-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
UDP-glucose 4, 6-dehydratase Activity Plays an Important Role in Maintaining Cell Wall Integrity and Virulence of
Candida albicans, a human fungal pathogen, undergoes morphogenetic changes that are associated with virulence. We report here that GAL102 in C. albicans encodes a homolog of dTDP-glucose 4,6-dehydratase, an enzyme that affects cell wall properties as well as virulence of many pathogenic bacteria. We found that GAL102 deletion leads to greater sensitivity to antifungal drugs and cell wall destabilizing agents like Calcofluor white and Congo red. The mutant also formed biofilms consisting mainly of hyphal cells that show less turgor. The NMR analysis of cell wall mannans of gal102 deletion strain revealed that a major constituent of mannan is missing and the phosphomannan component known to affect virulence is greatly reduced. We also observed that there was a substantial reduction in the expression of genes involved in biofilm formation but increase in the expression of genes encoding glycosylphosphatidylinositol-anchored proteins in the mutant. These, along with altered mannosylation of cell wall proteins together might be responsible for multiple phenotypes displayed by the mutant. Finally, the mutant was unable to grow in the presence of resident peritoneal macrophages and elicited a weak pro-inflammatory cytokine response in vitro. Similarly, this mutant elicited a poor serum pro-inflammatory cytokine response as judged by IFNγ and TNFα levels and showed reduced virulence in a mouse model of systemic candidiasis. Importantly, an Ala substitution for a conserved Lys residue in the active site motif YXXXK, that abrogates the enzyme activity also showed reduced virulence and increased filamentation similar to the gal102 deletion strain. Since inactivating the enzyme encoded by GAL102 makes the cells sensitive to antifungal drugs and reduces its virulence, it can serve as a potential drug target in combination therapies for C. albicans and related pathogens.
Published in the journal: UDP-glucose 4, 6-dehydratase Activity Plays an Important Role in Maintaining Cell Wall Integrity and Virulence of. PLoS Pathog 7(11): e32767. doi:10.1371/journal.ppat.1002384
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002384Summary
Candida albicans, a human fungal pathogen, undergoes morphogenetic changes that are associated with virulence. We report here that GAL102 in C. albicans encodes a homolog of dTDP-glucose 4,6-dehydratase, an enzyme that affects cell wall properties as well as virulence of many pathogenic bacteria. We found that GAL102 deletion leads to greater sensitivity to antifungal drugs and cell wall destabilizing agents like Calcofluor white and Congo red. The mutant also formed biofilms consisting mainly of hyphal cells that show less turgor. The NMR analysis of cell wall mannans of gal102 deletion strain revealed that a major constituent of mannan is missing and the phosphomannan component known to affect virulence is greatly reduced. We also observed that there was a substantial reduction in the expression of genes involved in biofilm formation but increase in the expression of genes encoding glycosylphosphatidylinositol-anchored proteins in the mutant. These, along with altered mannosylation of cell wall proteins together might be responsible for multiple phenotypes displayed by the mutant. Finally, the mutant was unable to grow in the presence of resident peritoneal macrophages and elicited a weak pro-inflammatory cytokine response in vitro. Similarly, this mutant elicited a poor serum pro-inflammatory cytokine response as judged by IFNγ and TNFα levels and showed reduced virulence in a mouse model of systemic candidiasis. Importantly, an Ala substitution for a conserved Lys residue in the active site motif YXXXK, that abrogates the enzyme activity also showed reduced virulence and increased filamentation similar to the gal102 deletion strain. Since inactivating the enzyme encoded by GAL102 makes the cells sensitive to antifungal drugs and reduces its virulence, it can serve as a potential drug target in combination therapies for C. albicans and related pathogens.
Introduction
Candida albicans is a polymorphic fungus that causes infection of skin, nail, mucous membrane in healthy individuals and can lead to more severe infections of the vital organs in case of immune-compromised patients leading to death [1]. It is capable of growing in both yeast and hyphal forms and the yeast to hyphal transition has been reported to play a key role in virulence [2]. Environmental cues such as temperature, pH, serum, nutrient deprivation on solid media, etc. are known to trigger yeast to hyphal transition and in vitro studies have led to the identification of some of the key regulators such as CPH1, EFG1, INT1, PRA1, RBF1, TUP1, UME6 etc. [3]–[5]. In general, mutants of some of these regulators show reduced hyphal morphology that correlates with reduced virulence, suggesting a direct correlation of the hyphal form with virulence of C. albicans [6]. Surprisingly, mutations in genes like TUP1 and NRG1 (global repressors of hyphal morphology) also show reduced virulence in spite of increased hyphal morphology [7]. This observation has raised questions about the validity of the direct correlation of hyphal morphology and virulence [8]. The morphological forms also differ in the cell wall composition [9]. Cell wall is the first cell organelle that comes in contact with the host and plays an important role in determining the outcome of the host pathogen interaction. Therefore, alterations in the cell wall composition and the associated transcriptional program, than the shape of the cell, per se, that might be crucial to virulence of C. albicans.
It has been reported that the cell wall architecture and virulence of microbial pathogens can be affected by presence of Galactose, a sugar that can act as a sole carbon source for many pathogens [10]. The effect of galactose on the biofilm development of C. albicans has been extensively studied. The rate of formation of biofilm is higher in the presence of galactose [11]. Further, galactose contributes to 3% of the dry weight of extra-cellular polymeric material of C. albicans biofilm [12]. Most organisms are able to metabolize galactose i.e. convert β-D-galactose to glucose 1-phosphate through four enzymes of the Leloir pathway which have been well characterized in E. coli and Saccharomyces cerevisiae [13]. The epimerase which catalyzes the third step in the Leloir pathway can clearly be an enzyme that may have a role beyond galactose metabolism, in that, the reversible reaction could be employed to generate galactose during growth on glucose as the sole carbon source. Indeed, various phenotypes are associated with mutations in this gene in different organisms [14], [15]. In S. cerevisiae, lack of Gal10p prevents growth on galactose as the sole carbon source [16] although, it has no reported effect on growth in the absence of galactose. Previous work from our laboratory has shown that CaGAL10 (orf19.3672) encodes a true homolog of UDP-galactose-4-epimerase and can functionally complement the S. cerevisiae gal10Δ [17]. We have shown that the ultrastructure of the biofilm of Cagal10 mutant is distinctly different from that of the wild type [17]. In Arabidopsis, the impairment results in root-specific phenotypes, including increased root hair elongation, decreased root length, and root epidermal bulging etc. [18]. In humans, impairment of galactose epimerase causes one of two clinically distinct forms of galactosemia, an autosomal recessive epimerase-deficiency syndrome [19].
The orf19.3674 which has been annotated as CaGAL102 in the Candida Genome Database, encodes a protein very similar to the epimerase domain of the CaGal10p. We have previously shown that the full length CaGal10p as well as its epimerase domain alone complements the S. cerevisiae gal10 deletion [17]. However, in light of a report [20] that Cryptococcus neoformans has two functional paralogs of GAL10, we asked the following questions: Does CaGAL102 encode a functional galactose epimerase? If yes, what is the significance of two Gal10 paralogs in C. albicans? If it is not a functional epimerase what role does it play in C. albicans?
In the present study, we aimed to determine the role of this putative galactose epimerase in C. albicans using multiple experimental approaches. We found that the GAL102 actually encodes a UDP-glucose 4,6-dehydratase activity and its loss affects the composition of cell wall mannans. The mutant cells lacking the activity also showed several defects in phenotypes associated with virulence of C. albicans. The mutant cells elicited a differential cytokine response from host immune system cells in vitro as well as in vivo, which reflected in attenuated virulence in mouse model of systemic candidiasis. These observations are in agreement with the reports that cell wall mannans contribute to host responses elicited by fungal pathogens [21], [22]. Our observations present a clear evidence for an enzyme activity that affects composition of cell wall mannans, cell morphology and contributes to virulence in C. albicans.
Results
A putative paralog of UDP-galactose 4 - epimerase encoded by GAL102 in C. albicans genome does not function as a galactose epimerase
Orf19.3674/GAL102 in the Candida Genome Database has been annotated as a UDP-galactose 4-epimerase. Multiple sequence alignment of Gal102p with UDP-galactose 4-epimerase (Gal10p) homologs using the software ClustalW showed that the active site residues in the epimerase domain are highly conserved (Figure 1A). Incidentally, CaGal10 shows 52% identity and ∼ 70% similarity to ScGal10p but Gal102p shows only 27% identity and 47% similarity. Most striking is the conservation of the YXXXK motif wherein the conserved tyrosine residue plays an important role during catalysis facilitating the transfer of a hydride from C4 of the sugar to C4 of NADH leading to a 4′-ketopyranose intermediate and NADH [23].
Fig. 1. Putative Gal10 paralog does not encode functional Galactose epimerase. 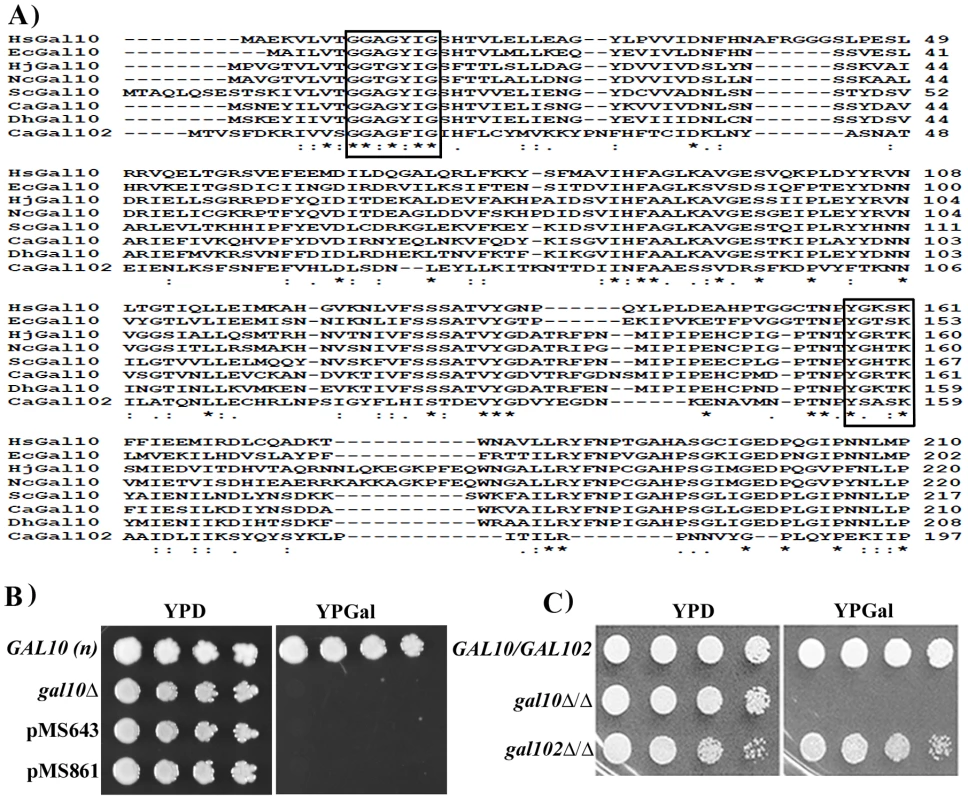
A) Multiple sequence alignment of Gal10 homologs from E. coli (Ec), N. crassa (Nc), H. jecorina (Hj), S. cerevisiae (Sc), C. albicans (Ca), D. hansenii (Dh), H. sapiens (Hs). The residues known to play a crucial role in catalysis or in NAD binding are shown in boxes. Identical residues are marked as * and similar residues are marked as ‘:’ B) S. cerevisiae gal10 deletion strain transformed with pPS189 (empty vector) or, pMS643 (PTEF1-Gal102), pMS861 (PTEF1-Gal102*) were spotted on YPD and YPGal plates and incubated at 30°C. Plates were photographed next day. WT S. cerevisiae strain BY4741 (GAL10) was spotted as control. C) WT SC5314 (GAL102/GAL102), Cagal10Δ/Δ and gal102Δ/Δ cells were spotted on YPD and YPGal plates and checked for growth after 1 day. Most ORFs from C. albicans expressed in S. cerevisiae are able to complement loss of function mutants of corresponding homologs. To test whether GAL102 encoded protein has UDP-galactose 4-epimerase activity we sub-cloned the ORF under the constitutive TEF1 promoter on a multi-copy plasmid (pMS643). C. albicans uses an alternative genetic code with CUG codon encoding for Serine in place of Leucine of the universal genetic code. We replaced the only CUG codon with UCG to introduce Serine at the 314 position in the recombinant Gal102 ORF (pMS861) and then tested its ability to complement the S. cerevisiae gal10 mutant strain PJB5. The gal10 mutant is unable to grow in the presence of galactose as the sole carbon source. Transformants expressing the open reading frame grew on plates containing glucose as the sole carbon source but not in the presence of galactose (Figure 1B). We tested the presence of this protein in cells transformed with pMS861 using Western blot analysis to confirm that the failure to complement was not due to poor or lack of expression of the heterologous protein (Figure S1A).
