-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
siRNA–Mediated Methylation of Telomeres
Chromosome termini form a specialized type of heterochromatin that is important for chromosome stability. The recent discovery of telomeric RNA transcripts in yeast and vertebrates raised the question of whether RNA–based mechanisms are involved in the formation of telomeric heterochromatin. In this study, we performed detailed analysis of chromatin structure and RNA transcription at chromosome termini in Arabidopsis. Arabidopsis telomeres display features of intermediate heterochromatin that does not extensively spread to subtelomeric regions which encode transcriptionally active genes. We also found telomeric repeat–containing transcripts arising from telomeres and centromeric loci, a portion of which are processed into small interfering RNAs. These telomeric siRNAs contribute to the maintenance of telomeric chromatin through promoting methylation of asymmetric cytosines in telomeric (CCCTAAA)n repeats. The formation of telomeric siRNAs and methylation of telomeres relies on the RNA–dependent DNA methylation pathway. The loss of telomeric DNA methylation in rdr2 mutants is accompanied by only a modest effect on histone heterochromatic marks, indicating that maintenance of telomeric heterochromatin in Arabidopsis is reinforced by several independent mechanisms. In conclusion, this study provides evidence for an siRNA–directed mechanism of chromatin maintenance at telomeres in Arabidopsis.
Published in the journal: siRNA–Mediated Methylation of Telomeres. PLoS Genet 6(6): e32767. doi:10.1371/journal.pgen.1000986
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000986Summary
Chromosome termini form a specialized type of heterochromatin that is important for chromosome stability. The recent discovery of telomeric RNA transcripts in yeast and vertebrates raised the question of whether RNA–based mechanisms are involved in the formation of telomeric heterochromatin. In this study, we performed detailed analysis of chromatin structure and RNA transcription at chromosome termini in Arabidopsis. Arabidopsis telomeres display features of intermediate heterochromatin that does not extensively spread to subtelomeric regions which encode transcriptionally active genes. We also found telomeric repeat–containing transcripts arising from telomeres and centromeric loci, a portion of which are processed into small interfering RNAs. These telomeric siRNAs contribute to the maintenance of telomeric chromatin through promoting methylation of asymmetric cytosines in telomeric (CCCTAAA)n repeats. The formation of telomeric siRNAs and methylation of telomeres relies on the RNA–dependent DNA methylation pathway. The loss of telomeric DNA methylation in rdr2 mutants is accompanied by only a modest effect on histone heterochromatic marks, indicating that maintenance of telomeric heterochromatin in Arabidopsis is reinforced by several independent mechanisms. In conclusion, this study provides evidence for an siRNA–directed mechanism of chromatin maintenance at telomeres in Arabidopsis.
Introduction
Telomeres safeguard the stability of eukaryotic chromosomes by protecting natural chromosome ends from triggering DNA damage responses. Chromosome termini consist of telomeric and subtelomeric repeats that are bound by a specific set of telomere binding proteins as well as nucleosomes that exhibit features of pericentric heterochromatin [1]. These regions are usually devoid of functional genes, and transgenes integrated in the vicinity of telomeres are subjected to transcriptional silencing, a phenomenon known as telomere position effect [2]. Studies in mammals indicate that telomeric heterochromatin plays an important function in chromosome end protection and telomere length regulation. Inactivation of the SIRT6 histone deacetylase in human cells causes hyperacetylation of telomeric histone H3, telomere dysfunction and premature cell senescence [3]. Deficiency in histone methyltransferases or the retinoblastoma tumor suppressor leads to disruption of telomeric heterochromatin and aberrant telomere elongation in mouse cells [4]–[6]. Another important hallmark of heterochromatin in mammals is DNA methylation. Although vertebrate telomeric DNA does not appear to be methylated due to the lack of canonical CG sites, subtelomeric repeats are heavily methylated [7]. Interestingly, inactivation of DNA methyltransferases in mouse cells decreases 5-methylcytosine at subtelomeres and leads to increased telomeric recombination, without a concomitant change in histone modifications [7]. These data indicate a functional interaction between subtelomeric and telomeric chromatin.
Heterochromatin was thought to be transcriptionally inactive, but this view has been challenged by discoveries of numerous non-coding (nc) transcripts derived from heterochromatic loci. Some of these transcripts directly contribute to the assembly of heterochromatin at defined chromosomal domains and their biogenesis is vital for processes such as X chromosome inactivation, genomic imprinting, transposon silencing and centromere function [8]. Thus, it is not surprising that although telomeres possess marks of repressive heterochromatin, they are not transcriptionally silent. Recent studies revealed the presence of telomeric repeat-containing RNAs (TERRA) that are transcribed from subtelomeric regions in yeast and vertebrates [9]–[11]. TERRA are removed from telomeres either through Rat1p-dependent degradation in budding yeast or through non-sense mediated RNA decay (NMD) in human; deficiencies in these RNA processing pathways have dramatic effects on telomere maintenance [9], [10]. Hypomethylation of subtelomeric regions in mammalian cells lacking DNA methyltransferases leads to the overproduction of TERRA [11], [12]. This suggests that the epigenetic status of subtelomeres and telomeres influences TERRA expression.
The discovery of TERRA raised the question of whether ncRNAs contribute to the establishment of telomeric heterochromatin. This hypothesis gained support in a recent study in which downregulation of TERRA by exogenous short interfering RNAs (siRNAs) in human cell lines led to depletion of histone heterochromatic modification from telomeres [13]. In many organisms, RNA-mediated chromatin silencing relies on small RNA molecules that guide effector complexes to target sites [8], [14]. However, involvement of small RNAs in chromatin formation at canonical telomeres has not been shown yet. In this study, we investigate chromatin organization and transcription at chromosome ends in the model plant Arabidopsis thaliana. We detect the presence of transcripts containing telomeric repeats and show that some of these transcripts are processed into ∼24 nt siRNAs. These transcripts are produced from telomeres as well as from intrachromosomal telomeric loci that are mainly located at centromeres. The 24 nt siRNAs are generated through the RNA-dependent DNA methylation (RdDM) pathway, which is a plant-specific mechanism that utilizes siRNAs to guide DNA methyltransferases to asymmetric cytosines (CNN) [15], [16]. We demonstrate that RdDM is responsible for methylation of telomeric DNA that contains cytosines exclusively in asymmetric sequence contexts and hence for reinforcement of heterochromatic marks at telomeres.
Results
Chromatin organization at Arabidopsis chromosome termini
Gene organization at chromosome ends in Arabidopsis appears to be unique. In contrast to the majority of organisms with known telomere/subtelomere sequences, 8 of the 10 Arabidopsis subtelomeres have no repetitive DNA, and predicted genes are annotated in the immediate vicinity of telomeres [17] (Figure 1A). We experimentally confirmed that sequences annotated as chromosome ends are indeed associated with telomeres for 7 chromosome arms with the exception of the right arm of chromosome 3 [18]. The two remaining chromosome termini contain clusters of ribosomal RNA genes (NORs) [19]. We performed reverse transcription (RT) PCR analysis to verify that all the predicted terminal genes are expressed and that they do not represent pseudogenes (Figure 1B). The genes showed distinct tissue-specific expression patterns and the size of the RT-PCR products corresponded to the predicted size of the spliced mRNAs. There was no obvious correlation between the level of expression and promoter distance from telomeres, and even the At2g48160 gene, with a promoter immediately adjacent to telomeric DNA, was robustly expressed. These data indicate that, in contrast to yeast and mammals, Arabidopsis telomeres do not silence genes located in their vicinity.
