Detection panel for identification of twelve hemorrhagic viruses using real-time RT-PCR
Real time RT-PCR panel pro identifikaci dvanácti virů hemoragických horeček
Úvod:
Virové hemoragické horečky jsou způsobovány zástupci čtyř virových čeledí a patří mezi choroby s velmi vysokou smrtností. Pro jejich diagnostiku není dostupný žádný komerční kit.
Výsledky:
Vytvořili jsme real time RT-PCR panel pro detekci a identifikaci virů Ebola, Marburg, Lassa, Guanarito, Machupo, Sabiá, Seoul, Puumala, Hantaan, virus krymsko-konžské hemoragické horečky a horečky Rift Valley. Protokol funguje na principu jednotných reakčních podmínek a byl testován na různých PCR přístrojích jak kapilárních, tak i destičkových včetně polní verze termocykleru Ruggedized Advanced Pathogen Identification Device (R.A.P.I.D.) (Idaho Technology, Inc.).
Závěr:
V kombinaci s adaptovanými protokoly z již publikovaných prací představujeme jednoduchý detekční systém pro rychlou identifikaci původců virových hemoragických horeček z čeledí filoviru, arenavirů a bunyavirů s dostatečnou senzitivitou a specificitou pro použití laboratořích prvního kontaktu a pro diagnostiku v polních podmínkách.
Klíčová slova:
hemoragické horečky – filoviry – arenaviry – real time RT-PCR –diagnostika
Authors:
M. Fajfr 1,2; V. Neubauerová 3; P. Pajer 3; P. Kubíčková 3; D. Růžek 4,5
Authors‘ workplace:
Faculty of Military Health Sciences, University of Defense, Hradec Králové
1; Institute of Clinical Microbiology, University Hospital in Hradec Králové, Hradec Králové
2; Central Military Institute of Health, Army of the Czech Republic, Prague
3; Institute of Parasitology, Biology Centre of the Academy of Sciences of the Czech Republic, České Budějovice
4; Department of Virology, Veterinary Research Institute, Brno
5
Published in:
Epidemiol. Mikrobiol. Imunol. 63, 2014, č. 3, s. 238-244
Category:
Review articles, original papers, case report
Overview
Background:
Viral hemorrhagic fevers are caused by viruses from four viral families and develop diseases with high fatality rates. However, no commercial diagnostic assay for these pathogens is available.
Findings:
We developed real-time RT-PCR assays for viruses Ebola, Marburg, Lassa, Guanarito, Machupo, Junin, Sabiá, Seoul, Puumala, Hantaan, Crimean-Congo hemorrhagic fever virus and Rift Valley fever virus. The assays were optimized for identical reaction conditions and can be performed using several types of real-time PCR instruments, both capillary and plate, including a portable Ruggedized Advanced Pathogen Identification Device (R.A.P.I.D.) (Idaho Technology, Inc.).
Conclusions:
In combination with primers and probes from previously published studies, we present a simple system for rapid identification of hemorrhagic filoviruses, arenaviruses and bunyaviruses with sufficient sensitivity for first contact laboratory and diagnosis under field conditions.
Keywords:
hemorrhagic fever – filovirus – arenavirus – real-time RT-PCR – detection
INTRODUCTION
Viral hemorrhagic fevers (VHFs) represent a wide group of diseases caused by viral pathogens, which belong to four different viral families – Filoviridae, Arenaviridae, Flaviviridae, and Bunyaviridae [1]. VHFs are serious diseases with high mortality, which mostly occur near the equatorial belt in the African or American continents. However, viruses causing VHFs can be imported into non-endemic countries, and have also a potential to be misused as aerobiological weapons. Also units potentially dispatched in endemic areas during UN, EU or NATO missions could be at risk.
Real-time RT-PCR assays for detection of various VHF species, and corresponding strains of Ebola virus [2–6], Marburg virus [2, 3, 6], Lassa virus [6, 7], Junin virus [8], hemorrhagic bunyaviruses [9–11] and other VHFs were developed. However, comprehensive panels of real-time RT-PCR assays for VHF detection are rare. Trombley et al. [12] developed a detection system for specific and absolute quantification of multiple hemorrhagic filoviruses, arenaviruses, and New World Hantaviruses based on a panel of 48 TaqMan-based PCR assays. A much simpler system is, however, needed for use under field conditions in mobile labs or fields hospitals. Drosten et al. [6] presented a principle of universal cycling profiles used in panels for detection of 7 different hemorrhagic viruses.