We further disrupted the GAL102 gene using a PCR based strategy as described earlier [24] and generated multiple isolate/mutants through independent transformations. We confirmed the deletion and the absence of the transcript by Southern and Northern analysis respectively (Figure S1B). We tested these mutants for various phenotypes and have presented here representative data from one of these isolates. We found that the null mutant gal102Δ/Δ had growth rate similar to that of the WT in normal rich media (YPD) at 30°C (data not shown). When spotted on YP glucose or YP galactose plates, the gal102Δ/Δ strain exhibited no growth defect (Figure 1C) while the gal10Δ/Δ mutant strain could not grow on galactose-containing medium [17]. This implied that the GAL102 gene does not play any significant role in galactose metabolism in C. albicans. It has been reported that the two paralogs of GAL10 in C. neoformans affect growth on galactose at different temperatures [20]. We saw no such difference in growth under suboptimal conditions such as high temperature or low galactose concentration in the absence of GAL102 (data not shown).
GAL102 encodes a nucleotide sugar dehydratase
Since the deletion of GAL102 showed no effect on galactose metabolism and associated phenotypes, we repeated the homology search through NCBI databases. We found that Gal102p showed 32% identity to dTDP-glucose 4,6-dehydratase (RmlB; Acc. No. AAB88398) of E. coli and 40% identity to the N-terminus of RHM2 (1–370 amino acids) of A. thaliana. The latter has been biochemically shown to have UDP-glucose 4,6-dehydratase activity. The dTDP/UDP-glucose 4,6-dehydratase enzyme is known to be involved in rhamnose biosynthesis in bacteria as well as in plants [25], [26]. Figure 2A shows the ClustalW alignment of Gal102p sequence with several other fungal homologs as well as those from E. coli and A. thaliana.
Fig. 2. GAL102 encodes a dTDP-glucose 4, 6-dehydratase. 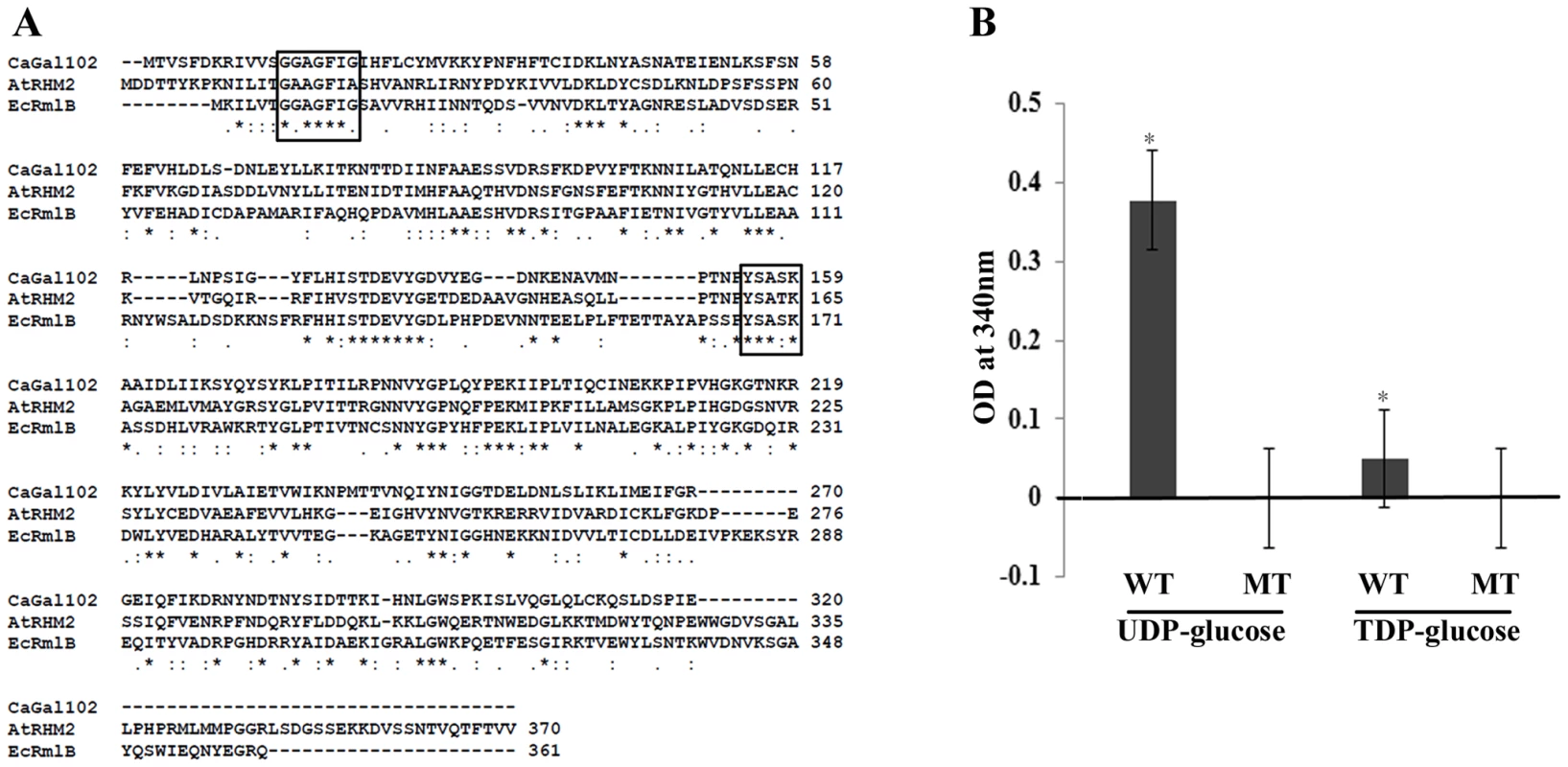
A) The sequence alignment of fungal homologs of dTDP-glucose 4, 6-dehydratase with Gal102 was generated using ClustalW. The homologs from E. coli and A. thaliana used for the multiple sequence alignments are shown. The conserved NAD binding motif and the active site YXXXK motif are boxed. B) Enzymatic activity of purified wildtype Gal102 (WT) and the catalytic mutant K159A (MT) was tested with UDP glucose, dTDP glucose as substrates. The activity was measured as NAD-NADH conversion by monitoring increase in A340. All the substrates and cofactor NAD were used at 3 mM concentration. The reactions were carried out at 37°C for 2 hr. Experiment was done in triplicate with 20 µg of recombinant protein expressed in E. coli and purified using Ni-NTA column per reaction. *p<0.05. The dTDP/UDP-glucose 4,6-dehydratase enzyme is a member of the SDR (short chain dehydratase/reductase) family of proteins, which have a highly conserved YXXXK motif, crucial for the enzyme activity. In case of A. thaliana, the RHM2K165A mutation completely abolishes UDP-glucose 4,6-dehydratase activity [26]. Lysine at position 159 in Gal102 lies within the putative catalytic motif, YXXXK. Therefore, we mutated it to alanine (AAA to GCA codon) as described in materials and methods section. We expressed the codon optimized Gal102p (as described above) and its mutant derivative Gal102pK159A in E. coli and purified the recombinant proteins. We assayed for the dehydratase activity by measuring NAD - NADH conversion at 340 nm using either UDP glucose or dTDP glucose as substrates. After 2 hr the native Gal102p showed significant activity with UDP glucose as the substrate but only very low activity with dTDP glucose as the substrate. This activity could not be detected with the same amount of Gal102K159A mutant protein (Figure 2B). This confirmed that Gal102p actually encodes a UDP-glucose dehydratase activity and that the lysine (Lys 159) in the conserved motif YXXXK, is essential for the activity of the enzyme.
Deletion of GAL102 leads to altered cell morphology and reduced cell wall integrity
We observed that the cells of the gal102Δ/Δ strain were more elongated when grown in normal rich media (YPD) at 30°C as compared to the WT (SC5314) strain (Figure 3A). Thus, the deletion of GAL102 has an effect on cell morphology which is also reflected in the wrinkled colony morphology (Figure S1C). When grown under hyphae inducing conditions, like growth in Lee's or Spider medium, the mutant showed much more enhanced filamentation as compared to the WT (SC5314).
Fig. 3. Deletion of GAL102 affects cell morphology and cell wall integrity. 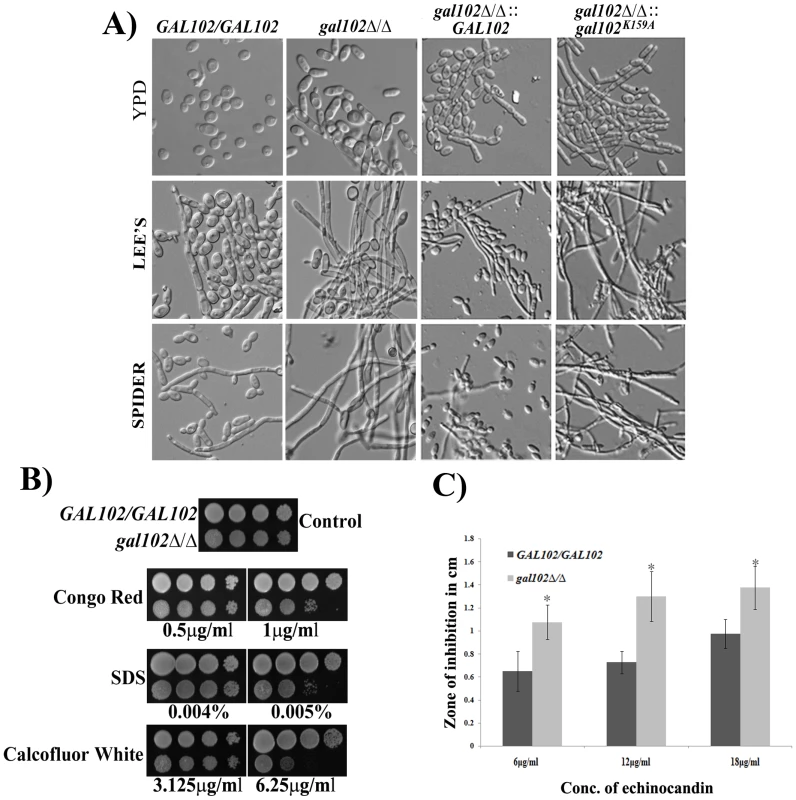
A) Cells of GAL102/GAL102, gal102Δ/Δ, WT reintegrant and the K159A mutant reintegrant in gal102Δ/Δ strain were grown overnight at 30°C in YPD supplemented with uridine or Lee's or Spider medium. Cells were photographed using Olympus BX51 microscope with Nomarsky optics at 400X magnification. B) The gal102Δ/Δ and the GAL102/GAL102 strain were incubated in YPD containing the indicated cell wall damaging agents at the indicated concentrations. C) The gal102Δ/Δ and the WT DAY286 strain were mixed with top agar and poured on top of YPD plates. Various concentration of Echinocandin (5–15 µg) was added to the wells and photographs were taken after 48 hr of incubation at 37°C. The results from 3 different plates were used to measure the diameter of zone of inhibition and plotted in the bar graph (lower panel). Grey bars represent WT while the black bars represent mutant. *p<0.05. To establish that the loss of enzyme activity of the Gal102p is responsible for the phenotypes associated with the deletion, we replaced the gal102::HIS1 allele with either WT GAL102 or gal102K159A allele. All these strains thus have URA3 gene integrated at the GAL102 locus whose expression from non-native locus has been reported to be associated with attenuation of virulence. The morphology of these marker matched reintegrant strains was then tested in hyphal inducing media like Lee's medium and Spider medium. It was observed that (Figure 3A) WT GAL102 integrant at the gal102::HIS1 locus reduced the extent of filamentation close to that of the WT (SC5314) strain. In contrast, the catalytically inactive gal102K159A integrant at the same locus continued to show high filamentation phenotype. This shows that the morphological phenotypes associated with the deletion are indeed due to the lack of the glucose dehydratase activity associated with the protein.
