-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
from the Aphid : A Missing Link from Facultative to Obligate Insect Endosymbiont
The genome sequencing of Buchnera aphidicola BCc from the aphid Cinara cedri, which is the smallest known Buchnera genome, revealed that this bacterium had lost its symbiotic role, as it was not able to synthesize tryptophan and riboflavin. Moreover, the biosynthesis of tryptophan is shared with the endosymbiont Serratia symbiotica SCc, which coexists with B. aphidicola in this aphid. The whole-genome sequencing of S. symbiotica SCc reveals an endosymbiont in a stage of genome reduction that is closer to an obligate endosymbiont, such as B. aphidicola from Acyrthosiphon pisum, than to another S. symbiotica, which is a facultative endosymbiont in this aphid, and presents much less gene decay. The comparison between both S. symbiotica enables us to propose an evolutionary scenario of the transition from facultative to obligate endosymbiont. Metabolic inferences of B. aphidicola BCc and S. symbiotica SCc reveal that most of the functions carried out by B. aphidicola in A. pisum are now either conserved in B. aphidicola BCc or taken over by S. symbiotica. In addition, there are several cases of metabolic complementation giving functional stability to the whole consortium and evolutionary preservation of the actors involved.
Published in the journal: from the Aphid : A Missing Link from Facultative to Obligate Insect Endosymbiont. PLoS Genet 7(11): e32767. doi:10.1371/journal.pgen.1002357
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002357Summary
The genome sequencing of Buchnera aphidicola BCc from the aphid Cinara cedri, which is the smallest known Buchnera genome, revealed that this bacterium had lost its symbiotic role, as it was not able to synthesize tryptophan and riboflavin. Moreover, the biosynthesis of tryptophan is shared with the endosymbiont Serratia symbiotica SCc, which coexists with B. aphidicola in this aphid. The whole-genome sequencing of S. symbiotica SCc reveals an endosymbiont in a stage of genome reduction that is closer to an obligate endosymbiont, such as B. aphidicola from Acyrthosiphon pisum, than to another S. symbiotica, which is a facultative endosymbiont in this aphid, and presents much less gene decay. The comparison between both S. symbiotica enables us to propose an evolutionary scenario of the transition from facultative to obligate endosymbiont. Metabolic inferences of B. aphidicola BCc and S. symbiotica SCc reveal that most of the functions carried out by B. aphidicola in A. pisum are now either conserved in B. aphidicola BCc or taken over by S. symbiotica. In addition, there are several cases of metabolic complementation giving functional stability to the whole consortium and evolutionary preservation of the actors involved.
Introduction
Symbiotic associations are widespread in insects, being particularly well studied in aphids [1], [2]. They feed on phloem sap, which has unbalanced nitrogen/carbon content and is deficient in a number of nutrients that the aphids cannot synthesize and that are provided by Buchnera aphidicola, their primary endosymbiont [3], [4]. In addition to B. aphidicola, some aphid populations harbor additional facultative (or secondary symbionts) that are not required for growth or reproduction [5], which are sometimes transmitted horizontally [6]–[8]. Three main facultative symbionts have been found in aphids, i.e Hamiltonella defensa, Regiella insecticola and Serratia symbiotica [9]. Although their presence is not necessary for the maintenance of the aphid-Buchnera association, several studies have demonstrated that they can provide certain benefits to their hosts, such as influencing interactions with the host's natural enemies, or defense against environmental heat stress (revised in [5]). Most of the experimental studies on facultative symbionts in aphids have involved members of the subfamily Aphidinae, mainly in Acyrthosiphon pisum. In addition, the genome of the three above mentioned facultative symbionts from A. pisum have been sequenced [10]–[12]. These sequences have revealed that all three bacteria have lost the ability to synthesize most of the essential amino acids, although they retain active uptake mechanisms to import them. Thus, it seems that while B. aphidicola does not need the help of these facultative symbionts for host survival, they are dependent on B. aphidicola for amino acid provision when they infect A. pisum.
Furthermore, in the cedar aphid Cinara cedri, a member of the Lachninae subfamily, it was found that the co-existing endosymbiont Serratia symbiotica SCc was necessary for the survival of the C. cedri consortium. The genome of B. aphidicola BCc, the obligate endosymbiont of C. cedri with only 416 kb is the smallest B. aphidicola among all Buchnera genomes described, and one of the smallest bacterial genomes sequenced so far. Functional genome analysis revealed that with 362 genes, B. aphidicola BCc is able to support cellular life. However, its symbiotic role has been questioned because, contrary to other Buchnera, it was unable to fulfill some symbiotic functions [13]. Thus, it was postulated that the nutrients that B. aphidicola BCc cannot synthesize could be supplied by S. symbiotica SCc. Moreover, it was reported that this bacterium has characteristics of an obligate symbiont which differ from the other S. symbiotica described so far [7]. Microscopic analysis of C. cedri demonstrated that S. symbiotica SCc is confined in a second type of bacteriocytes whose presence is as abundant and homogeneous as B. aphidicola BCc [14]. In addition, both endosymbionts were found to be involved in tryptophan biosynthesis, supplying this essential amino acid to both their host and themselves [15]. Regarding the situation in the subfamily Lachninae, most members of the subfamily were found to have a massive presence of secondary symbionts, mainly S. symbiotica [7], [8]. Phylogenetic studies of these symbionts in all aphids from different subfamilies whose presence was reported, showed the existence of two clades, A and B, of S. symbiotica hypothetically playing two different roles: clade A is composed of facultative endosymbionts, whereas in clade B they would be obligate endosymbionts. Interestingly, S. symbiotica from A. pisum (herein S. symbiotica SAp) whose genome has recently been sequenced [12] belongs to clade A, whereas in this work we report a S. symbiotica genome belonging to clade B.
In the present study, we have carried out the genome sequencing and metabolic analysis of S. symbiotica SCc, the third partner of the C. cedri consortium. This bacterium has suffered an important genome size reduction to become a co-obligate symbiont. The comparative genomics of S. symbiotica SCc with S. symbiotica SAp and other obligate and facultative symbiotic bacteria, as well as with free-living Serratia relatives, mainly S. proteamaculans and the genetic and metabolic information retrieved from the genome sequence of A. pisum [16] and derived studies [17]–[19], provide an evolutionary scenario of how a symbiotic bacterial consortium is established.
Results
Genome of S. symbiotica SCc strain
General and specific features of the S. symbiotica SCc genome (CP002295) reflect its lifestyle as a host-restricted, mutualistic symbiont that invades host cells. The moderately reduced genome consists of a 1,762.765 bp circular chromosome with average G+C content of 29.22% (Table 1 and Figure S1). This chromosome size represents a 67.7% reduction compared to the free-living bacterium S. proteamaculans (CP000826) and a 36.8% reduction compared to S. symbiotica SAp (AENX00000000) [12]. A total of 711 putative genes have been described, with 672 protein coding genes (CDS), 36 tRNAs, 3 rRNAs and one tmRNA. It is worth mentioning that the ribosomal genes 23S and 5S are located on a chromosomal region separated from the 16S rDNA gene, a situation already detected in other obligate endosymbionts. However, S. symbiotica SAp, H. defensa HAp (CP001277.1) and S. proteamaculans have more than one copy of the ribosomal gene in an operon structure. Also, the number of genes coding for tRNA are closer to B. aphidicola BAp (NC_011833) than to facultative or free-living bacteria. Finally, 58 readily identifiable pseudogenes were present, which is a number closer to the one observed in primary than in secondary symbionts (Table S1).
Tab. 1. Comparison of <i>S. symbiotica</i> SCc genome features to those of obligate (<i>B. aphidicola</i> BCc and <i>B. aphidicola</i> BAp, from <i>C. cedri</i> and <i>A. pisum</i>, respectively) and facultative (<i>S. symbiotica</i> SAp, and <i>H. defensa</i> HAp, both from <i>A. pisum</i>) endosymbionts, and a free-living bacterial genome (<i>S. proteamaculans</i>). 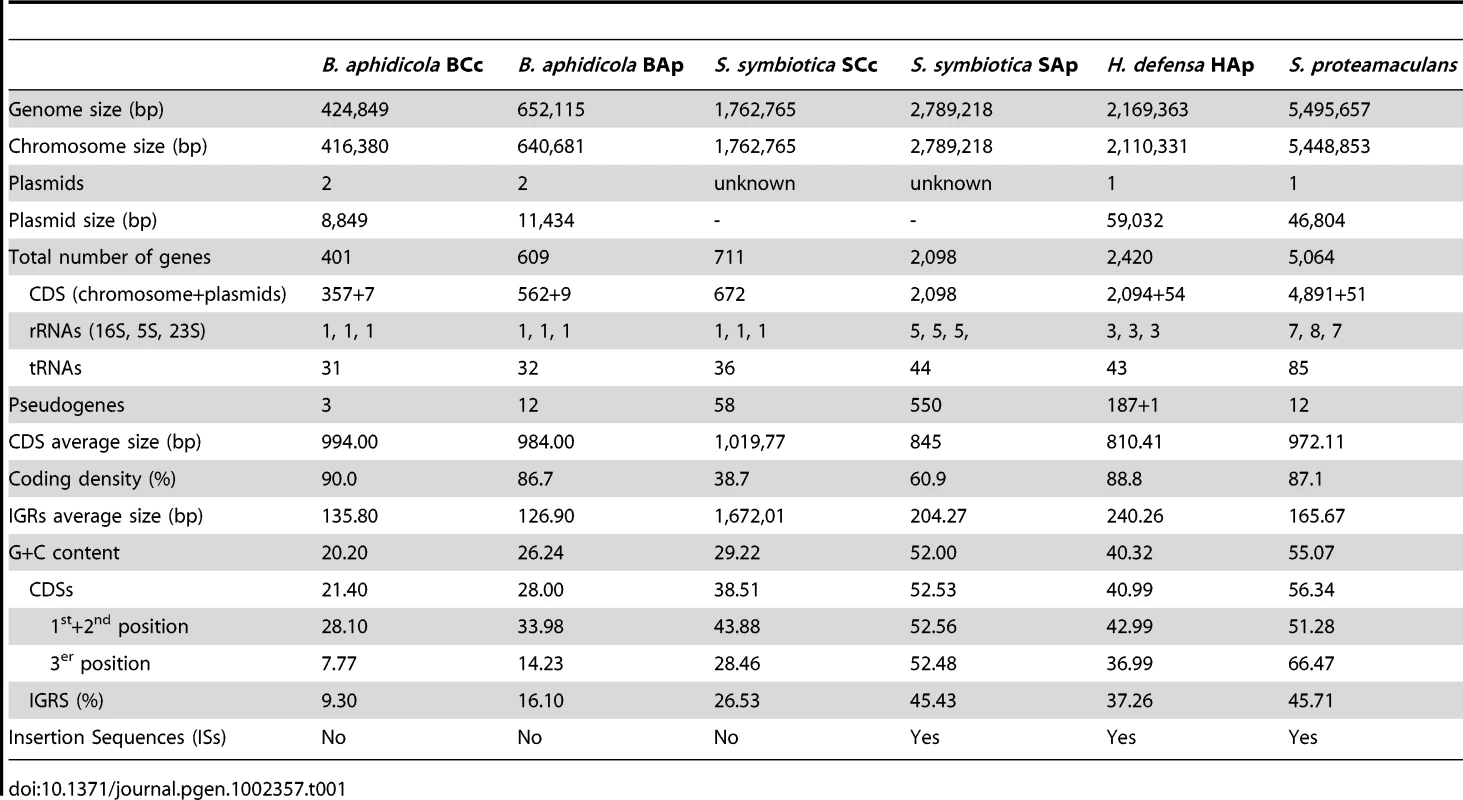
The origin of replication was located between the genes gidA and atpB. The overall coding density is 38.7%, the lowest among insect endosymbionts described so far, including facultative symbionts of aphids, like H. defensa HAp (88.8%), R. insecticola RAp (ACYF00000000) (71.0%) or S. symbiotica SAp (60.9%) [10]–[12]. A very interesting feature of this genome is the average length of the intergenic regions (IGRs) (1,672 bp), which is much higher than in the other selected species (Table 1). A detailed analysis of these IGRs indicates that they do not show any traces of homology to coding regions from other bacteria. Due to the fact that protein-coding regions (CDSs) were found to be more G+C rich than non-coding regions [20], we decided to analyze the GC content distribution of S. symbiotica SCc and compare it with selected bacteria. We found a striking two-peak distribution of the genome GC content in S. symbiotica SCc, instead of the one peak found in any of the other selected organism [21] (Figure S2). To analyze where this two-peak distribution could originate, we took both S. symbiotica and plotted their CDSs and IGRs GC distribution separately. In S. symbiotica SCc, the IGRs mean GC content (27%) was found to fall very far from that of CDSs (38.74%), which contrasts with the case of S. symbiotica SAp where both IGRs and CDSs mean GC content only differed by 7% (Figure S3). In addition, the great number of pseudogenes in S. symbiotica SAp (550) also gave a similar GC mean of 51%. This points towards the last stages of genomic degradation of S. symbiotica SCc IGRs by displaying no evident homology with any known gene and displaying a high A+T content, a common feature arising in many bacterial endosymbionts in advanced stages of genome reduction. Finally, this genome has lost all the insertion sequences (IS) that are characteristic of free-living bacteria and facultative symbionts, as also observed in other bacterial genomes with long-term insect host associations [1], [2].
Functional analysis of the predicted protein-coding genes
The protein genes of S. symbiotica SCc were classified according to COG categories [22] and compared with those of selected symbionts and free-living bacteria (Figure S4). The most relevant result is that S. symbiotica SCc has retained genes devoted to systems for which B. aphidicola BCc (NC_008513) was especially impaired compared with other Buchnera, such as biosynthesis of nucleotides, cofactors, lipid transport and metabolism, and cell envelope biogenesis. The only category in which it is clearly underrepresented is in amino acid metabolism (1.2% and 11.8% in S. symbiotica SCc and B. aphidicola BCc, respectively), which suggests the absolute metabolic dependence of S. symbiotica SCc on B. aphidicola BCc. Accordingly, S. symbiotica SCc possess many amino acid transport systems. Additionally, it has also preserved a wide range of transporters for other metabolites.
Metabolic pathway reconstruction
S. symbiotica SCc has preserved all the steps of glycolysis as well as pentose phosphate pathway. Contrary to S. symbiotica SAp but similar to Buchnera spp, it has lost a functional TCA cycle, preserving only the genes sucC and sucD. These two genes may have been retained to produce succinyl CoA, necessary for lysine biosynthesis. As in other endosymbiont, acetyl-CoA could be used to produce acetate and ATP via the products of the genes ackA and pta, and conserve energy under oxygen-limiting conditions.
For most of the other pathways, one must postulate the involvement of one or even the two other members of the consortium, i.e. B. aphidicola BCc and the aphid (see Figure 1 for a summary of the shared metabolism). This is the case of the purine metabolism, where S. symbiotica SCc can only synthesize AIR from PRPP, but needs an external uptake of IMP to obtain AMP, GMP and XMP (Figure 2). Additionally, S. symbiotica SCc could salvage nitrogen bases from nucleotides or nucleosides that, when in excess, could in turn be transformed and eliminated as uric acid excretion by the aphid metabolism. This role is taken by B. aphidicola BAp in A. pisum [23]. On the other hand, S. symbiotica SCc possesses the complete machinery for pyrimidine biosynthesis. This is in clear contrast with the situation in S. symbiotica SAp where the purine de novo synthesis is complete, but to obtain pyrimidines it requires the nucleoside import, either from the aphid of from B. aphidicola BAp [12], [23].
