-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
Sugars are important nutrients for many animals, but are also proposed to contribute to overnutrition-derived metabolic diseases in humans. Understanding the genetic factors governing dietary sugar tolerance therefore has profound biological and medical significance. Paralogous Mondo transcription factors ChREBP and MondoA, with their common binding partner Mlx, are key sensors of intracellular glucose flux in mammals. Here we report analysis of the in vivo function of Drosophila melanogaster Mlx and its binding partner Mondo (ChREBP) in respect to tolerance to dietary sugars. Larvae lacking mlx or having reduced mondo expression show strikingly reduced survival on a diet with moderate or high levels of sucrose, glucose, and fructose. mlx null mutants display widespread changes in lipid and phospholipid profiles, signs of amino acid catabolism, as well as strongly elevated circulating glucose levels. Systematic loss-of-function analysis of Mlx target genes reveals that circulating glucose levels and dietary sugar tolerance can be genetically uncoupled: Krüppel-like transcription factor Cabut and carbonyl detoxifying enzyme Aldehyde dehydrogenase type III are essential for dietary sugar tolerance, but display no influence on circulating glucose levels. On the other hand, Phosphofructokinase 2, a regulator of the glycolysis pathway, is needed for both dietary sugar tolerance and maintenance of circulating glucose homeostasis. Furthermore, we show evidence that fatty acid synthesis, which is a highly conserved Mondo-Mlx-regulated process, does not promote dietary sugar tolerance. In contrast, survival of larvae with reduced fatty acid synthase expression is sugar-dependent. Our data demonstrate that the transcriptional network regulated by Mondo-Mlx is a critical determinant of the healthful dietary spectrum allowing Drosophila to exploit sugar-rich nutrient sources.
Published in the journal: Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in. PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003438
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003438Summary
Sugars are important nutrients for many animals, but are also proposed to contribute to overnutrition-derived metabolic diseases in humans. Understanding the genetic factors governing dietary sugar tolerance therefore has profound biological and medical significance. Paralogous Mondo transcription factors ChREBP and MondoA, with their common binding partner Mlx, are key sensors of intracellular glucose flux in mammals. Here we report analysis of the in vivo function of Drosophila melanogaster Mlx and its binding partner Mondo (ChREBP) in respect to tolerance to dietary sugars. Larvae lacking mlx or having reduced mondo expression show strikingly reduced survival on a diet with moderate or high levels of sucrose, glucose, and fructose. mlx null mutants display widespread changes in lipid and phospholipid profiles, signs of amino acid catabolism, as well as strongly elevated circulating glucose levels. Systematic loss-of-function analysis of Mlx target genes reveals that circulating glucose levels and dietary sugar tolerance can be genetically uncoupled: Krüppel-like transcription factor Cabut and carbonyl detoxifying enzyme Aldehyde dehydrogenase type III are essential for dietary sugar tolerance, but display no influence on circulating glucose levels. On the other hand, Phosphofructokinase 2, a regulator of the glycolysis pathway, is needed for both dietary sugar tolerance and maintenance of circulating glucose homeostasis. Furthermore, we show evidence that fatty acid synthesis, which is a highly conserved Mondo-Mlx-regulated process, does not promote dietary sugar tolerance. In contrast, survival of larvae with reduced fatty acid synthase expression is sugar-dependent. Our data demonstrate that the transcriptional network regulated by Mondo-Mlx is a critical determinant of the healthful dietary spectrum allowing Drosophila to exploit sugar-rich nutrient sources.
Introduction
Mono - and disaccharides, i.e. sugars, are an important source of nutritional energy, but animal species display marked differences in the degree of sugar utilization and tolerance. While the diet of carnivores is typically low in sugars, nectarivores, like hummingbirds, feed primarily on sugar-rich nectar [1], [2]. Sugars from fruits and honey have been part of the ancestral human diet. However, the large quantities of refined sugars consumed by modern humans far exceed those available in natural sources [3]. In fact, it has been proposed that excessive added sugar in the diet, especially fructose, might contribute to the development of metabolic syndrome [3]–[5]. Yet the genetic factors governing the delicate balance between healthful dietary sugar utilization and the sugar overload-induced metabolic disturbance are poorly understood.
Drosophila is a well-suited model for exploring the physiological consequences of sugar intake. Drosophila melanogaster is a generalist fruit breeder naturally performing well on a broad range of dietary sugars [6]. However, excessive intake of sugars has been shown to cause diabetes-like metabolic changes in D. melanogaster, including insulin resistance, elevated circulating glucose and increased adiposity [7], [8]. Dietary sugars have also been shown to shorten Drosophila lifespan [9]. The sugar-induced insulin resistance has been attributed to the JNK-regulated lipocalin Neural Lazarillo [8]. Moreover, high sugar induced gene expression has been previously analysed [7], [10]. However, beyond these observations, the functional interactions between genotype and dietary sugar have remained poorly understood.
Elevated systemic glucose levels cause cellular stress and tissue damage [11], [12]. Animals therefore rapidly adapt their metabolism to fluctuating sugar intake, maintaining circulating glucose levels constant. A postprandial increase in circulating glucose triggers the release of insulin, which induces the rapid uptake of excess glucose by metabolic tissues including muscle, adipose tissue, and liver [13]. Intracellular glucose is immediately converted into glucose-6-phosphate and further metabolized into glycogen and lipids or catabolised to release energy. Metabolic tissues are exposed to large variations in the flux of intracellular glucose and therefore need to regulate their metabolism accordingly.
The basic helix-loop-helix transcription factor paralogs ChREBP (Carbohydrate Response Element Binding Protein) and MondoA act together with their common binding partner Mlx (Max-like protein X) to mediate transcriptional responses to intracellular glucose in mammals [14]. The ChREBP/MondoA-Mlx complex is activated by glucose-6-phosphate and other phosphorylated hexoses, and regulates gene expression by binding to target promoters containing a carbohydrate response element (ChoRE) [15]–[19]. ChREBP and MondoA regulate the majority of the global glucose-induced transcriptional responses and many of their target genes encode enzymes in glycolytic and lipogenic pathways [15], [16], [20], [21]. ChREBP and MondoA play differential tissue-specific roles in mammals: ChREBP functions in the liver, adipose tissue and pancreatic beta cells [22]–[26], while MondoA is predominantly expressed in the skeletal muscle [27].
Of the mammalian ChREBP/MondoA-Mlx complex, the role of ChREBP has been studied in a physiological setting using loss-of-function mice. While ChREBP is nonessential in terms of survival, the mutant mice display a number of metabolic phenotypes, including elevated plasma glucose and liver glycogen as well as reduced adiposity [25], [28]. ChREBP−/− mice survive poorly on a diet with high levels of sugars, but the underlying reasons have remained unexplored [25]. ChREBP is known to regulate a range of metabolic genes, including those involved in de novo lipogenesis [15], [21]. Which target genes contribute to the various physiological phenotypes and what is the causal interrelationship between the physiological phenotypes, are questions that require powerful genetics and are therefore challenging to address in vivo in mammals. Moreover, existence of another Mondo paralog, MondoA, might mask some phenotypes in the ChREBP−/− mouse. To better understand the physiological roles of the Mondo/ChREBP/-Mlx complex and its target genes, we have explored their role in Drosophila melanogaster. The Drosophila genome encodes one ortholog for each of ChREBP/MondoA and Mlx, which we call Mondo (alternative identifiers: CG18362, Mlx interactor, ChREBP) and Mlx (alternative identifiers: CG3350, Bigmax), respectively [27], [29], [30].
We have generated mlx null mutant flies, which displayed lethality in the late pupal stage. D. melanogaster larvae can normally utilize high levels of dietary sugars [6], but loss of Mlx or knockdown of Mondo caused striking intolerance towards sucrose, glucose and fructose. The mlx null mutant larvae also displayed extensive metabolic changes, with strongly elevated circulating glucose, signs of amino acid catabolism and altered lipid and phospholipid profiles. Systematic functional analysis of Mlx-regulated genes revealed three genes contributing to dietary sugar tolerance: cabut, encoding a Krüppel-like transcription factor, phosphofructokinase 2, a regulator of the glycolytic pathway, and Aldehyde dehydrogenase type III, which is linked to detoxification of reactive aldehydes.