In the Czech Republic there are no methods available for detection of hemorrhagic fever viruses. The problems of VHF diagnosis in the Czech Republic are partially solved by the National reference laboratory for arboviruses, which is able to detect only a few members – dengue virus and hantaviruses. Due to the fact that no commercial assays have been available, in house assays are only one possibility for diagnosis of VHFs. Here, we present a simple, convenient, and rapid detection system for identification of 12 hemorrhagic filoviruses, arenaviruses and bunyaviruses, which is designed to be suitable especially for field applications. The presented panel uses separate reverse transcription steps and a panel of single-plex real-time PCR with universal cycling profiles.
MATERIAL AND METHODS
Virus and RNA preparation
Purified viral RNA of Ebola Zaire virus (strain Gabon 2003), Ebola Sudan virus (strain Gulu), Marburg virus (strain Popp), Lassa virus (strain Josiah), Machupo virus (strain Carvallo), Junin virus (strain XJ), Sabiá virus (strain SPH114202), Guanarito virus (strain INH-95551) and Crimean-Congo hemorrhagic fever virus (strain AFGN09-2990) were purchased from the Bernhard Nocht Institute (Hamburg, Germany). Purified viral RNAs of Seoul virus (strain R22), Puumala virus (strain CG18-20), Hantaan virus (strain 76/118) and Rift Valley fever virus (strain 1.48) were purchased from the National Collection of Pathogenic Viruses (Health Protection Agency, Porton Down, UK). RNA concentration was measured using Qubit fluorometer (Invitrogen). cDNA was prepared using a Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacture’s manual. Nucleic acids from 24 viruses and 26 bacterial species were used as controls for specificity evaluation.
Primers and probe design
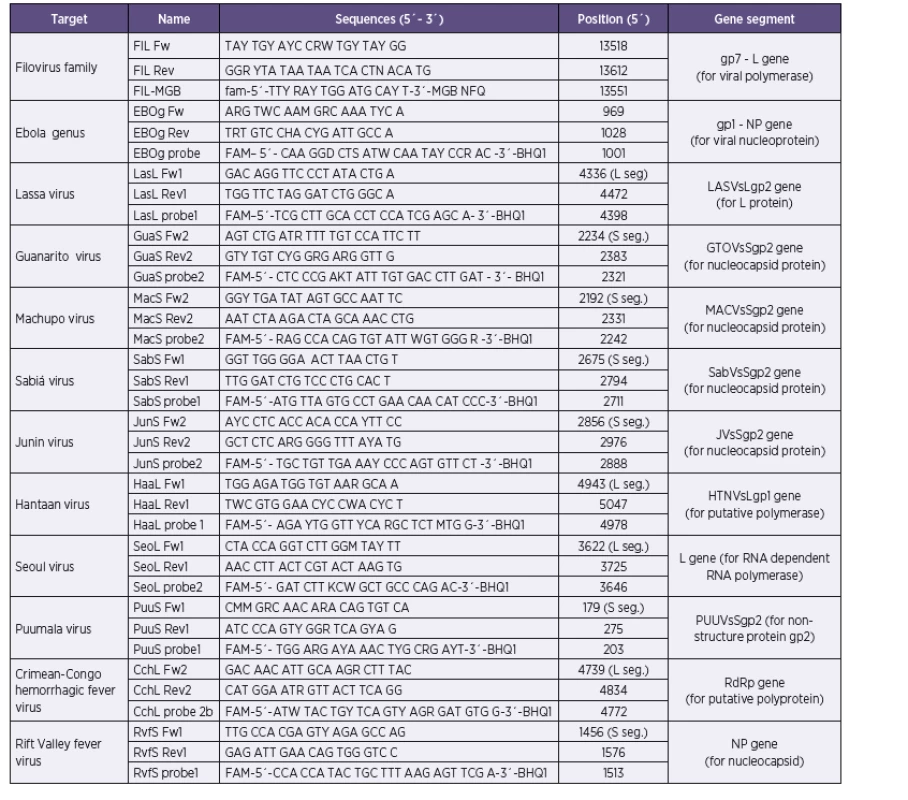
Our approach combines the use of previously published primer sequences, modified primer sequences, as well as newly designed primers (Table 1). Conservative segments of viral genomes suitable for primer and/or probe target design were identified in alignments of all relevant sequences available in the GenBank database in BLAST software (www.ncbi.nlm.nih.gov) and BioEdit (www.mbio.ncsu.edu/BioEdit/bioedit/html). The search for target sequences was performed either manually or using LightCycler Probe Design Software, version 1.0 (Idaho Technologies). All probes were conjugated with 6-carboxyfluorescein (FAM) and Black Hole Quencher 1 (BHQ1). According to project assignment were design protocols for Ebola, Marburg, Lassa, Junin, Machupo, Guanarito, Sabia, Crimean-Congo hemorrhagic fever, Rift Valley fever, Hantaan, Seoul and Puumala viruses. Dobrava/Belgrade virus was not in assignment of research project.
Real-time PCR
SYBR Green I real-time PCR was performed using LightCycler Fast Start DNA Master Plus SYBR Green I Kit (Roche). Briefly, the reaction mixture contained 3 µl of the cDNA template, 1 µl of each primer (0.5µM), 4 µl of the master mix, and 11 µl of PCR grade water. Reaction conditions were as follows: 95 °C for 10 minutes (1 cycle); 95 °C for 15 sec, 55 °C for 15 sec and 72 °C for 25 sec (35–45 cycles); 40 °C for 30 sec (1 cycle). For melting curve analysis, a temperature range from 95 °C to 55 °C was chosen.
TaqMan real-time PCR was done using LightCycler FastStart DNA Master Plus HybProbe Kit (Roche). Briefly, the reaction mix contained 1–3 µl of cDNA template, 1 µl of each primer and probe (0.5 µM), 4 µl of master mix, and PCR grade water made up to a total volume of 20 µl. Reaction conditions were: 95 °C for 10 minutes (1 cycle); 95 °C for 15 sec, 55 °C for 15 sec (52 °C for 20 sec for primers/probes sets from Panning et al., 2007) and 72 °C for 25 sec (45 cycles); 40 °C for 30 sec (1 cycle). A single fluorescence was taken at the end of each annealing step.
All reactions were performed using LightCycler 1.5 (Roche) and the data were analysed by LightCycler software, ver. 3.5 (Roche). The universality of the optimized reaction protocols was also tested using R.A.P.I.D. (Idaho Technology) and Chromo4 (Bio-Rad), real-time PCR thermocyclers at three workplaces. To test the detection limit, 10-fold dilutions of RNA template were analyzed. For each dilution at least 15 repeating reactions were used for average detection limit determination.
RESULTS
The published primers and probes were adapted to our filoviral family-specific real-time RT-PCR protocol. This reaction was based on the use of a previously published primer combination (FiloA2.2, 2.3, 2.4, FiloB, Filo B-Ra) with SYBR Green I marking [13], as well as a newly designed in-house assay (primers FIL Fw and FIL Rev combined with a probe FIL-MGB with minor groove binding (MGB)). The in-house protocol was optimized for use with SYBR Green I as well as TaqMan probe. Both protocols showed comparable results, the detection limit were 1–2 pg/1 µl. Due to the use of MGB probes in our in-house assay, the annealing temperature was decreased (48 °C) unlike all other protocols used in this study. With the purpose of keeping the reaction conditions the same in all reactions in our workflow, we prefer the use of primers by Panning et al. [13] with SYBR Green I as the method of choice. Marburg virus-specific assay is based on a previously published protocol by Panning et al. [13]. This method uses probe (FAMMBG) in combination with three primers (FiloA2.2, 2.3, 2.4, FiloB, Filo B-Ra). For detection of Ebola virus, a new combination of primers (EBOg Fw; EBOg Rev) and probe (EBOg probe) (see Table 1) was designed enabling the detection of major human Ebola viruses (Zaire and Sudan) and theoretically also the newly emerged Bundibugyo Ebola virus (tested only in silico). Detection limits were 80 fg RNA/1 µl of Ebola Zaire and 1,3 pg RNA/1 µl of Ebola Sudan.
For hemorrhagic bunyaviruses, new primer-probe sets for all chosen five members were designed. During the bio-analytic study it was shown that it was not possible to design a family specific assay. The detection limits for Seoul, Hantaan, Puumala viruses, causative agents of hemorrhagic fever with renal syndrome, were 60–100 fg/1 µl using SYBR Green I or 6–10 fg/1 µl with TaqMan probes. For Crimean-Congo hemorrhagic virus (CCHFV), the detection limit was 33 fg/1 µl for TaqMan and 100 fg/µl for SYBR Green I. For Rift Valley fever virus the detection limit was 80 fg/1 µl. No cross reactivity was observed with any of the tested control viral or bacterial pathogens. In all SYBR Green I assays worse detection limits were obtained in comparison with TaqMan probe use. Although highly degenerate primers were used, the melting curves were possible to evaluate (Figure 1).
Arenaviridae family specific assay is based on a previously published protocol by Vieth et al. [14, 15]. Arenaviruses are divided into two genetically very different groups, Old World arenaviruses and New World arenaviruses. Family specific primers are designed to discriminate these two groups. Both primer combinations exhibited nonspecific fluorescence in negative controls starting from cycle No. 34–36 in reactions with SYBR Green I, which is in accordance with the initial publication [14, 15]. Therefore, the number of cycles in our reactions was decreased to 35. The detection limit for reaction with standard results was 50–100 fg RNA/1 µl. The reactions were highly specific, no amplification was observed if nucleic acids from control viruses or bacteria were used. For detection of hemorrhagic arenaviruses, we designed assay for Lassa, Guanarito, Machupo, Junin, and Sabia viruses (see Table 1). These assays were developed for use with SYBR Green I or TaqMan probes. In reactions with SYBR Green I, the detection limit was 70–100 fg RNA/1 µl. If TaqMan probes were used, the detection limit was 7–10 fg RNA/1 µl.
All assays designed in this study or adapted from previously published protocols were optimized to identical reaction conditions to simplify the overall three-step procedure of hemorrhagic filovirus, arenavirus and bunyavirus detection (Diagram 1): reverse transcription – viral family specific Real time PCR (available only for filoviruses and arenaviruses) – virus specific Real time PCR. The suitability of our assays was tested using real-time PCR cyclers from different companies. Comparable results were obtained from all tested PCR cyclers.
DISCUSSION
The presented assay represents a compact real-time reverse transcription-PCR panel for detection of hemorrhagic filoviruses, arenaviruses and bunyaviruses designed especially for field conditions. Our system consisted of adopted and newly designed protocols which were able to detect on the first level the viral family (using SYBR Green I, not applicable for bunyaviruses) and on the second the pathogen itself (using SYBR Green I or TaqMan probes). For the high genetic variability of Bunyaviridae members our research team was not able to develop a family specific assay. The same problems with bunyavirus sequence variability have been published in previous studies. The principle of identical reaction condition was preferred, as was recommended by Drosten et al. [6]. The ideal method for VHF detection should be multiplex PCR reactions with high sensitivity and the possibility of multiple pathogen detection during one reaction run. However, it is quite difficult to design and optimize such an assay [16, 17]. Also the perspective method MassTaq PCR (combinations of Mass spectrometry and PCR) had the same drawbacks for widely routine use even though it theoretically allows the detection of 32 different pathogens [18]. On the contrary, presented protocols were very simple. The presented assay based on multiple single-plex PCR reactions allows adaptation of the method according to the actual conditions, for one sample or for multiple unknown samples. Analogous principles have been used in various commercial kits (like Bio threats screen kits (Idaho Technologies®). The assay modular principle (according to viral family) allows also the using of suitable protocols according to actual situations, like epidemiological data (place of stay, e.g .a patient infected during a stay in Africa – the protocols for New World arenaviruses should be removed from the assay used) or patient symptoms (type of bleeding, severity of case, etc.).