We tested the sensitivity of gal102Δ/Δ strain to various cell wall damaging agents, e.g. SDS, Congo red and Calcofluor white as well as an antifungal antibiotic echinocandin. The mutant cells showed greater sensitivity to the cell wall damaging agents as compared to the WT (SC5314) strain (Figure 3B). The increased sensitivity to several cell wall damaging agents suggests that the cell wall composition of the mutant is altered in such a way as to affect the integrity of the cell wall. The antibiotic echinocandin specifically showed larger zones of inhibition as compared to the WT, signifying greater sensitivity of the mutant to this antibiotic supporting the observed cell wall integrity defect A bar graph consolidating three independent measurements of zone of inhibition at the indicated concentration underscores the significance of the observation (Figure 3C).
Deletion of GAL102 leads to altered biochemical composition of the cell wall mannans
The above results indicated that the cell wall composition may be altered significantly in the gal102Δ/Δ mutant. Since the product of the Gal102 catalyzed reaction is a nucleotide sugar which might act as a sugar donor in cell wall protein glycosylation, we surmised, that the most likely target of the deletion was cell wall mannans which form the outermost layer of the C. albicans cell wall [9]. We analyzed the mannan composition of the mutant cell wall and compared it to that of the WT (SC5314). Earlier reports on NMR analysis of mannans from C. albicans cells have revealed differences among the yeast form and the hyphal form [9]. To ascribe the difference in mannan composition to the lack of Gal102 activity and not to the elongated cell morphology associated changes, we isolated mannans as described [9] from both the WT and the mutant cells grown at 25 °C at which the gal102Δ/Δ cells were much less elongated (Figure S1D). In our studies we performed 1H NMR and 31P NMR to study presence/absence of linkages among the mannans, and TOCSY, ROESY and HSQC to study the details of gross defects in specific bonding. The anomeric region of the 1D 1H-NMR spectra of mannans from WT [(i)], and gal102Δ/Δ [(ii)] are shown (Figure 4A).
Fig. 4. Deletion of the GAL102 affects cell wall mannan composition. 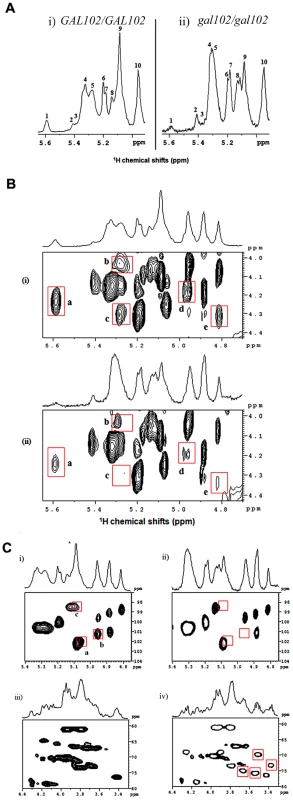
A) Anomeric region of 1H-NMR spectra of mannans from the WT (i) and gal102Δ/Δ (ii) of C. albicans recorded at 40°C temperature on a 500 MHz NMR spectrometer equipped with a triple resonance probe. Important changes that appear in the spectra for WT and the mutant samples are highlighted (and peaks are labeled with numbers). B) 2D 1H-1H TOCSY spectra of mannans from the WT (i) and the gal102Δ/Δ (ii) recorded on a 500 MHz NMR spectrometer equipped with a triple resonance probe. The mixing time for the TOCSY was fixed at 100 ms. The data was acquired at 40°C. Highlighted contours with red colors indicate the peaks of interest for comparison. C) Anomeric region of the 2D 1H-13C HSQC spectra of mannans from the WT (i) and gal102Δ/Δ (ii) recorded at 40°C on a 500 MHz NMR spectrometer equipped with a triple resonance probe. The panels (iii) and (iv) show 2D 1H-13C HSQC spectra of the WT and gal102Δ/Δ strain in the C-2, C-3 and C-6 proton/carbon region of the spectra. Highlighted contours with red box indicate the appearance of the new peaks in the spectra for mutant sample. A close observation of the corresponding proton NMR spectra revealed interesting differences in the structural assembly of mannan from the mutant as compared to WT. Both peaks #1 and #9 showed drastic reduction in the intensity. The former represents the loss or reduction of the entire β-1,2 link branched chain mannose units attached through phosphodiester linkage, and the latter, indicates the loss of considerable length of the α-1,6 backbone moiety. The internal change after the loss of α-1,6 backbone is reflected in the downfield shift of the peak 5 (merged into peak 4). The small, peak 3 which accounts for α-1,2 linked mannans in the acid-labile terminus is missing in the mutant. The mutant showed no change in the intensity of peak 10 suggesting the retention of the β-1,2 linked units (in the side-chain of acid resistant - terminus). The analysis of 31P NMR spectra (Figure S2) showed that the phosphodiester moiety in the side chain is absent in the mutant spectra implying that such linkage is either missing or substantially reduced.
A comparison of 2-dimensional 1H-1H-total correlation spectra (2D-TOCSY) of WT and the gal102Δ/Δ samples revealed significant difference in the spectra (Figure 4B). The reduced peak a indicated the loss of phosphodiester linkage with the β-1,2 mannan moiety. Peaks b and c come from the correlations between β 1 protons on the PO4-linked mannans (in the acid-labile region) to their respective C-6 and C-2 protons which are reduced in the spectra of the mutant samples. Additionally, the loss of intensity in peaks d and e indicates lack or substantial loss of part of the α-1,6 backbone mannan skeleton (revealing a reduction in the size of the α-1,6 backbone) which was further supported by the analysis of 2D 1H-1H ROESY (2-dimensional 1H-1H Rotating-frame Overhauser Effect Spectroscopy) spectra for both WT and the mutant (Figure S3).
A detailed analysis of a 2-dimensional 1H-13C-Heteronuclear Single Quantum Coherence (2D 1H-13C-HSQC) spectrum of WT and mutant samples revealed an interesting feature [Figure 4C (i) and (ii)]. Peak a in Figure 4C (i) corresponds to α-1 proton (δH 5.04 ppm; with its attached carbon, δC 101.73 ppm) of the 1,3,6-trisubstituted mannan moiety in the α-1,3 linkage of the acid-resistant terminus. The absence of this peak in the spectra for mutant reveals that the mutation leads to the loss of α-1,3 mannan linkage in the acid-resistant terminus. Peak b belongs to α/β proton of the terminal mannan β-1 unit (extreme left part of the α-1,6 backbone, see Figure 5) attached to α-1,2 mannose side-chain moiety missing in the mutant spectra, demonstrating the loss of entire α-1,2 side-chain in the acid-resistant terminal region of the mannan assembly. The significance of peak c has not been ascertained yet.
Fig. 5. The mutant mannan composition is significantly different from that of the WT and is qualitatively distinct from the other known mannosylation mutants. 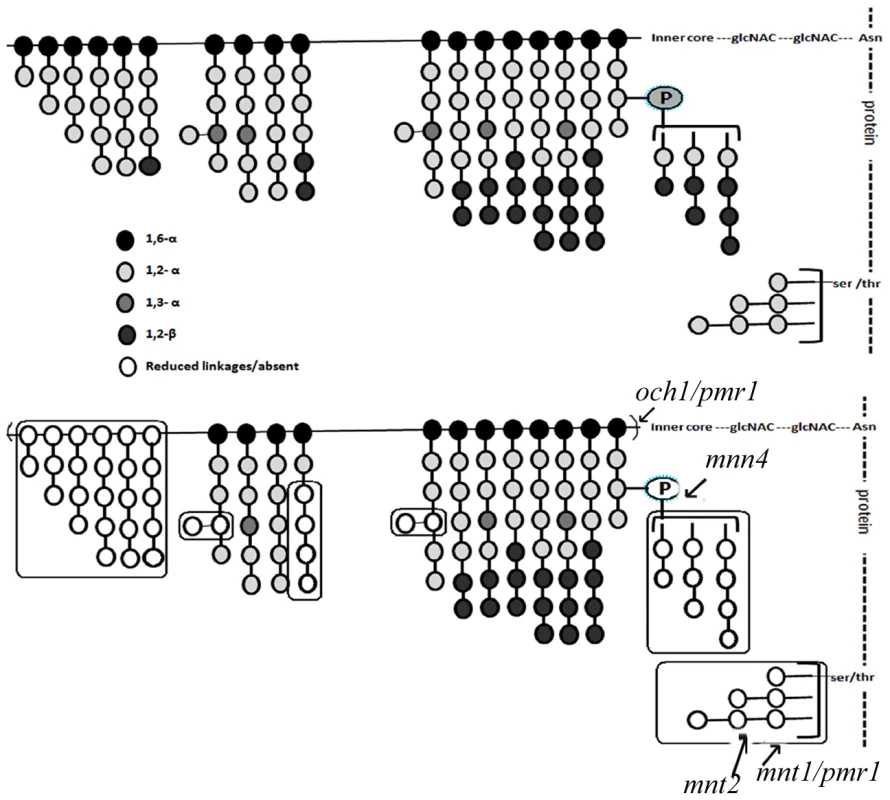
Top panel represents the mannan profile of WT (SC5314) strain, based on Netea et al. [22] illustration. Bottom panel, represents the mannan profile of gal102Δ/Δ. Effects of other known mannosylation mutants on the mannan structure is also as represented by Netea et al. [22]. These are indicated to allow comparison with gal102 mutant. The various linkages between the mannosyl residues have been indicated by the shading of the residues. The legend for the link and the shade intensity is given at the bottom of the figure. The boxes highlight the most significant differences in the mannan profile between the WT and the mutant linkages that are either reduced or absent (denoted by empty circles) in the mutant. The proton up-field region of the HSQC spectra [Figure 4C (iii) and (iv)] indicates loss of a part of the α-1,6 backbone mannan assembly and highlighted peaks with red boxes indicate the appearance of new peaks in the spectra for mutant. These new peaks are attributed to C-2, C-3 and C-6 protons/carbons of the β-1,2 mannose side-chain units in the acid-resistant terminus. The sharp and narrower peaks appearing in the anomeric proton regions of the HSQC spectra demonstrate reduction in chain length and other branching side-chains in the mutant sample. Thus, these observations offer details of the structure of the mannan assembly in C. albicans which corroborate published reports [9] and highlight the changes occurring in it upon deletion of GAL102.
Based on the above 1D as well as 2D NMR analyses we propose the mutant mannan structure to be much different than the WT mannan as shown in Figure 5. The upper panel refers to the WT mannan structure as accepted in the literature [22]. Our study clearly shows that the length of the mutant mannan is substantially reduced and it lacks specific linkages in the branches including the phospho-mannans (Figure 5, lower panel). Significance of these alterations in relation to the other known mannosylation defective mutants is discussed below (see discussion section).
Genome-wide differential gene expression in gal102Δ/Δ strain shows distinct reduction in expression of genes involved in cell wall morphogenesis and virulence
Since the mutant cells showed such distinct phenotypes as elongated cell morphology, and reduced cell wall integrity, we tested the genome wide expression profile of the mutant and compared it to that of the WT. Both WT and gal102Δ/Δ mutant were grown at 30°C in YPD till A600 = 1.2 and RNA was isolated from cells, labeled and used to probe the Agilent custom microarrays as described (see Materials and Methods).
In agreement with the filamentous morphology, the expression profile of the mutant cells showed several hyphal specific genes like HWP1 and a transcriptional regulator UME6 expressed at high levels while yeast phase specific genes like YWP1 were down regulated. Differential expression of some of the genes like HWP1, YWP1 and GAL102 was validated using semi-quantitative RT PCR (data not shown). The levels of transcripts encoding several (characterized as well as putative) GPI-anchored proteins, e.g. ALS7, PGA31, PGA37, RBR1 etc. were high in the mutant. While a negative regulator of hyphal morphology NRG1 was also found to be up-regulated (Table 1); several genes known to play a role in biofilm formation, along with genes like CHK1, CRK1, RRH2, SOD4 etc. that contribute to virulence of C. albicans, were significantly down-regulated (Tables S1, S2 in Text S1). While the gene expression profiling showed genes being expressed as expected from the observed phenotypes, it also provided hints that some of the other attributes associated with virulence such as biofilm formation and survival in disseminated candidiasis mouse model may be altered in the mutant. Apart from the GPI anchored protein coding genes and those involved in biofilm formation, several classes of genes also showed differential expression such as those encoding proteins intrinsic to membrane (GO:31224), α1,3 mannosyl transferase activity (GO:331), etc. Several genes associated with cell wall (GO: 5618) were also highly overexpressed.