Results
Mondo-Mlx complex is essential for sugar tolerance
To study the physiological role of Drosophila Mlx, we generated a mutant allele using imprecise P-element (P{XP}bigmaxd07258) excision. We recovered one mutant allele, mlx1, which lacked the entire coding region of mlx as well as 17 C-terminal amino acids of the neighbouring gene CG3368 (Figure 1A). As controls, we recovered lines from which the P-element had been excised precisely, leaving mlx intact. If not differently stated, the precise excision line is used as a control throughout the study. As expected, mlx1 mutant larvae expressed neither Mlx protein nor mRNA (Figure 1B; Figure S1A). The mlx1 mutants displayed lethality at the late pupal (pharate) stage (Figure S1B), and only a small number of adult flies could be recovered. mlx1 mutant flies also displayed a modest developmental delay when raised on our standard fly food (Figure S1C).
Fig. 1. Drosophila larvae lacking functional Mlx display strong intolerance to dietary sugars. 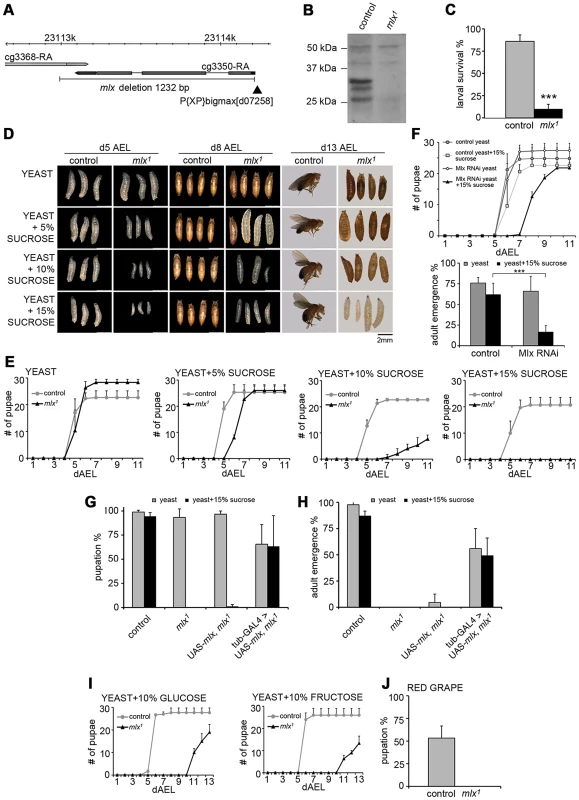
(A) Schematic presentation of mlx1 mutant locus. P-element is indicated with black arrowhead. (B) Western blot from control and mlx1 mutant larval lysates using anti-Mlx antibodies. (C) Survival of control and mlx1 mutant larvae after 4 days on a 20% sucrose-only diet. (D) Images of control and mlx1 mutants on 20% yeast diet supplemented with 0–15% sucrose after 5, 8, and 13 days after egg laying (dAEL). (E) Pupation kinetics of control and mlx1 mutants on a 20% yeast diet supplemented with 0–15% sucrose. (F) Pupation kinetics and survival of control (tub-GAL4>) and Mlx RNAi (tub-GAL4>Mlx RNAi) larvae on 20% yeast diet with or without 15% sucrose. (G) Rescue of mlx1 mutant pupation on high sugar diet by ubiquitous (tub-GAL4) transgenic expression of mlx. (H) Rescue of adult viability of mlx1 mutants by ubiquitous (tub-GAL4) transgenic expression of mlx. (I) Pupation kinetics of control and mlx1 mutants on a 20% yeast diet supplemented with 10% glucose or fructose. (J) Pupation % of control and mlx1 mutant larvae on pieces of red grape with baker's yeast inoculum. Error bars represent ± SEM (*** p<0.001). Experiments with defined nutrients revealed that mlx1 mutant larvae failed to survive on a diet with 20% sucrose as the sole nutrient source (Figure 1C). To test if the mutant lethality was due to either the inability to utilize carbohydrates as energy source or intolerance towards sucrose, we supplemented protein-rich diet (20% yeast) with increasing levels of sucrose. While mlx1 mutant larvae developed with similar kinetics as control animals on a high protein/low sugar diet, increasing the sucrose concentration gradually slowed down larval development of mlx1 mutants (Figure 1D,E). At higher sucrose levels, mlx1 mutants failed to pupate and died as larvae, while control animals displayed no apparent change in pupation kinetics with respect to 0–15% sucrose.
To confirm that the observed phenotypes were due to loss of mlx function, we used RNAi-mediated knockdown and transgenic rescue. Ubiquitous knockdown of Mlx by RNAi led to significantly slower pupation, and increased pupal lethality on protein rich food supplemented with 15% sucrose, while displaying no visible phenotype in the absence of added sucrose (Figure 1F). Driver line without RNAi was used as a control. Moreover, sugar intolerance and pupal lethality of the mlx1 mutants were efficiently rescued by ubiquitous expression of transgenic mlx (tub-GAL4>UAS-mlx) (Figure 1G, 1H). To further rule out the possibility that sugar intolerance was due to disturbed function of the neighbouring gene CG3368, we used RNAi for ubiquitous knockdown. CG3368 was efficiently silenced, but no sugar intolerance was observed (Figure S1D–S1F). Thus, mlx gene function is essential for tolerance to dietary sucrose.
To test for specific intolerance towards glucose or fructose, we supplemented the protein-rich food with 10% of either monosaccharide. Both caused clear developmental delays of mlx1 mutants (Figure 1I). Drosophila melanogaster is a dietary generalist, feeding on micro-organisms on decaying fruits and vegetables that have varying sugar content. To test whether the sugar intolerance of mlx1 mutants was relevant within the natural spectrum of D. melanogaster's diet, we allowed larvae to develop on pieces of red grape with baker's yeast inoculum. Indeed, mlx1 mutants were unable to pupate in these conditions, while >50% of the control larvae reached the pupal stage (Figure 1J).
In mammals, Mlx forms a functional complex with the Mondo paralogs, MondoA and ChREBP. We tested if the heterodimeric function of Mlx is conserved in Drosophila and essential for the sugar tolerance. Drosophila Mlx co-immunoprecipitated with Mondo when expressed in Drosophila S2 cells, suggesting heterodimeric function (Figure 2A). Notably, Mlx from S2 cells runs as two distinct bands (Figure 2A), which correspond to the two upper bands present in the in vivo sample (Figure 1B and Figure S2). The nature of these bands has remained unclear, no alternative splicing has been reported and both bands are resistant to alkaline phosphatase treatment (not shown). Ubiquitous RNAi knockdown of Mondo (tub-GAL4>Mondo RNAi) led to delayed pupation (Figure 2B) and reduced pupal survival on high sugar diet (20% yeast-15% sucrose) (Figure 2C). In sum, the biochemical and genetic evidence implies conservation of the Mondo-Mlx heterodimer function in Drosophila.
Fig. 2. Evidence for functional Mondo-Mlx heterodimer. 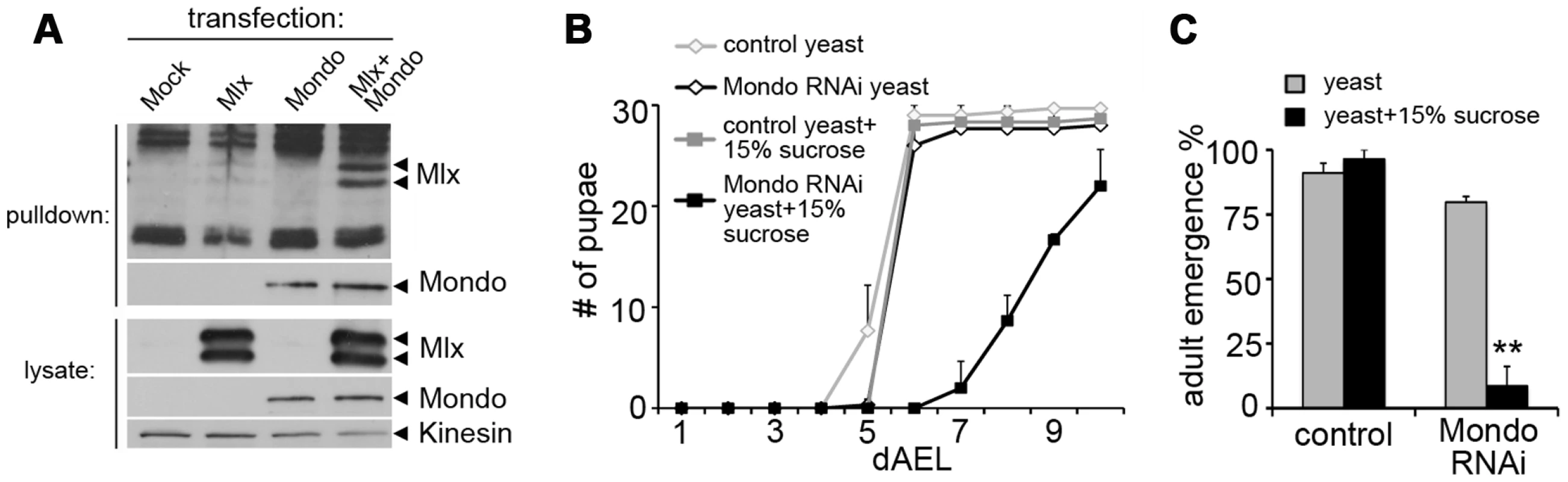
(A) Co-purification of Mlx upon pulldown of Strep-tagged Mondo from S2 cells. Mlx was detected on Western blot using anti-Mlx antibodies. Mondo-Strep-V5 was detected using anti-V5 antibodies. Kinesin was used as loading control. (B) Pupation kinetics of control (tub-GAL4>) and Mondo RNAi (tub-GAL4>Mondo RNAi) larvae on 20% yeast diet with or without 15% sucrose. (C) Survival into adulthood of control (tub-GAL4>) and Mondo RNAi (tub-GAL4>Mondo RNAi) animals grown on 20% yeast diet with or without 15% sucrose. Error bars represent ± SEM (** p<0.01). Changes in lipid, amino acid, and carbohydrate metabolism in mlx mutants
The diet-dependent phenotype of mlx1 mutants led us to perform a comprehensive survey of their metabolic status using mass-spectrometry based lipidomics and metabolomics. Lipidomics analysis revealed significant downregulation of key phospholipid groups, such as phosphatidylethanolamines (PE) and lysophosphatidylcholines (LysoPC) (Figure 3A). Total triglyceride (TG) levels showed a lower trend in mlx1 mutants, but the difference to the controls was not statistically significant (Figure 3B). However, mlx1 mutants showed significant enrichment in triglyceride species with long fatty acid tails (Figure 3B). At the same time, mlx1 mutants showed strong downregulation of certain fatty acids, such as myristoleic acid and lauric acid (Figure 3C). On the other hand, ceramide (Cer) levels were elevated in mlx1 mutants compared to controls (Figure 3D). Together, mlx1 mutants display signs of severely altered lipid and phospholipid metabolism. In addition to the changes in lipid profiles, total amino acid levels were significantly reduced in mlx1 mutants (Figure 3E), while concentration of urea was dramatically increased (Figure 3F). This implies that mlx1 mutants might catabolise amino acids for energy.
Fig. 3. Changes in lipid and amino acid profiles in mlx1 mutants. 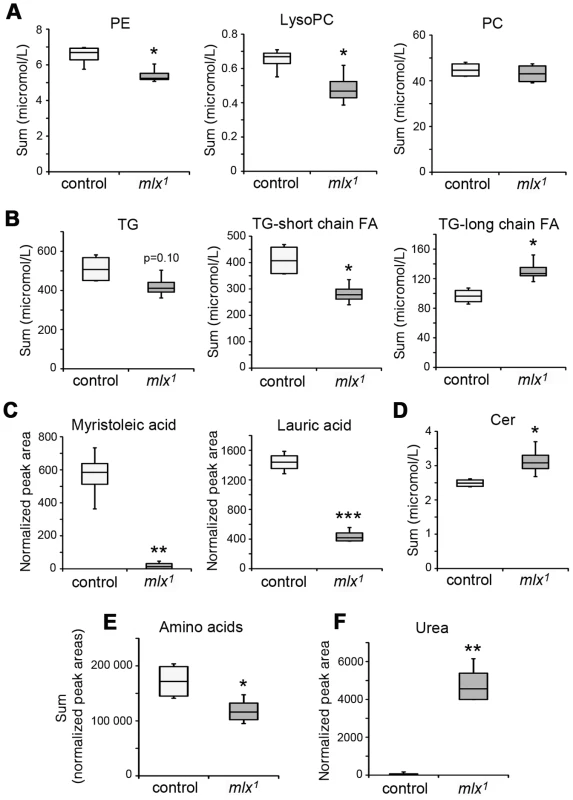
(A) Phospholipid levels in control and mlx1 mutant larvae. PE: phosphatidylethanolamine; LysoPC: lysophosphatidylcholine; PC: phosphatidylcholine. (B) Total triglyceride levels as well as triglycerides divided into two groups based on total fatty acid length. For TG-long chain FA group, total fatty acid carbon number >48. (C) Levels of myristoleic acid and lauric acid were strongly reduced in mlx1 mutant larvae. (D) Ceramide levels in control and mlx1 mutant larvae. (E) Total levels of amino acids in control and mlx1 mutant larvae. (F) Urea levels in control and mlx1 mutant larvae. Error bars represent ± SEM (*p<0.05; ** p<0.01; *** p<0.001). The box contains the middle 50% of the data, the two ends of the boxes represent the upper and the lower quartile of the data. The median is represented by the line in the center of the rectangular box. The vertical lines mark the minimum and maximum data values. To study changes in glucose metabolism, we measured glucose and trehalose levels in the hemolymph of larvae raised on diets with varying sucrose content. Trehalose is a disaccharide released by gluconeogenesis and glycogenolysis in insects [31]. The levels of circulating glucose were moderately elevated in mlx1 mutant larvae raised on a low-sugar diet (20% yeast) (Figure 4A). However, increasing the dietary sucrose to 5%, which still sustains larval development of mlx1 mutants, led to a prominent increase of circulating glucose in mlx1 mutants while remaining constant in control animals (Figure 4A). Trehalose levels were also significantly elevated in mlx1 mutants, but unlike glucose, trehalose levels were little affected by the dietary sucrose levels (Figure 4B). Furthermore, glycogen levels were significantly elevated in mlx1 mutants (Figure 4C), indicating that glucose catabolism, not cellular glucose intake, is limiting glucose clearance from circulation in mlx1 mutants.
Fig. 4. Elevated glucose, trehalose, and glycogen levels in the absence of Mondo-Mlx. 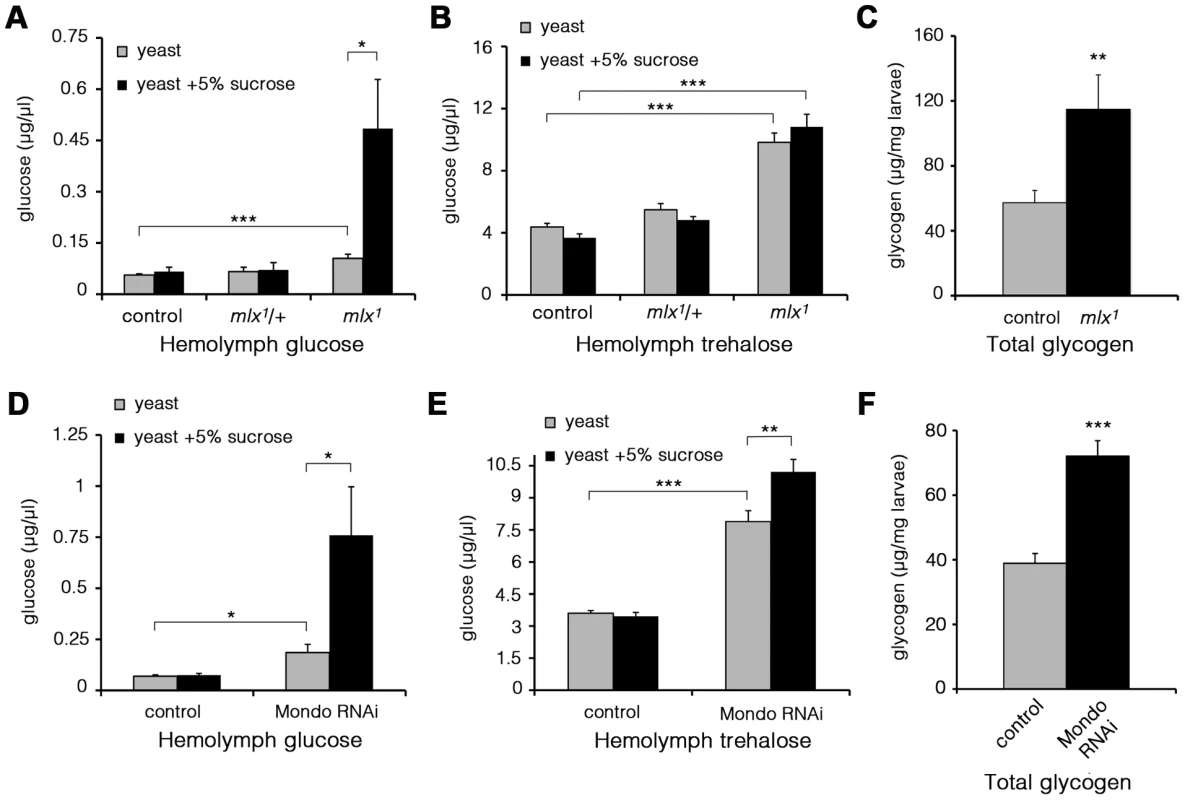
Hemolymph glucose (A) and trehalose (B) levels in control and mlx1 mutant larvae on a 20% yeast diet with or without 5% sucrose. (C) Glycogen levels in control and mlx1 mutant larvae on a 20% yeast-5% sucrose diet. Hemolymph glucose (D) and trehalose (E) levels in control (tub-GAL4>) and Mondo RNAi (tub-GAL4>Mondo RNAi) larvae grown on a 20% yeast-5% sucrose diet. (F) Glycogen levels in control (tub-GAL4>) and Mondo RNAi (tub-GAL4>Mondo RNAi) larvae on 20% yeast-5% sucrose diet. Error bars represent ± SEM (* p<0.05; ** p<0.01; *** p<0.001). To verify that the elevated glucose levels were due to loss of Mlx function, we performed a transgenic rescue, which normalized circulating glucose levels (Figure S3A). Further, RNAi-mediated knockdown of Mlx led to a clear increase in circulating glucose, trehalose and glycogen (Figure S3B–S3D). In line with the expectation of heterodimeric function for Mondo and Mlx proteins, Mondo RNAi knockdown led to a prominent increase in circulating glucose and trehalose (Figure 4D,E). Also the glycogen levels were significantly increased in Mondo RNAi larvae (Figure 4F). Knockdown of CG3368, the neighbouring gene of mlx, had no influence on circulating glucose or trehalose (Figure S3B, S3C). In conclusion, mlx null mutant and mondo knockdown larvae display a reduced capacity to utilize circulating glucose leading to poor homeostatic adaptation to elevated dietary sugar.
Mlx is functionally important in the fat body
As mammalian MondoA-Mlx and ChREBP-Mlx appear to display tissue-specific functions, we wanted to determine the functionally important tissues for Drosophila Mondo-Mlx. We first analyzed their mRNA expression using quantitative RT-PCR. Expression of Mondo and Mlx were highly correlated; highest levels were detected in the fat body, gut and Malpighian tubules (Figure 5A). To functionally dissect the contribution of different tissues to the sugar sensitivity, we used tissue-specific transgenic rescue. Restoring Mlx expression in neurons (Elav-GAL4) or muscle (Mef2-GAL4) did not significantly improve the sugar tolerance or survival of mlx1 mutants (Figure 5B). However, targeted expression in the fat body with two independent driver lines (ppl - and r4-GAL4) efficiently rescued survival on high sugar diet. Moreover, rescue of Mlx in the fat body, but not in muscle, was sufficient to normalize the levels of circulating glucose in mlx1 mutants (Figure 5C). Notably, transgenic Mlx expression by tub-GAL4 (Figure 1G) or Mef2-GAL4 (Figure 5B) caused moderately reduced survival, which was also observed in the presence of intact endogenous mlx (Figure S4).
Fig. 5. Mlx is functionally important in the fat body. 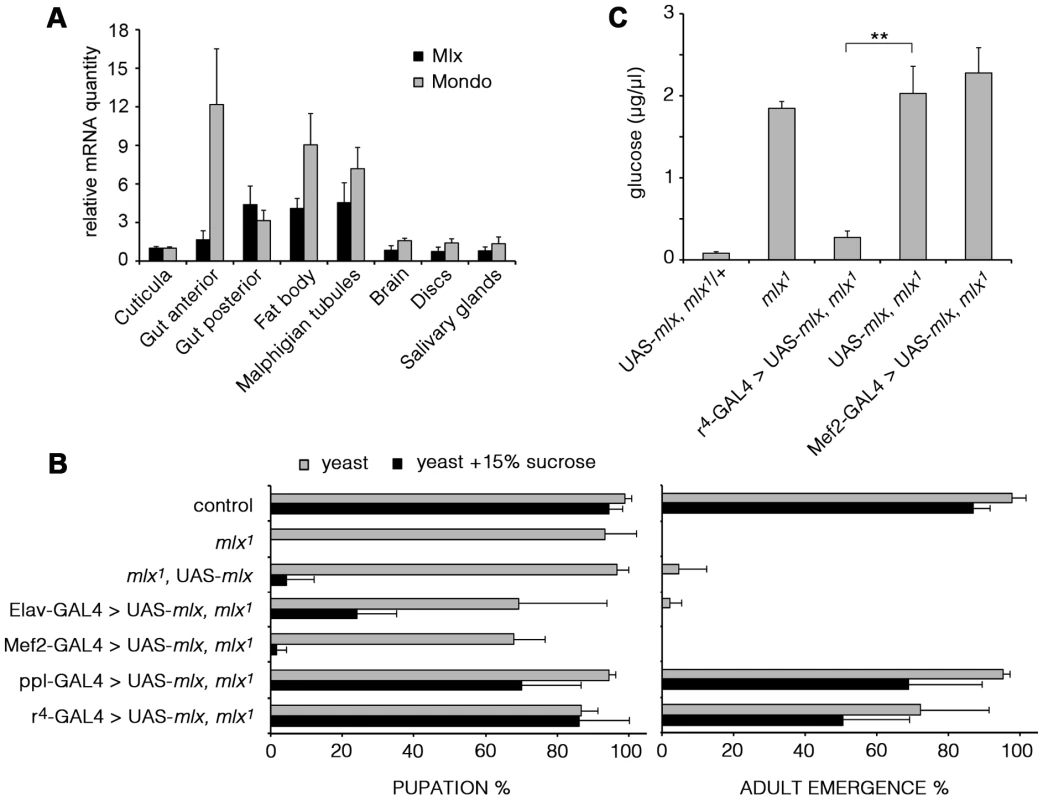
(A) mRNA expression of mondo and mlx in dissected tissues of 3rd instar larvae measured by quantitative RT-PCR. rp49 was used as a reference gene. (B) Tissue specific rescue of pupation and adult emergence of the mlx1 mutants using neuronal (Elav), muscle (Mef2), or fat body (ppl and r4) –specific GAL4 drivers. (C) Fat body –specific (r4-GAL4) expression of transgenic mlx rescues the elevated circulating glucose in mlx1 mutants. Error bars represent ± SEM (** p<0.01). Mlx regulates metabolic genes and a secondary transcriptional effector
To identify Mlx target genes, we performed a microarray gene expression profiling specifically in the fat body. Comparing gene expression between mlx1 mutant and control fat bodies from 3rd instar prewandering larvae raised on a moderate sucrose level diet (20% yeast-5% sucrose) revealed 97 down - and 96 up-regulated genes (>2-fold change and adjusted p-value<0.05) (Figure 6A; Table S1). As expected for a deletion mutant, mlx was identified as the most strongly downregulated gene on the microarray (Figure 6A). Gene Set Enrichment Analysis (GSEA) revealed a significant enrichment of KEGG categories involved in metabolic regulation (Figure 6B). For example, KEGG categories of fatty acid metabolism and nitrogen metabolism were strongly downregulated (Figure 6C), which is in good agreement with the metabolomics data (Figure 3). Many of the genes downregulated in mlx1 mutant fat body also showed reduced expression during earlier larval stages in whole larval samples (Figure 6D). Mlx-regulated genes include several key metabolic genes, such as glycerol-3-phosphate dehydrogenase-1 (Gpdh, CG9042), stearoyl-CoA 9-desaturase-1 (desat1, CG5887), Glutamine synthetase-1 (Gs1, CG2718), and 3-hydroxybutyrate dehydrogenase (sro, CG12068). The mean expression levels of three known ChREBP and MondoA targets in mammals, fatty acid synthase (Fas, CG3523), acetyl-CoA carboxylase (ACC, CG11198) and phosphofructokinase 2 (PFK2, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, CG3400), [20], [32], were also reduced, although they did not pass our strict microarray cut-offs (Figure 6D). Interestingly, one of the most strongly downregulated genes in mlx1 mutant fat body was that encoding the Krüppel-like transcription factor Cabut (cbt; CG4427) (Figure 6A, 6D).
Fig. 6. Mlx-regulated gene expression program. 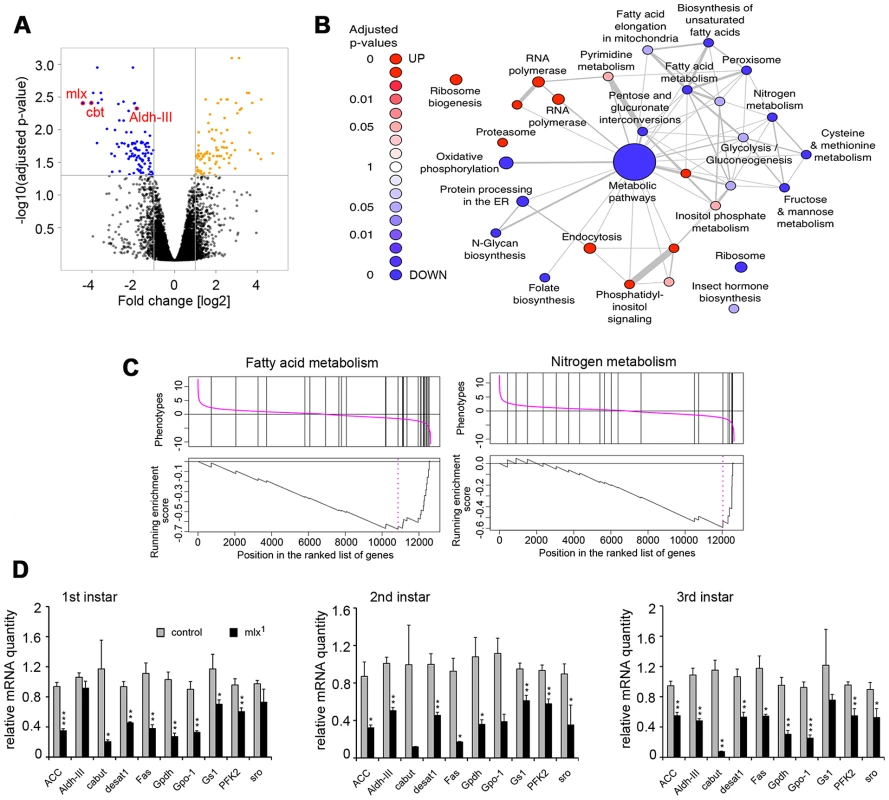
(A) Volcano plot indicating genes differentially expressed between control and mlx1 mutant fat bodies. Significant changes (p<0.05 and log2 fold changes>±1) are indicated in orange (upregulated in mlx1) and blue (downregulated in mlx1). (B) Results of a gene-set enrichment analysis (GSEA) of KEGG pathways. Red: upregulated set in mlx1 mutant, blue: downregulated set. The size of nodes illustrates the size of gene sets and the width of edges denotes the overlap between two gene sets. (C) GSEA charts for KEGG categories “Fatty acid metabolism” and “Nitrogen metabolism”. (D) Developmental stage-specific mRNA expression (quantitative-RT-PCR) of selected genes downregulated in mlx1 mutant in the microarray. Expression levels were normalized using Actin 42A as a reference gene. Error bars represent ± SEM (* p<0.05; ** p<0.01; *** p<0.001). Evidence for a transcriptional network mediating sugar tolerance
To test in an unbiased way if any of the genes downregulated in mlx1 mutants had an essential role in maintaining organismal sugar tolerance, we systematically targeted 103 candidate genes by RNAi (Table S2). Intriguingly, ubiquitous knockdown of two Mlx-regulated genes identified on the microarray led to significant sugar intolerance. Transcription factor Cabut was among the most highly Mlx-regulated genes in the microarray. Ubiquitous knockdown of Cabut expression by RNAi during the larval stage caused a modest delay of pupation on low sugar diet. However, on high sugar diet (20% yeast-15% sucrose) Cabut knockdown led to prominent developmental delay and impaired survival (Figure 7A). This suggests that Mondo-Mlx activates a hierarchical transcriptional network to regulate dietary sugar tolerance with Cabut as an essential downstream effector.
Fig. 7. Functional analysis of Mlx-regulated genes. 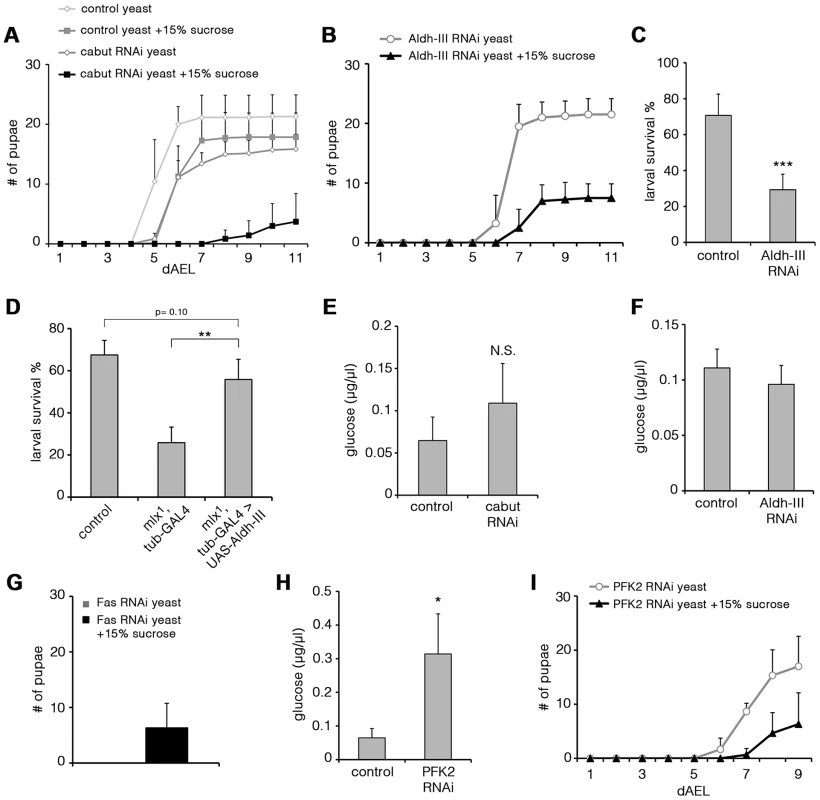
(A) Pupation kinetics of control (tubts-GAL4>) and Cabut RNAi (tubts-GAL4>Cabut RNAi) larvae on 20% yeast diet with or without 15% sucrose. To avoid early larval lethality, we used temperature sensitive GAL80 with tub-GAL4 system to induce RNAi expression during larval stage. Larvae were maintained in +29°C after collection. Cabut knockdown causes a moderate delay also in the low sugar (20% yeast) diet, but addition of 15% substantially reduces survival and delays pupation further. (B) Pupation kinetics of Aldh-III RNAi (tub-GAL4>Aldh-III RNAi) larvae on 20% yeast diet with or without 15% sucrose. (C) Survival of control (tub-GAL4>) and Aldh-III RNAi (tub-GAL4>Aldh-III RNAi) larvae after 6 days on 20% sucrose-only diet. (D) Survival of control, mlx1, tub-GAL4 and mlx1, tub-GAL4>UAS-Aldh-III larvae after 4 days on a 20% sucrose-only diet. (E) Hemolymph glucose levels in control (tubts-GAL4>) and Cabut RNAi (tubts-GAL4>Cabut RNAi) larvae on 20% yeast-5% sucrose. (F) Hemolymph glucose levels in control (tub-GAL4>) and Aldh-III RNAi (tub-GAL4>Aldh-III RNAi) larvae on 20% yeast-5% sucrose. (G) Total pupation of Fas RNAi (tub-GAL4>Fas RNAi) larvae on 20% yeast diet with or without 15% sucrose. (H) Hemolymph glucose levels in control (tub-GAL4>) and PFK2 RNAi (tub-GAL4>PFK2 RNAi) larvae on 20% yeast-5% sucrose. (I) Pupation kinetics of PFK2 RNAi (tub-GAL4>PFK2 RNAi) larvae on 20% yeast diet with or without 15% sucrose. Error bars represent ± SEM (* p<0.05; ** p<0.01; *** p<0.001). Uncoupling of dietary sugar tolerance and circulating glucose
Another Mlx-regulated gene, which caused sugar intolerance upon ubiquitous knockdown, was Aldehyde dehydrogenase type III (Aldh-III, CG11140; Figure 7B). Most Aldh-III knockdown animals reached the pharate stage on a low sugar diet, but died during early pupal stages on a high-sugar diet (Figure S5A). Similar early pupal lethality was observed in mlx1 mutants on sugar concentrations that allowed pupation (data not shown). Survival on a 20% sucrose-only diet was also significantly reduced upon Aldh-III knockdown (Figure 7C). We also tested whether restoring Aldh-III activity by transgenic expression would be sufficient to rescue impaired mlx1 mutant survival on high sugar diet. While transgenic expression of Aldh-III did not rescue mlx1 mutant pupation on 20% yeast-15% sucrose diet (data not shown), larval survival of mlx1 mutants on 20% sucrose-only diet was significantly improved by transgenic Aldh-III (Figure 7D). The same was true when Aldh-III expression was rescued only in the fat body (Figure S5B). In conclusion, Aldh-III is essential and sufficient for providing dietary sugar tolerance.
We also explored whether the sugar intolerance observed upon knockdown of Cabut and Aldh-III was associated with elevated circulating glucose levels. Surprisingly, knockdown of either gene did not result in a significant increase in circulating glucose (Figure 7E, 7F). This implies that impaired clearance of circulating glucose is not an essential prerequisite for intolerance to dietary sugars. Instead, these two Mondo-Mlx-regulated parameters can be uncoupled at the level of the downstream target genes. Based on these phenotypes, the Cabut-dependent branch of the transcriptional network mediates only a subset of Mondo-Mlx functions. We also studied the possibility that Cabut could be a direct regulator of Aldh-III, however the mRNA levels of Aldh-III were unchanged in Cabut RNAi larvae (Figure S5C). Also the mRNA levels of Mondo and Mlx were unchanced in Cabut RNAi larvae, indicating that Cabut is not a feedback regulator of Mondo-Mlx. Notably, it is possible that Cabut is regulating a subset of common genes with Mondo-Mlx.
Interference of fatty acid synthesis makes larvae dependent on sugar-rich diet
Perhaps the best established function of mammalian ChREBP-Mlx is promotion of de novo lipogenesis in response to high carbohydrate intake [14]. This function is mediated through upregulation of lipogenic genes and it appears to be conserved in Drosophila (Figure 6D; [33]). Two key targets of Mondo-Mlx involved in de novo lipogenesis are acetyl-CoA carboxylase (ACC) and fatty acid synthase (Fas). Thus exploring their function in respect to dietary sugar will reveal whether de novo lipogenesis is functionally coupled to sugar tolerance. While knockdown of ACC was embryonic lethal (data not shown), Fas knockdown animals displayed some degree of survival until pupal stage. Strikingly, Fas knockdown larvae displayed early larval lethality on high protein diet (20% yeast paste), but diet supplementation with 15% sucrose partially rescued the lethality allowing pupation (Figure 7G). Thus, Mondo-Mlx-mediated regulation of fatty acid synthesis is a non-essential function for dietary sugar tolerance. In contrast, high dietary sugars promote survival upon compromised fatty acid synthesis.
Phosphofructokinase 2 contributes to circulating glucose and sugar tolerance
As our systematic analysis of sugar tolerance genes did not reveal a mechanism by which Mondo-Mlx maintains low circulating glucose, we analyzed glucose levels on additional Mondo-Mlx targets that have a metabolic function. Phosphofructokinase 2 (PFK2, Pfrx, CG3400) synthesizes and breaks down fructose-2,6-bisphosphate, which is an allosteric activator of phosphofructokinase 1 and hence promotes glycolysis. PFK2 expression was downregulated in mlx1 mutants (Figure 6D). Knockdown of PFK2 led to elevated circulating glucose, showing that PFK2 is a Mondo-Mlx downstream target, which contributes to circulating glucose levels (Figure 7H). Interestingly, PFK2 knockdown also reduced pupation on high sugar diet (20% yeast-15% sucrose) (Figure 7I), implying that Mondo-Mlx-mediated activation of the glycolytic pathway contributes to dietary sugar tolerance.
Discussion
Our study demonstrates that interfering with the function of the Mondo-Mlx complex severely affects Drosophila energy metabolism, rendering animals highly intolerant to sugars in their diet. The sugar intolerance is likely due to a combined effect of several downstream effectors, since our systematic loss-of-function analysis revealed three Mlx target genes that are essential for survival on high sugar diet. Sugar tolerance is influenced by glycolysis, which also contributes to clearance of glucose from circulation. However, circulating glucose levels and sugar tolerance are phenotypes that can be uncoupled, as in the case of two Mlx targets, Cabut and Aldh-III, which only contribute to sugar tolerance.
Mondo-Mlx shows a high degree of functional conservation between flies and mammals, as orthologs of many Drosophila Mlx-regulated genes are known targets of ChREBP/MondoA-Mlx in mammals. These include glycerol-3-phosphate dehydrogenase-1, stearoyl-CoA 9-desaturase-1, fatty acid synthase, acetyl-CoA carboxylase, and phosphofructokinase 2 [15], [21], [25]. Drosophila Mlx displayed an essential role in the fat body, which is the counterpart of mammalian adipose tissue and liver. Thus, it is conceivable that the liver and adipose tissue-specific ChREBP-Mlx, instead of the muscle-specific MondoA-Mlx, represents the ancestral function of the heterodimer.
This study identifies Aldh-III as a novel gene contributing to dietary sugar tolerance. Aldh-III is the ortholog of mammalian Aldh3 family. Aldehyde dehydrogenases (Aldhs) are highly conserved NAD(P)+ -dependent enzymes that oxidize aldehydes to the corresponding carboxylic acid and act on a broad range of substrates [34]. Aldehydes are highly reactive compounds forming adducts with nucleic acids and proteins, thus disturbing cellular functions. Reactive aldehydes can originate from exogenous sources or be products of cellular metabolism. Aldhs have been shown to provide protection against a number of ectopic stresses, including toxic chemicals, heat stress, and UV irradiation [34]–[37]. One of the best-established functions of Aldh proteins is neutralization of acetaldehyde, a toxic metabolite of ethanol. Acetaldehyde elimination is mainly mediated by Aldh2 [34], [38]. Polymorphism in the Aldh2 gene is common is Asian populations and it leads to poor ethanol tolerance [39]. Our finding that another member of the Aldh family, Aldh-III, is essential for dietary sugar tolerance is intriguing, since there are multiple parallels between the hepatic pathophysiologies related to excessive ethanol and fructose consumption in humans [3]. Of note, we observed that Aldh-III is sufficient in rescuing the sugar intolerance of mlx1 mutants in the fat body, the insect counterpart of liver, suggesting that metabolic stress-induced dysfunction of the fat body contributes to sugar intolerance. Future studies should be aimed at understanding the generation of sugar-derived reactive aldehydes and their molecular targets.
We also provide evidence that PFK2 expression is positively regulated by Mondo-Mlx in Drosophila and that the PFK2 levels are critical in managing circulating glucose levels and providing sugar tolerance. Thus, our data suggests that regulation of PFK2 gene expression might be a suitable strategy to manage hyperglycemia in diabetes. Elevated PFK2 expression has also been associated with the high rate of glycolytic flux in neoplastic tumors [40]. Exploring the contribution of Mondo and Mlx proteins in this setting is therefore warranted. In addition to being regulated transcriptionally, PFK2 activity in mammals is known to be posttranslationally regulated by insulin signalling through protein kinase AKT [41], [42]. It will be interesting to learn, what is the contribution of insulin signalling pathway activity on the dietary sugar tolerance.
The finding that Cabut is an essential secondary effector of Mondo-Mlx is intriguing. In fact, cabut expression has been reported to respond to other metabolism-related signals: it is upregulated upon inhibition of TOR complex 1 signalling [43], which is likely mediated by activation of the Forkhead transcription factor, FoxA [44]. While the developmental role of Cabut has been studied [45], [46], its metabolic functions have remained unexplored. Our finding showing that Cabut plays an essential metabolic role in providing dietary sugar tolerance implies this topic deserves an in-depth survey in the future. Notably, the closest mammalian homologs of Cabut, Klf-10 and Klf-11, have been linked to metabolic regulation. Mutations in the klf-11 locus are associated with risk of diabetes [47], while Klf-10 appears to negatively regulate lipogenic genes in hepatocytes [48]. klf-10 expression is regulated both by circadian signals [49] as well as by ChREBP upon high glucose [48]. Thus, Klf-10 might be the functional ortholog of Cabut. Future studies should be aimed at identifying the Cabut target genes involved in its metabolic functions.
Dietary sugar tolerance displays a wide natural variation, even in closely related animal species. For example, two Drosophila species, D. melanogaster and D. mojavensis, have strikingly different tolerance to dietary sugars [6]. In contrast to the fruit generalist D. melanogaster, D. mojavensis is a cactus breeder, which does not naturally encounter high levels of simple sugars and displays poor survival on sugar-rich diet [6]. Based on our data, it is possible to hypothesize that Mondo-Mlx-regulated transcriptional network contributes to the natural variation in sugar tolerance. Genetic differences in sugar tolerance are also observed in humans. For example, hereditary fructose intolerance (HFI) is caused by mutations in the aldolase-B gene [50]. A fructose-restricted diet renders HFI relatively benign, but ingestion of fructose or sucrose leads to strong symptoms, including nausea and vomiting as well as a risk of liver and kidney damage. It will be important to explore whether genetic changes in the ChREBP/MondoA-Mlx network influence individual's risk for sugar overload-induced metabolic disturbance.
This study highlights the usefulness of Drosophila as a model for systematically exploring the genetic factors defining the range of healthful nutrient intake. Notably, the sugar intolerance in mlx1 mutants is not a pleiotropic consequence of a generally disturbed energy metabolism. Knockdown of other key transcriptional metabolic regulators, such as SREBP, did not cause notable sugar intolerance (unpublished observation). The availability of genome-scale reagents, including in vivo RNAi lines, offers the possibility for the systematic dissection of genes contributing to dietary sugar tolerance. These genes might include novel members of the Mondo-Mlx-regulated genetic network, but they will also uncover whether parallel regulatory pathways are involved. The Drosophila model is also particularly useful in dissecting the function of transcription factors that are master regulators of several gene groups that contribute to distinct physiological outputs. While this study has focused on uncovering those Mondo-Mlx targets that contribute to sugar tolerance and circulating glucose levels, uncovering the downstream effectors behind the other metabolic phenotypes of mlx1 mutant fly awaits for future studies.
Materials and Methods
Fly stocks
P{XP}bigmaxd07258 were obtained from Bloomington stock center. For generating UAS-mlx flies, the coding region of mlx cDNA was amplified by PCR and cloned into pUAST vector using BglII and XbaI restriction sites. FLAG-tag was incorporated into the C-terminus. Aldh-III coding region was cloned into pUAST vector using NotI and XhoI restriction sites. RNAi lines were obtained from Vienna Drosophila RNAi Center and from NIG-FLY Stock Center. The following GAL4 driver lines were used in this study: tub-GAL4 [51], ppl-GAL4 [52], r4-GAL4 [53], Elav-GAL4 [54] and Mef2-GAL4 [55].
Fly food
In standard conditions flies were maintained at 25°C on medium containing agar 0.6% (w/v), semolina 3.2% (w/v), malt 6.5% (w/v), dry baker's yeast 1.8% (w/v), propionic acid 0.7% (v/v) and Nipagin (methylparaben) 2.4% (v/v). For defined nutrient studies, larvae were grown on food containing 20% (w/v) dry baker's yeast, 0.5% (w/v) agar, and 2.5% (v/v) Nipagin (methylparaben) in PBS supplemented with varying concentrations of sucrose, glucose or fructose. 1st instar larvae were collected from apple juice plates (apple juice 33.33% (v/v), agar 1.75% (w/v), sugar 2.5% (w/v) and Nipagin (methylparaben) 2.0% (v/v)) and larvae were grown at controlled density (30 larvae per vial).
Mlx antibodies
For generating anti-Mlx antibodies full-length Drosophila mlx cDNA was cloned into pGEX-4T2. Recombinant GST-Mlx was purified using Glutathione-agarose (Sigma). Anti-Mlx antiserum was raised by immunizing a guinea pig (Storkbio Ltd).
Cell culture and transfections
Drosophila S2 cells were grown at 25°C in standard Shields and Sang M3 medium (Sigma) containing 2% of fetal bovine serum (Gibco), 1× insect medium supplement (Sigma) and penicillin/streptomycin (Gibco). The transfections were performed using Effectene (Qiagen), according to manufacturer's protocol. Expression of transfected genes was induced with 1.2 µM CuSO4 24 h post-transfection.
Pulldown and Western blotting
For detecting endogenous Mlx in vivo, 3rd instar prewandering larvae were homogenized in Laemmli sample buffer and boiled for 5 min. Samples were resolved on SDS-PAGE and detected by Western blotting using anti-Mlx antibodies. For the pulldown experiment, cells were lysed in IP lysis buffer (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0,1% NP40) and lysates were cleared by centrifugation. Lysate protein concentration was adjusted to 1 µg/µl. 1 ml of lysate was incubated o/n with 25 µl Strep-Tactin beads (IBA). The beads were washed 5 times with IP lysis buffer, after which Laemmli sample buffer was added and samples were boiled for 5 min. Pulldown and lysate samples were resolved on SDS-PAGE, transferred to nitrocellulose and analyzed by Western blotting using anti-V5 (Invitrogen), anti-Mlx and anti-Kinesin (Cytoskeleton).
Metabolomic analyses of small polar metabolites and lipids
Metabolomics was performed using prewandering 3rd instar larvae grown on 20% yeast-5% sucrose diet. Analysis was done in four biological replicas. Data was processed using Guineu [56] and MZmine 2 [57] software packages for small polar metabolites and molecular lipids, respectively. Detailed description is available in Protocol S1.
Metabolic assays
Metabolite assays were done using prewandering 3rd instar larvae grown on 20% yeast or 20% yeast-5% sucrose diet. All analyses were done at least in four biological replicas. Glucose, trehalose and glycogen measurements were conducted as described [58], [59].
Microarray and quantitative RT–PCR
Staged 3rd instar prewandering control and mlx1 mutant larvae were grown on a 20% yeast-5% sucrose diet. RNA was extracted from 3–4 larval fat bodies per sample in four biological replicas using the Nucleospin RNA XS kit (Macherey-Nagel). The Amino Allyl MessageAmp II aRNA Amplification kit (Ambion) was used for aRNA synthesis. Hybridization to Agilent Drosophila Gene Expression Microarray, 4×44K was performed according to the manufacturer's instructions. Data normalization and analysis was performed using R, taking advantage of packages from the Bioconductor repository. A full documentation of the data analysis is available in Protocol S1. For quantitative RT-PCR, the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) with random hexamer primers was used for first strand cDNA synthesis. PCR was performed using Maxima SYBR Green qPCR Master Mix (2X) (Fermentas) and analyzed on StepOnePlus (Applied Biosystems) real-time PCR system. Primer sequences are available as Table S3.
All expression profiling data is available under accession E-MTAB-699 at the ArrayExpress repository.
Statistics
Statistical significance for each experiment (excluding metabolomics and microarray) was determined with unpaired Student's t-test with unequal variance. All quantitative data are presented as mean ± SEM for a minimum of three independent biological replicates.
Supporting Information
Zdroje
1. TsaharE, del RioCM, AradZ, JoyJP, IzhakiI (2005) Are the low protein requirements of nectarivorous birds the consequence of their sugary and watery diet? A test with an omnivore. Physiol Biochem Zool 78 : 239–245.
2. WelchKCJr, SuarezRK (2007) Oxidation rate and turnover of ingested sugar in hovering Anna's (Calypte anna) and rufous (Selasphorus rufus) hummingbirds. J Exp Biol 210 : 2154–2162.
3. LustigRH, SchmidtLA, BrindisCD (2012) Public health: The toxic truth about sugar. Nature 482 : 27–29.
4. TappyL, LeKA, TranC, PaquotN (2010) Fructose and metabolic diseases: new findings, new questions. Nutrition 26 : 1044–1049.
5. StanhopeKL (2012) Role of fructose-containing sugars in the epidemics of obesity and metabolic syndrome. Annu Rev Med 63 : 329–343.
6. MatzkinLM, JohnsonS, PaightC, BozinovicG, MarkowTA (2011) Dietary protein and sugar differentially affect development and metabolic pools in ecologically diverse Drosophila. J Nutr 141 : 1127–1133.
7. MusselmanLP, FinkJL, NarzinskiK, RamachandranPV, HathiramaniSS, et al. (2011) A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech 4 : 842–849.
8. PascoMY, LeopoldP (2012) High sugar-induced insulin resistance in Drosophila relies on the lipocalin Neural Lazarillo. PLoS ONE 7: e36583 doi:10.1371/journal.pone.0036583.
9. MairW, PiperMD, PartridgeL (2005) Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol 3: e223 doi:10.1371/journal.pbio.0030223.
10. ZinkeI, SchutzCS, KatzenbergerJD, BauerM, PankratzMJ (2002) Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J 21 : 6162–6173.
11. GiaccariA, SoriceG, MuscogiuriG (2009) Glucose toxicity: the leading actor in the pathogenesis and clinical history of type 2 diabetes - mechanisms and potentials for treatment. Nutr Metab Cardiovasc Dis 19 : 365–377.
12. SinghDK, WinocourP, FarringtonK (2011) Oxidative stress in early diabetic nephropathy: fueling the fire. Nat Rev Endocrinol 7 : 176–184.
13. SaltielAR, KahnCR (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414 : 799–806.
14. HavulaE, HietakangasV (2012) Glucose sensing by ChREBP/MondoA-Mlx transcription factors. Semin Cell Dev Biol
15. MaL, RobinsonLN, TowleHC (2006) ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem 281 : 28721–28730.
16. StoltzmanCA, PetersonCW, BreenKT, MuoioDM, BillinAN, et al. (2008) Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci U S A 105 : 6912–6917.
17. LiMV, ChenW, HarmanceyRN, Nuotio-AntarAM, ImamuraM, et al. (2010) Glucose-6-phosphate mediates activation of the carbohydrate responsive binding protein (ChREBP). Biochem Biophys Res Commun 395 : 395–400.
18. DentinR, Tomas-CobosL, FoufelleF, LeopoldJ, GirardJ, et al. (2012) Glucose 6-phosphate, rather than xylulose 5-phosphate, is required for the activation of ChREBP in response to glucose in the liver. J Hepatol 56 : 199–209.
19. StoltzmanCA, KaadigeMR, PetersonCW, AyerDE (2011) MondoA senses non-glucose sugars: regulation of thioredoxin-interacting protein (TXNIP) and the hexose transport curb. J Biol Chem 286 : 38027–38034.
20. SansCL, SatterwhiteDJ, StoltzmanCA, BreenKT, AyerDE (2006) MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol 26 : 4863–4871.
21. JeongYS, KimD, LeeYS, KimHJ, HanJY, et al. (2011) Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS ONE 6: e22544 doi:10.1371/journal.pone.0022544.
22. YamashitaH, TakenoshitaM, SakuraiM, BruickRK, HenzelWJ, et al. (2001) A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci U S A 98 : 9116–9121.
23. WangH, WollheimCB (2002) ChREBP rather than USF2 regulates glucose stimulation of endogenous L-pyruvate kinase expression in insulin-secreting cells. J Biol Chem 277 : 32746–32752.
24. HeZ, JiangT, WangZ, LeviM, LiJ (2004) Modulation of carbohydrate response element-binding protein gene expression in 3T3-L1 adipocytes and rat adipose tissue. Am J Physiol Endocrinol Metab 287: E424–430.
25. IizukaK, BruickRK, LiangG, HortonJD, UyedaK (2004) Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci U S A 101 : 7281–7286.
26. DentinR, BenhamedF, HainaultI, FauveauV, FoufelleF, et al. (2006) Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes 55 : 2159–2170.
27. BillinAN, EilersAL, CoulterKL, LoganJS, AyerDE (2000) MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol 20 : 8845–8854.
28. HermanMA, PeroniOD, VilloriaJ, SchonMR, AbumradNA, et al. (2012) A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 484 : 333–338.
29. PeyrefitteS, KahnD, HaenlinM (2001) New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix-loop-helix genes. Mech Dev 104 : 99–104.
30. McFerrinLG, AtchleyWR (2011) Evolution of the Max and Mlx networks in animals. Genome Biol Evol 3 : 915–937.
31. BeckerA, SchloderP, SteeleJE, WegenerG (1996) The regulation of trehalose metabolism in insects. Experientia 52 : 433–439.
32. IshiiS, IizukaK, MillerBC, UyedaK (2004) Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc Natl Acad Sci U S A 101 : 15597–15602.
33. SassuED, McDermottJE, KeysBJ, EsmaeiliM, KeeneAC, et al. (2012) Mio/dChREBP coordinately increases fat mass by regulating lipid synthesis and feeding behavior in Drosophila. Biochem Biophys Res Commun
34. SinghS, BrockerC, KoppakaV, ChenY, JacksonBC, et al. (2012) Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med 56C: 89–101.
35. TownsendAJ, Leone-KablerS, HaynesRL, WuY, SzwedaL, et al. (2001) Selective protection by stably transfected human ALDH3A1 (but not human ALDH1A1) against toxicity of aliphatic aldehydes in V79 cells. Chem Biol Interact 130–132 : 261–273.
36. PappaA, ChenC, KoutalosY, TownsendAJ, VasiliouV (2003) Aldh3a1 protects human corneal epithelial cells from ultraviolet - and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med 34 : 1178–1189.
37. LassenN, PappaA, BlackWJ, JesterJV, DayBJ, et al. (2006) Antioxidant function of corneal ALDH3A1 in cultured stromal fibroblasts. Free Radic Biol Med 41 : 1459–1469.
38. YuHS, OyamaT, IsseT, KitakawaK, OgawaM, et al. (2009) Characteristics of aldehyde dehydrogenase 2 (Aldh2) knockout mice. Toxicol Mech Methods 19 : 535–540.
39. CrabbDW, EdenbergHJ, BosronWF, LiTK (1989) Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest 83 : 314–316.
40. YalcinA, TelangS, ClemB, ChesneyJ (2009) Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp Mol Pathol 86 : 174–179.
41. DeprezJ, VertommenD, AlessiDR, HueL, RiderMH (1997) Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem 272 : 17269–17275.
42. MoutonV, ToussaintL, VertommenD, GueuningMA, MaisinL, et al. (2010) Heart 6-phosphofructo-2-kinase activation by insulin requires PKB (protein kinase B), but not SGK3 (serum - and glucocorticoid-induced protein kinase 3). Biochem J 431 : 267–275.
43. GuertinDA, GunturKV, BellGW, ThoreenCC, SabatiniDM (2006) Functional genomics identifies TOR-regulated genes that control growth and division. Curr Biol 16 : 958–970.
44. BulowMH, AebersoldR, PankratzMJ, JungerMA (2010) The Drosophila FoxA ortholog Fork head regulates growth and gene expression downstream of Target of rapamycin. PLoS ONE 5: e15171 doi:10.1371/journal.pone.0015171.
45. Munoz-DescalzoS, TerolJ, ParicioN (2005) Cabut, a C2H2 zinc finger transcription factor, is required during Drosophila dorsal closure downstream of JNK signaling. Dev Biol 287 : 168–179.
46. RodriguezI (2011) Drosophila TIEG is a modulator of different signalling pathways involved in wing patterning and cell proliferation. PLoS ONE 6: e18418 doi:10.1371/journal.pone.0018418.
47. NeveB, Fernandez-ZapicoME, Ashkenazi-KatalanV, DinaC, HamidYH, et al. (2005) Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci U S A 102 : 4807–4812.
48. IizukaK, TakedaJ, HorikawaY (2011) Kruppel-like factor-10 is directly regulated by carbohydrate response element-binding protein in rat primary hepatocytes. Biochem Biophys Res Commun 412 : 638–643.
49. GuillaumondF, Grechez-CassiauA, SubramaniamM, BrangoloS, Peteri-BrunbackB, et al. (2010) Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol 30 : 3059–3070.
50. BouteldjaN, TimsonDJ (2010) The biochemical basis of hereditary fructose intolerance. J Inherit Metab Dis 33 : 105–112.
51. LeeT, LuoL (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22 : 451–461.
52. ZinkeI, KirchnerC, ChaoLC, TetzlaffMT, PankratzMJ (1999) Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development 126 : 5275–5284.
53. LeeG, ParkJH (2004) Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 167 : 311–323.
54. LuoL, LiaoYJ, JanLY, JanYN (1994) Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev 8 : 1787–1802.
55. RanganayakuluG, ElliottDA, HarveyRP, OlsonEN (1998) Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development 125 : 3037–3048.
56. CastilloS, MattilaI, MiettinenJ, OresicM, HyotylainenT (2011) Data analysis tool for comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry. Anal Chem 83 : 3058–3067.
57. PluskalT, CastilloS, Villar-BrionesA, OresicM (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11 : 395.
58. ZhangW, ThompsonBJ, HietakangasV, CohenSM (2011) MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet 7: e1002429 doi:10.1371/journal.pgen.1002429.
59. ParrouJL, FrancoisJ (1997) A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem 248 : 186–188.
Štítky
Genetika Reprodukčná medicína
Článek The G4 GenomeČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 4- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



