-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Evidence for the Robustness of Protein Complexes to Inter-Species Hybridization
Despite the tremendous efforts devoted to the identification of genetic incompatibilities underlying hybrid sterility and inviability, little is known about the effect of inter-species hybridization at the protein interactome level. Here, we develop a screening platform for the comparison of protein–protein interactions (PPIs) among closely related species and their hybrids. We examine in vivo the architecture of protein complexes in two yeast species (Saccharomyces cerevisiae and Saccharomyces kudriavzevii) that diverged 5–20 million years ago and in their F1 hybrids. We focus on 24 proteins of two large complexes: the RNA polymerase II and the nuclear pore complex (NPC), which show contrasting patterns of molecular evolution. We found that, with the exception of one PPI in the NPC sub-complex, PPIs were highly conserved between species, regardless of protein divergence. Unexpectedly, we found that the architecture of the complexes in F1 hybrids could not be distinguished from that of the parental species. Our results suggest that the conservation of PPIs in hybrids likely results from the slow evolution taking place on the very few protein residues involved in the interaction or that protein complexes are inherently robust and may accommodate protein divergence up to the level that is observed among closely related species.
Published in the journal: Evidence for the Robustness of Protein Complexes to Inter-Species Hybridization. PLoS Genet 8(12): e32767. doi:10.1371/journal.pgen.1003161
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003161Summary
Despite the tremendous efforts devoted to the identification of genetic incompatibilities underlying hybrid sterility and inviability, little is known about the effect of inter-species hybridization at the protein interactome level. Here, we develop a screening platform for the comparison of protein–protein interactions (PPIs) among closely related species and their hybrids. We examine in vivo the architecture of protein complexes in two yeast species (Saccharomyces cerevisiae and Saccharomyces kudriavzevii) that diverged 5–20 million years ago and in their F1 hybrids. We focus on 24 proteins of two large complexes: the RNA polymerase II and the nuclear pore complex (NPC), which show contrasting patterns of molecular evolution. We found that, with the exception of one PPI in the NPC sub-complex, PPIs were highly conserved between species, regardless of protein divergence. Unexpectedly, we found that the architecture of the complexes in F1 hybrids could not be distinguished from that of the parental species. Our results suggest that the conservation of PPIs in hybrids likely results from the slow evolution taking place on the very few protein residues involved in the interaction or that protein complexes are inherently robust and may accommodate protein divergence up to the level that is observed among closely related species.
Introduction
Identifying the molecular processes underlying species incompatibilities is a fundamental issue in evolutionary biology. Charles Darwin first suggested that the inviability and sterility often occurring after species hybridization could result from mechanisms other than the differences observed between parents and morphological disorders in the descendants [1]. Several theoretical models have since been developed to address how differences leading to incompatibilities may accumulate between species [2]–[6] and genetic mapping of such incompatibilities [7]–[10] has uncovered some molecular mechanisms by which they may take place. In theory, any system in which molecules or genes interact with each other to perform a function and that co-evolve within species, could give rise to such incompatibilities. Most studies on molecular incompatibilities have so far been performed using genetic approaches to identify the genes involved in hybrid lethality and sterility, for instance, in Drosophila [7]–[9] or in hybrid sterility in yeasts [10]. In a cell, all processes are controlled and mediated by protein-protein interactions (PPIs). Stable PPIs form protein complexes that are the workhorses of the cell and perform fundamental tasks such as DNA replication, repair and transcription, cellular transport or cytoskeleton architecture [11]–[16]. More transient interactions are involved, for instance, in cell signaling events such as phosphorylation cascades [17]. Proteins are suggested to co-evolve within species over-time as a response of selection to maintain these interactions [18]–[20], and this co-evolution is so important that it can be used to infer PPIs [20]. Each pairwise PPI therefore represents an opportunity for incompatibilities to accumulate between closely related species, although there is limited empirical evidence for this phenomenon [21]. Whether protein divergence can result in incompatibilities in protein complexes in hybrids is largely unknown because we lack experimental systems to directly measure physical interactions in living cells in closely related species and in inter-species hybrids.
Here we develop such a system using the budding yeast Saccharomyces cerevisiae (Scer) and its closely related species belonging to the Saccharomyces sensu stricto group [22], [23]. As its interactome has been intensely studied, we reasoned that the budding yeast would be an appropriate reference for protein interaction network (PIN) comparisons among species and in hybrids between species. More than 70,000 physical interactions have been reported in major protein interaction databases such as the BioGRID [24] for this species. Another reason why Scer is a particularly good model to study the implication of genetic incompatibilities in hybrids is the availability of closely related species that are genetically tractable [23] and that can be mated with each other. Indeed, F1 hybrids among Saccharomyces species are viable and are found in the wild [25] and can be readily produced in the laboratory [26]. However, they show strong incompatibilities, as they are almost entirely sterile [27], [28], which makes standard genetic mapping approaches inefficient to identify incompatibilities systematically.
Several techniques are available for interactome mapping and could be applied to the screening of abnormal PPIs directly in viable F1 hybrids. A technique that would be particularly powerful would integrate all factors that affect PPIs such as protein abundance and localization. It would allow detecting PPIs in vivo, among proteins expressed at their endogenous levels and at their normal cellular localization. One such method has recently been developed for the budding yeast and is based on the Protein-fragment Complementation Assay (PCA) technology [13], [29]. This technique detects structural relationships among proteins, as the C-termini of both proteins need to be within 80 Angstroms of each other for PPI detection [13]. The signal detected also reflects the amount of protein complexes formed [30]. Here, we set out to adapt the PCA based on the dihydrofolate reductase (DHFR), recently developed for Scer [13], to other species of the Saccharomyces sensu stricto group, which covers the same range of molecular divergence as that between birds and mammals [22]. Using this tool, we directly test whether 5–20 millions years of evolution result in protein incompatibility between species, i.e. in gain or loss of PPIs in the hybrid background as compared to the parental species.
We identified two protein complexes that can serve as models, namely the nuclear pore complex (NPC) and the RNA polymerase II (RNApII). We selected these two complexes because they have distinct functions and contrasting patterns of evolution. The NPC performs selective molecule transport across the nuclear envelope and its overall structure is conserved among eukaryotes [31], although the precise organization diverges among distantly related species [32]. The RNApII is a core enzyme that performs DNA transcription and its structure is fully conserved in eukaryotes [33]. The structure of these complexes has either been well established or extensively modeled. In Scer, the NPC is a circular assembly of about 500 proteins (nucleoporins) arranged in eight identical substructures, each constituted of about 30 nucleoporins, most of them being duplicated in a substructure [34]. The RNApII is composed of 12 interacting subunits [35]. These complexes were at least partially detected in vivo in Scer by DHFR-PCA [13] and in agreement with structural information provided by other techniques [24]. A number of characteristics make these complexes appropriate models to investigate the impact of hybridization on PPIs. First, proteins belonging to the same complex evolve at equivalent rates in Saccharomyces species (Figure 1), reflecting the potential coevolution of subunits within NPC and RNApII, as expected from previous reports [36]. Further, the NPC proteins are at the high end of the spectrum of divergence among the Saccharomyces species, whereas RNApII proteins show stronger conservation (Figure 1). Second, the NPC and RNApII represent contrasted situations because their respective functions require different levels of complex architecture flexibility. For instance, in Scer, most of RNApII subunits are essential for cell viability [37] and all are required for transcription [35], whereas about one third of nucleoporin genes are non-essential [37] and many NPC proteins perform redundant structural functions and are thus likely interchangeable [34]. Also, the Scer NPC includes several recent genes which originated during the whole yeast genome duplication [38]. All these observations suggest that PPIs in the NPC would be more likely to diverge among species. A lack of changes in the architecture of the NPC between species or in the hybrid would suggest that protein complexes can be robust to protein divergence.
Fig. 1. Protein divergence in the RNApII (black) and the NPC (grey) in the Saccharomyces sensu stricto group. 
Evolutionary trees were drawn for all proteins of a complex and are on the same scale (proportion of different amino acids). Distribution of protein divergence between Scer and Skud was calculated from multiple-sequence alignments available for 5261 orthologous proteins [23]. Arrows indicate protein divergence covered by the two complexes. Results
The DHFR-PCA permits detection of PPIs across the Saccharomyces sensu stricto species complexes
We first tested whether we could detect PPIs using the DHFR-PCA in S. paradoxus (Spar), S. uvarum (Suva) and S. kudriavzevii (Skud) and determined what would be the optimal experimental conditions for measuring PPIs in vivo. The DHFR-PCA assay is based on the reconstitution of the DHFR enzyme (see Figure S1A–S1B). In order to measure the signal to noise ratio of this assay in the four species, we engineered a strong PPI reporter by fusing the DHFR fragments (F[1], [2] and F[3]) downstream of the coding sequence of homodimerizing residues of the GCN4 parallel coiled-coil leucine zipper [39]. Overall, we observed the strongest signal-to-noise ratio (growth of positive/negative controls) in Scer and Skud and in conditions that are standard for the Scer DHFR-PCA (Figure 2A). We thereafter focused our study on Scer and Skud to ensure optimal conditions to detect PPIs in these species and in their hybrids. The two species show an average of 16.4% pairwise protein divergence (Figure 1), which is in the same order of magnitude than what it is observed between birds and mammals [22]. Because no hybridizable species presents such a protein divergence level in plants or animals, Scer and Skud offer a conservative system to measure how hybrid protein complexes form in inter-species crosses.
Fig. 2. Optimization of the DHFR-PCA in four Saccharomyces species. 
(A) Control diploid strains were grown in presence of DMSO (control) and methotrexate (MTX). In the positive controls (+), two zipper protein fragments (white) were fused via a linker (grey) to complementary protein fragments of the MTX-resistant DHFR (black). Zipper fragments strongly interact, reconstituting the DHFR activity and allowing growth on MTX medium. In the negative controls (−), the absence of interaction (L alone) prevents the DHFR reconstitution and strain growth. Optimal signal was found in Scer and Skud. (B) High density arrays were used to screen PPIs in Scer and Skud. Each plate contained crosses between 24 MATa and 16 MATα strains in three non adjacent replicates. Here, the interaction between Nup159 and Nup116 reconstitutes the DHFR (+) and allows growth of the colony in both species. Nup159 and Nup60 do not interact directly (−) and strains do not grow on MTX medium. Comparison of protein complexes between species and their hybrids
We set out to compare the architecture of the NPC (15 subunits considered) and the RNApII (9 subunits considered) between Scer, Skud and their F1 hybrids. The 24 subunits corresponded to a subset of Scer proteins, which were showed to be involved in at least one PPI in the considered complexes in Tarrassov et al. [13]. Not all Scer strains were available in the DHFR-PCA collection so we reconstructed them by homologous recombination as described in Tarrassov et al. [13]. We then attempted to construct all of the equivalent strains (24 in MATa and 24 in MATα) in Skud by targeting the orthologous genes [40]. All strains were obtained for Scer; 23 and 17 strains were obtained for, respectively, MATa and MATα strains in Skud (Table S1). We then performed a comprehensive screen of PPIs within and between the NPC and RNApII, within species and their hybrids. We crossed all Scer and Skud MATa strains with all Scer and Skud MATα strains. Most of interactions between two proteins P1 and P2 were testable in two reciprocal ways (P1 in MATa×P2 in MATα vs. P2 in MATa×P1 in MATα; Figure S2A), which was the case of 276 PPIs. The 24 remaining homomeric interactions (P1 in MATa×P1 in MATα) were also tested. We compared these PPIs between Scer, Skud and the inter-specific hybrids, namely hybrid 1 (Scer MATa×Skud MATα) and hybrid 2 (Skud MATa×Scer MATα), giving a total of 1927 possible combinations (Table S2). In order to evaluate the interaction between P1 from Scer and P2 from Skud, P1-P2 was measured in hybrid 1 and P2-P1 in hybrid 2 (Figure S2A). All combinations P1-P2 and P2-P1 were replicated three times. PPIs were measured by estimating colony size in pixels on high quality images of plates (Figure 2B). The DHFR-PCA signal was highly reproducible and the reproducibility was similar for the intra and inter-specific crosses (r = 0.90 to 0.97, p<0.001; Figure S3). Triplicates were averaged, giving a mean growth signal index (SI) for all downstream analyses (data available in Dataset S1). Because SI values associated to a particular protein were sometimes lower when the protein was tagged in MATa or MATα, potentially generating false positive or hide true interactions, we corrected SI values for these haploid strain effects (see Methods and Figures S2B–S2F, S4, S5, S6, S7, data available in Dataset S2).