The detection limits of our protocols 50–100 fg RNA/1 µl for family specific assay and tens fg RNA/1 µl for species specific reactions are sensitive enough in the first step (field) diagnosis of clinical or environmental samples. Previously published protocols worked with similar sensitivity 10–100 fg/1 µl [16]. The sensitivity of our protocols matched approximately from 102 to 104 copy/ml. The viral load in patients sera during the acute phase of viral hemorrhagic fever caused for example by Ebolavirus was measured 105–107/ml in surviving patients and 108–1010/ml in dead patients [19]. Other viruses causing hemorrhagic fevers should be found in serum with a quantity near 106/ml – for CCHFV 7,7 x 105/ml or for Lassa virus 106/ml [16]. As follows from previous data, the most used clinical material for PCR diagnosis of VHF was serum/plasma (majority of VHF) or cerebrospinal fluid (arenaviruses) [16]. Our SYBR Green I protocols for virus-specific reactions had lower specificity in comparison with TaqMan probe protocols, which is generally known [6, 16], but with the advantage of markedly lower cost. Based on these findings, SYBR Green I procedure was recommended only in viral family specific assays (adopted protocols), in accordance with other data [13,15]. For virus specific reaction, we used more specific TaqMan probes. A novel method for detection of 9 viral pathogens from a VHF group with high sensitivity (from 10-5 to 10-1 PFU/PCR) and specificity with the ability of product quantification was recently published [12]. Our protocols have worse detection limits, but we use strictly only one primers/probe set for one pathogen. Sensitivity of the assay is decreased by the use of high degenerate bases in the primers/probes. Trombley et al. chose high sensitivity with results in using of 48 sets of primers/probe (TaqMan and TaqMan-MGB) for detection of 9 pathogens [12]. Using of this amount of primers/probe sets could be highly inconvenient in routine diagnosis especially in field conditions as was mentioned by the authors themselves. Such an assay is more suitable for application in epidemiological studies or for antiviral treatment effectiveness studies.

In the Czech Republic there were presented from VHF only sporadic cases of HFRS (hemorrhagic fever with renal syndrome) caused by Dobrava/Belgrade virus [20]. During the serological survey were also found seropositivity against Hantaan, Puumala and Seoul [21]. Very similar results were also published from other European countries, where hantaviruses were detected annually with seroprevalence from 1 % to 9 % in North parts of Europe [22]. In European countries were also sporadically detected other VHF members, like Crimean-Congo hemorrhagic fever virus [23, 24] or some cases of dengue fever from Madeira [25]. But the risk of importation of some other VHF from Africa or South America is still real and the health care system must be prepared for its identification [16].
CONCLUSION
Our method was designed as a simple protocol suitable especially for field conditions even though of lower sensitivity. The assay can be used for first examination in areas of VHF presence. The protocol optimizations for R.A.P.I.D. cycler (Idaho Technology, Inc.) allow its use by mobile research or counter-epidemic teams. The sensitivity of the presented assay is sufficient for initial rapid examination. The assay design also allows two modifications according to actual demand: using one sample-multiple pathogens or multiple sample-one pathogen. As is recommended in WHO or CDC manuals, the confirmation tests in certificated laboratories are required for final evaluation of viral hemorrhagic fever cases.
The whole PCR VHF assay (with PCR assay for VHF flaviviruses, data not published) was in 2013 authorised for use in the Czech Army.
List of abbreviations
BHQ1, Black Hole Quencher 1; FAM, 6-carboxyfluorescein; R.A.P.I.D., Ruggedized Advanced Pathogen Identification Device; MGB, minor groove binding.
Competing interests
The authors declare that they have no competing interests.
Acknowledgement
We acknowledge financial support by grant OVUVZU2008002 of the Ministry of Defense of the Czech Republic, grant Z60220518 and the AdmireVet project No. CZ.1.05/2.1.00/01.006 (ED006/01/01) of the Ministry of Education, Youth and Sports of the Czech Republic.