Fig. 1. Inferred metabolism of amino acids, nucleotides, membrane compounds, cofactors, and vitamins of the C. cedri-consortium. 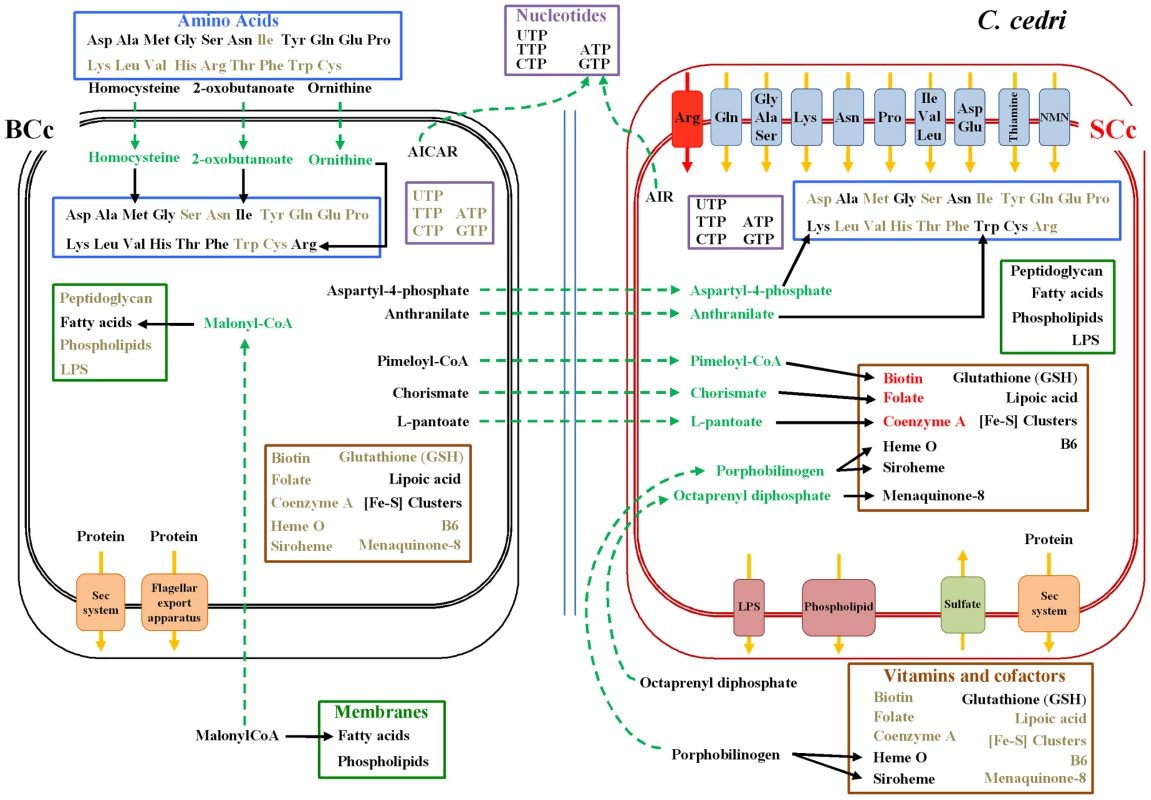
B. aphidicola BCc is represented by three concentric black rectangles and S. symbiotica SCc by three red ones. The two internal rectangles represent internal and external bacterial membranes and the external rectangle represents the vesicle of the eukaryotic cell. The two blue lines represent the membrane in the bacteriocytes. Metabolites are black when synthesized, grey when not synthesized, red when their synthesis is not clear, and green for intermediates exchanged between partners. Black lines indicate intact pathways. Blue, pink and green squares on the membranes represent transporters (red indicating non-functional system). Orange boxes correspond to secretion systems. Fig. 2. Outline of the putative synthesis of nucleotides by the consortium (C. cedri, B. aphidicola BCc, and S. symbiotica SCc). 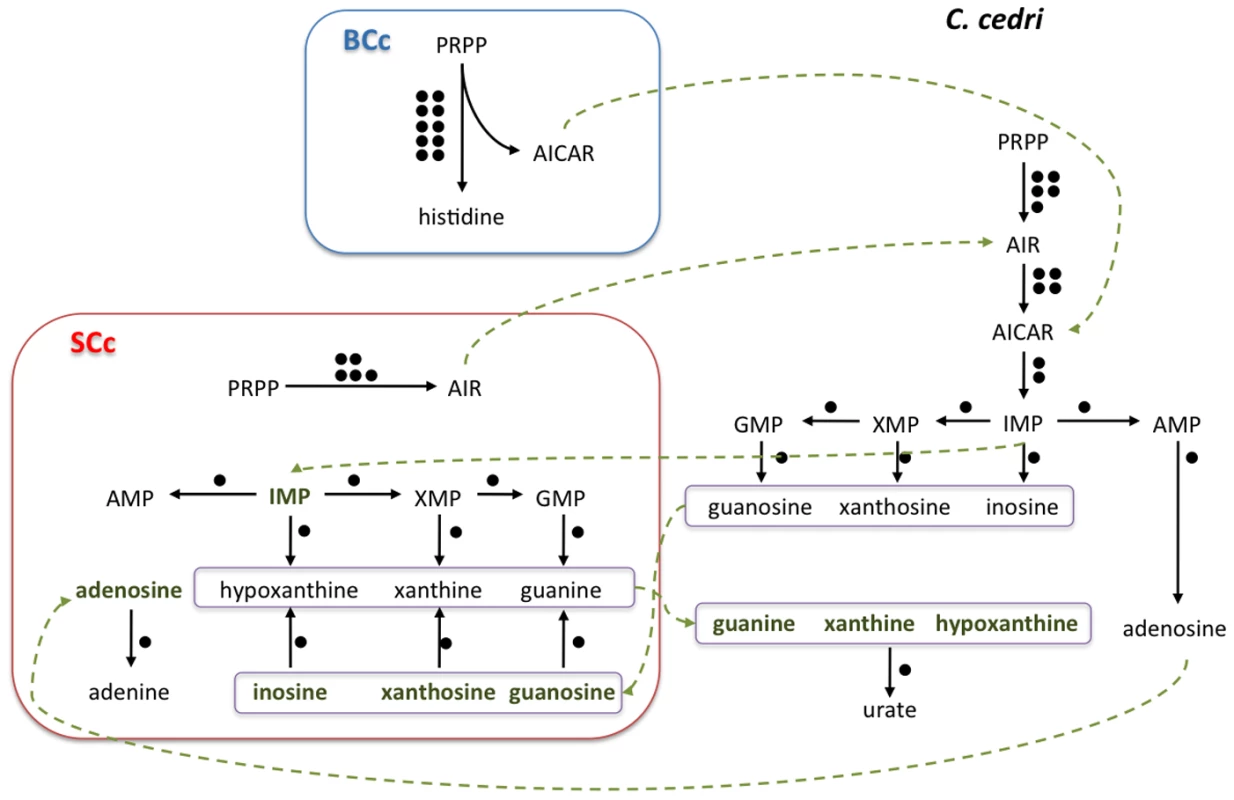
The number of genes involved in each pathway is shown as black circles beside them. The intermediate metabolites are colored in green. Green arrows indicate the movement of intermediary metabolites. In the case of aphid, the genes are postulated. Most secondary endosymbionts retain the pathways for the synthesis of non-essential amino acids. However, S. symbiotica SCc has only preserved the pathways for alanine, cysteine and asparagine (Table 2). Regarding essential amino acid biosynthesis, S. symbiotica SCc has retained the ability to synthesize lysine and tryptophan provided that Buchnera metabolism supplies the respective precursors, i.e., aspartyl-4-phosphate and anthranilate (Figure 1). The latter situation (described elsewhere [15]) is similar in S. symbiotica SAp, which would also require exogenous anthranilate to synthesize tryptophan. However, in A. pisum, B. aphidicola BAp can provide the tryptophan as it possesses the complete pathway. A striking result relates the case of the non-essential amino acids serine and cysteine and the essential ones isoleucine and methionine, which, as shown in Figure 3, is necessary to postulate the metabolic complementation of all three members of the consortium to be synthesized. The aphid would produce serine from glicerate-3-p and then, S. symbiotica SCc could make cysteine. In turn, B. aphidicola BCc can provide threonine to the aphid to obtain the precursor of isoleucine. This is also similar in A. pisum and B. aphidicola BAp (17–19). Finally, B. aphidicola BCc could synthesize methionine, isoleucine and arginine with the external supply of homocysteine, 2-oxobutanoate and ornithine, respectively. We postulate that they come from the aphid, as might be the case in B. aphidicola BAp for methionine and isoleucine biosynthesis [17], [19].
Fig. 3. Outline of the putative synthesis of the amino acids serine, cysteine, isoleucine, and methionine by the consortium (C. cedri, B. aphidicola BCc, and S. symbiotica SCc). 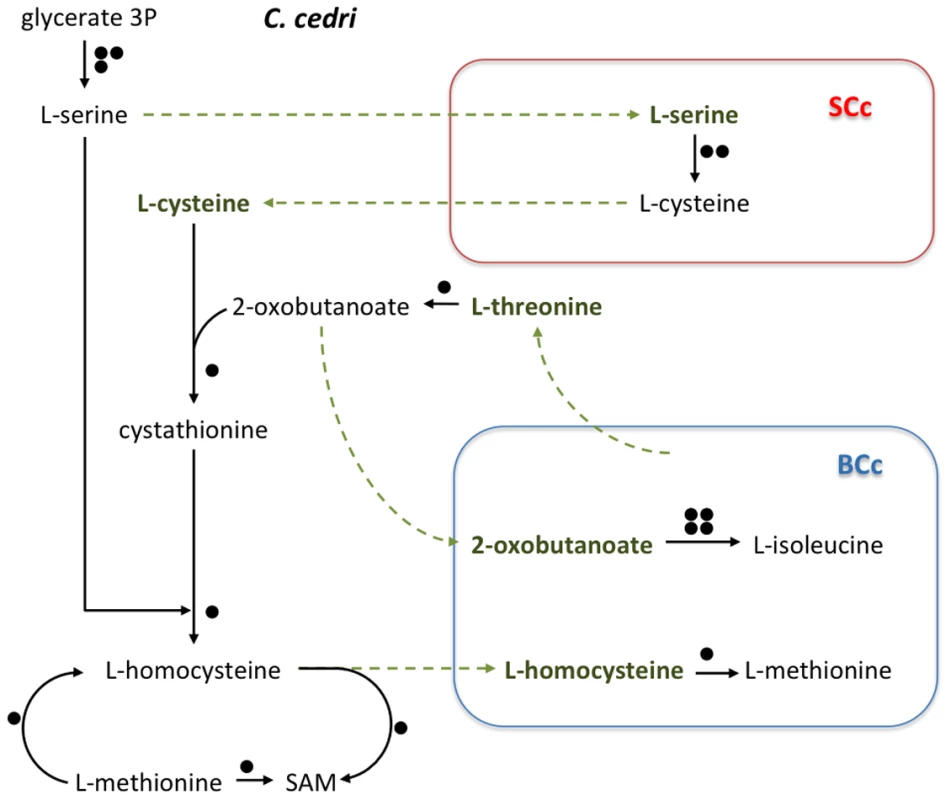
Rest as in Figure 2. Tab. 2. Metabolic capacity of amino acids biosynthesis by selected endosymbionts and free-living bacteria. 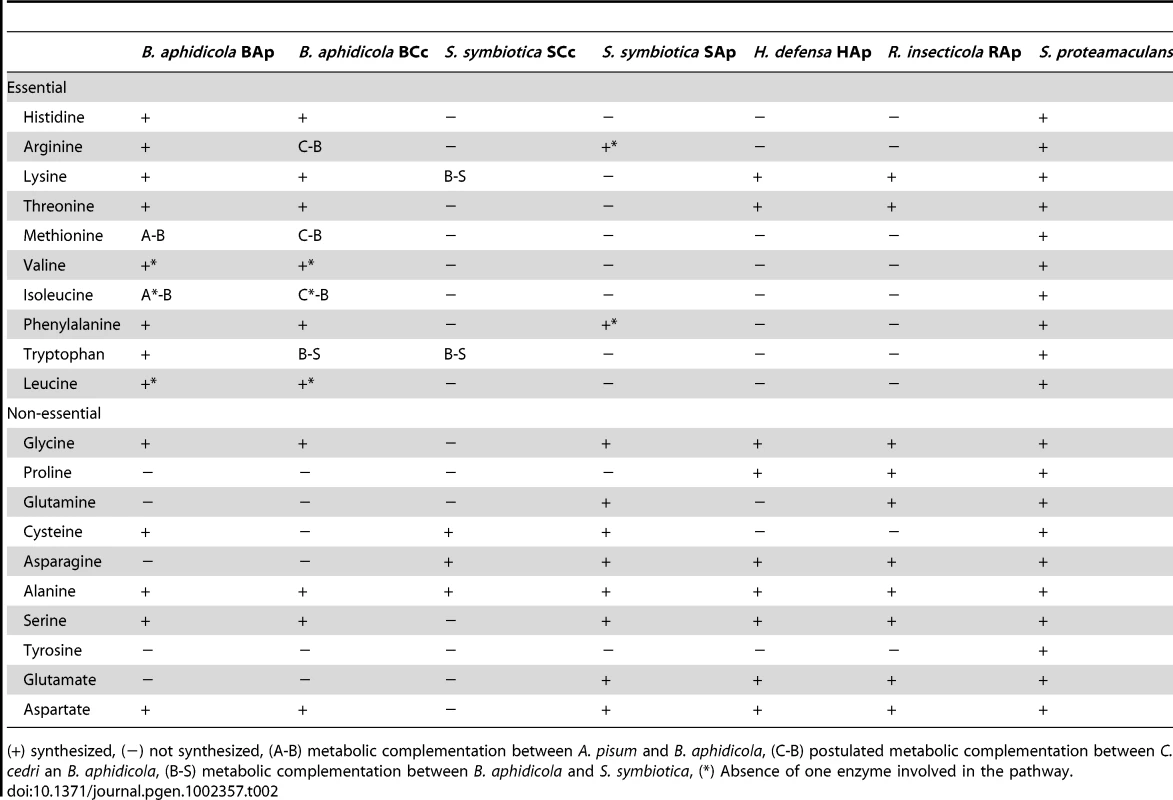
(+) synthesized, (−) not synthesized, (A-B) metabolic complementation between A. pisum and B. aphidicola, (C-B) postulated metabolic complementation between C. cedri an B. aphidicola, (B-S) metabolic complementation between B. aphidicola and S. symbiotica, (*) Absence of one enzyme involved in the pathway. With regard to cofactors and vitamins, genome sequencing has revealed that S. symbiotica SCc is capable of synthesizing the same metabolites as B. aphidicola BAp as well as vitamin B6 (Table 3) although for biotin, folate and CoA, S. symbiotica SCc would require the provision of the respective precursors from B. aphidicola BCc, i.e. pimeloyl CoA, chorismate and L-pantoate (Figure 1). Clearly, S. symbiotica SCc has taken over these functions, which have been completely lost in B. aphidicola BCc. Moreover, S. symbiotica SCc could synthesize heme group in collaboration with the aphid, which must provide the porphobilinogen. This differs hugely from S. symbiotica SAp, which has preserved only four pathways (Table 3).
Tab. 3. Metabolic capacity of cofactor and vitamin synthesis by selected endosymbionts and free-living bacteria. 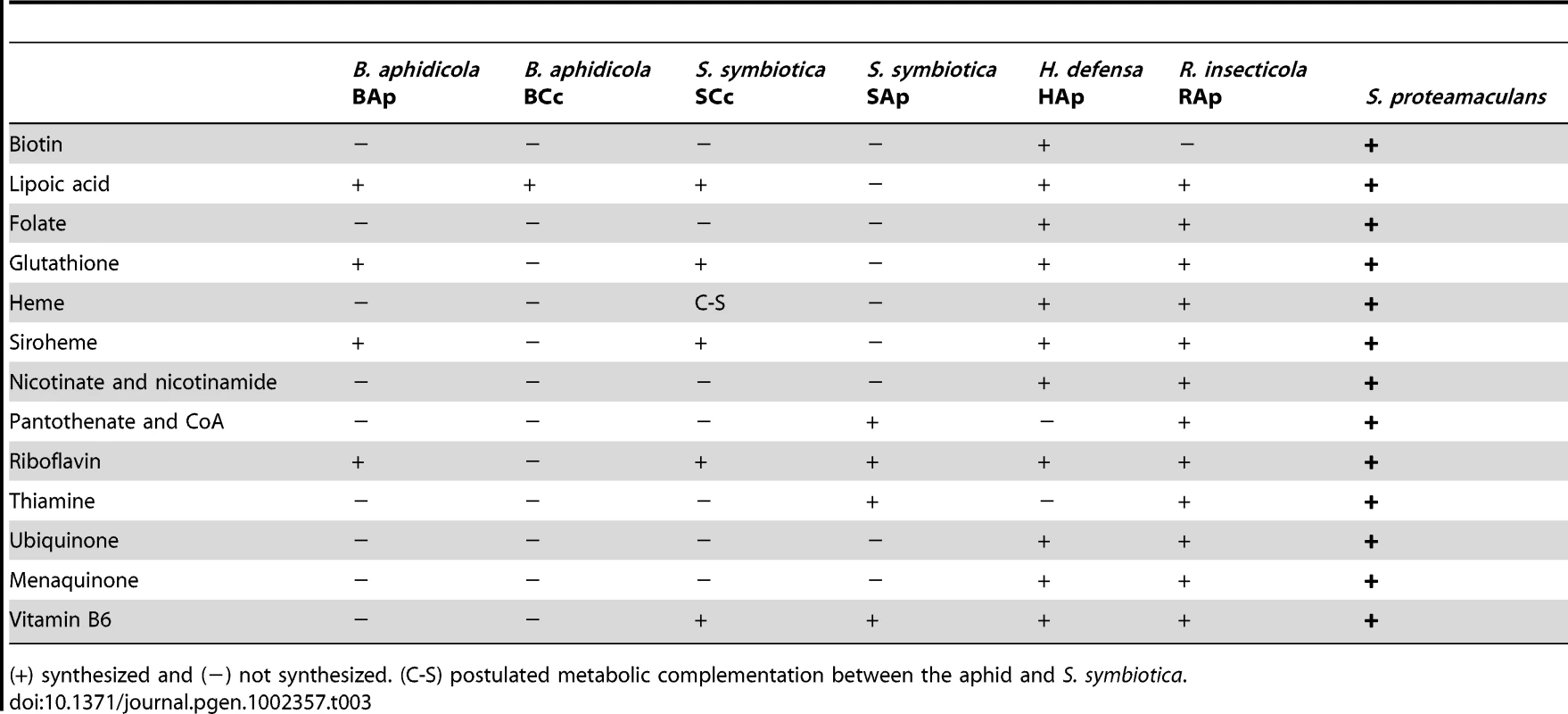
(+) synthesized and (−) not synthesized. (C-S) postulated metabolic complementation between the aphid and S. symbiotica. Cell wall and membranes
The genome sequencing of S. symbiotica SCc revealed that it retains the ability to synthesize peptidoglycan and liposaccharides to make its well-structured and complex membranes (Figure 4). This contrasts with its B. aphidicola partner, which has lost all the genes related to these functions [13]. Although S. symbiotica SCc retains the ability to synthesize these compounds, they are macromolecules and it is unlikely that they can enter B. aphidicola BCc. However, as can be seen in Figure S5, both bacteria maintain all three expected membranes, the two gram-negative and the external bacteriocyte-derived membrane.
Fig. 4. Outline of the putative synthesis of fatty acids, phospholipids (cardiolipin, phosphatidyl-ethanolamine, dyacilglycerol prolipoprotein), peptidoglycan, and lipopolysaccharide by the consortium (C. cedri, B. aphidicoa BCc, and S. symbiotica SCc). 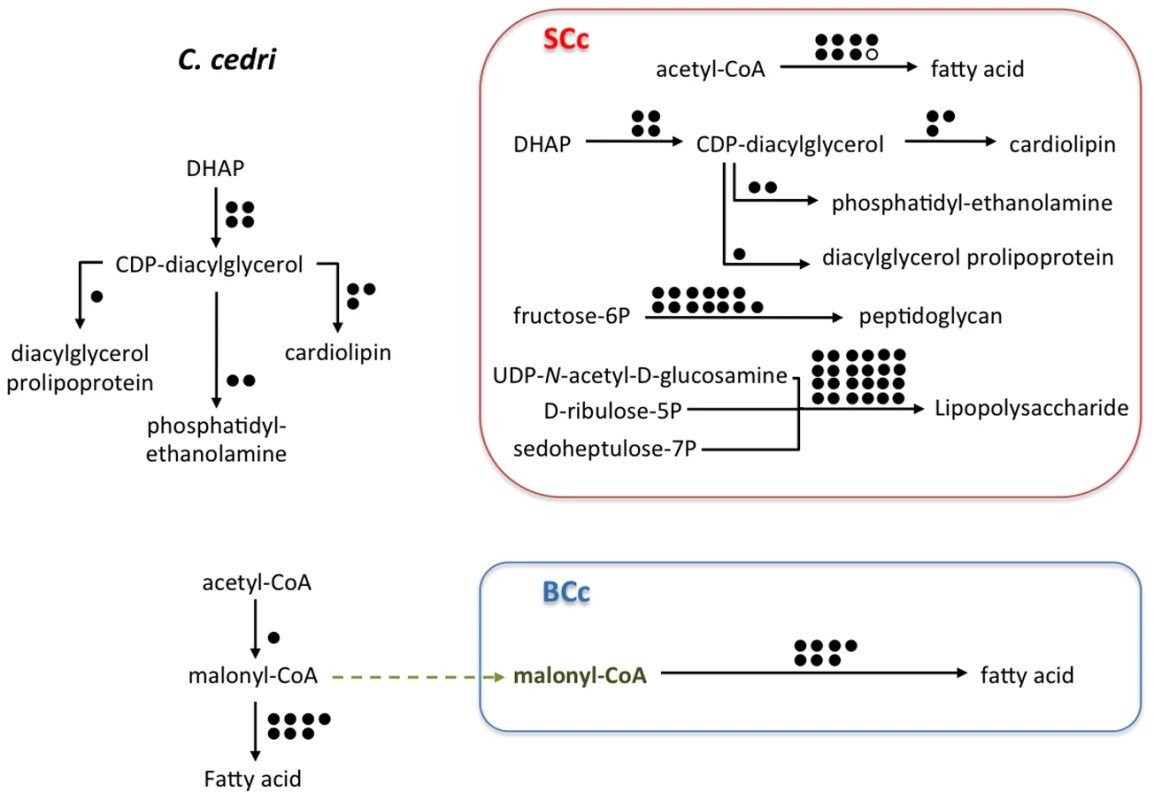
White circles representing absent genes. Rest as in Figure 2. Pseudogenes, absent genes, and genome degradation
To gain insight into the pseudogenization process undergone by S. symbiotica SCc and S. symbiotica SAp in their respective lineages, we have compared the state of the annotated pseudogenes in both Serratia and the free-living S. proteamaculans (see Table S1 for details). From the 58 pseudogenes found in S. symbiotica SCc, two (tuf and bamA) have a duplicated functional copy. From the other 56, eighteen are also inactive genes (nine pseudogenes and nine absent genes) in S. symbiotica SAp, whereas 38 are active copies. Regarding the 311 chosen pseudogenes in S. symbiotica SAp (see Materials and Methods), as expected, most are absent in S. symbiotica SCc, and some are also absent in S proteamaculans, thus being strain specific A very interesting result is that sixteen of the S. symbiotica SAp pseudogenes are putatively active genes in S. symbiotica SCc (Table S1), thus indicating differential degradation fates in both Serratia lineages. Moreover, S. symbiotica SCc possesses 20 CDSs that are totally absent in S. symbiotica SAp.
Synteny plots of Serratia species
In order to further compare the two intracellular Serratia, we performed the analysis of the synteny between both bacteria and also a comparison with free-living relatives. The results are shown in Figure 5 and clearly display the great number of rearrangements that occurred when the bacterium adopted an intracellular lifestyle, as is the case for both S. symbiotica compared to S. proteamaculans. The most interesting result is the comparison between S. symbiotica SCc and S. symbiotica SAp (panel D) where a series of rearrangements are found even in the biggest contigs, which suggest a past history of active mobile elements in S. symbiotica SCc, which are already unidentifiable in the current genome but still present in the S. symbiotica SAp.
Fig. 5. Synteny plots. 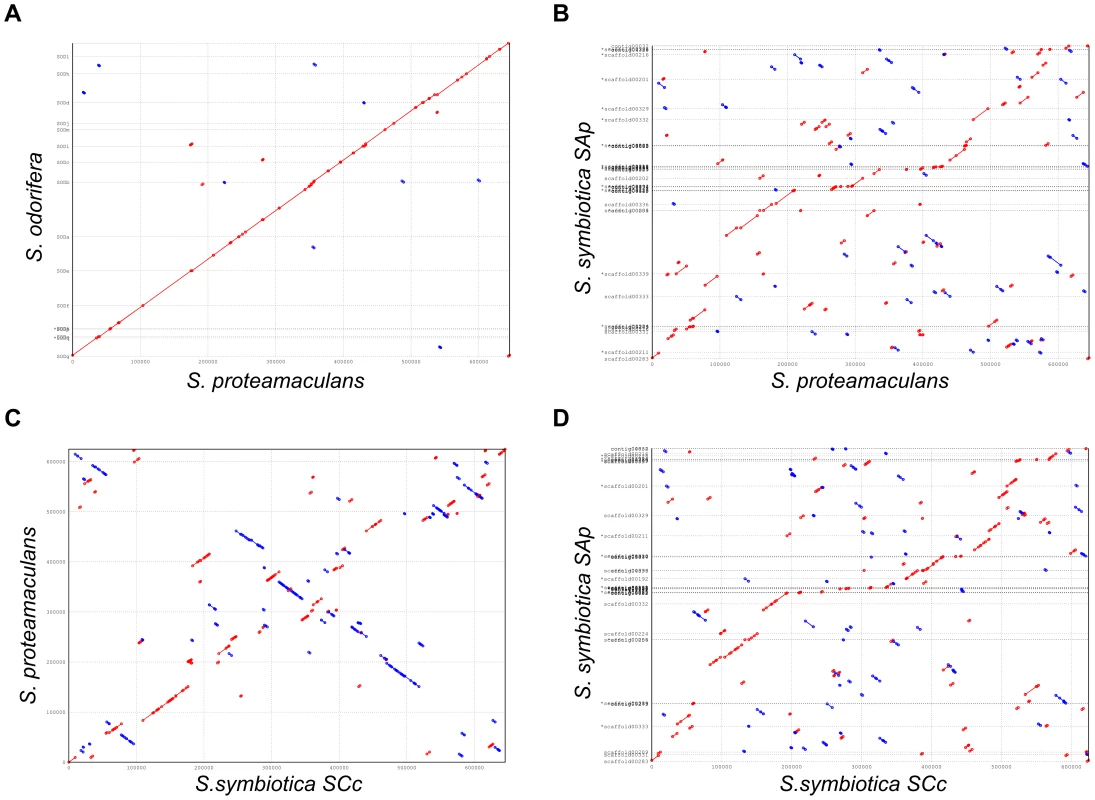
Dot plots displaying syinteny between different species of Serratia in the shared groups of genes. (A) S. proteamaculans is taken as reference against S. odorifera. (B) S. proteamaculans is taken as reference against S. symbiotica SAp. (C) S. symbiotica SCc is taken as reference against S. proteamaculans and (D) S. symbiotica SCc is taken as reference against S. symbiotica SAp. Red dots, direct match; blue dots, reverse match. Discussion
Symbioses involving prokaryotes living in close relationships with insects have been widely studied from the genomic perspective [1], [2]. In the process towards host accommodation, symbionts experience a series of major genetic and phenotypic changes that can be detected by comparison with free-living relatives. Several scenarios could account for the evolution of symbiotic associations, from the first stages of free-living bacteria, through facultative symbiosis, to obligate symbionts. Of particular relevance is the association formed by the coexistence of several symbionts in a given host. Aphids are a good model to dissect the different stages of the integration process undertaken by the different symbionts coexisting therein. At present, the genome of B. aphidicola from five aphid species, belonging to different aphid lineages have been sequenced providing information of the last steps leading to obligate endosymbiosis [13], [24]–[27]. On the other hand, the genome of three facultative endosymbionts from the aphid A. pisum are also available [10]–[12]. They are in the early stages of transition from a free-living to symbiotic lifestyle, with S. symbiotica SAp probably representing the earliest stage of all three [12].
Our work indicates that S. symbiotica from C. cedri is a good candidate for a missing link between a facultative and an obligate insect endosymbionts. For comparative purposes, the two most relevant genomes are B. aphidicola BAp, the Buchnera with the biggest genome that does not need a second symbiont for aphid survival [17], [18], and S. symbiotica SAp because it is a Serratia symbiont, but in a much earlier step of the integration process [12].
Many features of the S. symbiotica SCc genome, such as the A+T content, the number of genes, the loss of recA gene, as well as the total absence of ISs or other mobile DNA still present in all the facultative symbionts analyzed so far, are indicative of an obligate endosymbiont. It is worth mentioning that in S. symbiotica SAp, there are still a certain number of ISs, although because the genome sequence is incomplete, the exact number is not known. Moreover, transposases, plasmid-associated genes, and phage-associated genes can make up to 4% of the total number of genes [12]. On the other hand, S. symbiotica SCc has lost all the genes involved in bacterial pathogenesis that are still retained in S. symbiotica SAp. However, the S. symbiotica SCc genome size (1,763 kb) is intermediate between the two A. pisum symbionts, the obligate B. aphidicola (641 kb) and the facultative S. symbiotica SAp (ca. 2,789 kb), with non-coding DNA comprising a huge part of the genome. In fact, the coding density is extremely low (more than two times lower than that of B. aphidicola BCc), whereas the average size of the intergenic regions is extremely high (more than seven-fold that of H. defensa). According to our knowledge of prokaryotic genomes, these regions must correspond to ancient genes. However, in contrast with its related and recent symbiont S. symbiotica SAp, which has around 550 pseudogenes, in S. symbiotica SCc only 58 pseudogenes could be clearly identified [12]. These data support the postulated gradual process of genome degradation of the pseudogenes, ending up in their total disappearance in obligate bacterial endosymbionts [28]–[30]. In fact, if we substitute the size of the intergenic region in S. symbiotica SCc (1,672 bp on average) for the size of these regions in B. aphidicola BCc (135.8 bp on average), the chromosomal length would be 771,075 bp, a reduction of 43.7% and in the range of other obligate endosymbionts published so far (reviewed in [1], [2]).
The functional annotation of the S. symbiotica SCc genome indicated that its main symbiotic role would be the metabolism of cofactors, vitamins and nucleotides, whereas in B. aphidicola BCc it would be that of amino acid provider. However, the inferred metabolism of both endosymbionts has revealed a strong interdependence and a fine tuning of different biosynthetic pathways which, in some cases, probably also involves metabolic complementation with the aphid, as shown to occur in A. pisum [16]–[19], [23]. Overall, it seems that B. aphidicola BCc and S. symbiotica SCc in C. cedri jointly perform the metabolic functions that B. aphidicola BAp performs in A. pisum.
Another interesting feature relates to cell morphology. When S. symbotica SCc was first reported, its spherical morphology at the microscopic level was surprising [14], similar to the shape cells of B. aphidicola (Figure S5C), and different to the rod-shaped bacteria observed in S. symbiotica SAp [31] and in S. symbiotica from C.tujafilina [7]. These last two Serratia could be present in different locations in some individuals of the population, whereas S. symbiotica SCc are confined to their own bacteriocytes and occur in all individuals and at the same density as B. aphidicola BCc [14]. However, S. symbiotica SCc, like S. symbiotica SAp, has retained the genes involved in bacillary morphology (mreB, mreC, mreD, mrdB). These genes have been lost in all B. aphidicola genomes sequenced so far. At present, it is not clear whether these genes are being expressed or not, although the observed morphology is unexpected. The possible role played by the intracellular environment cannot be ruled out, possibly exerting some kind of effect on the morphology if those genes are expressed [31].
In summary, all the data presented (diversity in symbiont morphology, distribution and function) correlate with the existence of two different clades of S. symbiotica in aphids, at least, as also indicated by the phylogenetic analyses [7], [8]. The analysis of the synteny between S. symbiotica SCc and S. symbiotica SAp and the comparison with free-living Serratia indicate the great and different number of rearrangements undergone when the two bacteria adopted an intracellular lifestyle (Figure 5).
The comparison of the genome of all three secondary endosymbionts of A. pisum, H. defensa, S. symbiotica and R. insecticola, provides some clues to the scenario of how the C. cedri consortium came into being. These three bacteria, despite being facultative, could be retained by the aphid because they provide certain benefits to the host under particular conditions (for a review see [5]). Specifically, S. symbiotica SAp is involved in defense against environmental heat stress [32]–[34]. Due to the inactivation of some of their biosynthetic pathways, such as those related to the essential amino acid biosynthesis, over time, these bacteria have become dependent on the presence of Buchnera, and thus preserve active uptake mechanisms for their provision. On the other hand, as B. aphidicola is still undergoing a genome reduction process, some symbiotic functions may be lost and taken over by the second endosymbiont. When this happens, the consortium is established. The different agents involved in tryptophan biosynthesis in A. pisum and C. cedri is an amazing example of evolution towards the establishment of a consortium. In all the B. aphidicola strains, the two first genes of the tryptophan pathway, trpE and trpG, coding for anthranilate synthase, are either on a plasmid or in the chromosome, but always separated from the rest of the genes on the chromosome. Both S. symbiotica have lost these two genes, but preserve the other genes of the pathway (trpABCD), implying Buchnera dependence for anthranilate provision. The main difference between both systems involves the obligate endosymbiont. In A. pisum, Buchnera can make tryptophan autonomously because it possesses the complete pathway, whereas in C. cedri, Buchnera has lost the trpABCD genes, which are present in Serratia. This example could be enough to seal a consortium. Another case of metabolic collaboration between the two endosymbionts is the biosynthesis of lysine from aspartate. This pathway is complete in B. aphidicola BAp [19] whereas in B. aphidicola BCc only the first step, catalyzed by aspartokinase (thrA), takes place, whereas the other eight steps occur in S. symbiotica SCc (Figure 1). Moreover, additional cases of metabolic complementation might also exist during the synthesis of biotin, folate, and CoA in S. symbiotica SCc.
Finally, the fact that 36 active genes in S. symbiotica SCc are either pseudogenes, or absent genes in S. symbiotica SAp point towards different genome degradation processes in both Serratia. Such processes are context-dependent, i.e., the consequence of the different gene repertoire of the other agents when the association started, particularly the different genome composition of B. aphidicola in A. pisum or C. cedri.
In summary, here we report a missing link in the evolution from a facultative to an obligate endosymbiont. This is the case of S. symbiotica SCc when compared with S. symbiotica SAp, two different endosymbionts belonging to the same genus but in two different stages of the integration process leading to intracellular lifestyle: S. symbiotica SAp, a recently acquire facultative symbiont, and S. symbiotica SCc a recent co-obligate endosymbiont. We also gain insights into the establishment of a bacterial consortium between two co-obligate symbionts in aphids, B. aphidicola BAp and S. symbiotica SCc.
Materials and Methods
Aphid collection and total DNA extraction
C. cedri aphids were collected in Valencia, Spain. An enriched fraction of bacteriocytes was obtained as in [35], and then used to extract total DNA following a CTAB (Cetyltrimethylammonium bromide) method [36].
Genome sequencing and assembling
The complete genome sequence of S. symbiotica SCc was obtained using single and paired-end shotgun reads from 454 pyrosequencing method (454 Life Science, Lifesequencing). The sequencing run generated 831,450 reads that assembled into 108,723 contigs using the GS De novo Assembler (version 1.1.03.24). Contigs expected to belong to the Serratia genome were identified by BLASTX searches against the GenBank non-redundant database [37], and reads associated with these contigs were extracted and reassembled to generate the S. symbiotica SCc genome. Reassembly produced 15 contigs. The order and orientation of some of the 15 contigs were predicted using the pair-ends information. All contig joins were confirmed using PCR amplification followed by Sanger sequencing. The tool Gap4.8b1 from Staden Package [38] was used for the total assembling of the Sanger sequences. This resulted in a single 1,762,765 bp contig with an average 454 (both single and paired-ends) coverage of 25.90×.
Gene annotation and pseudogene prediction
The protein coding sequences (CDSs) were identified with the GLIMMER v3.02 program [39]. The ARTEMIS [40] program was used to check for the start and stop codons. Final annotation was performed using BLASTP comparison [37]. The tRNAscan [41] program was used to predict tRNAs, as well as other small RNAs, like tmRNA, the RNA component of the RNase P. Signal Recognition Particle RNA was identified by programs like SRPscan [42], as well as consulting the Rfam database [43]. Intergenic regions (IGRs) were manually analyzed by BLASTX and BLASTN to locate pseudogenes that were not found by GLIMMER. Then they were reanalyzed with Rfam, Pfam and NCBIs BLASTX [37], [43], [44] against the non-redundant database to look for any trace of coding fragments. Once the genome was finally annotated, the size of the genome, genes and intergenic regions was determined with ARTEMIS [40]. GC content was calculated by the on-line tool GeeCee (http://srs.nchc.org.tw/emboss-bin/emboss.pl?_action=input&_app=geecee).
Genome GC difference and CDSs and IGRs GC content analysis
The nucleotide sequences from complete genomes, or from the contigs when the closed genome was unavailable, as occurred in S. symbiotica SAp and R. insecticola, were recovered from both S. symbiotica (SCc and SAp), S. proteamaculans (as a free-living Serratia representative), H. defensa and R. insecticola (as facultative aphid endosymbionts representatives), and B. aphidicola BCc (as a primary endosymbiont representative). Genomic GC difference was calculated as described in [21].
The IGRs and CDSs nucleotide FASTA files were extracted for both S. symbiotica, and GC content was calculated for each sequence.
Inferred metabolism
The ORFs orthologous to known genes in other species were catalogued based on non-redundant classification schemes, such as COG (Clusters of Orthologous Groups of Proteins). The metabolic network was reconstructed using the automatic annotator server from KAAS-KEEG [45]. According to our genome annotation, each pathway was examined checking the BRENDA [46] and EcoCyc databases [47].
Electron microscopy
C. cedri adult aphids were dissected (the head was removed) under a microscope in 0.9% NaCl fixed in 2% paraformaldehyde-2,5% glutaraldehyde in 0.2 M phosphate buffer (PB) for 24 h, and washed several times in 0.1 M PB. They were then post fixed in 2% osmium tetroxide in 0.1 M PB for 90 min in darkness, dehydrated in ethanol, and embedded in araldite (Durcupan, Fluka). Semithin sections (1.5 µm) were cut with a diamond knife, and stained with toluidine blue (Nikon Eclipse E800). Ultrathin (0.05 µm) sections were cut with a diamond knife, stained with lead citrate, and examined under transmission electron microscope (JEOLJEM1010).
Analysis of presence/absence of pseudogens
Due to the fact that the S. symbiotica SAp genome is incompleted (12), CDSs that were present in more than one sequence (i.e. they had a gap spanning two contigs in a scaffold), as well as the pseudogenes without a name assigned or annotated as phage, transposase, integrase or hypothetical, were excluded from the analysis. This resulted in 311 pseudogenes and 2041 CDSs for this organism. First, the 58 pseudogenes present in S. symbiotica SCc were matched with their counterparts in S. symbiotica SAp and S. proteamaculans genomes (in both pseudogene and CDS databases for each organism). The CDSs were grouped by presence of same annotation or by BLAST (using pseudogenes as query and CDSs as subjects, with an e-value cut-off of 1e−03) and checked manually for function annotation. Also, BLASTX was run using pseudogenes as query against CDSs from all three Serratia and hits with a Bit Score Ratio > = 30 were selected and manually checked. Finally, genes not detected in S. proteamaculans (both against S. symbiotica SCc and S. symbiotica SAp CDSs and pseudogenes) were manually searched on the KEGG orthology database and in selected cases using BLAST with the pseudogene sequence against nr (restricted to Serratia taxonomy). The S. symbiotica SAp pseudogenes that did not match any S. symbiotica SCc pseudogenes in the analysis described above were matched to their CDS or pseudogene counterparts in both S. symbiotica (SCc and SAp) and S. proteamaculans in a similar fashion. COG categories from all pseudogenes, when not available, were obtained from both NCBI and KEGG. Plots were done using R [48].
Synteny plots
Protein coding genes from S. symbiotica SAp, S. odorifera 4Rx13 (ADBX00000000) and S. proteamaculans 568 were downloaded from Genbank. S. symbiotica SAp CDSs that were in more than one sequence were omitted. We then used BLAST with an e-value cut-off of 1e-05 and 70% match cut-off. The results were clustered using MCL [49]. Common genes from all different Serratia species were extracted from each nucleotide FASTA file and ordered by contigs (when not in a single one), and Promer from the Mummer package [50] was used to plot the comparisons. S. odorifera 4Rx13 was used to exemplify the contig rearrangement algorithm due to the low number of contigs present in the genome annotation.
Supporting Information
Zdroje
1. MoyaAPeretoJGilRLatorreA 2008 Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet 9 218 229
2. MoranNAMcCutcheonJPNakabachiA 2008 Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42 165 190
3. BaumannP 2005 Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59 155 189
4. DouglasAE 1998 Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43 17 37
5. OliverKMDegnanPHBurkeGRMoranNA 2010 Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55 247 266
6. RussellJAMoranNA 2005 Horizontal transfer of bacterial symbionts: heritability and fitness effects in a novel aphid host. Appl Environ Microbiol 71 7987 7994
7. LamelasAPérez-BrocalVGómez-ValeroLGosalbesMJMoyaA 2008 Evolution of the secondary symbiont “Candidatus Serratia symbiotica” in aphid species of the subfamily Lachninae. Appl Environ Microbiol 74 4236 4240
8. BurkeGRNormarkBBFavretCMoranNA 2009 Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl Environ Microbiol 75 5328 5335
9. MoranNARussellJAKogaRFukatsuT 2005 Evolutionary Relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol 71 3302 3310
10. DegnanPHLeonardoTECassBNHurwitzBSternD 2009 Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol 12 2060 2069
11. DegnanPHYuYSisnerosNWingRAMoranNA 2009 Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A 106 9063 9068
12. BurkeGRMoranNA 2011 Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol 3 195 208
13. Pérez-BrocalVGilRRamosSLamelasAPostigoM 2006 A small microbial genome: The end of a long symbiotic relationship? Science 314 312 313
14. Gómez-ValeroLSoriano-NavarroMPérez-BrocalVHeddiAMoyaA 2004 Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the Aphid Cinara cedri. J Bacteriol 186 6626 6633
15. GosalbesMJLamelasAMoyaALatorreA 2008 The striking case of tryptophan provision in the cedar aphid Cinara cedri. J Bacteriol 190 6026 6029
16. ConsortiumTIAG 2010 Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol 8 e1000313 doi:10.1371/journal.pbio.1000313
17. WilsonACCAshtonPDCalevroFCharlesHColellaS 2010 Genomic insight into the amino acid relations of the pea aphid Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol 19 249 258
18. HansenAKMoranNA 2011 Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A 108 2849 2854
19. ShigenobuSWilsonACC 2011 Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont. Cell Mol Life Sci 68 1297 1309
20. BohlinJSkjerveEUsseryDW 2008 Investigations of oligonucleotide usage variance within and between prokaryotes. PLoS Comput Biol 4 e1000057 doi:10.1371/journal.pcbi.1000057
21. BohlinJSnipenLHardySPKristoffersenaBLagesenK 2010 Analysis of intra-genomic GC conent homogeneity within prokaryotes. BMC Genomics 11 464
22. TatusovRLFedorovaNDJacksonJDJacobsARKiryutinB 2003 The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4 41
23. RamseyJSMacDonaldSJJanderGNakabachiAThomasGH 2010 Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol 19 Suppl 2 241 248
24. ShigenobuSWatanabeHHattoriMSakakiYIshikawaH 2000 Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407 81 86
25. TamasIKlassonLCanbackBNaslundAKErikssonAS 2002 50 million years of genomic stasis in endosymbiotic bacteria. Science 296 2376 2379
26. van HamRCKamerbeekJPalaciosCRausellCAbascalF 2003 Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci U S A 100 581 586
27. LamelasAGosalbesMJMoyaALatorreA 2011 New clues about the evolutionary history of metabolic losses in bacterial endosymbionts, provided by the genome of Buchnera aphidicola from the aphid Cinara tujafilina. Appl Environ Microbiol 77 4446 4454
28. SilvaFJLatorreAMoyaA 2001 Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet 17 615 618
29. BeldaEMoyaABentleySSilvaFJ Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 11 449
30. ColeSTEiglmeierKParkhillJJamesKDThomsonNR 2001 Massive gene decay in the leprosy bacillus. Nature 409 1007 1011
31. FukatsuTNikohNKawaiRKogaR 2000 The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: homoptera). Appl Environ Microbiol 66 2748 2758
32. ChenD-QMontllorCBPurcellAH 2000 Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol Exp Appl 95 315 323
33. MontllorCBMaxmenAPurcellAH 2002 Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27 189 195
34. RussellJAMoranNA 2006 Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc Biol Sci 273 603 610
35. GilRSilvaFJZientzEDelmotteFGonzalez-CandelasF 2003 The genome sequence of Blochmannia floridanus: Comparative analysis of reduced genomes. Proc Natl Acad Sci U S A 100 9388 9393
36. AusubelFM 1999 Short protocols in molecular biology: a compendium of methods from Current protocols in molecular biology. 44th edn New York Wiley
37. AltschulSFMaddenTLSchafferAAZhangJZhangZ 1997 Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389 3402
38. StadenRBealKFBonfieldJK 2000 The Staden package. Methods Mol Biol 132 115 130
39. DelcherALHarmonDKasifSWhiteOSalzbergSL 1999 Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27 4636 4641
40. RutherfordKParkhillJCrookJHorsnellTRiceP 2000 Artemis: sequence visualization and annotation. Bioinformatics 16 944 945
41. LoweTMEddySR 1997 tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25 955 964
42. RegaliaMRosenbladMASamuelssonT 2002 Prediction of signal recognition particle RNA genes. Nucleic Acids Res 30 3368 3377
43. Griffiths-JonesSMoxonSMarshallMKhannaA 2005 Rfam: Annotating Non-Coding RNAs in Complete Genomes. Nucleic Acids Res 33 D121 D141, 2005
44. BatemanACoinLDurbinRFinnRDHollichV 2004 The Pfam protein families database. Nucleic Acids Res 32 D138 D141
45. MoriyaYItohMOkudaSYoshizawaACKanehisaM 2007 KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35 W182 185
46. ChangAScheerMGroteASchomburgISchomburgD 2009 BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res 37 D588 592
47. CaspiRFoersterHFulcherCAKaipaPKrummenackerM 2008 The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res 36 D623 631
48. RDC-Team 2010 R: A language for statistical computing. R Foundation For Statistical Computing, Vienna, Austria, 2.11.1
49. Van DongenS 2000 Graph clustering by flow simulation [PhD Thesis] Utrecht, The Netherlands University of Utrecht
50. KurtzSPhillippyADelcherALSmootMShumwayM 2004 Versatile and open software for comparing large genomes. Genome Biol 5 R12
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- De Novo Origins of Human Genes
- Duplication Hotspots Are Associated with Late-Replicating Regions of the Genome
- De Novo Origin of Human Protein-Coding Genes
- Cyclin D/CDK4 and Cyclin E/CDK2 Induce Distinct Cell Cycle Re-Entry Programs in Differentiated Muscle Cells
- Short Day–Mediated Cessation of Growth Requires the Downregulation of AINTEGUMENTALIKE1 Transcription Factor in Hybrid Aspen
- Physiological IRE-1-XBP-1 and PEK-1 Signaling in Larval Development and Immunity
- Role of Pirh2 in Mediating the Regulation of p53 and c-Myc
- Signatures of Environmental Genetic Adaptation Pinpoint Pathogens as the Main Selective Pressure through Human Evolution
- FOXO Regulates Organ-Specific Phenotypic Plasticity In
- Heritable Epigenetic Variation among Maize Inbreds
- Foxn1 Regulates Lineage Progression in Cortical and Medullary Thymic Epithelial Cells But Is Dispensable for Medullary Sublineage Divergence
- Attenuation of the Sensing Capabilities of PhoQ in Transition to Obligate Insect–Bacterial Association
- A Novel Protein LZTFL1 Regulates Ciliary Trafficking of the BBSome and Smoothened
- Activation of Bmp2-Smad1 Signal and Its Regulation by Coordinated Alteration of H3K27 Trimethylation in -Induced Senescence
- Histone H3K56 Acetylation, CAF1, and Rtt106 Coordinate Nucleosome Assembly and Stability of Advancing Replication Forks
- The SUN Protein Mps3 Is Required for Spindle Pole Body Insertion into the Nuclear Membrane and Nuclear Envelope Homeostasis
- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- Effect of Host Species on the Distribution of Mutational Fitness Effects for an RNA Virus
- Pch2 Acts through Xrs2 and Tel1/ATM to Modulate Interhomolog Bias and Checkpoint Function during Meiosis
- SOX9 Governs Differentiation Stage-Specific Gene Expression in Growth Plate Chondrocytes via Direct Concomitant Transactivation and Repression
- from the Aphid : A Missing Link from Facultative to Obligate Insect Endosymbiont
- Recessive Antimorphic Alleles Overcome Functionally Redundant Loci to Reveal Function in Flowers and Meristems
- Over-Expression of DSCAM and COL6A2 Cooperatively Generates Congenital Heart Defects
- Consequences of Eukaryotic Enhancer Architecture for Gene Expression Dynamics, Development, and Fitness
- Distinct Genetic Architectures for Male and Female Inflorescence Traits of Maize
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- For Male , Sperm Activation Is a “Just-in-Time” Event
- PcG Complexes Set the Stage for Epigenetic Inheritance of Gene Silencing in Early S Phase before Replication
- The Gene Contains Hotspots for L1 Endonuclease-Dependent Insertion
- Relative Burden of Large CNVs on a Range of Neurodevelopmental Phenotypes
- Multiple Means to the Same End: The Genetic Basis of Acquired Stress Resistance in Yeast
- Genome-Wide Crossover Distribution in Meiosis Reveals Sex-Specific Patterns along Chromosomes
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
- Homologs of Retinoblastoma-Associated Protein 46/48 Associate with a Histone Deacetylase to Act Redundantly in Chromatin Silencing
- Genetic Interaction Maps in Reveal Functional Crosstalk among Cell Envelope Biogenesis Pathways
- The ERI-6/7 Helicase Acts at the First Stage of an siRNA Amplification Pathway That Targets Recent Gene Duplications
- PBX1 Genomic Pioneer Function Drives ERα Signaling Underlying Progression in Breast Cancer
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- De Novo Origins of Human Genes
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