Tab. 1. The list of representative genes differentially expressed in C. albicans gal102Δ/Δ as compared to the WT. 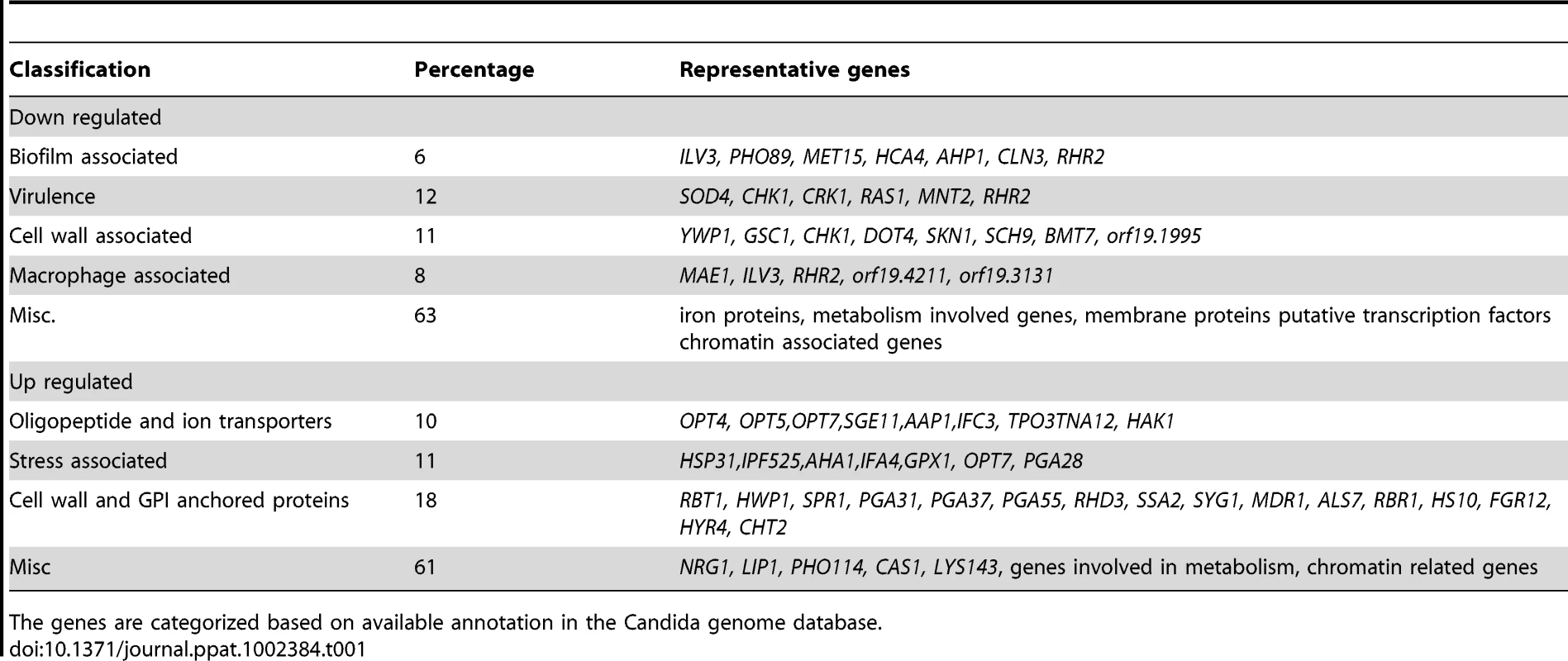
The genes are categorized based on available annotation in the Candida genome database. Macrophages inhibit the growth of gal102Δ/Δ strain in vitro
Since the cell wall mannans constitute the outermost layer of the C. albicans cell wall we expected that the altered mannan composition might alter interactions with the host cells. The outcome of the interaction between phagocytic cells such as macrophages and pathogens such as C. albicans can be studied using cell viability assays in vitro [27]. To study the response of macrophages to gal102Δ/Δ strain, both WT and gal102Δ/Δ mutant cells were incubated with mouse resident peritoneal macrophages and their growth was monitored at indicated time points. At 6 hr, there was no difference in the growth as measured by the CFU numbers, in the presence or absence of macrophages. However, by 18 hr there was a slight reduction in the growth of gal102Δ/Δ strain in the presence of macrophages and by 30 hr there was a substantial reduction in the proliferation relative to cultures without macrophages. The WT strain, on the other hand, proliferated to the same extent in the presence or absence of macrophages (Figure 6A). This inhibition of growth of the gal102Δ/Δ strain in the presence of macrophages is illustrated in micrographs (Figure 6B). These observations demonstrate that resident macrophages suppressed the proliferation of the gal102Δ/Δ strain but not WT C. albicans.
Fig. 6. Macrophages suppress the growth of gal102Δ/Δ in vitro. 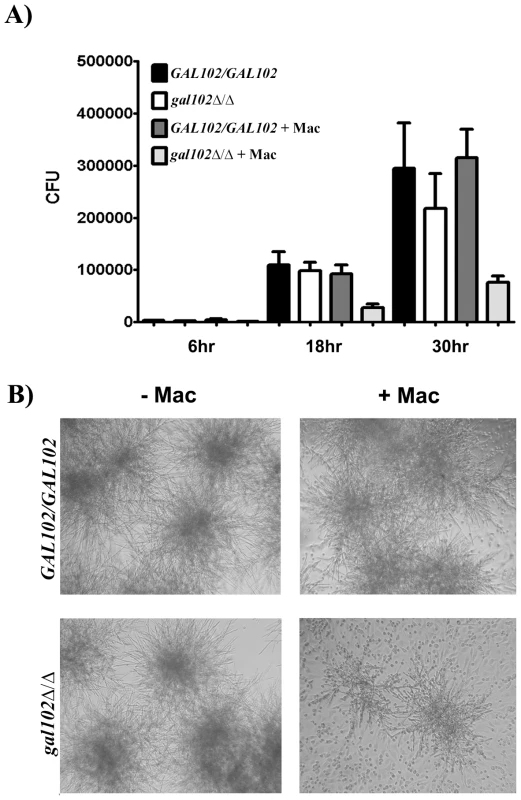
A) Live C. albicans (WT or gal102Δ/Δ) were cultured in the absence or presence of adherent resident peritoneal macrophages for different time periods. Following incubation, C. albicans were washed and plated onto YPD agar plates. Data are pooled from at least two separate experiments with a total of three experiments (mean ± SD), *p < 0.05, t test. B) Representative micrographs depicting growth of WT and gal102Δ/Δ strains of Candida in the absence (left panel) or presence (right panel) of resident peritoneal macrophages for 18 hr is shown. To determine cytokine response mediated by WT and gal102Δ/Δ in vitro, live and heat-killed C. albicans strains were incubated with resident peritoneal macrophages for varying time points. Incubation of macrophages with live WT Candida induced 3–4 fold higher levels of TNFα and IFNγ compared to the gal102Δ/Δ strain and the amounts steadily increased from 6 to 30 hr (Figure S4A and B; left panel). In case of heat-killed Candida, the levels were reduced but differences in the cytokine amounts between WT and gal102Δ/Δ were still significant (Figure S4A and B; right panel) and reflected the same pattern as that induced by the live cells. Interestingly, the gal102Δ/Δ strain induced higher amounts of IL4 production compared to that induced by WT (Figure S4C). These observations demonstrate that peritoneal macrophages elicit distinct cytokine patterns in response to WT and the gal102Δ/Δ strains.
Deletion of GAL102 leads to changes in biofilm characteristics
The problem in treating Candida infection becomes more complicated when it adheres to host surface or to the inner walls of catheters in patients and forms a biofilm, which is resistant to a variety of antifungal agents [28]. The biofilm formed by the WT strain is made up of both the yeast and the hyphal forms and the mutants that are unable to show yeast to hyphal transition, do not form effective biofilms [28]. The ability of gal102Δ/Δ, which shows predominantly elongated cell morphology, to form biofilm was determined using protocols described previously [17]. The WT SC5314 cells formed biofilm which consisted of a mixture of yeast form and filamentous cells which appear healthy and exhibit turgor (Figure 7). Although the ability of the gal102Δ/Δ to form biofilm was not affected, the ultrastructure observed under a scanning electron microscope was quite different (Figure 7). In case of the gal102Δ/Δ mutant strain, the biofilm consisted mainly of elongated cells that showed relatively less turgor, which could reflect weakened structure of the cell wall. The WT reintegrant biofilm showed presence of much higher proportion of yeast form cells as observed in case of biofilms formed by DAY286 WT strain (data not shown). The mutant reintegrant showed biofilm comparable to that of the gal102Δ/Δ strain.
Fig. 7. Biofilm structure and diffusion properties are seriously compromised in C. albicans lacking GAL102. 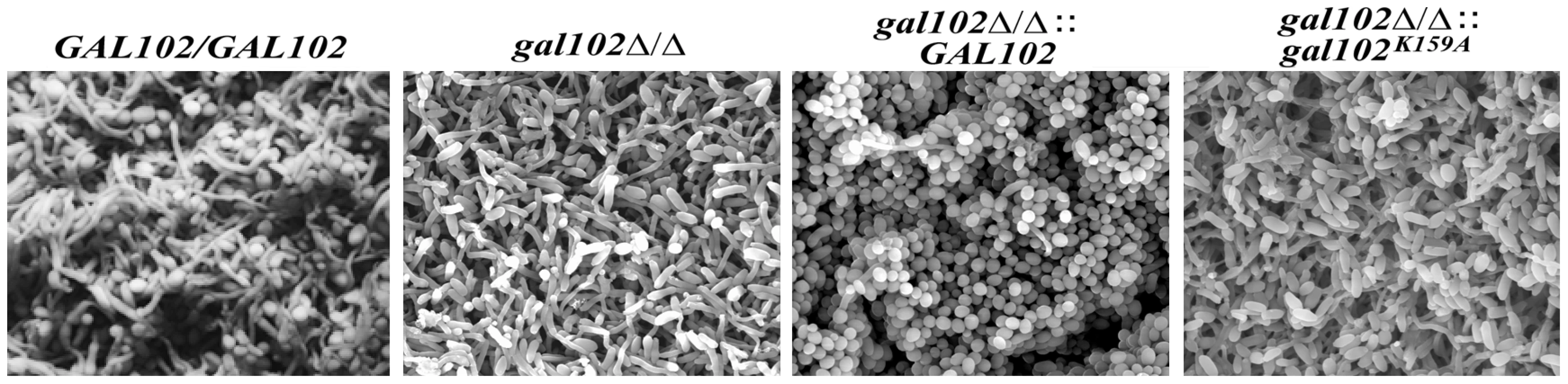
The biofilms of WT (SC5314), gal102Δ/Δ, and WT and the K159A mutant reintegrant in gal102Δ/Δ strain were allowed to be developed as described in the Materials and Methods section. The biofilms were observed under a scanning electron microscope after appropriate treatment. Representative pictures at 1500X magnification are shown. The gal102Δ/Δ strain is less virulent and causes much less organ damage than the WT in a mouse model of systemic candidiasis
To test the effect of alteration in the cell wall mannan composition on virulence of the gal102Δ/Δ, we performed intravenous injection into the lateral tail vein of female BALB/c mice with the WT and the gal102Δ/Δ strain. To ensure that high filamentation of gal102Δ/Δ strain does not lead to clogging of the tail vein during injection, or lead to inaccurate estimation of cell numbers we grew both WT and gal102Δ/Δ strain at 25°C. As mentioned earlier, at this temperature gal102Δ/Δ strain exhibited much less elongated cell morphology (Figure S1D). The resulting survival curves demonstrate that virulence of gal102Δ/Δ strain is greatly reduced. At 5×106 cells of WT C. albicans per animal, 50% of the mice died within 3–4 days and the rest by 6 days (Figure 8A). However, ∼80% or more of mice injected with the gal102Δ/Δ strain survived the entire 30 day duration of observation (data not shown). As mentioned above, the gal102Δ/Δ and the WT and the gal102K159A reintegrant strains, all have URA3 gene integrated at the gal102 locus. We tested the virulence of these genotype matched strains in a mouse model of systemic candidiasis test as before. The clinical isolate SC5314 which is routinely used as the standard for virulence studies showed that it kills the host with faster kinetics than the DAY286, a strain with URA3 reintegrated at the ARG4 locus. This is consistent with earlier reports that the DAY286 strain is ∼70% as virulent as compared to SC5314 [29]. As seen in the survival curve (Figure 8A), the WT reintegrant showed virulence levels comparable to the WT DAY286 strain while the mutant gal102K159A reintegrant strain, as expected, showed low virulence, comparable to the gal102Δ/Δ parent strain. These results clearly show that the deletion/inactivation of GAL102, is responsible for the alteration in morphology as well as compromised virulence and not the expression of URA3 gene from a non-native locus [30].
Fig. 8. Infection of mice with gal102Δ/Δ leads to enhanced survival and reduced fungal tissue burden. 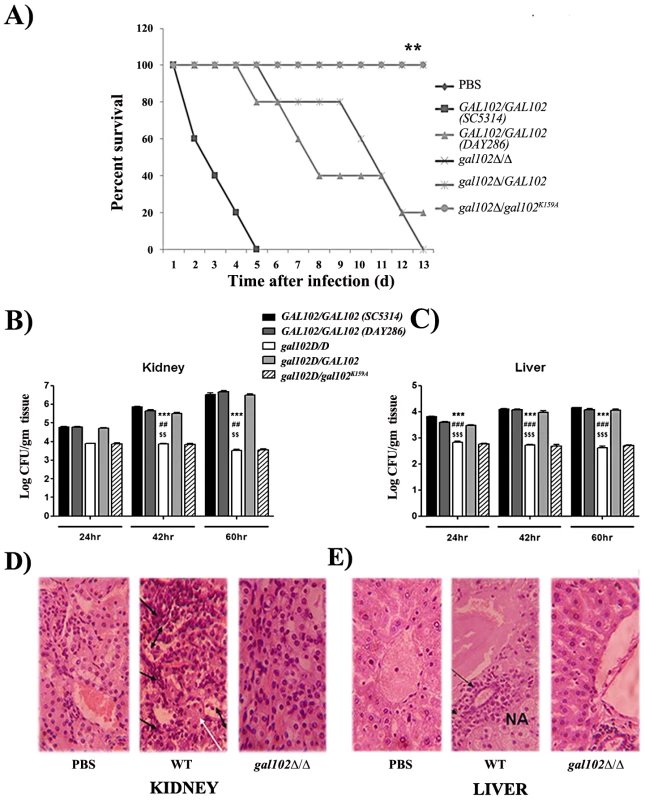
Mice were infected intravenously with 5×106 cells of WT (SC5314 and DAY286), gal102Δ/Δ, and the GAL102 or the gal102K159A mutant reintegrants in gal102Δ/Δ strain. Mice were sacrificed at 24, 42 and 60 hr after infection and disease progression was assessed. A) The survival curve and statistical differences between strains were analyzed by Prism software using the Log-rank (Mantel-Cox) test. Data from one experiment using five mice is shown and is representative of three identical, separate experiments. **p < 0.01, t test. (B and C) Subgroups of three to five mice were sacrificed at 24, 42 and 60 hr after infection Quantitative fungal burden in kidney and liver was measured by serial dilution and expressed as log CFU/gram tissue. Data are represented as mean ± standard errors of the means from three separate experiments with three to five mice per strain. ***p<0.0001, ##p<0.01 and $$p<0.01 compared with SC5314, DAY286 and WTRI (for (gal102Δ/Δ) injected mice) in kidney at 42 and 60 hr. ***p<0.0001, ###p<0.0001 and $$$p<0.0001 compared with SC5314, DAY286 and WTRI (for (gal102Δ/Δ) injected mice) in liver at 24, 42 and 60 hr. (D and E) Kidney and liver tissue sections were dissected from mice sacrificed at 60 hr, stained with hematoxylin & eosin and histological changes were observed. Sections from PBS treated mice were used as control. A representative histopathological examination is shown (magnification 400X for both panels). Black arrows indicate inflammatory cells, NA denotes necrotic areas in liver and white arrows indicate tubular casts and double headed arrows indicate sloughing of the tubular epithelial cells in the kidney. To evaluate whether reduced virulence of gal102Δ/Δ strain was due to its reduced multiplication in vivo, in a parallel set of experiment mice infected with both WT and mutant were sacrificed and fungal colonization of both, kidney and liver, the major target organs in systemic candidiasis, was determined. Increased fungal burden was observed in WT-infected mice from 24 hr to 60 hr after initiation of infection. On the other hand, CFUs recovered from the kidney and liver of mice infected with the gal102Δ/Δ strain were significantly lower than the WT and steadily decreased with time (Figure 8B and C). Both the reintegrants showed expected phenotypes similar to the WT and gal102Δ/Δ strains respectively. The Candida cells isolated from kidneys of mice infected with the gal102Δ/Δ strain clearly showed the wrinkled colony morphology characteristic of the mutant strain (data not shown) confirming the presence of the mutant in the infected hosts. The other markers of kidney and liver damage namely elevated levels of serum urea [31] and of the enzyme alanine aminotransferase (ALT) [32] were also monitored in case of mice infected with either the WT (SC5314) or the gal102Δ/Δ mutant and corroborated the damage to these organs by the WT but not the gal102Δ/Δ strain (Figure S5A and B). The liver sections from the control (PBS-treated) mice revealed normal hepatic architecture whereas mice infected with WT C. albicans showed necrotic patches. Moreover, the kidney sections of WT infected mice showed damage probably due to an excessive inflammatory response. In WT kidney, massive sloughing of the tubular epithelial cells into tubular lumen and deposition of tubular casts was observed compared to PBS and gal102Δ/Δ. Intra-tubular infiltration of inflammatory cells was also high in WT compared to PBS and gal102Δ/Δ. In WT infected liver, significant infiltration of the inflammatory cells around the peri-arterial space was observed compared to the PBS and gal102Δ/Δ. The hepatocytes around the peri-arterial space also exhibited nuclear pycnosis and hydrophobic degeneration in WT compared to PBS and gal102Δ/Δ group. However, liver of mice infected with the mutant strain did not display any necrotic regions or infiltration in the kidney by cells involved in inflammatory response and was comparable to PBS treated mice (Figure 8D and E; Figure S6A and B). These observations demonstrate that WT infection causes organ damage which is not seen upon infection with gal102Δ/Δ.
The reduced virulence of gal102Δ/Δ is associated with its inability to elicit a strong pro-inflammatory response
The observation that the gal102Δ/Δ cells displayed reduced virulence led us to study their ability to elicit cytokine production. Mice were injected (as above) with equal number of cells of WT (SC5314 and DAY286), gal102Δ/Δ, and the GAL102 or the gal102K159A mutant reintegrants in gal102Δ/Δ strain and sacrificed at various intervals. The amounts of different cytokines in serum were monitored by ELISA. We observed that the amounts of the pro-inflammatory cytokines, TNFα and IFNγ were significantly higher in mice infected with WT than those infected with gal102Δ/Δ strain (Figure 9A and B). It is interesting that IFNγ amounts were initially high and decreased later; however TNFα was induced early and remained high for the WT. At the later time points, the lowered IFNγ amounts correlated with increase in the anti-inflammatory cytokine IL-4. In striking contrast, infection with the gal102Δ/Δ strain was characterized by a rapid and increased production of IL-4 (Figure 9C). These experiments demonstrate that infection with the gal102Δ/Δ strain resulted in an altered cytokine response in the host. The WT cells elicited high levels of pro-inflammatory cytokines, such as TNFα and IFNγ, whereas these cells elicited much higher levels of anti-inflammatory cytokine response, e.g. IL-4 (Figure 9A–C). This was also true with the WT and the mutant reintegrant indicating that the activity of the enzyme was restored in the WT reintegrant and lack of which in the mutant reintegrant is responsible for the observed opposing effects on cytokine production.
Fig. 9. Reduced virulence is associated with the inability of gal102Δ/Δ to elicit pro-inflammatory cytokine response in the host. 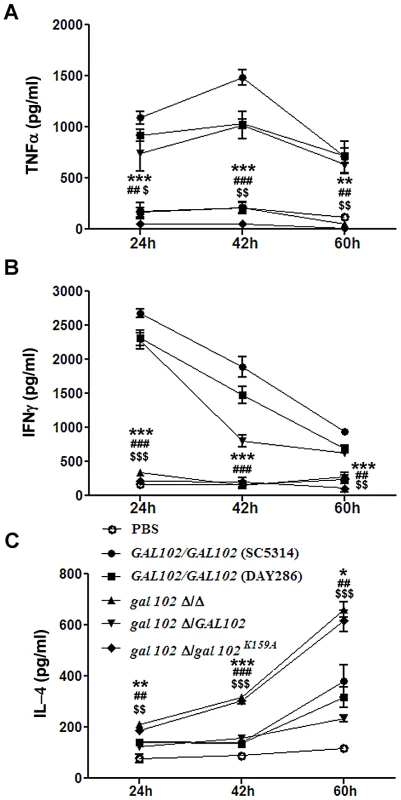
(A, B and C) Mice were infected intravenously with WT (SC5314 and DAY286), gal102Δ/Δ, and the GAL102 or the gal102K159A mutant reintegrants in gal102Δ/Δ strains and were sacrificed at different time points for measurement of serum cytokine amounts. TNFα (A), IFNγ(B) and IL-4 (C) amounts were determined by ELISA. Sera from PBS treated mice were taken as control. Data are represented as means ± standard errors of the means from three separate experiments with sera of five mice. ***p<0.001, **p<0.01, *p<0.05; ###p<0.001, ##p<0.01, #p<0.05 and $$$p<0.001, $$p<0.01, $p<0.05 compared with SC5314, DAY286 and WTRI (for (gal102Δ/Δ) treated). Discussion
We initiated this work following the observation that C. albicans genome carries two genes encoding putative paralogs of UDP-galactose epimerase enzyme. We had earlier shown that CaGAL10 gene encodes a UDP-galactose epimerase [17]. During the course of this study we found that although the gene in C. albicans was annotated as UDP-galactose epimerase (CaGAL102), the gene does not appear to encode a functional galactose epimerase but an enzyme with UDP-glucose 4,6-dehydratase activity. The RHM2 gene of A. thaliana, is the only other eukaryotic homolog which has been demonstrated to have this activity [26]. Prokaryotic homologs of this enzyme typically use dTDP-glucose as the substrate [25]. The dTDP/UDP-glucose 4,6-dehydratase enzyme homologs control cell wall composition in bacteria and plants and in both cases the enzyme functions in the rhamnose (and/or related sugar) biosynthetic pathway. Rhamnose, is a component of the cell wall of these organisms and inhibition of its biosynthesis leads to cell wall integrity defects. In pathogenic bacteria the defect in cell wall integrity leads to loss of virulence. In A. thaliana mutants, it is known to lead to root morphology defects [33]. However, in the genomes of very few fungal species has the homolog of this enzyme been reported and is clearly absent in the filamentous fungi, e.g. Aspergillus. The homolog in some of the above fungal species has been annotated as a putative dTDP-glucose 4,6-dehydratase without any experimental evidence and in C. albicans it was annotated incorrectly as galactose epimerase as evidenced from our present study. Since the presence of rhamnose has not been reported in any of these fungal species, and the homologs of downstream enzymes in the rhamnose biosynthetic pathway are absent in their genomes, we addressed the following questions with C. albicans, an important fungal pathogen as the representative species: a) what is the significance of presence of this gene in C. albicans? b) Does it affect cell wall biogenesis and c) is it relevant to its virulence?
The inactivation of UDP-glucose dehydratase drastically affects composition of cell wall mannan
Cell wall is the major organelle of a pathogen that comes in contact with the host cells and plays an important role in the outcome of the host-pathogen interaction. Many studies have revealed that the major components of the fungal cell wall such as mannans, glycans and proteins all contribute to morphology as well as the host response elicited by the pathogen. The mutants of C. albicans which lack specific components of mannan, the outermost layer of the cell wall, show reduced virulence indicating subtle contributions of individual components to virulence [34]. The two morphological forms of C. albicans, the yeast and the hyphae, are known to differ in the proteins associated with the cell wall, as well as the mannans which greatly contribute to the host response [34]. Several mutants have been studied which show alterations in various features of the cell wall. It has been reported that the och1 mutant is defective in outer, branched N-linked glycans [35]. The mnt1 mnt2 mutant lacks all the 4 terminal O-linked α1,2-mannosyl residues, but has normal N-linked mannan [36]. The pmr1 mutant has defects in both N - and O-linked mannosylation [37] and the mnn4 mutant lacks phosphomannan [38]. While the gal102 mutant lacked the phosphomannan like the mnn4 mutant, it showed substantially reduced levels of the O-linked mannosyl residues and had less drastic effect on the N-linked mannan as compared to the pmr1 mutant. Thus, the gal102 mutant displayed a mannan profile that is distinct from any of these above mentioned known mannosylation defective mutants (Figure 5).
Alteration in cell wall composition is associated with differential expression of several cell wall protein coding genes
Cell wall has been considered a dynamic rather than a static assemblage of macromolecules and recent studies have indicated that the Candida cells exhibit altered mannoprotein composition in response to environmental stimuli [39]. Our genome-wide differential gene expression studies of the mutant showed that a large number of hypha-specific genes were upregulated while yeast form-specific Ywp1 protein was clearly downregulated which is commensurate with the morphology of the cells and increased tendency of the mutant cells to show hyper-filamentation. Among the classes of downregulated genes, the most significant were membrane protein encoding genes as well as those involved in biofilm formation. Interestingly, among the over expressed genes under the categories like cell wall (p = 4×10−8) hyphal cell wall, carbohydrate metabolism etc. were much higher and most significant. Some of the genes encoding cell-surface proteins such as ALS2, ALS9 which are known to be expressed during infection were also found to be downregulated while ALS6/ALS7 appear to be upregulated. It has been suggested that in response to cell wall integrity defects some compensatory mechanisms cause increase in expression of several cell wall proteins [40]. In the gal102Δ/Δ mutant the genes coding for GPI-anchored cell wall proteins showed increased expression levels. It has been known from work based on S. cerevisiae system as well, that expression of GPI-anchored proteins is drastically altered when cell wall synthesis is defective [40]. It has been suggested through many studies that cell wall is an important organelle in fungi, whose complex structure is carefully managed by the cell and altered according to various environmental and internal cues. The cell wall composition in turn affects survival of the fungal cell in the host as it elicits specific response from host immune system.
Altered cell wall composition of the gal102 mutant induces altered host cytokine response which correlates with reduced virulence
Mammalian hosts can respond to the presence of a pathogen by producing a variety of inflammatory molecules which help curtail the infection. These can be categorized into two broad groups: those involved in pro-inflammatory responses (e.g. TNFα & IFNγ) and anti-inflammatory responses (e.g. IL4 & IL13). The balance of pro - and anti-inflammatory response is thought to be important for the outcome of fungal infections. TNFα, a pro-inflammatory cytokine is a key mediator in protecting against disseminated candidiasis, as demonstrated by the fact that the lack of TNFα worsens the course of disseminated candidiasis [41]. Furthermore, IFNγ another pro-inflammatory cytokine is thought to play a dual role during candidiasis. On one hand, it is required for host resistance as shown in studies using Ifnγ−/− mice [42]; on the other hand, excess IFNγ production during infection leads to host damage leading in turn, to increased susceptibility to C. albicans [43]. Most importantly, dendritic cells pulsed (ex vivo) with yeast cells, when transferred into mice, protect them from challenge with C. albicans [44]. This protection was not observed with dendritic cells pulsed with hyphal cells. These studies clearly underscore the importance of pro-inflammatory cytokines in the host resistance to C. albicans infection. There are conflicting reports on the role of IL-4 during candidiasis. One report involving intra-peritoneal challenge with C. albicans blastoconidia showed that IL-4 was not involved in resistance [42]. However, the unrestrained IFNγ response during early stages of C. albicans infection in Il4−/− mice, leads to resistance and only at later stages are the Il4−/− mice susceptible due to failure in mounting a strong Th1 response. In fact, treatment with exogenous IL4 boosts immunity against C. albicans [45].
The gal102 mutant showed virulence defect in disseminated mouse model system and the same reflected in its inability to grow in the presence of macrophages in vitro. In vitro as well as in vivo studies showed that the mutant was unable to induce pro-inflammatory cytokine production in contrast to that observed with WT C. albicans infection. This was indeed surprising since reduced pro-inflammatory cytokine response could be expected to increase survival of the pathogen in the host. It is well known that C. albicans yeast and the hyphal forms elicit different responses from the host immune system with the yeast form inducing pro-inflammatory cytokines, and the hyphal form inducing anti-inflammatory cytokines [34], [44]. The high pro-inflammatory cytokine response is required for host defence; however excessive inflammatory responses cause damage to the host [46], [47]. The better survival and growth of the WT pathogen in the host leads to increased production of pro-inflammatory cytokines, resulting in excessive tissue damage. On the other hand, it is possible that the defective cell wall of gal102 mutant renders these cells susceptible to killing by host cells (Figure 6) and at the same time lower levels of pro-inflammatory cytokines induced by the mutant cause reduced tissue damage (Figure 8D and E) resulting in increased host survival (Figure 8A). The gal102 mutant induces increased production of IL4, an anti-inflammatory cytokine which is responsible for the increased killing and reduced growth of mutant cells (Figure 9). These results clearly underscore the important roles of pro - and anti-inflammatory cytokines, which have major implications in terms of immunity against fungal pathogens.
In C. albicans marker gene expression has been shown to be drastically affected depending on the locus of integration. Among the most favorite of the marker genes viz. URA3 gene in C. albicans has been shown to affect virulence attributes depending on its chromosomal location [30]. Re-integration of the WT ORF and the catalytically inactive mutant ORF in the native locus allowed us to compare the phenotypes of the deletion mutant irrespective of the URA3 expression levels since all the three strains, i.e. gal102Δ/Δ and the WT or mutant reintegrant carried the URA3 marker at the GAL102 locus. The WT reintegrant showed rescue of most phenotypes tested e.g. filamentous morphology, virulence attributes and the cytokine response elicited by the mutant, while the catalytically inactive mutant with only a single lysine to alanine change in the YXXXK motif showed phenotypes akin to the parent gal102Δ/Δ mutant.
UDP glucose 4,6-dehydratase activity although present in non-pathogenic S. pombe affects virulence of C. albicans
In summary, our characterization of gal102, a putative paralog of UDP-galactose 4-epimerase in C. albicans, has led to the conclusion that it is not a galactose epimerase. Although sequence analysis reveals that it is a member of the Short chain dehydratase reductase (SDR) family, whose members have a conserved, glycine rich, NAD/NADP-binding motif as well as a catalytic YXXXK motif [48]. The present study for the first time has presented evidence that a fungal homolog actually possesses the UDP-glucose 4,6-dehydratase activity. Yet, the homologs of enzymes participating in the subsequent steps involved in rhamnose biosynthesis have not been identified and the product rhamnose has not been detected in C. albicans or other related fungi. This raises a distinct possibility that Gal102p may not be involved in the rhamnose biosynthetic pathway in these fungi. It is known that nucleotide sugar moieties are used as sugar donors in glycosylation reactions. Since we have observed significant differences in the mannosylation profile of the mutant, it is possible that this enzyme functions upstream of a variety of glycosylation reactions involved in cell wall protein mannosylation in C. albicans and many of the related fungal pathogens. We have shown here that in C. albicans, the absence of the activity affects cell wall mannans, cell wall integrity and morphology with consequences on survival in the host and virulence. The product of this enzyme activity likely acts as a donor of sugar residues in the synthesis of mannans and might affect GPI anchoring of proteins which are known to affect cell wall integrity and morphology of cells [49], [50]. We speculate that loss of cell wall integrity might trigger over-expression of several GPI proteins as a salvage response. The mutant with reduced cell wall integrity is also more susceptible to extracellular killing in the host as compared to WT cells. At the same time altered mannan composition elicits weaker pro-inflammatory cytokine response reducing host tissue damage and further allowing host to survive longer to be able to overcome the infecting mutant cells. The WT cells, on the other hand, can cause greater mortality due to the increased tissue damage induced by high levels of pro-inflammatory cytokines. Hence, in spite of the elongated cell morphology, the gal102 mutant remains avirulent. Perhaps, it's not the cell morphology, but the composition of the cell wall that plays a key role in virulence.
Interestingly, CaGAL102 was not included among the 674 target genes studied by Noble et al. [51] as those likely to be involved in virulence of C. albicans. It turns out that this study had excluded genes that had sequence homologs in either S. cerevisiae or S. pombe, two of the well-studied non-pathogenic yeasts. CaGal102p is highly homologous to the protein encoded by the S. pombe gene SPBPB2B2.11 and recent phylogenetic analysis of GAL cluster has indicated that S. pombe might have acquired the entire GAL cluster including CaGAL102 homolog from a Candida species [52]. Hence it is not surprising that the above interesting study [51] missed CaGAL102 as one of the genes affecting virulence of C. albicans. It is also well known that genes whose products contribute to virulence can have homologs in related non-pathogenic microorganisms and, thus, are not necessarily unique to the genomes of pathogens. Assigning the direct role of the biochemical activity of CaGal102p in virulence of the fungal pathogens will require further in-depth studies through biochemical, genetic and molecular analyses.
Materials and Methods
Animal ethics statement
All experiments involving mice were conducted in compliance of the Ministry of Environment and Forests Act on breeding of and experiments on animals (control and supervision) rules, 1998. Mice were housed in the Central animal facility of IISc and studies were performed as per the guidelines laid out by the institutional animal ethics committee, IISc (permit no. CAF/Ethics/174/2009). The Central animal facility is accredited to the Ministry of Environment and Forests, Government of India. The guidelines followed are approved in consultation with the Committee for the purpose and control and supervision of experiments on animals (CPCSEA) can be seen in detail in the following document: http://envfor.nic.in/divisions/awd/cpcsea_laboratory.pdf.
Strains and growth medium
The C. albicans strains used in this study are listed in Table 2. Standard growth media were used for C. albicans. Details of composition are provided in supplementary methods section. Prior to challenge in the animal model, a suspension was made by transferring to YPD broth and incubating at 30°C with shaking for a period of 16–24 hr. The S. cerevisiae strain was transformed with the plasmid constructs listed in Table 3 to generate the appropriate strains assayed for various phenotypes in this study.
Tab. 2. List of <i>C. albicans</i> and <i>S. cerevisiae</i> strains. 
In all assays such as mouse infection, etc. where number of cells is critical, the cells were counted using hemocytometer.
Mice
Female BALB/c mice, aged 6–8 weeks and weighing ∼18–25 g, were obtained from the Central Animal Facility, Indian Institute of Science, Bangalore. All mice received care according to institutional guidelines and were maintained under controlled conditions and fed with a standard diet.
DNA manipulation
GAL102 (http://www.candidagenome.org/) was amplified with the CaputGAL10(f) and CaputGAL10(r) primers. The 963 bp fragment was cloned in pGEM-Teasy vector (Promega) to construct pVM602. pVM603 (pGAL1-ORF19.3674) was constructed by cloning of pYES2 (Invitrogen). pMS643 (pTEF1-ORF19.3674) was constructed by cloning BamHI/XhoI insert from pVM603 in the same sites in pPS189. The construct thus obtained was used to transform S. cerevisiae gal10 deletion strain, PJB5.Codon optimized CaGAL102 was also cloned in the same sites in pPS189 (pMS861) and transformed in pJB5. A list of all the plasmids generated during this study is provided in Table 3.
To generate K159A mutation in Gal102, the first 490 bp of the wildtype open reading frame was amplified using Pfu (Fermentas) with primers CaputGAL10(f) and Gal102 _int _r (Table 4) harbouring the TTT to TGC change in the primer. The remaining 473 bp was amplified using Pfu (Fermentas) using primers Gal102 _int _f harbouring the AAA to GCA change and CaputGAL10(r). The final product was generated by overlap PCR using Pfu using primers CaputGAL10(f) and CaputGAL10(r) and cloned in pBSKS (Promega) in EcoRV site thus generating plasmid pMS833 The clone had K159A mutation in the Gal102 open reading frame and this was confirmed by sequencing. The Gal102 K159A open reading frame was released as a BamHI-XhoI fragment from pMS833 and subcloned in pSJ821 in BglII-XhoI sites thus generating plasmid pMS834.
In order to express CaGal102 in E. coli, the only CUG codon in Gal102 (seventh codon from the C - terminus) was converted to other serine encoding codon to compensate for the alternative genetic code in C. albicans. The ORF was amplified using Caput Gal10 (f) and a reverse primer Gal102 CTG* r using Pfu polymerase (Fermentas) and cloned in pBSKS. The mutant CaGal102 hereafter referred to as Gal102CTG*,was further subcloned in pET32(a) to generate plasmid pMS836.To express the catalytic mutant in E. coli, the mutant ORF was PCR amplified using primers Ca put Gal10 (f) and Gal102 CTG* r from pMS833 (described above) and cloned in pET32(a) thus generating plasmid pMS838 (Table 3).
Transformation of C. albicans and S. cerevisiae
The lithium acetate procedure as described previously was used to transform C. albicans [53] and S. cerevisiae [54]. For Nourseothricin based plasmids transformation was done by electroporation and selected on plates containing 200 µg/ml of nourseothricin at 30°C [55].
Total protein extraction and Western Blot Analysis
Protein extracts were prepared from PJB5 cells transformed with pPS189 and pMS643 by glass bead lysis method. Protein samples were quantified using Bradford Reagent (Sigma) and approximately 50 µg of total protein was electrophoresed in 12.5% SDS-PAGE transferred to ImmobilonTM-P Transfer Membrane (Amersham), and probed with rabbit polyclonal 1° antibodies raised against bacterially expressed orf19.3674p. The blots were probed with HRP-conjugated anti-rabbit 2° antibody (Sigma) and developed using Western Lighting TMChemiluminescence Reagent plus (Perkin Elmer) and X-ray film reagents.
RNA isolation, RT PCR and microarray analysis
Strains were grown in YPD and YPGal at 30°C. Overnight saturated cultures were diluted in their respective media to an A600 of 0.1 and were allowed to grow until the A600 reached 1.2 following which total RNA was extracted as described below. WT SC5314 cells were induced to form hyphae using various hyphae inducing media described above. RNA was isolated by hot phenol method using liquid nitrogen [56]. RNA samples were quantified by measuring the absorbance at 260 nm and equal amounts of RNA were run on a 1% agarose-formaldehyde gel to check the quality. Total RNA was used for Genome wide expression profiling through Genotypic technology. Agilent's C. albicans custom arrays (AMADID 16356) were used and hybridized with the cDNAs that were labeled using the Agilent's RNA linear amplification kit following manufacturer's protocol. Two biological replicates were used for both WT and mutant NA samples and single color hybridization was performed on four independent arrays. The differentially expressed genes were identified using Genespring software based normalization methods (Typically LOWESS method was applied for array wide normalization and only mutant/WT ratio of >2 or <0.5 was considered as significant differential expression). The entire data has been deposited at the GEO database and can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20883. (Text S2).
Biofilm production and biofilm permeability assay
Biofilms were produced as described [57] and biofilm permeability assay was performed as described [17]. The concentration of echinocandin used for this assay was 300 µg/ml and 600 µg/ml.
Scanning electron microscopy
Biofilm developed was fixed with 4% (v/v) formaldehyde made in 1X phosphate buffer (pH 7.5) for 15 min followed by washing with 1X phosphate buffer. Then the biofilm was treated with 1% osmium tetroxide made in 1X phosphate buffer for 1 hr in dark. Then the subsequent dehydration steps with ethanol were carried out as follows: 70% for 10 min, 95% for 10 min, 100% for 20 min, followed by air drying in a desiccator overnight prior to gold coating in a sputter.
Spot assay for analyzing sensitivity to cell wall damaging agents
Cultures were grown in 5 ml of YPD medium until the exponential phase and then diluted to an optical density at 600 nm (OD600) of 0.5. Each undiluted culture and its four 10 fold serial dilutions were spotted onto YPD plates containing the following: Calcoflour White (10–40 µg/ml), sodium dodecyl sulfate (SDS; 0.005–0.04%), Congo red (5–30 µg/ml). Growth differences were recorded after incubation of the plates at 37°C for 72 hr.
Preparation of mannans
Candida strains were grown at 30°C for 36 hr with shaking in YPD medium containing 1% yeast extract, 2% peptone and 2% dextrose. Mannan was extracted from the cells with water at 120°C in an autoclave for 3 hr [9]. Briefly, after centrifugation the supernatant was treated with equal volume of Fehling's reagent for a short term precipitation. The copper-mannan complex was dissolved in 3N HCl and further precipitated drop-wise in Methanol: Acetic acid (8∶1) solvent. The carbohydrate was dried, dissolved in water, purified and then lyophilized till further use.
NMR spectroscopy of cell wall mannans
The mannans obtained so from the above mentioned procedure were then subjected to NMR analysis (all NMR studies were carried at the NMR Facility, Piramal Life Sciences Ltd., Mumbai, India). All NMR spectra were recorded for a solution sample of each (purified and lyophilized) mannan in 700 µl of deuterium oxide (D2O) at 40°C using BrukerAvance 500 MHz spectrometer equipped with z-gradient triple resonance (1H, 13C, X) probe. Acetone (δH 2.17 ppm) was used as an internal standard for the spectral referencing. The 2D TOCSY spectra (pulse, mlevphpr) were recorded containing 256 increments, consisting of 64 scans. The mixing time for each spectrum was set to 100 ms. 2D 1H-13C HSQC (pulse, hsqcetgp)) spectra were composed of 256 increments consisting of 64 scans. 2D ROESY spectra were acquired using a pulse sequence roesyphpr, with 256 increments consisting of 128 scans. 1D NMR spectra were recorded suing a standard pulse sequence, zg30. The 31P NMR spectra were recorded on Bruker 300 MHz spectrometer equipped with a BBO probe at the Piramal Life Sciences Ltd. research facility, Mumbai. Orthophosphoric acid was used as an external standard (δ31P 7 ppm) for 31P NMR. All spectra were processed and analyzed using Topsin (version 2.1, Bruker, March 2007) program package.
Co-culture studies with peritoneal macrophages and C. albicans
Cells were isolated from the peritoneal cavity of mice by flushing with 5 ml of ice-cold 0.32 M sucrose. 2×105 cells/well were seeded in 96-well flat-bottom tissue culture plates in 100 µl/well culture medium (RPMI 1640, 5% FBS, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 10 µg/ml gentamycin, 10% nonessential amino acids, 10% HEPES, 10% sodium pyruvate, 50 µM 2-ME). After incubation for 2 hr at 37°C with 5% CO2, wells were washed with medium to remove non-adherent cells and adherent cells were used for subsequent co-culture studies with C. albicans strains. Adherent resident peritoneal macrophages were incubated with 100 µl of various strains of live or heat-killed (30 min at 75°C) C. albicans cells at a concentration 104 cells/ml. Supernatants were harvested at different time points (6, 18 and 30 hr) and stored at –80°C until analysis of cytokines. The experiments were performed in triplicate with samples from three mice. The differences between strains were analyzed by using the Student's t test, and the level of significance was set at p < 0.05.
Infection of mice with C. albicans strains
Age-matched mice were challenged intravenously with 5×106 C. albicans WT (SC5314/DAY286), gal102Δ/Δ, or WT or gal102K159A re-integrants in gal102Δ/Δ per mouse via lateral tail vein injection in 100 µl PBS. Control mice were injected with 100 µl PBS as vehicle control. Cohorts of five mice per C. albicans strain were inoculated for survival study. Mice were monitored daily for signs of morbidity (weight loss, ruffled fur, hunched appearance, and decreased activity).
Tissue burden assay and histological examination
For the tissue burden assays same dose was injected as above (five mice per strain per experiment) and mice were sacrificed at varying time points (24, 42 and 60 hr) post-infection. Kidney and liver from individual mice (three to five mice per group) were removed aseptically, weighed and homogenized in 1X PBS buffer using homogenizer. CFU was determined by plating serial dilutions on YPD agar medium. The colonies were counted after 24 hr at 37°C, and results were expressed as log CFU/g tissue. These experiments were performed thrice with 5/6 mice per group. For histological examinations the kidney and liver tissues were dissected, fixed in 10% neutral formalin buffer and embedded in paraffin wax. The sections were mounted on slides and stained with haematoxylin-eosin (H&E). Examination was performed under a light microscope and photographs were taken by Nikon camera fitted to the microscope.
Cytokine assays
The mice injected with WT (SC5314/DAY286), gal102Δ/Δ, or WT or gal102K159A re-integrants in gal102Δ/Δ or PBS was sacrificed at 24, 42 and 60 hr post-infection. Blood was collected by cardiac puncture, allowed to clot at room temperature and centrifuged (10,000 rpm for 10 min) to separate serum for subsequent assays. Cytokine concentrations were measured by commercial ELISA kits from eBioscience, San Diego, USA according to the manufacturer's instructions using 96 well plates. The serum samples were diluted 1∶5 and the concentrations of cytokines were determined, using a standard curve. The linear range of detection for TNFα, IFNγ and IL-4 was 31.25–1,000 pg/mL, 62.5–2,000 pg/mL and 15.125–500 pg/mL respectively. This experiment was performed thrice with 5 mice per group-per time point.
UDP-glucose 4,6 - dehydratase activity assay
The enzyme activity was measured as described [26]. Briefly, the recombinant protein expressed in E. coli was purified using Ni-NTA column and 20 µg of protein was incubated at 37°C for 2 hr in reaction buffer containing 50 mM Tris-Cl (pH-7.6). At various time points the A340 nm was measured. The assay was repeated with two independent purified enzyme preps. The results include data from three replicates.
Statistical analysis
Data were analyzed using commercial software (GraphPad Prism 5 Software). Differences in C. albicans colonization of tissues and cytokine levels between two groups were analyzed by Student's t test and comparison between the survival curves were done using the Logrank test.
Supporting Information
Zdroje
1. BastidasRJHeitmanJ 2009 Trimorphic stepping stones pave the way to fungal virulence. Proc Natl Acad Sci U S A 106 351 352
2. WhitewayMBachewichC 2007 Morphogenesis in Candida albicans. Annu Rev Microbiol 61 529 553
3. ErnstJF 2000 Transcription factors in Candida albicans environmental control of morphogenesis. Microbiol 146 1763 1774
4. DhillonNKSharmaSKhullerGK 2003 Signaling through protein kinases and transcriptional regulators in Candida albicans. Crit Rev Microbiol 29 259 275
5. CarlislePLBanerjeeMLazzellAMonteagudoCLo'pez-RibotJL 2009 Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci U S A 106 599 604
6. LiuH 2002 Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int J Med Microbiol 292 299 311
7. García-SánchezSMavorALRussellCLArgimonSDennisonP 2005 Global roles of Ssn6 in Tup1 - and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol Biol Cell 16 2913 2925
8. EckertSEShethCCMuhlschlegelFA 2007 Regulation of Morphogenesis in Candida albicans. EnfertCHubeB Candida comparative and functional genomics 263 292
9. ShibataNSuzukiAKobayashHOkawaY 2007 Chemical structure of the cell wall mannan of Candida albicans serotype A and its difference in yeast and hyphal forms. Biochem J 404 365 372
10. RoperJRGütherMLMilneKGFergusonMAJ 2002 Galactose metabolism is essential for the African sleeping sickness parasite Trypanosoma brucei. Proc Natl Acad Sci USA 99 5884 5889
11. JinYSamaranayakeLPSamaranayakeYYipHK 2004 Biofilm formation of Candida albicans is variably affected by saliva and dietary sugars. Arch Oral Biol 49 789 798
12. BaillieGSDouglasLJ 2000 Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J Antimicrob Chemotherpy 46 397 403
13. HoldenHMRaymentIThodenJB 2003 Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem 278 43885 43888
14. BhatPJMurthyTV 2001 Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose mediated signal transduction. Mol Microbiol 40 1059 1066
15. BarberCRostiJRawatAFindlayKRobertsK 2006 Distinct properties of the five UDP-D-glucose/UDP - D -galactose 4-epimerase isoforms of Arabidopsis thaliana. J Biol Chem 281 17276 17285
16. SeibothBKaraVaLSandorEKubicekC 2002 The Hypocrea jecorina gal10 (uridine 50-diphosphate-glucose 4-epimerase-encoding) gene differs from yeast homologs in structure, genomic organization and expression. Gene 295 143 149
17. SinghVSomisettyVMysoreLRSadhalePP 2007 The key enzyme in galactose metabolism, UDPgalactose-4-epimerase, affects cell-wall integrity and morphology in Candida albicans even in the absence of galactose. Fungal Genet Biol 44 563 574
18. WubbenMJ2ndRodermelSRBaumTJ 2004 Mutation of a UDP-glucose-4-epimerase alters nematode susceptibility and ethylene responses in Arabidopsis roots. Plant J 40 712 724
19. QuimbyBBAlanoAAlmashanuSDeSandroAMCowanTM 1997 Characterization of two mutations associated with epimerase - deficiency galactosemia, by use of a yeast expression system for human UDP galactose-4-epimerase. Am J Hum Genet 61 590 598
20. MoyrandFLafontaineIFontaineTJanbonG 2008 UGE1 and UGE2 regulate the UDP-Glucose/UDP-Galactose equilibrium in Cryptococcus neoformans. Eukaryotic Cell 7 2069 2077
