-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
Vertebrate mesendoderm specification requires the Nodal signaling pathway and its transcriptional effector FoxH1. However, loss of FoxH1 in several species does not reliably cause the full range of loss-of-Nodal phenotypes, indicating that Nodal signals through additional transcription factors during early development. We investigated the FoxH1-dependent and -independent roles of Nodal signaling during mesendoderm patterning using a novel recessive zebrafish FoxH1 mutation called midway, which produces a C-terminally truncated FoxH1 protein lacking the Smad-interaction domain but retaining DNA–binding capability. Using a combination of gel shift assays, Nodal overexpression experiments, and genetic epistasis analyses, we demonstrate that midway more accurately represents a complete loss of FoxH1-dependent Nodal signaling than the existing zebrafish FoxH1 mutant schmalspur. Maternal-zygotic midway mutants lack notochords, in agreement with FoxH1 loss in other organisms, but retain near wild-type expression of markers of endoderm and various nonaxial mesoderm fates, including paraxial and intermediate mesoderm and blood precursors. We found that the activity of the T-box transcription factor Eomesodermin accounts for specification of these tissues in midway embryos. Inhibition of Eomesodermin in midway mutants severely reduces the specification of these tissues and effectively phenocopies the defects seen upon complete loss of Nodal signaling. Our results indicate that the specific combinations of transcription factors available for signal transduction play critical and separable roles in determining Nodal pathway output during mesendoderm patterning. Our findings also offer novel insights into the co-evolution of the Nodal signaling pathway, the notochord specification program, and the chordate branch of the deuterostome family of animals.
Published in the journal: Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin. PLoS Genet 7(5): e32767. doi:10.1371/journal.pgen.1002072
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002072Summary
Vertebrate mesendoderm specification requires the Nodal signaling pathway and its transcriptional effector FoxH1. However, loss of FoxH1 in several species does not reliably cause the full range of loss-of-Nodal phenotypes, indicating that Nodal signals through additional transcription factors during early development. We investigated the FoxH1-dependent and -independent roles of Nodal signaling during mesendoderm patterning using a novel recessive zebrafish FoxH1 mutation called midway, which produces a C-terminally truncated FoxH1 protein lacking the Smad-interaction domain but retaining DNA–binding capability. Using a combination of gel shift assays, Nodal overexpression experiments, and genetic epistasis analyses, we demonstrate that midway more accurately represents a complete loss of FoxH1-dependent Nodal signaling than the existing zebrafish FoxH1 mutant schmalspur. Maternal-zygotic midway mutants lack notochords, in agreement with FoxH1 loss in other organisms, but retain near wild-type expression of markers of endoderm and various nonaxial mesoderm fates, including paraxial and intermediate mesoderm and blood precursors. We found that the activity of the T-box transcription factor Eomesodermin accounts for specification of these tissues in midway embryos. Inhibition of Eomesodermin in midway mutants severely reduces the specification of these tissues and effectively phenocopies the defects seen upon complete loss of Nodal signaling. Our results indicate that the specific combinations of transcription factors available for signal transduction play critical and separable roles in determining Nodal pathway output during mesendoderm patterning. Our findings also offer novel insights into the co-evolution of the Nodal signaling pathway, the notochord specification program, and the chordate branch of the deuterostome family of animals.
Introduction
The Nodal signaling pathway performs several key steps during vertebrate development. Nodal signals are required for the initial specification and animal-vegetal patterning of mesoderm and endoderm. Nodal is also crucial for induction of the dorsal organizer, a specialized tissue that secretes a host of signals to pattern mesodermal fates along the dorsal-ventral axis and to induce the neuroectoderm [1]. During gastrulation, Nodal signals are maintained in the notochord and prechordal plate, the dorso-axial derivatives of the organizer. These structures are crucial for patterning the neural tube and brain, events which also involve Nodal signals. Finally, asymmetric Nodal activation during somitogenesis governs the laterality of organs such as the gut and heart, and asymmetric lobe development of mammalian lungs. The dependence of the embryo on proper Nodal signaling is evidenced clearly in zebrafish by double mutants for the Nodal homologs cyclops and squint and by maternal-zygotic (MZ) one-eyed pinhead (oep) mutants. These mutants, which entirely lack either the two early zebrafish Nodals or the essential extracellular EGF-CFC coreceptor, respectively, exhibit no Nodal signaling and consequently a near-complete loss of mesoderm, an absence of endoderm, and a severe disruption in neural patterning [2], [3].
Many components of the Nodal pathway have been identified and characterized in addition to the EGF-CFC coreceptor [4], [5]. The Nodal family of ligands belongs to the activin-like subgroup of the TGF-ß superfamily and shares many signaling components with other activin-like pathways. These common components include the type I and II activin receptors, Alk4 and ActRIIA/B respectively, the receptor-activated Smads, Smad2/3, and the effector Smad, Smad4. The Xenopus FoxH1 gene encodes the first transcription factor found to bind to activated Smads in response to activin-like signaling [6]. A Forkhead-family transcription factor conserved across vertebrate species, FoxH1 activates several Nodal targets, including nodal homologues themselves, the lefty Nodal inhibitors, and several mesendoderm-specific transcription factors, including goosecoid (gsc), no tail (ntl)/brachyury, the zebrafish floating head (flh) gene, and certain members of the Mix/Bix family of paired-like homeodomain factors [7]. Loss of FoxH1 function, through a targeted knockout in mouse or morpholino knockdown in Xenopus, causes a significant reduction in head structures and a complete loss of axial mesoderm [8]–[11]. These defects are similar to, but less severe than, those caused by a complete loss of Nodal signaling, suggesting that other transcription factors can also activate Nodal targets.
Two alleles of the zebrafish mutant schmalspur (sur) were independently isolated from two ENU mutagenesis screens [12]–[14]. The sur alleles were mapped to the FoxH1 locus and found to encode single-nucleotide substitutions ten bases apart from each other, leading to an Arg→His (FoxH1m768) or a Lys→Asn (FoxH1ty68b) at the beginning of the Forkhead DNA-binding domain [15], [16]. Due to the mutations' positions in the FoxH1 polypeptide and the failure of both mutant proteins to activate a luciferase reporter linked to FoxH1 binding sites [17], the sur alleles have been assumed to represent null mutations of FoxH1. However, embryos possessing both maternal and zygotic sur alleles display only mild versions of FoxH1 loss-of-function phenotypes observed in other organisms, including variable deficiencies in axial mesoderm and floor plate, as well as variable degrees of synopthalmia/cyclopia [15], [16]. The relatively mild defects of MZsur compared to FoxH1 loss in Xenopus and mouse led to the speculation that another Smad-interacting transcription factor, such as the zebrafish Mixer homologue bonnie and clyde (bon), can partially compensate for the sur mutation [18].
In this study we describe a novel mutation in zebrafish FoxH1, named midway (mid). This mutation causes highly penetrant defects in axial mesoderm specification that are significantly stronger than those of sur. Early molecular markers for, and later morphogenesis of, axial mesoderm are severely reduced or absent in MZmid embryos. These phenotypes more closely resemble loss of FoxH1 function in other organisms, suggesting that FoxH1 has a conserved role in axial development among all vertebrate species. Furthermore, investigation into the differences between the MZmid phenotypes and those caused by a complete loss of Nodal signaling provides new insights into the functions of the Nodal pathway during mesendoderm induction and patterning. FoxH1 function is required for notochord formation but is dispensable for most nonaxial mesoderm fates, which appear to rely on Eomesodermin (Eomes) activity for their earliest specification. Early endoderm induction also does not strictly require FoxH1, instead depending on Eomes and Bon. All three transcription factors contribute to gene expression in the organizer/prechordal plate and subsequent anterior neural development. Our results lead to a model in which the roles of Nodal during early development are partially distinct and separable according to the transcription factor or factors used by the responding cells.
Results
The midway (mid) allele encodes a novel mutation of FoxH1
The FoxH1Pr1 allele (which we refer to as midway) was isolated as a spontaneously occurring recessive mutation exhibiting a ventral body curvature at 24 hours post-fertilization (hpf; data not shown). Initial morphological analysis revealed that mid homozygotes failed to undergo cardiac jogging (data not shown), prompting the name midway and suggesting that the mid mutation perturbs the process of left-right patterning. RNA in situ hybridization analysis for southpaw, a Nodal homologue that is the earliest left-right asymmetrically expressed gene known in zebrafish [19], revealed a complete absence of expression in the lateral plate mesoderm (data not shown). These phenotypes closely resembled those caused by the sur alleles of the FoxH1 gene [20]–[22].
Bulked segregant analysis followed by genetic mapping by recombination frequency [23] supported the identity of mid as an allele of FoxH1. mid mapped to a roughly 10-cM interval on chromosome 12, defined by SSLP markers z27025 and z11549, that included the FoxH1 locus. The phenotypes and mapping data prompted a complementation analysis between mid and sur (FoxH1m768, the strain we used for all subsequent experiments involving sur) heterozygotes. These matings consistently produced clutches in which roughly 25% of the embryos exhibited ventral body curvature similar to homozygotes of either allele, indicating that mid and sur are in the same complementation group (data not shown). As final confirmation of the identity of the mid locus, FoxH1 mRNA transcribed from a pCS2 expression vector containing the full-length FoxH1 cDNA was microinjected into mid heterozygote incross progeny at the one-cell stage. Injection of 10 pg FoxH1 mRNA reduced the occurrence of ventral body curvature from 23% in uninjected clutches (n = 162) to 5% (n = 313). The phenotypes, sur complementation failure, mapping results, and rescue injections together indicate that the mid mutation lies within the FoxH1 gene.
To identify the molecular lesion in the mid allele, we sequenced the genomic FoxH1 locus in mid mutants. Importantly, the missense mutations of the two sur alleles were not present in the mid locus, distinguishing the mid and sur lesions at the molecular level. We discovered a two-nucleotide insertion at the beginning of the Smad-interaction domain (SID) which causes a frameshift at residue 337 of the 472-amino acid polypeptide. This frameshift causes a truncation that would eliminate all but the most N-terminal three amino acids of the SID, presumably prohibiting the resulting truncated FoxH1 protein from mediating any Smad-transduced transcriptional responses (Figure 1A–1C).
Fig. 1. MZmid mutants lack notochords. 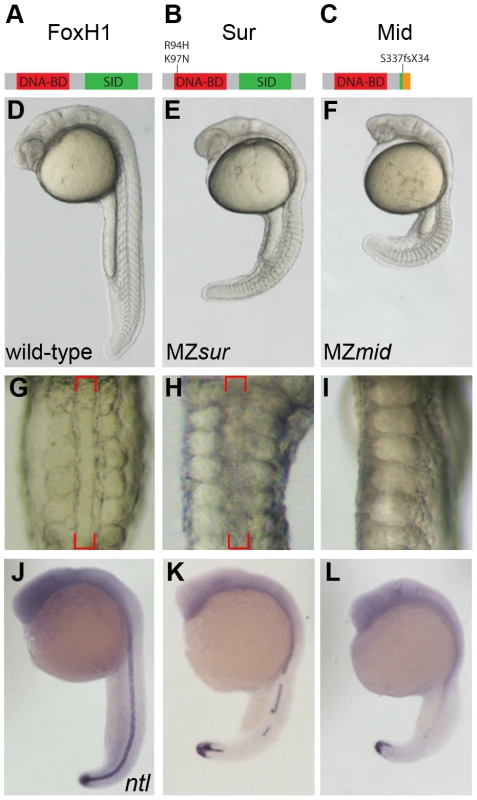
(A–C) Protein diagrams of FoxH1 alleles. (D–F) 24 hour post-fertilization (hpf) images of live wild-type and maternal-zygotic FoxH1 mutant embryos. (G–I) Dorsal zooms (2.5× relative magnification) of embryos in D–F at the level of the yolk extension. Red brackets mark the notochord; MZmid mutants lack this structure and exhibit midline-fused somites. (J–L) RNA in situ hybridization for ntl expression in 24 hpf wild-type and maternal-zygotic FoxH1 mutants. Axial mesoderm induction is differentially disrupted in FoxH1 mutants
FoxH1 is supplied to oocytes as a maternal mRNA, and MZsur embryos display stronger phenotypes than their zygotic counterparts (Zsur). These defects include variable deficiencies in axial mesoderm-derived tissues, particularly the notochord and prechordal plate, the latter leading to variable synopthalmia [15], [16]. We therefore wanted to compare the severity of MZmid phenotypes to the defects observed in MZsur mutants. To do so, we rescued mid homozygous embryos by FoxH1 mRNA injection and genotyped them as adults to verify the identities of the homozygous mutants. The genotyped mutants were then mated with each other to produce clutches consisting exclusively of MZmid embryos.
In contrast to the variable defects of MZsur mutants, MZmid embryos consistently display a highly penetrant absence of notochord and full cyclopia, hallmarks of Nodal signaling deficits (172/172; Figure 1D–1I). The loss of a morphological notochord is corroborated by a complete absence of the notochord marker ntl at 24 hpf in the midlines of MZmid mutants (71/72), whereas midline ntl expression was observed in a discontinuous pattern in a majority of MZsur embryos (35/51) (Figure 1J–1L), with the remainder exhibiting strong continuous midline ntl expression. Similar results were seen for other notochord markers, such as flh, sonic hedgehog (shh), and collagen2a (col2a) (data not shown).
To determine how early these defects were first apparent in MZmid mutants, we compared dorsal mesoderm marker expression in MZmid and MZsur mutants at 50% epiboly and 90% epiboly (Figure 2). At 50% epiboly, all MZsur and MZmid embryos display a significant thinning of the ntl-expressing dorsal margin (MZsur n = 81; MZmid n = 87) and a significant reduction of gsc expression (MZsur n = 60; MZmid n = 68) (Figure 2A–2I). However, at 90% epiboly, MZsur embryos show significant, though abnormal, midline expression of ntl (40/40) and gsc (34/34), whereas MZmid embryos almost completely lack midline ntl expression (63/63) and have less gsc expression (44/44) than MZsur (Figure 2M–2O). Other gastrulation-stage notochord markers, including axial, flh, shh, and lefty1, are also reduced in MZsur but almost completely absent in the midlines of MZmid (data not shown). Note that the endodermal marker cas is retained in both MZsur and MZmid (see below). These early phenotypes indicate a defect in the initial specification of the chordamesoderm in FoxH1 mutants. Therefore, our results not only strongly suggest that mid represents a stronger loss of FoxH1 function than sur, they also reveal an absolute requirement for FoxH1 in zebrafish axial specification.
Fig. 2. Nodal-dependent tissue specification is differentially disrupted in FoxH1 mutants. 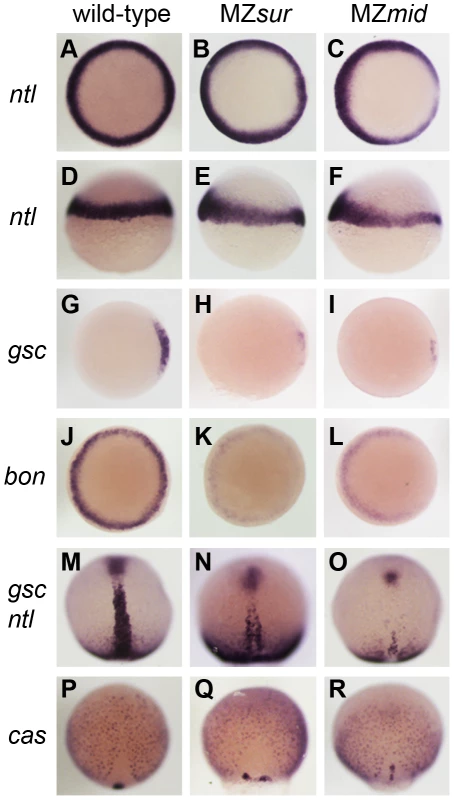
(A–L) Mesendoderm marker expression in wild-type and maternal-zygotic FoxH1 mutants at 40–50% epiboly. Dorsal is to the right; A–C, G–L are animal views, D–F are lateral views. Note the dorsal reduction of ntl expression in MZsur (E) and MZmid (F). (M–R) Axial mesoderm and endoderm marker expression at 90% epiboly, viewed dorsally with anterior up. FoxH1 mutants respond differently to Activin-like signaling
The presence of a notochord-like structure in MZsur mutants suggests that these embryos retain some ability to transduce Nodal signals in a FoxH1-dependent manner. To test this idea, we mated rescued sur homozygotes with rescued mid homozygotes. Regardless of whether the male or female adult was the sur mutant, the progeny of these crosses largely resembled MZsur embryos (Figure 3A). About 80% of these embryos (n = 375) display a morphological structure similar to the irregular notochords of MZsur mutants, with the remainder lacking a recognizable midline structure. This result was also observed when MZmid embryos were injected with RNA encoding the sur allele of FoxH1 (80% MZsur phenotype, n = 30). These observations support the hypothesis that the sur mutation of FoxH1 represents a hypomorphic allele and not a null.
Fig. 3. sur retains more Nodal transduction capability than mid. 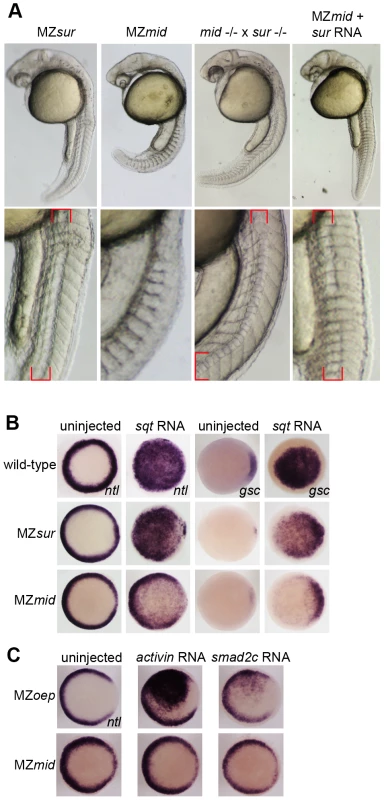
(A) Genetic interactions between the sur and mid alleles, demonstrating the ability of sur to partially rescue the loss of notochord caused by mid. Lower panels are enlargements showing the structures of the notochords at 24 hpf; red brackets indicate notochord domains. (B) Nodal overexpression in maternal-zygotic FoxH1 mutants. 50 pg sqt RNA was injected into wild-type, MZsur, and MZmid embryos at the one-cell stage, and embryos were assayed for Nodal target gene expression at 30–40% epiboly. Note the greater ability of MZsur embryos to respond to ectopic sqt compared to the MZmid response. (C) The mid mutation perturbs activin-like signaling. MZoep and MZmid embryos were injected with RNA encoding either a Xenopus activin homologue (2.5 pg) or an activated form of mouse Smad2 (100 pg). Responses were assayed by observing ntl expression at 30–40% epiboly. To directly test the ability of FoxH1 mutants to transduce Nodal signals, we performed an overexpression assay using RNA encoding the Nodal homologue squint (sqt; Figure 3B). Wild-type and mutant embryos were injected with 50 pg of sqt RNA at the one-cell stage and fixed at 30–50% epiboly to examine their ability to upregulate expression of ntl and gsc in a Nodal-dependent manner. Wild-type embryos responded by ubiquitously activating both ntl (81/81) and gsc (85/87) expression. MZsur embryos consistently upregulated both targets ubiquitously, similarly to wild-type (ntl 78/79; gsc 39/39). However, while ntl was activated throughout the animal pole in MZmid, the activation was much weaker than in either wild-type or MZsur, with the endogenous marginal expression pattern plainly visible (74/74). Upregulation of gsc was largely confined to the dorsal margin, where it is normally expressed, with little or no ventral or animal activation detected (32/37 dorsal expansion only, 4/37 weak animal activation, 1/37 no upregulation). These results suggest that while MZmid mutants are able to respond to ectopic Nodal, presumably through at least one other transcription factor, the response is much weaker and/or restricted to endogenous marker expression domains.