Fig. 1. Expression of Arabidopsis chromosome-terminal genes. 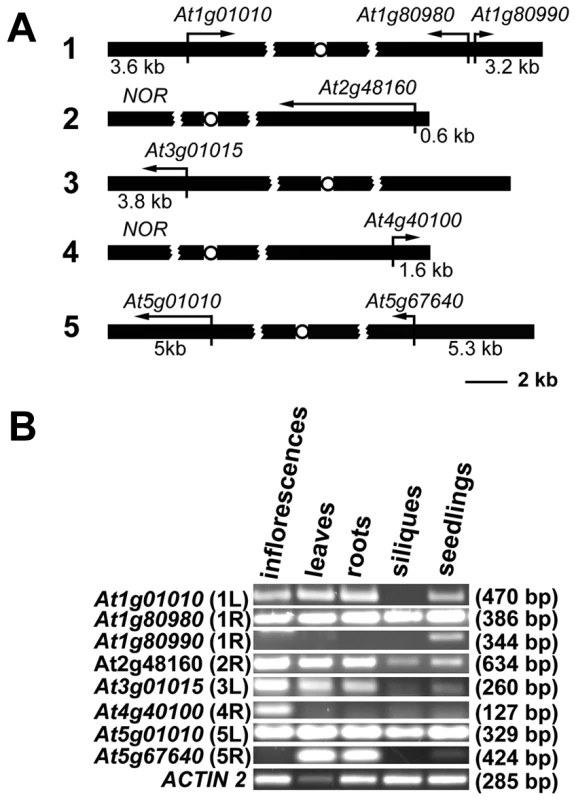
(A) A diagram of gene arrangement at the ends of five Arabidopsis chromosomes. Arrows illustrate the relative size and direction of transcripts of annotated terminal genes. The distance of the predicted ATG codon from the telomere is indicated. (B) Expression of the terminal genes in different tissues of wild-type plants assayed by RT-PCR. The size of the PCR products is indicated in parenthesis. The high transcriptional activity near telomeres raised questions about the chromatin structure of chromosome termini in Arabidopsis. We investigated the distribution of histone modification marks typical for plant euchromatin (tri-methylation of histone H3 at Lys4, H3K4me3) and heterochromatin (di-methylation of H3 at Lys9, H3K9me2; and mono-methylation of H3 at Lys27, H3K27me1) at telomere-associated regions by chromatin immunoprecipitation (ChIP). The ∼600 bp region immediately adjacent to the telomere on the right arm of chromosome 2 (2R) represents the promoter of the At2g48160 gene (Figure 2A) and carries typical euchromatic histone marks (Figure 2B). The H3K4me3 euchromatin mark was also dominant at the promoter of the At1g01010 gene that is located ∼3.5 kb from the telomere on the left arm of chromosome 1 (region 1L-3, Figure 2A and 2B), although we could detect a weak H3K27me1 signal that is usually typical of heterochromatin. Histone heterochromatic marks (H3K9me2 and H3K27me1) became more pronounced at the 1L-2 and 1L-1 regions that are located on the same arm ∼1.5 kb and 1 kb from the telomere, respectively (Figure 2A and 2B). The 1L telomere contains a recent 104 bp insertion of mitochondrial DNA embedded within the centromere-proximal region of telomeric repeats [20] (Figure 2A). Using this insertion to design primers that span the centromere-proximal part of the 1L telomere (1L-0, Figure 2A), we were able to demonstrate that this region also displays heterochromatin marks (Figure 2B). Nevertheless, the 1L-0 region still possessed clearly detectable H3K4me3, which is atypical of classical heterochromatin where the H3K4me3 modification is strongly reduced in comparison to H3K27me1 and H3K9me2. A similar histone-modification pattern was also observed in telomere-adjacent regions of five other chromosome arms (Figure 2B).
Fig. 2. Chromatin structure of Arabidopsis chromosome termini. 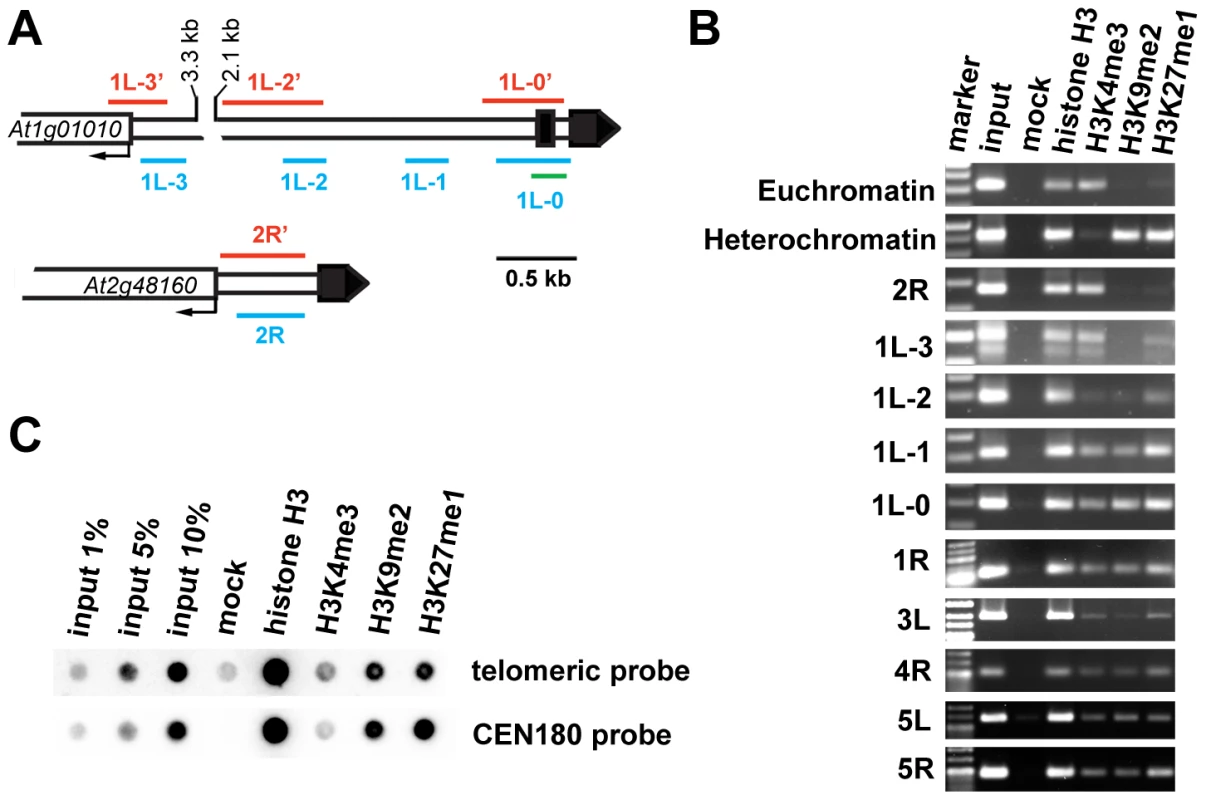
(A) Schematic diagram of the 1L and 2R chromosome ends. Black boxes represent telomeric DNA. The red and blue bars span regions analyzed by bisulfite sequencing and ChIP PCR, respectively. The green bar indicates the region analyzed by ChIP qPCR. (B) ChIP PCR data showing the distribution of methylated histones at unique regions immediately adjacent to telomeres at the indicated chromosome arms. A euchromatic fragment of the MEE51 gene and a heterochromatic gypsy-like retrotransposon (At4g03770) were amplified from the ChIP fractions as a control. (C) Immunoprecipitated DNA analyzed by sequential dot-blot hybridization with a telomeric probe and the centromeric CEN180 probe. To further examine chromatin at telomeres, we analyzed ChIP fractions by dot-blot hybridization with a telomeric probe (Figure 2C). The Arabidopsis genome is enriched for intrachromosomal degenerated telomeric repeats that are mainly localized at centromeres (Figure S1). To specifically assay for chromatin at telomeres, we used stringent hybridization conditions at which the centromere-derived signal is eliminated to less than 2% of the total telomeric signal (Figure S1). We readily detected H3K27me1 and H3K9me2 modifications, and a weaker but still clearly detectable H3K4me3 signal. This hybridization pattern was reminiscent of the results obtained by ChIP analysis of telomere-adjacent regions by PCR (Figure 2B). Thus, our ChIP data show that Arabidopsis telomeres form chromatin that is enriched for H3K9me2 and H3K27me1 heterochromatic marks, but still retains the euchromatic H3K4me3 modification. We found that the heterochromatin marks extend ∼1.5 kb into the subtelomeric region of 1L. A survey of a high-resolution genome-wide map of H3K9me2 distribution indicates that H3K9me2 also spreads up to 1.5 kb from telomeres at chromosome arms 1R, 3L, 4R and 5L [21] (http://epigenomics.mcdb.ucla.edu/H3K9m2/). However, detecting the prominent H3K4me3 signal side by side with the heterochromatic marks (Figure 2B and 2C) strongly indicates that Arabidopsis telomeres exhibit features of intermediate heterochromatin that is characterized by retention of opposing histone H3 methylation marks [22].
