Cirkulární RNA hsa-circ-0006203 – hsa-circ-0004872 jako nové biomarkery detekce karcinomu dutiny ústní
Cirkulární RNA hsa-circ-0006203 – hsa-circ-0004872 jako nové biomarkery detekce karcinomu dutiny ústní
Východiska: Spinocelulární karcinom dutiny ústní (oral squamous cell carcinoma – OSCC) je nejběžnějším typem zhoubných nádorů v oblasti hlavy a krku. Je známo, že cirkulární RNA (circRNA) hraje důležitou roli v karcinogenezi různých typů zhoubných nádorů. Role circRNA při OSCC však zůstává nejasná.
Materiál a metody: Byly získány tkáně OSCC a přilehlé normální tkáně k detekci exprese circRNA sekvenováním nové generace (next generation sequencing – NGS) a byly vybrány tkáně OSCC k ověření circRNAs různého významu pomocí kvantitativní polymerázové řetězové reakce s reverzní transkriptázou (reverse transcriptase-quantitative polymerase chain reaction – RT-qPCR). Pro další zkoumání role hsa-circ-0006203 – hsa-circ-0004872 byl vytvořen design primeru a byla provedena RT-PCR. Hladiny exprese byly detekovány pomocí RT-qPCR.
Výsledky: Výsledky NGS ukázaly, že u OSCC byly výrazně exprimovány circRNA, přičemž dvě circRNAs byly exprimovány výrazně odlišným způsobem. hsa-circ-0006203 – hsa-circ-0004872 byly ve vzorcích tkáně OSCC výrazně downregulována, což statisticky korelovalo s jejich patologickou diferenciací.
Závěr: Souhrnně lze říci, že výsledky prezentované studie odhalily zvýšené množství circRNA ve tkáních OSCC a pokud je nám známo, náš tým jako první zkoumal regulační roli sítě hsa-circ-0006203 – hsa-circ-0004872 u OSCC. Výsledky ukázaly, že hsa-circ-0006203 – hsa-circ-0004872 může být případným biomarkerem OSCC.
Klíčová slova:
biomarker – karcinom dutiny ústní – cirkulární RNA – hsa-circ-0006203 – hsa-circ-0004872
Authors:
H. Ghaffari Monfared; G. Taheri Sangsari; F. Jamshidian
Authors‘ workplace:
Department of Biology, East Tehran Branch, Islamic Azad University, Tehran, Iran.
Published in:
Klin Onkol 2023; 36(5): 378-381
Category:
Original Articles
doi:
https://doi.org/10.48095/ccko2023378
Overview
Background: Oral squamous cell carcinoma (OSCC) is the most common cancer of the head and neck region. The circular RNA (circRNA) is known to serve an important role in the carcinogenesis of different types of cancer. However, the circRNA role of OSCC remains unclear.
Material and methods: OSCC tissues and adjacent normal tissues were obtained to detect circRNAs expression by the next generation sequencing (NGS), and OSCC tissues were selected to verify the differentially significant circRNAs by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). To further investigate the role of hsa-circ-0006203 – hsa-circ-0004872, the primer design and RT-PCR were performed. The expression levels were detected by RT-qPCR.
Results: The NGS results demonstrated that circRNAs were abundantly expressed in OSCC, and two circRNAs were significantly differentially expressed. hsa-circ-0006203 – hsa-circ-0004872 were significantly downregulated in OSCC tissue samples and was statistically correlated with pathological differentiation.
Conclusion: In summary, the results of the present study revealed that OSCC tissues have abundant circRNAs and, to the best of our knowledge, it was our team who firstly explore the regulatory role of the hsa-circ-0006203 – hsa-circ-0004872 network in OSCC. The results indicated that hsa-circ-0006203 – hsa-circ-0004872 may be a potential biomarker for OSCC.
Keywords:
oral cancer – biomarker – circular RNA – hsa-circ-0006203 – hsa-circ-0004872
Introduction
Oral squamous cell carcinoma (OSCC), the most frequently occurring oral malignancy, is the sixth most prevalent cancer worldwide and the third most common carcinoma in some developing countries [1,2]. OSCC is one of the most frequently occurring head-neck tumors and accounts for ~80% of all cases [3]. In addition, ~600,000 new clinical cases are diagnosed annually and the age at the time of diagnosis is becoming lower each year [4]. Although the treatments modalities have improved, the mortality rate has not decreased significantly and the 5-year survival rate of OSCC is only ~63% [5]. Cancers including OSCC have been widely regarded as gene-related diseases; however, the precise molecular mechanisms and genetic basis of OSCC carcinogenesis remain largely unclear. Therefore, it is imperative to identify therapeutic targets to improve OSCC diagnosis and treatment.
Recently, circular RNAs (circRNAs) have been considered to be a special type of noncoding RNA, which are widespread and diverse in mammals [6]. Unlike linear RNA, circRNA contains covalently closed loop structures and has neither 5’caps nor 3’tails; therefore, the majority of circRNA cannot be degraded by RNA exonucleases or RNase R, thereby maintaining an improved stability compared with linear transcripts [6]. Emerging evidence has revealed various functions of circRNA such as competing endogenous RNAs, miRNA sponges, or transcriptional regulators [7]. CircRNA has also been considered to serve a crucial role in cancers, and due to its functions, circRNAs may serve as a potential target for tumor therapy or diagnostic biomarkers [8]. For example, CDR1as and circ-FOXO3 sponge microRNA (miR) regulate the development of breast carcinoma; additionally, circ-ITCH has an inhibitory effect on esophageal squamous cell carcinoma by suppressing the Wnt/b-catenin pathway [8]. However, the role of circRNA in the initiation and progression of OSCC has not been completely elucidated. In the present study, circRNA expression profiles were analyzed through the new generation sequencing (NGS), followed by RT-qPCR to verify the significantly expressed circRNAs in the OSCC tissues. hsa-circ-0008309 was identified to be downregulated in the cancer tissues. We explored the functions of hsa-circ-0006203 – hsa-circ-0004872 binding sites. These networks were further investigated in OSCC cell lines. In summary, the data provided a novel basis for circRNA functional research in OSCC and indicated that hsa-circ-0006203 – hsa-circ-0004872 may be novel potential biomarkers in OSCC research.
Material and methods
Patients and samples
A total of 60 tumor samples, 30 normal tissues, 20 benign tissues, and 10 malignant tissues from patients with oral cancer were obtained from the tumor bank of Imam Khomeini Hospital Complex, Tehran University of Medical Sciences. These tissues were prepared for NGS and validation by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR). Patients had not undergone additional treatments prior to surgery and all OSCC tissues were confirmed by strict pathological examination. The age of patients ranged from 29–78 years, and the median age at the time of diagnosis was 54 years. The male to female ratio was 34 : 11. The tissues were obtained from the tongue, gingiva, and floor of the mouth. Clinical and pathological characteristics of the patients were based on the most recent World Health Organization (WHO) classification and UICC tumor-node-metastasis (TNM) classification. Written informed consent from all patients was obtained, and the study was approved by the Medical Ethics Committee of Imam Khomeini Hospital Complex, Tehran University of Medical Sciences. All methods were performed in accordance with the relevant guidelines and regulations.
RNA extraction
Samples obtained from surgical specimens were immediately frozen using liquid nitrogen. Total RNA was extracted from frozen tissues using TRIzol® reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s protocol. The quality and quantity of the RNA were evaluated at a 260/280 ratio using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc.).
RT-qPCR
The reaction mixture (20 μL) containing 1 μg of total RNA was reverse transcribed into cDNA with the PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China). The mixture was incubated at 37 °C for 15 min and 85 °C for 5 s to acquire cDNA. RT-qPCR was conducted with a Roche Applied Science LightCycler® 96 Real-Time PCR System (Roche Diagnostics, Indianapolis, Indiana, USA) in accordance with the manufacturer’s protocol. The reaction mixture comprised of 2 μL cDNA, 5 μL SYBR® Premix Ex Taq™ II (Takara, China) and 1 μL primers (reverse and forward) with RNase-Free water to a final volume of 10 μL. The thermocycling conditions were as follows: 95 °C for 3 min and amplified by 40 cycles of denaturing at 95 °C for 10 s and 60 °C for 30 s. b-actin was used as an internal standard. Melting curves were produced to check product purity and the expression levels of circRNAs were detected by using the 2-DDCq method. The Cq value was the fractional cycle number at which the fluorescence exceeded the given threshold. Primer sequences are listed as follows: hsa-circ-0004872: F: TCAAGATCTGTGACTTTGGCCT, R: GGTGCTCAAAGGGGCTGATT. circ0006203: F: TCCCCATCACAAGAAGACCTGA, R: TGGTGTAGCCCTTTGGAGTCA.
Quantifying the extracted RNA
To determine the amount and purity of the extracted RNA, the nanodrop device (Thermo scientific-Nanodrop 2000) was used by the method of determining the optical density (OD: Optical Density). The samples had a light absorption ratio of 260–280 nm in the range of 1.8–2.2 and a light absorption ratio of 260.230 between 1.7 and 1.9, which was suitable for cDNA production.
Qualitative evaluation of extracted RNAs
After extracting the genetic material or amplifying a fragment of a specific gene product, the next step was to separate it into constituent fragments and identify each fragment. Agarose gel was used to separate nucleic acids. The size of the molecules in each band could be well determined by comparing it to bands made from nucleic acid fragments of a certain size, which were electrophoresed simultaneously with the samples.
cDNA synthesis extracted from RNA, reverse transcription reaction
In this study, a Thermo fisher kit was used to make cDNA. All steps were performed on ice. For each sample, the following materials were added to the nuclease-free microtube according to the protocol. First, to equalize the concentrations, we brought all the extracted RNAs to 1000 ng/μL after reading the amount of RNA concentration by the nanodrop device. It was poured into 0.2 mL sterile microtubes, then the microtubes were incubated at 65 °C with the aim of opening the nucleotide strands for 5 min, then immediately placed on ice to cool. After cooling (to prevent premature enzyme action), 10 µL of RT Master mix was added in PCR at 25 °C for 10 min (primer binding), 42 °C for 60 min (cDNA synthesis) and 70 °C for 10 min (enzyme inactivation).
Statistical analysis
Data were described and the relationship between them was assessed using specific statistical tests by SPSS program and at a significant level (P-value < 0.05): 1) Kolomogorov-Smirnov one-sample test was used to measure the normality of the data; 2) independent sample t-test was used to determine the significant level of mean Dcts between the two tumor groups and tumor margins; 3) one-way ANOVA test was used to evaluate the difference in expression levels of MAPK1 gene in different tumor sizes.
Results
Description of tumor and normal samples collected in this study
Thirty samples of oral cancer tissue and 30 healthy samples were used. Sixteen patients were classified in grade I, 13 patients in grade II and 1 patient in grade X. There were 3 patients with a tumor size of < 2 cm and 18 patients with a tumor size of 2–5 cm and 9 patients with a tumor size of > 5 cm.
Determining the quality and quantity of extracted RNA
After RNA extraction of the samples, in order for the extracted RNA to be true, the amount of RNA obtained was measured by a nanodrop device. The value of the wavelength ratio of 260–280 nm with acceptable values1.8–2.2 was considered. Therefore, 1.5% agarose gel was loaded to evaluate the quality of the extracted RNA. Based on Fig. 1, the 18 s and 28 s bands were well visible, which indicated the accuracy of the extraction.
Real time PCR reaction results
Using synthesized cDNAs, real time PCR was performed for all samples and ACTB reference gene. After performing the reactions, CT samples were extracted from real time PCR analysis. Fig. 2, 3 and 4 show the hsa-circ-0004872, hsa-circ - 0006203 and MAPK1 gene proliferation curves, respectively.
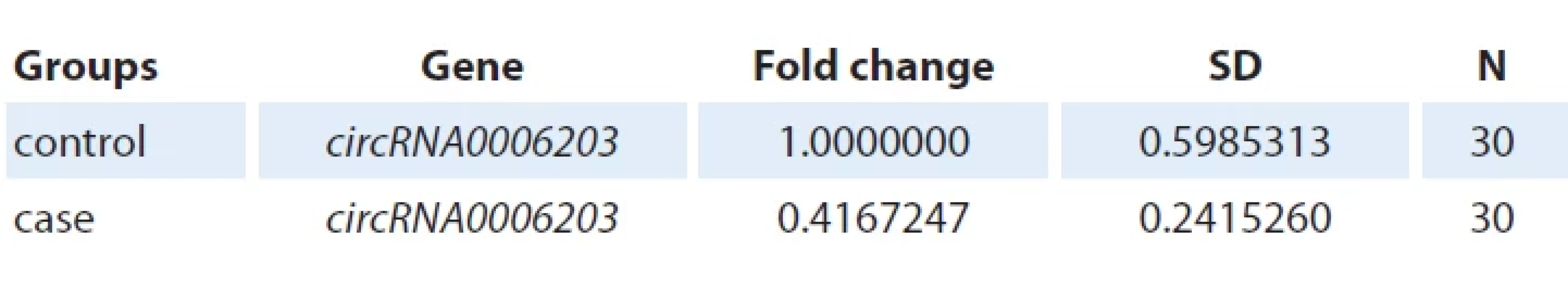
Evaluation of circRNA0006203 gene expression change between control and case groups
In Fig. 5 and tab. 1 and 2, the value of P = 0.00000 indicates a significant effect of case group on the expression of circRNA0006203 gene. The expression of circRNA0006203 gene in the case group compared to the control group shows a significant decrease: fold change = = 0.4167 and P = 0.00000.
Discussion
CircRNA was previously considered to be a noise from aberrant RNA splicing [9,10]. However, it was recently identified to have the potential as miR sponges and ideal biomarkers in various diseases. In addition, a large number of studies have indicated that various circRNAs were associated with the initiation and progression of cancers [11]; however, there is little evidence regarding the role of circRNA in OSCC.

In the present study, a total of 11,942 circRNAs were identified, demonstrating that circRNAs were in abundant existence in the OSCC samples (N = 8). Furthermore, 16 circRNAs were identified to be significantly differentially expressed in the OSCC samples through analysis. The results suggest that circRNA may serve an important role in OSCC, and these significantly differentially expressed circRNAs were subsequently validated in the 45 pairs of OSCC samples. As a result, hsa-circ-0006203 – hsa-circ-0004872 was demonstrated to be significantly downregulated in OSCC tissues (N = 16). Notably, compared with normal tissues, the hsa-circ-0006203 – hsa-circ-0004872 expression level was revealed to be 2.010811 times in OSCC tissues by NGS. The levels of hsa-circ-0006203 – hsa-circ-0004872 in certain samples were downregulated in the 45 pairs of OSCC tissues. These results indicated that hsa - -circ-0006203 – hsa-circ-0004872 expression level of each patient was not consistent and varied between individuals. The expression of hsa-circ-0006203 – hsa-circ-0004872 was downregulated in tumor tissues and was, therefore, more likely to act as a tumor suppressor. The ROC analysis indicated that the hsa-circ-0006203 – hsa-circ-0004872 expression level exhibited a diagnostic role in distinguishing OSCC tissues from adjacent normal tissues. Taken together, we hypothesized that hsa-circ-0006203 – hsa-circ-0004872 has a potential effect on OSCC. Compared with linear RNA, circRNA has been reported to have an increased number of miR binding sites and may regulate gene expression by acting as miR sponges, thereby regulating linear RNA transcription and protein production. Consequently, the present study examined whether hsa-circ-0006203 – hsa-circ-0004872 had the potential to affect the miRs in OSCC.
Conclusion
In summary, the present study revealed that the OSCC tissues have abundant circRNAs and identified that hsa - -circ-0006203 – hsa-circ-0004872 were significantly downregulated in OSCC tissues. Herein is an offered novel approach to explore the role of circRNA by various analyses. Furthermore, the regulatory role of hsa-circ-0006203 – hsa-circ-0004872 network was identified in OSCC. These results highlighted the possibility that hsa-circ-0006203 – hsa--circ-0004872 could serve as a potential target for OSCC. The functions and mechanisms of hsa-circ-0006203 – hsa--circ-0004872 in OSCC should continue to be extensively investigated.
For fi gures 1–5, see the online version of the article at www.linkos.cz.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
This paper has not been published elsewhere in whole or in part. All authors have read and approved the content and agree to submit for consideration for publication in the journal. There is no any ethical/legal conflicts involved in the article.
Conflicts of interest
The authors declare that they have no conflicts of interest.
The authors declare that they have no potential conflicts of interest concerning drugs, products, or services used in the study.
Autoři deklarují, že v souvislosti s předmětem studie nemají žádné komerční zájmy.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do
biomedicínských časopisů.
Dr. Faranak Jamshidian
Department of Biology
East Tehran Branch, Islamic Azad University
Tehran, Iran
e-mail: faranak.jamshidian@gmail.com
Submitted/Obdrženo: 13. 7. 2022
Accepted/Přijato: 13. 9. 2022
Sources
1. Rosebush MS, Rao SK, Samant S et al. Oral cancer: enduring characteristics and emerging trends. J Tenn Dent Assoc 2011; 91 (2): 24–27.
2. Casiglia J, Woo SB. A comprehensive review of oral cancer. Gen Dent 2001; 49 (1): 72–82.
3. Sun ZJ, Chen G, Zhang W et al. Mammalian target of rapamycin pathway promotes tumor-induced angiogenesis in adenoid cystic carcinoma: its suppression by isoliquiritigenin through dual activation of c-Jun NH2-terminal kinase and inhibition of extracellular signal-regulated kinase. J Pharmacol Exp Ther 2010; 334 (2): 500–512. doi: 10.1124/jpet.110.167692.
4. He KF, Zhang L, Huang CF et al. CD163+ tumor-associated macrophages correlated with poor prognosis and cancer stem cells in oral squamous cell carcinoma. Biomed Res Int 2014; 2014 : 838632. doi: 10.1155/2014/838632.
5. Miller KD, Siegel RL, Lin CC et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016; 66 (4): 271–289. doi: 10.3322/caac.21349.
6. Li Y, Zheng Q, Bao C et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25 (8): 981–984. doi: 10.1038/cr.2015.82.
7. Qu S, Yang X, Li X et al. Circular RNA: a new star of non--coding RNAs. Cancer Lett 2015; 365 (2): 141–148. doi: 10.1016/j.canlet.2015.06.003.
8. Memczak S, Jens M, Elefsinioti A et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495 (7441): 333–338. doi: 10.1038/nature11 928.
9. Zhang H, Cai K, Wang J et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014; 32 (11): 2858–2868. doi: 10.1002/stem.1795.
10. Yang W, Du WW, Li X et al. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 2016; 35 (30): 3919–3931. doi: 10.1038/onc.2015.460.
11. Li F, Zhang L, Li W et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015; 6 (8): 6001–6013. doi: 10.18632/oncotarget.3469.
Labels
Paediatric clinical oncology Surgery Clinical oncologyArticle was published in
Clinical Oncology

2023 Issue 5
- Possibilities of Using Metamizole in the Treatment of Acute Primary Headaches
- Metamizole at a Glance and in Practice – Effective Non-Opioid Analgesic for All Ages
- Metamizole vs. Tramadol in Postoperative Analgesia
- Spasmolytic Effect of Metamizole
- Metamizole in perioperative treatment in children under 14 years – results of a questionnaire survey from practice
-
All articles in this issue
- Editorial
- Využití botulotoxinu při léčbě nežádoucích účinků radioterapie
- Transformace indolentního folikulární lymfomu v difuzní velkobuněčný B-lymfom – molekulární podstata „nádorové agresivity“
- Jak neznámý je karcinom neznámého primárního zdroje metastazující do krčních uzlin?
- Stereotaktická radioterapie v léčbě časného stadia nemalobuněčného karcinomu plic
- Cirkulární RNA hsa-circ-0006203 – hsa-circ-0004872 jako nové biomarkery detekce karcinomu dutiny ústní
- Léčba dospělých pacientů s akutní lymfoblastovou leukemií v České republice v letech 2007–2020
- Alveolárny sarkóm mäkkých tkanív u dieťaťa – opis prípadu
- Raritní případ uroteliálního karcinomu metastazujícího do stěny žlučníku s projevy akutní cholecystitidy
- Aktuality z odborného tisku
- Nová možnost kombinované léčby pro pacienty s metastatickým renální karcinomem v 1. linii léčby: lenvatinib s pembrolizumabem – výsledky studie CLEAR
- Jubilant doc. MUDr. Bohuslav Konopásek, CSc.
- Životní jubileum první dámy české onkologie
- Spomienka na akademika MUDr. Viliama Ujházyho, DrSc. (1926–2023)
- Clinical Oncology
- Journal archive
- Current issue
- About the journal
Most read in this issue
- Transformace indolentního folikulární lymfomu v difuzní velkobuněčný B-lymfom – molekulární podstata „nádorové agresivity“
- Léčba dospělých pacientů s akutní lymfoblastovou leukemií v České republice v letech 2007–2020
- Stereotaktická radioterapie v léčbě časného stadia nemalobuněčného karcinomu plic
- Využití botulotoxinu při léčbě nežádoucích účinků radioterapie
