-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
DNA sequences capable of adopting non-canonical secondary structures have been associated with gross-chromosomal rearrangements in humans and model organisms. Previously, we have shown that long inverted repeats that form hairpin and cruciform structures and triplex-forming GAA/TTC repeats induce the formation of double-strand breaks which trigger genome instability in yeast. In this study, we demonstrate that breakage at both inverted repeats and GAA/TTC repeats is augmented by defects in DNA replication. Increased fragility is associated with increased mutation levels in the reporter genes located as far as 8 kb from both sides of the repeats. The increase in mutations was dependent on the presence of inverted or GAA/TTC repeats and activity of the translesion polymerase Polζ. Mutagenesis induced by inverted repeats also required Sae2 which opens hairpin-capped breaks and initiates end resection. The amount of breakage at the repeats is an important determinant of mutations as a perfect palindromic sequence with inherently increased fragility was also found to elevate mutation rates even in replication-proficient strains. We hypothesize that the underlying mechanism for mutagenesis induced by fragile motifs involves the formation of long single-stranded regions in the broken chromosome, invasion of the undamaged sister chromatid for repair, and faulty DNA synthesis employing Polζ. These data demonstrate that repeat-mediated breaks pose a dual threat to eukaryotic genome integrity by inducing chromosomal aberrations as well as mutations in flanking genes.
Published in the journal: Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in. PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003551
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003551Summary
DNA sequences capable of adopting non-canonical secondary structures have been associated with gross-chromosomal rearrangements in humans and model organisms. Previously, we have shown that long inverted repeats that form hairpin and cruciform structures and triplex-forming GAA/TTC repeats induce the formation of double-strand breaks which trigger genome instability in yeast. In this study, we demonstrate that breakage at both inverted repeats and GAA/TTC repeats is augmented by defects in DNA replication. Increased fragility is associated with increased mutation levels in the reporter genes located as far as 8 kb from both sides of the repeats. The increase in mutations was dependent on the presence of inverted or GAA/TTC repeats and activity of the translesion polymerase Polζ. Mutagenesis induced by inverted repeats also required Sae2 which opens hairpin-capped breaks and initiates end resection. The amount of breakage at the repeats is an important determinant of mutations as a perfect palindromic sequence with inherently increased fragility was also found to elevate mutation rates even in replication-proficient strains. We hypothesize that the underlying mechanism for mutagenesis induced by fragile motifs involves the formation of long single-stranded regions in the broken chromosome, invasion of the undamaged sister chromatid for repair, and faulty DNA synthesis employing Polζ. These data demonstrate that repeat-mediated breaks pose a dual threat to eukaryotic genome integrity by inducing chromosomal aberrations as well as mutations in flanking genes.
Introduction
Chromosomal instability and mutagenesis are two fundamental processes that alter prokaryotic and eukaryotic genomes. The deleterious consequences of excessive DNA perturbations are hereditary diseases and cancer in humans (reviewed in [1], [2], [3]). At the same time, a fine balance between acquiring genetic changes and restoring original DNA content is paramount for organismal development, adaptation, polymorphism and evolution (for example [4], [5], [6]).
Double-strand breaks (DSBs) in DNA are a driving force for both chromosomal instability and accumulation of mutations. DSBs are a well-established source of a variety of chromosomal aberrations including translocations and copy number variations [7], [8]. It has also become evident from studies in bacteria and yeast that DSB formation and repair are associated with an increased level of mutations, even during homologous recombination which was considered to be an error-free process. In E.coli, the role of DSB formation in the induction of mutagenesis was first inferred based on the requirement of RecA and RecBCD for the occurrence of adaptive mutations in the LacZ gene [9] and was later directly demonstrated by using I-SceI endonuclease-induced breaks [10]. In yeast, elevated levels of base substitutions and frame shift mutations were shown to be due to DSB repair in meiosis [11] and as a result of induction of DSBs in mitotically-dividing cells as shown in gene conversion (GC) [12], [13], break-induced replication (BIR) [14] and single-strand annealing (SSA) assays [15]. The proposed mechanism for break-induced mutagenesis, surmised from these studies, involves the formation of long regions of single-stranded DNA (ssDNA) as a result of DSB end resection. Mutations arise during error-prone synthesis either across the damaged ssDNA template or during synthesis following invasion into the undamaged donor strand. There are two lines of evidence supporting this mechanism. First, Yang et al. [15] have shown in yeast, that single-stranded DNA is drastically more prone to the accumulation of mutations with and without treatment with DNA damaging agents than double-stranded DNA. Second, in several studies, mutagenesis was shown to be fully or partially dependent on highly inaccurate translesion polymerases (TLS). The bacterial TLS polymerase, DinB is responsible for 85% of the mutations triggered by DSB repair during adaptive mutagenesis [16]. In yeast, depending on the assay and nature of mutations, DSB-induced mutagenesis is either completely (SSA [15]), partially (GC next to the DSB site and BIR [12], [13], [14]) or not (classical GC assay [17]) attributed to the activity of the error-prone TLS polymerase, Polζ.
Problems encountered by DNA replication machinery are a major source of spontaneous chromosomal breakage in eukaryotes, estimated to be approximately 10 DSBs per cell cycle in human cells (reviewed in [18]). Certain chromosomal regions, the fragile sites, often containing secondary structure-forming repeats, are susceptible to breakage especially under conditions of replication stress [19]. The mutagenic potential of replication-associated breaks has not been studied in detail. It is also unknown what the level of breaks during replication should be for mutagenesis to be manifested. The latter is important considering the fact that in previous studies mutagenesis was detected under conditions of extremely high level of DSBs, reaching up to 100% as seen in the case of site-specific endonucleases. Whether fragile motifs on their own or under conditions of replication stress could be a potent endogenous source of mutations remains to be established.
In this study, we investigate the mutagenic potential of two sequence motifs, inverted repeats and GAA/TTC tracts, which are natural chromosomal fragile sites [20], [21] under conditions of unperturbed and compromised replication. Long inverted repeats can adopt non-B DNA secondary structures such as hairpins and cruciforms owing to their internal symmetry [22]. They are a potent source of genome rearrangements in both prokaryotes and eukaryotes including humans [23]–[26]. We have previously demonstrated that in yeast a 320 bp Alu-quasi-palindrome triggers gross chromosomal rearrangements by inducing special type of DSBs that have hairpin-capped termini [21], [26]. The hairpin ends are a substrate for opening and processing by Sae2 and the Mre11/Rad50/Xrs2 (MRX) complex. In Δmre11, Δrad50, Δxrs2, or Δsae2 mutants, the resection of broken ends is completely blocked, giving rise to inverted dimers. GAA/TTC tracts adopt another kind of non-canonical DNA structure, namely, H-DNA or triplex DNA (reviewed in [27]). The triplex secondary structure is a driving force for the expansions of GAA tracts, a phenomenon responsible for Friedreich's ataxia in humans [28]. Triplex-adopting sequences, including GAA/TTC repeats, are also responsible for breakage and induction of recombination and rearrangements in bacteria, yeast and humans [20], [29]–[34]. Using yeast as an experimental system, we previously demonstrated that triplex structure-imposed replication problems can contribute to breakage at long GAA/TTC tracts [20]. At the same time, GAA-mediated breaks can occur in non-dividing cells where transcription is an important determinant of DSBs [35], [36]. H-DNA forming sequences are mutagenic in yeast and mammalian systems [34], [37]–[39], albeit, direct evidence that repeat-induced fragility is the reason for mutagenesis in the vicinity of the repeats remains to be found.
In this work, we demonstrate that increased break formation at the location of inverted repeats causes mutagenesis at distances up to 8 kb away from the DSB site. The accumulation of mutations requires the Sae2 protein, indicating that resection and generation of long ssDNA is a critical parameter for this phenomenon. We have found that error–prone synthesis involving the translesion polymerase Polζ during repair is primarily responsible for the observed mutagenesis. We also show that in replication-deficient strains the triplex-adopting GAA/TTC repeats are associated with hypermutability at distant loci, suggesting that a similar mechanism of mutagenesis can operate at repeat-associated chromosomal break sites under conditions of replication stress. These data demonstrate that secondary structure-mediated breaks pose a dual threat to eukaryotic genome integrity by inducing chromosomal aberrations and mutations extending to distant chromosomal sites. It is conceivable that the mechanisms of DSB-induced mutagenesis uncovered in this study are also relevant to human evolution, polymorphism and tumorigenesis.
Results
Experimental system
The experimental system used to assess the mutagenic potential of fragile inverted and GAA/TTC repeats in this study is based on the GCR assay described in [20], [26]. Briefly, the LYS2 gene containing the fragile motifs was inserted 43 kb from the telomere on the left arm of chromosome V in haploid yeast strains (Figure 1). There are no essential genes between the left telomere and the LYS2 gene. CAN1 is located 8 kb telomere-proximal to the LYS2 gene. The insertion of ADE2 between CAN1 and LYS2 allows for the differentiation between two types of events on media containing canavanine and low amounts of adenine. Breakage at the location of structure-forming repeats leads to the loss of the terminal 43 kb of the chromosomal arm containing both CAN1 and ADE2, resulting in canavanine-resistant red-colored colonies (CanRAde−). On the other hand, mutations in CAN1 are manifested as white-colored canavanine-resistant colonies (CanRAde+) (Figure 1). The correlation between colony color and the requirement of adenine for growth was verified by replica plating the CanR colonies to media lacking adenine. The three fragile motifs inserted into LYS2 were 100% homologous inverted Alu repeats, 320 bp each with a 12 bp spacer (Alu-IRs); 100% homologous IS50 palindromic repeats (IS50-PAL) , 1.3 kb each; and 230 repeats of GAA/TTC in the orientation wherein the GAA sequence is the template for the lagging strand synthesis.
Fig. 1. Experimental system to study fragile motif-induced mutagenesis. 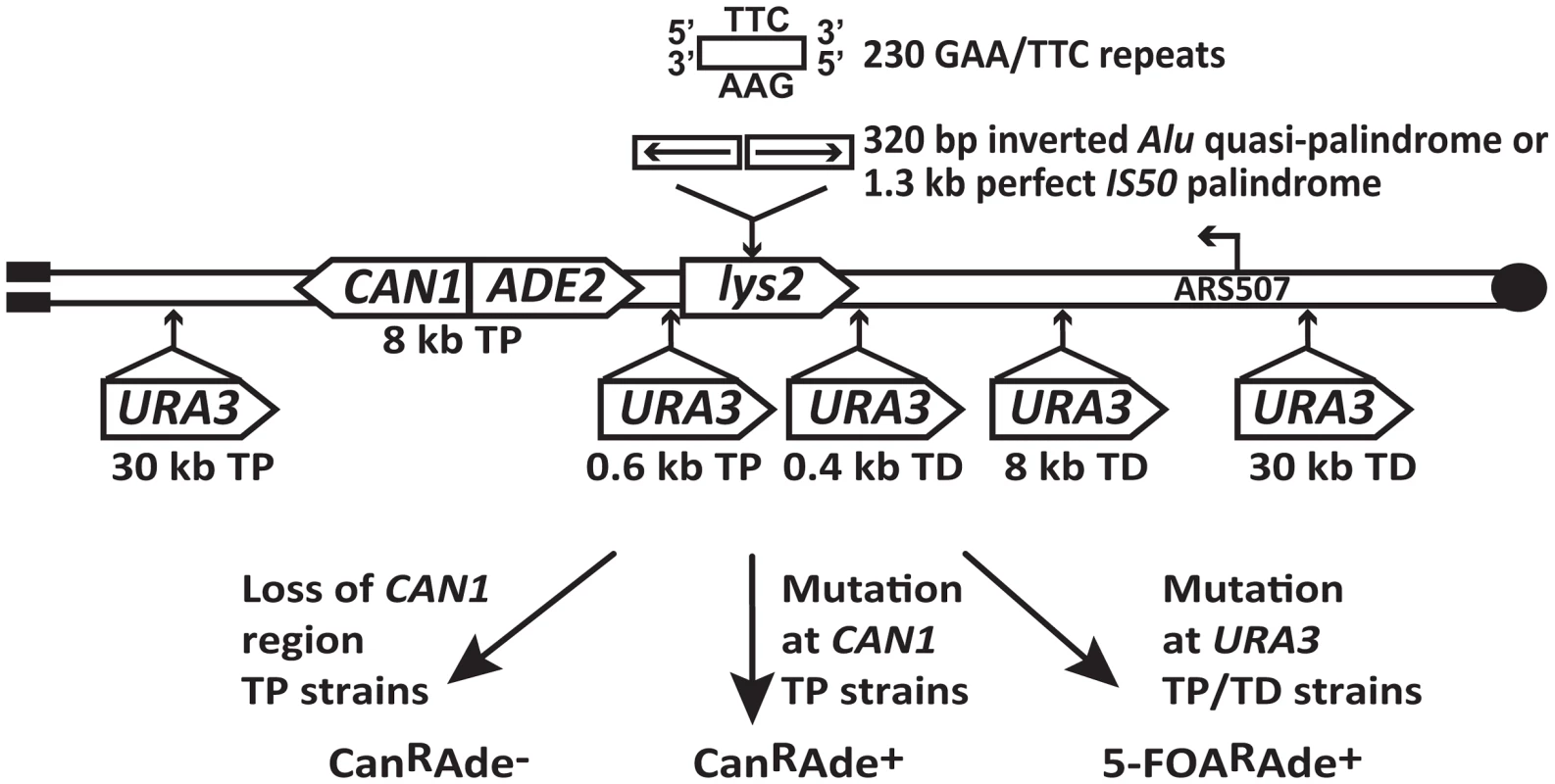
Alu-quasi-palindrome, IS50-palindrome or GAA/TTC repeats were inserted into LYS2 gene on the left arm of chromosome V. Positions of CAN1 and URA3 reporters located telomere-proximal (TP) or telomere-distal (TD) to the repeat insertion are shown. The position of the ARS507 and the direction of replication fork migrating through the repeat region are indicated. Breakage at the location of secondary-structure-adopting repeats can lead to loss of 43 kb telomere-proximal deletion resulting in red-colored CanRAde− clones. Mutations in CAN1 reporter will yield white-colored CanRAde+ isolates. Mutations in URA3 gene will give rise to colonies resistant to medium containing 5-fluoorotic acid (5-FOAR). To estimate how far mutagenesis can extend from the break site, URA3 was inserted into chromosome V telomere-proximal (TP) 0.6 kb and 30 kb away from the repeats, and telomere-distal (TD) 0.4 kb, 8 kb and 30 kb away from the repeats (Figure 1). Mutations in URA3 were measured on 5-fluoroorotic acid-containing media lacking adenine (5-FOAR Ade+), allowing us to preferably select these events in contrast to GCRs that give rise to 5-FOAR Ade− colonies.
A defect in DNA replication leads to increased Alu-quasipalindrome-induced breakage and mutagenesis at the CAN1 locus
Mutation levels in the CAN1 locus in wild-type strains with inverted Alu-quasipalindrome are not different from strains that lack the sequence motif. We wanted to determine whether addition of replication stress will enhance the fragility potential of these repeats and increase mutagenesis. In a screen for mutants that exhibit an increased level of hairpin-capped DSBs we identified the pol3-P664L allele that affects the functions of replicative polymerase δ responsible for synthesis of the lagging strand [40]. The P664L mutation is located in the polymerase domain of Polδ [41] and the yeast strains carrying this mutant allele exhibit temperature-sensitive growth at 37°C (data not shown). The rate of CAN1 region loss in strains containing Alu-quasipalindrome was 40-fold higher in pol3-P664L mutants than in wild-type (Table 1). Moreover, pol3-P664L strains with Alu-IRs exhibited elevated levels of mutagenesis in CAN1 loci located 8 kb away from the DSB site. Notably, the mutagenesis was completely dependent on the presence of fragile motifs, suggesting that the mutator phenotype is not a feature of the pol3 allele but rather is a consequence of increased breakage. It is important to note that in pol3-P664L strains without the Alu-quasi-palindrome, the relative rate of arm loss was nearly 3-fold higher than in wild-type strains. However, the fragility due to deficiency in Polδ is not high enough to induce mutagenesis.
Tab. 1. Polζ- and Sae2-dependent mutagenesis by Alu-quasi-palindrome in replication mutants. 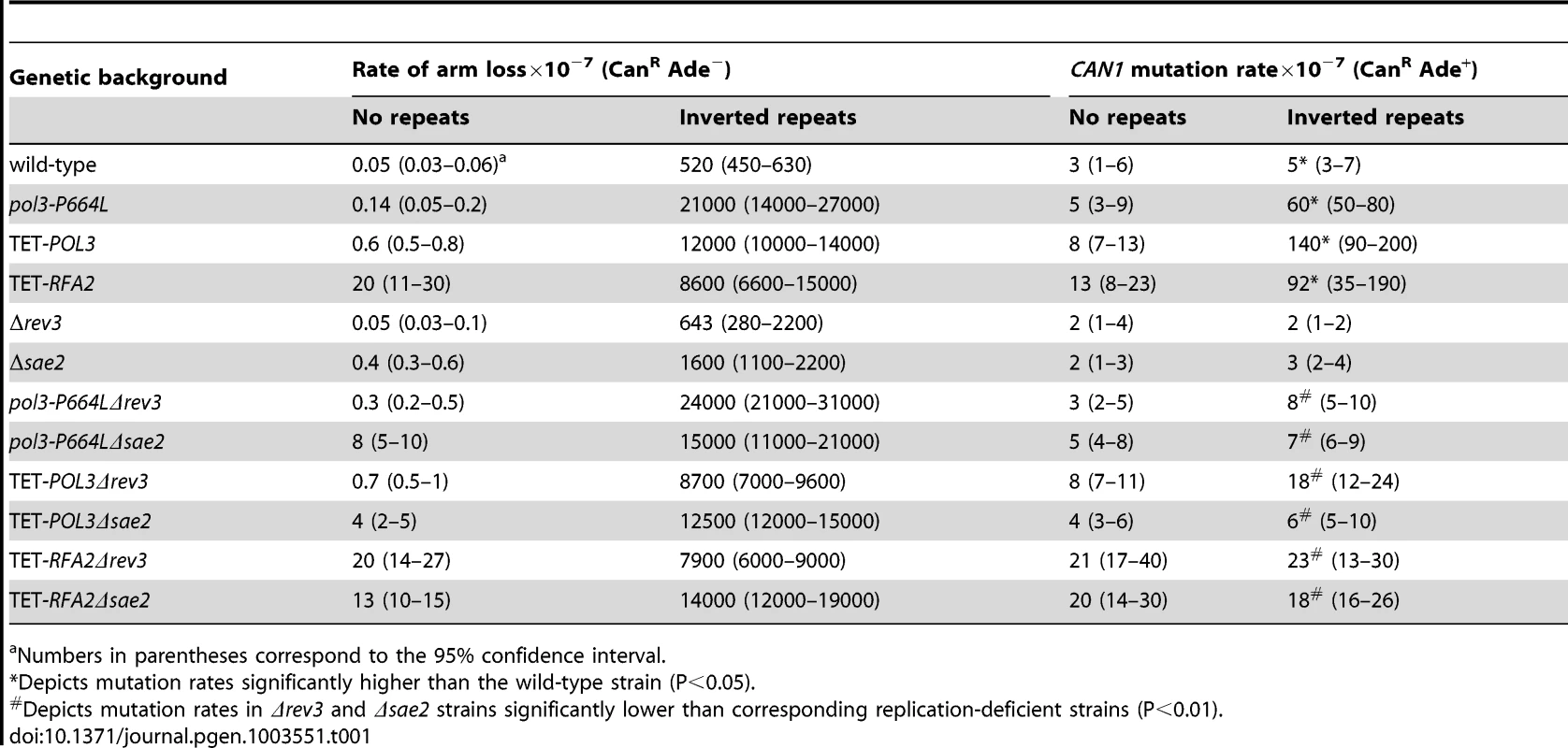
Numbers in parentheses correspond to the 95% confidence interval. A similar increase in Alu-IR-dependent fragility and mutagenesis in CAN1 gene was observed in strains where the POL3 expression was under the control of a tetracycline-repressible promoter (tetO7) [42]. Belli et al., 1998 [42] showed that tetO7–driven expression of genes in the presence of the antibiotic leads to a reduction in protein levels in comparison to conditions when genes were expressed from their native promoters. Western blotting analysis of c-Myc-tagged Pol3 revealed that upon treatment of cells with doxycycline the protein level was indeed ∼10 fold decreased in comparison with the wild-type level (Figure 2). Hence, we refer to TET-POL3 as a mutant allele and all further tests were carried out in the presence of doxycycline (see Material and Methods).
Fig. 2. Analysis of protein levels of Pol3 and Rfa2 in the wild-type and tetracycline downregulatable strains. 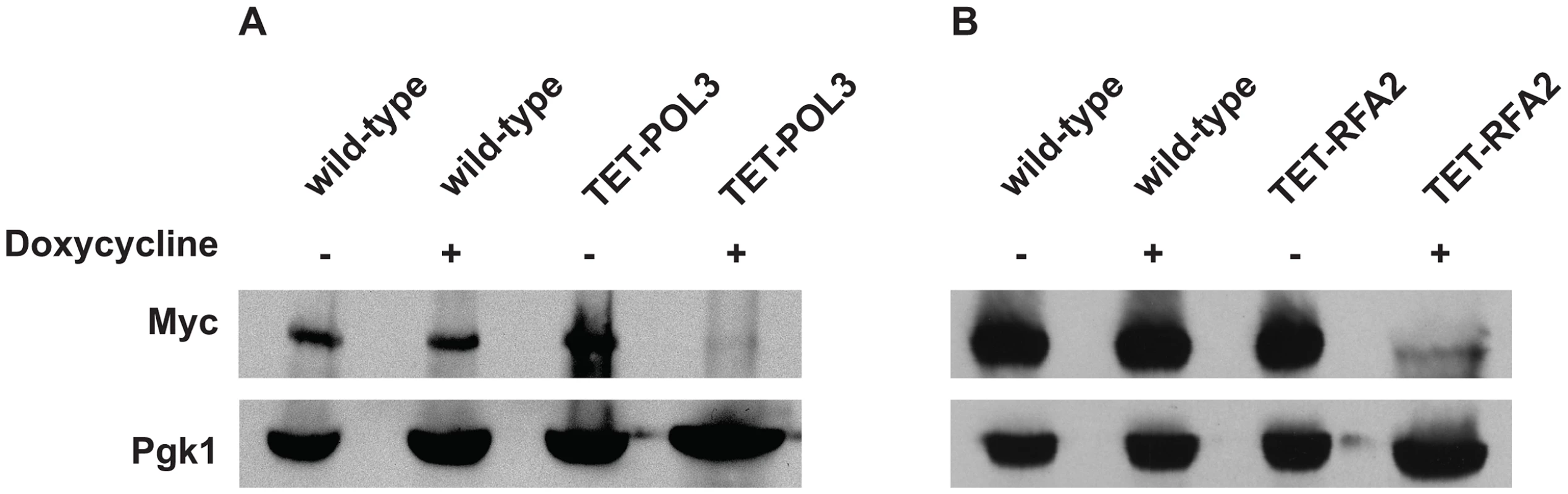
(A) Effect of downregulation of Pol3. (B) Effect of downregulation of Rfa2. Pol3 and Rfa2 were c-Myc tagged at the C-terminus in the wild-type, TET-POL3 and TET-RFA2 strains respectively. Proteins were extracted with (+) or without (−) treatment with doxycycline. Pol3 and Rfa2 were detected by Western blot with anti-c-MYC antibody. The protein levels were compared against Pgk1 levels (detected with anti-Pgk1 specific antibody) which acted as the loading control. Upon treatment with doxycycline, Pol3 expression was lowered 10 fold (average of 9, 10 and 12) and Rfa2 expression was lowered 4-fold (average of 4.1, 4.4 and 3.7). We also replaced the native promoter of another replication gene, RFA2, that encodes one of the subunits of the single-stranded DNA-binding protein participating in DNA replication and repair, with the tetO7 promoter. Upon downregulation with doxycycline, the expression of Rfa2 was ∼4-fold lower than the wild-type level (Figure 2). Similar to the TET-POL3 strain, the TET-RFA2 strain exhibited increased levels of arm loss and mutagenesis (Table 1).
It is important to note that neither the pol3-P664L strain nor the TET-POL3 and TET-RFA2 strains grown in the presence of doxycycline at chosen concentrations showed sensitivity to DNA damaging agents such as MMS and camptotechin, indicating that they are proficient in DNA repair (Figure S1).
Overall, these data show that mutations at distant loci require the presence of fragile motifs and are dependent on the amount of replication-associated breaks.
1.3 kb perfect palindrome induces mutagenesis at the CAN1 locus even in the wild-type strains
We addressed directly whether a fragile site can induce mutations at distant loci in replication-proficient strains. This experiment also helps to distinguish which is the key factor in mutagenesis, the level of breakage or repair of broken molecules by faulty replication proteins. The 1.3 kb long IS50 palindrome was found to induce mutagenesis in CAN1 when replication was unimpaired (3-fold). The increased length of the interacting arms and the lack of a spacer between them likely create a problem even for intact replication machinery and render this motif highly fragile with a 14-fold increase in GCR rates as compared to the Alu-IR strain (Table 2). Consistently, using Southern hybridization, we estimated the level of breakage at this palindrome to be 4.8% (average of 4.6%, 5% and 4.9%) which is ∼3-times higher than in strains carrying the Alu-quasi-palindrome (1.6%, average of 1.4%, 1.5% and 1.9%) (Figure 3). Taking into account that a deficiency in Pol3 causes a 7-fold increase in Alu-IR-mediated breakage (11%, average of 11%, 10% and 11%) and a 12-fold increase in mutagenesis, it is evident that the levels of DSB formation and not DSB repair by defective replication proteins are the important determinant of mutagenesis.