Identification of telomeric DNA–containing transcripts and siRNAs
We next asked whether Arabidopsis telomeres are transcribed by assaying for the presence of TERRA by Northern hybridization with a CCCTAAA probe. We readily detected two types of TERRA: heterogeneous transcripts which ranged from high molecular weight strands that migrated at the limits of gel resolution to hundreds of nucleotides, and several distinct bands (Figure 3A). We also detected antisense telomeric transcripts (ARRET) that gave a similar hybridization pattern as the TERRA by the complementary TTTAGGG probe (Figure 3A). These signals disappeared after pretreatment of the samples with RNaseA (Figure 3B and data not shown) demonstrating that they do not represent remnants of DNA in RNA preparations. Expression of TERRA varied between RNA samples extracted from different tissues of Arabidopsis (Figure 3C). Interestingly, remarkable variation in expression was also detected between different Arabidopsis accessions, as the levels of TERRA in seedlings of Zur and Ws ecotypes were almost two orders of magnitude higher than in Col and Ler (Figure 3C). Arabidopsis TERRA and ARRET can originate at telomeres or arise from transcription of degenerated intrachromosomal telomeric sequences localized at centromeric regions (Figure S1). The bulk of centromeric DNA consists of 177–179 bp satellite repeats (CEN180), a subset of which is transcribed [23]. Sequential hybridization of a Northern blot with probes detecting TERRA and CEN180 resulted in an almost identical hybridization pattern, characterized by five distinct bands (Figure 3A). Hybridization of the blots with probes detecting sequences immediately adjacent to telomeres did not produce any detectable signal (data not shown). These results suggest that TERRA and ARRET transcripts detected by Northern analysis mainly arise from centromeric regions that contain remnants of telomeric DNA and not from the transcription of telomeres.
Fig. 3. Identification of TERRA and ARRET transcripts in Arabidopsis. 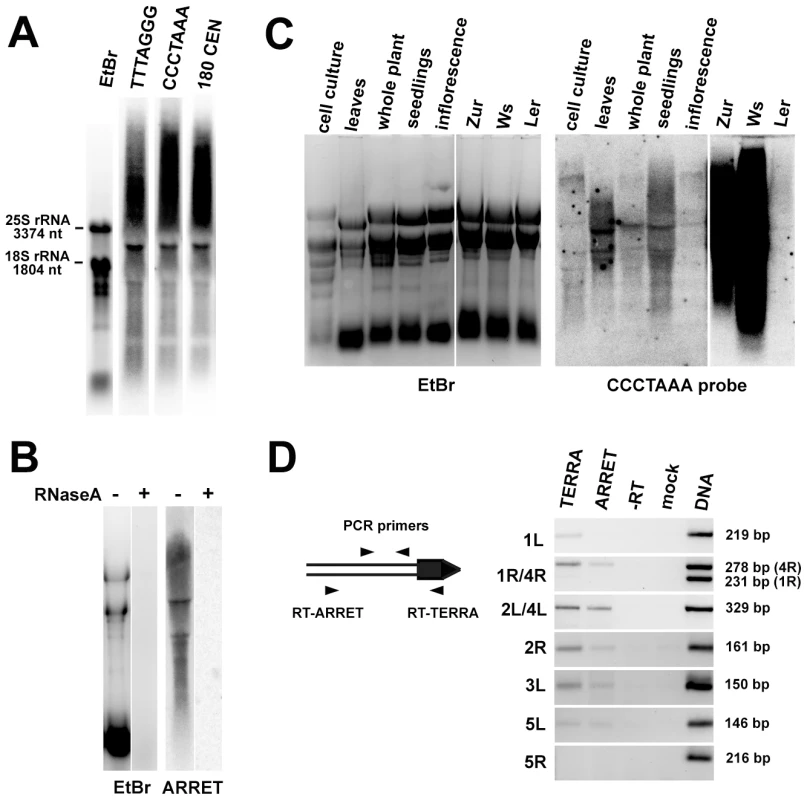
(A) Northern blot analysis of wild-type RNA that was hybridized with a strand-specific TTTAGGG probe, stripped after exposure further sequentially rehybridized with CCCTAAA and the centromeric CEN180 probe. The gel stained with ethidium bromide (EtBr) is shown as a loading control. (B) Sensitivity of ARRET transcripts to RNaseA. (C) Northern blot detection of TERRA in different tissues of wild-type Col plants and in seedlings of the Ws, Zur and Ler ecotypes. The left and right parts of the membrane were exposed for 1 day and 2 h, respectively. (D) Detection of telomere-derived TERRA and ARRET transcripts by RT-PCR. The diagram outlines the strategy used for strand-specific RT-PCR at a hypothetical chromosome end (the telomere is indicated as a black box). The size of the expected PCR product for each telomere is indicated. As chromosome ends 1R and 4R contain a stretch of sequence homology, one set of primers was used to assay for the expression at both telomeres in one reaction. The resulting chromosome-end-specific products can be distinguished by their size. It is currently unknown whether the subtelomere sequence at the NOR-bearing chromosome end represents the 2L or 4L telomere. ARRET transcripts at this arm were not analyzed because they correspond to the nascent 45S rRNA. To examine whether telomeres are transcribed at levels non-detectable by Northern hybridization, we analyzed expression of subtelomeric regions adjacent to telomeric DNA by strand-specific RT-PCR in flowers. We could distinguish expression of TERRA and ARRET by using either telomeric or subtelomeric arm-specific primers for reverse transcription (Figure 3D). We detected expression of both TERRA and ARRET at four out of eight analyzed chromosome arms. We failed to detect any transcription at chromosome arms 1R and 5R. Interestingly, only the TERRA but not ARRET transcripts were detected at 1L. The RT-PCR data demonstrate that at least five Arabidopsis telomeres are indeed transcribed, albeit at a low level. To gain further insights into telomere transcription, we cloned a ∼500 nt promoter of the At2g48160 gene, which is located next to the telomere (Figure 1), in front of a reporter β-glucuronidase (GUS) gene in both sense and antisense orientations. We could detect GUS transcripts in transgenic plants carrying both constructs, although the expression in the antisense direction was much weaker than in the sense orientation (Figure S2). This experiment further supports the idea that telomere adjacent regions can drive transcription into a telomere.
The presence of centromeric and telomeric TERRA and ARRET indicated that telomeric transcripts are able to form partially double stranded (ds) intermediates that could be processed by a Dicer into siRNA. In support of this hypothesis, siRNAs corresponding to both strands of telomeric DNA were detected in wild-type plants (Figure 4A). We estimate the size of the telomeric C-rich strand siRNAs (C-siRNA) to be 24–25 nt, and the size of G-siRNAs to be 23–24 nt (Figure S3). The formation of 24 nt siRNAs in Arabidopsis is mediated by RNA-processing enzymes of the RdDM pathway [24]. This pathway is specific to plants and mediates methylation of cytosine residues in an asymmetric sequence context (CNN). The absence of telomeric 23–25 siRNAs in plants lacking RNA-dependent RNA polymerase 2 (RDR2), Dicer-like 3 (DCL3) or subunits of RNA Polymerase IV (NRPD1 or NRPD2) and their reduction in two other RdDM mutants (drd1 and nrpe1) further demonstrated that telomeric siRNAs belong to the category of 24 nt heterochromatic siRNAs (Figure 4A). These siRNAs are usually derived from heterochromatic loci and form the most abundant fraction of plant small RNAs [25], [26]. They typically associate with Argonaute 4 (AGO4) that is part of the effector complex that, together with Polymerase V, mediates CNN methylation [27], [28]. To determine whether telomeric siRNAs associate with AGO4, we surveyed published datasets containing ∼600,000 Argonaute (AGO1, AGO2, AGO4 and AGO5)-bound small RNAs [29]. We identified a total of 133 small RNAs containing at least 12 nucleotides with a perfect telomeric repeat (Table S1). As expected, the majority of these small RNAs were associated with AGO4 (Figure 4B). Surprisingly, the AGO4-associated telomeric siRNAs were almost exclusively G-siRNAs and only a few C-siRNAs containing no more than 14 nt of the CCCTAAA repeat sequence were found in the dataset (Figure 4C). Since the levels of total G - and C-siRNAs are similar (Figure 4A), this bias may be caused by a selective incorporation of the G-siRNAs into the AGO4 complex.