We defined a threshold (t) above which SI would represent a PPI by comparing the distributions of SI measured among (SA) and within (SW) the NPC and RNApII (Figure S8 and S9), assuming that among complex crosses represent background growth of the colonies, which is well supported by the almost complete absence of physical interactions reported between NPC and RNApII proteins [24]. As all SA values fell below t≈1.4, we considered SW values above 1.4 to be a conservative threshold (Figure S8 and S9). In Scer, 39 out of 54 interactions (corresponding to 82 SW values with SW>t before reciprocal redundant combinations were collapsed) were concordant with physical interactions already reported in this species by independent methods (Figure S10A). Also, SW values measured in Scer were positively correlated to the number of times a PPI was reported in BioGRID (r = 0.55, p<0.001; Figure S10B). In Scer, PPIs were mostly detected between proteins for which C-termini are close to each other in the case of RNApII (Figure S10C) or corresponded to the architecture of NPC previously defined (Figure S10D). We observed a strong and highly significant correlation between SI values measured in the two species (r = 0.93, p<0.001; Figure 3A), suggesting a high degree of conservation in the protein complexes. Most interactions were shared between species, and SI remained highly correlated between species when only considering these interactions (r = 0.87, p<0.001). Among the 54 PPIs observed in Scer, 44 were comparable with Skud and 36 physical interactions were common to both species (8 in the RNApII, 28 in the NPC, Figure 3A–3C), corresponding to 50 significant SI values before reciprocal redundant combinations were collapsed. The remaining unshared PPIs in Scer and Skud (eight and four PPIs respectively) were all close to the detection threshold t and further examination by regression analysis showed that most of the differences were not significant (Figure 3D). This was the case for three very weak PPI signals found in RNApII in Scer but not in Skud (Figure 3B and Figure S10C). Other differences involved nucleoporins Nup1, Nup2 and Nup60 belonging to the subcomplex of the nuclear pore basket, with respectively one and three PPIs specific to Scer and Skud (Figure 3D). The structure of this subcomplex is poorly known [41] mainly because its location at the nuclear side makes unsuccessful the use of most of PPIs detections methods, which was in agreement with weak signals we found for these interactions. Thus, like the differences we found in RNApII, these variations have to be carefully interpreted. Other variations in PPIs involved proteins from the outer ring NPC subcomplex. Four PPIs were absent in Skud and three of them involved interactors of Nup145; Nup120, Nup82 and Nup100 (Figure 3D). However, the only statistically significant difference we found was the absence of Nup120-Nup145 interaction in Skud, which we discuss in-depth below (Figure 3D).
Fig. 3. The NPC and RNApII networks are largely conserved between Scer and Skud. 
(A) Comparison of SI values (strains growth signal index; log10) between species. Grey dotted lines indicate SI threshold values (t). SI values were considered to correspond to interactions when greater than t in both species (purple) or specific to one species when greater than t only in Scer (red), or only in Skud (blue). Black dotted lines indicate 5%, 1% and 0.1% threshold above which SI residuals significantly deviate from the Scer-Skud regression (black line). (B) Overlapped networks of Scer and Skud. Only SI above t and comparable interactions are represented. Line width is proportional to SI values measured between proteins in Scer (red) and Skud (blue) in the NPC (left) and the RNApII (right). Interactions appear in purple when Scer and Skud SI values overlap. Different degrees of purple depend on whether the interaction could be tested in reciprocal ways or not. (C) Venn diagram indicating the overlap of PPIs detected by PCA of other methods (BioGRID) in Scer, and by PCA in Skud. Reciprocal combinations of PPIs were collapsed. (D) Representation of PPIs shared (purple lines) or unique to Scer (red lines) or Skud (blue lines) in the NPC. Only proteins involving divergences in PPIs are showed. Only the difference in the Nup120-Nup145 PPI was significant (**: p<0.01). Protein complex architecture in the hybrid background
We assessed the extent to which the integrity of the protein complexes was preserved in hybrids by comparing PPIs in their parental background to PPIs in F1 hybrids. In hybrids, protein complexes can be formed with subunits from the two species. Incompatibilities between subunits could directly disrupt interactions between the two proteins of interest or indirectly affect the architecture of the complex and disrupt or lead to novel, spurious, unexpected interactions [42]. We considered that an interaction was disrupted or gained in hybrids when its residual value resulting from the species/hybrid comparison was significantly different from the distribution of all residual values pooled together (Figure S2E–S2F). Overall, we found that all interactions that were conserved between parental species were also seen in the hybrids, i.e. no interaction was disrupted specifically in the hybrid. We also found no instance of hybrid-specific interactions. For instance, we found that interactions involving Nup82 in the Scer background were fully conserved in hybrids between the SkudNup82 and Scer NPC subunits (Figure 4A–4B). Similar observations were made for Rpb3 in RNApII (Figure 4C–4D). Additionally, SI values from species and hybrids were strongly correlated in most of cases (FigureS S4, S5, S6, S7), suggesting that inter-species PPIs could not be differentiated from within species PPIs. We observed the same pattern for almost all comparisons between hybrids and Scer (Figure S8) and Skud (Figure S9).
Fig. 4. The NPC and RNApII are robust to hybridization. 
SI values compared between Scer (red) or Skud (blue) and Scer-Skud hybrids (green). Examples of proteins Nup82 (A–B) and Nup145 (E–F) in the NPC and Rpb3 (C–D) in the RNApII. Segment width is proportional to SI (dotted line if SI<t). For each comparison, networks on left show interactions measured between the protein of interest tagged in MATa (center) and other proteins of the same complex tagged in MATα. Networks on right show the reciprocal interactions. The protein of interest comes from Scer (red ring) or Skud (blue ring) and is in the species background (outer ring) or in the hybrid background (inner ring). Asterisks indicate whether the SI value measured in hybrid is significantly different from that measured in species (*: p<0.05; **: p<0.01; ***: p<0.001). Only the absence of ScerNup120-SkudNup145 is significant in reciprocal comparisons (E). Protein names were blurred when strains were unavailable. We found one exception for the interaction between proteins Nup120 and Nup145, which are also involved in few divergent interactions between Scer and Skud. The Nup120-Nup145 interaction is present in Scer but is undetectable in Skud (Figure 3D), neither in hybrids between the ScerNup120 and the SkudNup145 (Figure 4E). Conversely, the SkudNup120-ScerNup145 interaction was observed (Figure 4F), suggesting that the absence of interaction in Skud is due to changes in the amino acid sequence of Nup145 rather than Nup120. In order to determine whether the Nup145-Nup120 interaction was lost in Skud or gained in Scer, we repeated the PCA experiment between Nup145 and Nup120 by including Suva, which diverged early in the Saccharomyces sensu scricto group. As shown in Figure 5A, we found that the Nup145-Nup120 interaction was present in Suva, suggesting that the absence of interaction in Skud is a lost rather than a gain in Scer. Figure 5B shows three protein domains of Nup145 (namely I, II and III) essential for its interaction with Nup120 and Nup85 that have been recently mapped in Scer using deletion analysis [43]. Domains I and II also overlap a 900 bp region that is the only essential fragment of the gene [44]. We examined Nup85 and showed that it strongly interacted with Nup145 and Nup120 in three species and their hybrids, suggesting that the inability of Skud-Nup145 to interact was unlikely affected by the DHFR fragment and that the effect was specific to its interaction with Nup120 (Figure 5C). We thus looked at whether amino acid differences in Nup145 between Skud and Scer could affect the three interaction domains. Because the yeast Nup145 protein is self-cleaved in vivo into two distinct but functional protein fragments, Nup145N and Nup145C [45], and because the DHFR fragments are fused to the C-terminus of proteins, we focused on the Nup145C fragment.
Fig. 5. The absence of Nup120-Nup145C interaction in Skud is likely a PPI loss. 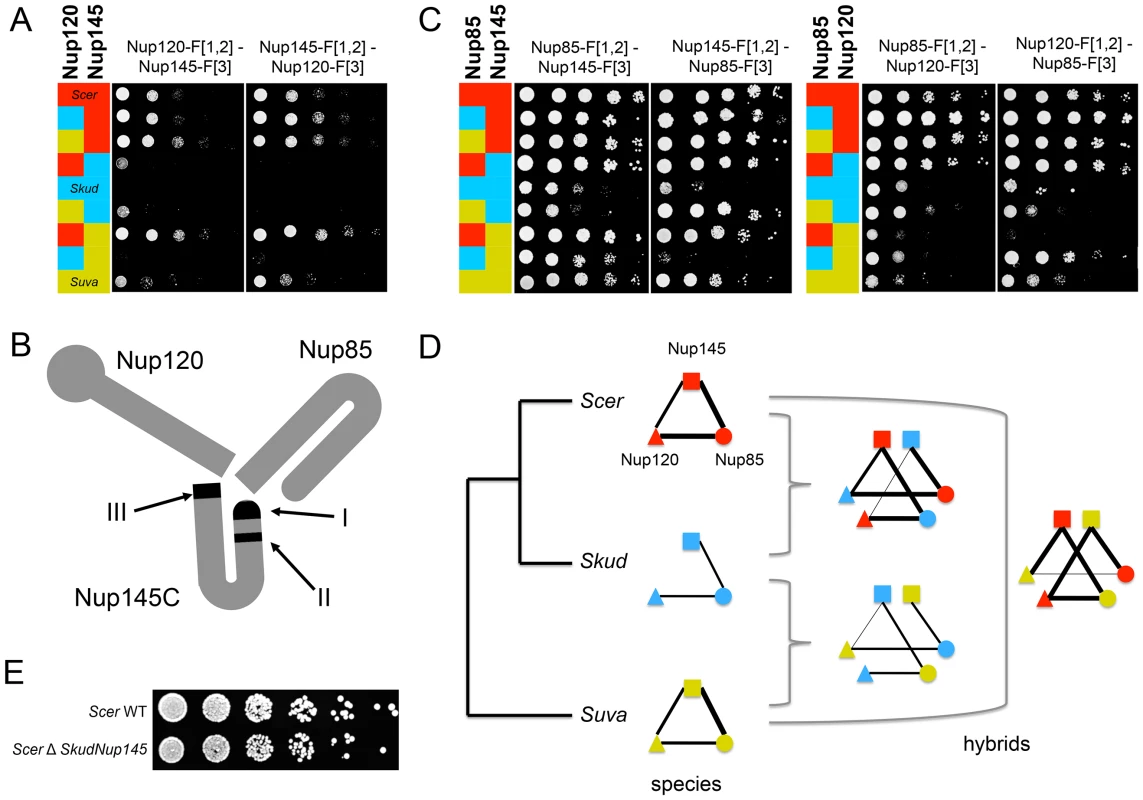
(A) Spot assays on Methotrexate medium (six days of growth at 30°C) to dissect the interaction in Scer (red), Skud (blue), Suva (yellow) and their hybrids. The interaction between Nup145C from Scer and Nup120 from Scer, Skud or Suva is detected, whereas it is lost when Nup145C comes from Skud. The interaction was also absent when it involved Nup120 from Skud and Nup145 from Suva. (B) Schematic structure of the Scer Nup120-Nup85-Nup145 sub-complex adapted from Fernandez-Martinez et al. [43]. The three interacting domains are indicated in black. (C) Interactions between Nup85 and Nup145C or Nup120 in three species and their hybrids confirm that not all Nup145C interactions are lost. (D) Evolutionary tree of Scer, Skud and Suva and schematic representation of Nup145-Nup120-Nup85 interactions in species and hybrids according to spot assays (A–C), revealing several other loss of interaction in hybrids with Suva. Line width is proportional to the number of spot growth observed for each interaction. (E) Similar growth for BY4741 Scer wild type (WT) and modified strains (ΔSkud-NUP145) suggest that Skud-Nup145 complements the absence of Scer-Nup145 (YPD medium, two days of growth at 30°C), as Nup145 is essential for growth in Scer. We found 81 amino acid (aa) changes unique to Skud Nup145C sequence (i.e. changes accumulated since the Scer-Skud divergence), with 27 of them located in three domains involved in Nup45C-Nup120 interaction [43], thus representing changes that could potentially affect the interaction (Figure S11A). Among these changes, seven were concentrated in a very short region (13 aa length) of the main interaction domain (domain I) and resulted in three important aa polarity inversion (Figure S11B). The fact that domain I is also located at the N-terminal end of Nup145C, that it concentrates several hypervariable regions, three phosphorylated sites [46], [47] and most of disordered regions of Nup145C [48], strongly suggests that this is the main interacting domain of the protein. Hence, several aa changes we observed in this domain between Scer and Skud are good candidates to explain the loss of interaction in Skud, since these changes could potentially enhanced a modification of the Nup145C N-terminal activity that subsequently caused this loss. We also found a 8 aa insertion at the C-terminal of the Scer Nup145, in the domain III (Figure S11), that may thus potentially affect the activity of this domain, while the absence of this insertion in Skud and Suva was not concordant with the absence of Nup145-Nup120 interaction in Skud only.
Interestingly, the interaction between Nup145C from Suva and Nup120 from Skud was also lost, as the interaction between Nup120 from Suva and Nup85 from Skud (Figure 5C–5D), suggesting that the architecture of this subcomplex, while conserved between Scer and Suva, may be modified in hybrids of Suva due to changes that occurred in this lineage. Many other variations in Nup145C sequence, like the 8 amino acids insertion in Scer, but also post-translational modifications and changes in regulation, may modulate these differences in PPIs we observed. Because Nup145 is an essential gene [37], we could expect that these interaction modifications would lead to incompatibilities in hybrids. However, we did not know whether the hybrid Nup145C-Nup120 interaction is essential for the cell, nor if incompatibilities between Nup145C and Nup120 actually affect the hybrid fitness, since parental proteins are all present in the diploid hybrid and thus parental interactions could occur and perform the function if hybrid interactions fail. In order to test if the loss of interactions could lead to an incompatibility between Scer and Skud, we constructed a haploid strain of Scer in which we replaced Nup145 by its homologue from Skud (see Text S1). If the lack of interaction were causing an incompatibility, we would expect Skud-Nup145 to not be able to complement the loss of Scer-Nup145. Then we compared the relative growth of modified strains and wild types (WT) on enriched YPD medium, to directly test whether SkudNup145 complemented ScerNup145. We observed a normal and similar growth for the WT and modified Scer strains, suggesting that, while essential, Nup145 from Scer, can be complemented by Nup145 from Skud (Figure 5E).
Discussion
Investigating the role of protein-protein incompatibilities among closely related species is an underexplored but fundamental issue in evolutionary biology. Because interacting proteins are known to co-evolve within species [18]–[20], such incompatibilities are expected to accumulate as species diverge. Very few cases of such incompatibilities have been reported in the literature. One instance has recently been reported in the Proliferating Cell Nuclear Antigen (PCNA) protein complex between two groups of yeasts species [42]. However, the species investigated are distantly related (300 My), such that the question of whether protein incompatibilities can accumulate fast enough to result in inter-species incompatibilities during or just after a speciation event and thus cause inter-species incompatibilities is still an open one. Here, we adapted tools developed for the model species Scer to study PPIs in vivo in the closely related species Skud. We examined how two protein complexes with contrasting evolutionary properties could assemble in the hybrid between Scer and Skud. Given that the divergence time between Skud and Scer represents about 78% of the age of the Saccharomyces genus [23], with a molecular divergence range that is equivalent to molecular divergence between birds and mammals [22], Saccharomyces species cover the higher end of the spectrum of molecular divergence that can be observed among species that can be hybridized. As a result of this high divergence, F1 haploid recombinant hybrids of Scer and Skud are unviable [27], [28], while F1 diploid hybrids are viable [25], [26]. The inviability of F1 haploid hybrids makes the use of standard genetic mapping techniques to locate incompatible loci in the genome impossible. However, PCA allows us to examine potential incompatibilities resulting from PPIs perturbations directly in diploid F1 at the molecular and network levels.
Our results indicate that neither of our model protein complexes, the NPC nor the RNApII, is significantly perturbed in the hybrid between species. Our results are unintuitive since these two complexes have highly contrasting evolutionary histories in the two Saccharomyces species. Scer-Skud protein divergence within the RNApII never exceeds 3%, whereas nucleoporins diverge in the range of 7–41% (Figure 1), suggesting that unlike the RNApII where all proteins are essential for DNA transcription [35], the structure of the NPC is highly flexible [49]. The high conservation of PPIs is thus consistent with the evolutionary history of RNApII but not NPC. For instance, Nup82 is a key protein of the NPC that is involved in PPIs with almost all nucleoporins we tested. Despite the high protein divergence between Scer and Skud Nup82 (26%), we found that all these interactions were indistinguishable and highly correlated between species. This result likely illustrates that only few protein residues contribute to PPIs, as has been previously shown in many protein complexes of known structure [50]. In the NPC, very little detailed structural data is available, most of it from human [51], [52] and Scer [43], [53]. For instance, the Nup82-Nup116-Nup159 sub-complex structure was solved in Scer [53]. The Nup116-Nup82 interaction that we found to be the strongest in the NPC for Scer and Skud involves 23 Nup82 residues, among which 21 are conserved between these two species. This observation suggests that at least part of the robustness of protein complexes to inter-species hybridization results from the fact that few sites are involved and those are conserved, while the rest of the protein is free to evolve. The evolution of PPIs has recently been investigated for orthologous proteins in distant Eukaryotes [54]. While the high level of divergence between studied organisms restricted the number of investigable PPIs, these works provided strong evidences that PPIs evolve much slower than proteins, particularly for orthologous proteins which genes underwent no duplication event during their evolutionary history. Our works based on the in vivo detection of PPIs through a same method in two divergent species but with comparable level of organization, confirmed the high level of PPIs conservation at a lower scale of evolutionary time.
The few differences we observed for PPIs in hybrids were not significant in most cases (Figure 4), with the exception of one particular interaction occurring in Scer but not in Skud between nucleoporins Nup120 and Nup145C. By comparing PPIs signals in Scer, Skud, Suva and hybrids, we deduced that this interaction change was likely a lost in Skud rather than a gain in Scer (Figure 5A). Nup145C and Nup120 belong to a NPC heptameric sub-complex that has a partially known structure in Scer (Figure 5B). Fernandez-Martinez et al. showed that three Nup145C protein regions disrupted the Nup145C-Nup120 interaction when deleted [43]. Interestingly, one region that likely corresponds to the functional domain of Nup145C contains a high concentration of amino acids changes proper to Skud, potentially leading to substantial changes in the domain functionality (Figure S11), enforcing the hypothesis that these amino acid changes may had an impact on Nup145C interactions with other proteins. The complicated pattern of PPIs loss we observed in hybrids of Suva also suggests that the Nup145C-Nup120-Nup85 subcomplex accumulated several other differences in amino acids, in post-translational modifications or in protein regulation during Suva divergence with other Saccharomyces species (Figure 5D). Such changes, without affecting the functionality of the subcomplex in parental species, may increase the probability of protein incompatibilities to occur in hybrids. Our observations, based on evolutionary protein divergence, thus corroborate the work of Fernandez-Martinez et al. based on experimental protein domain mapping [43]. These results show that protein complexes such as the NPC diverge among closely related species. Because the tools we developed for the comparison of PINs among species are also suitable for high throughput screening of PPIs, they provide a powerful method to identify these changes at the whole interactome level.
The loss or gain of PPIs in hybrids as compared to parental species could potentially affect the functional organization of hybrid complexes, and thus lead to incompatibilities. However, PPI changes we found in the Scer-Skud hybrid were mostly associated with differences we already identified between parents, and were always congruent with the architecture of the complex. Because we demonstrated that the Skud gene coding for protein Nup145C, losing its interactions in hybrids, was interchangeable in function with its ortholog in Scer (Figure 5E), one can support that in this particular case, PPIs changes in hybrids had no effect on hybrid viability. It is interesting to note that the gene coding for the Nup145C is orthologous to Nup96 of Drosophila simulans, which was shown to be involved in male hybrid inviability by interacting with genes located on the X chromosome of D. melanogaster [55]. While the precise mechanism underlying this incompatibility is unknown, studies suggest that it likely occurs at the protein-protein interaction level, and that the strong coevolution acting on interacting nucleoporins drove the divergence between D. melanogaster and D. simulans Nup96 ortholog, the latter potentially becoming incapable of some vital interaction in the hybrid NPC [56].
Few genes were found to have a strong effect on yeast hybrid sterility [10] and because most of genetic incompatibilities are rather caused by interactions involving many genes [28], [57], [58], they potentially involve many PPI perturbations in hybrids. However, we found that two large protein complexes in yeasts Skud and Scer were highly robust to hybridization and thus unlikely to be involved in hybrid incompatibilities. Some works provided indirect evidence about the potential implication of PPIs loss in hybrids inviability due to variations found in natural populations of S. cerevisiae [59] and S. paradoxus [60], suggesting that even within-species protein variation is enough to generate PPIs perturbations in hybrids. Skud and Scer diverged 5–20 My ago and underwent contrasting evolutionary pressures. Further studies will be needed to examine other types of PPIs, such as smaller protein complexes and transient, signaling interactions. Incompatibilities may also accumulate in parts of PINs that underlie species-specific traits that would result from the contrasting ecological and metabolic characteristics of these two species: difference in growth temperature [61], in carbon source preference [62] and in alcohol metabolism [23]. The tools we developed here could first be used to identify these species-specific networks and then be harnessed to specifically address these questions.
Methods
Adaptation of the DHFR-PCA method to S. paradoxus, S. kudriavzevii, and S. uvarum
The DHFR-PCA method was developed for Scer strains BY4741 and BY4742 [13]. Haploid strains of other species used or constructed in this study are detailed in Table S3 and were constructed as described in Text S1, using oligonucleotides described in Table S4. In order to optimize PCA conditions for Spar, Skud and Suva and allow comparisons with Scer, we tested different growth conditions using control diploid strains able to produce a strong signal and with the same molecular interactions for the four species (Figure S1C). In each species, we transformed the MATa and MATα strains with plasmids p41-ZL-DHFR[1,2] and p41-ZL-DHFR[3] respectively (construction detailed in Text S1). These plasmids express interacting leucine zipper moieties that strongly dimerize and thus lead to a strong signal in PCA. Negative controls consisted of plasmids expressing the linkers and DHFR fragments alone (p41-Linker-DHFR[1,2] and p41-Linker-DHFR[3]) [13] (Figure S1C). We then tested different PCA conditions (temperature and methotrexate concentration) using spot dilution assays. Transformations, crosses, diploid selection and PCA were performed as described in Text S1. Plates were manually analyzed with Adobe Photoshop from digital images and cell growth was estimated from pixel intensities of spot dilutions after six days of incubation (Figure S12).
Construction of strains for the DHFR-PCA in the NPC and RNApII complexes
All Skud MATa (FM1109) and Skud MATα (FM1110) strains for PPI screening (RNApII, 9 proteins; NPC, 15 proteins; see Figure S1A and Text S1 for details of constructions) were constructed by homologous recombination, by using primers described in Table S5. We completed the DHFR collection for Scer [13] since some strains were missing for these complexes and/or incorrectly tagged (Table S1). We confirmed all strains by PCR-sequencing across the 3′ end of the coding region of the genes and the fragment of the DHFR-PCA cassette encompassing the entire linker and the 5′ region of the DHFR fragments. All strains were reconfirmed by PCR a second time before screening and those that did not show PCR amplification were discarded.
Screening of PPIs in the NPC and RNApII complexes
We performed all possible crosses among the 24 MATa-DHFR F[1,2] and 24 MATα-DHFR F[3] strains constructed and confirmed for the NPC and the RNApII for a maximum of 576 pairwise combinations per species. We additionally made all possible hybrid crosses between Scer and Skud, given two types of hybrid diploids per combination: hybrid 1 (MATa Scer crossed with MATα Skud) and hybrid 2 (MATa Skud crossed with MATα Scer). Each possible combination was independently repeated three times (triplicates) on the same plate at random positions (Figure S13). Haploid strains were crossed on solid YPD medium and resulting diploid strains were selected two successive times on YPD with antibiotics using a robot-handled pin tool (see Text S1 and Figure S13). Diploid cells were transferred onto 1536-array plates of solid synthetic medium with methotrexate 200 µg/mL and incubated for five days at 30°C. Digital image analyses were performed using a custom script implemented in the software ImageJ 1.45 m (http://rsbweb.nih.gov/ij/).
Correction of SI values and regression analysis
Uncorrected SI values measured in species were highly correlated to those measured in hybrids (r = 0.87 to 0.91, p<0.001; Figures S4, S5, S6, S7). However, SI values associated to a particular protein in a species were sometimes biased between MATa and MATα strains. For instance, SI values associated to proteins Rpb8 and Nsp1 of Scer tagged in MATα were always lower than SI measured in MATa (Figures S4 and S6), whereas we observed the reverse for protein Rpb10 (Figures S5 and S7). In most of regressions tested, SI values associated to a protein tagged in a mating-type remained significantly correlated between species and hybrid, while differing from SI values associated to the opposed mating-type. We thus performed a correction of SI values by considering independently each protein tagged in a particular strain (Figure S2B–S2F). For each set of SI values associated to a haploid strain, we performed a correction only if the correlation species/hybrid was significant (Figure S2C), which was not the case for only two comparisons (Figure S7A). Then, we fitted the regression so that mean SI was equal in species and hybrid (Figure S2D). Corrections could be realized in two different ways: by increasing SI for biased strains, with the risk to increase the signal background, or by decreasing SI for unbiased strains, with the risk to lose PPI signals in the background. We choose the second way, which was the most conservative one. Then we grouped all corrected SI values together (Figure S2E) and obtained a significant increase of correlations quality for comparisons between species and hybrids (r = 0.94 to 0.97, p<0.001; Figures S4, S5, S6, S7). For downstream analysis, we considered that a SI value was different between species and hybrid or among species when the associated residual value significantly deviate from the distribution of all residual values grouped together (Figure S2F).
Supporting Information
Zdroje
1. Darwin C (1859) On the origin of species by means of natural selection. London: J. Murray. ix: , 1, 502 p.
2. MullerHJ, PontecorvoG (1940) Recombinants between Drosophila species the F-1 hybrids of which are sterile. Nature 146 : 199–200.
3. Bateson (1909) Discussion on the Influence of Heredity on Disease, with special Reference to Tuberculosis, Cancer, and Diseases of the Nervous System: Introductory Address. Proc R Soc Med 2 : 22–30.
4. DobzhanskyT (1940) Speciation as a stage in evolutionary divergence. American Naturalist 74 : 312–321.
5. GourbiereS, MalletJ (2010) Are species real? The shape of the species boundary with exponential failure, reinforcement, and the “missing snowball”. Evolution 64 : 1–24.
6. OrrHA, TurelliM (2001) The evolution of postzygotic isolation: Accumulating Dobzhansky-Muller incompatibilities. Evolution 55 : 1085–1094.
7. AraripeLO, MontenegroH, LemosB, HartlDL (2010) Fine-scale genetic mapping of a hybrid sterility factor between Drosophila simulans and D. mauritiana: the varied and elusive functions of “speciation genes”. BMC Evol Biol 10 : 385.
8. BarbashDA (2007) Nup96-dependent hybrid lethality occurs in a subset of species from the simulans clade of Drosophila. Genetics 176 : 543–552.
9. BrideauNJ, FloresHA, WangJ, MaheshwariS, WangX, et al. (2006) Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314 : 1292–1295.
10. LeeHY, ChouJY, CheongL, ChangNH, YangSY, et al. (2008) Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135 : 1065–1073.
11. Arabidopsis-Interactome-Mapping-Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333 : 601–607.
12. GuruharshaKG, RualJF, ZhaiB, MintserisJ, VaidyaP, et al. (2011) A protein complex network of Drosophila melanogaster. Cell 147 : 690–703.
13. TarassovK, MessierV, LandryCR, RadinovicS, Serna MolinaMM, et al. (2008) An in vivo map of the yeast protein interactome. Science 320 : 1465–1470.
14. ChoudharyC, KumarC, GnadF, NielsenML, RehmanM, et al. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325 : 834–840.
15. CollinsSR, MillerKM, MaasNL, RoguevA, FillinghamJ, et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 : 806–810.
16. GuoW, SacherM, BarrowmanJ, Ferro-NovickS, NovickP (2000) Protein complexes in transport vesicle targeting. Trends Cell Biol 10 : 251–255.
17. LemmonMA, SchlessingerJ (2010) Cell signaling by receptor tyrosine kinases. Cell 141 : 1117–1134.
18. LovellSC, RobertsonDL (2010) An integrated view of molecular coevolution in protein-protein interactions. Mol Biol Evol 27 : 2567–2575.
19. PazosF, ValenciaA (2008) Protein co-evolution, co-adaptation and interactions. Embo J 27 : 2648–2655.
20. RamaniAK, MarcotteEM (2003) Exploiting the co-evolution of interacting proteins to discover interaction specificity. J Mol Biol 327 : 273–284.
21. HarrisonJS, BurtonRS (2006) Tracing hybrid incompatibilities to single amino acid substitutions. Mol Biol Evol 23 : 559–564.
22. DujonB (2006) Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet 22 : 375–387.
23. ScannellDR, ZillOA, RokasA, PayenC, DunhamMJ, et al. (2011) The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. Genes Genomes Genetics 1 : 11–25.
24. StarkC, BreitkreutzBJ, Chatr-AryamontriA, BoucherL, OughtredR, et al. (2011) The BioGRID Interaction Database: 2011 update. Nucleic Acids Res 39: D698–704.
25. GonzalezSS, GalloL, ClimentMA, BarrioE, QuerolA (2007) Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int J Food Microbiol 116 : 11–18.
26. SniegowskiPD, DombrowskiPG, FingermanE (2002) Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res 1 : 299–306.
27. GreigD, BortsRH, LouisEJ, TravisanoM (2002) Epistasis and hybrid sterility in Saccharomyces. P R Soc B 269 : 1167–1171.
28. XuM, HeX (2011) Genetic incompatibility dampens hybrid fertility more than hybrid viability: yeast as a case study. PLoS ONE 6: e18341 doi:10.1371/journal.pone.0018341.
29. MichnickSW, EarPH, LandryC, MalleshaiahMK, MessierV (2010) A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods in Enzymology 470 : 335–368.
30. FreschiL, Torres-QuirozF, DubéAK, LandryCR (2012) qPCA: a scalable assay to measure the perturbation of protein-protein interactions in living cells. Molecular BioSystems DOI: 10.1039/C2MB25265A.
31. NeumannN, LundinD, PooleAM (2010) Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE 5: e13241 doi:10.1371/journal.pone.0013241.
32. BaptesteE, CharleboisRL, MacLeodD, BrochierC (2005) The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol 6: R85.
33. WoychikNA (1998) Fractions to functions: RNA polymerase II thirty years later. Cold Spring Harb Symp Quant Biol 63 : 311–317.
34. AlberF, DokudovskayaS, VeenhoffLM, ZhangW, KipperJ, et al. (2007) The molecular architecture of the nuclear pore complex. Nature 450 : 695–701.
35. CramerP, BushnellDA, FuJ, GnattAL, Maier-DavisB, et al. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288 : 640–649.
36. FraserHB, HirshAE, SteinmetzLM, ScharfeC, FeldmanMW (2002) Evolutionary rate in the protein interaction network. Science 296 : 750–752.
37. WinzelerEA, ShoemakerDD, AstromoffA, LiangH, AndersonK, et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 : 901–906.
38. KellisM, BirrenBW, LanderES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428 : 617–624.
39. EarPH, MichnickSW (2009) A general life-death selection strategy for dissecting protein functions. Nat Methods 6 : 813–816.
40. HittingerCT, GoncalvesP, SampaioJP, DoverJ, JohnstonM, et al. (2010) Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature 464 : 54–58.
41. HoelzA, DeblerEW, BlobelG (2011) The structure of the nuclear pore complex. Annu Rev Biochem 80 : 613–643.
42. ZamirL, ZaretskyM, FridmanY, Ner-GaonH, RubinE, et al. (2012) Tight coevolution of proliferating cell nuclear antigen (PCNA)-partner interaction networks in fungi leads to interspecies network incompatibility. Proc Natl Acad Sci U S A 109: E406–414.
43. Fernandez-MartinezJ, PhillipsJ, SekedatMD, Diaz-AvalosR, Velazquez-MurielJ, et al. (2012) Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol 196 : 419–434.
44. EmtageJL, BucciM, WatkinsJL, WenteSR (1997) Defining the essential functional regions of the nucleoporin Nup145p. J Cell Sci 110 (Pt 7) 911–925.
45. TeixeiraMT, FabreE, DujonB (1999) Self-catalyzed cleavage of the yeast nucleoporin Nup145p precursor. J Biol Chem 274 : 32439–32444.
46. FicarroSB, McClelandML, StukenbergPT, BurkeDJ, RossMM, et al. (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol 20 : 301–305.
47. SmolkaMB, AlbuquerqueCP, ChenSH, ZhouH (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A 104 : 10364–10369.
48. MalmstromL, RiffleM, StraussCE, ChivianD, DavisTN, et al. (2007) Superfamily assignments for the yeast proteome through integration of structure prediction with the gene ontology. PLoS Biol 5: e76 doi:10.1371/journal.pbio.0050076.
49. RoutMP, AitchisonJD, SupraptoA, HjertaasK, ZhaoY, et al. (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148 : 635–651.
50. LevyED (2010) A simple definition of structural regions in proteins and its use in analyzing interface evolution. J Mol Biol 403 : 660–670.
51. MelcakI, HoelzA, BlobelG (2007) Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science 315 : 1729–1732.
52. BoehmerT, JeudyS, BerkeIC, SchwartzTU (2008) Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell 30 : 721–731.
53. YoshidaK, SeoHS, DeblerEW, BlobelG, HoelzA (2011) Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc Natl Acad Sci U S A 108 : 16571–16576.
54. QianW, HeX, ChanE, XuH, ZhangJ (2011) Measuring the evolutionary rate of protein-protein interaction. Proc Natl Acad Sci U S A 108 : 8725–8730.
55. PresgravesDC, BalagopalanL, AbmayrSM, OrrHA (2003) Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423 : 715–719.
56. PresgravesDC, StephanW (2007) Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol Biol Evol 24 : 306–314.
57. GreigD (2007) A screen for recessive speciation genes expressed in the gametes of F1 hybrid yeast. PLoS Genet 3: e21 doi:10.1371/journal.pgen.0030021.
58. KaoKC, SchwartzK, SherlockG (2010) A genome-wide analysis reveals no nuclear dobzhansky-muller pairs of determinants of speciation between S. cerevisiae and S. paradoxus, but suggests more complex incompatibilities. PLoS Genet 6: e1001038 doi:10.1371/journal.pgen.1001038.
59. HeckJA, ArguesoJL, GemiciZ, ReevesRG, BernardA, et al. (2006) Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc Natl Acad Sci U S A 103 : 3256–3261.
60. LitiG, HaricharanS, CubillosFA, TierneyAL, SharpS, et al. (2009) Segregating YKU80 and TLC1 alleles underlying natural variation in telomere properties in wild yeast. PLoS Genet 5: e1000659 doi:10.1371/journal.pgen.1000659.
61. Arroyo-LopezFN, OrlicS, QuerolA, BarrioE (2009) Effects of temperature, pH and sugar concentration on the growth parameters of Saccharomyces cerevisiae, S. kudriavzevii and their interspecific hybrid. Int J Food Microbiol 131 : 120–127.
62. HittingerCT, RokasA, CarrollSB (2004) Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc Natl Acad Sci U S A 101 : 14144–14149.
63. WardJJ, SodhiJS, McGuffinLJ, BuxtonBF, JonesDT (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337 : 635–645.
Štítky
Genetika Reprodukčná medicína
Článek Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction DiseaseČlánek Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination SchemesČlánek Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- A Mystery Unraveled: Essentiality of RNase III in Is Caused by Resident Prophages
- Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction Disease
- Insertion/Deletion Polymorphisms in the Promoter Are a Risk Factor for Bladder Exstrophy Epispadias Complex
- Mi2β Is Required for γ-Globin Gene Silencing: Temporal Assembly of a GATA-1-FOG-1-Mi2 Repressor Complex in β-YAC Transgenic Mice
- Dissection of a Quantitative Trait Locus for PR Interval Duration Identifies as a Novel Modulator of Cardiac Conduction
- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Construction of a Global Pain Systems Network Highlights Phospholipid Signaling as a Regulator of Heat Nociception
- Genes Contributing to Pain Sensitivity in the Normal Population: An Exome Sequencing Study
- Identification of , a Locus Controlling Dominant Resistance to Autoimmune Orchitis, as Kinesin Family Member 1C
- ATX1-Generated H3K4me3 Is Required for Efficient Elongation of Transcription, Not Initiation, at ATX1-Regulated Genes
- Dynamic and Differential Regulation of Stem Cell Factor FoxD3 in the Neural Crest Is Encrypted in the Genome
- Identification of Novel Type 2 Diabetes Candidate Genes Involved in the Crosstalk between the Mitochondrial and the Insulin Signaling Systems
- The Genetic Architecture of Adaptations to High Altitude in Ethiopia
- Population Genomics of the Endosymbiont in
- Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination Schemes
- Testicular Differentiation Occurs in Absence of R-spondin1 and Sox9 in Mouse Sex Reversals
- A Yeast GSK-3 Kinase Mck1 Promotes Cdc6 Degradation to Inhibit DNA Re-Replication
- Genetic Adaptation Associated with Genome-Doubling in Autotetraploid
- The Essential Function of RNase III Is to Silence Foreign Toxin Genes
- Long-Range Regulatory Polymorphisms Affecting a GABA Receptor Constitute a Quantitative Trait Locus (QTL) for Social Behavior in
- A New Isolation with Migration Model along Complete Genomes Infers Very Different Divergence Processes among Closely Related Great Ape Species
- Chromosome Fragile Sites in Harbor Matrix Attachment Regions That May Be Associated with Ancestral Chromosome Rearrangement Events
- Genome-Wide Association Study Implicates Testis-Sperm Specific as a Susceptibility Locus for Impaired Acrosome Reaction in Stallions
- A Mechanism of Gene Amplification Driven by Small DNA Fragments
- Base Damage within Single-Strand DNA Underlies Hypermutability Induced by a Ubiquitous Environmental Agent
- Integrative Analysis of a Cross-Loci Regulation Network Identifies as a Gene Regulating Insulin Secretion from Pancreatic Islets
- Reciprocal Signaling between the Ectoderm and a Mesendodermal Left-Right Organizer Directs Left-Right Determination in the Sea Urchin Embryo
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Genome-Wide Fine-Scale Recombination Rate Variation in
- Deciphering the Transcriptional-Regulatory Network of Flocculation in
- On Lung Function and Interactions Using Genome-Wide Data
- Genome-Wide Joint Meta-Analysis of SNP and SNP-by-Smoking Interaction Identifies Novel Loci for Pulmonary Function
- The Effective Population Size of Malaria Mosquitoes: Large Impact of Vector Control
- Recessive Mutations in Implicate β-III Spectrin in Both Cognitive and Motor Development
- An Excess of Gene Expression Divergence on the X Chromosome in Embryos: Implications for the Faster-X Hypothesis
- Reduced Life- and Healthspan in Mice Carrying a Mono-Allelic MVA Mutation
- Natural Variation at the MATE Transporter Locus Reveals Cross-Talk between Fe Homeostasis and Zn Tolerance in
- Histone Deacetylase HDA6 Is Functionally Associated with AS1 in Repression of Genes in
- A Framework for the Establishment of a Cnidarian Gene Regulatory Network for “Endomesoderm” Specification: The Inputs of ß-Catenin/TCF Signaling
- A Polycomb Group Protein Is Retained at Specific Sites on Chromatin in Mitosis
- Diapause Formation and Downregulation of Insulin-Like Signaling via DAF-16/FOXO Delays Axonal Degeneration and Neuronal Loss
- Genes That Act Downstream of Sensory Neurons to Influence Longevity, Dauer Formation, and Pathogen Responses in
- A Genome-Wide RNAi Screen Reveals MAP Kinase Phosphatases as Key ERK Pathway Regulators during Embryonic Stem Cell Differentiation
- Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing–Based Approach
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
- Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
- Systems Genetic Analysis of Osteoblast-Lineage Cells
- Population Genomics of Inversion Polymorphisms in
- Spreading of Heterochromatin Is Limited to Specific Families of Maize Retrotransposons
- DNA Topoisomerases Maintain Promoters in a State Competent for Transcriptional Activation in
- A Histone Deacetylase Adjusts Transcription Kinetics at Coding Sequences during Morphogenesis
- Approaching the Functional Annotation of Fungal Virulence Factors Using Cross-Species Genetic Interaction Profiling
- Evidence for the Robustness of Protein Complexes to Inter-Species Hybridization
- Systematic Identification of Rhythmic Genes Reveals as a New Element in the Circadian Clockwork
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