21. PoulainDJouaultT 2004 Candida albicans cell wall glycans, host receptors and responses: elements for a decisive crosstalk. Curr Opin Microbial 7 342 349
22. NeteaMGGowNAMunroCABatesSCollinsC 2006 Immune sensing of Candida albicans requires cooperative recognition of glucans and mannans by lectin and Toll-like receptors. J Clin Invest 116 1642 1650
23. ThodenJBHoldenHM 2005 The molecular architecture of galactose mutarotase/UDP-galactose-4-epimerase from Saccharomyces cerevisiae. J Biol Chem 280 21900 21907
24. GolaSMartinRWaltherADünklerAWendlandJ 2003 New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20 1339 1347
25. MaYSternRJSchermanMSVissaVDYanW 2001 Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-Rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-Glucose to dTDP-Rhamnose. Antimicrob Agents Chemother 45 1407 1416
26. OkaTNemotoTJigamiY 2007 Functional analysis of Arabidopsis thaliana RHM2/MUM4 a multidomain protein involved in UDP-D glucose to UDP-L-rhamnose conversion. J Biol Chem 282 5389 5403
27. MarcilAHarcusDThomasDYWhitewayM 2002 Candida albicans killing by RAW 264.7 mouse macrophage cells: effects of Candida genotype, infection ratios, and gamma interferon treatment. Infect Immun 70 6319 6329
28. BlankenshipJRMitchellAP 2006 How to build a biofilm, a fungal perspective. Curr Opin Microbial 9 588 594
29. SanchezAAJohnstonDAMyersCEdwardsJEMitchellAP 2004 Relationship between Candida albicans virulence during experimental hematogenously disseminated infection and endothelial cell damage In Vitro. Infect Immun 72 598 601
30. ChengSNguyenMHZhangZJiaHHandfieldM 2003 Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect Immun 71 6101 6103
31. LeunkRDMoonRJ 1979 Physiological and metabolic alterations accompanying systemic candidiasis in mice. Infect Immun 26 1035 1041
32. KretschmarMHubeBBertschTSanglardDMerkerR 1999 Germ tubes and proteinase activity contribute to virulence of Candida albicans in murine peritonitis. Infect Immun 67 6637 6642
33. DietALinkBSeifertGJSchellenbergBWagnerU 2006 The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18 1630 1641
34. TorosantucciAChianiPCassoneA 2000 Differential chemokine response of human monocytes to yeast and fungal forms of Candida albicans and its relation to the beta-1,6 glucan of the fungal cell wall. J Leuko Biol 68 923 932
35. BatesSHughesHBMunroCAThomasWPHMacCallumDM 2006 Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J Biol Chem 281 90 98
36. MunroCABatesSBuurmanETHughesHBMacCallumDM 2005 Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J Biol Chem 280 1051 1060
37. BatesSMacCallumDMBertramGMunroCAHughesHB 2005 Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J Biol Chem 280 23408 23415
38. HobsonRPMunroCABatesSMacCallumDMCutlerJE 2004 Loss of cell wall mannosyl phosphate in Candida albicans does not influence macrophage recognition. J Biol Chem 279 39628 39635
39. KruppaMGreeneRRNossILowmanDWWilliamsDL 2011 C. albicans increases cell wall mannoprotein, but not mannan, in response to blood, serum and cultivation at physiological temperature. Glycobiol 21 1173 1180
40. MunroCASelvagginiSdeBruijnIWalkerLLenardonMD 2007 The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in C. albicans. Mol Microbiol 63 1399 1413
41. LouieABaltchALSmithRPFrankeMARitzWJ 1994 Tumor necrosis factor alpha has a protective role in a murine model of systemic candidiasis. Infect Immun 62 2761 2772
42. KáposztaRTreePMaródiLGordonS 1998 Characteristics of invasive candidiasis in gamma interferon - and interleukin-4-deficient mice: role of macrophages in host defense against Candida albicans. Infect Immun 66 1708 1717
43. LavigneLMSchopfLRChungCLMaylorRSypekJP 1998 The role of recombinant murine IL-12 and IFN-gamma in the pathogenesis of a murine systemic Candida albicans infection. J Immunol 160 284 292
44. d'OstianiCFDel SeroGBacciAMontagnoliCSprecaA 2000 Dendritic cells discriminate between yeasts and hyphae of the fungus Candida albicans. Implications for initiation of T helper cell immunity in vitro and in vivo. J Exp Med 191 1661 1674
45. MencacciADel SeroGCenciEd'Ostiani CFBacciA 1998 Endogenous interleukin 4 is required for development of protective CD4+ T helper type 1 cell responses to Candida albicans. J Exp Med 187 307 317
46. SahaBNandiD 2009 Farnesyl transferase inhibitors reduce ras activation and ameliorate acetaminophen-induced liver injury in mice. Hepatology 50 1547 1557
47. SahaBPrasannaSJChandrasekarBNandiD 2010 Gene modulation and immunoregulatory roles of Interferon gamma. Cytokine 50 1 14
48. AllardSTMBeisKGiraudM-FHegemanADGroMssJW 2002 Toward a structural understanding of the dehydratase mechanism. Structure 10 81 92
49. Moreno-RuizEOrtuGde GrootPWJCottierFLoussertC 2009 The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiol 155 2004 2020
50. RichardMIbata-OmbettaSDromerFBordon-PallierFJouaultT 2002 Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol Microbiol 44 841 853
51. NobleSMFrenchSKohnLAChenVJohnsonAD 2010 Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genetics 42 590 598
52. SlotJCRokasA 2010 Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc Natl Acad Sci USA 107 10136 10141
53. WaltherAWendlandJ 2003 An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet 42 339 343
54. ShermanFFinkGRHicksJ 1986 Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY Cold Spring Harbor Laboratory
55. ReuBOVikAKolterRMorschhauserJ 2004 The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341 119 127
56. VijayaraghavanUCompanyMAbelsonJ 1989 Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev 3 1206 1216
57. SamaranayakeYHYeJYauJYCheungBPSamaranayakeLP 2005 In vitro method to study antifungal perfusion in Candida biofilms. J Clin Microbiol 43 818 825
58. GillumAMTsayEYKirschDR 1984 Isolation of the Candida albicans gene for orotidine-5-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198 179 182
59. FonziWAIrwinMY 1993 Isogenic Strain Construction and Gene Mapping in Candida albicans. Genetics 134 717 728
60. MehtaDVKabirABhatPJ 1999 Expression of human inositol monophosphatase suppresses galactose toxicity in Saccharomyces cerevisiae: possible implications in galactosemia. Biochim Biophys Acta 1454 217 226
61. MumbergDMullerRFunkM 1995 Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156 119 22
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Microbial Spy Games and Host Response: Roles of a Small Molecule in Communication with Other SpeciesČlánek Sequence-Based Analysis Uncovers an Abundance of Non-Coding RNA in the Total Transcriptome ofČlánek The Splicing Factor Proline-Glutamine Rich (SFPQ/PSF) Is Involved in Influenza Virus Transcription
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 11- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Microbial Spy Games and Host Response: Roles of a Small Molecule in Communication with Other Species
- Simple Rapid Near-Patient Diagnostics for Tuberculosis Remain Elusive—Is a “Treat-to-Test” Strategy More Realistic?
- Ultra-Efficient PrP Amplification Highlights Potentialities and Pitfalls of PMCA Technology
- Assessing Predicted HIV-1 Replicative Capacity in a Clinical Setting
- Inhibition of IL-10 Production by Maternal Antibodies against Group B Streptococcus GAPDH Confers Immunity to Offspring by Favoring Neutrophil Recruitment
- Anti-filarial Activity of Antibiotic Therapy Is Due to Extensive Apoptosis after Depletion from Filarial Nematodes
- West Nile Virus Experimental Evolution and the Trade-off Hypothesis
- Shedding Light on the Elusive Role of Endothelial Cells in Cytomegalovirus Dissemination
- Galactosaminogalactan, a New Immunosuppressive Polysaccharide of
- Fatal Prion Disease in a Mouse Model of Genetic E200K Creutzfeldt-Jakob Disease
- BST2/Tetherin Enhances Entry of Human Cytomegalovirus
- Metagenomic Analysis of Fever, Thrombocytopenia and Leukopenia Syndrome (FTLS) in Henan Province, China: Discovery of a New Bunyavirus
- Neurons are MHC Class I-Dependent Targets for CD8 T Cells upon Neurotropic Viral Infection
- Sap Transporter Mediated Import and Subsequent Degradation of Antimicrobial Peptides in
- A Molecular Mechanism for Bacterial Susceptibility to Zinc
- Genomic Transition to Pathogenicity in Chytrid Fungi
- Evolution of Multidrug Resistance during Infection Involves Mutation of the Essential Two Component Regulator WalKR
- ChemR23 Dampens Lung Inflammation and Enhances Anti-viral Immunity in a Mouse Model of Acute Viral Pneumonia
- SH3 Domain-Mediated Recruitment of Host Cell Amphiphysins by Alphavirus nsP3 Promotes Viral RNA Replication
- A Gammaherpesvirus Cooperates with Interferon-alpha/beta-Induced IRF2 to Halt Viral Replication, Control Reactivation, and Minimize Host Lethality
- Early Secreted Antigen ESAT-6 of Promotes Protective T Helper 17 Cell Responses in a Toll-Like Receptor-2-dependent Manner
- CD4 T Cell Immunity Is Critical for the Control of Simian Varicella Virus Infection in a Nonhuman Primate Model of VZV Infection
- The Role of the P2X Receptor in Infectious Diseases
- Down-Regulation of Shadoo in Prion Infections Traces a Pre-Clinical Event Inversely Related to PrP Accumulation
- Cross-Reactive T Cells Are Involved in Rapid Clearance of 2009 Pandemic H1N1 Influenza Virus in Nonhuman Primates
- Single Molecule Analysis of Replicated DNA Reveals the Usage of Multiple KSHV Genome Regions for Latent Replication
- The Critical Role of Notch Ligand Delta-like 1 in the Pathogenesis of Influenza A Virus (H1N1) Infection
- Sequence-Based Analysis Uncovers an Abundance of Non-Coding RNA in the Total Transcriptome of
- Murine Gamma Herpesvirus 68 Hijacks MAVS and IKKβ to Abrogate NFκB Activation and Antiviral Cytokine Production
- EBV Tegument Protein BNRF1 Disrupts DAXX-ATRX to Activate Viral Early Gene Transcription
- SAG101 Forms a Ternary Complex with EDS1 and PAD4 and Is Required for Resistance Signaling against Turnip Crinkle Virus
- Multiple Candidate Effectors from the Oomycete Pathogen Suppress Host Plant Immunity
- Rab7A Is Required for Efficient Production of Infectious HIV-1
- A TNF-Regulated Recombinatorial Macrophage Immune Receptor Implicated in Granuloma Formation in Tuberculosis
- Novel Anti-bacterial Activities of β-defensin 1 in Human Platelets: Suppression of Pathogen Growth and Signaling of Neutrophil Extracellular Trap Formation
- CD11b, Ly6G Cells Produce Type I Interferon and Exhibit Tissue Protective Properties Following Peripheral Virus Infection
- The Splicing Factor Proline-Glutamine Rich (SFPQ/PSF) Is Involved in Influenza Virus Transcription
- A Kinase Chaperones Hepatitis B Virus Capsid Assembly and Captures Capsid Dynamics
- Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Pathovar
- Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler
- The Pore-Forming Toxin Listeriolysin O Mediates a Novel Entry Pathway of into Human Hepatocytes
- The Human Herpesvirus-7 (HHV-7) U21 Immunoevasin Subverts NK-Mediated Cytoxicity through Modulation of MICA and MICB
- Avirulence Effector Avr3b is a Secreted NADH and ADP-ribose Pyrophosphorylase that Modulates Plant Immunity
- Murid Herpesvirus-4 Exploits Dendritic Cells to Infect B Cells
- Unique Type I Interferon Responses Determine the Functional Fate of Migratory Lung Dendritic Cells during Influenza Virus Infection
- Evolution of a Species-Specific Determinant within Human CRM1 that Regulates the Post-transcriptional Phases of HIV-1 Replication
- Transcriptome Analysis of Transgenic Mosquitoes with Altered Immunity
- Antibody Evasion by a Gammaherpesvirus O-Glycan Shield
- UDP-glucose 4, 6-dehydratase Activity Plays an Important Role in Maintaining Cell Wall Integrity and Virulence of
- Protease-Resistant Prions Selectively Decrease Shadoo Protein
- A LysM and SH3-Domain Containing Region of the p60 Protein Stimulates Accessory Cells to Promote Activation of Host NK Cells
- Deletion of AIF Ortholog Promotes Chromosome Aneuploidy and Fluconazole-Resistance in a Metacaspase-Independent Manner
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Multiple Candidate Effectors from the Oomycete Pathogen Suppress Host Plant Immunity
- The Splicing Factor Proline-Glutamine Rich (SFPQ/PSF) Is Involved in Influenza Virus Transcription
- A TNF-Regulated Recombinatorial Macrophage Immune Receptor Implicated in Granuloma Formation in Tuberculosis
- SH3 Domain-Mediated Recruitment of Host Cell Amphiphysins by Alphavirus nsP3 Promotes Viral RNA Replication
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy