-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Endemic Dengue Associated with the Co-Circulation of Multiple Viral Lineages and Localized Density-Dependent Transmission
Dengue is one of the most important infectious diseases of humans and has spread throughout much of the tropical and subtropical world. Despite this widespread dispersal, the determinants of dengue transmission in endemic populations are not well understood, although essential for virus control. To address this issue we performed a phylogeographic analysis of 751 complete genome sequences of dengue 1 virus (DENV-1) sampled from both rural (Dong Thap) and urban (Ho Chi Minh City) populations in southern Viet Nam during the period 2003–2008. We show that DENV-1 in Viet Nam exhibits strong spatial clustering, with likely importation from Cambodia on multiple occasions. Notably, multiple lineages of DENV-1 co-circulated in Ho Chi Minh City. That these lineages emerged at approximately the same time and dispersed over similar spatial regions suggests that they are of broadly equivalent fitness. We also observed an important relationship between the density of the human host population and the dispersion rate of dengue, such that DENV-1 tends to move from urban to rural populations, and that densely populated regions within Ho Chi Minh City act as major transmission foci. Despite these fluid dynamics, the dispersion rates of DENV-1 are relatively low, particularly in Ho Chi Minh City where the virus moves less than an average of 20 km/year. These low rates suggest a major role for mosquito-mediated dispersal, such that DENV-1 does not need to move great distances to infect a new host when there are abundant susceptibles, and imply that control measures should be directed toward the most densely populated urban environments.
Published in the journal: Endemic Dengue Associated with the Co-Circulation of Multiple Viral Lineages and Localized Density-Dependent Transmission. PLoS Pathog 7(6): e32767. doi:10.1371/journal.ppat.1002064
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002064Summary
Dengue is one of the most important infectious diseases of humans and has spread throughout much of the tropical and subtropical world. Despite this widespread dispersal, the determinants of dengue transmission in endemic populations are not well understood, although essential for virus control. To address this issue we performed a phylogeographic analysis of 751 complete genome sequences of dengue 1 virus (DENV-1) sampled from both rural (Dong Thap) and urban (Ho Chi Minh City) populations in southern Viet Nam during the period 2003–2008. We show that DENV-1 in Viet Nam exhibits strong spatial clustering, with likely importation from Cambodia on multiple occasions. Notably, multiple lineages of DENV-1 co-circulated in Ho Chi Minh City. That these lineages emerged at approximately the same time and dispersed over similar spatial regions suggests that they are of broadly equivalent fitness. We also observed an important relationship between the density of the human host population and the dispersion rate of dengue, such that DENV-1 tends to move from urban to rural populations, and that densely populated regions within Ho Chi Minh City act as major transmission foci. Despite these fluid dynamics, the dispersion rates of DENV-1 are relatively low, particularly in Ho Chi Minh City where the virus moves less than an average of 20 km/year. These low rates suggest a major role for mosquito-mediated dispersal, such that DENV-1 does not need to move great distances to infect a new host when there are abundant susceptibles, and imply that control measures should be directed toward the most densely populated urban environments.
Introduction
Dengue is the most important mosquito-borne viral disease of humans, annually responsible for approximately 40 million cases and some 20,000 deaths in tropical and subtropical regions [1]. Dengue is caused by one of four single-stranded positive-sense RNA viruses (DENV-1 to DENV-4, also referred to as serotypes) of the genus Flavivirus (family Flaviviridae). Despite the large burden of dengue disease, and considerable research effort, there are currently no licensed vaccines or specific therapies. The challenge of effective and safe dengue vaccination is increased by the possibility that imperfect cross-protective vaccination could enhance DENV infection, or even virulence [2], and that lineages within individual DEN viruses, particularly different ‘genotypes’, may also differ in antigenicity [3]–[6]. In addition, the population dynamics of DENV within individual localities are complex, involving the birth-and-death of viral lineages that may also differ in both virulence and fitness [7]–[13], as well as the intricate patterns of gene flow, at both the local and international scales [7], [14], [15].
DENV transmission among humans is largely caused by the urban adapted and anthropophillic Aedes aegypti mosquito. Spatial and temporal patterns of dengue prevalence are likely driven by multiple factors including the immune status of human hosts [16], their age [17], [18], virus traits [13], [19], [20], the mosquito vector, and environmental variables including aspects of climate such as levels of precipitation [21], [22]. Human movement must also be an important, but poorly understood, contributor to viral transmission dynamics, and is obviously responsible for the increasingly widespread and complex distribution of the four DEN viruses at the global scale. On a local scale, how much DENV transmission within a specific population is due to the local movement of infected human hosts rather than of mosquitoes is unclear. Understanding the spatial and temporal dynamics of dengue transmission in endemic dengue populations is therefore central to the rational deployment of vector control activities and the design of intervention strategies. In this respect it is critical to determine the spatial structure of DENV within endemic populations, the rate at which DENV lineages diffuse through space (particularly in the face of a partially immune population), whether specific lineages are spreading more rapidly than others and indicative of enhanced fitness, and the likely contribution of mosquitoes and humans to local transmission patterns.
To address these questions we employed a fine-scale molecular approach to characterize the virus population dynamics of a recent DENV-1 outbreak in southern Viet Nam, a region of high dengue endemicity. Between 2006–2008 the estimated incidence of DENV-1 infection in the southern twenty provinces of Viet Nam ranged from 86–190 cases/100,000 [13], markedly higher than during the preceding six-year period when it ranged from 1–28 cases/100,000. The causes of this increased incidence are unknown.
To determine the patterns and dynamics of dengue transmission we utilized an expansive data set of DENV-1 whole genome sequences sampled prior to and during the peak in DENV-1 prevalence over a period of six years (2003–2008). We inferred the dynamics of viral transmission within individual communities, between communities, and between neighboring countries, using recently developed Bayesian phylogenetic methods that utilize both the temporal and spatial information of the sampled sequences. Uniquely, these time-calibrated phylogenetic methods provide the ability to reveal the complex interplay of spatial, genetic and epidemiological dynamics at the local, regional and global scales, and have the ability to consider individual viral lineages, whereas epidemiological approaches based on the analysis of incidence data are at best only able to distinguish among the four DEN viruses.
Results
Phylogeography of DENV-1 in South East Asia
We determined the consensus DENV-1 genome sequence (minimum sequence from nt 70–10,400) in acute plasma samples collected from 751 hospitalized patients in urban Ho Chi Minh City (HCMC) (n = 575 sampled between 2003–2008) and rural Dong Thap Province in the Mekong Delta region (n = 176 sampled between 2006–2007). The majority of viruses were sampled from 2006 to 2008 during which DENV-1 was the most prevalent serotype in circulation (Figure S1).
To determine the evolutionary relationships of DENV-1 in Viet Nam in the context of surrounding countries we analyzed the envelope (E) gene sequences from these locations (Figure 1A). The 751 DENV-1 sequences sampled from Viet Nam fell into one of five clades within the broader Genotype I cluster of viruses [23]. Four of the five clades consistently clustered within the diversity of Cambodian viruses with good support (posterior probability ranging from 0.81 to 1.0). This phylogeographic evidence, coupled with Cambodia and Viet Nam's shared border, is compatible with Cambodia acting as the major source of Vietnamese DENV-1. A caveat to this is the lack of contemporaneous DENV-1 sequences from nearby Thailand, which has previously been shown to harbor substantial DENV diversity and importation into Viet Nam [13]. Clearly, wider sampling in both time and space is needed to test this hypothesis.
Fig. 1. Maximum Clade Credibility (MCC) trees from the discrete phylogeography analysis of DENV-1. 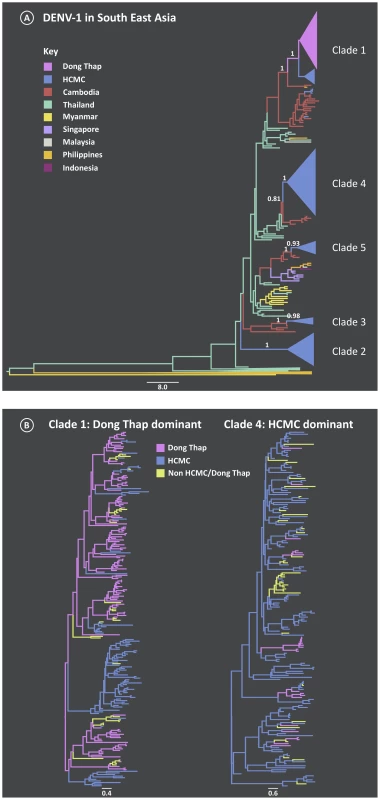
A) South East Asia tree reconstructed from the E gene, where the branches are colored by location of viral samples. The number above the branch indicates the posterior probability support for the Vietnamese clades and with nearest sister clades; B) An in-depth look at Vietnamese clades 1 and 4 which were found to be Dong Thap dominant and HCMC dominant, respectively. The branches are colored by sampling location (Dong Thap, HCMC or Non- HCMC and Dong Thap). The majority of the clades largely comprised viruses from HCMC, with the exception of clade 1, which was found to be Dong Thap dominant. The timing of these inferred introductions were gauged from the age of the most recent common ancestor (TMRCA) of each clade (Table 1). The period in which these different viral clades emerged in southern Viet Nam ranged from late 2001 to mid-2005. Apart from clade 1, which was found to be the most recent introduction, the mean ages of clades 2–5 did not differ significantly, suggesting that different viral lineages were imported over short or similar time-scales, and then co-circulated. These clades were chosen for more detailed phylogeographic analysis. Finally, genome-wide rates of nucleotide substitution – at ∼1×10−3 nucleotide substitutions per site, per year (Table 1) – were the same among clades and highly consistent with those previously determined for DENV [24], [25].
Tab. 1. Rate of nucleotide substitution of DENV-1 for each clade in Viet Nam, and the inferred time of the most recent common ancestor (TMRCA). 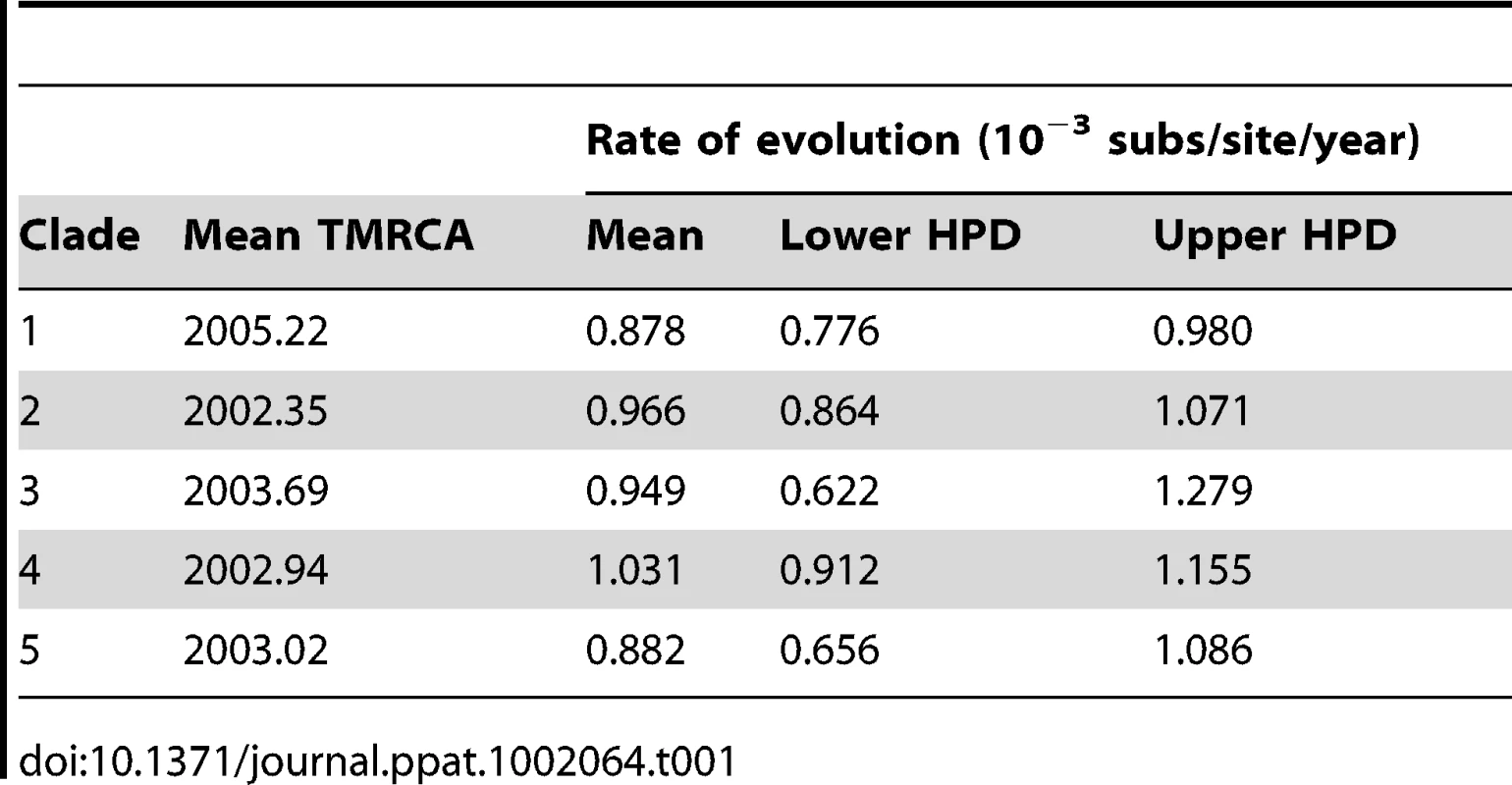
Viral migration between Dong Thap and HCMC
For the clades identified as being within Viet Nam, a discrete spatial model [26] was employed to reveal the migration between the sampling locations. The results are shown in Figure 1B, in which branches are colored by the most probable state location. In four of the five clades HCMC was the most likely viral source, with viruses exported to the rural area of Dong Thap. The non-HCMC isolates in these clades were interspersed among the HCMC sampled isolates, which strongly suggested that the DENV-1 epidemics in southern Viet Nam mainly emerged first in HCMC. The exception was clade 1, which was dominated by Dong Thap viruses and where Dong Thap was inferred to be the most likely place of origin. Moreover, the HCMC viruses in clade 1 did not form a monophyletic group, supporting the view that clade 1 viruses were imported into HCMC on multiple occasions from Dong Thap.
To determine whether the viral migration rates varied between urban and rural epidemics, we compared the spatial dynamics between clades 1 and 4 (Table 2). When focusing on the number of transitions from the inferred source location, a symmetrical pattern was observed between the two clades. For instance, the transmission rate between HCMC and Dong Thap was higher in the HCMC dominant clade 4, while for the reverse direction (Dong Thap to HCMC) it was greater in Dong Thap dominant clade 1. Hence, once a virus became established in a location, rural or urban, the rate of viral exportation was found to be greater than the rate of viral importation.
Tab. 2. Migration between HCMC and Dong Thap (DT) in clade 1 and clade 4, the number of transitions between each state along a branch in the tree using the robust counting method. 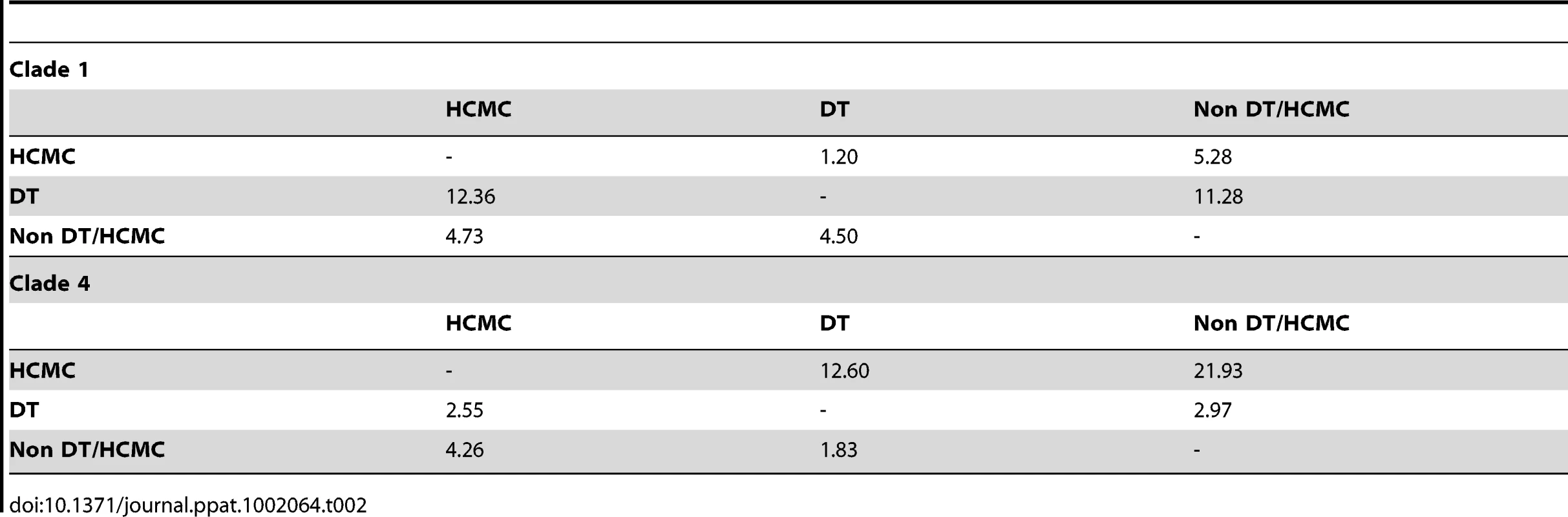
DENV-1 dispersion in urban and rural locales
The geographical coordinates of the patient's residential address in HCMC (n = 381) or Dong Thap Province (n = 175) was known for 556 cases and this information was employed to reconstruct the fine-scaled dispersion of the individual viral lineages within the sampling areas using a continuous spatial diffusion model with non-homogenous dispersion rates [27]. The average viral dispersion rate (km/year) was calculated for each clade, and separately for HCMC and/or Dong Thap data subsets, as if the epidemic in these regions derived from a single introduction (Table 2). We define virus dispersion rate as a measure of how quickly a virus lineage spreads geographically, given the inferred root location and final sampling locations. Even though we only had one estimate of the average dispersion rate of DENV-1 in Dong Thap, a clear disparity was observed when compared to the rates from HCMC lineages (Table 3). Specifically, the viral lineages from clade 1 in Dong Thap spread approximately 2–3 times faster than any lineage from HCMC. This is indicative of a fundamental difference in the epidemiological dynamics of DENV-1 in the two areas.
Tab. 3. Rate of virus dispersion of DENV-1 and mean age of each clade in the different localities in Viet Nam. 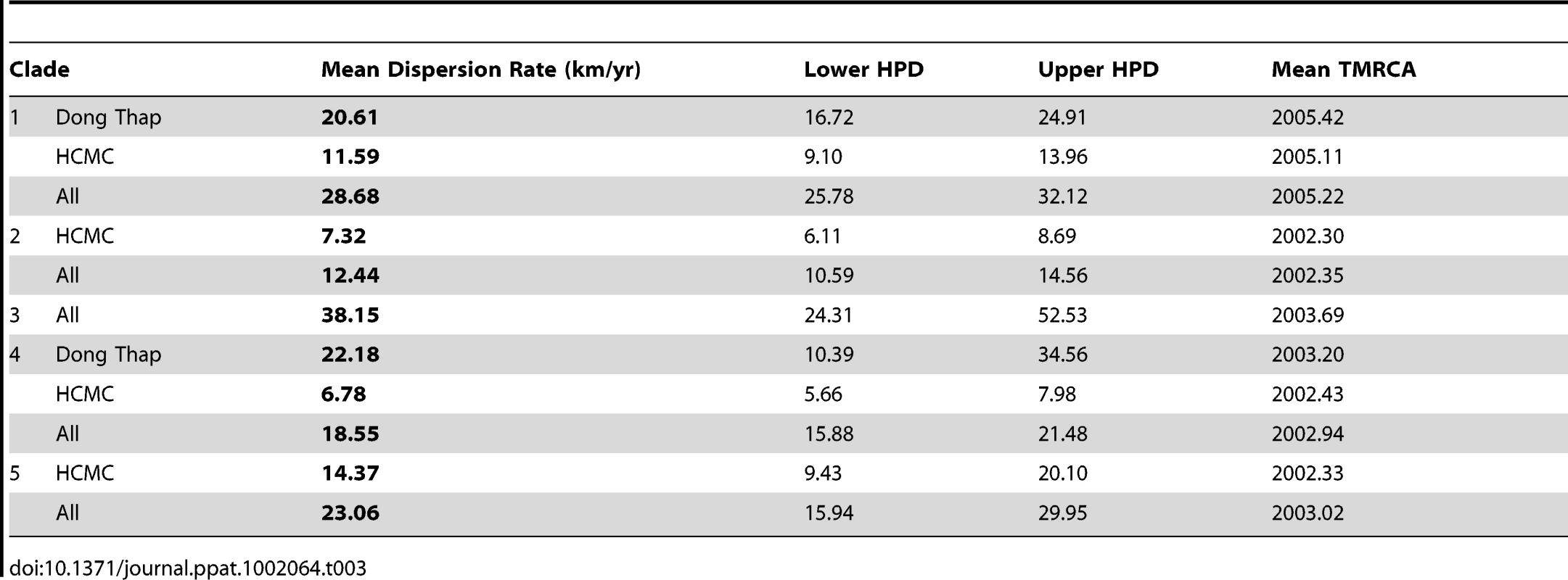
A further dissection of the dispersion rates through time in HCMC (clades 2, 4 and 5) and Dong Thap (clade 1) revealed interesting patterns in the rate of viral spread in the two locations. In HCMC (Figure 2A and B), the monthly incidence of DENV-1 showed a similar trend as in Dong Thap, with corresponding regular fluctuations and an increasing overall trend. However, there was no clear association between genetic diversity, incidence, and dispersion rate observed in the urban environment demonstrated by the roughly horizontal relationship in Figure 2B and the overlapping 95% HPD (highest posterior density) intervals. Hence, although the DENV-1 clades were introduced independently into HCMC, they had spread at similar and effectively constant rates. For Dong Thap, clade 1 was the only one clearly derived from a distinct single importation and of a sufficient size for analysis. The dispersion rate of DENV-1 appeared to be associated with the fluctuations in genetic diversity and monthly incidence in Dong Thap (Figure 2C and D). The two peaks in relative genetic diversity of clade 1 in Dong Thap coincided with the two major peaks in the monthly incidence, indicating that DENV-1 epidemic in Dong Thap is largely driven by this lineage.
Fig. 2. The genetic diversity, dengue incidence rate, and dispersion rates of DENV-1 in HCMC and Dong Thap. 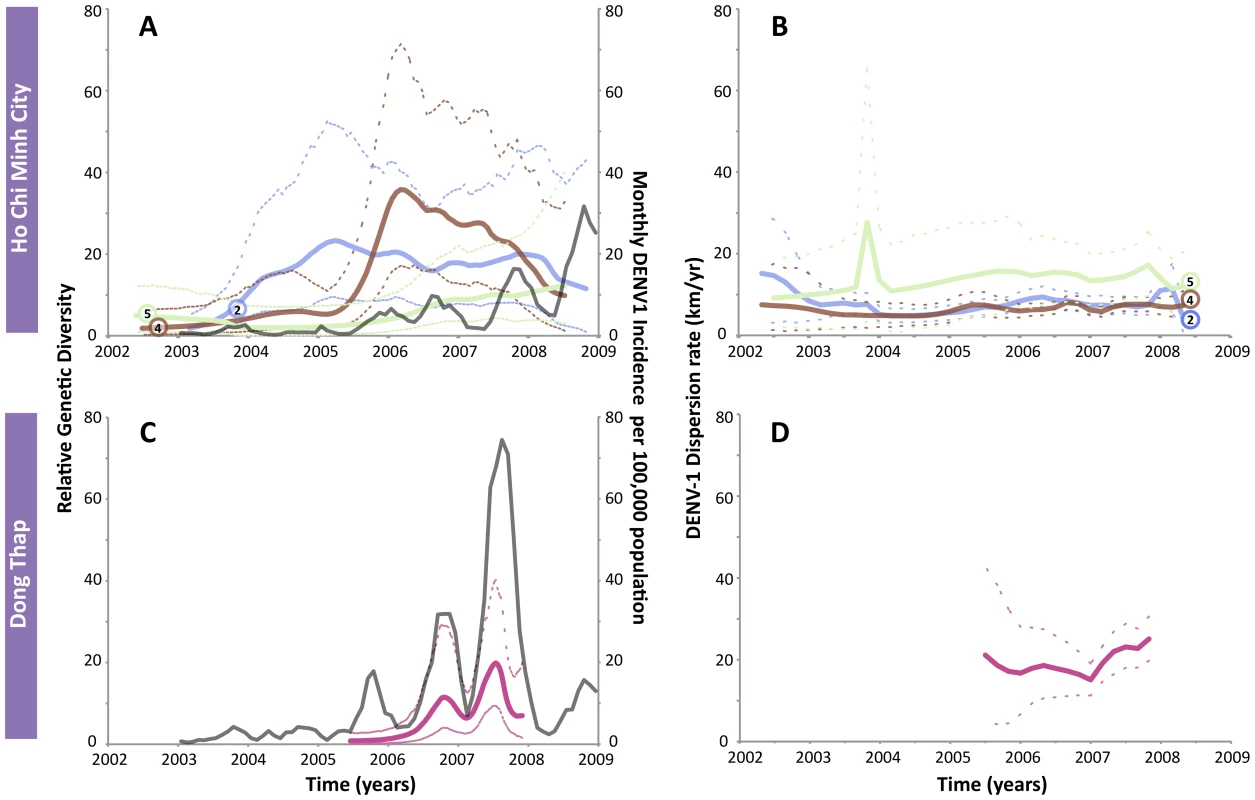
The top row shows the results of three dominant lineages in HCMC (clades 2, 4 and 5), while the bottom row shows the results of clade 1 in Dong Thap. Clade 1 is not included in HCMC since it was identified as having multiple origins from Dong Thap. A) Relative genetic diversity of the main clades in HCMC, superimposed with the incidence rate of DENV-1 in HCMC (grey). B) Dispersion rates of the three lineages in HCMC estimated in 6-month intervals from 2003–2009. C) and D) show the results of clade 1 in Dong Thap, relative genetic diversity and incidence rate (grey) and dispersion rate, respectively. To investigate whether these dispersion rate estimates in HCMC were simply a reflection of the geographic constraint of our samples, they were re-estimated by randomizing the tip location for each clade (Table 4). The results indicated what the maximum dispersion rate could be given the sampled locations, which were found to be 2–3 times greater than the empirical estimates, with wide HPD intervals (Table 4). The spatial reconstruction of the viral spread at different stages of the epidemics showed that these viral lineages had co-circulated in the same place at the same time (Figure 3). This observation is of fundamental importance as it suggests that the number of susceptible hosts to DENV-1 had not been saturated in HCMC, and could potentially have supported additional DENV-1 lineages in this area.
Fig. 3. The dispersion of the main clades in HCMC from 2003–2008 estimated from the continuous diffusion phylogeography process. 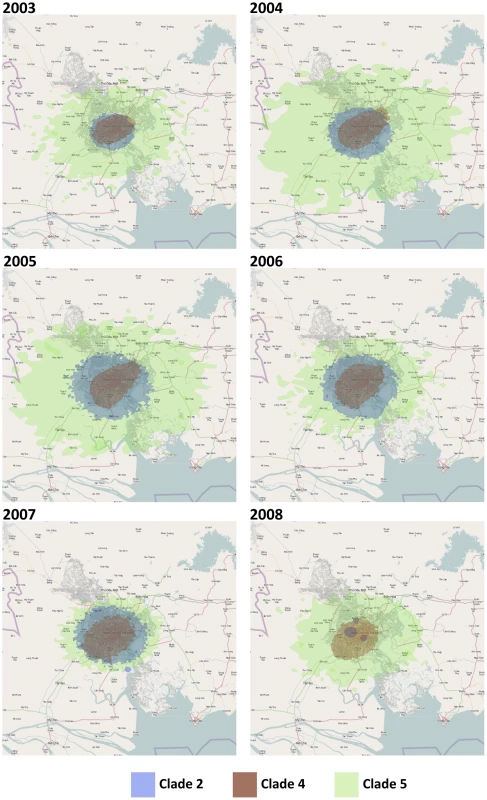
The different clades are color coded (clade 2 = blue, clade 4 = red, and clade 5 = green). All three clades appear to emerge from similar parts of HCMC and continue to co-exist in same geographic space at the same time. Map Data © 2011 OpenStreetMap Contributors, CC-BY-SA. Tab. 4. Results from randomizing locations at the tips to test the upper limits of the dispersion rates of DENV-1 in HCMC, Viet Nam. 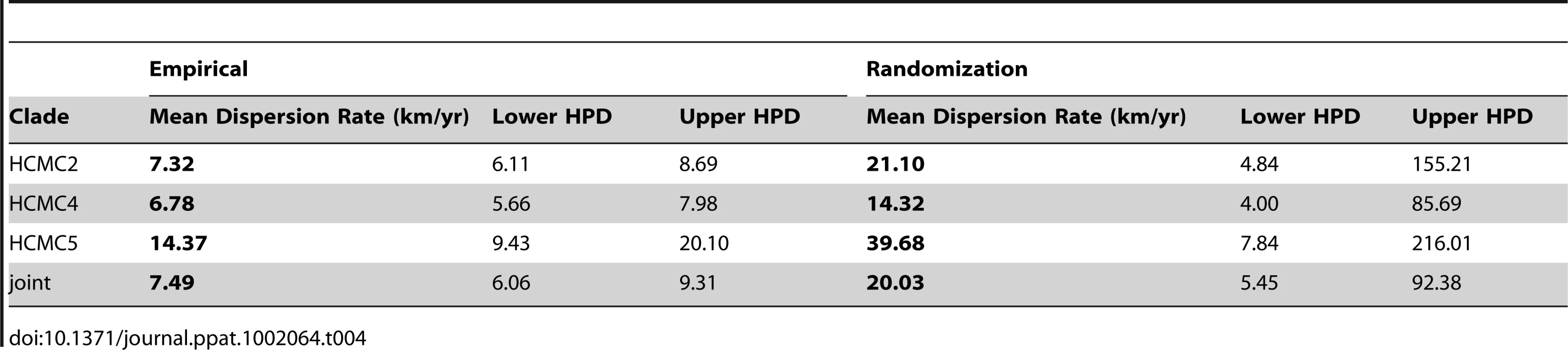
Population density and transmission routes
To determine whether transmission routes within HCMC varied according to population density, we employed a non-reversible discrete phylogeography model applied to district level data. Importantly, the more densely populated inner city districts (above 30,000 people per km2) were found to contribute significantly to DENV-1 transmission compared to the suburban districts (Figure 4). Moreover, the most densely populated region, District 5, had the highest number of connections, providing compelling evidence that this area might be a major hub in the city.
Fig. 4. Results from a non-reversible discrete phylogeography analysis of HCMC clades at the district level (21 states). 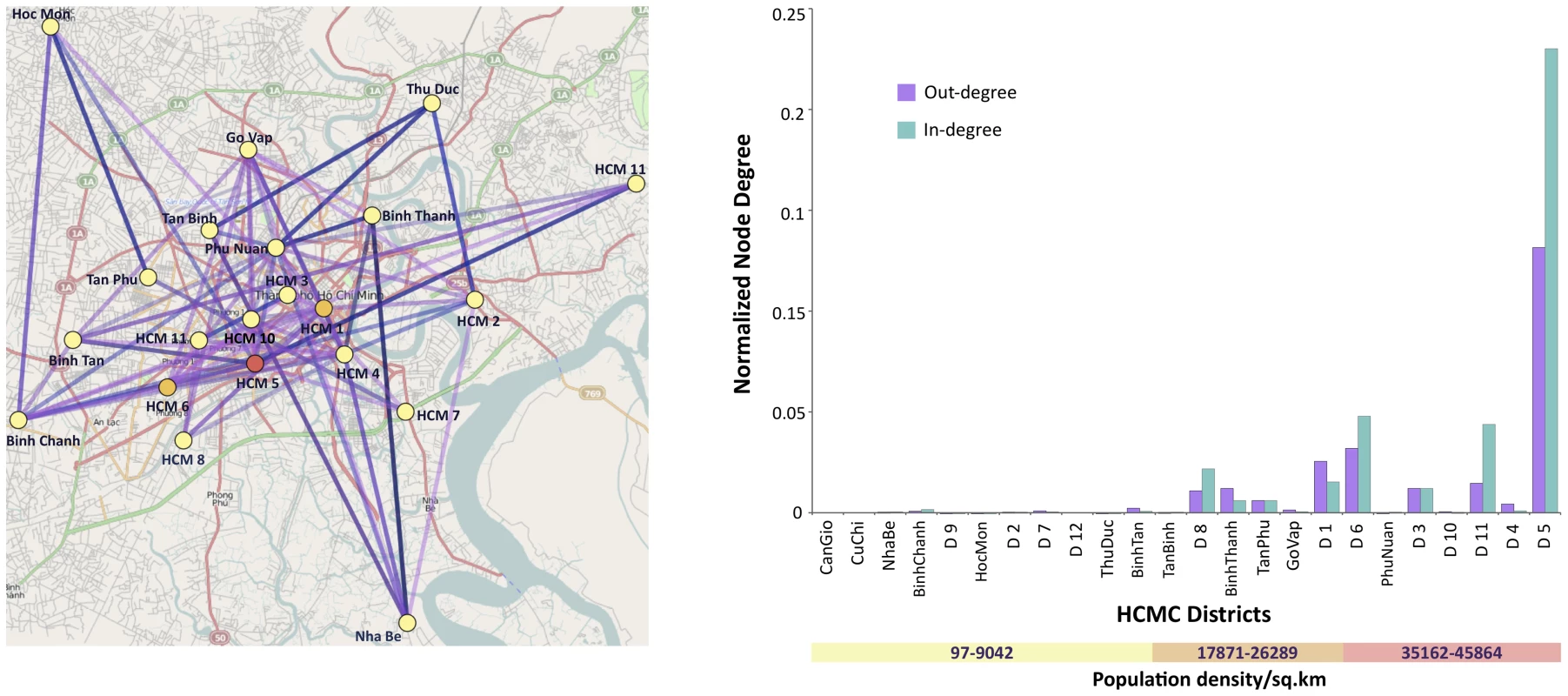
The significant connections at Bayes factor 3 are shown on the map of HCMC. The districts are colored by the number of normalized out-degree connections, where yellow, orange and red indicate low, intermediate, and high, respectively. The number of significant connections to and from a location (district) is represented by the bar chart in terms of in-degree and out-degree respectively. The districts along the x-axis are ordered by increasing population density. Map Data © 2011 OpenStreetMap Contributors, CC-BY-SA. Discussion
At the scale of South-East Asia, the observation that there is a strong clustering by country indicates that there is a far higher level of DENV-1 gene flow within than between countries. Such a phylogeographic pattern is compatible with relatively short transmission distances for DENV as a whole, including that meditated by mosquitoes. This rather limited spatial movement also sits in marked contrast to that observed in respiratory borne pathogens such as influenza, where there is relatively little clustering by place of isolation even on a global scale [28].
Each of the five clades of DENV-1 we identify has a very recent common ancestry, dating only shortly before the appearance of that clade. Given that dengue is endemic in southern Viet Nam, with DENV-1 circulating there for at least 23 years [29], such recent common ancestry suggests that there is a rapid and continual turnover of viral lineages, as has been increasingly described for this and other DEN viruses [8]–[11], [30], [31]. Less clear is whether these instances of lineage turnover are due to fitness differences between the lineages in question, such that natural selection is preferentially able to favor one lineage over another, or whether there is simply a stochastic die-off. That the three major clades we detect in HCMC co-circulate in the same spatial region with overlapping ranges, and possess broadly equivalent levels of relative genetic diversity, suggests that they are of similar fitness and hence that there is little, if any, competition between them. Consistent with this, we did not observe differences in early plasma viremia levels between patients infected with viruses belonging to the different clades (Figure S2). Indeed, we suggest below that HCMC is likely characterized by a large number of susceptible hosts, which would in turn reduce the extent of selective competition between lineages. More generally, these results indicate that although a specific viral serotype may appear to be endemic in a specific geographic region for an extended period, this does not mean that the same viral clades are involved throughout this period.
A striking result from this study is that the ‘virus dispersion rates’ we estimate appear to be very low, and particularly in HCMC where mean rates were universally <20 km/year. Such low rates are especially noteworthy given the rapidity and geographic scale with which DENV-1 re-emerged as the dominant serotype in southern Viet Nam [13]. We therefore interpret these low rates to mean that urban centers like HCMC are characterized by sufficiently high numbers of susceptible hosts such that the virus does not have to move very far to infect a new host. Such a notion is supported by the fact that higher virus dispersion rates are observed in Dong Thap, which is characterized by an approximately ten-fold lower population density (495 persons/km2) relative to HCMC (3024 persons/km2), although more estimates are clearly needed from this locality. In addition, the highest levels of viral movement were found in and out of the most densely populated region of HCMC (District 5), suggesting that this well-connected locality acts as a focal point for dengue dispersion within the city. Hence, it is not that DENV-1 moves slowly at a spatial scale in HCMC, but rather that it does not have to move far geographically to continue its transmission.
Although our sample of genome sequences is biased toward those from HCMC, our analysis indicates that DENV-1 generally diffuses from HCMC to Dong Thap. Again, this observation is suggestive of a gravity model of viral transmission, in which spatial diffusion occurs over a gradient of population density, and is compatible with our observation that dispersion rates are associated with the numbers of susceptible hosts. A similar gravity-dependent pattern of virus dispersion was recently suggested for DENV-2 in Viet Nam [14], although the use of a strictly reversible phylogeographic model in that case meant that directionality could not be ascertained with certainty. Combined, these studies strongly suggest that the density of the human host population plays a fundamental role in determining the transmission dynamics of endemic dengue.
Typically, adult A. aegypti mosquitoes travel short distances of less than ∼100 m during their average life-span of a few weeks [32]–[34]. The very short distances traveled by DENV-1, particularly in HCMC is consistent with mosquitoes, rather than humans, being responsible for the majority of the spatial spread in HCMC, which is again in part a function of the high density of susceptible hosts. A similarly limited movement of dengue has been reported by recent studies that focused on smaller geographic areas, reflecting the restricted spatial range of mosquito vectors, and corroborating the highly focal pattern of DENV transmission observed in HCMC [15], [35]. It is also notable that the geographical range of the three major clades in HCMC changed little from 2003–2008. As such, the full geographic range of these clades is established very early on as the virus is able to spread rapidly through a susceptible host population. Upon the introduction of a new dengue serotype into Iquitos, Peru, it was noted that early-confirmed cases were scattered throughout the city, suggesting a rapid establishment of the virus when entering a completely naïve population [36]. This observation gives added weight to our conclusion that the dispersion rates of DENV-1 in southern Viet Nam are largely a function of the availability of susceptible hosts.
These results have a number of important implications for the future control of dengue. Most generally, that DENV tends to spread relatively slowly on a spatial scale (such that DENV phylogenies exhibit a strong spatial structure both nationally and internationally) suggests that any future vaccine escape or drug resistance mutations would also spread relatively slowly. In addition, that the dispersion rates of DENV appear to largely reflect the density of human host population, including movement from Ho Chi Minh City to Dong Thap, suggests that future control measures, including mosquito spraying, should be directed toward the densest host populations and preferentially to urban over rural areas.
Methods
Patient population
The dengue patients from whom DENV whole genome sequences were determined were enrolled in one of two prospective studies at the Hospital for Tropical Diseases in Ho Chi Minh City, Viet Nam or at Dong Thap Hospital, Dong Thap Province, Viet Nam. The median age of these patients was 12 years (interquartile range 7–17 years) and 51% were male. Serological investigations (IgM and IgG capture ELISAs) were performed using paired plasma samples using methods described previously [37]. DENV serotype and viraemia levels were determined using an internally-controlled real-time RT-PCR assay that has been described previously [38].
Genomic sequencing
Viral genomes were sequenced using the Broad Institute's capillary sequencing (Applied Biosystems) directed amplification viral sequencing pipeline http://www.broadinstitute.org/scientific-community/science/projects/viral-genomics-initiative). This sequencing effort was part of the Broad Institute's Genome Resources in Dengue Consortium (GRID) project. Viral RNA was isolated from diagnostic plasma samples (QIAmp viral RNA mini kit, Qiagen) and the RNA genome reverse transcribed to cDNA with superscript III reverse transcriptase (Invitrogen), random hexamers (Roche) and a specific oligonucleotide targeting the 3′ end of the target genome sequences (nt 10868 to 10890, AGAACCTGTTGATTCAACAGCAC). cDNA was then amplified using a high fidelity DNA polymerase (pfu Ultra II, Stratagene) and a pool of specific primers to produce 14 overlapping amplicons of 1.5 to 2 kb in size for a physical coverage of 2-fold across the target genome (nt 40 to 10649). Amplicons were then sequenced in the forward and reverse direction using primer panels consisting of 96 specific primer pairs, tailed with M13 forward and reverse primer sequence, that produce 500–700 bp amplicons from the target viral genome. Amplicons were then sequenced in the forward and reverse direction using M13 primer. Total coverage delivered post amplification and sequencing was 8-fold. Resulting sequence reads were assembled de novo using the Broad Institute's AV454 assembly algorithm (Henn et al. 2011. in review) and a reference-based annotation algorithm.
All whole genome sequences newly determined here have been deposited in GenBank and assigned accession numbers (Table S1).
Phylogeographic analyses of DENV-1 in Southeast Asia and Viet Nam
A data set of DENV-1 sequences was collated to include isolates from countries in Southeast Asia that were likely linked to Viet Nam via migration. An alignment of the envelope (E) gene (1485 nt) was assembled for the Southeast Asian and Vietnamese isolates (n = 134 and 751, respectively) to include the broadest range of locations. An initial neighbor-joining tree was constructed in PAUP* [39], using a HKY85 nucleotide substitution model with gamma-distributed rates. This allowed us to make an initial identification of the major clades of DENV-1 in Viet Nam. These Vietnamese isolates were then subsampled (n = 101) to explore their phylogeography in context of the South East Asian isolates. Isolation dates for the South East Asia data set were obtained from GenBank annotations and via personal communication. Where specific dates were not available in terms of day and month, a mid-point of the year of isolation was used.
The spatial dynamics of DENV-1 in Southeast Asia were investigated with a discrete diffusion model [26] using Bayesian Monte Carlo Markov Chain (MCMC) method implemented in BEAST [40]. The phylogeography analysis was executed with a codon-structured SDR06 substitution model [41], a relaxed uncorrelated lognormal clock [42] and a Gaussian Markov Random Field (GMRF) coalescent prior [43] over the unknown phylogeny. The discrete diffusion model used the country of isolation of the sampled sequences to reconstruct the ancestral location states of the internal nodes from the posterior time-scaled tree distribution. The MCMC was run for 50 million generations, sampling every 5000th state, and executed multiple times to ensure adequate mixing and stationarity had been achieved.
Viral transmission between Ho Chi Minh City and Dong Thap province
Major clades of Vietnamese DENV-1 identified from the broad-scale South East Asian analysis were selected for further study to examine the spatial and temporal variation in Viet Nam. In clades with appreciable numbers of sequences from Dong Thap and HCMC, isolates from these locations were analyzed independently to gauge the regional variation in viral transmission patterns. For the fine-scale analysis, a continuous diffusion model based on a lognormal relaxed random walk [27] was employed to model the DENV-1 spatial dynamics in Viet Nam. For each isolate, the specific sample date and location information in terms of the longitude and latitude of the patient's household were used. Isolates that were identical in sample date and location information were down-sampled so as to reduce the potentially biasing effect of over-sampling of epidemiologically-linked cases. The MCMC runs were evaluated as previously described, and the chain lengths ranged from 50 to 100 million generations, and were sampled regularly to yield 10,000 trees from the posterior distribution.
The viral dispersion rates (km/yr) for each data set were calculated across the tree (i.e. total straight-line distance travelled divided by the total time) and biannually to consider the spatial heterogeneity in a time-scaled framework. Plots of relative genetic diversity over time were reconstructed using the GMRF coalescent prior to reveal the association between the genetic diversity of each group in terms of their evolutionary history [43].
Further discrete phylogeography analyses were performed with the robust counting method [44], [45] to determine the extent of viral migration between Dong Thap and HCMC and whether this varied when the lineage originated in a rural or urban area. In this case, the discrete states were represented by either the isolate being sampled from HCMC, Dong-Thap or neither (non-Dong Thap or HCMC).
Viral migration within Ho Chi Minh City
For the limiting case of a freely mixing (non-spatially structured) epidemic in HCMC, dispersion rates were estimated whilst randomizing the tip locations during the tree proposal in the MCMC, whilst co-estimating the rates for each independent lineage and the joint DENV-1 diffusion rate. To determine the viral transmission network within HCMC, a non-reversible discrete phylogeography model was applied to all the HCMC isolates, using the district of isolation for the discrete states. The analysis was performed and evaluated as described above with the addition of implementing Bayesian Stochastic Search Variable selection (BSSVS) to identify significant transition rates between locations [26]. The transition rates supported by a Bayes factor of at least 3 were examined further by looking at the number of in-degree and out-degree per district. The number of connections was normalized by the number of samples from the source location in order to reduce the bias from under-represented locations in our data set.
Ethics statement
Patients (or their parents/guardians) gave written informed consent to participate in each of the studies. The study protocols were approved by the Hospital for Tropical Diseases and the Oxford University Tropical Research Ethical Committee.
Supporting Information
Zdroje
1. Pediatric Dengue Vaccine Initiative. Global burden of dengue. http://www.pdvi.org/about_dengue/GBD.asp. Accessed 30 November 2009
2. GandonSMackinnonMJNeeSReadAF 2001 Imperfect vaccines and the evolution of pathogen virulence. Nature 414 751 756
3. WebsterDPFarrarJRowland-JonesS 2009 Progress towards a dengue vaccine. Lancet Infect Dis 9 678 687
4. WahalaWMDonaldsonEFde AlwisRAccavitti-LoperMABaricRS 2010 Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog 6 e1000821
5. ShresthaBBrienJDSukupolvi-PettySAustinSKEdelingMA 2010 The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog 6 e1000823
6. Sukupolvi-PettySAustinSKEngleMBrienJDDowdKA 2010 Structure and Function Analysis of Therapeutic Monoclonal Antibodies against Dengue Virus Type 2. J Virol 84 9227 9239
7. HolmesECTwiddySS 2003 The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol 3 19 28
8. MesserWBGublerDJHarrisESivananthanKde SilvaAM 2003 Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis 9 800 809
9. Myat ThuHLowryKJiangLHlaingTHolmesEC 2005 Lineage extinction and replacement in dengue type 1 virus populations are due to stochastic events rather than to natural selection. Virology 336 163 172
10. Rico-HesseRHarrisonLMSalasRATovarDNisalakA 1997 Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 230 244 251
11. BennettSNHolmesECChirivellaMRodriguezDMBeltranM 2006 Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J Gen Virol 87 885 893
12. McElroyKSantiagoGLennonNBirrenBHennM 2011 Endurance, refuge, and reemergence of dengue virus type II, Puerto Rico, 1986–2007. Emerg Infect Dis 17 64 71
13. HangVTTHolmesECDuongVNguyenTQTranTH 2010 Emergence of the Asian 1 genotype of dengue virus serotype 2 in viet nam: in vivo fitness advantage and lineage replacement in South-East Asia. PLoS Negl Trop Dis 4 e757
14. RabaaMATy HangVTWillsBFarrarJSimmonsCP 2010 Phylogeography of recently emerged DENV-2 in southern Viet Nam. PLoS Negl Trop Dis 4 e766
15. SchreiberMJHolmesECOngSHSohHSHLiuW 2009 Genomic Epidemiology of a Dengue Virus Epidemic in Urban Singapore. J Virol 83 4163 4173
16. EndyTPNisalakAChunsuttitwatSVaughnDWGreenS 2004 Relationship of preexisting dengue virus (DV) neutralizing antibody levels to viremia and severity of disease in a prospective cohort study of DV infection in Thailand. J Infect Dis 189 990 1000
17. NagaoYKoelleK 2008 Decreases in dengue transmission may act to increase the incidence of dengue hemorrhagic fever. Proc Natl Acad Sci U S A 105 2238 2243
18. AndersKLNguyetNMVan Vinh ChauNHungNTThuyTT 2011 Epidemiological Factors Associated with Dengue Shock Syndrome and Mortality in Hospitalized Dengue Patients in Ho Chi Minh City, Vietnam. Am J Trop Med Hyg 84 127 134
19. ColognaRArmstrongPMRico-HesseR 2005 Selection for virulent dengue viruses occurs in humans and mosquitoes. J Virol 79 853 859
20. LibratyDHEndyTPHoungHSGreenSKalayanaroojS 2002 Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis 185 1213 1221
21. ScottTWAmerasinghePHMorrisonACLorenzLHClarkGG 2000 Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: blood feeding frequency. J Med Entomol 37 89 101
22. Scott TWMA 2003 Aedes aegypti density and the risk of dengue virus transmission. Takken WST Ecological aspects for application of genetically modified mosquitoes Dordrecht, The Netherlands FRONTIS 187 206
23. ZhangCMammenMPJrChinnawirotpisanPKlungthongCRodpraditP 2005 Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J Virol 79 15123 15130
24. TwiddySSHolmesECRambautA 2003 Inferring the rate and time-scale of dengue virus evolution. Mol Biol Evol 20 122 129
25. LanciottiRSGublerDJTrentDW 1997 Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol 78 2279 2286
26. LemeyPRambautADrummondAJSuchardMA 2009 Bayesian Phylogeography Finds Its Roots. Plos Comput Biol 5 e1000520
27. LemeyPRambautAWelchJJSuchardMA 2010 Phylogeography Takes a Relaxed Random Walk in Continuous Space and Time. Mol Biol Evol 27 1877 1885
28. RussellCAJonesTCBarrIGCoxNJGartenRJ 2008 The global circulation of seasonal influenza A (H3N2) viruses. Science 320 340 346
29. HaDQNinhTU 2000 Virological Surveillance of Dengue Haemorrhagic Fever in Viet Nam, 1987-1999. Dengue Bulletin 24 18 23
30. SittisombutNSistayanarainACardosaMJSalminenMDamrongdachakulS 1997 Possible occurrence of a genetic bottleneck in dengue serotype 2 viruses between the 1980 and 1987 epidemic seasons in Bangkok, Thailand. Am J Trop Med Hyg 57 100 108
31. WittkeVRobbTEThuHMNisalakANimmannityaS 2002 Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 301 148 156
32. RussellRCWebbCEWilliamsCRRitchieSA 2005 Mark-release-recapture study to measure dispersal of the mosquito Aedes aegypti in Cairns, Queensland, Australia. Med Vet Entomol 19 451 457
33. ReiterPAmadorMAAndersonRAClarkGG 1995 Short report: dispersal of Aedes aegypti in an urban area after blood feeding as demonstrated by rubidium-marked eggs. Am J Trop Med Hyg 52 177 179
34. HarringtonLCScottTWLerdthusneeKColemanRCCosteroA 2005 Dispersal of the dengue vector Aedes aegypti within and between rural communities. Am J Trop Med Hyg 72 209 220
35. BalmasedaAStandishKMercadoJCMatuteJCTellezY 2010 Trends in Patterns of Dengue Transmission over 4 Years in a Pediatric Cohort Study in Nicaragua. J Infect Dis 201 5 14
36. MorrisonACMinnickSLRochaCForsheyBMStoddardST 2010 Epidemiology of Dengue Virus in Iquitos, Peru 1999 to 2005: Interepidemic and Epidemic Patterns of Transmission. PLoS Negl Trop Dis 4 e670
37. HangVTNguyetNMTrungDTTricouVYoksanS 2009 Diagnostic Accuracy of NS1 ELISA and Lateral Flow Rapid Tests for Dengue Sensitivity, Specificity and Relationship to Viraemia and Antibody Responses. PLoS Negl Trop Dis 3 e360
38. SimmonsCPDolocekCChauTNBGriffithsMDungNTP 2007 Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue haemorrhagic fever. J Infect Dis 195 1097 107
39. SwoffordD 2003 PAUP*.Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. ed. Sinauer Associates. Sunderland, Mass
40. DrummondAJRambautA 2007 BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7 214
41. ShapiroBRambautADrummondAJ 2006 Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23 7 9
42. DrummondAJHoSYWPhillipsMJRambautA 2006 Relaxed phylogenetics and dating with confidence. PLoS Biol 4 699 710
43. MininVNBloomquistEWSuchardMA 2008 Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 25 1459 1471
44. MininVNSuchardMA 2008 Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol 56 391 412
45. O'BrienJDMininVNSuchardMA 2009 Learning to count: robust estimates for labeled distances between molecular sequences. Mol Biol Evol 26 801 814
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Glycosaminoglycan Binding Facilitates Entry of a Bacterial Pathogen into Central Nervous SystemsČlánek The SV40 Late Protein VP4 Is a Viroporin that Forms Pores to Disrupt Membranes for Viral ReleaseČlánek Pathogen Recognition Receptor Signaling Accelerates Phosphorylation-Dependent Degradation of IFNAR1Článek High Affinity Nanobodies against the VSG Are Potent Trypanolytic Agents that Block Endocytosis
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 6- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus Is the Determinant of Genome Attachment
- Evolutionary Analysis of Inter-Farm Transmission Dynamics in a Highly Pathogenic Avian Influenza Epidemic
- Glycosaminoglycan Binding Facilitates Entry of a Bacterial Pathogen into Central Nervous Systems
- Endemic Dengue Associated with the Co-Circulation of Multiple Viral Lineages and Localized Density-Dependent Transmission
- The SV40 Late Protein VP4 Is a Viroporin that Forms Pores to Disrupt Membranes for Viral Release
- The -glycan Glycoprotein Deglycosylation Complex (Gpd) from Deglycosylates Human IgG
- The Lipid Transfer Protein CERT Interacts with the Inclusion Protein IncD and Participates to ER- Inclusion Membrane Contact Sites
- Induction of Noxa-Mediated Apoptosis by Modified Vaccinia Virus Ankara Depends on Viral Recognition by Cytosolic Helicases, Leading to IRF-3/IFN-β-Dependent Induction of Pro-Apoptotic Noxa
- Environmental Constraints Guide Migration of Malaria Parasites during Transmission
- HIV-1 Efficient Entry in Inner Foreskin Is Mediated by Elevated CCL5/RANTES that Recruits T Cells and Fuels Conjugate Formation with Langerhans Cells
- Kupffer Cells Hasten Resolution of Liver Immunopathology in Mouse Models of Viral Hepatitis
- Highly Pathogenic Avian Influenza Virus H5N1 Infects Alveolar Macrophages without Virus Production or Excessive TNF-Alpha Induction
- Uses Host Triacylglycerol to Accumulate Lipid Droplets and Acquires a Dormancy-Like Phenotype in Lipid-Loaded Macrophages
- Compensatory T Cell Responses in IRG-Deficient Mice Prevent Sustained Infections
- Detection of Inferred CCR5- and CXCR4-Using HIV-1 Variants and Evolutionary Intermediates Using Ultra-Deep Pyrosequencing
- A Dynamic Landscape for Antibody Binding Modulates Antibody-Mediated Neutralization of West Nile Virus
- HSV-2 Infection of Dendritic Cells Amplifies a Highly Susceptible HIV-1 Cell Target
- “Rational Vaccine Design” for HIV Should Take into Account the Adaptive Potential of Polyreactive Antibodies
- Impact of Endofungal Bacteria on Infection Biology, Food Safety, and Drug Development
- Infection Reduces B Lymphopoiesis in Bone Marrow and Truncates Compensatory Splenic Lymphopoiesis through Transitional B-Cell Apoptosis
- The Intrinsic Antiviral Defense to Incoming HSV-1 Genomes Includes Specific DNA Repair Proteins and Is Counteracted by the Viral Protein ICP0
- Molecular Interactions that Enable Movement of the Lyme Disease Agent from the Tick Gut into the Hemolymph
- Pathogen Recognition Receptor Signaling Accelerates Phosphorylation-Dependent Degradation of IFNAR1
- A Freeze Frame View of Vesicular Stomatitis Virus Transcription Defines a Minimal Length of RNA for 5′ Processing
- High Affinity Nanobodies against the VSG Are Potent Trypanolytic Agents that Block Endocytosis
- Tipping the Balance: Secreted Oxalic Acid Suppresses Host Defenses by Manipulating the Host Redox Environment
- Bacteria-Induced Dscam Isoforms of the Crustacean,
- Structural and Mechanistic Studies of Measles Virus Illuminate Paramyxovirus Entry
- Insertion of an Esterase Gene into a Specific Locust Pathogen () Enables It to Infect Caterpillars
- A Role for TLR4 in Infection and the Recognition of Surface Layer Proteins
- The Binding of Triclosan to SmeT, the Repressor of the Multidrug Efflux Pump SmeDEF, Induces Antibiotic Resistance in
- Low CCR7-Mediated Migration of Human Monocyte Derived Dendritic Cells in Response to Human Respiratory Syncytial Virus and Human Metapneumovirus
- HIV/SIV Infection Primes Monocytes and Dendritic Cells for Apoptosis
- Sporangiospore Size Dimorphism Is Linked to Virulence of
- Productive Parvovirus B19 Infection of Primary Human Erythroid Progenitor Cells at Hypoxia Is Regulated by STAT5A and MEK Signaling but not HIFα
- Identification of DNA-Damage DNA-Binding Protein 1 as a Conditional Essential Factor for Cytomegalovirus Replication in Interferon-γ-Stimulated Cells
- The Influenza Virus Protein PB1-F2 Inhibits the Induction of Type I Interferon at the Level of the MAVS Adaptor Protein
- Passively Administered Pooled Human Immunoglobulins Exert IL-10 Dependent Anti-Inflammatory Effects that Protect against Fatal HSV Encephalitis
- Infection of Induces Antifungal Immune Defenses
- Merozoite Invasion Is Inhibited by Antibodies that Target the PfRh2a and b Binding Domains
- Cyclic di-GMP is Essential for the Survival of the Lyme Disease Spirochete in Ticks
- Cross-Neutralizing Antibodies to Pandemic 2009 H1N1 and Recent Seasonal H1N1 Influenza A Strains Influenced by a Mutation in Hemagglutinin Subunit 2
- Spatial Dynamics of Human-Origin H1 Influenza A Virus in North American Swine
- Coronavirus Gene 7 Counteracts Host Defenses and Modulates Virus Virulence
- Clathrin Facilitates the Morphogenesis of Retrovirus Particles
- Contribution of Intrinsic Reactivity of the HIV-1 Envelope Glycoproteins to CD4-Independent Infection and Global Inhibitor Sensitivity
- Functional Analysis of Host Factors that Mediate the Intracellular Lifestyle of
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- High Affinity Nanobodies against the VSG Are Potent Trypanolytic Agents that Block Endocytosis
- Structural and Mechanistic Studies of Measles Virus Illuminate Paramyxovirus Entry
- Sporangiospore Size Dimorphism Is Linked to Virulence of
- The Binding of Triclosan to SmeT, the Repressor of the Multidrug Efflux Pump SmeDEF, Induces Antibiotic Resistance in
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