Fig. 3. Inverted repeat and GAA/TTC-induced DSB detection in wild-type and mutant strains. 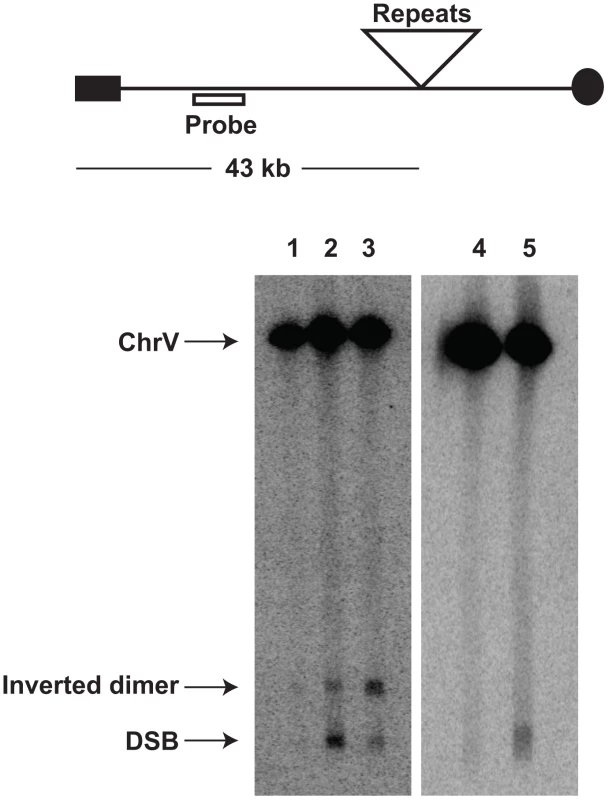
Upper panel depicts the relative positions of the inverted repeats and the probe (open rectangle) used. For the detection of inverted repeat-mediated breaks Δsae2 strains were used as in these mutants the hairpin-capped breaks are not opened and resection is abolished [21]. As a consequence, inverted dimer molecules accumulate in Δsae2 mutants as previously demonstrated. Contour-clamped homogeneous electric field gel electrophoresis and Southern hybridization were used to highlight the intact chromosome V and the broken fragment. Lanes 1, 2 and 3 depict the Alu-IR, pol3-P664L Alu-IR, and IS50-IR strains respectively. Lanes 4 and 5 depict GAA/TTC(230) and TET-POL3 GAA/TTC(230) strains respectively. Intact chromosome V, DSB fragments and inverted dimers (in the case of inverted repeats) are indicated. Tab. 2. Polζ- and Sae2-dependent mutagenesis by IS50-perfect palindrome. 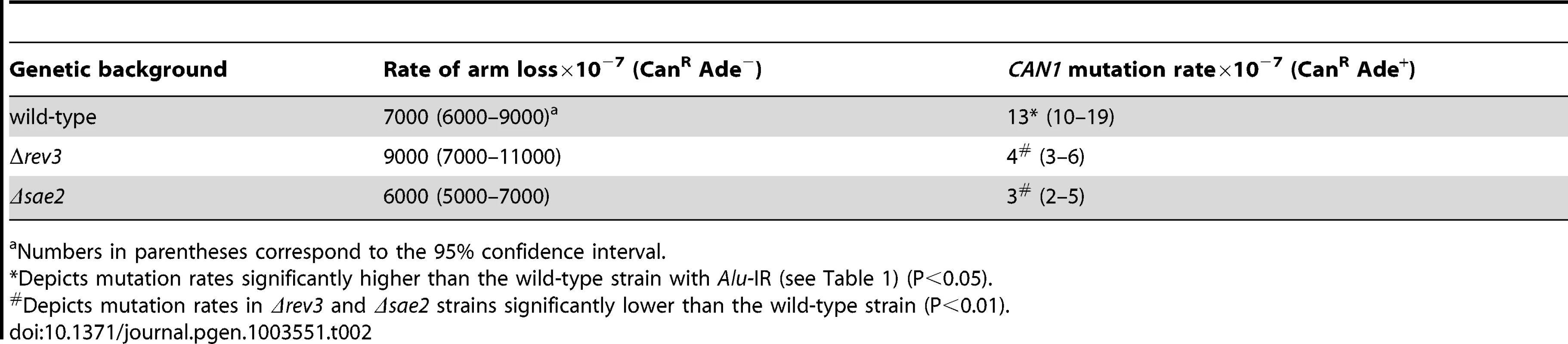
Numbers in parentheses correspond to the 95% confidence interval. Mutagenesis by inverted repeats depends on the distance of the reporter from the DSB site and on the activity of the Sae2 protein
Previously, we have shown that Alu-IRs induce DSBs that have hairpin-capped termini [21]. The resection and repair of these DSBs requires the hairpin-opening activity of Sae2 and the Mre11 nuclease [43]. To test if ssDNA generated as a result of 5′-3′DSB end resection is a critical requirement for repeat-induced mutagenesis, the SAE2 gene was disrupted in pol3-P664L, TET-POL3 and TET-RFA2 Alu-IR strains. The level of CAN1 mutagenesis in pol3-P664LΔsae2, TET-POL3Δsae2 and TET-RFA2Δsae2 mutants was reduced to levels observed in strains without inverted Alus (Table 1), indicating that mutations are indeed a consequence of DSB resection. Similarly, in the strains carrying IS50 repeats, mutation rates declined upon deletion of SAE2 (Table 2).
To determine to what distance the DSB-associated mutagenesis can spread on either side of the fragile site, we inserted the URA3 reporter 0.4, 8 and 30 kb telomere-distal (TD) and 0.6 and 30 kb telomere-proximal (TP) to Alu-IRs in pol3-P664L strains (Figure 1). The average length of ssDNA generated via DSB end resection in yeast varies from 2 kb to 10 kb [44]. This predicts that mutations in URA3 situated past 10 kb should diminish. Consistently, although mutation rates at 0.4 kb, 0.6 kb and 8 kb were approximately the same (10–15-fold higher than in wild-type strain), at 30 kb the rate of ura3 mutations significantly decreased in TP and TD constructs (Table 3).
Tab. 3. Mutagenesis by fragile Alu-IRs depends on the distance of reporter from the DSB site. 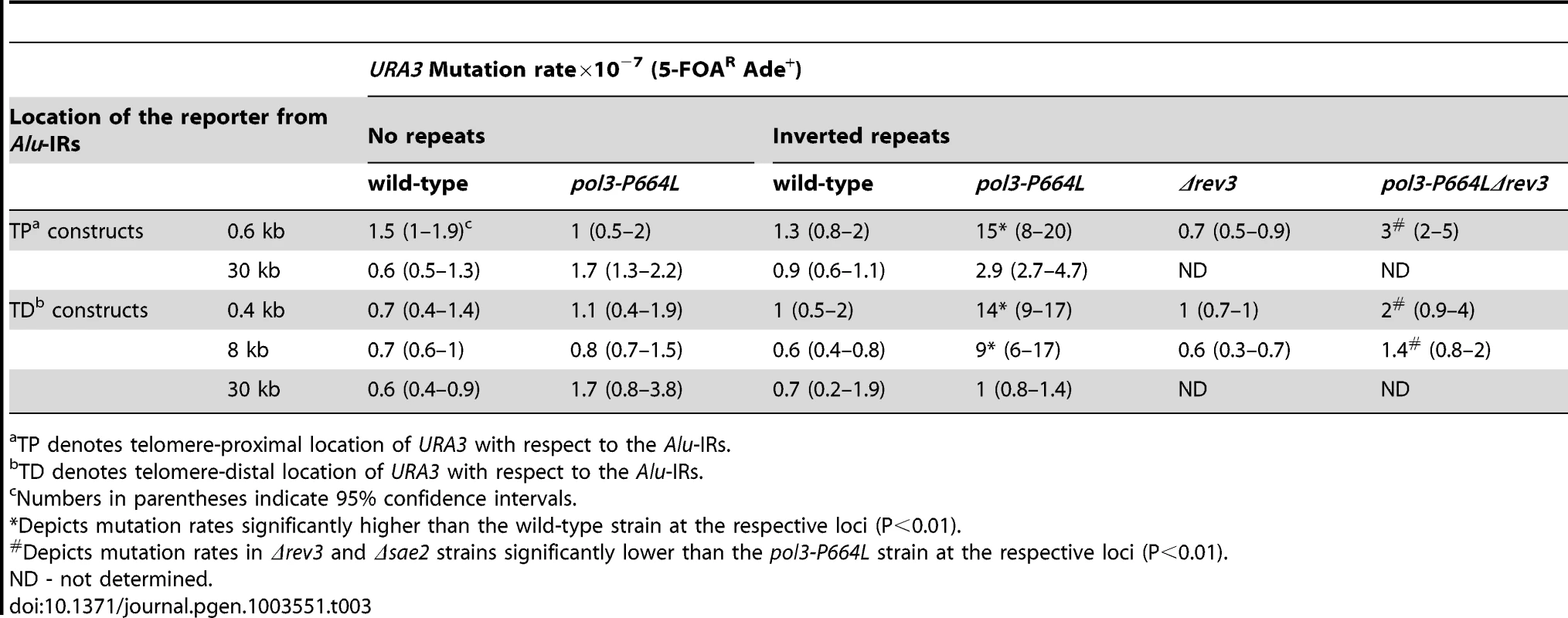
TP denotes telomere-proximal location of URA3 with respect to the Alu-IRs. The dependence of the efficiency of mutagenesis on the activity of Sae2 and the distance of the reporter from DSB site demonstrates that ssDNA is an intermediate for the occurrence of mutations.
Increase in mutagenesis observed in replication-deficient and –proficient strains is mostly attributed to the activity of Polζ translesion polymerase
Holbeck and Strathern, [45] and Rattray et al. [12] showed that Polζ translesion synthesis activity is required for the generation of base substitutions in a reporter located 0.3 kb from the site of an HO-endonuclease-induced break. To assess if mutagenesis induced by fragile motifs depends on translesion synthesis, we disrupted the REV3 gene encoding the catalytic subunit of Polζ [46] in wild-type, pol3-P664L, TET-POL3 and TET-RFA2 strains with Alu-IRs (Table 1). REV3 disruption in replication-defective strains brought the mutation level in the CAN1 reporter to almost the level observed in the wild-type strain. In replication-proficient strains carrying Δrev3, only a modest 2-fold decrease in CAN1 mutation rate was observed. Augmented mutation rates in the strains with IS50 repeats were also dependent on the activity of Polζ (Table 2).
To gain further insight into the spectrum of mutations generated at distant loci as a result of DSB formation by inverted repeats, we sequenced 22–31 independent CanRAde+ isolates from wild-type, pol3-P664L and pol3-P664LΔrev3 strains, respectively. In the wild-type strain with Alu-IRs, 85% of the mutations were base substitutions and 15% were single base deletions (Table 4 and Table S1.). A similar mutation spectrum was also observed in other studies [47], [48]. This correlates with the lack of increase of CAN1 mutagenesis in the wild-type Alu-IR strain (Table 1), indicating that the observed mutations in replication-proficient strains were a result of spontaneous mutagenesis rather than secondary structure-induced DSBs. In the pol3-P664L strain the mutation spectrum was changed. There was a significant increase in the frequencies of base substitutions, particularly G∶C→T∶A and G∶C→C∶G transversions characteristic of Polζ errors during spontaneous mutagenesis [49] (Table 4 and Table S2). Increases in deletions ranging from 1 to 5 bp and complex mutations (two or more changes in a run of 10 bp) were also observed. These types of changes were also previously attributed to the TLS activity of Polζ [48], [50]. A similar mutation spectrum was also seen for CanRAde+ clones from strains containing the IS50-perfect palindrome (Table 4 and Table S5). Since mutagenesis observed in these strains requires the activity of Polζ, it is likely that error-prone synthesis by the TLS polymerase during DSB repair causes base substitutions as well as deletions and complex mutations. Consistently, errors that could be assigned to the activity of Polζ were suppressed in pol3-P664LΔrev3 strains (Table 4 and Table S3). We also uncovered large deletions (up to 39 bp) and a duplication of 27 bp flanked by short direct repeats in pol3 mutants with or without Alu-IRs. This is most probably attributed to the defective Polδ. Notably, pol3-P664L strains that lack fragile motifs also exhibited complex mutations (Table 4 and Table S4). Taking into account that fragility in pol3-P664L without Alu-IRs is low, it can be inferred that these changes reflect mutations arising during DNA replication carried out by a faulty DNA polymerase (a process that also might require TLS polymerases [51]) rather than a consequence of error-prone synthesis during DSB repair.
Tab. 4. Mutation spectra in CAN1 reporter. 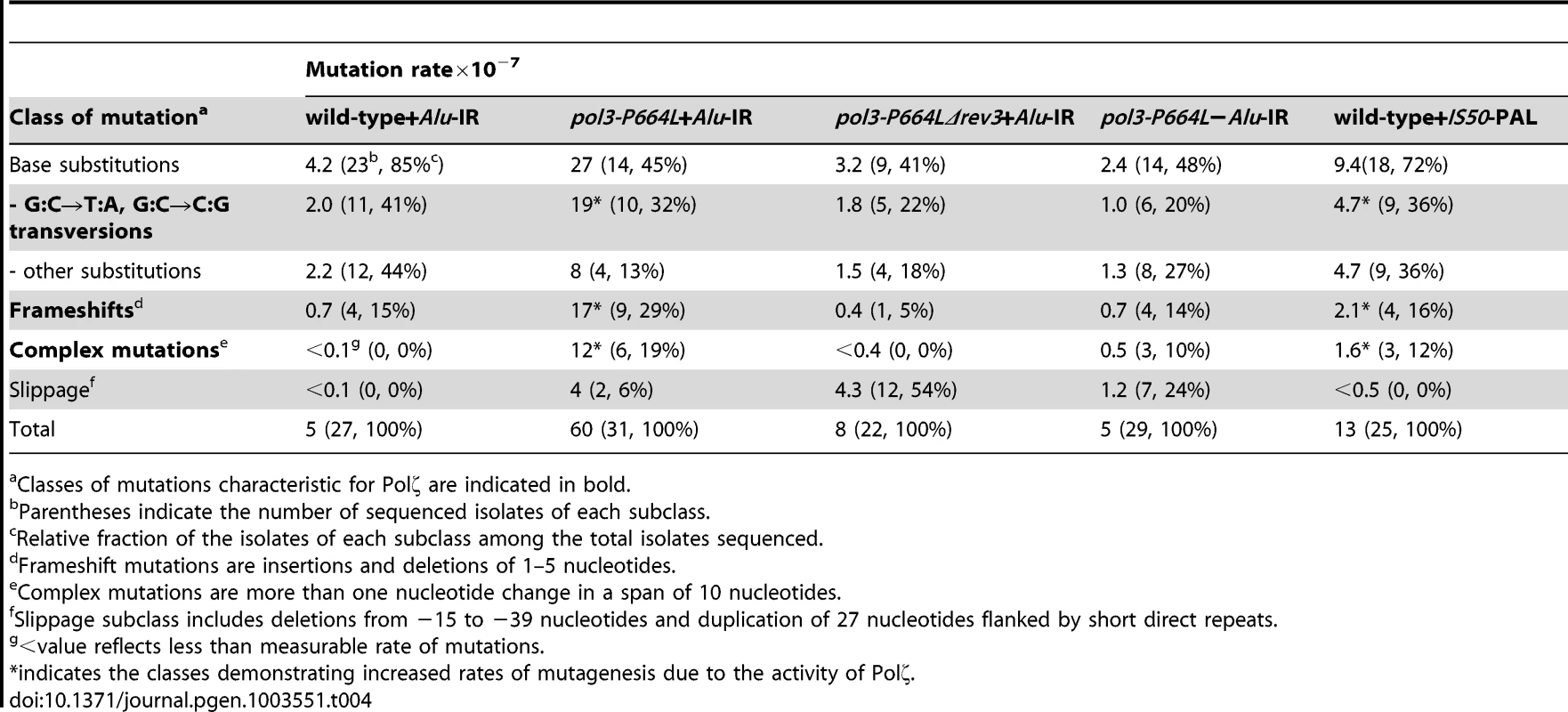
Classes of mutations characteristic for Polζ are indicated in bold. Overall, analysis of mutation spectra in wild-type and replication-deficient strains is in agreement with genetic analysis and supports the conclusion that repeat-mediated mutations are generated by error-prone Polζ and do not occur due to faulty synthesis by replicative polymerases.
GAA/TTC fragile motif also induces mutagenesis at distant chromosomal loci that is partly Polζ dependent
To determine if DSB-induced mutagenesis can be observed at another fragile motif, we assessed CAN1 mutation rate in strains carrying 230 repeats of the triplex-adopting GAA/TTC (Figure 1). Although the rate of CanR mutations was unaltered in wild-type strains, a 4-fold increase in mutagenesis was detected in pol3-P664L and TET-POL3 strains (Table 5). The level of DSB formation at GAA/TTC repeats in the TET-POL3 strain was estimated to be 3.3% (average of 3.1%, 3.3% and 3.6%, Figure 3). This is a minimal estimation of GAA/TTC-mediated DSBs since, unlike the situation with palindromic sequences, resection of the broken fragments cannot be prevented by SAE2 disruption and a proportion of degraded DSBs are excluded from detection.