Fig. 4. Detection of telomeric siRNAs. 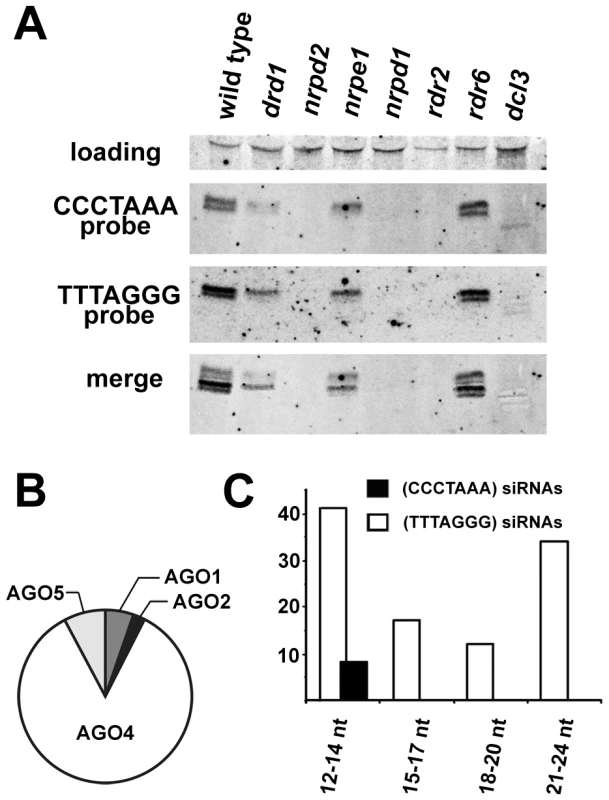
(A) Northern analysis of small RNAs from wild-type and the indicated RdDM mutants. The membrane was hybridized with a CCCTAAA probe, stripped and rehybridized with the TTTAGGG probe. Electronically merged autoradiograms show faster migration of the C-siRNA that is due to a sequence bias towards pyrimidines. The loading control represents a large RNA that hybridizes to the TTTAGGG probe. (B) Distribution of siRNAs containing at least 12 nt of telomeric sequence in different Argonaute complexes. (C) Distribution of AGO4-associated C- and G-siRNAs according to the extent of homology to telomeric sequence. The total number of siRNAs is indicated on the y-axis. As TERRA transcripts are produced from telomeres as well as from centromere-located telomeric DNA, the siRNAs may be of either telomeric or centromeric origin. To determine whether telomere-derived transcripts are processed into siRNAs, we aligned Argonaute-associated siRNAs with telomere-adjacent sequences. We found abundant siRNAs corresponding to both strands of subtelomeric DNA at chromosome arms 1L, 1R, 3L, 4R and 5L (Figure 5, Table S2). Since these regions are formed by unique sequences, the origin of the siRNAs can be unambiguously traced to these loci. Interestingly, AGO4-associated siRNAs were particularly enriched at the chromosome ends that also exhibited expression of TERRA and ARRET (1L, 3L, 4R, 5L; Figure 5). These data strongly argue that telomeric TERRA and/or ARRET are processed into siRNAs.
Fig. 5. Distribution of Argonaute-associated siRNAs in telomere-adjacent regions. 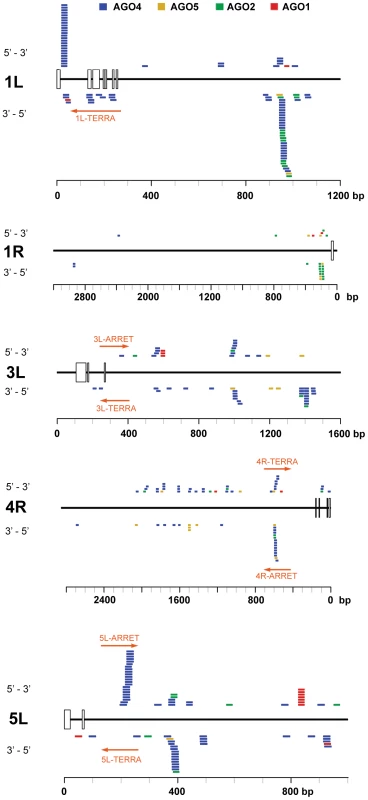
The diagrams represent subtelomeric regions at the indicated chromosome arms. The rulers indicate the distance from a continuous array of telomeric repeats, open boxes mark positions of telomeric repeats intermingled within subtelomeric sequences. Only chromosome arms containing siRNAs detected within 5 kb from telomeres are shown. The position and orientation of AGO-associated siRNAs is indicated by colored bars. Only siRNAs that aligned to unique locations are included (Table S2). The TERRA and ARRET transcripts detected by strand-specific RT-PCR are depicted by a red arrow. The RdDM pathway mediates methylation of telomeric DNA
Plants can methylate cytosines in all sequence contexts, and DNA methylation at asymmetric positions relies largely on 24 nt siRNAs and on the RdDM pathway. The presence of telomeric siRNAs prompted us to ask whether telomeric DNA, which contains cytosines exclusively in the CNN context, can be methylated. We took advantage of the unique insertion in the 1L telomere that allowed us to design primers spanning 13 CCCTAAA repeats located in the centromere-proximal part of the 1L telomere (region 1L-0'; Figure 2A). Bisulfite sequencing of the 1L-0' region in wild-type plants revealed that over 40% of cytosines in these telomeric repeats are methylated (Figure 6). In contrast, the 1L and 2R subtelomeric regions are devoid of DNA methylation (Figure S4). The telomeric methylation in 1L-0' is non-randomly distributed, with preferential enrichment at the third cytosine in the CCCTAAA sequence (Figure 6A and 6B). A similar observation was recently made through whole genome bisulfite sequencing that also revealed methylation of telomeric repeats, albeit at a lower total frequency than reported here [30]. The level of 5-methylcytosine in all sequence contexts was dramatically reduced in rdr2 mutants, arguing that methylation of the 1L-0' region primarily depends on the RdDM mechanism (Figure 6A and 6C).
Fig. 6. Methylation of telomeric DNA in wild-type and RdDM-deficient plants. 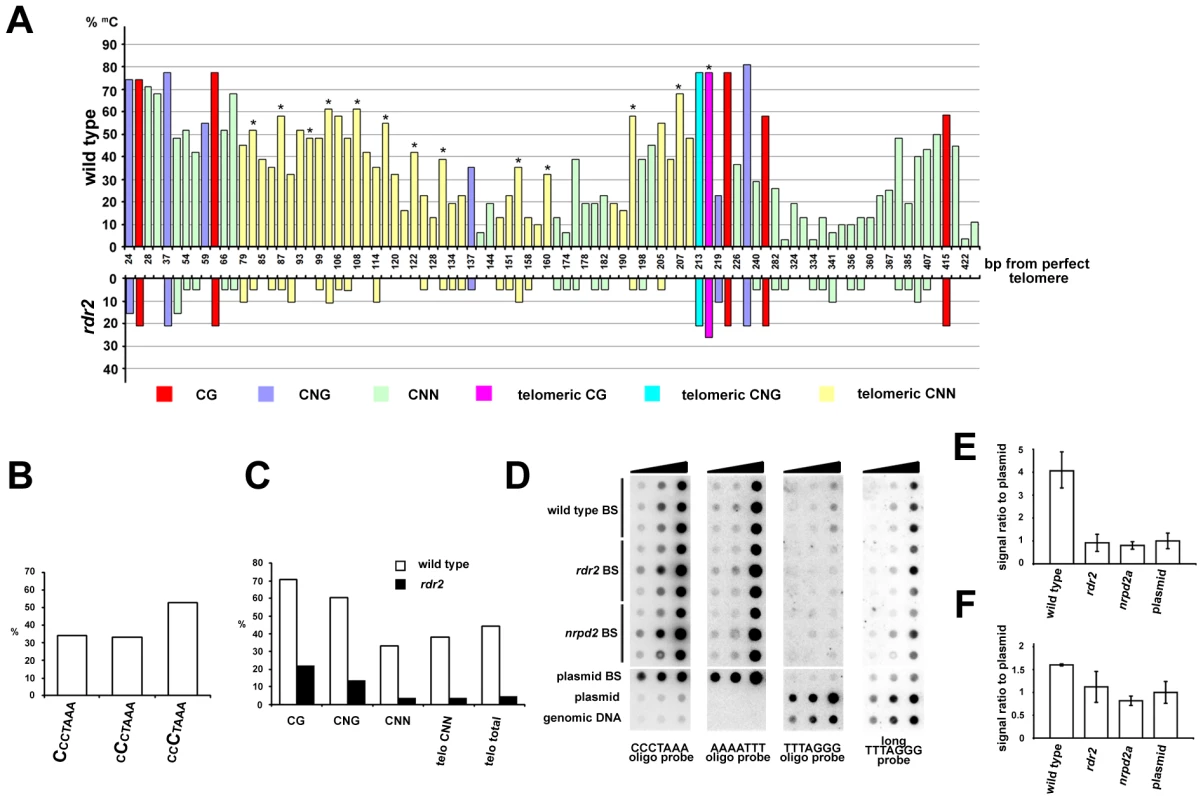
(A) The chart shows the frequency and distribution of cytosine methylation in the 1L-0' region. In total, 30 clones from five independently treated wild-type samples and 19 clones from three independent rdr2 samples were analyzed. Asterisks indicate the third cytosine in the CCCTAAA sequence. (B) The proportion of methylated cytosines in the 1L-0' region of wild-type plants depending on the position within the telomeric repeat as determined by bisulfite sequencing. (C) Frequency of cytosine methylation in the whole 1L-0' region according to sequence context in wild-type and rdr2 plants. (D) Cytosine methylation in bulk telomeric DNA assayed by dot-blot hybridization. Bisulfite-treated (BS) genomic DNA was spotted onto a membrane (∼7, 33 and 200 ng from each sample) and sequentially hybridized with AAAATTT, TTTAGGG and CCCTAAA probes. Untreated wild-type genomic DNA, and untreated and bisulfite-treated (BS) plasmids carrying ∼750 nt of Arabidopsis telomeric DNA were used as controls. (E,F) Quantification of signals obtained with oligo (E) and long (F) TTTAGGG probes. The signal intensity of non-converted DNA (obtained with the TTTAGGG probe) was normalized to the amount of telomeric DNA determined from hybridization with the CCCTAAA probe. The signal from BS-treated plasmid served to determine background hybridization of the probes to fully converted non-methylated telomeric DNA. Error bars represent standard deviation (N = 3). We next examined whether cytosine methylation and its dependence on the RdDM pathway is a general feature of telomeric DNA. We sequentially hybridized bisulfite-treated total genomic DNA to oligonucleotide probes that first detected fully converted telomeric DNA (probe AAAATTT), then unconverted, and thus completely methylated DNA (probe TTTAGGG), and finally the complementary cytosine-free strand (probe CCCTAAA) as a control for loading (Figure 6D). A strong hybridization AAATTTT signal suggested that the bulk of telomeric DNA is only weakly methylated. Nevertheless, a portion of wild-type DNA was resistant to bisulfite conversion as hybridization with the TTTAGGG oligo probe showed a signal that was ∼4-fold higher than a background signal from a corresponding amount of non-methylated bisulfite-converted telomeric DNA cloned in a plasmid (Figure 6D and 6E). These data further indicate the presence of some heavily methylated CCCTAAA sequences in wild-type plants. Importantly, this CCCTAAA signal was reduced to a background level in rdr2 and nrpd2a mutants (Figure 6D and 6E). To further investigate whether methylation occurs at telomeres, we performed high-stringency hybridization of the bisulfite-converted samples with a long telomeric TTTAGGG probe (Figure 6C). Under these conditions, converted plasmid-cloned telomeric DNA produces a high background hybridization signal that is likely caused by sufficiently stable interactions between longer fragments of the (TTTTAAA)n converted telomeric DNA and the (TTTAGGG)n probe. Nevertheless, wild-type DNA samples still produced a signal that was significantly higher than the background hybridization (Figure 6F). These data, together with the bisulfite sequencing of the 1L-0' telomeric region, strongly argue that DNA methylation is a general characteristic of Arabidopsis telomeres and that its maintenance requires the RdDM pathway.
Loss of DNA methylation is often accompanied by chromatin remodeling. However, the decrease in telomeric DNA methylation did not result in a significant loss of heterochromatic histone marks, and both H3K9me2 and H3K27me1 remained enriched at the bulk of telomeric DNA in rdr2 mutants (Figure 7A). However, analysis of histone modifications at the 1L-0' locus by ChIP and quantitative PCR (Figure 7B and 7C) showed a decrease in H3K9me2 and H3K27me1 (Figure 7C) in rdr2 mutants. These data indicate that although the RdDM-dependent mechanism is not solely responsible for heterochromatin formation at telomeres, it contributes to its maintenance by mediating methylation of telomeric DNA, thereby reinforcing heterochromatic histone modifications.
Fig. 7. Telomeric heterochromatin in RdDM mutants. 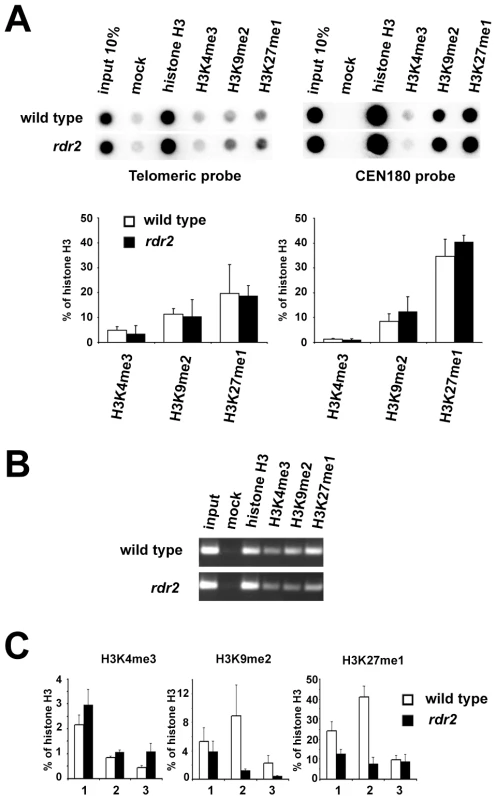
(A) Representative ChIP data of wild-type and rdr2 plants. Chromatin was immunoprecipitated with antibodies against histone H3, H3K4me3, H3K9me2 and H3K27me1, blotted onto a membrane and hybridized with the TTTAGGG probe. The same membrane was stripped after exposure and rehybridized with the centromeric CEN180 probe. Data from three independent ChIP experiments were used for quantification. Signals were normalized to mock. (B) Analysis of ChIP fractions from wild-type and rdr2 plants by PCR with primers spanning the 1L-0 locus. (C) qPCR analysis of H3K4me3, H3K9me2 and H3K27me1 at the 1L-0 locus in wild-type and rdr2 plants. Each value represents an average of three qPCR measurements normalized to histone H3 occupancy for each ChIP fraction. The results of three independent pairwise ChIP experiments (1, 2 and 3) are presented. Disruption of telomeric heterochromatin or demethylation of subtelomeric sequences leads to increased telomere elongation and recombination in mouse [7]. Our analysis of telomere length and intrachromatid recombination at chromosome ends did not reveal any differences between RdDM mutants and wild-type plants (Figure S5 and Figure S6). This observation further corroborates our finding that despite reduced DNA methylation, the bulk of telomeric chromatin in rdr2 mutants still retains heterochromatic features.
Discussion
Heterochromatin is a universal characteristic of chromosome termini in a variety of organisms, including yeast, flies and mammals. Subtelomeric regions in these organisms are gene-poor and enriched for middle to highly repetitive sequences that contribute to the formation of a heritably repressed chromatin structure at chromosome termini that shares similarities with pericentromeric heterochromatin [1], [31], [32]. Nevertheless, some aspects of chromatin organization appear to be unique at telomeres as telomeric chromatin in humans and plants display unusually short nucleosomal spacing (∼160 nt) in comparison with the ∼180 nt periodicity at the bulk of chromatin [33]–[35].
In contrast to many other organisms, telomeres in Arabidopsis are directly adjacent to transcriptionally active genes. This situation is more similar to silenced transposons inserted in gene-rich regions than to pericentromeric heterochromatin. This is also reflected in the organization of telomeric chromatin that exhibits features of intermediate heterochromatin that is characterized by the presence of both active and repressive histone H3 marks. Such chromatin was described to be associated with some Arabidopsis transposons and transgenic loci [22], [36]. Chromatin analysis of the 1L subtelomere demonstrates that repressive histone H3 modifications are most pronounced immediately next to telomeres and that their presence gradually recedes with growing distance from telomeres. Data on whole-genome distribution of H3K9me2 indicate that this also holds true for telomere-associated regions of several other chromosome arms [21]. These data infer that repressive histone marks are primarily established at telomeres and spread only a limited distance within adjacent subtelomeric sequences. The existence of such relatively small clusters of repressive chromatin (2–5 kb) next to otherwise large gene-rich regions suggests a functional importance for the heterochromatization of telomeres in Arabidopsis. It further suggests the existence of mechanisms that specifically maintain repressive histone modifications at telomeres. Assembly of heterochromatin at chromosome ends in budding yeast is partially dependent on tethering Sir proteins to telomeres via the Rap1 telomere-binding protein [31]. Human SIRT6 histone deacetylase preferentially associates with telomeres, although how it is recruited to chromosome termini is not known [3]. A recent study in mice overexpressing TRF2 indicates that, similar to the situation in yeast, heterochromatin formation at telomeres in mammals may also involve telomere-binding proteins [37].
The discovery of TERRA provides another attractive model that involves targeting the chromatin remodeling machinery to chromosome termini through ncRNA [38], [39]. This suggestion was recently corroborated by the finding that downregulation of TERRA by RNAi in human cells causes a decrease in histone H3K9 methylation [13]. It was proposed that TERRA facilitates heterochromatin formation by stabilizing interactions between heterochromatin factors and telomeric DNA. In this study, we demonstrate expression of telomeric transcripts in Arabidopsis and describe a mechanism by which telomeric repeats-containing RNAs affect telomeric chromatin through siRNA.
In contrast to the situation in mammals, where only UUAGGG telomeric transcripts were detected [10], [11], both telomeric strands appear to be transcribed from some telomeres in Arabidopsis. This indicates that canonical telomeric DNA may, under certain circumstances, act as a promoter and initiate transcription. Two lines of observations further corroborate the link between transcription and telomeric DNA in Arabidopsis. Firstly, short stretches of a telomeric sequence were found in numerous Arabidopsis promoters and it has been shown that these interstitial telomere motifs are required for transcription [40]. Secondly, several transcription factors have been identified in Arabidopsis that specifically bind to telomeric DNA in electromobility shift assays (reviewed in [41]). Thus, it is possible that some of these transcription factors localize to telomeres and promote their expression.
In addition to transcripts that originated at telomeres, we detected TERRA and ARRET that are apparently generated by transcription of centromere-associated telomeric loci. We cannot currently determine the exact identity of telomere - or centromere-derived TERRA/ARRET that is processed by DCL3 and degraded to telomeric siRNAs. The requirement of RDR2 for siRNA formation indicates that the predicted dsRNA intermediate is not a simple annealing product of complementary TERRA and ARRET, but is dependent on additional RNA-dependent RNA synthesis. Thus, even relatively low level transcripts can yield significant amounts of siRNA. In fact, direct detection of precursor transcripts in the RdDM pathway has been so far reported only in a special transgene system [42]. In plants, heterochromatic siRNAs serve to guide DNA methylases to specific asymmetric CNN positions in a mechanism that relies on AGO4 [28]. Interestingly, AGO4 appears to retain telomeric G-siRNAs, and not the complementary C-siRNAs, although these data should still be verified by Northern analysis of AGO4 co-immunoprecipitated siRNAs. It is unknown whether the bias towards G-siRNAs is of biological significance, but it is interesting that the AGO4 complex appears to specifically retain siRNAs complementary to the telomeric strand to be methylated.
Our data, showing methylation of bulk telomeric DNA as well as heavy methylation of the centromere-proximal region of the 1L telomere, together with data from whole genome bisulfite sequencing [30], argue that telomeric heterochromatin in Arabidopsis is not only defined by histone modifications, but also by DNA methylation. Although mammalian telomeres lack CG sites, and are, thus, believed to be unmethylated, at least two proteins linked to DNA methylation (SMCHD1, MBD3) have been found in purified fractions of human telomeric chromatin [43]. Additionally, the recent discovery of CNN and CNG methylation in human embryonic stem cells warrants the re-examination of DNA methylation at human telomeres [44].
We demonstrate that the maintenance of telomeric DNA methylation depends, to a large extent, on heterochromatic siRNA and the RdDM machinery. Intriguingly, loss of telomeric DNA methylation only has a slight effect on histone methylation at bulk telomeres, indicating that assembly of Arabidopsis telomeric heterochromatin relies on several reinforcing mechanisms that recruit histone methyltransferases such as SUVH4 to telomeres [45]. Loss of DNA methylation has a more profound effect on histone methylation at the centromere-proximal part of the 1L telomere. This indicates that RdDM may play a role in maintaining heterochromatin at the boundary between telomeres and adjacent euchromatic genes. The involvement of siRNA in modulation of telomeric heterochromatin may not be restricted to plants. Our data in Arabidopsis are reminiscent of the situation in fission yeast where heterochromatin in subtelomeric regions is established by two independent pathways, one of which relies on the telomere-binding protein Taz1, while the other involves RNA-induced transcriptional silencing (RITS) [46]. However, in contrast to the situation in Arabidopsis where siRNA targets canonical telomeric repeats, RITS in fission yeast is directed at centromere-like sequences that are located ∼15 kb from telomeres. In humans, TERRA has been proposed to act as a scaffold, reinforcing interactions between telomere-binding proteins and heterochromatin factors such as ORC1 and HP 1 [13]. Nevertheless, human TERRA could also promote heterochromatin formation through an siRNA-mediated pathway. This notion is supported by the observation that enrichment of Argonaute-1 at human telomeres is correlated with increased H3K9 methylation and HP1 association [47], and by the discovery of telomere-derived human siRNAs [48].
Materials and Methods
Plant material and growth conditions
Arabidopsis mutants carrying the following alleles were used in this study: dcl3-1 (dcl3), rdr2-1 (rdr2), nrpd1a-4 (nrpd1), nrpd1b-1 (nrpe1), sgs2-1 (rdr6), drd1-1 (drd1) and nrpd2a-1 (nrpd2). Plants were grown in soil under long-day conditions (16 h light/8 h dark) at 22°C.
RNA analyses
Total RNA was extracted using TriReagent solution (Sigma). For Northern blot analysis, 10 µg aliquots were separated on 1.2% formaldehyde agarose gels, blotted onto a nylon membrane and hybridized with [32P] 5′ end-labeled (TTTAGGG)4 (TTTAGGG probe) or (TAAACCC)4 (CCCTAAA probe) oligonucleotides. Oligo hybridizations were carried out at 55°C as previously described [49]. Centromeric transcripts were detected by hybridization with a [32P]-labeled CEN180 repeat unit amplified from Arabidopsis genomic DNA using primers CEN1 and CEN2 (Table S3). For RT-PCR analyses, ∼2 µg of total RNA was reverse transcribed by using oligo dT for gene expression. The (CCCTAAA)3 oligo or subtelomere-specific primers (Table S3) were used for RT of TERRA and ARRET, respectively. The respective cDNAs were amplified by 25–35 cycles of PCR with specific primers (Table S3). Small RNAs were isolated from inflorescences using the mirVana miRNA isolation kit (Ambion), separated on 15% polyacrylamide gels and electroblotted onto a nylon membrane. Telomeric siRNAs were detected by hybridization with either (TTTAGGG)4 or (TAAACCC)4 oligo probes in ULTRAhyb-Oligo hybridization buffer (Ambion) at 42°C. The artificial 25 and 23 nt siRNAs were synthesized by in vitro transcription using T7 RNA polymerase (MBI). The T7-TOP oligonucleotide (10 µM) was annealed to a template oligonucleotide (10 µM) as indicated in Figure S3. In vitro transcription was carried out with 30U of T7 RNA polymerase (MBI) and the annealed oligos (0.5 µM) in 50 µL of 1× Transcription buffer (MBI) supplemented with NTPs (10 mM) and RiboLock RNase inhibitors (MBI) for 60 min at 37°C. 25 µL of the reaction was separated on a 15% polyacrylamide gel, electroblotted onto a nylon membrane and analyzed by Southern hybridization.
Analysis of DNA methylation
Genomic DNA was extracted from 4 week old plants with the DNAeasy Plant Maxi Kit (Qiagen). Bisulfite modification was performed using the EpiTect Bisulphite Kit (Qiagen) according to the manufacturer's instructions. The completeness of the conversion was tested by PCR amplification of a non-methylated genomic region [50]. Modified DNA was used as a template for PCR amplification with the primers indicated in Table S3. The PCR products were cloned into the pCR2.1 TOPO cloning vector (Invitrogen) and sequenced using a BigDye terminator and an ABI310 sequencer (Applied Biosystems). The sequence of the clones was analyzed with the software CyMATE [50]. The efficiency of cytosine conversion in the 1L-0' region in these samples was further controlled by either spiking genomic DNA with a bacterial plasmid containing a region that partially overlaps with 1L-0' or by sequence analysis of other genomic loci that are devoid of 5-methylcytosines. For methylation analysis at bulk telomeric DNA, bisulfite-modified genomic DNA was transferred onto a nylon membrane by vacuum-blotting. As a control, a bisulfite-modified plasmid containing 750 bp of plant non-methylated telomeric DNA was blotted onto the membrane in an amount that roughly corresponded to the total amount of telomeric DNA present in genomic samples (1 ng of the plasmid contained telomeric DNA equivalent to ∼260 ng of genomic DNA). The membrane was hybridized with the [32P] 5′ end-labeled (TTTAAAA)4 oligo (AAAATTT probe) in a standard hybridization buffer [49] at 40°C. The membrane was washed twice for 10 min at 40°C in 2× SSC followed by a 40 min wash in 1× SSC at 40°C. The membrane was exposed to a Kodak Phosphor screen (Biorad) and scanned with Molecular Imager FX (Biorad). The membrane was then stripped and sequentially rehybridized with the TTTAGGG and CCCTAAA oligo probes at 55°C as described [49]. The final rehybridization was performed at 65°C with a strand-specific (TTTAGGG)n probe that was obtained by labeling of a 750 bp fragment of telomeric DNA with α-[32P]-GTP. The signals were quantified using QuantityOne software (Biorad).
Chromatin immunoprecipitation
Chromatin isolation and immunoprecipitation were performed as described [51] using antibodies against histone H3 (Abcam; cat. no. ab1791), H3K9me2 (Abcam; cat. no. ab1220), H3K4me3 (Abcam; cat. no. ab8580) and H3K27me1 (provided by Thomas Jenuwein). The DNA was column-purified from immunoprecipitated chromatin and concentrated in 50 µl of elution buffer. For dot-blot analysis, 40 µl of the DNA was blotted onto a nylon membrane and analyzed by hybridization with a [32P]-labeled 750 bp (TTTAGGG)n probe. For PCR analysis, 1 µl of the eluted DNA was amplified by 30 cycles of PCR with the primers specified in Table S1. Quantitative PCR analysis of the 1L-0 region was performed using the iQ5 Real Time PCR detection system (Biorad) and a 2× SensiMix Plus SyBR Kit (PeqLab).
Analysis of Argonaute-associated siRNAs
The sequences of Argonaute-associated siRNAs were retrieved from the NCBI (accession number GSE10036). The individual AGO datasets were searched for the presence of siRNAs containing a string of at least 12 nucleotides of Arabidopsis telomeric repeats of any possible permutation. Telomeric siRNAs were copied into an Excel table and manually annotated. Subtelomeric siRNAs were identified by attempting to align all Argonaute-associated siRNAs to an ∼15 kb sequence from the ends of each Arabidopsis chromosome using the publicly available program SOAP [52]. The subtelomeric sequences were derived from the sequences of whole chromosomes available in TAIR, and from cloned fragments of telomere-associated sequences deposited in the Gene Bank (AB033278 and AM177017). Perfectly matching siRNA alignments were retained, and plotted using R.
Cytology
Mitotic chromosomes prepared from pistils of wild-type plants were subjected to fluorescence in situ hybridization (FISH) with a Cy3-conjugated (CCCTAAA)2 PNA probe (Metabion) as previously described [53]. Chromosomes were examined using a Zeiss Axioscope fluorescence microscope equipped with a CCD camera.
Telomere analyses
The PETRA assay was carried out with genomic DNA extracted from a fifth generation tert mutant plant [54] according to the published protocol [18]. Terminal restriction fragment analysis was performed as described [49], [55]. Analysis of intrachromatid telomeric recombination was performed by the t-circle amplification assay [56]. DNA extracted from Arabidopsis ku70 [57] mutants was used as a positive control.
Supporting Information
Zdroje
1. BlascoMA
2007 The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8 299 309
2. OttavianiA
GilsonE
MagdinierF
2008 Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie 90 93 107
3. MichishitaE
McCordRA
BerberE
KioiM
Padilla-NashH
2008 SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452 492 496
4. Garcia-CaoM
O'SullivanR
PetersAH
JenuweinT
BlascoMA
2004 Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36 94 99
5. GonzaloS
Garcia-CaoM
FragaMF
SchottaG
PetersAH
2005 Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 7 420 428
6. JonesB
SuH
BhatA
LeiH
BajkoJ
2008 The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4 e1000190 doi:10.1371/journal.pgen.1000190
7. GonzaloS
JacoI
FragaMF
ChenT
LiE
2006 DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8 416 424
8. ZaratieguiM
IrvineDV
MartienssenRA
2007 Noncoding RNAs and gene silencing. Cell 128 763 776
9. LukeB
PanzaA
RedonS
IglesiasN
LiZ
2008 The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32 465 477
10. AzzalinCM
ReichenbachP
KhoriauliL
GiulottoE
LingnerJ
2007 Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318 798 801
11. SchoeftnerS
BlascoMA
2008 Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10 228 236
12. YehezkelS
SegevY
Viegas-PequignotE
SkoreckiK
SeligS
2008 Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet 17 2776 2789
13. DengZ
NorseenJ
WiedmerA
RiethmanH
LiebermanPM
2009 TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35 403 413
14. MoazedD
2009 Small RNAs in transcriptional gene silencing and genome defence. Nature 457 413 420
15. PikaardCS
HaagJR
ReamT
WierzbickiAT
2008 Roles of RNA polymerase IV in gene silencing. Trends Plant Sci 13 390 397
16. MatzkeM
KannoT
DaxingerL
HuettelB
MatzkeAJ
2009 RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol
17. 2000 Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796 815
18. HeacockM
SpanglerE
RihaK
PuizinaJ
ShippenDE
2004 Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. Embo J 23 2304 2313
19. CopenhaverGP
PikaardCS
1996 RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J 9 259 272
20. KuoHF
OlsenKM
RichardsEJ
2006 Natural variation in a subtelomeric region of Arabidopsis: implications for the genomic dynamics of a chromosome end. Genetics 173 401 417
21. BernatavichuteYV
ZhangX
CokusS
PellegriniM
JacobsenSE
2008 Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE 3 e3156 doi:10.1371/journal.pone.0003156
22. HabuY
MathieuO
TariqM
ProbstAV
SmathajittC
2006 Epigenetic regulation of transcription in intermediate heterochromatin. EMBO Rep 7 1279 1284
23. MayBP
LippmanZB
FangY
SpectorDL
MartienssenRA
2005 Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet 1 e79 doi:10.1371/journal.pgen.0010079
24. HuettelB
KannoT
DaxingerL
BucherE
van der WindenJ
2007 RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta 1769 358 374
25. MosherRA
SchwachF
StudholmeD
BaulcombeDC
2008 PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A 105 3145 3150
26. ZhangX
HendersonIR
LuC
GreenPJ
JacobsenSE
2007 Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A 104 4536 4541
27. HeXJ
HsuYF
ZhuS
WierzbickiAT
PontesO
2009 An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4 - and RNA-binding protein. Cell 137 498 508
28. WierzbickiAT
ReamTS
HaagJR
PikaardCS
2009 RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41 630 634
29. MiS
CaiT
HuY
ChenY
HodgesE
2008 Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 116 127
30. CokusSJ
FengS
ZhangX
ChenZ
MerrimanB
2008 Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452 215 219
31. PerrodS
GasserSM
2003 Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell Mol Life Sci 60 2303 2318
32. PrydeFE
GorhamHC
LouisEJ
1997 Chromosome ends: all the same under their caps. Curr Opin Genet Dev 7 822 828
33. TommerupH
DousmanisA
de LangeT
1994 Unusual chromatin in human telomeres. Mol Cell Biol 14 5777 5785
34. FajkusJ
KovarikA
KralovicsR
BezdekM
1995 Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol Gen Genet 247 633 638
35. SykorovaE
FajkusJ
ItoM
FukuiK
2001 Transition between two forms of heterochromatin at plant subtelomeres. Chromosome Res 9 309 323
36. LippmanZ
MayB
YordanC
SingerT
MartienssenR
2003 Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1 e67 doi:10.1371/journal.pbio.0000067
37. BenettiR
SchoeftnerS
MunozP
BlascoMA
2008 Role of TRF2 in the assembly of telomeric chromatin. Cell Cycle 7 3461 3468
38. HorardB
GilsonE
2008 Telomeric RNA enters the game. Nat Cell Biol 10 113 115
39. LukeB
LingnerJ
2009 TERRA: telomeric repeat-containing RNA. Embo J 28 2503 2510
40. TremousaygueD
ManevskiA
BardetC
LescureN
LescureB
1999 Plant interstitial telomere motifs participate in the control of gene expression in root meristems. Plant J 20 553 561
41. ZellingerB
RihaK
2007 Composition of plant telomeres. Biochim Biophys Acta 1769 399 409
42. DaxingerL
KannoT
BucherE
van der WindenJ
NaumannU
2009 A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. Embo J 28 48 57
43. DejardinJ
KingstonRE
2009 Purification of proteins associated with specific genomic Loci. Cell 136 175 186
44. ListerR
PelizzolaM
DowenRH
HawkinsRD
HonG
2009 Human DNA methylomes at base resolution show widespread epigenomic differences. Nature
45. GrafiG
Ben-MeirH
AviviY
MosheM
DahanY
2007 Histone methylation controls telomerase-independent telomere lengthening in cells undergoing dedifferentiation. Dev Biol 306 838 846
46. KanohJ
SadaieM
UranoT
IshikawaF
2005 Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15 1808 1819
47. HoCY
MurnaneJP
YeungAK
NgHK
LoAW
2008 Telomeres acquire distinct heterochromatin characteristics during siRNA-induced RNA interference in mouse cells. Curr Biol 18 183 187
48. CaoF
LiX
HiewS
BradyH
LiuY
2009 Dicer independent small RNAs associate with telomeric heterochromatin. Rna 15 1274 1281
49. RihaK
FajkusJ
SirokyJ
VyskotB
1998 Developmental control of telomere lengths and telomerase activity in plants. Plant Cell 10 1691 1698
50. HetzlJ
FoersterAM
RaidlG
Mittelsten ScheidO
2007 CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 51 526 536
51. LawrenceRJ
EarleyK
PontesO
SilvaM
ChenZJ
2004 A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13 599 609
52. LiR
LiYR
KristiansenK
WangJ
2008 SOAP: short oligonucleotide aligment program. Bioinformatics 24 713
53. AkimchevaS
ZellingerB
RihaK
2008 Genome stability in Arabidopsis cells exhibiting alternative lengthening of telomeres. Cytogenet Genome Res 122 388 395
54. RihaK
McKnightTD
GriffingLR
ShippenDE
2001 Living with genome instability: plant responses to telomere dysfunction. Science 291 1797 1800
55. FitzgeraldMS
RihaK
GaoF
RenS
McKnightTD
1999 Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci U S A 96 14813 14818
56. ZellingerB
AkimchevaS
PuizinaJ
SchiratoM
RihaK
2007 Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell 27 163 169
57. RihaK
WatsonJM
ParkeyJ
ShippenDE
2002 Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. Embo J 21 2819 2826
Štítky
Genetika Reprodukčná medicína
Článek The Met 470 Allele Is Associated with Lower Birth Rates in Fertile Men from a Population IsolateČlánek Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2010 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Translational Selection Is Ubiquitous in Prokaryotes
- Whole-Genome Sequencing of a Single Proband Together with Linkage Analysis Identifies a Mendelian Disease Gene
- Web-Based, Participant-Driven Studies Yield Novel Genetic Associations for Common Traits
- The Met 470 Allele Is Associated with Lower Birth Rates in Fertile Men from a Population Isolate
- Contributions of Status and Allele Expression, But Not Copy Number Variation, to the Control of SIVmac251 Replication in Indian-Origin Rhesus Monkeys
- A Genome-Wide Association Study of Optic Disc Parameters
- Amplification of a Cytochrome P450 Gene Is Associated with Resistance to Neonicotinoid Insecticides in the Aphid
- The Transcription Factor REST Is Lost in Aggressive Breast Cancer
- 14-3-3 Mediates Histone Cross-Talk during Transcription Elongation in
- Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
- Use of Genome-Wide Expression Data to Mine the “Gray Zone” of GWA Studies Leads to Novel Candidate Obesity Genes
- Genome-Wide RNAi Screen Identifies Multiple Regulators of HIF–Dependent Transcription in Hypoxia
- The CYCLIN-A CYCA1;2/TAM Is Required for the Meiosis I to Meiosis II Transition and Cooperates with OSD1 for the Prophase to First Meiotic Division Transition
- Inactivation of hnRNP K by Expanded Intronic AUUCU Repeat Induces Apoptosis Via Translocation of PKCδ to Mitochondria in Spinocerebellar Ataxia 10
- Mice with Alopecia, Osteoporosis, and Systemic Amyloidosis Due to Mutation in , a Gene Coding for Palmitoyl Acyltransferase
- siRNA–Mediated Methylation of Telomeres
- Chromosome 4 Replicates in Two Phases That Correlate with Chromatin State
- Dynamic Switch of Negative Feedback Regulation in Akt–TOR Signaling
- On the Use of Variance per Genotype as a Tool to Identify Quantitative Trait Interaction Effects: A Report from the Women's Genome Health Study
- and Deficiency Cooperate in the Progression of Mouse Prostate Tumourigenesis
- An Integration of Genome-Wide Association Study and Gene Expression Profiling to Prioritize the Discovery of Novel Susceptibility Loci for Osteoporosis-Related Traits
- Consent and Internet-Enabled Human Genomics
- Understanding Adaptation in Large Populations
- Identification of a Functional Genetic Variant at 16q12.1 for Breast Cancer Risk: Results from the Asia Breast Cancer Consortium
- Evidence that Adaptation in Is Not Limited by Mutation at Single Sites
- The IG-DMR and the -DMR at Human Chromosome 14q32.2: Hierarchical Interaction and Distinct Functional Properties as Imprinting Control Centers
- Cushing's Syndrome and Fetal Features Resurgence in Adrenal Cortex–Specific Knockout Mice
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Web-Based, Participant-Driven Studies Yield Novel Genetic Associations for Common Traits
- The IG-DMR and the -DMR at Human Chromosome 14q32.2: Hierarchical Interaction and Distinct Functional Properties as Imprinting Control Centers
- Amplification of a Cytochrome P450 Gene Is Associated with Resistance to Neonicotinoid Insecticides in the Aphid
- Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



