-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Negative Regulation of Notch Signaling by Xylose
The Notch signaling pathway controls a large number of processes during animal development and adult homeostasis. One of the conserved post-translational modifications of the Notch receptors is the addition of an O-linked glucose to epidermal growth factor-like (EGF) repeats with a C-X-S-X-(P/A)-C motif by Protein O-glucosyltransferase 1 (POGLUT1; Rumi in Drosophila). Genetic experiments in flies and mice, and in vivo structure-function analysis in flies indicate that O-glucose residues promote Notch signaling. The O-glucose residues on mammalian Notch1 and Notch2 proteins are efficiently extended by the addition of one or two xylose residues through the function of specific mammalian xylosyltransferases. However, the contribution of xylosylation to Notch signaling is not known. Here, we identify the Drosophila enzyme Shams responsible for the addition of xylose to O-glucose on EGF repeats. Surprisingly, loss - and gain-of-function experiments strongly suggest that xylose negatively regulates Notch signaling, opposite to the role played by glucose residues. Mass spectrometric analysis of Drosophila Notch indicates that addition of xylose to O-glucosylated Notch EGF repeats is limited to EGF14–20. A Notch transgene with mutations in the O-glucosylation sites of Notch EGF16–20 recapitulates the shams loss-of-function phenotypes, and suppresses the phenotypes caused by the overexpression of human xylosyltransferases. Antibody staining in animals with decreased Notch xylosylation indicates that xylose residues on EGF16–20 negatively regulate the surface expression of the Notch receptor. Our studies uncover a specific role for xylose in the regulation of the Drosophila Notch signaling, and suggest a previously unrecognized regulatory role for EGF16–20 of Notch.
Published in the journal: Negative Regulation of Notch Signaling by Xylose. PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003547
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003547Summary
The Notch signaling pathway controls a large number of processes during animal development and adult homeostasis. One of the conserved post-translational modifications of the Notch receptors is the addition of an O-linked glucose to epidermal growth factor-like (EGF) repeats with a C-X-S-X-(P/A)-C motif by Protein O-glucosyltransferase 1 (POGLUT1; Rumi in Drosophila). Genetic experiments in flies and mice, and in vivo structure-function analysis in flies indicate that O-glucose residues promote Notch signaling. The O-glucose residues on mammalian Notch1 and Notch2 proteins are efficiently extended by the addition of one or two xylose residues through the function of specific mammalian xylosyltransferases. However, the contribution of xylosylation to Notch signaling is not known. Here, we identify the Drosophila enzyme Shams responsible for the addition of xylose to O-glucose on EGF repeats. Surprisingly, loss - and gain-of-function experiments strongly suggest that xylose negatively regulates Notch signaling, opposite to the role played by glucose residues. Mass spectrometric analysis of Drosophila Notch indicates that addition of xylose to O-glucosylated Notch EGF repeats is limited to EGF14–20. A Notch transgene with mutations in the O-glucosylation sites of Notch EGF16–20 recapitulates the shams loss-of-function phenotypes, and suppresses the phenotypes caused by the overexpression of human xylosyltransferases. Antibody staining in animals with decreased Notch xylosylation indicates that xylose residues on EGF16–20 negatively regulate the surface expression of the Notch receptor. Our studies uncover a specific role for xylose in the regulation of the Drosophila Notch signaling, and suggest a previously unrecognized regulatory role for EGF16–20 of Notch.
Introduction
Notch signaling is a juxtacrine cell-cell communication pathway with broad roles in animal development and adult tissue homeostasis [1], [2]. Both gain - and loss-of-function mutations in Notch pathway components cause human disease [3]–[5], and therapeutic approaches to alter the activity of Notch signaling are a subject of intense research and development [6]. The extracellular domains of Notch receptors contain a large number (up to 36) of EGF repeats. Each EGF repeat contains six cysteine residues, which are linked to each other via three disulfide bonds [7]. Of the several forms of O-linked carbohydrates found on Notch EGF repeats [8]–[10], two have been shown to be required for Notch signaling in both flies and mammals: O-fucose and O-glucose [11]–[16]. Addition of N-acetylglucosamine (GlcNAc) to O-fucose by Fringe glycosyltransferases modulates Notch signaling in several contexts [17], [18].
O-linked glucose is attached to serine residues in the consensus sequence C1-X-S-X-(P/A)-C2 by the protein O-glucosyltransferase Rumi/POGLUT1 [8], [14], [19]. Rumi is required for both fly and mammalian Notch signaling at a step downstream of ligand-binding [14], [15], [20]. In vivo structure-function analyses indicate that all of the 18 Rumi target sequences in Drosophila Notch (dNotch) contribute to Notch activation, with O-glucose sites on EGF10-15 playing a more important role than others [20]. O-glucose can serve as a substrate for additional sugar modifications to generate xylose-xylose-glucose trisaccharides [8], [19], [21]. The human enzymes responsible for the addition of xylose to O-glucosylated Notch EGF repeats have recently been identified: glucoside xylosyltransferase (GXYLT)1 and GXYLT2 add the first xylose and xyloside xylosyltransferase (XXYLT1) adds the second (Figure 1A) [22], [23]. Thus far, no functional studies have been performed to analyze the role of xylosylation in Notch signaling.
Fig. 1. Shams functions as a glucoside xylosyltransferase on Notch. 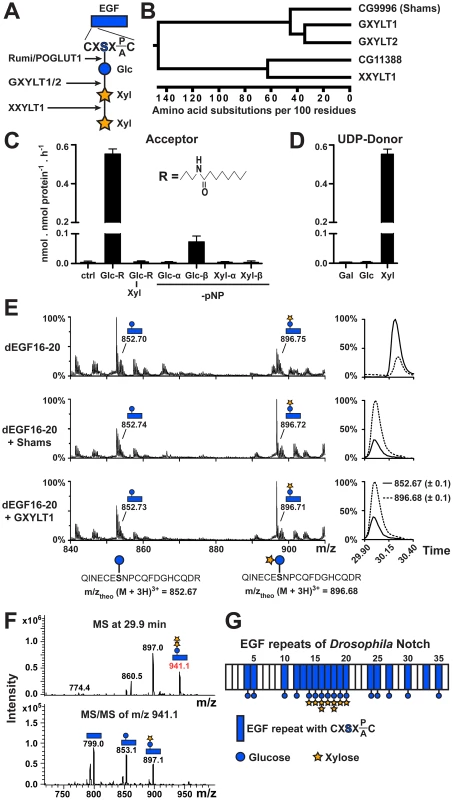
(A) Schematic of the xylose-xylose-glucose trisaccharide attached to the serine (S) residue in the consensus sequence on an EGF repeat and the glycosyltransferases involved in its generation. (B) Phylogenetic tree of human GXYLT1/2 and XXYLT1, and their Drosophila homologs CG9996 (Shams) and CG11388 based on the Clustal W algorithm. (C) Xylosyltransferase assays using UDP-[14C]xylose donor and synthetic lipophilic acceptors to determine acceptor specificity of Shams. R represents the drawn hydrophobic aglycon; pNP, para-nitrophenol. (D) Donor substrate specificity using Glc-R as substrate. (E) Mass spectrometric analysis of a glycosylated peptide of Drosophila Notch (d) EGF16–20 expressed in Sf9 insect cells shows an increase in the ratio of disaccharide- versus monosaccharide-modified form after incubation with Shams or human GXYLT1 and UDP-xylose in vitro. Extracted ion chromatograms on the right indicate that the ratio between xylosylated and non-xylosylated peptides (dashed and solid lines, respectively) is inverted by Shams and GXYLT1. (F) Mass spectrometry demonstrates the presence of O-glucose trisaccharide on a peptide from EGF16 (639QINECESNPCQFDGHCQDR657). Top and bottom panels show MS and MS/MS spectra, respectively. (G) Schematic representation of sites of dNotch xylosylation identified by mass spectrometry (spectra shown in Figure S2). The most elongated glycan structure detected on each EGF repeat is shown, but the shorter forms can also exist. Here, we identify the Drosophila glucoside xylosyltransferase which adds xylose to O-glucosylated EGF repeats and show that this enzyme negatively regulates Drosophila Notch signaling in certain contexts. We use a combination of mass spectrometry, genetic and in vivo mutational studies to show that the functionally important sites of xylosylation reside in EGF repeats 16–20 of Notch, and that xylose negatively regulates the surface expression of Notch. Given that O-glucose positively regulates Notch signaling in all contexts studied so far [20], negative regulation of Notch signaling by xylosylated O-glucose glycans provides an example of how the strength of a signaling pathway can be fine-tuned by stepwise addition of carbohydrate molecules to a receptor.
Results
Drosophila CG9996 (shams) Encodes a Glucoside Xylosyltransferase which Adds Xylose to O-glucosylated Notch EGF Repeats
Using homology searches, we identified two novel Drosophila proteins (CG9996 and CG11388) homologous to human Notch xylosyltransferases. Sequence comparison between these two fly proteins and human Notch xylosyltransferases indicated that CG9996 is the only close fly homolog of human GXYLT1 and GXYLT2, whereas CG11388 bears much more sequence identity with XXYLT1 (Figure 1B and Figure S1). We named CG9996 Shams, a companion and muse for the poet Rumi. To test whether Shams can add xylose to glucose similar to its human homologs, we first performed in vitro glycosyltransferase assays by using purified, recombinant Shams and synthetic lipophilic acceptors (Figure 1C). We found that Shams can indeed add xylose specifically to a synthetic acceptor harboring a glucose residue (Glc-β1-R) but not to one harboring a xylose-glucose disaccharide (Xyl-α1,3-Glc-β1-R) (Figure 1C). To further examine the substrate specificity of Shams, we used glucose or xylose residues attached to para-nitrophenol (pNP) in α - or β-linkage as the acceptor for the xylosyltransferase activity of Shams and found that Shams can only transfer xylose to Glc-β1-pNP (Figure 1C). We also performed similar assays to determine the donor substrate specificity of Shams and found that Shams is able to transfer xylose, but not glucose or galactose, to the Glc-β1-R acceptor (Figure 1D). To examine whether Shams is able to add xylose to O-glucosylated EGF repeats of Notch, we assayed purified Shams using a fragment of Drosophila Notch (EGF16–20) harboring several O-glucosylation sites, expressed in and purified from Sf9 cells. Mass spectrometric analysis of a glycosylated peptide of EGF16 showed that Sf9 cells produce a mixture of glucose and xylose-glucose in a ratio of about 3 to 1 at this site. Sf9 cells apparently highly glucosylate EGF16, but show limited xylosylation capacity. In vitro, both Shams and human GXYLT1 can transfer xylose to O-glucosylated EGF16, changing the ratio in favor of the xylosylated form (Figure 1E). Taken together, these experiments show that Shams functions as a glucoside xylosyltransferase capable of adding the first xylose to O-glucose on Notch EGF repeats, similar to its human homologs GXYLT1 and GXYLT2 [23].
Addition of Xylose to O-glucose Occurs on a Subset of Notch EGF Repeats in Drosophila
Mass spectrometry on mouse Notch1 expressed in several mammalian cell lines has shown that xylose-xylose-glucose trisaccharide is the dominant form of O-glucose glycans on all mouse Notch1 EGF repeats harboring an O-glucosylation site, although the stoichiometry varies among different EGF repeats [8], [19]. To determine the distribution of xylosylated O-glucose glycans on Drosophila Notch, we performed systematic mass spectrometric analyses of Drosophila Notch expressed in Drosophila S2 cells. We find that while O-glucose is found at all predicted sites analyzed, xylose is only detected on EGF14–20, and xylose-xylose-glucose trisaccharides are only detected at EGF16 and EGF18 (Figure 1F and 1G and Figure S2). Therefore, unlike mammalian Notch1, addition of xylose to O-glucose appears to be limited to a subset of EGF repeats of the Drosophila Notch.
shams Mutations Promote Notch Signaling
To examine the role of xylosylation in Drosophila Notch signaling, we performed genetic experiments on two independent alleles of shams (Figure 2A). Flies homozygous for the piggyBac insertion shamse01256 (shamsPB/PB) are viable at 25°C and do not exhibit any adult phenotypes besides a loss of the posterior cross-vein in 20% of the flies (Figure 2C; compare to 2B). When raised at 30°C, 56% of shamsPB/PB flies lose the distal portion of the L5 wing vein (Figure 2D), similar to the phenotype observed in gain-of-function Abruptex alleles of Notch (NAx) [24]. Precise excision of this piggyBac insertion fully reverts the phenotype, indicating that the observed loss of the wing vein is due to the insertion (Figure 2E). We also generated a null allele lacking 97% of the Shams coding region by using FLP/FRT-mediated recombination on two piggyBac insertions in the region (Figure 2A) [25]. Animals homozygous or hemizygous for the null allele shamsΔ34 survive to adulthood and exhibit a 100% penetrant loss of the L5 wing vein at 25°C (Figure 2F). At 30°C, shamsΔ34/Df animals are semi-lethal and all the escapers exhibit partial loss of multiple wing veins (Figure 2G). The wing vein loss phenotype can be rescued by overexpression of shams cDNA (Figure 2H and 2I), by providing a shamsgt-wt genomic transgene that contains the shams locus (Figure 2J), or by an HA-tagged version of the shams genomic transgene (shamsgt-wt-HA, data not shown). However, even though the shamsΔ34 allele also affects CG11836 (Figure 2A), a genomic transgene containing this gene (CG11836gt-wt) does not rescue the wing vein phenotypes of the shamsΔ34/Df animals (Figure 2K). Of note, each of the shamsgt-wt and CG11836gt-wt genomic transgenes partially rescues the semi-lethality of these animals, indicating that both transgenes are functional. These observations indicate that loss of shams results in a wing vein loss phenotype. shamsΔ34/Df animals raised at 30°C also lose the ocellar and postvertical bristles in the head (Figure 2M; compare to 2L) similar to NAx alleles [26]. Again, this phenotype can be fully suppressed by a shams genomic transgene (Figure 2N). Together, these data indicate that loss of shams results in phenotypes reminiscent of Notch gain-of-function phenotypes.
Fig. 2. Mutations in shams result in the loss of wing veins and head bristles. 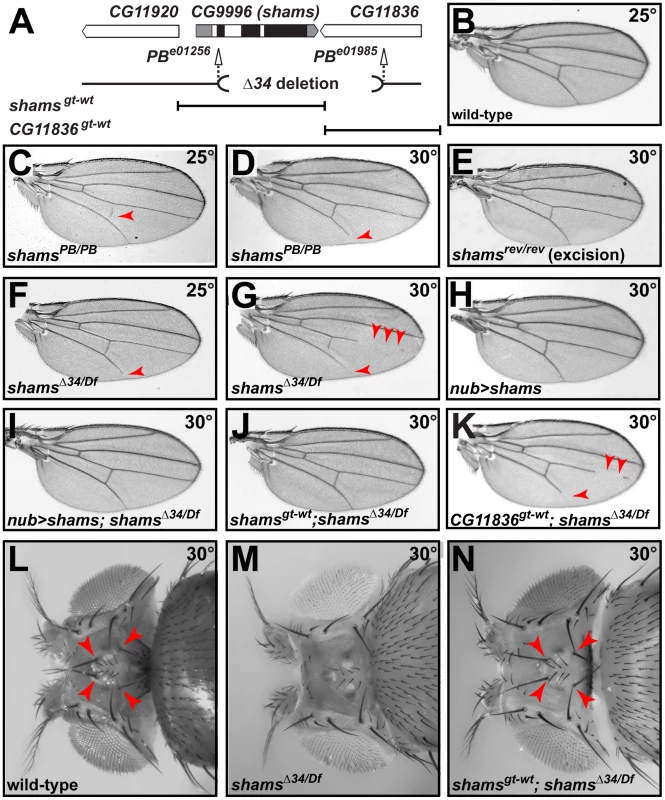
(A) Schematic representation of the genomic region containing shams (CG9996) and its neighboring genes, shams alleles, and the shams and CG11836 rescue transgene. Black boxes indicate the coding parts of exons. (B) Adult wing of a wild-type fly. (C,D) shamsPB/PB (e01256) mutants raised at 25°C exhibit a partially penetrant loss of the posterior cross-vein (C) and at 30°C lose the distal portion of the L5 wing vein (arrowheads) (D). (E) Precise excision of the piggyBac insertion results in animals with normal wing veins. (F,G) Adult wings of shamsΔ34/Df(3R)BSC494 flies raised at 25°C lose wing vein material at the distal end of L5 (F) and at 30°C exhibit substantial loss of L4, L5, and posterior cross-vein (G). (H,I) Overexpression of shams with nubbin-GAL4 does not cause any phenotypes in the wing (H), but rescues the wing vein loss in shamsΔ34/Df(3R)BSC494 animals (I). (J) shamsgt-wt rescues shamsΔ34/Df(3R)BSC494 wing defects. (K) A genomic rescue transgene harboring CG11836 does not rescue the wing vein phenotype of shamsΔ34/Df(3R)BSC494 animals. (L) Wild-type adult heads have two ocellar bristles and two post-vertical bristles (arrowheads). (M) In shamsΔ34/Df(3R)BSC494 mutants raised at 30°C, ocellar and post-vertical bristles are lost. (N) This bristle phenotype is rescued by shamsgt-wt (arrowheads). To examine whether the observed phenotypes are indeed due to increased Notch signaling, we performed genetic interaction studies. As reported previously, loss of one copy of Notch results in wing margin defects and wing vein expansion (Figure 3A) [27]. Homozygosity for the shamsPB allele suppresses the N55e11/+ haploinsufficient phenotypes (Figure 3B and Figure S3). In a reciprocal experiment, we find that shamsPB enhances the wing vein loss observed in the NAx-E2 dominant gain-of-function allele (Figure 3C and 3D). Together, these observations indicate that Shams decreases the activity of Notch, potentially by adding xylose to O-glucose residues on Notch EGF repeats.
Fig. 3. Xylosyltransferases inhibit Drosophila Notch signaling. 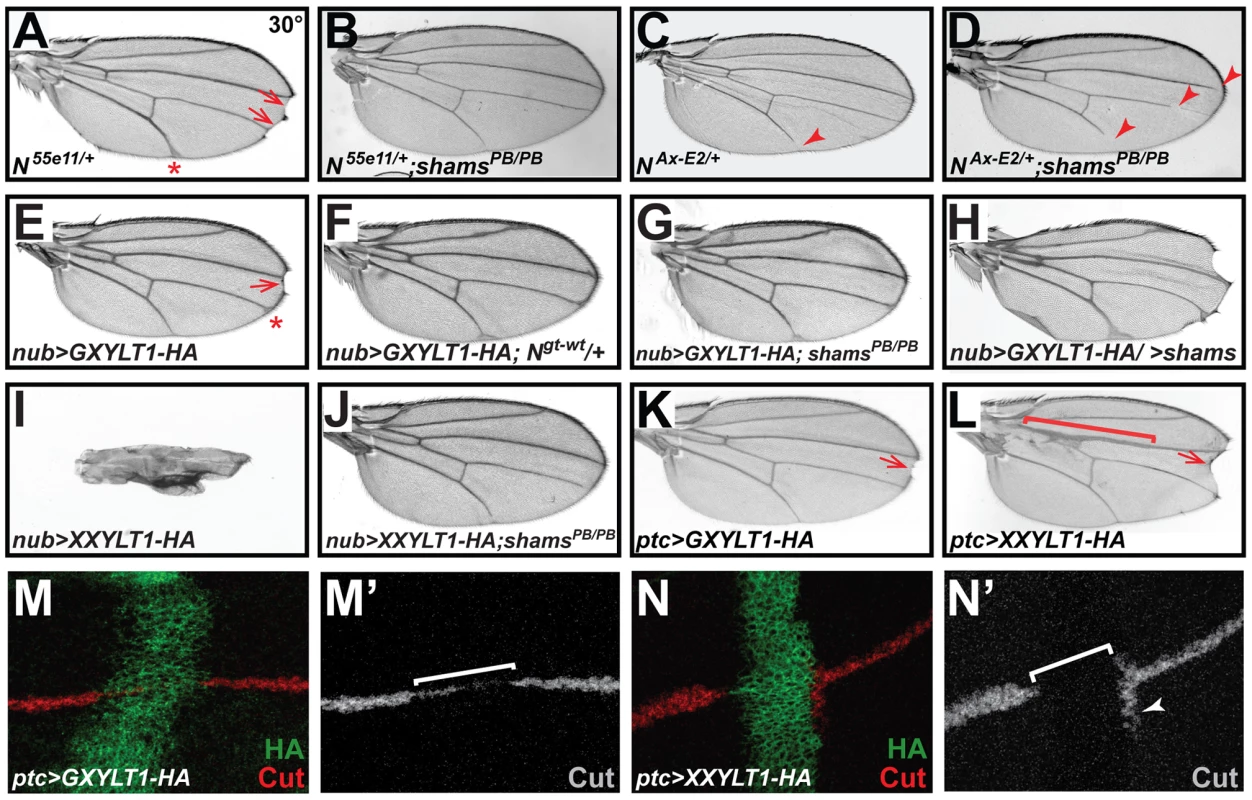
All animals were raised at 30°C. (A) N55e11/+ animals show wing vein thickening (asterisk) and margin defects (arrows). (B) shamsPB/PB suppresses the N55e11/+ phenotypes. (C) The Abruptex mutant NAx-E2/+ exhibits loss of wing vein at distal L5 (arrowhead) and occasionally L2. (D) shamsPB/PB enhances the NAx-E2/+ phenotype. (E) Wing-specific overexpression of HA-tagged human GXYLT1 by nubbin-GAL4 (nub>GXYLT1-HA) induces wing vein thickening (asterisk) and wing margin scalloping (arrow). (F) An additional copy of Notch suppresses the wing margin defect in nub>GXYLT1-HA. (G) The wing margin defect of nub>GXYLT1-HA flies is suppressed in a shamsPB/PB background. Note that GXYLT1-HA rescues the L5 wing vein loss phenotype of shamsPB/PB (compare to Figure 2D). (H) Co-overexpression of GXYLT1-HA and Shams results in the enhancement of the wing margin loss and wing vein expansion. (I) Overexpression of human XXYLT1-HA in the wing results in severe wing vein expansion and a complete loss of wing margin. (J) shamsPB/PB fully suppresses these phenotypes, but XXYLT1-HA overexpression does not suppress the L5 vein loss phenotype of shams PB/PB, indicating that the XXYLT1-HA phenotypes are strictly mediated by its enzymatic activity. (K) Overexpression of GXYLT1-HA by patched-GAL4 results in a mild wing margin loss in the patched domain (arrow). (L) Overexpression of XXYLT1-HA by patched-GAL4 results in wing vein thickening (bracket) and a more pronounced wing margin loss (arrow) in the patched domain. (M–N′) Double staining of wing imaginal discs by antibodies against HA (green) and the Notch downstream target Cut (red in M,N; gray in M′,N′). Note that the loss of Cut is less severe upon GXYLT1-HA overexpression (M,M′) compared to that resulting from XXYLT1-HA overexpression (N,N′). Overexpression of Human Xylosyltransferases Inhibits Notch Signaling
To examine the effects of increased xylosylation on Notch signaling, we overexpressed an HA-tagged version of human GXYLT1 in developing fly wings. We observed margin scalloping in 40% of adult wings at 30°C (Figure 3E) and a mild wing vein expansion at 25°C and 30°C (Figure 3E and Figure S4). Increasing the Notch gene dosage suppressed the wing margin loss caused by GXYLT1-HA overexpression (Figure 3F), indicating that the phenotype is due to decreased Notch signaling. Overexpression of GXYLT1-HA in a shamsPB/PB background resulted in the mutual suppression of the wing scalloping and vein loss phenotypes observed in each genotype (Figure 3G; compare to Figure 3E and Figure 2D). Simultaneous overexpression of GXYLT1-HA and Shams resulted in an enhancement of the wing vein and margin defects (Figure 3H). Together, these data indicate that Shams and GXYLT1 are functionally homologous, although GXYLT1 seems to be more potent than Shams (see Figure 2H), and suggest that increased xylosylation negatively regulates Notch signaling. Ectopic expression of human XXYLT1-HA resulted in a dramatic loss of margin and thickening of the wing veins, consistent with severe loss of Notch signaling (Figure 3I and Figure S4). In accordance with their enzymatic functions (Figure 1) [22], the XXYLT1-HA overexpression phenotypes are completely suppressed in a shamsPB/PB background with 100% penetrance (Figure 3J; n = 10). This indicates that the observed phenotypes are due to the enzymatic activity of XXYLT1, as they are dependent on Shams to generate xylose-glucose disaccharide substrates. The data also agree with the homology searches which indicate that Shams is the only GXYLT in flies.
To more directly show that the phenotypes caused by overexpressing human xylosyltransferases are due to a loss of Notch signaling, we overexpressed these enzymes along the antero-posterior axis of the developing wing discs by using the patched (ptc)-GAL4 driver. Again, both enzymes showed phenotypes compatible with loss of Notch signaling in the adult wings, with XXYLT1-HA phenotypes being stronger than the GXYLT1-HA phenotypes (Figure 3K and 3L). Overexpression of XXYLT1-HA resulted in loss of wing margin tissue and a collapse of the L3 and L4 wing veins without affecting proliferation or apoptosis (Figure 3L and S5A–D′). These phenotypes are specific to XXYLT1 overexpression, because they are fully rescued in a shams background (Figure S5E). Antibody staining of the third instar wing imaginal discs in these animals showed that the Notch downstream target Cut is either decreased (GXYLT1-HA) or lost (XXYLT1-HA) in the ptc-GAL4 domain upon human xylosyltransferase overexpression (Figure 3M–3N′). Altogether, these observations indicate that xylosylation negatively regulates Notch signaling.
EGF Repeats 16–20 of the Drosophila Notch Harbor the Functionally Important Sites of Xylosylation
Notch transgenes harboring serine-to-alanine mutations in all or most O-glucosylation sites show a temperature-sensitive loss of Notch signaling, similar to rumi animals [14], [20]. However, when smaller subsets of the O-glucosylation sites are mutated and the animals are raised at 25°C or lower, the negative effects of loss of O-glucose on Notch is significantly decreased [20]. If loss of xylose on specific EGF repeats results in increased Notch signaling, these mutations should recapitulate the shams mutant phenotypes, as loss of O-glucose precludes the addition of xylose. To test this, we generated animals that lack endogenous Notch but are rescued by one copy of Notch genomic transgenes carrying mutations in various subsets of O-glucosylation sites (Figure 4A) [20]. Serine-to-alanine mutations in EGF10–15 or EGF24–35 did not result in loss of wing vein, similar to a wild-type Notch transgene (Figure 4B, 4C and 4E). However, O-glucose mutations in EGF16–20 resulted in a partial loss of wing veins L2, L4, and L5 (Figure 4D), similar to but somewhat stronger than the shams null phenotypes (Figure 2F and 2G). N54l9/Y; Ngt-16_20/+ animals also exhibited head bristle defects similar to shams mutants (Figure S6). These observations nicely match our mass spectrometry data (Figure 1G) and indicate that xylosylation of EGF16–20 plays a negative regulatory role in Drosophila Notch signaling.
Fig. 4. Xylosylation of EGF16–20 negatively regulates Drosophila Notch signaling in vivo. 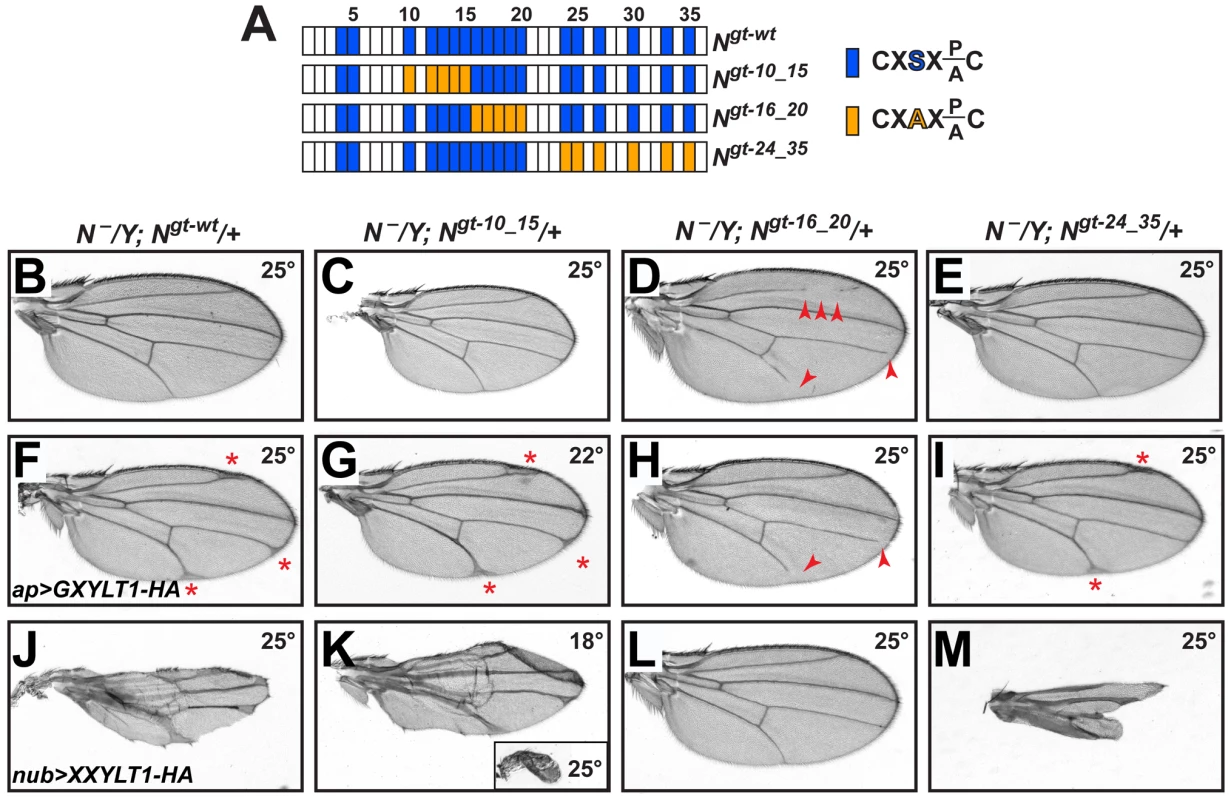
(A) Schematic of the EGF repeats of wild-type and mutant Notch genomic transgenes. Blue boxes show EGF repeats with a consensus O-glucosylation site; orange boxes denote EGF repeats with a serine-to-alanine mutation in the O-glucosylation site, which prevents the addition of O-glucose and therefore xylose. (B–E) N−/Y; Ngt-wt/+, N−/Y; Ngt-10_15/+, and N−/Y; Ngt-24_35/+ males exhibit no wing vein loss, but N−/Y; Ngt-16_20/+ males (D) exhibit loss of L2, L4 and L5 veins (arrowheads). (F) At 25°C, N−/Y; Ngt-wt/+ males expressing GXYLT1-HA in the apterous-GAL4 domain show thickening of the distal wing veins. (G) In a N−/Y; Ngt-10_15/+ background, ap>GXYLT1-HA becomes lethal at 25°C, and is not suppressed at 22°C. (H) In a N−/Y; Ngt-16_20/+ background, the ap>GXYLT1-HA phenotype is fully suppressed. Note the presence of wing vein loss. (I) Ngt-24_35 does not suppress the ap>GXYLT1-HA phenotype. (J) At 25°C, N−/Y; Ngt-wt/+ males expressing nub>XXYLT1-HA show severe wing vein and margin defects. (K) The phenotypes are dramatically enhanced in N−/Y; Ngt-10_15/+ males raised at 25°C (inset) and are comparable to (J) when raised at 18°C. (L,M) The nub>XXYLT1-HA phenotypes are fully suppressed in N−/Y; Ngt-16_20/+ males (L), but are enhanced in N−/Y; Ngt-24_35/+ males (M; compare to J). All wings, including the inset in M, are shown to scale. One prediction from the above conclusion is that mutations in EGF16–20 should suppress the loss of Notch signaling caused by the overexpression of human xylosyltransferases. To test this, we overexpressed GXYLT1-HA and XXYLT1-HA in genetic backgrounds lacking endogenous Notch and rescued by one copy of a wild-type or O-glucose mutant Notch transgenes. Overexpression of these enzymes in animals with one copy of a wild-type Notch transgene raised at 25°C results in phenotypes very similar to their overexpression in a wild-type background raised at the same temperature (Figure 4F and 4J and Figure S4). Among the mutant Notch transgenes, Ngt-16_20 is the only one which could fully suppress both GXYLT1-HA and XXYLT1-HA overexpression phenotypes (Figure 4G–4I and 4K–4M; n = 10 for each genotype;100% penetrance). Of note, in Ngt-10_15 and Ngt-24_35 backgrounds the XXYLT1-HA overexpression phenotype is even enhanced, most likely due to the negative effect of loss of these O-glucose residues on Notch signaling (Figure 4K and 4M) [20]. Altogether, these data indicate that Shams regulates Notch signaling via an enzymatic mechanism, and that the O-glucosylation sites in Drosophila Notch EGF16–20 are the biologically-relevant targets of xylosylation by Shams.
Notch Surface Expression in the Pupal Wing Is Increased upon Loss of Shams or Mutating O-glucose Sites in EGF16–20 of Notch
To examine the effects of loss of shams on Notch localization, we performed Notch surface staining on larval wing imaginal discs and pupal wings harboring shams mutant clones. Loss of shams does not affect the surface expression of Notch in third instar wing imaginal disc (Figure S7A–A″). However, more Notch protein is present in and at the surface of shams mutant cells in the pupal wing (Figure 5A and 5A′ and Figure S7B–B′). We also sought to determine whether mutating the O-glucose sites on EGF16–20 of Notch results in increased cell surface levels of Notch. To this end, we generated Mosaic Analysis with a Repressible Cell Marker (MARCM) clones [28] of a protein-null allele of Notch in a background harboring one copy of the wild-type or an EGF16–20 mutant Notch, similar to what we have described before for other mutant Notch transgenes [20]. In these animals, heterozygous cells have both endogenous Notch and a copy of our transgene, but cells in the mutant clones only harbor a copy of the transgene. Accordingly, the level of Notch expressed from the wild-type transgene in the clones is less than that in the heterozygous cells (Figure 5B and 5B′), in agreement with our previous report [20]. Clones of Notchgt-16_20 in larval wing disc do not show an increase in Notch surface expression (data not shown). However, in the pupal wing, the level of surface Notch expressed from the Notchgt-16_20 transgene in the clones is significantly increased compared to that expressed from the wild-type Notchgt-wt transgene (Figure 5C and 5C′; compare to 5B′). One potential explanation for the difference between the effects of loss of Notch xylosylation in pupae versus larvae could be different levels of Shams expression at these stages. Indeed, Western blot confirmed higher Shams expression in pupae compared to third instar larvae (Figure 5D). In agreement with a role for xylose in surface expression of Notch, overexpression of human XXYLT1 results in a significant decrease and overexpression of human GXYLT1 results in a mild and partially penetrant decrease in Notch surface expression in the larval wing imaginal discs (Figure S8). Altogether, these observations suggest that addition of xylose residues to EGF16–20 of Notch decreases the availability of Notch at the cell surface.
Fig. 5. Increased surface expression of Notch in shams and Notchgt-16_20 clones in the pupal wing. 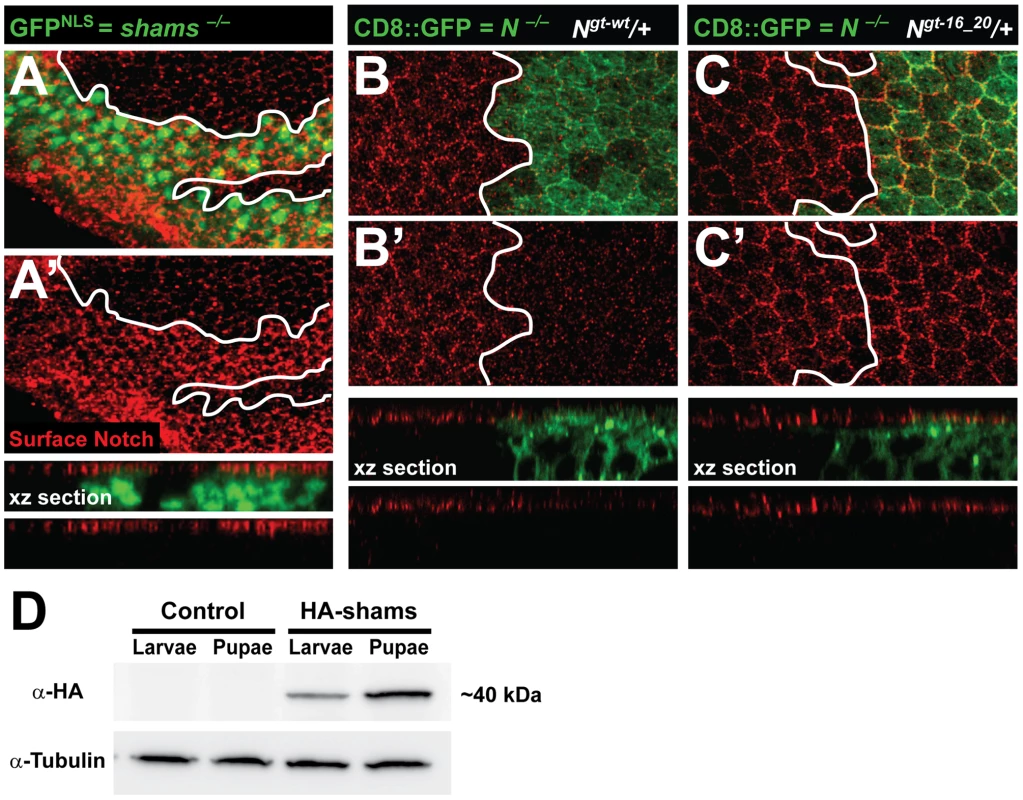
All animals were raised at 30°C. (A,A′) Shown are confocal images from a pupal wing at 22–24 hours after puparium formation (APF) with a MARCM clone of shamsΔ34 marked by nuclear GFP (GFPNLS). Surface expression of Notch is shown in red. Note, also in the xz section, that the Notch surface level at this stage is increased in shams mutant cells. (B–C′) Shown are confocal images of pupal wings around 22 hours APF from animals harboring MARCM clones of the Notch54l9 protein-null allele (marked by CD8::GFP) with one copy of either a wild-type Notch transgene (B,B′) or a Notch transgene with O-glucose mutations in EGF16–20 (C,C′). The only source of Notch in the clones is the Notch transgene. Note, also in xz sections, that the level of surface Notch in clones harboring the Notchgt-16_20 is increased compared to that in clones harboring Notchgt-wt. (D) Anti-HA Western blot on larval and pupal protein extracts from animals harboring one copy of an HA-tagged Shams genomic transgene (HA-Shams; shamsgt-wt-HA-attVK22) or attVK22 control animals. Tubulin was used as loading control. Pupal extracts show relatively higher levels of HA-Shams. Discussion
Our data indicate that there are significant differences between the ways O-glucose monosaccharides and their extended (xylosylated) form regulate Drosophila Notch signaling. First, O-glucosylation promotes Notch signaling [14], [20], but xylosylation inhibits Notch signaling. Secondly, loss of O-glucosylation affects Notch signaling in all contexts studied so far [14], [20], [29], but loss of xylosylation only affects Notch signaling in certain contexts, i.e. wing vein development and head bristle formation. Finally, O-glucose residues on all EGF repeats contribute to the Notch signal strength in redundant and/or additive fashions [20], but xylose residues seem to be only required on a subset of Notch EGF repeats. In other words, unlike glucose residues which function globally on the Notch extracellular domain to promote Notch signaling in various contexts [14], [20], [29], xylose residues function locally on a specific region of Notch to decrease signaling in certain contexts. Thus, our data show that the strength of the Notch signaling pathway can be fine-tuned by altering the distribution and relative levels of O-glucose monosaccharide and their xylose-containing extended forms on the Notch receptor. Since xylose-xylose-glucose glycans on EGF repeats are the only known glycans in animals with a terminal xylose [30], [31], our data strongly suggest that at least in Drosophila, terminal xylose residues play a fairly specific role in regulating the Notch signaling pathway.
Our observations provide compelling evidence that the negative effects of xylosyltransferases on Drosophila Notch signaling are primarily mediated via their enzymatic activity. First, serine-to-alanine mutations in the O-glucosylation sites in EGF16–20 of the Drosophila Notch recapitulate the wing vein and head bristle phenotypes of shams. Since loss of the protein O-glucosyltransferase Rumi results in loss of Notch signaling [14], [20], the observed gain of Notch signaling phenotypes cannot be due to loss of O-glucose from these EGF repeats, but are very likely due to the loss of xylose normally attached to the O-glucose. Secondly, mutating the O-glucose sites of EGF16–20 fully suppresses the Notch loss-of-function phenotypes caused by GXYLT1 and XXYLT1 overexpression, strongly suggesting that these phenotypes result from the addition of xylose by these enzymes to O-glucose on these EGF repeats. Lastly, although the Notch loss-of-function phenotypes of XXYLT1 overexpression are much more severe than those caused by GXYLT1 overexpression, loss of shams fully suppresses the XXYLT1 overexpression phenotypes but only partially suppresses the GXYLT1 overexpression phenotype. These data fully match the enzymatic activities of these proteins: GXYLT1 and Shams both add the first xylose to O-glucose and function in parallel, but XXYLT1 adds the second xylose and therefore completely depends on the activity of GXYLT1/Shams.
A hallmark of Notch receptors is the presence of many EGF repeats in their extracellular domain. However, the functional importance of only a handful of Notch EGF repeats has been elucidated. Specifically, EGF11 and 12 are necessary for ligand-binding [32], EGF 24, 25, 27 and 29 negatively regulate Notch signaling, likely through opposing ligand binding [33], and EGF8 is required for binding of Notch to Serrate but not Delta [34]. By showing that EGF16–20 (or a subset of them) negatively regulate Drosophila Notch signaling in a xylose-dependent manner, our data assign a function to these EGF repeats. Loss of shams or loss of xylosylation on EGF16–20 affects surface expression of Notch in the pupal wing and results in defects in wing vein formation, a process primarily regulated in the pupal stage. In contrast, loss of shams does not affect Notch expression in the larval wing discs, or the wing margin formation, which primarily occurs at the larval stage. Together, these observations suggest that Notch signaling in the pupal wing is more sensitive to loss of xylosylation compared to the larval wing disc, likely because of the relatively higher levels of Shams expression in the pupal stage. Nevertheless, given the loss of Notch signaling observed upon GXYLT1 and XXYLT1 overexpression, the larval wing discs likely have the machinery required to recognize and respond to xylose on Notch. Increased availability of Notch at the cell surface of shams and Notchgt-16_20 mutant clones in the pupal wing suggests a molecular mechanism for the role of xylose in regulating Notch signaling. However, it remains to be seen whether other steps of Notch signaling, including ligand-binding and cis-inhibition are also affected by the loss of xylose.
Although cell type variability exists, all O-glucosylated EGF repeats of the mouse Notch1 harbor xylose-xylose-glucose trisaccharides at high stoichiometry [19]. Our previous work [14] and current mass spectrometry experiments indicate that all of the EGF repeats of the Drosophila Notch with a consensus Rumi target motif analyzed thus far (16 out of 18) harbor O-glucose. However, we find that xylose is only added to a subset of O-glucosylated Notch EGF repeats in Drosophila. This difference might result from different efficiency of the mammalian versus Drosophila xylosyltransferases in vivo despite their similar in vitro levels of activity (Figure 1E). Indeed, overexpression of human GXYLT1, but not its Drosophila homolog Shams, results in Notch loss-of-function phenotypes in the wing, suggesting that GXYLT1 is more potent in adding xylose to Notch in vivo. Moreover, the difference between the distribution of (xylose)-xylose-glucose saccharides on fly and mammalian Notch could in part be due to the presence of two GXYLT enzymes in mammals instead of only one (Shams) in flies. Regardless of the mechanisms underlying the observed differences, it will be of great interest to determine whether alteration of the level or distribution of xylose-xylose-glucose saccharides on Notch receptors can modulate the strength of mammalian Notch signaling as well.
Materials and Methods
Drosophila Strains and Genetics
The following strains were used in this study: y w, y w; D/TM6, Tb1, w; nocSco/CyO, w; nocSco/CyO; TM3, Sb/TM6, Tb1, w; CyO, P{FRT(w+)Tub-PBac\T}2/wgSp-1, y w hsFLP; Dr/TM3, apterous-GAL4, patched-GAL4, y w N54l9 FRT19A/FM7, N55e11/FM7c, NAx-E2, Df(3R)BSC494/TM6C, Sb1, w; PBac{RB}CG999601256, attVK22, UAS-CG8::GFP (Bloomington Stock Center), w; PBac{RB}CG11836e01985 (Exelixis), shamsrev, shamsΔ34/TM6, Tb1, shamsgt-wt-attVK22, shamsgt-wt-HA-attVK22, CG11836gt-wt-attVK22, UASattB-shams-HA-VK22, UASattB-GXYLT1-HA-VK22, UASattB-XXYLT1-HA-VK22, Ubx-FLP FRT19A/FM7; Act-GAL4 UAS-CD8::GFP/CyO (this study), Ngt-wt-attVK22, Ngt-10_15-attVK22, Ngt-16_20-attVK22, Ngt-24_35-attVK22 [20], y w Ubx-FLP tub-GAL4 UAS-GFPnls-6X-Myc; FRT82B y+ tub-GAL80/TM6, Ubx [14], vas-int-ZH-2A; attVK22 [35], nubbin-GAL4 (Georg Halder). All crosses were performed on standard media and incubated at described temperatures.
To test the effect of Notch mutations on the adult wing, w; Ngt-wt, Ngt-10_15, Ngt-16_20, or Ngt-24_35 males were crossed to N54l9/FM7, B females. B+ male progeny were scored for wing and head bristle defects. To determine the effect of Notch mutations on GXYLT1-HA and XXYLT1-HA overexpression in the wing, B+, Cy+ male progeny were scored from apterous-GAL4 UAS-GXYLT1-HA/CyO or nubbin-GAL4 UAS-XXYLT1-HA/CyO males crossed with N54l9/FM7; Ngt-wt/+, N54l9/FM7; Ngt-10_15/+, N54l9/FM7; Ngt-16_20/+, or N54l9/FM7; Ngt-24_35/+ females. To generate MARCM clones of Notch harboring a Notch genomic transgene, Ubx-FLP FRT19A/Y; Act-GAL4 UAS-CD8::GFP/CyO males were crossed to Notch54l9/FM7; Ngt-wt/+ or N54l9/FM7; Ngt-16_20/+ females. All MARCM crosses were set at room temperature and transferred to 30°C at L1–L2 instar stage. For further details on Drosophila genetics and other techniques used in this study, please see Text S1.
Glycosyltransferase Assays
For recombinant expression of Shams (CG9996) the predicted C-terminal lumenal domain starting from Gln23 was amplified from Drosophila w1118 cDNA and cloned into the pFast-Bac1 vector (Invitrogen) encoding the HBM-secretion signal followed by the Protein A coding sequence, as described for human GXYLT1 [23]. Constructs were expressed in Sf9 insect cells using the Bac-to-Bac System (Invitrogen) and secreted proteins were purified by IgG-Sepahrose-6 Fast Flow beads (GE-Healthcare) as described [23]. Activity assays were performed on bead-coupled enzyme in the presence of radiolabeled UDP-sugars as described before [23], except that samples were incubated for 2 h at 27°C. To measure activity on EGF repeats, the Notch EGF16–20 fragment was expressed in Sf9 insect cells, purified by Nickel affinity chromatography, and used in an in vitro assay followed by mass spectrometric analysis as described [22].
O-Glucose Site Mapping on Drosophila Notch EGF1–36 Expressed in S2 Cells
A construct encoding EGF1–36 from Drosophila Notch with a C-terminal 3X-FLAG tag (EGF1–36-FLAG3, generously provided by Dr. Ken Irvine) was expressed in Drosophila S2 cells. EGF1–36-FLAG3 was purified from medium, reduced and alkylated, and subjected to in-gel protease digests as described [36]. O-Glucose modified glycopeptides were identified by neutral loss of the glycans during collision-induced dissociation (CID) using nano-LC-MS/MS as described [19].
Dissections, Staining, Image Acquisition and Processing
Dissection and staining were performed by using standard methods. For surface staining, third instar larval imaginal discs and pupal wings were dissected and incubated with anti-Notch antibody in the absence of detergent. Antibodies used were mouse α-Cut (2B10) 1∶500, mouse anti-Notch (C458.2H) 1∶100, mouse α-Delta (C594.9B) 1∶100 (Developmental Studies Hybridoma Band), goat α-HA 1∶50 (GenScript), mouse α-phosphorylated Histone H3 (Ser10) 1∶100, rabbit α-Cleaved Caspase 3 (Asp175) 1∶50 (Cell Signaling), donkey-α-goat-Dylight549 1∶500, donkey-α-mouse-Cy5 1∶500, goat-α-mouse-Cy3 1∶500, donkey-α-Rabbit-Cy5 1∶500, donkey-α-Guinea Pig-Cy3 1∶500 (Jackson ImmunoResearch Laboratories). Confocal images were scanned using a Leica TCS-SP5 microscope and processed with Amira5.2.2. Images were processed with Adobe Photoshop CS5; Figures were assembled in Adobe Illustrator CS5.