Tab. 5. Mutagenesis in CAN1 reporter stimulated by GAA/TTC repeats. 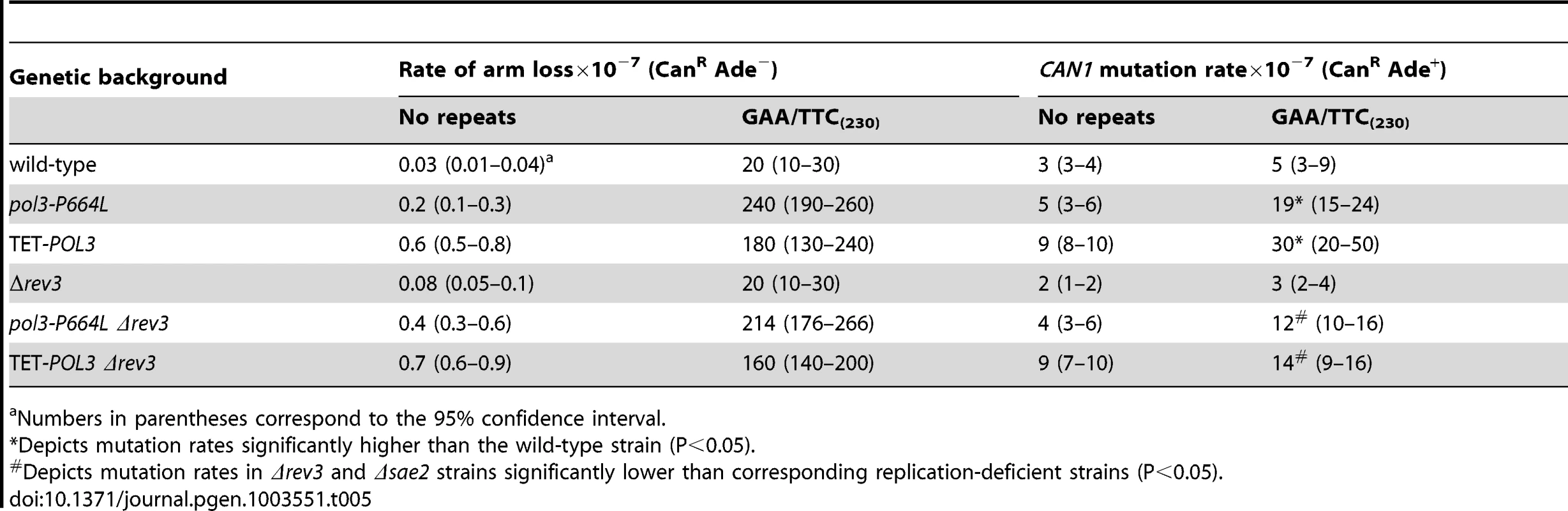
Numbers in parentheses correspond to the 95% confidence interval. Similar to Alu-IR-mediated mutagenesis, Polζ plays a role in the induction of mutations by GAA/TTC repeats. There was a mild but statistically significant reduction (2-fold) in mutagenesis in pol3-P664LΔrev3 versus pol3-P664L (p<0.05) and TET-POL3Δrev3 versus TET-POL3 (p<0.05) strains as determined using an unpaired t-test. Although it is difficult to evaluate the contribution of resection and long ssDNA to GAA/TTC-associated mutagenesis, the involvement of REV3 suggests that the mechanism underlying mutagenesis in the case of inverted repeats and GAA/TTC fragile sites can be similar.
Discussion
The induction of DSBs using site-specific endonucleases has been shown to drive mutagenesis [12]–[15]. This study demonstrates that natural chromosomal fragile sites comprising of sequence motifs that can adopt non-B DNA structures are also mutagenic. Under condition of replication stress, the mutagenesis can reach up to the levels caused by deficiency in the mismatch repair system [52]. We also show that the mutations are a consequence of error-prone repair of repeat-induced DSBs. Overall, we establish secondary structure-forming motifs as a potent source of endogenous mutagenesis and reveal the mechanism underlying this phenomenon.
In this study we found that when replication is compromised, Alu-quasi-palindrome promotes chromosomal fragility and mutagenesis at CAN1 and URA3 reporters located 8 kb from the break site. Mutations were also increased in strains with a perfect IS50-palindrome with inherently higher fragility even in replication-proficient strains. We have previously shown that inverted repeats induce hairpin-capped DSBs in replication-proficient strains [21]. We have found that in replication-defective mutants the DSBs mediated by the Alu-quasi-palindrome also have hairpin-capped termini (Y. Zhang, N. Saini, Z. Sheng, K.S. Lobachev, in preparation). The opening of the hairpins necessitates the nuclease activity of the MRX complex and Sae2. The requirement of Sae2 for mutagenesis at distant loci unequivocally demonstrates that mutations are a consequence of DSB formation (Table 1 and Table 2). Moreover, these data also implicate the formation of long ssDNA upon resection of DSB ends as the second step in repeat-mediated mutagenesis. ssDNA has been shown to be prone to accumulation of mutations during SSA or in a situation where the telomeres become uncapped [15]. Therefore, it is possible that hairpin-processing generates damaged ssDNA that can serve as a faulty template for synthesis during SSA or GC. Alternatively, the undamaged ssDNA can be involved in strand invasion and mutations could arise due to error-prone synthesis during homologous recombination as suggested in other studies [9], [13], [14]. Error-prone synthesis of the undamaged DNA template in replication deficient strains by Polζ was observed by Northam et al. [51].
Although we cannot determine whether mutagenesis is due to accumulation of damage in resected DNA or error-prone synthesis on undamaged template, our data point towards synthesis-dependent strand annealing (SDSA) as the underlying mechanism for mutagenesis (Figure 4). None of the analyzed CanR clones contained interstitial deletions and all of the clones retained intact Alu-IRs or IS50-palindrome (data not shown). This suggests that SSA is unlikely to operate during Alu-IR-mediated mutagenesis and alludes to a template-dependent repair process that involves the undamaged sister chromatid. Thus, we favor a scenario wherein hairpin-capped DSBs are induced in late S or G2 stage of the cell cycle. Upon hairpin opening by Sae2 and MRX, the 3′ end of the resected DSB invades the intact sister chromatid template. The requirement for invasion in mutagenesis is a likely step but ultimately cannot be proven by using rad51 or rad52 mutants for two reasons: these strains exhibit a mutator phenotype on their own [49] and Rad51 and Rad52 proteins are required for DSB formation at the Alu-IRs in replication-defective strains (Y. Zhang, N. Saini, Z. Sheng, K.S. Lobachev, in preparation). It is conceivable that the invasion event can proceed either as a BIR or as an SDSA event. SDSA is the most probable mechanism owing to the fact that mutations were observed in both TP and TD reporters and that reduced mutation rates were measured at reporters 30 kb from the break site (Table 3). It is important to note that SDSA preserves the original inverted repeats that can trigger additional rounds of breakage and associated mutagenesis. If extrapolated to humans, these observations identify secondary structure-forming repeats as a potent source of mutagenesis that can change the expression of flanking genes during the lifetime of healthy individuals even in the absence of exogenous damage.
Fig. 4. Model for mutagenesis induced by Alu-IRs and GAA/TTC repeats. 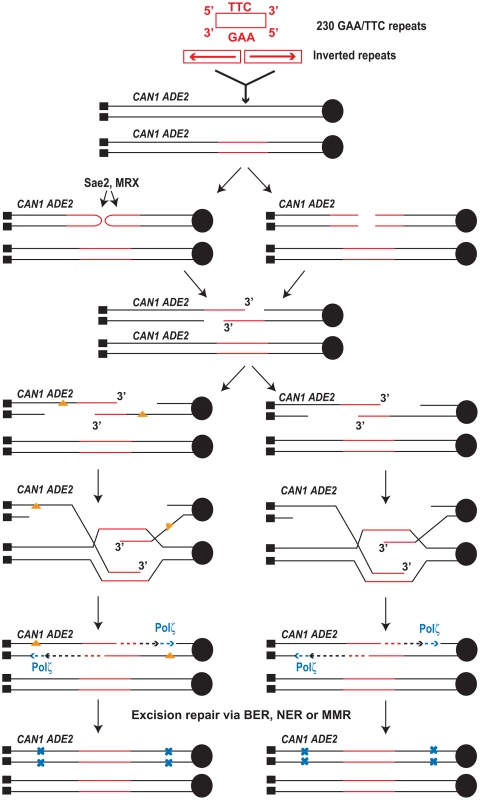
The inverted repeats and 230 repeats of GAA/TTC inserted into LYS2 are shown in red (not drawn to scale). Centromere (filled black circle) and telomeres (filled black squares) are also shown. Inverted repeats and GAA/TTC repeats trigger DSBs in late S or in G2 wherein the intact sister chromatid is present. The inverted repeats-induced hairpin-capped DSB are processed by Sae2 and the MRX complex (shown on the left). GAA/TTC tracts induce DSBs that have exposed 5′ and 3′ termini (shown on the right). Two scenarios for the accumulation of mutations are shown. On the left, ssDNA generated as a result of extensive resection can accumulate damages (orange triangles). Error-prone synthesis during the fill in reaction will lead to mutations (shown as blue x). On the right, errors can be made by Polζ during synthesis across the undamaged template. Mutations will be incorporated either due to the action of mismatch repair or in next round of DNA replication (not shown). Mutations generated in the reporter 8 kb away from DSB site were also strongly dependent on the activity of Polζ (Table 1 and Table 2) indicating that the error-prone translesion synthesis operates during SDSA. This is consistent with the Hirano and Sugimoto, 2006 study that showed that Mec1 kinase is needed to recruit the Polζ-Rev1 complex to the DSB site [53] and other studies where DSB-induced mutagenesis required Polζ [12]–[15]. Analysis of mutations in the CAN1 locus of the hyper-fragile strains revealed an increase in G∶C→T∶A and G∶C→C∶G transversions, frameshift and complex mutations that are signatures of Polζ (Table 4) [48]–[50].
In this study we also show that DSB-triggering long GAA/TTC repeats induce mutagenesis at distant loci, indicating that a similar underlying mechanism of mutagenesis described above for inverted repeats can operate for triplex-forming motifs. The requirement of Rev3 for mutagenesis is more evident for inverted repeats than for GAA/TTC repeats. It would be interesting to see if other TLS polymerases, Rev1 and Polη, besides Polζ operate in GAA/TTC-associated mutagenesis and to determine if the mutation spectra in GAA/TTC - and inverted repeat-containing strains differ. It is also important to note that in our experimental system, we observe mutagenesis by GAA/TTC tracts only under conditions of compromised replication wherein the repeat-mediated fragility is further increased. In other studies, mutagenesis is induced by GAA/TTC repeats in replication-proficient strains [37], [38]. These discrepancies might reflect the distance of the used reporter from the fragile motif. It is possible that at closer distances, SSA might be the predominant pathway for mutagenesis where mutations introduced by Polζ can be scored above the spontaneous level of mutagenesis, while SDSA requires higher frequencies of breakage and longer ssDNA. This can be checked experimentally in future studies. Our data are also in agreement with the recent study by Shah et al., 2012 wherein GAA/TTC-induced mutations in a closely juxtaposed reporter in Polδ mutants were dependent on Polζ [39].
Overall, this study demonstrates that fragile sequence motifs that are found in eukaryotic genomes, including humans, can be potent inducers of mutagenesis. Thus, secondary structure-adopting repeats can represent a dual threat to DNA stability by changing the structural organization of the genome and causing mutations. Recent studies linking the occurrence of mutations near chromosomal rearrangement break-points in primates and humans suggest that error-prone repair of DSBs can operate during speciation, evolution and tumorigenesis [54]–[57]. Thus it is likely that fragile and mutagenic non B-DNA-forming motifs are contributing factors to these processes.
Materials and Methods
Yeast strains
The yeast strains used for the analysis of the inverted repeat-induced mutagenesis were derivatives of the KT19 strain (MATa, bar1-Δ, his7-2, trp1-Δ, ura3-Δ, leu2-3,112, ade2-Δ, lys2-Δ, cup1-Δ, yhro54c-Δ, cup2-Δ, V34205::ADE2lys2::Alu-IRs, V29616::CUP1). GAA/TTC-mediated mutagenesis was measured in strains that were derivative of YKL36 (MATa, bar1-Δ, his3-Δ, trp1-Δ, ura3-Δ, leu2-Δ, ade2-Δ, lys2-Δ, V34205::ADE2lys2::(GAA)230). Alu-IRs and GAA repeats were inserted into the BamHI site and the IS50 palindrome was inserted into HpaI site in LYS2. The strains without repeats had an intact LYS2 gene. For measuring the distance dependence of repeat-induced mutagenesis, URA3 was amplified from pRS306 with flanking regions for the points of insertion into chromosome V. URA3 was inserted close to the repeats in lys2 (586 bp TP and 352 bp TD), ∼8 kb TD of the repeat locus between SGD coordinates 42096 and 42097 and ∼30 kb TP between SGD coordinates 11910 and 11911 and TD between coordinates 64686 and 64687 (Figure 1, Table S6). The pol3-P664L allele was created via site-directed mutagenesis using p170 [58]. The mutation P664L results in the appearance of the AseI site. The plasmid was digested with HpaI and the mutation was obtained using pop-in pop-out methodology. The mutant shows mild temperature sensitivity at 37°C. The tetracycline promoter construct was obtained from Euroscarf (pCM225). PCR was performed with primers carrying overhangs for RFA2 and POL3 promoter regions and one-step integration was used to replace the promoters for RFA2 and POL3 (Table S6). REV3 was replaced with the kanMX cassette in wild-type and pol3-P664L strain and with the hphMX cassette amplified from pAG32 in the TET-POL3 and TET-RFA2 strains [59]. SAE2 was disrupted with the kanMX cassette in the wild-type strains and with TRP1 in pol3-P664L, TET-POL3 and TET-RFA2 strains.
GCR and mutation rates estimations
Fluctuation tests were carried out to estimate mutation and GCR rates. The strains were allowed to grow on YPD agar for 3 days at 30°C. The TET-POL3 and TET-RFA2 strains were grown on YPD containing 2 µg/ml and 0.1 µg/ml doxycycline, respectively. At these chosen concentrations of doxycycline the proteins are downregulated leading to an increase in fragility without significantly affecting viability of the strains. 14 individual colonies were diluted in 0.25 ml water each and serial dilutions were made to approximately 1∶10000. The cultures were plated on YPD and on L-canavanine (60 mg/L) low adenine (5 mg/L) containing synthetic media in order to obtain approximately several hundred colonies per plate after incubating for 3 days at 30°C. White colonies on canavanine-containing media are indicative of mutations in CAN1 while red colonies depict GCR events. For mutation rate estimation at URA3, the cultures were appropriately diluted and plated on 5-FOA (1 g/L) containing synthetic media lacking adenine. Mutation rates and 95% confidence intervals were calculated as previously described [21].
DSB detection and quantification
Yeast cells were embedded into agarose plugs at a concentration of 2×109 cells/ml for detection of inverted-repeat-mediated DSBs and at a concentration of 8×109 cells/ml for detection of GAA/TTC-induced DSBs. The chromosomes were separated using contour-clamped homogeneous electric field gel electrophoresis as described previously [36] and transferred onto a nylon membrane. Southern hybridization was carried out using a probe specific to HPA3 that is telomere-proximal to the repeats. Densitometry analysis was performed using ImageJ (NIH) and the intensity of the broken product was normalized against the intact chromosome V.
Sequence analysis of mutations at CAN1
CAN1 mutants were obtained by plating approximately 30 individual colonies from two independent isolates for each strain on L-canavanine low adenine containing synthetic media. The CanRAde+ isolates were then streaked out on YPD to obtain single colonies from which DNA was extracted. PCR was carried out using primers 60 bp upstream and 158 bp downstream of CAN1. The PCR product was sequenced using 4 internal primers for CAN1 such that the entire gene would be covered at least twice during sequencing. The primers used for sequencing CAN1 are
can1-o1 : 5′CATCTACTGGTGGTGACAAAG3′;
can1-s1 : 5′GCCACGGTATTTCAAAGCTTGC3′;
can1-s2 : 5′GGCTCTTGGAACGGATTTTC3′;
can1-s3 : 5′TGTAGCCATTTCACCCAAGG3′. The sequencing results are depicted in Table 4 and Tables S1, S2, S3, S4, S5.
Estimation of Pol3 and Rfa2 expression
To quantify the changes in protein expression POL3 was tagged with 13 copies of c-Myc-epitope tag and RFA2 was tagged with 9 copies of the c-Myc-epitope tag at the C-terminus in both wild-type and tetracycline downregulatable strains. TET-POL3 was grown in the presence of 2 µg/ml doxycycline overnight, while TET-RFA2 was grown with 0.1 µg/ml doxycycline overnight. Total protein was extracted as previously described [60] and separated using SDS-polyacrylamide gel electrophoresis on an 8% gel. After electrophoresis the gel was blotted on a nitrocellulose membrane and probed with an antibody specific to c-Myc (Genscript) and an antibody for Pgk1 (Invitrogen). The membrane was further treated with anti-mouse secondary antibody (Genscript) and chemiluminescent detection was carried out using the protocol described by GE Healthcare. Densitometry analysis was performed using ImageJ (NIH) and the intensity of Pol3 and Rfa2 were normalized against the loading control Pgk1.
Supporting Information
Zdroje
1. CharamesGS, BapatB (2003) Genomic instability and cancer. Curr Mol Med 3 : 589–596.
2. HarperJW, ElledgeSJ (2007) The DNA damage response: ten years after. Mol Cell 28 : 739–745.
3. JacksonSP, BartekJ (2009) The DNA-damage response in human biology and disease. Nature 461 : 1071–1078.
4. PollardKS, SalamaSR, KingB, KernAD, DreszerT, et al. (2006) Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2: e168.
5. StamatoyannopoulosJA, AdzhubeiI, ThurmanRE, KryukovGV, MirkinSM, et al. (2009) Human mutation rate associated with DNA replication timing. Nat Genet 41 : 393–395.
6. ZhangF, GuW, HurlesME, LupskiJR (2009) Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10 : 451–481.
7. HastingsPJ, LupskiJR, RosenbergSM, IraG (2009) Mechanisms of change in gene copy number. Nat Rev Genet 10 : 551–564.
8. WymanC, KanaarR (2006) DNA double-strand break repair: all's well that ends well. Annu Rev Genet 40 : 363–383.
9. HarrisRS, LongerichS, RosenbergSM (1994) Recombination in adaptive mutation. Science 264 : 258–260.
10. PonderRG, FonvilleNC, RosenbergSM (2005) A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell 19 : 791–804.
11. MagniGE (1963) The Origin of Spontaneous Mutations during Meiosis. Proc Natl Acad Sci U S A 50 : 975–980.
12. RattrayAJ, ShaferBK, McGillCB, StrathernJN (2002) The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics 162 : 1063–1077.
13. StrathernJN, ShaferBK, McGillCB (1995) DNA synthesis errors associated with double-strand-break repair. Genetics 140 : 965–972.
14. DeemA, KeszthelyiA, BlackgroveT, VaylA, CoffeyB, et al. (2011) Break-induced replication is highly inaccurate. PLoS Biol 9: e1000594.
15. YangY, SterlingJ, StoriciF, ResnickMA, GordeninDA (2008) Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet 4: e1000264.
16. McKenzieGJ, LeePL, LombardoMJ, HastingsPJ, RosenbergSM (2001) SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell 7 : 571–579.
17. HicksWM, KimM, HaberJE (2010) Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329 : 82–85.
18. HaberJE (1999) DNA recombination: the replication connection. Trends Biochem Sci 24 : 271–275.
19. SchwartzM, ZlotorynskiE, KeremB (2006) The molecular basis of common and rare fragile sites. Cancer Lett 232 : 13–26.
20. KimHM, NarayananV, MieczkowskiPA, PetesTD, KrasilnikovaMM, et al. (2008) Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J 27 : 2896–2906.
21. LobachevKS, GordeninDA, ResnickMA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108 : 183–193.
22. Sinden RR (1994) DNA structure and Function. San Diego: Academic Press.
23. DarmonE, EykelenboomJK, LinckerF, JonesLH, WhiteM, et al. (2010) E. coli SbcCD and RecA control chromosomal rearrangement induced by an interrupted palindrome. Mol Cell 39 : 59–70.
24. EdelmannL, SpiteriE, KorenK, PulijaalV, BialerMG, et al. (2001) AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet 68 : 1–13.
25. KurahashiH, EmanuelBS (2001) Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet 10 : 2605–2617.
26. NarayananV, MieczkowskiPA, KimHM, PetesTD, LobachevKS (2006) The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125 : 1283–1296.
27. Frank-KamenetskiiMD, MirkinSM (1995) Triplex DNA structures. Annu Rev Biochem 64 : 65–95.
28. CampuzanoV, MonterminiL, MoltoMD, PianeseL, CosseeM, et al. (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271 : 1423–1427.
29. BlaszakRT, PotamanV, SindenRR, BisslerJJ (1999) DNA structural transitions within the PKD1 gene. Nucleic Acids Res 27 : 2610–2617.
30. NapieralaM, DereR, VetcherA, WellsRD (2004) Structure-dependent recombination hot spot activity of GAA.TTC sequences from intron 1 of the Friedreich's ataxia gene. J Biol Chem 279 : 6444–6454.
31. PatelHP, LuL, BlaszakRT, BisslerJJ (2004) PKD1 intron 21: triplex DNA formation and effect on replication. Nucleic Acids Res 32 : 1460–1468.
32. RaghavanSC, ChastainP, LeeJS, HegdeBG, HoustonS, et al. (2005) Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J Biol Chem 280 : 22749–22760.
33. RaghavanSC, SwansonPC, MaY, LieberMR (2005) Double-strand break formation by the RAG complex at the bcl-2 major breakpoint region and at other non-B DNA structures in vitro. Mol Cell Biol 25 : 5904–5919.
34. WangG, VasquezKM (2004) Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc Natl Acad Sci U S A 101 : 13448–13453.
35. TangW, DominskaM, GreenwellPW, HarvanekZ, LobachevKS, et al. (2011) Friedreich's ataxia (GAA)n*(TTC)n repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet 7: e1001270.
36. ZhangY, ShishkinAA, NishidaY, Marcinkowski-DesmondD, SainiN, VolkovKV, MirkinSM, LobachevKS (2012) Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol Cell 48 : 254–65.
37. ShishkinAA, VoineaguI, MateraR, CherngN, ChernetBT, et al. (2009) Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Mol Cell 35 : 82–92.
38. TangW, DominskaM, GawelM, GreenwellPW, PetesTD (2013) Genomic deletions and point mutations induced in Saccharomyces cerevisiae by the trinucleotide repeats (GAA.TTC) associated with Friedreich's ataxia. DNA Repair (Amst) 12 : 10–17.
39. ShahKA, ShishkinAA, VoineaguI, PavlovYI, ShcherbakovaPV, et al. (2012) Role of DNA polymerases in repeat-mediated genome instability. Cell Rep 2 : 1088–1095.
40. Nick McElhinnySA, GordeninDA, StithCM, BurgersPM, KunkelTA (2008) Division of labor at the eukaryotic replication fork. Mol Cell 30 : 137–144.
41. PavlovYI, ShcherbakovaPV, RogozinIB (2006) Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol 255 : 41–132.
42. BelliG, GariE, AldeaM, HerreroE (1998) Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14 : 1127–1138.
43. LengsfeldBM, RattrayAJ, BhaskaraV, GhirlandoR, PaullTT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28 : 638–651.
44. ZhuZ, ChungWH, ShimEY, LeeSE, IraG (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134 : 981–994.
45. HolbeckSL, StrathernJN (1997) A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147 : 1017–1024.
46. PrakashS, JohnsonRE, PrakashL (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74 : 317–353.
47. LangGI, MurrayAW (2008) Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178 : 67–82.
48. SakamotoAN, StoneJE, KisslingGE, McCullochSD, PavlovYI, et al. (2007) Mutator alleles of yeast DNA polymerase zeta. DNA Repair (Amst) 6 : 1829–1838.
49. EndoK, TagoY, DaigakuY, YamamotoK (2007) Error-free RAD52 pathway and error-prone REV3 pathway determines spontaneous mutagenesis in Saccharomyces cerevisiae.. Genes Genet Syst 82 : 35–42.
50. AbdulovicAL, MinesingerBK, Jinks-RobertsonS (2008) The effect of sequence context on spontaneous Polzeta-dependent mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res 36 : 2082–2093.
51. NorthamMR, RobinsonHA, KochenovaOV, ShcherbakovaPV (2010) Participation of DNA polymerase zeta in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics 184 : 27–42.
52. ChenC, MerrillBJ, LauPJ, HolmC, KolodnerRD (1999) Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol Cell Biol 19 : 7801–7815.
53. HiranoY, SugimotoK (2006) ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol 16 : 586–590.
54. BergerMF, LawrenceMS, DemichelisF, DrierY, CibulskisK, et al. (2011) The genomic complexity of primary human prostate cancer. Nature 470 : 214–220.
55. DeS, BabuMM (2010) A time-invariant principle of genome evolution. Proc Natl Acad Sci U S A 107 : 13004–13009.
56. RobertsSA, SterlingJ, ThompsonC, HarrisS, MavD, et al. (2012) Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 46 : 424–35.
57. DrierY, LawrenceMS, CarterSL, StewartC, GabrielSB, et al. (2013) Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res 23 : 228–235.
58. KokoskaRJ, StefanovicL, TranHT, ResnickMA, GordeninDA, et al. (1998) Destabilization of yeast micro - and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol Cell Biol 18 : 2779–88.
59. GoldsteinAL, McCuskerJH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553.
60. KnopM, SiegersK, PereiraG, ZachariaeW, WinsorB, et al. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15 : 963–72.
Štítky
Genetika Reprodukčná medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene ClusterČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