MZoep mutants, which lack the essential coreceptor of the Nodal pathway, have been reported to be entirely refractory to ectopically supplied Nodal. However, general Smad2/3-mediated TGFß signaling is attainable by bypassing the lack of the Nodal coreceptor. Injection of mRNA encoding either a Xenopus activin homologue (XactßB) or an activated truncation of mouse Smad2 (mSmad2c) into MZoep causes robust activation of the Nodal targets ntl and gsc, as all intracellular components of the pathway are intact [3]. Because the mid allele encodes a putative truncated form of FoxH1 that lacks a SID, we hypothesized that treatments eliciting a Nodal-like response in MZoep mutants would not be as effective in MZmid embryos. Indeed, the lowest doses of XactßB (2.5 pg) and mSmad2c (100 pg) we found to produce a reliable upregulation of ntl in MZoep mutants (66/71 and 8/21, respectively) had no effect in MZmid mutants (0/21 and 0/68, respectively; Figure 3C). A higher dose of XactßB did produce a response, but a much weaker one than observed in MZoep (data not shown). These results, together with the sqt injection data and the sur/mid trans-heterozygote phenotypes, indicate that mid encodes a stronger allele of FoxH1 than sur, and that it causes deficiencies in general Smad2/3-mediated TGFß signaling.
The above findings support the notion that the mid allele behaves in a recessive fashion and is a stronger loss-of-function allele than sur. However, if the mid locus is producing a signal transduction-incompetent truncation of the FoxH1 protein, it may act as a recessive antimorph, potentially by blocking promoter binding by other transcription factors with which FoxH1 shares targets. To directly test this potential dominant inhibition by Mid protein, we injected FoxH1, sur, or mid mRNA into wild-type embryos to observe the resulting overexpression defects. Phenotypes were categorized as mild, severe, or catastrophic and included defects in anterior and axial structures. (see Table S1 for more detailed descriptions, and Figure S1 for examples of the defects observed). Injection of 50 pg FoxH1 mRNA, five times the dose used to rescue sur or mid mutants, produced a range of defects, with only 25.6% of embryos (n = 297) appearing wild-type at 24 hpf. Injection of 50 pg sur mRNA yielded similar but somewhat stronger phenotypes, with only 17.6% of embryos (n = 136) appearing wild-type. In contrast, injection of 50 pg mid mRNA had a significantly weaker effect, with 85.6% of embryos (n = 167) appearing wild-type. These results suggest that, at identical doses, Mid protein overexpression has much less of an effect on development than either FoxH1 or Sur protein, and that physiological levels of Mid protein present in MZmid mutants are most likely not acting in a dominant fashion.
We also wished to determine if blocking production of Mid protein in MZmid mutants could alleviate any potential dominant effects of the mutant protein and produce embryos that resemble MZsur mutants. Alternatively, if Sur protein retains some partial signaling capability, then preventing its production should abrogate this function and produce embryos that resemble MZmid mutants. To block production of each of these mutant proteins, we injected MZsur and MZmid mutants with a morpholino targeting the translational start site of FoxH1 mRNA (gift of B. Feldman; [17]). However, the published phenotype caused by this morpholino is more severe than either of the two maternal-zygotic FoxH1 mutants. We therefore used a low dose of the morpholino to determine whether either of the mutants was sensitive to a partial depletion of mutant protein (Figure S2). Injection of 4 ng of FoxH1MO into MZsur mutants caused a significant increase in embryos lacking a notochord (115/187 injected vs. 10/155 uninjected). Injection into MZmid mutants never allowed formation of a structure resembling a notochord (0/182); in fact, a majority of these embryos (158/182) resembled their uninjected siblings. Together with the above overexpression data, these results strongly suggest that Mid protein does not act in a dominant fashion to disrupt axial development, and that Sur protein retains some capacity to transduce Nodal signals and allow for notochord formation.
Mid and Sur proteins differ in their DNA–binding abilities
The difference in allelic strength between mid and sur seems to contradict the current consensus that sur represents a null allele of FoxH1. Due to the location of the sur lesion, which is either of two amino acid substitutions at the N-terminal extreme of the Forkhead DNA-binding domain, it had been assumed that sur caused a total loss of DNA binding, leading to a complete failure of signal transduction through Sur protein [15], [16]. This assumption is supported by a failure of the sur mutation to activate a luciferase reporter driven by an activin response element [17]. However, the highly penetrant losses of notochord markers and structure caused by the mid mutation, and the partial rescue of these axial phenotypes by providing the sur allele in trans, suggest that Sur protein retains some ability to transduce Nodal signals, albeit below wild-type levels. We therefore investigated the DNA-binding capabilities of the wild-type and mutant versions of zebrafish FoxH1 protein.
We employed an electrophoretic mobility shift assay using a DNA probe derived from a putative FoxH1 binding site in the proximal promoter of the zebrafish gsc gene and in-vitro translated FoxH1, Sur, and Mid proteins N-terminally tagged with 6xHis and 3xHA epitopes (Figure 4A). Wild-type FoxH1 protein shifted the probe in a protein - and sequence-specific manner, as judged by effective competition by unlabeled probe, no competition by unlabeled mutated probe, and successful supershifting by an antibody against the HA epitope. However, Sur protein produced no detectable shift of the probe, suggesting that the sur mutation does indeed abolish DNA binding. Surprisingly, a qualitative analysis suggests that Mid protein binds to the probe much more strongly than wild-type protein. Because the mid mutation removes the C-terminal 25% of the FoxH1 polypeptide, we speculated that, in the absence of activated Smads in the nucleus, the C-terminus of wild-type FoxH1 may normally function to occlude the DNA-binding domain. If so, we hypothesized that the complete loss of binding we observed for the Sur protein may actually be a combination of two separate phenomena: a partial impairment in DNA binding caused by the sur mutation, and the wild-type C-terminal occlusion of the DNA-binding domain. Removal of the C-terminus from the Sur polypeptide may then reveal a weak DNA-binding ability of the Sur DNA-binding domain.
Fig. 4. DNA–binding activities of FoxH1 mutants. 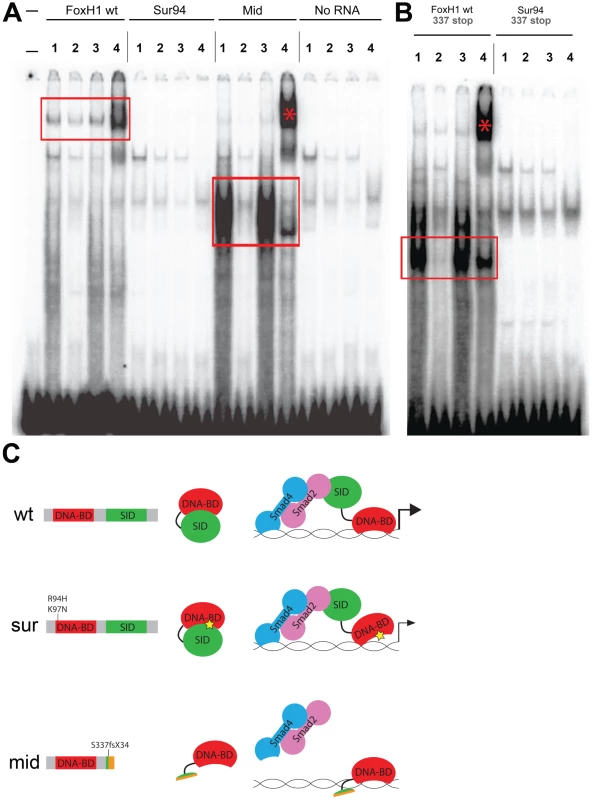
(A) Electrophoretic mobility shift assays using a FoxH1 binding site probe derived from the zebrafish gsc promoter and in vitro-translated epitope-tagged full-length proteins. FoxH1 and Mid protein bind the probe in a protein- and sequence-specific manner (red boxes and asterisk), while Sur protein shows no binding activity. Upper lane labels indicate the RNA translated for use in each binding reaction; individual lane numbers denote additions to the basic binding reactions (1: no additions; 2: 100-fold excess unlabeled competitor probe; 3: 100-fold excess mutated unlabeled competitor probe; 4: anti-HA antibody). (B) EMSAs using truncated FoxH1 and Sur proteins lacking the C-terminal SID. Truncated wild-type protein specifically binds the probe (red box), while truncated Sur protein does not. Lane markings are as described in (A) above. (C) Model for DNA-binding activities of wild-type and mutant FoxH1 proteins. Wild-type protein binds weakly to its recognition sites alone, but can bind strongly upon loss of its C-terminus, suggesting that Smad interaction may “open up” the conformation of the wild-type protein and allow for strong binding upon pathway activation. Sur protein is impaired in its DNA-binding ability, but may be weakly/transiently tethered to its recognition sequences by activated Smads or other unknown factors. Mid protein cannot interact with Smads and so cannot transduce Nodal signals, but can bind strongly to FoxH1 recognition sites. To determine whether the C-terminus of Sur protein was masking some partial ability of the mutated Forkhead domain to bind to its DNA recognition sequence, we generated C-terminally truncated versions of wild-type FoxH1 and Sur proteins (denoted by “337 stop”). We observed that the wild-type truncated protein now bound to the probe much more strongly than full-length FoxH1 and at a level comparable to Mid protein, supporting the idea that the C-terminus possesses some ability to partially inhibit DNA-binding by the Forkhead domain (Figure 4B). However, the truncated Sur protein still did not shift the probe, indicating that the sur mutation genuinely abolishes DNA-binding, even without a potential inhibitory activity from the C-terminus. This result is surprising given the weaker phenotypes and stronger ectopic-Nodal responses of MZsur compared to MZmid. However, it may be possible that interaction with activated Smads is enough to weakly tether the Sur protein to its target promoters long enough to activate some transcription (Figure 4C).
FoxH1 and Bon do not account for all zebrafish Nodal transduction
Since mid phenotypes more closely resemble those of FoxH1 loss in other organisms, we used MZmid and MZoep mutants as representatives to study the differences between loss of FoxH1 and loss of Nodal across species. MZmid embryos clearly develop more somites and are generally larger than MZoep mutants, and form a clear mid-hindbrain boundary (Figure 5B, 5F). Furthermore, early markers of endoderm specification that are lost in MZoep mutants are expressed in MZmid embryos: while significantly reduced, bon, the zebrafish Mixer homologue, is expressed prior to gastrulation (70/75), and casanova/sox32 (cas; n = 38), axial (n = 46), and sox17 (n = 34) are expressed abundantly in endoderm precursors of all late-gastrulation MZmid mutants analyzed (Figure 2L, 2R and data not shown). Because the oep mutation disrupts the Nodal pathway at a very early step in signal transduction (ligand-receptor binding) whereas the mid mutation affects a later step (transcriptional target activation), we wanted to confirm that the phenotypic differences between these two mutants was due to an otherwise intact Nodal pathway in MZmid embryos. We injected MZmid embryos with a mixture of morpholinos targeting the Nodal homologues sqt and cyclops (cyc) [24], which effectively phenocopy the defects of both MZoep mutants and cyc;sqt double mutants [3]. Knockdown of these Nodal homologues in MZmid efficiently phenocopies the MZoep defects (139/139), indicating that Nodal signals are active in MZmid (Figure 5E). Therefore, we hypothesized that at least one other transcription factor capable of transducing Nodal signals, presumably through interaction with activated Smad2/3, was present in MZmid embryos.
Fig. 5. Inhibiting Nodal signals in MZmid mutants. 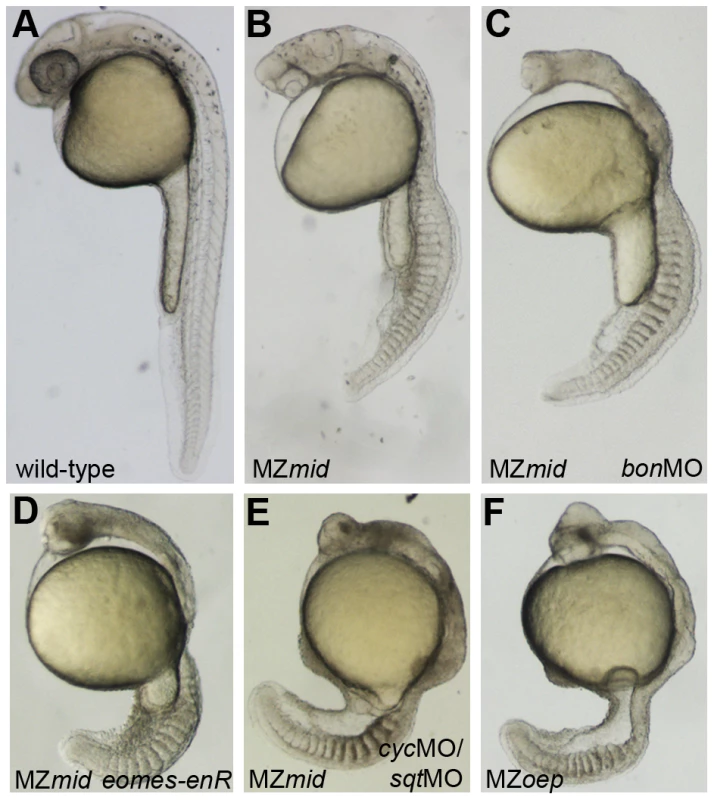
Wild-type (A), uninjected MZmid (B), and injected MZmid (C–E) embryos compared to a complete loss of Nodal signaling in MZoep (F) mutants. Inhibiting bon function (3 ng bonMO) in an MZmid background further impairs anterior development but does not affect tail development (C). Blocking eomes function (15–20 pg eomes-enR mRNA; D) resembles both a loss of the Nodal ligands cyc and sqt (8 ng each cycMO+sqtMO; E) and MZoep (F), indicating that Nodal signaling is occurring in MZmid mutants and is mediated by Eomes. Mixer/Bon has previously been proposed as a compensatory factor upon loss of FoxH1 function in MZsur mutants [18]. Bon was already known to be very important for proper endoderm specification in a Nodal-dependent manner [25], and could physically interact with phosphorylated Smad2 as part of the Nodal pathway [26]. However, loss of Bon function in MZsur mutants did not recapitulate the MZoep phenotype, leading to the assumption that another transcription factor was transducing Nodal signals in these embryos [18]. Since our results suggest that sur does not completely abolish FoxH1 function, it is possible that this previous study did not involve a complete loss of Nodal signaling through FoxH1 and Bon together. We therefore injected a morpholino targeting bon into MZmid mutants to attempt to phenocopy the MZoep phenotype. At a dose that reliably knocked down endoderm marker expression in wild-type embryos (data not shown), bonMO caused a further impairment in anterior neural patterning in MZmid mutants, as judged by the loss of a morphological mid-hindbrain barrier and a striking similarity to the anterior structures of MZoep embryos (77/79; Figure 5C). This effect was somewhat expected, given a prior study implicating bon in organizer specification, prechordal plate formation, and proper neural patterning [27]. However, outside of the head region, bonMO did not have much of an effect on a gross morphological level (77/79). The embryos developed the same numbers of somites and were generally the same size as their uninjected siblings. Therefore, in agreement with the aforementioned investigation into the interaction between the sur and bon mutations, we conclude that FoxH1 and Bon do not represent the entire complement of Nodal-transducing transcription factors in the early zebrafish embryo.
Eomes inhibition in MZmid recapitulates a total loss of Nodal signaling
An intriguing candidate for another Nodal responsive factor is Eomesodermin (Eomes), a T-box transcription factor repeatedly implicated in mesendoderm induction in several species. In Xenopus, Eomes is one of the earliest expressed mesoderm inducers, and its overexpression leads to ectopic activation of a number of Nodal signaling targets, such as Xbra (the Xenopus homologue of no tail) and gsc. Inhibition of Eomes causes gastrulation failure and mesoderm marker downregulation [28]. A more recent study showed that cooperative Nodal signaling and Eomesodermin function are required during Xenopus paraxial mesoderm induction [29]. In mouse, Eomes is required for proper prospective mesoderm ingression through the primitive streak and the consequent formation of the mesoderm germ layer [30], and also for the definitive endoderm lineage [31]. Zebrafish Eomes has similarly been implicated in the Nodal-dependent induction of dorsal mesoderm markers [32] and is required for endoderm specification [33]. Recently, Xenopus Eomes protein was shown to physically interact with phosphorylated Smad2, potentially placing it parallel to FoxH1 in the Nodal signaling pathway [34]. Eomes was therefore a prime candidate for mediating the Nodal-dependent mesendoderm induction observed in MZmid mutants.
In order to test this hypothesis, we wanted to inhibit Eomes function in an MZmid background. However, eomes morpholinos have limited effects on development, most likely due to the presence of maternally deposited Eomes protein in zebrafish oocytes [32]. We therefore employed a fusion of the DNA-binding domain of zebrafish EomesA and the transcriptional repressor domain of the Drosophila Engrailed protein (eomes-enR; gift of A. Bruce) in order to block endogenous Eomes function. This fusion was shown previously to inhibit dorsal mesoderm marker expression, and its effects on development could be rescued by coinjection of eomesA mRNA [32]. When injected into MZmid mutants at the one-cell stage, eomes-enR caused an impairment of anterior neural development similar to inhibition of bon function (78/122; Figure 5D). However, in contrast to bonMO, which had no discernible effect outside of the head region, injection of eomes-enR also caused a significant reduction in embryo length with a coincident decrease in somite number. These embryos closely resembled the MZoep phenotype at 24 hpf (84/122), suggesting that Eomes is, in fact, responsible for Nodal signaling functions observed in MZmid mutants.
To validate this result, we examined expression of a number of mesendoderm markers in MZmid mutants injected with eomes-enR (Figure 6). Repression of Eomes function in an MZmid background appears to exacerbate the preexisting dorsal mesoderm marker reduction at 50% epiboly. Residual dorsal ntl and flh expression in MZmid (38/38 and 38/38, respectively) were further downregulated by eomes-enR (51/51 and 47/51, respectively), whereas expression of these markers in wild-type embryos was only mildly affected, if at all, upon Eomes inhibition alone (11/54 and 7/33 with mild reductions, respectively). Intriguingly, blocking Eomes function in both wild-type (56/56) and MZmid (53/53) embryos strongly reduces or completely abolishes bon expression, whereas MZoep mutants exhibit very weak expression of bon prior to gastrulation (30/35; arrowheads in Figure 6), as has been reported previously [18], [35]. This result indicates that Eomes is required to initiate bon expression and may do so in a Nodal-independent manner. Importantly, we observed that repression of Eomes function did not affect the residual expression of cyc or sqt in MZmid mutants (Figure S3). All MZmid mutants analyzed showed marginal expression of cyc (59/59) and sqt (53/53) at 30–40% epiboly, and injection of the eomes-enR mRNA into MZmid did not perturb expression of either Nodal homologue (70/70 and 54/54 with unaffected cyc and sqt expression, respectively). While these results do not address a potential ability of Eomes to regulate Nodal expression during later stages of development, they indicate that Eomes is not involved in regulation of cyc or sqt during Nodal-dependent mesendoderm induction, as FoxH1 is [10], [15], [36]–[38].
