an APETALA2-like gene of arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8 155 168
17. MizukamiYFischerRL 2000 Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proceedings of the National Academy of Sciences of the United States of America 97 942 947
18. DewitteW 2002 Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79 92
19. HoriguchiGGonzalezNBeemsterGTInzeDTsukayaH 2009 Impact of segmental chromosomal duplications on leaf size in the grandifolia-D mutants of Arabidopsis thaliana. Plant Journal 60 122 133
20. ShigyoMHasebeMItoM 2006 Molecular evolution of the AP2 subfamily. Gene 366 256 265
21. Espinosa-RuizASaxenaSSchmidtJMellerowiczEMiskolcziP 2004 Differential stage-specific regulation of cyclin-dependent kinases during cambial dormancy in hybrid aspen. Plant Journal 38 603 615
22. RohdeAVanMontaguMInzeDBoerjanW 1997 Factors regulating the expression of cell cycle genes in individual buds of Populus. Planta 201 43 52
23. IbanezCKozarewaIJohanssonMOgrenERohdeA 2010 Circadian Clock Components Regulate Entry and Affect Exit of Seasonal Dormancy as Well as Winter Hardiness in Populus Trees. Plant Physiology 153 1823 1833
24. HarashimaHSchnittgerA 2010 The integration of cell division, growth and differentiation. Current Opinion in Plant Biology 13 66 74
25. InzeDVandepoeleKRaesJDe VeylderLRouzeP 2002 Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903 916
26. CockcroftCEMurrayJAH 1997 The role of Arabidopsis thaliana cyclin D2 in plant development and the cell cycle: Analysis of expression pattern. European Journal of Cell Biology 72 57 57
27. Riou-KhamlichiCHuntleyRJacqmardAMurrayJAH 1999 Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283 1541 1544
28. AbeMKobayashiYYamamotoSDaimonYYamaguchiA 2005 FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052 1056
29. GronlundABhaleraoRPKarlssonJ 2009 Modular gene expression in Poplar: a multilayer network approach. New Phytologist 181 315 322
30. Suarez-LopezPWheatleyKRobsonFOnouchiHValverdeF 2001 CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116 1120
31. JeanmouginFThompsonJDGibsonTJPlewniakFHigginsDG 1997 The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25 4876 4882
32. KarimiMInzeDDepickerA 2002 GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7 193 195
33. KonczCSchellJ 1986 The promoter of Tl-DNA Gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Molecular & General Genetics 204 383 396
34. NilssonOAldénTSitbonFAnthony LittleCChalupaV 1992 Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging. Transgenic research 1 209 220
35. VandesompeleJDe PreterKPattynFPoppeBVan RoyN 2002 Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3 -
36. FulopKPettko-SzandtnerAMagyarZMiskolcziPKondorosiE 2005 The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant Journal 42 810 820
37. MeskieneIBaudouinESchweighoferALiwoszAJonakC 2003 Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. Journal of Biological Chemistry 278 18945 18952
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- De Novo Origins of Human Genes
- Duplication Hotspots Are Associated with Late-Replicating Regions of the Genome
- De Novo Origin of Human Protein-Coding Genes
- Cyclin D/CDK4 and Cyclin E/CDK2 Induce Distinct Cell Cycle Re-Entry Programs in Differentiated Muscle Cells
- Short Day–Mediated Cessation of Growth Requires the Downregulation of AINTEGUMENTALIKE1 Transcription Factor in Hybrid Aspen
- Physiological IRE-1-XBP-1 and PEK-1 Signaling in Larval Development and Immunity
- Role of Pirh2 in Mediating the Regulation of p53 and c-Myc
- Signatures of Environmental Genetic Adaptation Pinpoint Pathogens as the Main Selective Pressure through Human Evolution
- FOXO Regulates Organ-Specific Phenotypic Plasticity In
- Heritable Epigenetic Variation among Maize Inbreds
- Foxn1 Regulates Lineage Progression in Cortical and Medullary Thymic Epithelial Cells But Is Dispensable for Medullary Sublineage Divergence
- Attenuation of the Sensing Capabilities of PhoQ in Transition to Obligate Insect–Bacterial Association
- A Novel Protein LZTFL1 Regulates Ciliary Trafficking of the BBSome and Smoothened
- Activation of Bmp2-Smad1 Signal and Its Regulation by Coordinated Alteration of H3K27 Trimethylation in -Induced Senescence
- Histone H3K56 Acetylation, CAF1, and Rtt106 Coordinate Nucleosome Assembly and Stability of Advancing Replication Forks
- The SUN Protein Mps3 Is Required for Spindle Pole Body Insertion into the Nuclear Membrane and Nuclear Envelope Homeostasis
- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- Effect of Host Species on the Distribution of Mutational Fitness Effects for an RNA Virus
- Pch2 Acts through Xrs2 and Tel1/ATM to Modulate Interhomolog Bias and Checkpoint Function during Meiosis
- SOX9 Governs Differentiation Stage-Specific Gene Expression in Growth Plate Chondrocytes via Direct Concomitant Transactivation and Repression
- from the Aphid : A Missing Link from Facultative to Obligate Insect Endosymbiont
- Recessive Antimorphic Alleles Overcome Functionally Redundant Loci to Reveal Function in Flowers and Meristems
- Over-Expression of DSCAM and COL6A2 Cooperatively Generates Congenital Heart Defects
- Consequences of Eukaryotic Enhancer Architecture for Gene Expression Dynamics, Development, and Fitness
- Distinct Genetic Architectures for Male and Female Inflorescence Traits of Maize
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- For Male , Sperm Activation Is a “Just-in-Time” Event
- PcG Complexes Set the Stage for Epigenetic Inheritance of Gene Silencing in Early S Phase before Replication
- The Gene Contains Hotspots for L1 Endonuclease-Dependent Insertion
- Relative Burden of Large CNVs on a Range of Neurodevelopmental Phenotypes
- Multiple Means to the Same End: The Genetic Basis of Acquired Stress Resistance in Yeast
- Genome-Wide Crossover Distribution in Meiosis Reveals Sex-Specific Patterns along Chromosomes
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
- Homologs of Retinoblastoma-Associated Protein 46/48 Associate with a Histone Deacetylase to Act Redundantly in Chromatin Silencing
- Genetic Interaction Maps in Reveal Functional Crosstalk among Cell Envelope Biogenesis Pathways
- The ERI-6/7 Helicase Acts at the First Stage of an siRNA Amplification Pathway That Targets Recent Gene Duplications
- PBX1 Genomic Pioneer Function Drives ERα Signaling Underlying Progression in Breast Cancer
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- De Novo Origins of Human Genes
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



