-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Potential anticancer agent hypericin and its model compound emodin: interaction with DNA
Authors: Jana Staničová 1,2; Valéria Verebová 1; Alena Strejčková 1
Authors place of work: The University of Veterinary Medicine & Pharmacy, Košice, Slovakia 1; First Faculty of Medicine, Charles University, Prague, Czech Republic 2
Published in the journal: Čes. slov. Farm., 2016; 65, 28-31
Category: 44<sup>th</sup> Conference drug synthesis and analysis
*Part 1: Čes. slov. Farm. 2015; 64, 200–225.
*Part 2: Čes. slov. Farm. 2015; 64, 269–304.Introduction
Hypericin (HYP) (Fig. 1a) is a natural photosensitizing pigment occurring in plants of the genus Hypericum. HYP under light illumination displays anti-proliferative and cytotoxic effects on many tumour cell lines1, 2). These properties together with minimal dark toxicity, tumour selectivity and high clearance from the host body make HYP a very promising agent in photodynamic therapy of cancer. Due to its prospective pharmaceutical utilization, this interesting molecule has been the subject of investigation of many scientific research groups3–6).
Fig. 1. Chemical structure of hypericin (a) and emodin (b) 
Our team has been dealing with the interaction of HYP and its derivatives (such as emodin, quinizarin, danthron, alizarin) with important bio macromolecules to better understand a drug’s mechanism of action.
The interaction of the drug with linear calf thymus DNA is the main aim of this contribution. The binding constant and interaction mode have been determined by spectrophotometric methods. The model compound of HYP, emodin (E) (Fig. 1b), was considered in our study for its structural similarity with the HYP molecule. In addition, emodin, a plant derived anthraquinone, was found to be a photosensitizer which possess anti-tumour, anti-bacterial, anti-viral, anti-inflammatory, and myorelaxing activities7, 8). Detailed knowledge of the interaction of HYP and E with the DNA at the molecular level is extremely important in determination of their distribution and therapeutic effect in the body.
Experimental methods
Hypericin was purchased from KRD (Slovakia), emodin and high polymerized calf thymus DNA were obtained from Sigma-Aldrich (Germany). Stock solutions of HYP (10–4 mol/l) and E (10–3 mol/l) were prepared in DMSO, which was acquired from Sigma-Aldrich (Germany). Calf thymus DNA (2 · 10–3 mol/l) was dissolved in TE buffer (10 mmol/l Tris and 1 mmol/l EDTA), pH = 7.4 by stirring at 4 °C.
Binding constants were determined by fluorescence (fluorimeter RF – 5301 PC Shimadzu) and absorption spectrophotometry (an absorption spectrophotometer UV-VIS 2401 PC Shimadzu) using equilibrium receptor - ligand binding analysis by Copeland9).
We used an influence of the drug on thermal denaturation of DNA to determine the mode of its interaction with the macromolecule. For this purpose, an absorption spectrophotometer Specord UV-VIS Analytik Jena connected with Peltier module was used. This experimental arrangement enables continuous increasing of temperature with step 1 °C per minute. Temperature range of measurement was 20–90 °C. Absorption curves of samples were recorded every 2 minutes. The samples were filtrated on MillexGV (0.22 ∝m) before measurement. Melting curves were fitted by Van’t Hoff equation:
where A absorbance, Amin minimal measured absorbance, Amax maximal measured absorbance, ΔH enthalpy of transition, R gas constant, T temperature, Tm melting temperature.
All spectra and figures were evaluated in Origin 6.0 and 8.0 and Grafit programs.
Results and discussion
Binding constants express the strength of interaction between macromolecules and ligands9). We used fluorescence spectroscopy for a measurement of dependence of HYP (10–6 mol/l) fluorescence intensity on DNA concentration (10–8 – 3 · 10–5 mol/l) to determine binding constants of the complexes HYP/DNA. Figure 2a shows experimental data fitted by Langmuir isotherm, which gives the equilibrium dissociation constant. Semi-logarithmic graphical representation of equilibrium binding analysis was also used in our calculation to find a more exact binding constant value (Fig. 2b). Considering both graphical approaches, the binding constant of the HYP/DNA complex has been appointed as 4.0 · 104 l/mol. Comparing with other drugs which are typical DNA intercalators such as actinomycin D (binding constant Kb = 1.9 · 106 l/mol)10) or mitoxantron (Kb = 3.9 · 105 l/mol)11) our obtained value does not point to an intercalation mode of the DNA – HYP interaction. In addition, the HYP derivatives danthron and quinizarin interacting with DNA by electrostatic interactions12) have similar binding constant as HYP13). Moreover, another model compound of HYP – emodin has been examined by fluorescence and absorption spectroscopy (see below) and the value of its measured binding constant and thermodynamic parameters point to a non-intercalative mode of interaction with DNA.