Fig. 6. Eomes inhibition enhances the Nodal signaling and mesendoderm deficiencies of MZmid. 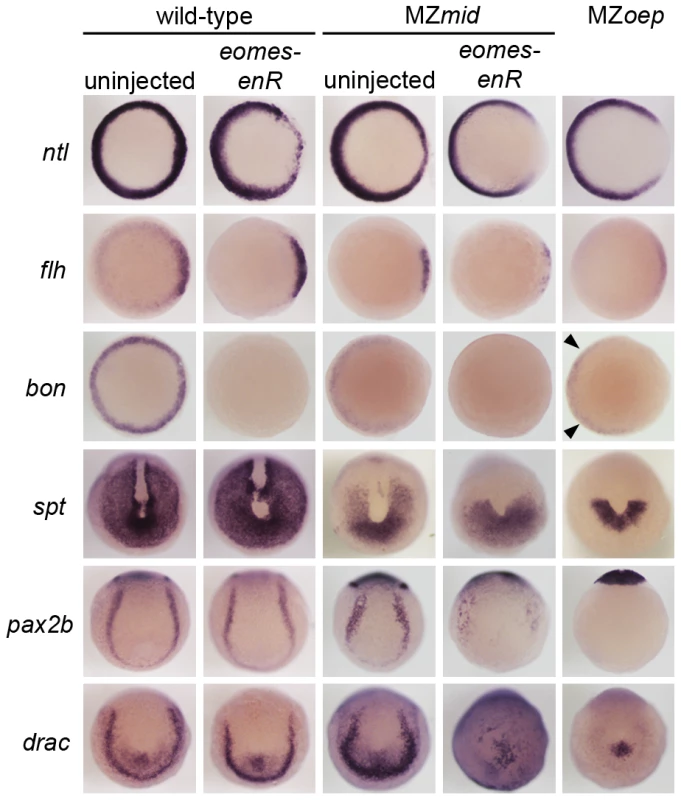
Markers for various populations of mesendoderm derivatives were analyzed in wild-type and MZmid embryos in which Eomes function was inhibited. Eomes inhibition in wild-type embryos has a minimal effect on expression of most markers, whereas in an MZmid background it has significant effects on mesendoderm-derived tissues, approaching MZoep levels for most markers. Arrowheads indicate Nodal-independent expression of bon in MZoep mutants. Because Eomes can regulate certain Nodal transcriptional targets, we investigated whether FoxH1 and Eomes shared roles in the morphological development of dorsal mesoderm-derived structures. First, MZmid mutants were injected with mRNA encoding a translational fusion of the Eomes DNA-binding domain and the VP16 transcriptional activator domain (gift of A. Bruce, [32]). This fusion was previously shown to upregulate several Nodal targets in a Nodal-dependent manner. When injected into MZmid mutants, eomes-VP16 caused a significant rescue of notochord development (Figure S4A; 102/201). While this result suggests that FoxH1 and Eomes both contribute to notochord development, it should be noted that this effect was observed upon ubiquitous overexpression of a constitutively active form of Eomes. To test whether Eomes normally contributes to notochord development, we injected wild-type embryos with eomes-enR. Importantly, we very rarely observed a loss of notochord in wild-type embryos injected with eomes-enR (Figure S4B; 2/156 with no notochord), although these embryos do display other severe morphological defects. This result demonstrates a certain level of specificity of the Eomes fusion proteins, as other T-box functions, like that of no tail in notochord development [39], are not disrupted by eomes-enR. It also suggests that properly regulated Eomes protein, expressed at endogenous locations and levels, does not play a significant role in notochord specification.
In addition to dorsal mesoderm and endoderm induction, which are well-known Nodal-dependent processes, other mesoderm-derived tissues present in MZmid but reduced or absent in MZoep are affected by Eomes repression (Figure 6). Because we observed a reduction in somite number in MZmid embryos injected with eomes-enR, we examined the expression of tbx16/spadetail (spt), a T-box factor known to be required for proper somitogenesis, especially for formation of the anterior (trunk) somites [40]. At the end of gastrulation, spt is expressed in the presomitic mesoderm at the vegetal pole of the embryo and excluded from the dorsal axis. MZmid mutants display a partial reduction in the spt expression domain, which resembles a vegetal “U” with expression reaching into the dorsal side of the embryo (53/53). However, eomes-enR further restricted the expression domain of spt (40/52) to one resembling that observed in MZoep embryos, which express spt only in a semicircular band of cells at the ventrovegetal extreme of the embryo (36/36). This further reduction of spt expression likely explains the significant loss of anterior somites observed later in MZmid mutants injected with eomes-enR. A similar reduction of expression is observed for pax2.1 (56/62) and draculin (39/39), markers of the intermediate mesoderm and blood precursors, respectively, which appear wild-type in uninjected MZmid embryos (65/65 wild-type for pax2b; 36/36 wild-type for draculin). While Eomes inhibition in wild-type embryos had little effect on expression of these markers (78/78 wild-type for pax2b; 47/47 wild-type for draculin), it drastically reduced their expression in an MZmid background. Based on the morphological and marker expression phenotypes in MZmid mutants upon inhibition of Eomes function, we propose that Eomes is the transcription factor responsible for the Nodal-dependent mesendoderm specification observed in embryos lacking Nodal transduction through FoxH1.
Discussion
Transcriptional regulation of Nodal signaling
The ability of many intercellular signaling pathways to function effectively depends on the transcription factors that regulate target gene expression. Our investigation into the early roles of Nodal signaling reveals distinct and biologically separable functions for the Nodal pathway during zebrafish development, contrary to the current understanding of Nodal-dependent mesendoderm specification. Through identifying a novel mutation in the Nodal signaling effector FoxH1 that removes its ability to bind to the Smad intracellular Nodal effectors, we have shown that certain functions of the pathway can be blocked without overtly perturbing its other roles. This functional segregation is achieved, at least in part, by the use of multiple transcription factors capable of interacting with Smad2 upon stimulation of the pathway (Figure 7). FoxH1 appears to be absolutely required for formation of the notochord and the most anterior trunk somites. Eomesodermin is responsible for much of the nonaxial mesoderm specification observed in MZmid mutants, although in wild-type embryos Eomes and FoxH1 cooperate in this function, since loss of either protein alone does not appreciably affect most nonaxial mesoderm marker expression. Eomes is also required for at least two steps of endoderm specification: the initiation of bon expression (Figure 6) and the assembly of a transcriptional activator complex, composed of Eomes, Bon, and Gata5, on the cas promoter [33]. Despite these partially separable roles of the Nodal responsive transcription factors, their activities appear to converge at the shield/organizer, which will later form the prechordal plate. All three proteins contribute to marker expression at the shield and to subsequent function of the prechordal plate in neural development, based on the loss of morphological neural structures (such as the mid-hindbrain boundary) in pairwise versus single loss of function situations (Figure 5). It is unclear whether Eomes' contribution to prechordal plate specification derives from direct target activation or through its activation of Bon. However, given the relatively minor neural patterning and morphology defects in bon mutants [27], and Eomes' reported ability to induce entire secondary axes when overexpressed in zebrafish [32], it is highly likely that it is making direct contributions to shield formation and function.
Fig. 7. Model for roles of Nodal signaling in early zebrafish embryos. 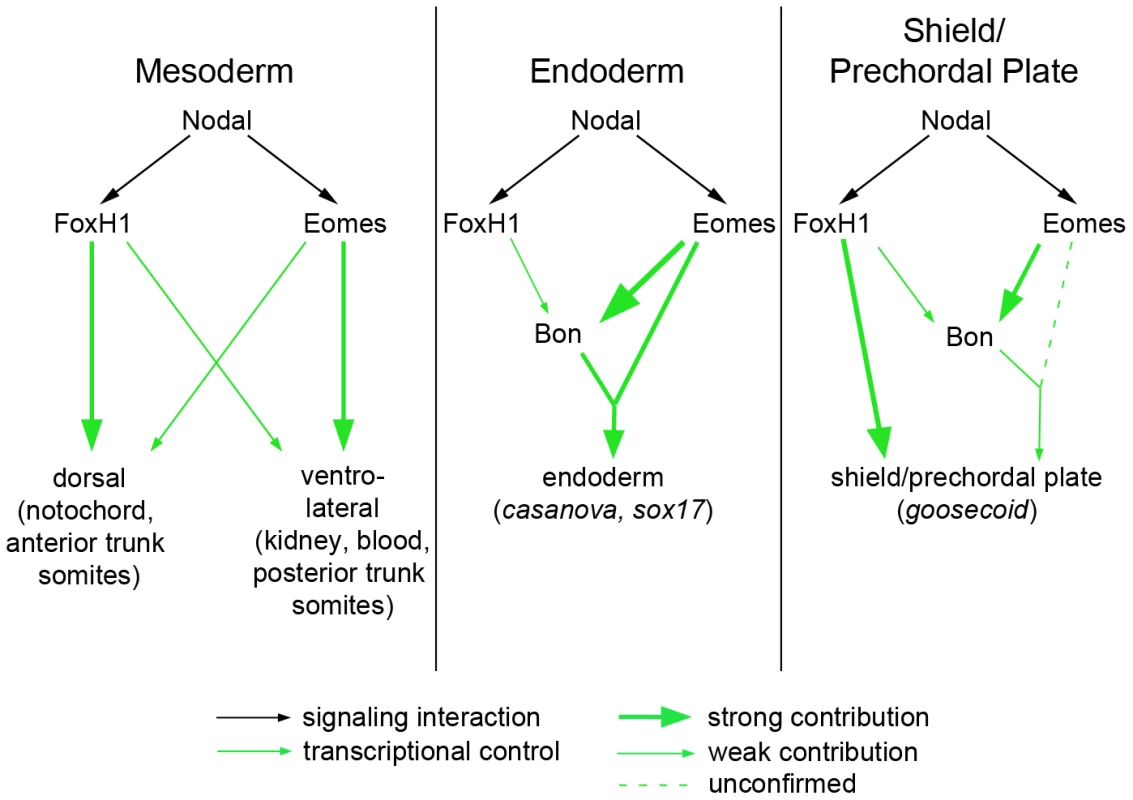
Nodal signaling functions are partially separable according to the transcription factors used by the pathway. (A) FoxH1 is absolutely required for notochord specification and for the most anterior trunk somites, while Eomes is essential for nonaxial mesoderm induction upon loss of FoxH1 function. These two factors may cooperate in a wild-type embryo for normal nonaxial mesoderm induction, as loss of either alone does not have a significant impact on early marker expression (Figure 6). (B) FoxH1 is not strictly required for the endoderm specification pathway, although it makes contributions to bon expression. Eomes is required both for bon initiation and for proper downstream endoderm marker expression, as has been demonstrated previously. (C) In the organizer, FoxH1, Eomes, and Bon function meet for full induction of marker expression and prechordal plate derivation. DNA–binding capabilities of wild-type and mutant FoxH1 proteins
We have also demonstrated that the previously characterized sur allele is not a complete loss of function, contrary to current assumptions. Based on the two mutants' phenotypes and relative abilities to transduce Nodal signals, we find that sur retains some in vivo activity and therefore represents a less deleterious mutation of FoxH1 than mid. Surprisingly, despite the evidence supporting sur's partial functionality, we find that the sur mutation genuinely abolishes binding to a target gene reporter element in a gel shift assay. If Sur protein cannot bind its target regulatory sequences in the genome, how can it activate target genes in response to Nodal? One possibility is that its interaction with activated Smads in the nucleus is enough to allow it to transiently interact with its target sequences through the weak DNA-binding ability of Smad4 (Figure 4C). In a similar vein, it should be noted that the broader environment of FoxH1 binding sites in the genome has not been well studied; there may be other factors with which FoxH1 normally interacts at its target sites to efficiently activate transcription in vivo. Sur protein may still be able to interact with these hypothetical factors, allowing for weak tethering of an otherwise active Sur-Smad complex to its genomic binding sites.
The observation that in vitro translated Mid protein can bind to a FoxH1 target probe, and with apparently greater affinity than full-length FoxH1, might suggest that the Mid protein could possess dominant negative or recessive antimorphic functions. A mutated transcription factor with tighter binding to its consensus site that is unable to carry out target gene transcription could theoretically compete with wild-type protein for consensus occupation. However, several of our findings indicate this is not the case. mid heterozygous fish are viable and produce phenotypically mutant progeny at a typical Mendelian frequency, and MZmid defects can be partially rescued by sur RNA injection. Furthermore, overexpression of mid mRNA results in developmental defects much less frequently than either FoxH1 or sur RNA, and morpholino knockdown of any endogenous Mid protein in MZmid mutants does not rescue notochord formation. Finally, we do not see alterations in the expression domains of certain genes whose promoters are bound by the FoxH1 protein, including cyc, sqt, and nonaxial ntl, arguing that the mutant FoxH1 protein in midway is not blocking these promoters. Given these results, dominant-negative and recessive antimorphic activities for Mid protein seem highly unlikely. Indeed, documented instances of inhibitory functions for FoxH1 involve recruitment of co-inhibitors. The Forkhead domain of mouse Foxh1 recruits Gsc to the murine Mixl1 promoter, allowing for proper histone deacetylase-mediated repression of the Mixl1 locus during early development [41]. Foxh1 can also inhibit the transcription of gsc itself by incorporating Smad3, rather than Smad2, into a regulatory complex at the murine gsc promoter in luciferase assays [42]. Therefore, it is possible that the mere presence of non-functional Mid protein at a FoxH1 target promoter does not strictly imply inhibition of that promoter. It seems more likely, based on our in vivo results, that an activated FoxH1-Smad2/4 complex has a higher affinity for its target promoters than do the truncated Mid protein or the full-length wild-type FoxH1 protein alone.
Our investigation into the biochemical properties of the wild-type and mutant FoxH1 proteins revealed a new potential regulatory mechanism for FoxH1 transcriptional activity. Removal of the C-terminal Smad interaction domain, either through artificial truncation or through the midway frameshift, allows qualitatively enhanced DNA binding of the truncated protein relative to the full-length wild-type protein. This apparent autoinhibitory function of the FoxH1 C-terminus is not an unprecedented phenomenon. Several transcription factors display autoinhibition activities between their DNA-binding domains and other regions of their polypeptides, including the Ets-1 protooncoprotein [43]–[46] and the cell-cycle transcription factor Swi4 [47]. In fact, the mechanism of Swi4 autoinhibition relief may be similar to what we propose for FoxH1. C-terminal occlusion of the Swi4 DNA binding domain is interrupted by interaction with Swi6, much as Smad2/4 binding to the FoxH1 C-terminus may serve to “open up” the polypeptide to reveal the Forkhead DNA-binding domain. A detailed structural and biochemical analysis of this autoinhibition activity will shed light on this potential mode of FoxH1 transcriptional regulation.
Potential Nodal-independent roles for FoxH1 in early development
A recent study investigated the functions of FoxH1 in early zebrafish development by use of morpholinos to knock down FoxH1 translation [17]. Their results indicated a very early, and potentially Nodal-independent, role for FoxH1 during epiboly. High doses of a FoxH1 translation-blocking morpholino caused embryos to developmentally freeze at about 50% epiboly, remaining temporally halted while their control-injected siblings progressed relatively normally through gastrulation and somitogenesis. The highly penetrant MZmid phenotype we have described here, which closely resembles loss of FoxH1 function in other organisms, is not nearly as severe as that of FoxH1 morphants. However, while the mid mutation removes FoxH1's ability to transduce Nodal signals via Smad interaction, it is possible that the Forkhead DNA-binding domain retains some function in MZmid embryos that is lost in the FoxH1 morphants. Indeed, certain studies have described Nodal-independent functions for the FoxH1 Forkead domain in other organisms. During anterior heart field development in mouse, the Forkhead domain of FoxH1 was shown to bind to Nkx2.5, and this interaction was required to fully activate a mef2c transcriptional reporter [48]. In light of this observation, and the aforementioned interaction with Gsc at the mouse Mixl1 promoter, it is possible that the Forkhead DNA-binding domain of FoxH1 retains some early developmental functions in MZmid embryos that are blocked by morpholino knockdown. Indeed, we occasionally observed a midline bifurcation phenotype in MZmid embryos injected with a low dose of FoxH1 MO (4/182). This defect is reminiscent of the low-penetrance midline bifurcation phenotype observed in MZsqt embryos [49] and ethanol-induced midline bifurcations in wild-type zebrafish embryos [50], both of which are attributed to impairments in gastrulation movements during early development. Given the Nodal-independent inhibition of cell motility described in the original FoxH1MO study, it seems likely that the Mid protein (i.e. the FoxH1 DNA-binding domain) can still act to promote cellular movements at the start of gastrulation. In further support of the Mid protein having some functionality, we also observed a few MZmid;FoxH1MO embryos that developed two eyes (5/182; see Figure S2 caption), a phenotype we never see in MZmid mutants. Therefore, we postulate that the Mid protein can still function endogenously to repress some targets that affect prechordal plate formation. In fact, it is known that mouse Foxh1, in cooperation with Gsc, can repress mixl1 (the mouse bon homologue). Normally this repression would occur simultaneously with strong Nodal signaling to allow for prechordal plate formation. It is intriguing to speculate that the Mid protein still binds to and represses this promoter in zebrafish, in cooperation with the small amount of Gsc being produced, and that this repression is not overcome since Nodal signaling is impaired. Upon FoxH1MO injection into MZmid mutants, this endogenous function of the FoxH1 DNA-binding domain is blocked, allowing for enough prechordal plate function to split the eye field on rare occasions. For these reasons, we believe that the mid allele is not a complete FoxH1 null mutation, but one that eliminates a specific function of the protein (the Smad-mediated Nodal signal transduction) while leaving other endogenous functions intact. Investigation into the phenotypic differences between MZmid mutants and FoxH1 morphants could uncover further novel roles for this transcription factor during early development. Thus, the midway mutation will serve as an important tool for understanding the Nodal dependent and independent roles of FoxH1 in development.
Evolution of the notochord specification program
The strict requirement of FoxH1 for notochord development, as observed in MZmid mutants, may shed some light on a long-standing evolutionary question: how did the notochord genetic program evolve? Homologues of many known players in notochord specification and development have been identified outside of the chordate lineage. Nodal pathway components are found in non-chordate deuterostomes [51], [52] and non-deuterostome bilaterians [53], and homologues of brachyury/ntl have been found in protostomes [54] and possibly in the last common ancestor of bilateria and sponges [55], [56]. Therefore, these factors alone cannot account for the evolution of the notochord, indicating that other evolutionary innovations must have arisen in order to harness this machinery for use in notochord development. Intriguingly, although the Forkhead superfamily is represented in all animal phyla [56] and even in fungi [57], members of the FoxH subfamily have not been identified outside of the chordate subgroups of the deuterostome lineage [56], [58]. FoxH homologues have been discovered in the tunicate Ciona intestinalis [58] and the cephalochordate Amphioxus [59], but are absent in echinoderm genomes [52], [60], suggesting that this Forkhead subfamily could have evolved concomitantly, and perhaps causally, with the chordate lineage. Indeed, a recent analysis of the regulation of “notochord-specific” genes in Ciona indicates that a cohort of these genes falls outside the transcriptional control of Brachyury [61]. It is tempting to consider that some of these genes, which may be essential for the formation of the notochord in Ciona and chordates, could be transcriptionally regulated by FoxH homologues.
This proposed recent evolution of the FoxH subfamily may help explain an apparent discrepancy between the MZmid phenotypes and the current model for Nodal-mediated mesendoderm induction. A host of experiments in various species [62] indicates that high levels of Nodal signals are required for endoderm and prechordal plate specification, tissues which are most sensitive to partial reduction of Nodal signaling. Lower levels are sufficient for specification of dorsal mesoderm fates such as notochord, which are relatively unperturbed upon partial loss of Nodal signals. In conflict with this model, MZmid embryos consistently lack notochords, as is seen in Foxh1 knockout mice [8], [10], while prechordal plate expression of gsc is reduced but present and early endoderm specification appears largely unaffected. If FoxH1 evolved relatively recently in deuterostomes, its unique function in notochord formation would have been superimposed upon the preexisting dose-dependent functions of the Nodal pathway in the differential specification of mesendoderm. Perhaps this late addition of the notochord genetic program causes it to fall outside of the Nodal dose-response model of mesendoderm induction. In chordates, then, endoderm and nonaxial mesoderm would be induced by higher and lower levels of Nodal signals, respectively, with FoxH1 serving mainly to properly modulate expression levels via Nodal feedback loops. The newly evolved axial mesoderm domain, however, would form through direct activation by FoxH1 of a notochord-specific gene network in a particular location of the embryo, relying on this specific activity of FoxH1 rather than the preexisting Nodal dose-response mechanism. This evolutionary layering of Nodal signaling responses in chordates may have been further complicated by divergences among the different vertebrate lineages, as loss of Foxh1 function in mice does lead to a failure of definitive endoderm induction [8], [10].
In sum, our current work demonstrates that FoxH1 plays a required conserved role in notochord specification through characterization of the novel FoxH1 mutant mid. We also identify Eomes as a Nodal-transducing factor that acts, directly and indirectly, in concert with FoxH1 to carry out all Nodal-dependent processes during early mesendoderm specification. Our results provide novel insights into the previously unappreciated separation of roles of the Nodal pathway based on the transcription factors available for signal transduction. We also shed light on the evolution of the genetic program that leads to development of the chordate animal lineage.
Materials and Methods
Ethics statement
All protocols for the care and use of zebrafish were approved by Princeton's Institutional Animal Use and Care Committee and the University Veterinarian.
Zebrafish strains
FoxH1m768 and oeptz257 were generated in the Boston and Tübingen ENU mutagenesis screens, respectively [63], [64]. FoxH1Pr1 was isolated as a spontaneous mutation by R. Burdine. Fish strains were maintained by outcrossing to various wild-type strains, including AB and WIK, and recessive pigment mutants including alb, gol, and leo.
Mapping, sequencing, and genotyping the midway locus
The mid locus was mapped to a 10 cM region of chromosome 12, first using a genome-wide panel of SSLP markers for low-resolution mapping and then using additional selected SSLP markers for finer mapping [23]. This region, defined by markers z27025 and z11549, covered the FoxH1 locus, prompting complementation matings between sur and mid heterozygotes. Upon complementation failure, the FoxH1 locus was sequenced in mid homozygotes, and a two-nucleotide insertion was discovered after nucleotide 1007 of the open reading frame.
A three-nucleotide deletion in the mid 3′UTR was used as a RFLP for genotyping purposes. A 324-bp region was amplified using the following primers: forward 5′ - CCAGTATGCCCTACAGAACGGACCTTCCC-3′; reverse 5′-CTGTACAACAGCTTGTTGCCAGGGC-3′. This product was digested with BtsCI, which cuts only the mutant fragment to produce a 200-bp band upon 3% agarose gel electrophoresis.
Plasmid construction
The complete FoxH1 cDNA clone was obtained from American Type Culture Collection (GenBank ID BC044340.1). The FoxH1 cDNA was subcloned into pCS2+ using the flanking XhoI sites to make the pCS2-FoxH1 expression plasmid. For the related sur and mid expression plasmids, the surm768 and mid mutations were engineered into pCS2-FoxH1 using the Stratagene QuikChange II Site-Directed Mutagenesis Kit (Agilent #200523) and verified by sequencing. To make the epitope-tagged expression constructs for EMSAs, the FoxH1, sur, and mid ORFs were PCR-amplified from the pCS2 expression vectors and ligated into a modified pET15b vector containing an expanded multiple cloning site and three copies of the hemagglutinin epitope tag in-frame downstream of the 6xHis sequence. The entire 6xHis-3HA-FoxH1 cassette of each pET15b plasmid was then PCR-amplified and ligated into pCS2. The mid ORF contained the entire FoxH1 ORF with the mid AT insertion; plasmids containing this ORF produced a polypeptide that was of the predicted smaller size compared to the wild-type and sur plasmids upon in vitro transcription/translation and Western blotting. Truncated FoxH1 and Sur expression plasmids were created by PCR-amplifying the corresponding ORFs from the full-length constructs using a reverse primer that replaced the serine at codon 337 with a stop codon (e.g. FoxH1-337X).
Whole-mount in situ hybridization
Antisense RNA probes were transcribed from linearized plasmid templates using DIG-labeled nucleotides and used in a standard protocol for whole-mount in situ hybridization [65]. Probes used were no tail [66], goosecoid [67], bonnie and clyde/mixer [68], axial/FoxA2/HNF3ß [69], casanova/sox32 [70], floating head/Znot [71], spadetail/tbx16 [40], pax2.1 [72], draculin [73], sonic hedgehog [74], lefty1/antivin [75], squint/ndr1 [2], [76], cyclops/ndr2 [77], southpaw/ndr3 [19], sox17 [35], and collagen2a [78].
Microinjections
In vitro-transcribed mRNAs were generated from linearized plasmid templates using the mMessage mMachine SP6 transcription kit (Ambion #AM1340). Template plasmids were: pCS2-FoxH1, pCS2-sur, pCS2-mid, pCS2-6xHis-3HA-FoxH1, pCS2-6xHis-3HA-sur (see Plasmid Construction above), pCS2-squint [2], pSP64T-XactßB [79], pCS-cytßgal-madr2(C) [80], and pENG-N-eomes and pVP16-N-eomes [32]. sur and mid homozygous embryos were rescued by injection of 10 pg of FoxH1 mRNA transcribed from pCS2-FoxH1, raised to adulthood, and genotyped as described above. Morpholinos for FoxH1, bonnie and clyde, squint, and cyclops were described previously [17], [18], [24], [27], [81], [82]. mRNAs and morpholinos were diluted in 10 mg/mL Phenol Red and injected in 500 pL drops into the yolks of 1–4 cell stage embryos.
Microscopy
Live embryo and in situ hybridization images were captured at 4× or 10× magnification using a ProgressC14 digital camera (Jenoptik) on a Leica MZFLIII microscope.
Electrophoretic mobility shift assays
Probe synthesis and labeling were performed as described [83]. The double-stranded FoxH1 binding site probe was derived from a 36-base pair sequence centered around a putative FoxH1 binding site in the zebrafish goosecoid proximal promoter (wild-type: 5′-TCAAATTAATTCTCAATACACAGATCGGTGGTTTTC-3′; mutant: 5′-TCAAATTAATTCTCAAGACCCAGATCGGTGGTTTTC-3′; underlined bases denote FoxH1 binding site).
pCS2-6xHis-3HA plasmids (see Plasmid construction above) were linearized with Asp718I and transcribed using SP6 RNA polymerase and a ribonucleotide mixture containing 7-methyl guanosine. mRNAs were subsequently used in in vitro translation reactions using a rabbit reticulocyte lysate system (Promega). Relative amounts of translated protein in each reaction were determined by Western blotting for the HA epitopes, and equal amounts of proteins were used in binding reactions as described [83] with the addition of 50 ng/µL each of poly(dI-dC)/poly(dI-dC) and poly(dA-dT)/poly(dA-dT).
Supporting Information
Zdroje
1. WengWStempleDL 2003 Nodal signaling and vertebrate germ layer formation. Birth Defects Res C Embryo Today 69 325 332
2. FeldmanBGatesMAEganESDouganSTRennebeckG 1998 Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature 395 181 185
3. GritsmanKZhangJChengSHeckscherETalbotWS 1999 The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 97 121 132
4. SchierAF 2009 Nodal morphogens. Cold Spring Harb Perspect Biol 1 a003459
5. ShenMM 2007 Nodal signaling: developmental roles and regulation. Development 134 1023 1034
6. ChenXRubockMJWhitmanM 1996 A transcriptional partner for MAD proteins in TGF-beta signalling. Nature 383 691 696
7. WhitmanM 2001 Nodal signaling in early vertebrate embryos: themes and variations. Dev Cell 1 605 617
8. HoodlessPAPyeMChazaudCLabbeEAttisanoL 2001 FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev 15 1257 1271
9. KofronMPuckHStandleyHWylieCOldR 2004 New roles for FoxH1 in patterning the early embryo. Development 131 5065 5078
10. YamamotoMMenoCSakaiYShiratoriHMochidaK 2001 The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev 15 1242 1256
11. HowellMInmanGJHillCS 2002 A novel Xenopus Smad-interacting forkhead transcription factor (XFast-3) cooperates with XFast-1 in regulating gastrulation movements. Development 129 2823 2834
12. HammerschmidtMPelegriFMullinsMCKaneDABrandM 1996 Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development 123 143 151
13. SchierAFNeuhaussSCHarveyMMalickiJSolnica-KrezelL 1996 Mutations affecting the development of the embryonic zebrafish brain. Development 123 165 178
14. Solnica-KrezelLStempleDLMountcastle-ShahERanginiZNeuhaussSC 1996 Mutations affecting cell fates and cellular rearrangements during gastrulation in zebrafish. Development 123 67 80
15. PogodaHMSolnica-KrezelLDrieverWMeyerD 2000 The zebrafish forkhead transcription factor FoxH1/Fast1 is a modulator of nodal signaling required for organizer formation. Curr Biol 10 1041 1049
16. SirotkinHIGatesMAKellyPDSchierAFTalbotWS 2000 Fast1 is required for the development of dorsal axial structures in zebrafish. Curr Biol 10 1051 1054
17. PeiWNoushmehrHCostaJOuspenskaiaMVElkahlounAG 2007 An early requirement for maternal FoxH1 during zebrafish gastrulation. Dev Biol 310 10 22
18. KunwarPSZimmermanSBennettJTChenYWhitmanM 2003 Mixer/Bon and FoxH1/Sur have overlapping and divergent roles in Nodal signaling and mesendoderm induction. Development 130 5589 5599
19. LongSAhmadNRebagliatiM 2003 The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 130 2303 2316
20. BisgroveBWEssnerJJYostHJ 2000 Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development 127 3567 3579
21. BrandMHeisenbergCPWargaRMPelegriFKarlstromRO 1996 Mutations affecting development of the midline and general body shape during zebrafish embryogenesis. Development 123 129 142
22. ChenJNvan EedenFJWarrenKSChinANusslein-VolhardC 1997 Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 124 4373 4382
23. LiaoECZonLI 1999 Simple sequence-length polymorphism analysis. Methods Cell Biol 60 181 183
24. FanXHagosEGXuBSiasCKawakamiK 2007 Nodal signals mediate interactions between the extra-embryonic and embryonic tissues in zebrafish. Dev Biol 310 363 378
25. KikuchiYTrinhLAReiterJFAlexanderJYelonD 2000 The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev 14 1279 1289
26. RandallRAGermainSInmanGJBatesPAHillCS 2002 Different Smad2 partners bind a common hydrophobic pocket in Smad2 via a defined proline-rich motif. EMBO J 21 145 156
27. TrinhLAMeyerDStainierDY 2003 The Mix family homeodomain gene bonnie and clyde functions with other components of the Nodal signaling pathway to regulate neural patterning in zebrafish. Development 130 4989 4998
28. RyanKGarrettNMitchellAGurdonJB 1996 Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell 87 989 1000
29. FukudaMTakahashiSHaramotoYOnumaYKimYJ 2010 Zygotic VegT is required for Xenopus paraxial mesoderm formation and is regulated by Nodal signaling and Eomesodermin. Int J Dev Biol 54 81 92
30. RussAPWattlerSColledgeWHAparicioSACarltonMB 2000 Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404 95 99
31. ArnoldSJHofmannUKBikoffEKRobertsonEJ 2008 Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135 501 511
32. BruceAEHowleyCZhouYVickersSLSilverLM 2003 The maternally expressed zebrafish T-box gene eomesodermin regulates organizer formation. Development 130 5503 5517
33. BjornsonCRGriffinKJFarrGH3rdTerashimaAHimedaC 2005 Eomesodermin is a localized maternal determinant required for endoderm induction in zebrafish. Dev Cell 9 523 533
34. PicozziPWangFCronkKRyanK 2009 Eomesodermin requires transforming growth factor-beta/activin signaling and binds Smad2 to activate mesodermal genes. J Biol Chem 284 2397 2408
35. AlexanderJStainierDY 1999 A molecular pathway leading to endoderm formation in zebrafish. Curr Biol 9 1147 1157
36. NorrisDPBrennanJBikoffEKRobertsonEJ 2002 The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development 129 3455 3468
37. OsadaSISaijohYFrischAYeoCYAdachiH 2000 Activin/nodal responsiveness and asymmetric expression of a Xenopus nodal-related gene converge on a FAST-regulated module in intron 1. Development 127 2503 2514
38. SaijohYAdachiHSakumaRYeoCYYashiroK 2000 Left-right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol Cell 5 35 47
39. HalpernMEHoRKWalkerCKimmelCB 1993 Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell 75 99 111
40. GriffinKJAmacherSLKimmelCBKimelmanD 1998 Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development 125 3379 3388
41. IzziLSilvestriCvon BothILabbeEZakinL 2007 Foxh1 recruits Gsc to negatively regulate Mixl1 expression during early mouse development. EMBO J 26 3132 3143
42. LabbeESilvestriCHoodlessPAWranaJLAttisanoL 1998 Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell 2 109 120
43. GravesBJCowleyDOGoetzTLPetersenJMJonsenMD 1998 Autoinhibition as a transcriptional regulatory mechanism. Cold Spring Harb Symp Quant Biol 63 621 629
44. HagmanJGrosschedlR 1992 An inhibitory carboxyl-terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc Natl Acad Sci U S A 89 8889 8893
45. LimFKrautNFramptomJGrafT 1992 DNA binding by c-Ets-1, but not v-Ets, is repressed by an intramolecular mechanism. EMBO J 11 643 652
46. WasylykCKerckaertJPWasylykB 1992 A novel modulator domain of Ets transcription factors. Genes Dev 6 965 974
47. BaetzKAndrewsB 1999 Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding. Mol Cell Biol 19 6729 6741
48. von BothISilvestriCErdemirTLickertHWallsJR 2004 Foxh1 is essential for development of the anterior heart field. Dev Cell 7 331 345
49. PeiWFeldmanB 2009 Identification of common and unique modifiers of zebrafish midline bifurcation and cyclopia. Dev Biol 326 201 211
50. ZhangYShaoMWangLLiuZGaoM 2010 Ethanol exposure affects cell movement during gastrulation and induces split axes in zebrafish embryos. Int J Dev Neurosci 28 283 288
51. CheaHKWrightCVSwallaBJ 2005 Nodal signaling and the evolution of deuterostome gastrulation. Dev Dyn 234 269 278
52. LaprazFRottingerEDubocVRangeRDuloquinL 2006 RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev Biol 300 132 152
53. GrandeCPatelNH 2009 Nodal signalling is involved in left-right asymmetry in snails. Nature 457 1007 1011
54. KispertAHerrmannBGLeptinMReuterR 1994 Homologs of the mouse Brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium, and Locusta. Genes Dev 8 2137 2150
55. AdellTGrebenjukVAWiensMMullerWE 2003 Isolation and characterization of two T-box genes from sponges, the phylogenetically oldest metazoan taxon. Dev Genes Evol 213 421 434
56. LarrouxCLukeGNKoopmanPRokhsarDSShimeldSM 2008 Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol 25 980 996
57. ZhuGMullerEGAmacherSLNorthropJLDavisTN 1993 A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the fork head family of DNA-binding proteins. Mol Cell Biol 13 1779 1787
58. MazetFYuJKLiberlesDAHollandLZShimeldSM 2003 Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene 316 79 89
59. YuJKMazetFChenYTHuangSWJungKC 2008 The Fox genes of Branchiostoma floridae. Dev Genes Evol 218 629 638
60. TuQBrownCTDavidsonEHOliveriP 2006 Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev Biol 300 49 62
61. KuglerJEPassamaneckYJFeldmanTGBehJRegnierTW 2008 Evolutionary conservation of vertebrate notochord genes in the ascidian Ciona intestinalis. Genesis 46 697 710
62. StainierDY 2002 A glimpse into the molecular entrails of endoderm formation. Genes Dev 16 893 907
63. DrieverWSolnica-KrezelLSchierAFNeuhaussSCMalickiJ 1996 A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123 37 46
64. HaffterPGranatoMBrandMMCHammerschmidtM 1996 The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123 1 36
65. ThisseCThisseB 2008 High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3 59 69
66. Schulte-MerkerSHoRKHerrmannBGNusslein-VolhardC 1992 The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 116 1021 1032
67. StachelSEGrunwaldDJMyersPZ 1993 Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development 117 1261 1274
68. AlexanderJRothenbergMHenryGLStainierDY 1999 casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol 215 343 357
69. StrahleUBladerPHenriqueDInghamPW 1993 Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev 7 1436 1446
70. KikuchiYAgathonAAlexanderJThisseCWaldronS 2001 casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev 15 1493 1505
71. TalbotWSTrevarrowBHalpernMEMelbyAEFarrG 1995 A homeobox gene essential for zebrafish notochord development. Nature 378 150 157
72. KraussSJohansenTKorzhVFjoseA 1991 Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development 113 1193 1206
73. HerbomelPThisseBThisseC 1999 Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126 3735 3745
74. KraussSConcordetJPInghamPW 1993 A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75 1431 1444
75. BisgroveBWEssnerJJYostHJ 1999 Regulation of midline development by antagonism of lefty and nodal signaling. Development 126 3253 3262
76. ErterCESolnica-KrezelLWrightCV 1998 Zebrafish nodal-related 2 encodes an early mesendodermal inducer signaling from the extraembryonic yolk syncytial layer. Dev Biol 204 361 372
77. RebagliatiMRToyamaRHaffterPDawidIB 1998 cyclops encodes a nodal-related factor involved in midline signaling. Proc Natl Acad Sci U S A 95 9932 9937
78. YanYLHattaKRigglemanBPostlethwaitJH 1995 Expression of a type II collagen gene in the zebrafish embryonic axis. Dev Dyn 203 363 376
79. SokolSChristianJLMoonRTMeltonDA 1991 Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell 67 741 752
80. BakerJCHarlandRM 1996 A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes Dev 10 1880 1889
81. FeldmanBStempleDL 2001 Morpholino phenocopies of sqt, oep, and ntl mutations. Genesis 30 175 177
82. KarlenSRebagliatiM 2001 A morpholino phenocopy of the cyclops mutation. Genesis 30 126 128
83. AokiTSchweinsbergSManassonJSchedlP 2008 A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila. Mol Cell Biol 28 1047 1060
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Structural and Functional Differences in the Long Non-Coding RNA in Mouse and Human
- Identification, Replication, and Functional Fine-Mapping of Expression Quantitative Trait Loci in Primary Human Liver Tissue
- A −436C>A Polymorphism in the Human Gene Promoter Associated with Severe Childhood Malaria
- A Decline in p38 MAPK Signaling Underlies Immunosenescence in
- The Operon Balances the Requirements for Vegetative Stability and Conjugative Transfer of Plasmid R388
- Novel and Conserved Protein Macoilin Is Required for Diverse Neuronal Functions in
- Ixr1 Is Required for the Expression of the Ribonucleotide Reductase Rnr1 and Maintenance of dNTP Pools
- Genome of Strain SmR1, a Specialized Diazotrophic Endophyte of Tropical Grasses
- A Deficiency of Ceramide Biosynthesis Causes Cerebellar Purkinje Cell Neurodegeneration and Lipofuscin Accumulation
- A Latent Pro-Survival Function for the Mir-290-295 Cluster in Mouse Embryonic Stem Cells
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time
- Prion Formation and Polyglutamine Aggregation Are Controlled by Two Classes of Genes
- Integrated Genome-Scale Prediction of Detrimental Mutations in Transcription Networks
- Post-Embryonic Nerve-Associated Precursors to Adult Pigment Cells: Genetic Requirements and Dynamics of Morphogenesis and Differentiation
- A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
- A Genetic and Structural Study of Genome Rearrangements Mediated by High Copy Repeat Ty1 Elements
- A Missense Mutation in Causes a Major QTL Effect on Ear Size in Pigs
- A Flurry of Folding Problems: An Interview with Susan Lindquist
- Meiotic Recombination Intermediates Are Resolved with Minimal Crossover Formation during Return-to-Growth, an Analogue of the Mitotic Cell Cycle
- A Nervous Origin for Fish Stripes
- The ISWI Chromatin Remodeler Organizes the hsrω ncRNA–Containing Omega Speckle Nuclear Compartments
- The Telomerase Subunit Est3 Binds Telomeres in a Cell Cycle– and Est1–Dependent Manner and Interacts Directly with Est1
- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Characterizing Genetic Risk at Known Prostate Cancer Susceptibility Loci in African Americans
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



