Sample Processing and Methodological Pitfalls in Multiple Myeloma Research
Zpracování vzorků a metodická úskalí ve výzkumu mnohočetného myelomu
V tomto úvodním článku pro ostatní metodiky využívané na našem pracovišti při studiu mnohočetného myelomu a monoklonálních gamapatií jsme se zaměřili na postupy vlastního zpracování biologického materiálu, principy separace buněk a nastavené algoritmy dalších postupů. Běžně používaná metodika magnetické separace buněk MACS je vhodná pouze pro vzorky se vstupní infiltrací plazmatickými buňkami > 5 %. Pro nízce zastoupené populace buněk pak využíváme výhradně metodu fluorescencí aktivované separace FACS. Izolované plazmatické buňky jsou dále využívány pro molekulárně biologické studie, pro cytogenetická vyšetření a k proteinovým analýzám. Dále se v této práci zmiňujeme o úskalích, která souvisejí s výzkumem mnohočetného myelomu, některá z nich již umíme překonat, s jinými se zatím neúspěšně potýkáme.
Klíčová slova:
mnohočetný myelom – monoklonální gamapatie – separace buněk – CD138
Tato práce byla podpořena výzkumnými projekty MŠMT ČR LC06027, MSM0021622434; granty IGA MZ ČR NS10207, NS10387, NS10406, NS10408, NT11154 a granty GAČR GAP304/10/1395, GP301/09/P457.
Autoři deklarují, že v souvislosti s předmětem studie nemají žádné komerční zájmy.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do bi omedicínských časopisů.
Authors:
A. Potáčová 1; J. Štossová 1; I. Burešová 1; L. Kovářová 1,2; M. Almaši 1,3
; M. Penka 2; R. Hájek 1,3,4
Authors‘ workplace:
Babak Myeloma Group, Department of Pathological Physiology, Faculty of Medicine, Masaryk University, Brno, Czech Republic
1; Department of Clinical Hematology, University Hospital Brno, Czech Republic
2; Laboratory of Experimental Hematology and Cell Immunotherapy, Department of Clinical Hematology, University Hospital Brno, Czech Republic
3; Department of Internal Medicine – Hematooncology, University Hospital Brno, Czech Republic
4
Published in:
Klin Onkol 2011; 24(Supplementum 1): 18-23
Overview
In this paper, initial processing of biological material, cell separation algorithms and other procedures are discussed. For samples with initial infiltration of plasma cells > 5%, CD138 MicroBeads and Auto-Magnetic-Activated Cell Sorting program are used. Fluorescence-Activated Cell Sorting is used exclusively for cell populations with low-abundance; these samples are detected using fluorescently labeled antibodies only. Isolated plasma cells are further processed for molecular biological studies, for cytogenetics and protein analyses. Furthermore, this work examines the pitfalls of research related to multiple myeloma; some of them we have overcome, while the others are still problematic.
Key words:
multiple myeloma – monoclonal gammopathy – cell separation – CD138
This work was supported by research grants of The Ministry of Education, Youth and Sports: LC06027, MSM0021622434; research projects of IGA of The Ministry of Health: NS10207, NS10387, NS10406, NS10408, NT11154 and grants of GACR GAP304/10/1395, GP301/09/P457.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
The Editorial Board declares that the manuscript met the ICMJE “uniform requirements” for biomedical papers.
Introduction
Laboratory studies of monoclonal gammopathies and multiple myeloma (MM) are based on separated plasma cells (PC) from bone marrow aspirates. Pathological populations in the bone marrow (BM) are heterogeneous and may contain less mature forms of B lymphocytes [1]. In addition, various types of monoclonal gammopathies or MM differ in PC infiltration of the bone marrow. For PC analysis by modern molecular biology approaches, it is necessary to obtain pure populations of cells of sufficient quantity and purity [2]; therefore, it is very important to find the optimal separation strategy. There are several options for separation of PC. Our department has been pursuing this issue for many years; our experiences and results have been previously published [3–7]. This work focuses on a brief overview of the principles of separation methods and describes our optimized algorithm, which is followed for sample processing.
Currently, we process samples not only from the Department of Internal Hematooncology (University Hospital Brno), but also samples of collaborating institutions throughout the Czech Republic and the Visegrad region. All patients have been informed about research purposes and signed informed consent forms approved by the ethical committees of all institutions.
Initial processing of clinical samples, their distribution between research teams and biobanking are carried out at the Laboratory of Experimental Hematology and Cellular Immunotherapy at the Department of Clinical Hematology, University Hospital Brno (LEHABI OKH FN Brno). This laboratory is specialized in initial processing of samples (plasma, serum collection) and in the immunomagnetic cell separation and determination of purity (autoMACS, cytospin). LEHABI closely cooperates with the flow cytometry laboratory at the OKH FN Brno. Fluorescence-activated cell sorting and other analyses are done at the Integrated Laboratories of Biomedical Technologies (ILBIT) at the University Campus Brno. More than 200 samples of bone marrow were processed in our laboratories every year. In 2010, the processed material doubled. So far, we have processed 556 bone marrow samples and more than 1,500 samples of peripheral blood. In the next chapters of this article, we focus on the principles and procedures for initial processing of biological material, cell separation algorithms and other procedures.
Initial Processing of Samples
Bone marrow samples (10–40 ml) are first mixed with the same volume of Iscove’s modified Dulbecco’s medium (Sigma--Aldrich) containing 100 U/mL heparin and 100 U/mL DNAse I (Roche Diagnostics). Bone marrow mononuclear cells (BMMC) are isolated by density centrifugation on Ficoll-Paque Plus (Scintila) at 400 g for 35 min at 20°C. Then, collected cells are washed twice with phosphate buffered saline (PBS) containing 2 mM EDTA (centrifugation at 300 g for 10 min at 20°C). In the prepared sample, percentage of CD138+ cells is measured by flow cytometry (using CD138/PE, Exbio), and the samples are processed based on a protocol (Fig. 1).

CD138+ Cells Separation
Separation techniques based on antibody binding to a surface marker are commonly used for PC sample enrichment. PC separation is mostly done by positive cell selection using anti-CD138 monoclonal antibody. According to the infiltration of PC in the bone marrow, we use either magnetic-activated cell sorting (MACS) and/or fluorescence-activated cell sorting (FACS). Both methods are optimized and used in relation to the percentage content of PC, to achieve very high purity of collected populations, as well as maximal yield of cells.
MACS separation = Magnetic--Activated Cell Sorting
Immunomagnetic separation of PC based on CD138 has been described by several groups [8–10]. This method is based on immunomagnetic labeling of target cells by monoclonal antibody coupled to the magnetic particle. Washed cells are labeled for 15 minutes in the refrigerator with CD138 Microbeads (10 µl per 10 × 106 cells). Then, labeled and washed cells are captured in separation column which is placed in a magnetic field in the autoMACS separator (Miltenyi Biotec). The entire process of sorting is automated – unlabeled cells pass through the column to the negative fraction. After the separation column is removed from the magnetic field, the target cells (= enriched positive fraction) are eluted (Fig. 2). General protocol of sample labeling and magnetic separation is available free at http://www.miltenyibiotec.com. Then, sorted cells are washed in PBS; yield and purity of the fractions are determined, and the samples are processed according to requirements of further analyses. For FISH analysis (interphase fluorescein in situ hybridization), the cells are suspended in warm potassium chloride (KCl) (37°C), incubated for 15 minutes at 37°C and centrifuged. Then, the cell pellet is fixed in fixative solution (Carnoy: 60% ethanol, 30% chloroform and 10% glacial acetic acid) and stored at –20°C. For ribonucleic acid (RNA) isolation, native cells are used, while for other analyses dry pellets are frozen (–196°C).

The established algorithm uses autoMACS separator for samples with more than 5% of PC in BMMC fraction. For samples with more than 10% of PC, program Possels is used. The Posseld2 program for special double selection is used for cases with PC infiltration of 5–10% [7]. In compliance with this protocol, we are able to separate highly purified cell fractions. The median PC purity is about 93% for possels and 84% for program posseld2. These results are acceptable for most types of cytogenetic and molecular analyses. So far, results of immunomagnetic separation for samples with very low PC percentage (< 5%) are not optimal.
FACS separation = Fluorescence--Activated Cell Sorting
For samples with less than 5% of PC, fluorescence-activated cell sorting (FACS) is used, especially for MGUS samples, where more phenotypically distinct populations of PC can be found [1]. We can select multiple surface markers and sort more subpopulations of cells. Fluorescence-based separation uses antibodies conjugated with fluorochrome(s) for the identification of target population; this analysis is performed on the cell sorter (in our laboratory we use FACSAria with two lasers, BD Biosciences). Main principle of separation is as follows [11,12]: Antibody-labeled cell suspension is formed into a narrow stream. A piezoelectric crystal in the nozzle holder causes the cell stream to break into individual droplets. The system is adjusted so that there is a low probability of more than one cell being in a droplet. Just before the stream breaks into droplets, the flow passes through the observation point where the fluorescence intensities of each cell are measured by the flow cytometer. At this point, the cells for sorting are selected. An electrical contact placed in the nozzle holder loads the abrupting stream at the moment of disruption of the droplet. The charged droplets containing selected cells move through the electrostatic field that diverts the droplets into containers based upon their charge. After separation of the stream, the droplets are discharged and the system is ready for the next cycle.
Determination of the PC phenotype foregoes own FACS separation. Antibodies: CD38/APC; CD138/PE; CD45/PerCP; CD56/FITC from Exbio and CD19/PC7 from Beckman Coulter are used for phenotypic determination. Samples are incubated for 15 minutes in the dark at 4°C. Then, different populations of PC are separated using various phenotypic markers: CD19 and CD56 (CD19+/CD56–; CD19–/CD56+; CD19–/CD56–; CD19+/CD56+). BMMC samples are incubated with appropriate amount of antibody (CD138/PE, CD56/FITC from Exbio; CD19/PC7 from Beckman Coulter; 30 minutes in the dark at 4°C). Cells are then washed with cold PBS and diluted to 5–10 × 106 cells/ ml with cold PBS enriched with 1% fetal calf albumin (Sigma). Separation of cells runs at 3–8 × 103 cells/s. Separated cells are collected into a tube with RPMI-1640 medium (Sigma) enriched with 20% bovine fetal serum.
In addition to FISH analysis, we are able to sort a very small number of cells on the microscopic slide covered by fetal calf albumin. As needed, KCl solution and/or the Carnoy fixative are added.
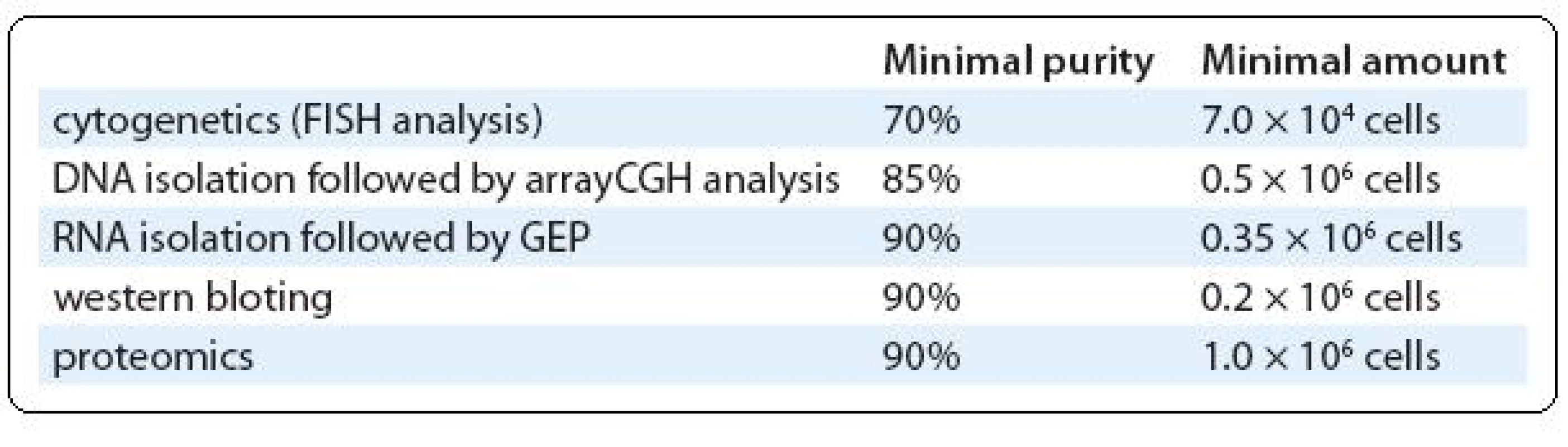
For a special group of samples with high cellularity (more than 30 × 106 cells) and/or very low infiltration of PC, we use combination of magnetic and fluorescence-based separations. Cells are labeled with CD138 antibody conjugated with fluorochrome (CD138/PE, Exbio) in the first step and with magnetically labeled antibody against used fluorochrome (anti-PE MicroBeads, Miltenyi) in the second step. So, these double labeled cells are separated in the magnetic system (we use VarioMACS – semi-automated magnetic separator from Miltenyi Biotec). Fluorochrome positive fraction is immediately sorted in the cell sorter to high purity. Magnetic pre-enrichment markedly reduces the cellularity of sorted sample and saves time. Moreover, more markers labeled with different fluorochromes can be used in the first step, allowing the combined separation of normal and abnormal PC. However, it is true that final yield of target cells is lower in comparison with one-step procedure. On the other hand, prolonged sorting lasting several hours will reduce the yield of sorted cells, probably because of lower viability of sorted cells. Further distribution of separated cells between research groups is done based on their purity and quantity (Tab. 1).
Biobanking
A biobank is a cryogenic storage facility used to archive biological samples for research and experimental purposes. Formation of myeloma database and biobank is a key input and a prerequisite for any further research applications. Since 2001, in our myeloma bank, samples from patients with different stages of multiple myeloma and MGUS have been collected. We archive samples of separated cells (positive and negative fractions), serum, plasma of peripheral blood and plasma of bone marrow. All reports of the frozen samples and their storage are maintained in a database of samples called Myelab. Currently, for subsequent research purposes, there are more than 730 separated PC samples, 2,500 samples of bone marrow plasma and more than 10,000 samples of plasma/serum from patients with MM and MGUS (Fig. 3) in the bank of myeloma samples. All samples are archived under codes. Samples of separated CD138+ cells are usually used immediately for consequent analyses (RNA isolation, FISH analysis). Other samples (positive and negative fraction) are frozen as pellets and stored in Dewars containers in liquid nitrogen (–190°C). Stable temperature conditions are protected by a security system in the cryobank. Long-term storage and archiving in the frozen state guarantees sufficient cell viability, which is important for their future use. Samples of plasma or serum are stored at –80°C in 0.5 ml aliquots.

Methodological Pitfalls in Multiple Myeloma Research
Is the CD138 Positive Cell “the real thing”?
PC separation from the bone marrow of patients with monoclonal gammopathy is a challenging methodological problem. Bone marrow is a complex mixture of different types of cells with highly variable PC abundance. For separation of PC from the bone marrow, we use surface marker CD138 (Syndecan-1). CD138 is a transmembrane heparan sulphate proteoglycan that is expressed by both normal and malignant PC of the bone marrow and PC in the peripheral blood of MM patients [13]. CD138 (as other syndecans) binds and modifies various growth factors, enzymes and extracellular matrix components and is shed constitutively by cultured cells as well as apoptotic cells [14,15]. CD138 positivity is typical not only for PC, but also for normal and neoplastic epithelial tissues, for a small subset of mesenchymal neoplasms, squamous cell carcinoma, renal cell carcinoma and prostate adenocarcinoma [16]. These cells may also be found in metastases in the bone marrow [17]. Thus, separated cells must be evaluated not only by their phenotype but by their morphology and clinical context. In contrast, there is also a much smaller CD138 – fraction with a strong clonogenic potential [13,15]. CD138 – fraction of cells has demonstrated some important differences from CD138+ cells (increased immaturity and greater proportion of cells in S phase). These results support the hypothesis that CD138 – cells have a greater proliferative potential [13]. Unfortunately, CD138 – cells are often not included in studies, because only CD138+ cells are isolated. In spite of this, CD138 was accepted as convenient and highly representative marker of PC selection. However, it is necessary to take into account all known facts and limits of this marker.
Methodological Pitfalls in Myeloma Research
For all consequent analyses of MM and MGUS samples, it is necessary to obtain sufficient amount of purified PC. Samples are used for: molecular diagnostics in MM with detailed look at new potentially prognostic factors such as centrosome amplification and abnormal expression of mitotic genes in B cells and PC; characterisation of genetic abnormalities on chromosome 1; proteomic and genomic analyses of resistance or sensitivity to anti-myeloma drugs; study of microenvironment and angiogenesis in MM; analysis of proliferative and self renewal potential of myeloma cells progenitors/precursors; for study of pathogenesis of extramedullary relapse in MM and other partial aims.
Abnormal clones of PC in many patients with multiple myeloma have a low proliferative activity and low mitotic activity [18,19]. These limitations have been overcome by the introduction of new molecular techniques, such as fluorescence in situ hybridisation (FISH) and comparative genomic hybridisation (CGH) [20]. In MM and MGUS research, one of the most fundamental problems remains the lack of sorted cells. Although we use the most modern methods, there is not always enough cells for all experiments. If all the research groups should receive a sufficient number of cells, we would have to separate at least 2.3 million of CD138+ cells. Only about 20% of all sorted samples fulfill this limit. It is still necessary to improve the methodological processes in terms of minimalization of cells demands (ideally to tens of thousands cells) or to focus on methods and analyses that work with samples of peripheral blood, easily available biological material, collection of which does not burden the patient and can be collected repeatedly. At present, we are only partially successful. We were able to reduce the number of required cells from initial one million to 0.35 × 106 for arrayCGH and qRT-PCR (where the quantity of DNA or RNA, respectively is more decisive than the absolute number of cells); we presume that this number will lowered when we optimize DNA/RNA amplification. Unfortunately, some methods, such as conventional proteomics – using two-dimensional gel electrophoresis (2-DE) followed by liquid nanochromatography coupled with mass spectrometry – still require at least one million cells. Although conventional 2-DE remains generally a fundamental tool in expression proteomics, it has significant limitations. It is very time consuming (analysis of one experiment may take, according to the scale, up to several months), as well as the low dynamic range and insufficient detection limit [21]. All of this and the impossibility of a large set of samples (patients) with consequent robust statistic analysis were the reasons that lead us to leave 2-DE and to focus on other methods (e. g. method of relative quantification of proteins using mass spectrometry: Isotope-Coded Protein Labeling). This approach uses different isotopic tags as an alternative to previously used conventional procedures based on separation of proteins using 2-DE and removes its limitations.
Another viable option for MM research is analysis of biomarkers from peripheral blood (or plasma and serum). For example, single nucleotide polymorphisms were successfully determined from peripheral blood [22]. Other promising prognostic markers may be some miRNAs. It was reported that these RNAs are stable in serum [23] and their extracellular presence is important for cell – cell communication and can be perspective indicator of cancer progression, multi-drug resistance or invasion and metastasis of tumors [24]. In proteomic research, we optimize antibody protein chip technique that allows us to detect 70 cytokines, chemokines, growth factors, matrix metalloproteinases and theirs inhibitors and other important potential markers of tumor-associated cases in one run. We expect that new knowledge and dynamically evolving methods will allow us to overcome the lack of CD138+ cells and open more topics and horizons to explore for our further research.
Conclusion
Research of CD138+ fraction of myeloma cells can probably answer many questions about diagnosis, prognosis or pathogenesis of MM or MGUS. The introduction of optimal separation strategy enabled us to obtain sufficient amount of highly purified CD138+ cells, which are required for subsequent experiments. Especially optimization of fluorescence-based separation opened the way to MGUS research – now we are able to reach highly pure populations of PC for sophisticated research applications (such as genomic analyses). Considering the fact that we will never have enough cells in MGUS for all wanted research applications, we will concentrate on biomarkers from easily available peripheral blood.
Acknowledgement
We are very grateful to the Department of Internal Medicine – Hematooncology for providing us with the bone marrow samples. We also thank all participating members of the Czech Myeloma Group, all patients, their caregivers, and referring physicians for making this work possible.
Prof.
MUDr. Roman Hájek, CSc.
Babak Myeloma Group
Department
of Pathological Physiology
Faculty
of Medicine
Masaryk University
Kamenice
5
625
00 Brno
Czech Republic
e-mail:
r.hajek@fnbrno.cz
Sources
1. Kovarova L, Buresova I, Buchler T et al. Phenotype of plasma cells in multiple myeloma and monoclonal gammopathy of undetermined significance. Neoplasma 2009; 56(6): 526–532.
2. Stranneheim H, Orre LM, Lehtiö J et al. A comparison between protein profiles of B cell subpopulations and mantle cell lymphoma cells. Proteome Sci 2009; 7 : 43.
3. Fišerová A, Hájek R, Doubek M et al. Imunomagnetická separace myelomových buněk. Klin Onkol 2001; 14(2): 46–50.
4. Čumová J, Burešová I, Kovářová L et al. Selekce plazmatických buněk. Klin Onkol 2008; 21 (Suppl 1): S190–S194.
5. Burešová I, Čumová J, Kovářová L et al. Srovnání selekce plazmatických buněk metodami MACS a FACS. Klin Onkol 2008; 21 (Suppl 1): S195–S197.
6. Burešová I, Kyjovská D, Kovářová L et al. Algoritmus separace plazmatických buněk ze vzorků kostní dřeně. Klin Onkol 2011; 24(1): 35–40.
7. Cumova J, Kovarova L, Potacova A et al. Optimization of immunomagnetic selection of myeloma cells from bone marrow using magnetic activated cell sorting. Int J Hematol 2010; 92(2): 314–319.
8. Draube A, Pfister R, Vockerodt M et al. Immunomagnetic enrichment of CD138 positive cells from weakly infiltrated myeloma patients samples enables the determination of the tumor clone specific IgH rearrangement. Ann Hematol 2001; 80(2): 83–89.
9. Horst A, Hunzelmann N, Arce S et al. Detection and characterization of plasma cells in peripheral blood: correlation of IgE+ plasma cell frequency with IgE serum titre. Clin Exp Immunol 2002; 130(3): 370–378.
10. Chen L, Li J, Xu W et al. Molecular cytogenetic aberrations in patients with multiple myeloma studied by interphase fluorescence in situ hybridization. Exp Oncol 2007; 29(2): 116–120.
11. Presentation of Joachim W. Ellwart Cell Sorting Facility Helmholtz Zentrum München German Research Center for Environmental Health Institute of Molecular Immunology (IMI) Workgroup Kremmer NMI Reutlingen. Available from: http://www.slidefinder.net/2/22jul08/1214224.
12. Bonner WA, Hulett HR, Sweet RG et al. Fluorescence activated cell sorting. Rev Sci Instrum 1972; 43(3): 404–409.
13. Reid S, Yang S, Brown R et al. Characterisation and relevance of CD138-negative plasma cells in plasma cell myeloma. Int J Lab Hematol 2010; 32 (6 Pt 1): e190–e196.
14. Jourdan M, Ferlin M, Legouffe E et al. The myeloma cell antigen syndecan-1 is lost by apoptotic myeloma cells. Br J Haematol 1998; 100(4): 637–646.
15. Fuhler GM, Baanstra M, Chesik D et al. Bone marrow stromal cell interaction reduces syndecan-1 expression and induces kinomic changes in myeloma cells. Exp Cell Res 2010; 316(11): 1816–1828.
16. Kambham N, Kong C, Longacre TA et al. Utility of syndecan-1 (CD138) expression in the diagnosis of undifferentiated malignant neoplasms: a tissue microarray study of 1,754 cases. Appl Immunohistochem Mol Morphol 2005; 13(4): 304–310.
17. O’Connell FP, Pinkus JL, Pinkus GS. CD138 (syndecan-1), a plasma cell marker immunohistochemical profile in hematopoietic and nonhematopoietic neoplasms. Am J Clin Pathol 2004; 121(2): 254–263.
18. Rao PH. Comparative genomic hybridization for analysis of changes in DNA copy number in multiple myeloma. Methods Mol Med 2005; 113 : 71–83.
19. Sawyer JR. Metaphase cytogenetic techniques in multiple myeloma. Methods Mol Biol 2011; 730 : 149–158.
20. Schilling G, Dierlamm J, Hossfeld DK. Prognostic impact of cytogenetic aberrations in patients with multiple myeloma or monoclonal gammopathy of unknown significance. Hematol Oncol 2005; 23(3–4): 102–107.
21. Petrak J, Ivanek R, Toman O et al. Déjà vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics 2008; 8(9): 1744–1749.
22. Almasi M, Sevcikova S, Slaby O et al. Association study of selected genetic polymorphisms and occurrence of venous thromboembolism in multiple myeloma patients treated with thalidomide. Clin Lymph Myel Leuk. Accepted for publication.
23. Chen X, Ba Y, Cai X et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18(10): 997–1006.
24. Muralidharan-Chari V, Clancy JW, Sedgwixk A et al. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 2010; 123 (Pt 10): 1603–1611.
Labels
Paediatric clinical oncology Surgery Clinical oncologyArticle was published in
Clinical Oncology

2011 Issue Supplementum 1
- Possibilities of Using Metamizole in the Treatment of Acute Primary Headaches
- Metamizole vs. Tramadol in Postoperative Analgesia
- Spasmolytic Effect of Metamizole
- Metamizole at a Glance and in Practice – Effective Non-Opioid Analgesic for All Ages
- Safety and Tolerance of Metamizole in Postoperative Analgesia in Children
-
All articles in this issue
- Editorial (CZ)
- Radiotherapeutic methods
- Multiple Myeloma
- Monoclonal Gammopathy of Undeterminated Significance: Introduction and Current Clinical Issues
- Sample Processing and Methodological Pitfalls in Multiple Myeloma Research
- Flow Cytometry in Monoclonal Gammopathies
- Flow Cytometric Phenotyping and Analysis of T Regulatory Cells in Multiple Myeloma Patients
- Genomics in Multiple Myeloma Research
- Polymorphisms Contribution to the Determination of Significant Risk of Specific Toxicities in Multiple Myeloma
- Oligonucleotide-based Array CGH as a Diagnostic Tool in Multiple Myeloma Patients
- Visualization of Numerical Centrosomal Abnormalities by Immunofluorescent Staining
- Impact of Nestin Analysis in Multiple Myeloma
- Editorial (EN)
- List of authors and reviewers
- Clinical Oncology
- Journal archive
- Current issue
- About the journal
Most read in this issue
- Multiple Myeloma
- Flow Cytometric Phenotyping and Analysis of T Regulatory Cells in Multiple Myeloma Patients
- Monoclonal Gammopathy of Undeterminated Significance: Introduction and Current Clinical Issues
- Flow Cytometry in Monoclonal Gammopathies