Western Blots
Third instar larvae and pupae aged 22 hours after puparium formation (APF) were collected from VK22 control animals or animals harboring one copy of the shamsgt-wt-HA genomic transgene. Protein extracts were generated using RIPA buffer (Boston BioProducts) and a Protease Inhibitor Cocktail (Roche). Protein extracts were separated on 12% acrylamide gel and transferred to PVDF membrane. Blots were probed with goat α-HA 1∶300 (GenScript), mouse α-Tubulin 1∶1000 (Santa Cruz Biotech), donkey-α-goat-HRP 1∶2000 and goat α-mouse-HRP 1∶2000 antibodies (Jackson ImmunoResearch Laboratories). Western blots were developed using Pierce ECL Western Blotting Substrates (Thermo Scientific) and imaged with an LAS4000 GE ImageQuant Imager. Three independent experiments showed the same result.
Supporting Information
Zdroje
1. Artavanis-TsakonasS, MuskavitchMA (2010) Notch: the past, the present, and the future. Curr Top Dev Biol 92 : 1–29.
2. KopanR, IlaganMX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137 : 216–233.
3. PentonAL, LeonardLD, SpinnerNB (2012) Notch signaling in human development and disease. Semin Cell Dev Biol 23 : 450–457.
4. LouviA, Artavanis-TsakonasS (2012) Notch and disease: A growing field. Semin Cell Dev Biol 23 : 473–480.
5. SouthAP, ChoRJ, AsterJC (2012) The double-edged sword of Notch signaling in cancer. Semin Cell Dev Biol 23 : 458–464.
6. AsterJC, BlacklowSC (2012) Targeting the Notch Pathway: Twists and Turns on the Road to Rational Therapeutics. J Clin Oncol 30 : 2418–2420.
7. HarrisRJ, SpellmanMW (1993) O-linked fucose and other post-translational modifications unique to EGF modules. Glycobiology 3 : 219–224.
8. MoloneyDJ, ShairLH, LuFM, XiaJ, LockeR, et al. (2000) Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem 275 : 9604–9611.
9. MatsuuraA, ItoM, SakaidaniY, KondoT, MurakamiK, et al. (2008) O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J Biol Chem 283 : 35486–35495.
10. TakeuchiH, Fernandez-ValdiviaRC, CaswellDS, Nita-LazarA, RanaNA, et al. (2011) Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc Natl Acad Sci U S A 108 : 16600–16605.
11. OkajimaT, IrvineKD (2002) Regulation of notch signaling by o-linked fucose. Cell 111 : 893–904.
12. ShiS, StanleyP (2003) Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci U S A 100 : 5234–5239.
13. SasamuraT, SasakiN, MiyashitaF, NakaoS, IshikawaHO, et al. (2003) neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development 130 : 4785–4795.
14. AcarM, Jafar-NejadH, TakeuchiH, RajanA, IbraniD, et al. (2008) Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132 : 247–258.
15. Fernandez-ValdiviaR, TakeuchiH, SamarghandiA, LopezM, LeonardiJ, et al. (2011) Regulation of the mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 138 : 1925–1934.
16. ZhouL, LiLW, YanQ, PetryniakB, ManY, et al. (2008) Notch-dependent control of myelopoiesis is regulated by fucosylation. Blood 112 : 308–319.
17. BrucknerK, PerezL, ClausenH, CohenS (2000) Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406 : 411–415.
18. MoloneyDJ, PaninVM, JohnstonSH, ChenJ, ShaoL, et al. (2000) Fringe is a glycosyltransferase that modifies Notch. Nature 406 : 369–375.
19. RanaNA, Nita-LazarA, TakeuchiH, KakudaS, LutherKB, et al. (2011) O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J Biol Chem 286 : 31623–31637.
20. LeonardiJ, Fernandez-ValdiviaR, LiYD, SimcoxAA, Jafar-NejadH (2011) Multiple O-glucosylation sites on Notch function as a buffer against temperature-dependent loss of signaling. Development 138 : 3569–3578.
21. WhitworthGE, ZandbergWF, ClarkT, VocadloDJ (2010) Mammalian Notch is modified by D-Xyl-alpha1-3-D-Xyl-alpha1-3-D-Glc-beta1-O-Ser: implementation of a method to study O-glucosylation. Glycobiology 20 : 287–299.
22. SethiMK, BuettnerFF, AshikovA, KrylovVB, TakeuchiH, et al. (2012) Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch. J Biol Chem 287 : 2739–2748.
23. SethiMK, BuettnerFF, KrylovVB, TakeuchiH, NifantievNE, et al. (2010) Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J Biol Chem 285 : 1582–1586.
24. de CelisJF, BarrioR, del ArcoA, Garcia-BellidoA (1993) Genetic and molecular characterization of a Notch mutation in its Delta - and Serrate-binding domain in Drosophila. Proc Natl Acad Sci U S A 90 : 4037–4041.
25. ParksAL, CookKR, BelvinM, DompeNA, FawcettR, et al. (2004) Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36 : 288–292.
26. RoyetJ, BouwmeesterT, CohenSM (1998) Notchless encodes a novel WD40-repeat-containing protein that modulates Notch signaling activity. EMBO J 17 : 7351–7360.
27. MohrOL (1919) Character Changes Caused by Mutation of an Entire Region of a Chromosome in Drosophila. Genetics 4 : 275–282.
28. LeeT, LuoL (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24 : 251–254.
29. PerdigotoCN, SchweisguthF, BardinAJ (2011) Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development 138 : 4585–4595.
30. BakkerH, OkaT, AshikovA, YadavA, BergerM, et al. (2009) Functional UDP-xylose transport across the endoplasmic reticulum/Golgi membrane in a Chinese hamster ovary cell mutant defective in UDP-xylose Synthase. J Biol Chem 284 : 2576–2583.
31. Freeze HH, Elbein AD (2009) Glycosylation Precursors. In: Varki A, Cummings, R. D., Esko JD, Freeze HH, Stanley P, et al.., editors. Essenstial of Glycobiology. New York: Cold Spring Harbor Laboratory Press. pp. 47–61.
32. RebayI, FlemingRJ, FehonRG, CherbasL, CherbasP, et al. (1991) Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67 : 687–699.
33. PeiZ, BakerNE (2008) Competition between Delta and the Abruptex domain of Notch. BMC Dev Biol 8 : 4.
34. YamamotoS, CharngWL, RanaNA, KakudaS, JaiswalM, et al. (2012) A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science 338 : 1229–1232.
35. VenkenKJ, HeY, HoskinsRA, BellenHJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 : 1747–1751.
36. XuA, HainesN, DlugoszM, RanaNA, TakeuchiH, et al. (2007) In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe. J Biol Chem 282 : 35153–35162.
Štítky
Genetika Reprodukčná medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene ClusterČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