Do redakce došlo dne 30. 1. 2014.
Adresa pro korespondenci:
Dr. Miroslav Fajfr
Ústav klinické mikrobiologie FN Hradec Králové
Sokolská 581
50005 Hradec Králové
e-mail: fajfrmiroslav@seznam.cz
Sources
1. Borio L, Inglesby T, Peters CJ, Schmaljohn AL, Hughes JM, Jahrling PB, et al. Hemorrhagic Fever Viruses as Biological Weapons (Medical and Public Health Management). JAMA, 2002;287(18):2391–2405.
2. Gibb TR, Norwood DA Jr, Woollen N, Henchal EA. Development and evaluation of a fluorogenic 5´ nuclease assay to detect and differentiate between Ebola virus subtype Zaire and Sudan. J Clin Microbiol, 2001;39(11):4125–4130.
3. Weidmann M, Mühlberger E, Hufert FT. Rapid detection protocol for filoviruses. J Clin Virol, 2004;30(1):94–99.
4. Towner JS, Rollin PE, Bausch DG, Sanchez A, Crary SM, Vincent M, et al. Rapid Diagnosis of Ebola Hemorrhagic Fever by Reverse Transcription-PCR in an Outbreak Setting and Assessment of Patient Viral Load as a Predictor of Outcome. J Virol, 2004;78(8):4330–4341.
5. Towner JS, Khristova ML, Sealy TK, Vincent MJ, Erickson BR, Bawiec DA, et al. Marburgvirus Genomics and Association with a Large Hemorrhagic Fever Outbreak in Angola. J Virol, 2006;80(13):6497–6516.
6. Drosten C, Göttig S, Schilling S, Asper M, Panning M, Schmitz H, Günther S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol, 2002;40(7):2323–2330.
7. Lunkenheimer K, Hufert FT, Schmitz H. Detection of Lassa virus RNA in specimens from patients with Lassa fever by using the polymerase chain reaction. J Clin Microbiol, 1990; 28 : 2689–2692.
8. Lozano ME, Enría D, Maiztegui JI, Grau O, Romanowski V. Rapid diagnosis of Argentine hemorrhagic fever by reverse transcriptase PCR-based essay. J Clin Microbiol, 1995; 33(5):1327–1332.
9. Aitichou M, Saleh SS, McElroy AK, et al. Identification of Dobrava, Hantaan, Seoul, and Puumala viruses by one-step real-time RT-PCR. J Virol Method, 2005;124(1–2):21–26.
10. Bird BH, Bawiec DA, Ksiazek TG, et al. Highly sensitive and broadly reactive quantitative reverse transcription-PCR assay for high-throughput detection of Rift Valley fever virus. J Clin Microbiol, 2007;45(11):3506–3513.
11. Garrison AR, Alakbarova S, Kulesh DA, et al. Development of a TaqMan®-Minor Groove Binding Protein Assay for the Detection and Quantification of Crimean-Congo Hemorrhagic Fever Virus. Am J Trop Med Hyg, 2007;77(3):514–520.
12. Trombley AR, Wachter L, Garrison J, Buckley-Beason VA, Jahrling J, Hensley LE, et al. Comprehensive panel of Real-time TaqManTM polymerase chain reaction assays for detection and absolute quantification of filoviruses, arenaviruses and New World hantaviruses. Am J Trop Med Hyg, 2010;82(5):954–960.
13. Panning M, Laue T, Olschlager S, Eickmann M, Becker S, Raith S, et al. Diagnostic Reverse-Transcription Polymerase Chain Reaction Kit for Filoviruses Based on the Strain Collections of all European Biosafety Level 4 Laboratories. J Inf Dis, 2007, 196(Suppl. 2):S199–204.
14. Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, et al. RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg, 2007;101(12):1253–1264.
15. Vieth S, Drosten C, Charrel R, Feldmann H, Günther S. Establishment of conventional and fluorescence resonance energy transfer-based real-time PCR assays for detection of pathogenic New World arenaviruses. J Clin Virol, 2005;32(3):229–235
16. Drosten C, Kummerer BM, Schmitz H, et al. Molecular diagnostics of viral hemorrhagic fevers. Antiviral Res, 2003;57(1–2):61–87.
17. Jabado OJ, Palacios G, Kapoor V, et al. Greene SCPrimer: a rapid comprehensive tool for designing degenerate primers from multiple sequence alignments. Nucleic Acids Res, 2006;34(22):6605–6611.
18. Palacios G, Briese T, Kapoor V et al. MassTag Polymerase Chain Reaction for Differential Diagnosis of Viral Hemorrhagic Fevers. Emerg Infect Dis, 2006;12(4):692–695.
19. Towner JS, Rollin PE, Bausch DG, et al. Rapid diagnosis of Ebola Hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral loads as a predictor of outcome. J Virol, 2004;78(8):4330–4341.
20. Papa A, Zelená H, Barnetová D, Petroušová L. Genetic detection of Dobrava/Belgrade virus in a Czech patient with Haemorrhagic fever with renal syndrome. Clin Microbiol Infect, 2010;16(8):1187–1190.
21. Vacková M, Douda P, Beran J, et al. Serologic detection of hantavirus antibodies. Epidemiol Mikrobiol Imunol, 2002;51(2):74–77.
22. Vapalathi O, Mustonen J, Lundkvist A, et al. Hantavirus in Europe. Lancet Infect Dis, 2003;3 : 653–661.
23. Fajs L, Jakupi X, Ahmeti S, et al. Molecular epidemiology of Crimean-Congo hemorrhagic fever virus in Kosovo. PLoS Negl Trop Dis, 2014;8(1):e2647.
24. Barr DA, Aitken C, Bell DJ, et al. First confirmed case of Crimean-Congo hemorrhagic fever in the UK. Lancet, 2013;282(9902):1458.
25. Frank C, Höhle M, Stark K, Lawrence J. More reason to dread rain on vacation? Dengue fever in 42 German and United Kingdom Madeira tourists during autumm 2012. Euro Surveill, 2013;18(14):20446.
Labels
Hygiene and epidemiology Medical virology Clinical microbiologyArticle was published in
Epidemiology, Microbiology, Immunology

2014 Issue 3
-
All articles in this issue
- Infections caused by human alpha herpes viruses
- Opposite trends in the incidence of gonorrhea and syphilis in the East Bohemian Region – 30 years of surveillance
- The current view of the diagnosis and management of amebiasis in the light of the authors’ own case reports
- Prevalence and characteristics of thermotolerant Campylobacter spp. in the human food chain
- Importance of vaccination against influenza in individuals with cardiovascular disease
- Evaluation of the importance of a ready-made, gentamicin-impregnated spacer in relation to bacteriological findings in patients with periprosthetic joint infections
- Q fever – an occupational disease leading to disability – case report
- Measles re-emerging in the Ústí Region
- Klinická a mikrobiologická charakteristika závažných infekcií spôsobených streptokokmi skupiny A
- Antibiofilm activity of agents for disinfection of skin, mucosa, and wound on microorganisms isolated from patients with catheter-related infections
- Enterohemorrhagic Escherichia coli as the cause of diarrhea in the Czech Republic, 1965–2013
- Resistance monitoring working group: Resistance to erythromycin, ciprofloxacin, and tetracycline in human isolates of Campylobacter spp. in the Czech Republic tested by the EUCAST standard method
- Occurrence and characteristic of Staphylococcus aureus in pork meat
- Identification of Mycobacterium spp. isolates using matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS)
- Occupational tuberculosis in Slovakia and in the Czech Republic
- Avidity of antiphospholipid antibodies – our current knowledge
- Detection panel for identification of twelve hemorrhagic viruses using real-time RT-PCR
- Epidemiology, Microbiology, Immunology
- Journal archive
- Current issue
- About the journal
Most read in this issue
- The current view of the diagnosis and management of amebiasis in the light of the authors’ own case reports
- Infections caused by human alpha herpes viruses
- Enterohemorrhagic Escherichia coli as the cause of diarrhea in the Czech Republic, 1965–2013
- Detection panel for identification of twelve hemorrhagic viruses using real-time RT-PCR