Fig. 2. Langmuir isotherm of HYP/DNA complex (a). L represents the concentration of DNA and B is normalized HYP fluorescence intensity (excitation wavelength: 560 nm). Concentrations are mentioned in the text. Semi-logarithmic representation of binding curve (b). Final binding constant was taken as an average of two ones obtained from a, and b. 
The most probable binding sites in the DNA structure for HYP molecule were found by Raman spectroscopy14). Vibrational spectra HYP in a complex with polynucleotides show that N7 of purines, preferentially guanine, is included into the interaction. These findings and the value of the binding constant lead us to the supposition of the groove binding mode of interaction.
Model compound of HYP emodin is a molecule which can serve for a better understanding of the binding effect of more complex drugs on macromolecules such as DNA or transport proteins. Fluorescence and absorption spectroscopy were used to measure the binding constant of E/DNA complex, which was 8.1 · 104 l/mol.
Fluorescence quenching of emodin as a result of adding DNA, formation of the non-fluorescence complex E/DNA and the value of the binding constant can lead us to assume that emodin interacts with DNA by a non-intercalative but groove binding mode of interaction.
Absorption analysis has confirmed our results obtained by fluorescence measurements. Hyperchromicity and small bathochromic shift of the absorption maximum (not shown) indicate that E and DNA molecules interact through electrostatic forces or hydrogen bindings15). Bond by electrostatic forces can be excluded since the value of the binding constant indicates that it is the interactions through hydrogen bonds that are associated with binding to the groove.
In addition, we proved our results by denaturation of pure DNA and E/DNA complexes in ratios 1/1, 2/1, 3/1 by absorption spectroscopy connected with a Peltier module. Melting curves that can be seen in Figure 3a show a destabilizing effect of the drug on calf thymus DNA. They were fitted (Fig. 3b) using Van’t Hoff equation to obtain main thermodynamic characteristics (Tm, ΔT, ΔH).
Fig. 3. Normalized melting curves of pure DNA and complexes E/DNA in ratios 1/1, 2/1, 3/1. DNA concentration: 3 · 10<sup>–5</sup> mol/l, emodin concentration: 3 · 10<sup>–5</sup> mol/l – 9 · 10<sup>–5</sup> mol/l (a). Illustration of Van‘t Hoff fit for complex E/DNA in ratio 1/1 (b). 
Melting temperature Tm is shifted from 67.5 °C (pure DNA) to 57 °C for the complex E/DNA in 2/1 concentration ratio (Table 1). Also Van’t Hoff enthalpy ΔH is decreasing from 388 kJ (pure DNA) to 126 kJ (E/DNA = 2/1), which means that lower energy is needed for denaturation of 50% base pairs of DNA in the complex with emodin. The changes in all thermodynamic parameters lead us to a claim that emodin destabilizes the calf thymus DNA molecule and this destabilization is associated with the groove binding interaction mode16). Our findings can be supported by the fact that an intercalative mode of interaction manifests in thermal stabilization of the DNA molecule due to drug binding16).
Tab. 1. Thermodynamic characteristics determined from the melting curves of pure DNA and the complexes E/DNA in concentration ratios 1/1, 2/1, 3/1 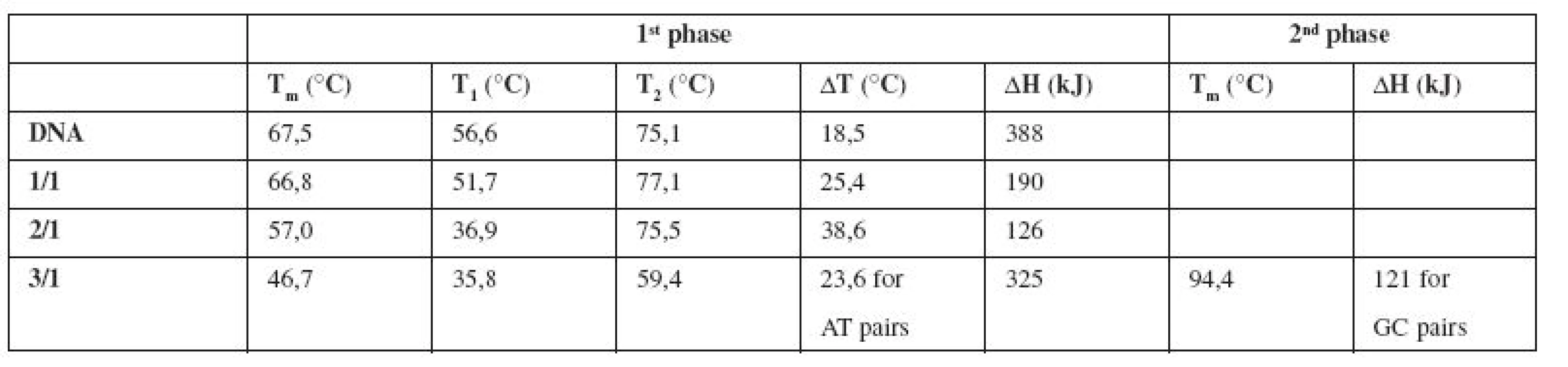
It is generally supposed that small molecules like emodin bind preferentially into the minor groove of DNA, especially to regions which are rich in AT base pairs17).
In our experiments the melting curve of the complex E/DNA in 3/1 (Fig. 3a, Table 1) shows a two-phase character with an expressive destabilization of AT regions in DNA.
Conclusion
The binding constant values of the potential anticancer drugs hypericin and emodin lead us to assume that these molecules do not bind into the DNA structure by an intercalative mode of interaction.
The emodin influence on denaturation of the DNA macromolecule points to the groove binding mode of interaction and the incorporation of the drug takes place probably into the DNA minor groove by hydrophobic or hydrogen interactions. Shahabadi et al.18) showed that the antidiabetic agent metformin with the binding constant of 8.3 · 104 l/mol binds into the minor groove in the DNA structure. These findings can support our results.
Considering the binding characteristics of emodin as a model compound of hypericin, we can suppose that hypericin binds into the DNA major groove. Results obtained by Kočišová14) and the largeness of the hypericin molecule lead us to this assumption.
This work was supported by grant KEGA No. 014 UVLF – 4/2013. Authors are pleased to thank dr. P. Miškovský and dr. R. Varhač for the possibility to use experimental equipment in their laboratories.
Conflict of interest: none.
doc. RNDr. Jana Staničová, PhD.
Department of Chemistry, Biochemistry & Biophysics
Institute of Biophysics, UVMP
Komenského 73,
040 01 Košice,
Slovak Republic
e-mail: jana.stanicova@uvlf.sk
Zdroje
1. Lopez-Bazzocchi I., Hudson J. B., Towers G. H. N. Antiviral activity of the photoactive plant pigment hypericin. Photochem. Photobiol. 1991; 54, 95–98.
2. Andreoni A., Colasanti A., Colasanti P., Mastrocinque M., Riccio P., Roberti G. Laser photosensitization of cells by hypericin. Photochem. Photobiol. 1994; 59, 529–533.
3. Miškovský P. Hypericin – A new antiviral and antitumor photosensitizer: mechanism of action and interaction with biological macromolecules. Current Drug Targets 2002; 3, 55–84.
4. Dougherty T. J., Gomer C. J., Henderson B. W., Jori G., Kessel D., Korbelik M., Peng Q. J. Photodynamic therapy. Natl. Cancer Inst. 1998; 90, 889–905.
5. Kiesslich T., Krammer B., Plaetzer K. Cellular mechanisms and prospective applications of hypericin in photodynamic therapy. Curr. Med. Chem. 2006; 13, 2189–20201.
6. Karioti A., Bilia A. R. Hypericins as potential leads for new therapeutics. Int. J. Mol. Sci. 2010; 11, 562–594.
7. Radovic J., Maksimovic-Ivanic D., Timotijevic G., Popadic S., Ramic Z., Trajkovic V., Miljkovic D., Stosic-Grujicic S., Mijatovic S. Cell-type dependent response of melanoma cells to aloe emodin. Food and chemical toxicology 2012; 50(9), 3181–3189.
8. Poliaček I., Stránsky A., Jakuš J., Baráni H., Tomori Z., Halašová E. Activity of the laryngeal abductor and adductor muscles during cough, expiration and aspiration reflexes in cats. Physiological Research 2003; 52(6), 749–762.
9. Copeland R. A. Enzymes – a practical introduction to structure, mechanism, and data analysis. 2nd edition. New York – Chichester – Weinheim – Brisbane – Singapore – Toronto: John Wiley & Sons INC 2000.
10. Ciolkowski M. L., Fang M. M., Lund M. E. A surface plasmon resonance method for detecting multiple modes of DNA-ligand interactions. J. Pharm. Biomed. Anal. 2000; 22, 6.
11. Agarwal S., Jangir D. K., Mehrotra R. Spectroscopis studies of the effects of anticancer drug mitoxantrone interaction with calf-thymus DNA. J. Photochem. Photobiol. B 2013; 120, 177–182.
12. Gholivand M. B., Kashanian S., Peyman H., Roshanfekr H. DNA-binding study of anthraquinone derivatives using Chemometrics methods. European Journal of Medicinal Chemistry 2011; 46, 2630–2638.
13. Verebová V., Adamčík J., Danko P., Podhradský D., Miškovský P., Staničová J.Anthraquinones quinizarin and danthron unwind negatively supercoiled DNA and lengthen linear DNA. Biochemical and Biophysical Research Communication 2014; 444, 50–55.
14. Kočišová E., Chinsky L., Miškovský P. Sequence specific interaction of the photoactive drug hypericin depends on the structural arrangement and the stability of the structure containing its specific 5 ‘ AG3 ‘ target: A resonance Raman spectroscopy study J. Biomol. Structure Dynamics 1999; 17(1), 51–59.
15. Kumar C. V., Turner R. S., Asuncion E. H. Groove binding of astyrylcyanine dye to the DNA double helix: the salt effect. Journal of Photochemistry and Photobiology A: Chemistry 1993; 74(2–3), 231–238.
16. Bi S., Zhang H., Qiao C., Sun Y., Liu C. Studies of interaction of emodin and DNA in the presence of ethidium bromide by spectroscopic method. Spectrochimica Acta A: Mol. Biomol. Spectrosc. 2008; 69(1), 123–129.
17. Lavery R., Pullman, B. Molecular electrostatic potential on the surface envelopes of macromolecules: B-DNA. International Journal of Quantum Chemistry 1981; 20(1), 259–272.
18. Shahabadi N., Heidari L. Binding studies of the antidiabetic drug, metformin to calf thymus DNA using multispectroscopic methods. Spectrochimica Acta A: Mol. Biomol. Spectrosc. 2012; 97, 406–410.
Štítky
Farmácia Farmakológia
Článok vyšiel v časopiseČeská a slovenská farmacie

2016 Číslo 1-
Všetky články tohto čísla
- Antimicrobial, antiparasitic and anticancer properties of Hibiscus sabdariffa (L.) and its phytochemicals: in vitro and in vivo studies
- Oral films as perspective dosage form
- Influence of the mesh of extrusion dies on the properties of cores intended for the preparation of pellets with controlled glucose release
- Potential anticancer agent hypericin and its model compound emodin: interaction with DNA
- The relationship of lipid imbalance and chronic inflammation mediated by PPAR
- Development of a spectrophotometric method for carmoisine determination in quality control of equipment cleaning
- Thin-layer chromatography application for the standardization of Sophora flowers
- 35. technologické dni a pohľad do histórie tohto podujatia
- Farmakognózia. Biologicky aktívne rastlinné metabolity a ich zdroje.
- Česká a slovenská farmacie
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Oral films as perspective dosage form
- Farmakognózia. Biologicky aktívne rastlinné metabolity a ich zdroje.
- Antimicrobial, antiparasitic and anticancer properties of Hibiscus sabdariffa (L.) and its phytochemicals: in vitro and in vivo studies
- The relationship of lipid imbalance and chronic inflammation mediated by PPAR
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy






