-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
Bdellovibrio bacteriovorus is a Delta-proteobacterium that oscillates between free-living growth and predation on Gram-negative bacteria including important pathogens of man, animals and plants. After entering the prey periplasm, killing the prey and replicating inside the prey bdelloplast, several motile B. bacteriovorus progeny cells emerge. The B. bacteriovorus HD100 genome encodes numerous proteins predicted to be involved in signalling via the secondary messenger cyclic di-GMP (c-di-GMP), which is known to affect bacterial lifestyle choices. We investigated the role of c-di-GMP signalling in B. bacteriovorus, focussing on the five GGDEF domain proteins that are predicted to function as diguanylyl cyclases initiating c-di-GMP signalling cascades. Inactivation of individual GGDEF domain genes resulted in remarkably distinct phenotypes. Deletion of dgcB (Bd0742) resulted in a predation impaired, obligately axenic mutant, while deletion of dgcC (Bd1434) resulted in the opposite, obligately predatory mutant. Deletion of dgcA (Bd0367) abolished gliding motility, producing bacteria capable of predatory invasion but unable to leave the exhausted prey. Complementation was achieved with wild type dgc genes, but not with GGAAF versions. Deletion of cdgA (Bd3125) substantially slowed predation; this was restored by wild type complementation. Deletion of dgcD (Bd3766) had no observable phenotype. In vitro assays showed that DgcA, DgcB, and DgcC were diguanylyl cyclases. CdgA lacks enzymatic activity but functions as a c-di-GMP receptor apparently in the DgcB pathway. Activity of DgcD was not detected. Deletion of DgcA strongly decreased the extractable c-di-GMP content of axenic Bdellovibrio cells. We show that c-di-GMP signalling pathways are essential for both the free-living and predatory lifestyles of B. bacteriovorus and that obligately predatory dgcC- can be made lacking a propensity to survive without predation of bacterial pathogens and thus possibly useful in anti-pathogen applications. In contrast to many studies in other bacteria, Bdellovibrio shows specificity and lack of overlap in c-di-GMP signalling pathways.
Published in the journal: Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in. PLoS Pathog 8(2): e32767. doi:10.1371/journal.ppat.1002493
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002493Summary
Bdellovibrio bacteriovorus is a Delta-proteobacterium that oscillates between free-living growth and predation on Gram-negative bacteria including important pathogens of man, animals and plants. After entering the prey periplasm, killing the prey and replicating inside the prey bdelloplast, several motile B. bacteriovorus progeny cells emerge. The B. bacteriovorus HD100 genome encodes numerous proteins predicted to be involved in signalling via the secondary messenger cyclic di-GMP (c-di-GMP), which is known to affect bacterial lifestyle choices. We investigated the role of c-di-GMP signalling in B. bacteriovorus, focussing on the five GGDEF domain proteins that are predicted to function as diguanylyl cyclases initiating c-di-GMP signalling cascades. Inactivation of individual GGDEF domain genes resulted in remarkably distinct phenotypes. Deletion of dgcB (Bd0742) resulted in a predation impaired, obligately axenic mutant, while deletion of dgcC (Bd1434) resulted in the opposite, obligately predatory mutant. Deletion of dgcA (Bd0367) abolished gliding motility, producing bacteria capable of predatory invasion but unable to leave the exhausted prey. Complementation was achieved with wild type dgc genes, but not with GGAAF versions. Deletion of cdgA (Bd3125) substantially slowed predation; this was restored by wild type complementation. Deletion of dgcD (Bd3766) had no observable phenotype. In vitro assays showed that DgcA, DgcB, and DgcC were diguanylyl cyclases. CdgA lacks enzymatic activity but functions as a c-di-GMP receptor apparently in the DgcB pathway. Activity of DgcD was not detected. Deletion of DgcA strongly decreased the extractable c-di-GMP content of axenic Bdellovibrio cells. We show that c-di-GMP signalling pathways are essential for both the free-living and predatory lifestyles of B. bacteriovorus and that obligately predatory dgcC- can be made lacking a propensity to survive without predation of bacterial pathogens and thus possibly useful in anti-pathogen applications. In contrast to many studies in other bacteria, Bdellovibrio shows specificity and lack of overlap in c-di-GMP signalling pathways.
Introduction
Predatory, “attack-phase” cells of Bdellovibrio bacteriovorus HD100 use flagellar motility in liquid environments; and gliding motility on solid surfaces, to encounter other Gram-negative bacteria [1], [2]. Prey bacteria include a wide range of pathogens of man, animals and plants, [3], [4] thus Bdellovibrio can be seen as “pathogens of pathogens” [5]. B. bacteriovorus attach to these prey and enter their periplasms, by mechanisms that remain to be fully understood. Once inside prey, B. bacteriovorus become non-motile and degrade prey macromolecules, using them for their own growth and replication (Figure S1 in Text S1). Growth occurs from both poles, giving rise to odd and even numbered progeny by synchronous septation of the elongated multi-nucleoid B. bacteriovorus filament, inside the infected, spherical, prey cell which is now called a “bdelloplast” [6]. At the end of predatory growth and septation, B. bacteriovorus induce motility once more, and use flagellar motility to emerge from prey in liquid media, or gliding motility to emerge from prey on solid surfaces, and move off, in a non-replicative, “attack phase” to seek more prey encounters. Cultures of B. bacteriovorus growing in this predatory or prey/host-dependent (HD) manner require entry to another prey cell to replicate as the “attack-phase” cells have replication suppressed (by as yet unknown control mechanisms) and do not grow using organic nutrients from the external media [7]. Attack phase cells are vibroid and 0.25 µm by 1.25 µm with a single polar sheathed flagellum, they attach to prey at the non-flagellar pole [8].
In the laboratory a small fraction: 1×10−7, of attack phase B. bacteriovorus populations can also be cultured axenically without prey, as so-called HI (host-independent) cultures on peptone-rich artificial media; here they grow and divide as if inside prey cells (Figure S1 in Text S1) [9]. Natural point mutations in the Bd0108 Type IVB pilus-like gene product are reported to account for the small fraction of predatory cells that can adopt this “wild type” HI phenotype [10]. Wild-type HI B. bacteriovorus HD100 convert readily back to HD cultures, via predatory invasion, if presented with prey cells in the absence of rich media. HI cells of B. bacteriovorus are typically longer than the attack phase HD cells, being 2–10 µm long but 0.25–0.3 µm in width; longer cells usually have a serpentine morphology and one or more flagella in polar or other cell sites.
Cyclic di-GMP is a bacterial second messenger that controls various processes. In the Proteobacteria, c-di-GMP controls a lifestyle transition between single, usually motile, cells and surface-attached multicellular communities (biofilms) [11], [12]. It also contributes to the switch between environmental and pathogenic lifestyles in other organisms [13]. C-di-GMP is synthesized by diguanylyl cyclases containing GGDEF domains, and degraded by phosphodiesterases containing EAL or HD-GYP domains. C-di-GMP acts via a variety of receptors (effector proteins) [14]. Among the most common receptors are PilZ domain proteins and a sub-group of GGDEF domain proteins containing so-called I-sites [15]. These sites are present in many diguanylyl cyclases where they function in feedback inhibition. However, a sub-group of the GGDEF domain proteins, which are enzymatically inactive, have evolved that bind c-di-GMP via the I-sites and function as c-di-GMP receptors.
B. bacteriovorus HD100 is predicted to have a high c-di-GMP “intelligence” because its 3.8Mb genome contains as many as 15 PilZ domain proteins representing putative c-di-GMP receptors [16], [17], [18]. It is peculiar that the number of predicted diguanylyl cyclases and phosphodiesterases is relatively low, i.e. 5 GGDEF, 1 EAL and 6 HD-GYP domain proteins, which facilitates analysis of c-di-GMP signalling cascades as genes encoding enzymes for c-di-GMP synthesis and degradation can be deleted and the resultant phenotypes tested.
In this study, we carried out a deletion analysis of genes encoding 5 GGDEF domain proteins that may initiate c-di-GMP signalling cascades. Unexpectedly, this analysis produced a discretely different phenotype for each GGDEF domain gene deletion strain. One GGDEF knockout strain was rendered obligately predatory, a phenotype that is very desirable for the future application of Bdellovibrio as anti-infective agents to kill pathogenic bacteria [5]. We discovered that individual c-di-GMP signalling pathways control each of the axenic and predatory lifestyles and B. bacteriovorus gliding and flagellar motility. The extreme degree of specificity and lack of the overlap among the signalling pathways involving a small diffusible molecule has not been previously observed in bacteria, or believed to be possible. The small (0.25 µm×1.25 µm) B. bacteriovorus cell size, which might have been thought to facilitate rapid c-di-GMP equilibration, did not cause nonspecific cross-activation of c-di-GMP cascades. While the mechanisms involving pathway separation are yet to be uncovered, unique cellular localizations of some diguanylyl cyclases and c-di-GMP receptors observed here may contribute to the lack of pathway cross-reactivity. The pathways include one degenerate GGDEF domain protein, with a GVNEF motif, which we found to be a c di-GMP receptor required for the rapid entry of B. bacteriovorus into prey cells. Thus c di-GMP signalling has evolved to control the predatory ability of this naturally invasive killer of pathogenic bacteria.
Results/Discussion
Bioinformatic analysis of the B. bacteriovorus GGDEF domain proteins
We analyzed the sequences of the 5 GGDEF domain proteins encoded in the B. bacteriovorus genome using Pfam (Figure S2 in Text S1) The sequences of GGDEF domains of four proteins were very similar to the consensus (Pfam database [19]) and did not contain residues predicted to interfere with the diguanylyl cyclase activity [20]. This suggested that these four proteins are likely enzymatically competent. We designated the B. bacteriovorus HD100 locus tags for these proteins [21] as encoding DgcA (Bd0367), DgcB (Bd0742), DgcC (Bd1434) and DgcD (Bd3766), where Dgc stands for diguanylyl cyclase. A genome sequence comparison revealed that DgcA, DgcB and DgcC are conserved between different Bdellovibrio strains, whereas gene Bd3766, encoding DgcD, is not conserved in the genome of another B. bacteriovorus strain tiberius (Hobley et al., in preparation).
Pfam predicts that [19] DgcA has an N-terminal receiver domain typical of bacterial response regulators (Figure S2 in Text S1), which implies that its activity is regulated by phosphorylation by an unknown histidine kinase. DgcB has an N-terminal forkhead domain, which may be involved in binding of a protein containing a phosphorylated serine or threonine residue [22]. DgcC contains a large N-terminal domain of unknown function. DgcD has a predicted periplasmic N-terminal domain containing 4 tetratricopeptide repeats, TPR domains, flanked by the transmembrane domains.
The GGDEF domain of the fifth protein, Bd3125, was clearly degenerate. Among others, it had two substitutions at key residues in the most conserved GGDEF (Gly-Gly-Asp-Glu-Phe) motif, the catalytic half site required for substrate, GTP, binding, i.e., GGDEF→GVNEF. Thus protein sequence analysis suggested that Bd3125 is not enzymatically competent. Each of the five proteins, including Bd3125, contains an RxxD motif 5 residues upstream of the GGDEF motif, which is a typical sequence and position for the I-site. For the four Dgc proteins, this site likely represents a site for feedback inhibition. Given the likely lack of diguanylyl cyclase activity of Bd3125 but the presence of the I-site, we predicted that it may function as a c-di-GMP receptor (effector) protein, and as such designated it CdgA.
Biochemical characterization of the B. bacteriovorus GGDEF domain proteins
To characterize activities of the GGDEF domain proteins, we first expressed them as fusions to the maltose-binding protein, MBP, in the highly motile strain, E. coli MG1655. For DgcD that contains a large transmembrane domain, the fusion was made directly to the GGDEF domain. Ko and co-workers, followed by Ryjenkov and co-workers have shown that elevated c-di-GMP levels inhibit E. coli motility in swim agar plates [23], [24]. Each MBP-fusion was tested for the effect it had on E. coli motility in swim agar plates. As expected, DgcA, DgcB and DgcC strongly inhibited E. coli swimming (Figure S3 in Text S1), consistent with their predicted diguanylyl cyclase activities. However, the MBP-fusion to the GGDEF domain of DgcD did not inhibit swimming, either because its GGDEF domain is enzymatically inactive, or, more likely, because it could not be activated in E. coli. Because the B. bacteriovorus dgcD knockout produced no observable phenotype, biochemical characterization of DgcD was not pursued. Expression of CdgA produced a significantly larger swim zone than that of the empty vector (Figure S3 in Text S1). Since CdgA cannot possibly act as a c-di-GMP degrading enzyme, we predict that it likely binds c-di-GMP and acts as a c-di-GMP sink, thus effectively decreasing the pool of available c-di-GMP in E. coli.
We purified the MBP-fusions to DgcA, DgcB, DgcC and CdgA and tested diguanylyl cyclase activities and c-di-GMP binding in vitro. As expected, DgcA, DgcB and DgcC proved to function as diguanylyl cyclases (Figure 1A–C). Because each of these proteins contains an N-terminal regulatory domain whose activation cannot be ensured in E. coli, it is likely that the observed activities of the purified fusions lie between their respective active and inactive states.
Fig. 1. Enzymatic activity of the MBP-fusion proteins. 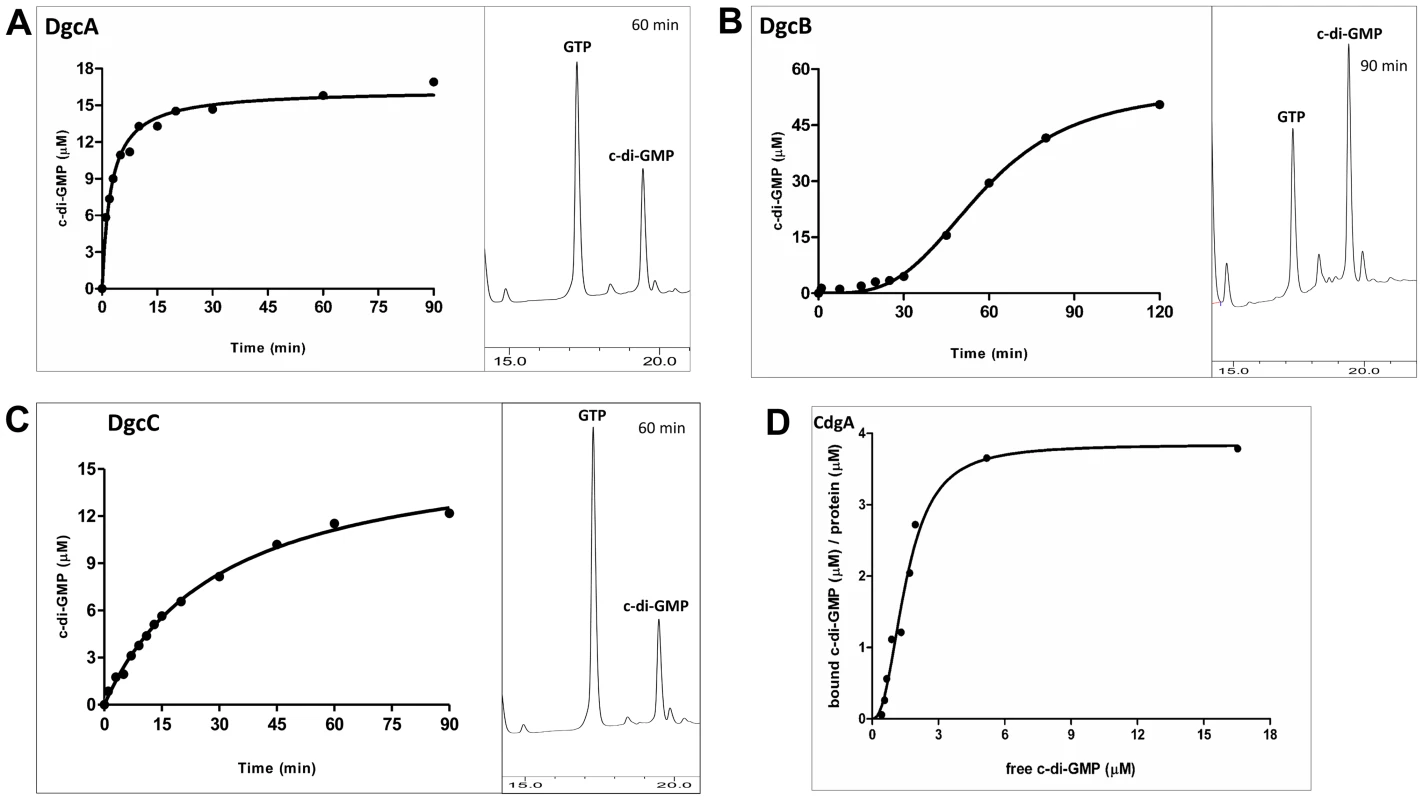
Enzymatic activity of the MBP-fusion to A: DgcA (Bd0367); B: DgcB (Bd0742); C: DgcC (Bd1434); the HPLC profiles show peaks of c-di-GMP (product) and GTP (substrate) at a single time point. D: Saturation plot of equilibrium binding between c-di-GMP and CdgA (Bd3125). Protein-nucleotide binding was examined by equilibrium dialysis in Dispo-Biodialyzer cassettes as described earlier [24]. It appears that CdgA binds 4 molecules of c-di-GMP, which is possible if CdgA were to form tetramers in the presence of c-di-GMP, similar to those described for c-di-GMP-inhibited state of diguanylate cyclases [50]. Among the three cyclases, DgcB is most sensitive to feedback inhibition by c-di-GMP (Figure 1B). DgcB purified from E. coli contained c-di-GMP at approximately 1∶1 protein:c-di-GMP molar ratio. No diguanylyl cyclase activity was observed following addition of the substrate, GTP, to the purified protein. Only after extended (approximately 30 minutes) incubation at 37°C, could the DgcB activity be detected, probably due to unfolding of the c-di-GMP-inhibited DgcB conformation and spontaneous formation of the enzymatically competent DgcB dimer [25].
The CdgA protein containing a degenerate GGDEF domain showed no diguanylyl cyclase activity, which is consistent with the sequence analysis and motility assays in E. coli (Figure S3 in Text S1). However, this protein bound c-di-GMP in vitro with an apparent Kd ∼2 µM (Figure 1D), as measured by equilibrium dialysis [26]. This value is well within the range of intracellular c-di-GMP concentrations observed in bacteria [12]. Therefore, CdgA is a c-di-GMP receptor (effector protein), which, according to the genetic analysis shown below, acts downstream of the diguanylyl cyclase DgcB.
DgcA signals control the motile escape of Bdellovibrio from prey after their digestion, controlling both swarming and swimming motility
An in-frame deletion of dgcA was achieved by conjugation of a suicide vector pK18mobsacB into B. bacteriovorus HD100 and subsequent sucrose suicide screening for gene replacement and loss of the plasmid (see materials and methods and Text S1; [2], [27]). The ΔdgcA mutant strain could not grow predatorily and did not form plaques (Figure 2Ai) on lawns of prey bacteria, thus it was isolated solely by axenic culturing on peptone-rich media. The ΔdgcA mutant was seen by phase contrast microscopy to be non-motile in liquid media.
Fig. 2. Predatory and axenic growth on agar plates. 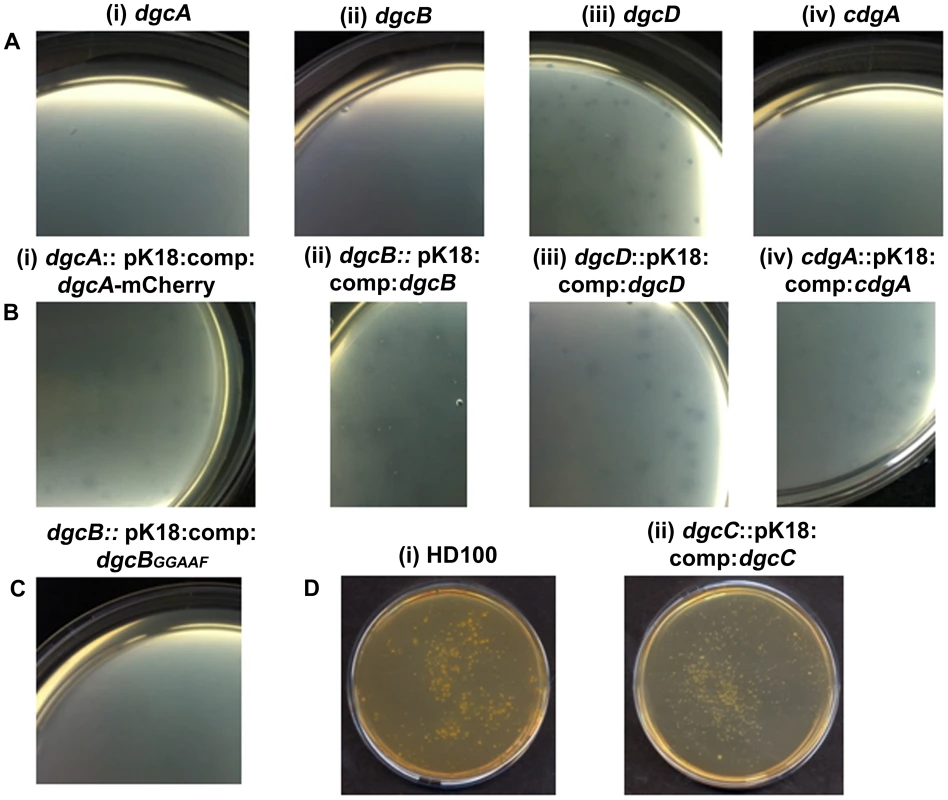
A: Bdellovibrio deletion strains plated onto lawns of prey (i) ΔdgcA (ii) ΔdgcB (iii) ΔdgcD and (iv) ΔcdgA. The ΔdgcD strain forms plaques in the prey lawns. B: Complementation by single crossover of wild-type and mCherry fusion proteins restores plaquing ability to each deletion mutant: (i) dgcA::pK18:comp:dgcA-mCherry; (ii) dgcB::pK18:comp:dgcB; (iii) dgcD::pK18:comp:dgcD; and (iv) cdgA::pK18:comp:cdgA. C: Plaquing is not restored to the ΔdgcB mutant when a GGAAF mutation is introduced into the complementation plasmid. D: Axenic growth by (i) wild-type Bdellovibrio; and (ii) the complemented ΔdgcC strain. Electron microscopy (Figure 3AI, ii) showed that although a membranous flagellar sheath was made, no functional flagellum was assembled within it. This appearance was reminiscent of a non-motile fliC3 flagellin mutant previously studied in Bdellovibrio [1], [28]. Studies with that fliC3 mutant had shown it would enter prey cells if applied directly onto them on a solid surface, and could use gliding motility to exit the bdelloplast after septation and lysis [1], [2]. Therefore cells of the ΔdgcA mutant were applied to E. coli prey (that were constitutively expressing mCherry) on a 1% agarose surface, the prey were invaded and the ΔdgcA mutant replicated and septated within them. However, after septation no induction of (flagellar or) gliding motility was seen in the ΔdgcA strain and the septated Bdellovibrio remained motionless inside the dead “shell” of the prey bdelloplast long after (as long as 120 minutes was measured without any change being observed) the prey mCherry fluorescence was dissipated by the lysis of the bdelloplast (Figure 4). In contrast, wild-type Bdellovibrio lyse bdelloplasts and immediately (after 10 minutes), emerge, using gliding motility if on an agarose surface. Thus the deletion of the dgcA gene rendered the cells effectively unable to be predatory, as they could not escape the prey remains to search for new prey. A DgcA-mCherry strain was seen to be fluorescent throughout the cell at all times through the predatory cycle (Figure 5Ai), and during Host-Independent growth (Figure 5Aii), not only at times when the cells would be motile (by either flagellar - or gliding - motility).
Fig. 3. Cell morphology of dgc and cdg deletion strains revealed by electron microscopy. 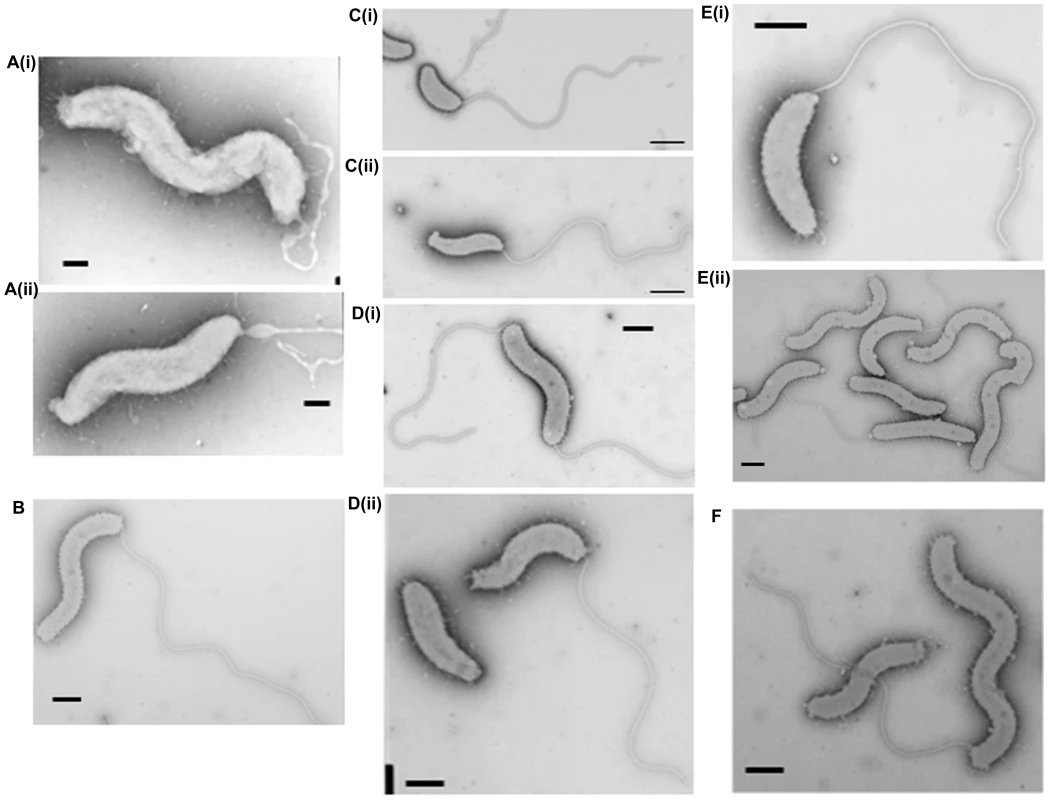
A(i) and (ii): HI ΔdgcA cells showed no functional flagella filaments, only empty membranous flagellar sheath. B: HI ΔdgcB cells showing typical cell morphologies. C: (i) ΔdgcC HD cells and (ii) typical wild-type HD cells, showing the cell size differences of the ΔdgcC mutant. D: (i) rare HI ΔdgcCsup populations have a higher than normal percentage (10%) of bipolarly flagellate cells, (ii) a wild-type Bdellovibrio strain HID13 has a mix of cells with no or one polar flagellum. E: (i) HD ΔdgcD cells and (ii) HI ΔdgcD cells, showing wild-type cell morphology and typical sheathed flagella. Some membrane blebbing was visible on the HI ΔdgcD cells but not in significant excess to that seen on other wild-type HI cells. F: the ΔcdgA HI cells have typical HI cell morphologies and typical levels of flagellated cells. All cells were stained with 1% PTA (pH 7.0), scale bars = 0.5 µm. Fig. 4. Predatory escape defect of the ΔdgcA cells from fluorescent E. coli prey. 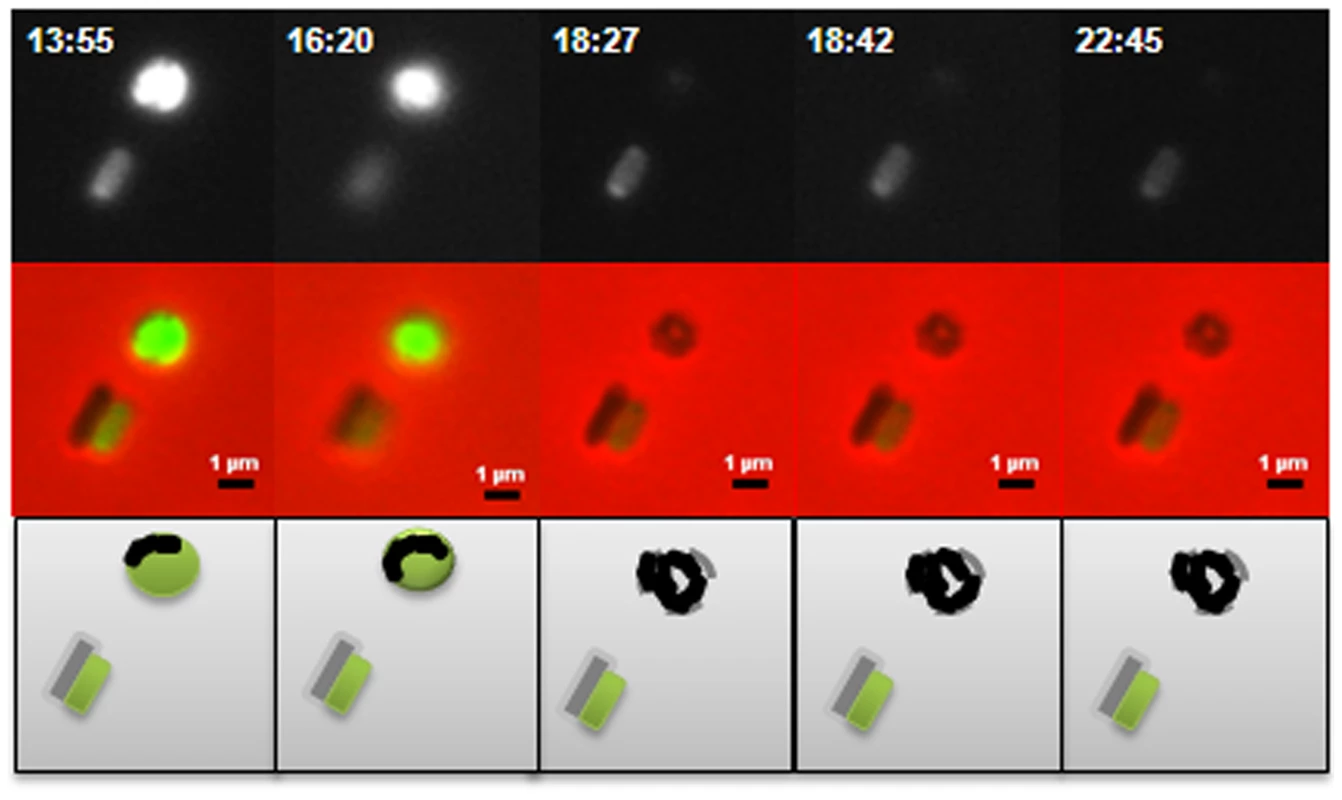
The ΔdgcA strain enters prey (not shown) grows inside the bdelloplast, septates and lyses the prey outer membrane, releasing the fluorescence of the prey cell, but does not escape the remnants of the prey, remaining there for over 4 hours, compared to 10 minutes for wild type; scale bars = 1 µm. Fig. 5. Location of fluorescently tagged Dgc and Cdg proteins in wild-type predatory and axenically growing cells. 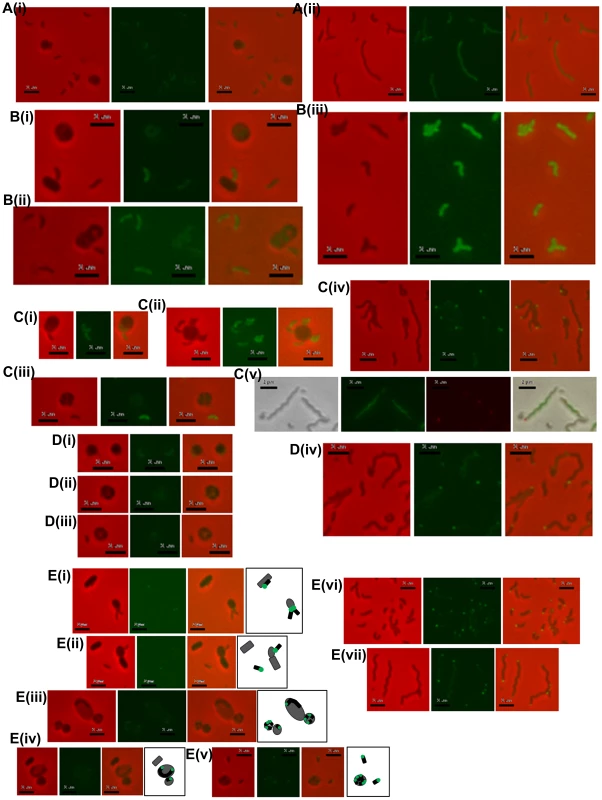
A: (i) HD wild-type Bdellovibrio expressing DgcA-mCherry, showing constitutive low level fluorescence in cells that have invaded prey or which are free swimming; (ii) HI axenic Bdellovibrio expressing DgcA-mCherry, again fluorescence is distributed throughout the cells. B: (i and ii) HD wild-type Bdellovibrio expressing DgcB-mCherry showing constitutive fluorescence throughout the cell at all points in the predatory lifecycle, in Bdellovibrio that have invaded prey or which are free swimming; (iii) HI axenic wild-type Bdellovibrio expressing DgcB-mCherry, again fluorescence is seen throughout the cells. C: HD wild-type Bdellovibrio expressing DgcC-mCherry showing constitutive fluorescence throughout both the (i) attack-phase cell and (ii) early in predation, but a dissipation of DgcC fluorescence (iii) later in predation and at septation inside prey; (iv) HI axenic wild-type Bdellovibrio expressing DgcC-mCherry, most cells contain some bright foci, typically at the poles of the cell, but some in the centre; (v) Dapi-stained HI cells with DgcC-mCherry, green fluorescence is the Dapi-stained DNA, the red foci are the DgcC-mCherry. DgcC-mCherry often forms mid-cell foci in areas without chromosomal DNA, suggestive of location at the potential septal points. D: (i, ii and iii) HD wild-type Bdellovibrio expressing DgcD-mCherry showing very weak fluorescence; (iv) HI axenic Bdellovibrio expressing DgcD-mCherry, faint fluorescence throughout the cells with some bright foci. E: (i – v) Fluorescent localization of CdgA during the predatory cycle with illustrative diagrams. During intracellular growth and division the CdgA remains at the poles (i, ii and iii) early in the predatory cycle, after prey invasion, the fluorescence is seen only at one pole, (iv) later, when the Bdellovibrio is growing (inside prey) from both poles, CdgA is seen at both poles (v) at septation each progeny CdgA-mCherry cell has both a fluorescent and a non-fluorescent pole; (vi and vii) HI cells containing CdgA-mCherry also show polar localization, (vi) smaller cells have a single fluorescent pole whilst (vii) longer cells (part of the natural variation in lengths of HI grown Bdellovibrio) typically have fluorescence at both poles, both of which may be active for prey-interaction (Figure S6 in Text S1). All scale bars = 2 µm. Complementation of the ΔdgcA mutant with a full length but C-terminally mCherry-tagged wild type dgcA gene was achieved by single recombination into the chromosome from suicide plasmid pK18mobsacB. Exconjugant Bdellovibrio from this complementation regained the ability to form plaques on E. coli prey lawns (Figure 2Bi) and typically formed 2×03 plaques per conjugation, compared to zero for the parental ΔdgcA mutant strain (Figure 2Ai). Gliding was restored to the ΔdgcA mutant by this complementation (Figure S4 in Text S1) but interestingly flagellar motility was not. However, we have previously shown that flagellar motility is not required for predation [1]. We were able to observe, by video microscopy, gliding cells of the complemented strain having lysed and escaped from the prey bdelloplasts. Unfortunately the full-length dgcA gene without mCherry tag proved difficult to isolate (possibly due to c-di-GMP effects in cloning strains of E. coli) so the phenotype of the complemented ΔdgcA strain was only discernable from the mCherry tagged dgcA gene. We acknowledge that the presence of the mCherry tag may interfere with protein-protein interactions that could be required for full complementation, so we cannot test the flagellar effect. However as gliding motility and predatory growth were restored by this dgcA-mcherry complementation, we conclude that the DgcA protein controls gliding motility and thus successful predatory growth on prey bacteria.
DgcB controls processes essential for Bdellovibrio entry into prey
Inactivation of dgcB gave a mutant strain which did not form plaques (Figure 2Aii) and which could only be isolated by HI axenic growth (Figure 3B) but which retained expression of wild-type flagella. Some 50% of axenically grown wild type HI Bdellovibrio have a flagellum but only 5% of those HI cells swim (in contrast to the 98% motility of predatory Bdellovibrio cultures). For the dgcB mutant HI cells 50% had flagella and 10–15% of the cells were motile, more than for the wild type HI strains. When 21 cultures, consisting of seven replicates of three independent isolates, of this strain were challenged with prey bacteria in liquid culture, versus 3 cultures of two wild type (WT) B. bacteriovorus HI strain controls, predatory invasion and prey bdelloplast formation was seen in the WT B. bacteriovorus HI strains after 9 hours and the prey cultures began to clear by prey-lysis. At this point no bdelloplasts were seen for the ΔdgcB mutant strains. At 24 hours of incubation there were no prey remaining in the WT HI cultures but the ΔdgcB mutant cultures were full of prey bacteria. In 4 of the 21 test cultures of the ΔdgcB mutant strains prey killing and bdelloplast formation were seen after an additional 120 hours incubation. In the remaining 17 cultures however, no bdelloplasts were seen despite prolonged further incubation. The 4 presumed suppressor strains were tested by PCR for the presence of the original ΔdgcB mutation and this was confirmed.
Repeating this experiment on two different occasions, in a different experimental setting, luminescent prey bacteria [29] were incubated in 96 well Optiplates (Porvair Biosciences) in a BMG luminescent plate reader with 18 different cultures of two different isolates of the ΔdgcB mutant (including the isolate that had given presumed suppressor strains before), were incubated for 72 hours. The predator:prey ratio in these latter experiments should have shown clearly, even low-levels (5% of total) of prey-killing by a drop in prey luminescence. No such drop in luminescence was seen in any of the 18 isolates, (data not shown) showing that the phenotype of the ΔdgcB mutant is non-predatory, but that rare suppressor mutants may be isolated (these suppressor strains are currently the subject of further study).
A DgcB:mCherry fusion protein expressed in a wild type DgcB background was constitutively brightly fluorescent in the cytoplasm of B. bacteriovorus cells whether they were growing predatorily (Figure 5 Bi-ii) or axenically (Figure 5 Biii). As the DgcB protein contains a predicted forkhead domain at the N terminus, we conclude that it may be activated via this domain when its c-di-GMP synthetic properties are biologically appropriate. This activation may be in response to prey sensing.
Complementation of the ΔdgcB mutant with the wild type dgcB gene was achieved by single recombination into the chromosome from suicide plasmid pK18mobsacB. A single complementation conjugation gave rise to 1.7×104 plaques on prey lawns (Figure 2Bii) in stark contrast to the ΔdgcB mutant itself which did not form plaques (Figure 2Aii) and had to be cultured axenically as HI strains (and in which only very rare suppressor strains emerged after many days of prey challenge in liquid cultures of those axenically growing HI ΔdgcB mutants).
Further evidence that the diguanylyl cyclase activity of the DgcB itself was important for predation came from the conjugation of a GGAAF variant into the ΔdgcB mutant instead of the wild type conjugation. The GGAAF plasmid did not restore plaquing ability (Figure 2C) to the ΔdgcB strain. Thus we conclude that the diguanylyl cyclase activity of the DgcB protein controls, via c-di-GMP signalling, processes that are required for the invasion of prey bacteria.
DgcC is required for the conversion of B. bacteriovorus to axenic growth
Inactivation of the dgcC gene produced attack phase Bdellovibrio cells that although normally flagellate, were smaller, but wider (Figure 3Ci) than wild type (Figure 3C ii) (Figure 6). The dgcC mutant was still predatory, and invaded and killed E. coli prey cells at wild type rates. However, the normal 1 in 107 conversion of predatory Bdellovibrio to axenically growing HI Bdellovibrio [9] was lost in this mutant and 4.5×1011 cells had to be applied to nutrient media to isolate and culture a rare single colony (Figure 3Di), growing axenically. This suggested a secondary mutation was required to allow the ΔdgcC mutant to grow axenically. Bacteria from such suppressor strains were frequently (10%) biflagellate, but not markedly different (within the natural variation of HI Bdellovibrio from wild-type HI strains) as exemplified by the wild-type strain HID13 (Figure 3Dii).
Fig. 6. Cell size comparisons of wild-type, ΔdgcC, dgcC wild-type GGEEF-complemented and dgcCGGAAF “complemented” Bdellovibrio strains. 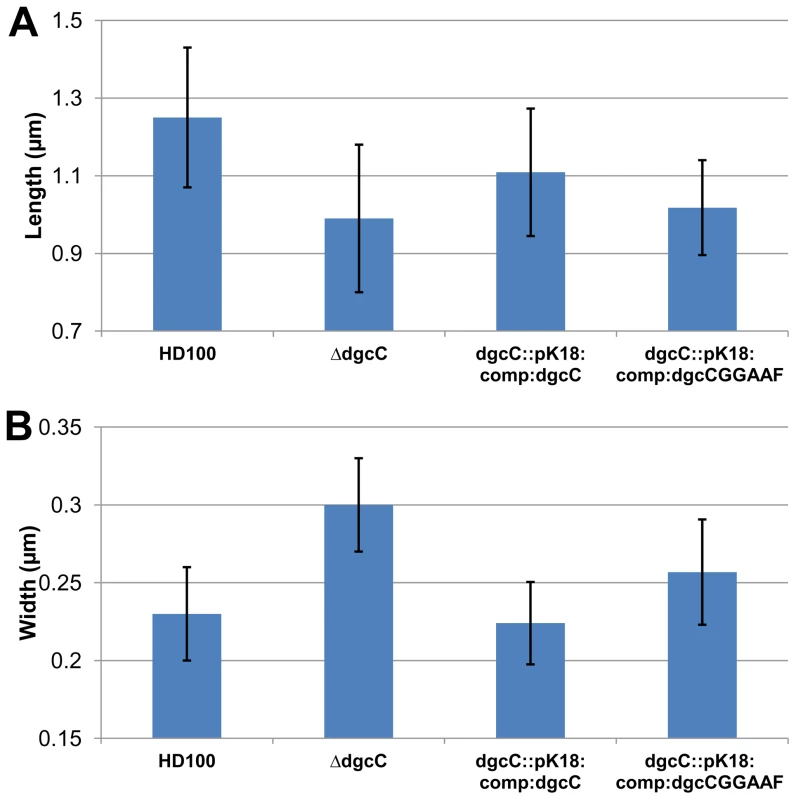
Histograms show the mean A: length and B: width measured from negatively strained electron microscope images of a minimum of 50 cells per strain. Error bars show one standard deviation above and below the mean. The lengths of each of the mutant and complemented strains was significantly shorter than wild-type (p<0.001) but for the widths, the strain complemented with the wildtype dgcC gene which was not significantly wider than wild type, but the mutant strain was significantly (p<0.001) wider; with the dgcCGGAAF complemented strain being of intermediate width. Fluorescent microscopy of DgcC-mCherry in B. bacteriovorus carrying a wild type copy of the dgcC gene, showed a cytoplasmic distribution in attack-phase B. bacteriovorus (Figure 5Ci) and only faint fluorescence when Bdellovibrio was elongating inside prey cells (Figure 5Cii), with fluorescence gone when the predatory cells were septating in the bdelloplast (Figure 5Ciii). Attack-phase cells of the fluorescent DgcC-mCherry strain (which also carried a wild type dgcC gene) readily converted (at wild type, 1 in 107 frequency), into axenically growing HI cultures. In these HI cells there were strong fluorescent foci of DgcC-mCherry expression at one pole of each cell (Figure 5Civ) and in 60% of dividing HI cells DgcC foci were seen associated with division points at non-Dapi-staining regions (Figure 5Cv). There were also fainter foci along the length of non-dividing longer HI cells (Figure 5Civ,v). There was no overall cytoplasmic distribution, in contrast to that of the DgcC-mCherry expression in predatory attack-phase cells (which do not divide outside of prey).
We propose that DgcC may be required to regulate predatory cell division and/or cytoskeletal regulation during predatory growth. This is because the ΔdgcC progeny produced were shorter and fatter than wild-type (Figure 3Ci, ii, Figure 6). Also it was noteworthy that DgcC-mCherry fluorescence was present when the Bdellovibrio were outside prey or in the early stages of invasion (Figure 5Ci,ii), but that it dissipated at septation (Figure 5Ciii)., suggesting DgcC turnover at septation. Wild-type attack-phase Bdellovibrio cannot replicate outside prey, due to an unknown control mechanism [7]. It is interesting, in support of this, that the conversion to axenic growth mode of Bdellovibrio was abolished in the DgcC mutant. Axenic and predatory division of Bdellovibrio does not have to use a completely overlapping set of cell division proteins, as the Bdellovibrio genome is large, and as predatory division is by synchronous septation of a long filament, yet in axenically growing Bdellovibrio budding events (of single cells from the tip of a long filament), binary fission events (of shorter serpentine HI cells) and other division patterns are seen.
One gene, Bd0108, at the hit locus has been reported by others as a hot spot for mutations which affect the ability of Bdellovibrio to convert from predatory to axenic growth [9], [10], [30]. Despite this the sequence of the Bd0108 gene in the ΔdgcC strain was found to be wild type, in addition the sequence of Bd0108 in the rare (1 in 4.5×1011) “HI suppressor of ΔdgcC” that grew was also wild type.
Complementation of the ΔdgcC mutant with the wild type dgcC gene was achieved by single recombination into the chromosome from suicide plasmid pK18mobsacB. Exconjugants from this complementation were predatory, as was the original ΔdgcC mutant strain, and the cells returned to the wild-type cell width dimension of 0.23 µm and almost returned to the wild-type length of 1.25 µm (Figure 6). However when a version of the dgcC gene, encoding GGAAF instead of wild-type GGEEF (to inactivate the catalytic site of the protein) was used in identical conjugation experiments, the exconjugant strains were not fully complemented in terms of cell size (Figure 6), showing intermediate size distributions of length and width.
The normal 1 in 107 conversion of wild type predatory Bdellovibrio to axenically growing HI Bdellovibrio (Figure 2Di) was restored to the ΔdgcC mutant by wild type dgcC complementation (Figure 2Dii) and colonies of axenically growing Bdellovibrio were readily isolated from nutrient media plates. Further experiments will determine whether the GGAAF encoding version of the dgcC plasmid restores the normal HI conversion rate.
DgcD protein has no obvious role in B. bacteriovorus behaviour
In contrast to the other GGDEF genes, an in-frame deletion of the dgcD gene did not cause an observable phenotype in the conditions that we measured and the mutant strains could be cultured in both HD (Figure 2Aiii) and HI growth modes without alteration in growth rates. Introduction of the wild type dgcD gene had no effects on growth (Figure 2Biii). They also retained wild type morphology and motility (Figure 3Ei, ii). Bright fluorescent foci of DgcD-mCherry were seen (Figure 5D iv) in wild type HI Bdellovibrio, but very weak constitutive fluorescence only, was seen in HD cells (Figure 5 D I,ii,iii). Sequencing of another B. bacteriovorus isolate in our laboratory (in preparation) has shown that while the other 4 GGDEF genes are conserved across strains, that dgcD is not. Due to the lack of a significant phenotype for the dgcD deletion and the lack of conservation among Bdellovibrio strains, we did not investigate the role of DgcD any further.
CdgA organizes processes at the Bdellovibrio “nose” that are crucial to rapid prey invasion
Exconjugants with an in-frame deletion of the cdgA gene were not predatory and did not form plaques on prey lawns (Figure 2A iv) and ΔcdgA Bdellovibrio could only be isolated in HI axenically grown plate cultures (Figure 3F). However, when the ΔcdgA mutant was offered to prey cells in mixed liquid cultures, prey invasion and bdelloplast formation did proceed, but slowly, i.e. 40–90 minutes for ΔcdgA versus 30–40 minutes for wild-type HD strains (Figure S5 in Text S1). This slow predation resulted in liquid cultures taking two days to clear the majority of prey cells as compared to around 16 hours for wild-type strains. This slow prey invasion, likely accounted for the failure of the ΔcdgA mutant to be efficiently cultured predatorily on plates of prey lawns when originally isolated. Furthermore, when the ΔcdgA mutant, that had been growing slowly predatorily in liquid cultures, was returned to prey lawns on agar plates, they again failed to form plaques. Complementation of the ΔcdgA mutant with the wild type cdgA gene was achieved by single recombination into the chromosome from suicide plasmid pK18mobsacB. Exconjugants from this complementation gave rise to >105 plaques per conjugation, counted on dilution plates of prey lawns (Figure 2B iv) in contrast to none from matched numbers of the original ΔcdgA strain (Figure 2A iv). Invasion of prey cells by the complemented strain, previously grown on prey, was found to occur after 30–40 minutes (a wild type speed) by timelapse microscopy (Figure S5 in Text S1).
Fluorescent protein localization of CdgA-mCherry, in a wild type CdgA-expressing B. bacteriovorus HD100, showed that it was expressed at the non-flagellar pole or prey-interacting “nose” of the predatory B. bacteriovorus cells and that this expression was seen when the Bdellovibrio were attached to the prey cell at the point of invasion (Figure 5E i, ii). After invasion a single fluorescent polar focus of CdgA-mCherry persisted whilst the Bdellovibrio began to elongate, initially from this single pole. Then a second CdgA-mCherry focus developed at the second pole - during the period in which the Bdellovibrio grows via bipolar elongation (Figure 5E iii, iv); [6]). Upon septation of the predatory Bdellovibrio, within the prey bdelloplast, a single bright CdgA-mCherry focus was seen at one pole of each septated Bdellovibrio cell, prior to release from prey (Figure 5E v).
In axenically-growing, longer, HI cells polar and sometimes bipolar expression of this protein was also seen (Figure 5E vi, vii). This pattern correlates with video-microscopy observations we have made where some unusually long HI Bdellovibrio wild-type cells, found by chance with each pole adjacent to a prey cell, simultaneously invade both prey cells at once. In such unusually long HI Bdellovibrio (which do not have the usual polar flagellum), it seems that each pole can be competent for prey entry (Figure S6 in Text S1).
Among mutations in the c-di-GMP signalling proteins, the ΔdgcB and ΔcdgA mutations were the only ones that impaired predatory growth, specifically at the prey entry stage, which suggests that DgcB and CdgA belong to the same regulatory cascade, where DgcB signals, via c-di-GMP synthesis, to CdgA. A more severe phenotype of the dgcB deletion versus the cdgA deletion implies that DgcB signals via more than a single target. The DgcB targets likely regulate activity/expression of host hydrolytic and prey modifying/degrading proteins involved in penetration of prey bacteria. It appears that diguanylyl cyclase activity of DgcB is activated via an encounter with prey, possibly via the forkhead domain of DgcB. Using the DgcB-mCherry fusion we determined that DgcB is apparently expressed at relatively high levels and located throughout the cell.
Analysis of c-di-GMP levels in axenically grown Δdgc and Δcdg mutant strains
The method described by Bobrov and co-workers [31] was used to determine the c-di-GMP levels in pure Bdellovibrio cells from the axenically grown ΔdgcA, B and ΔcdgA mutant HI strains, versus matched wet cell weights of wild type HI controls. A similar analysis was not possible for predatory strains due to possible contamination with prey-derived c-di-GMP. The mean level of c-di-GMP for wild type axenic HI B. bacteriovorus strains HID13 and HID26 was 1.4+/−0.1nM/mg wet weight of cells. For the ΔdgcA strain this value was considerably lower at 0.4nM/mg wet weight of cells, for the ΔdgcB strain the value was 1.5nM/mg wet weight of cells and for the ΔcdgA strain the value was slightly higher at 2.0nM/mg wet weight of cells. Thus the absence of the DgcA protein noticeably reduced the extractable c-di-GMP content of the B. bacteriovorus cells, consistent with the diguanylyl cyclase activity of DgcA. The absence of the CdgA protein slightly increased the extractable c-di-GMP content but the absence of the DgcB protein did not cause a significant difference to extractable c-di-GMP levels. The latter result is not surprising given the expectation that DgcB is activated when the Bdellovibrio are in contact with, or the immediate proximity of prey (which was not the case in the axenically grown cells used here.
Summary
In summary, (Figure 7) we have found that each of 4 conserved GGDEF proteins in B. bacteriovorus HD100 contributes non-overlapping regulatory controls to different aspects of the predatory or axenic life cycles of this bacterium, and that these can be seen in single GGDEF gene mutants with the remaining GGDEF genes intact. We found that DgcB controls predatory invasion of prey bacteria, by signalling to c-di-GMP receptors, probably including CdgA, a degenerate GGDEF protein located at the prey-interacting “nose” of Bdellovibrio. As DgcB may receive information that indicates proximity or attachment to prey being sensed by the Bdellovibrio, we are now searching for the signals that activate DgcB. This will allow us to understand signals that “tell” Bdellovibrio that prey are present and aid the targeting of Gram-negative pathogens for destruction in anti-infective settings.
Fig. 7. Summary of GGDEF domain protein activities in B. bacteriovorus HD100 lifestyles. 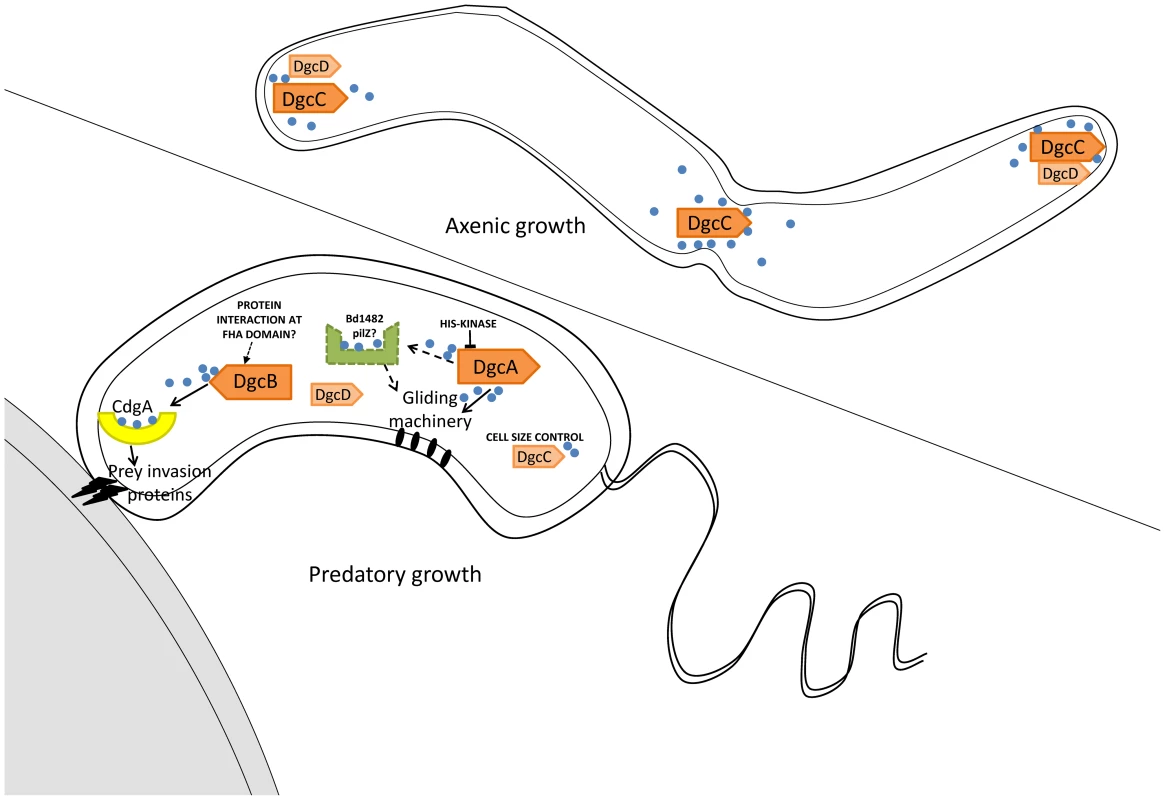
Arrows with solid lines summarise regulatory activities discovered by analysis of phenotypes of DgcA,B,C and CdgA mutant strains. Dotted arrows and lines indicate pathways inferred by association of a PilZ protein. Positions of DgcC and DgcD in the axenically growing Bdellovibrio were informed by mCherry tagging. The position of CdgA at the prey-interacting pole of the predatory Bdellovibrio was informed by mCherry tagging, with the remaining DgcA and DgcB mCherry proteins being found to be cytoplasmic in these cells. DgcA controls gliding motility (Figure 7) which is required for the Bdellovibrio to exit the exhausted prey debris after predation and to move off to regions where new prey encounters would be possible [1], [2]. To our knowledge this is the first case where c-di-GMP controls gliding motility, whereas c-di-GMP control of flagellar motility is better understood in other bacteria. As only gliding motility and predation, not flagellar synthesis, were found to be restored by the DgcA-mCherry complementation experiment, it may be that DgcA does not regulate flagellar synthesis in B. bacteriovorus HD100. DgcA has a response regulator domain at the N terminus and thus has some similarity to WspR which controls flagellar biogenesis versus pellicle formation in Pseudomonas [32], [33]. Flagellar biogenesis is not however required for B. bacteriovorus predation [1] and further studies on DgcA will define whether it also regulates flagellar biogenesis. We do note that, at the 5′ end of one of 4 operons of gliding motility genes of B. bacteriovorus [2], there is a gene, Bd1482, encoding a PilZ domain protein, which may be a candidate for receiving signals from DgcA to effect gliding and thus we have tentatively labelled this (Figure 7) as a candidate to receive c-di-GMP signals from DgcA. Gliding will be particularly important in the predation of pathogen biofilms where Bdellovibrio have been shown to be effective [34].
DgcC controls the transition between the predatory “attack phase” of Bdellovibrio when it is “locked into” a non-replicative phase, hunting prey; and the replicative, axenic, growth phase, on protein rich media. This is a mysterious transition [10], [35]. It is noteworthy that DgcC-mCherry forms foci in growing axenic cells and we hope that studying any proteins with which DgcC forms a complex may illuminate this control of non-predatory replication. The DgcC deletion mutant is the gateway to producing a useful, obligately predatory Bdellovibrio for application as a self-limiting living antibiotic.
We have revealed the impressive c-di-GMP intellect of this tiny bacterium and how it is employed in its unique intra-bacterial niche, and we can now see how this can be manipulated to use it as an anti-infective [18]. Studying the PilZ receptor domain proteins of Bdellovibrio, of which there are 15 [36], will dissect further, the stages by which predatory and axenic growth processes are regulated, and will reveal the cyclase - receptor relationships.
This study has contributed to our understanding of specificity of c-di-GMP signalling pathways; something where different models have been proposed from workers studying other bacteria. Some have suggested that little specificity exists between diguanylyl cyclases and their targets [37]; with all expressed diguanylyl cyclases contributing to the intracellular c-di-GMP pool that is sensed by all c-di-GMP receptors or targets. In an alternative, “c-di-GMP cloud” model [38], each diguanylyl cyclase has a specific target; little crosstalk exists between different c-di-GMP signalling pathways because c-di-GMP bursts are localized, and the spillover is prevented by c-di-GMP phosphodiesterases. Several studies have supported the later model by data showing that intracellular levels of c-di-GMP do not necessarily correlate with the observed phenotypes, that c-di-GMP is produced at unique cellular locations, and most directly, specific c-di-GMP targets were linked to individual diguanylyl cyclases [39]-[43]. However, B. bacteriovorus is the first bacterium in which the strikingly different phenotypes of the diguanylyl cyclase mutants shows an exclusive cyclase - c-di-GMP target(s) relationship. It is ironic that the task of proving the existence of c-di-GMP signalling pathway specificity has fallen onto one of the tiniest bacteria, where diffusion rates could have been envisioned to make c-di-GMP spillover unavoidable. The small size of Bdellovibrio cells makes the exclusivity point most convincing. While some bacteria may have ‘general purpose’ diguanylyl cyclases, and activation of parallel c-di-GMP targets is perhaps advantageous for some organisms, under certain circumstances; specific c-di-GMP signalling from discrete diguanylyl cyclases is clearly at play in Bdellovibrio control pathways, and as this specificity occurs in such small bacteria; it should also be considered in others.
Materials and Methods
Strains and growth conditions
Bdellovibrio and E. coli strains are listed in Table S1 (in Text S1). Predatory B. bacteriovorus strains were routinely grown in Ca/HEPES with E. coli S17-1 as prey as previously described [1], [44]. Host-Independent (HI) Bdellovibrio were isolated and grown in PY broth as previously described [44], [45]. Kanamycin (25 µg ml−1 for E. coli and 50 µg ml−1 for Bdellovibrio), ampicillin (50 µg ml−1) and isopropyl-β-D-1-thiogalactopyranoside (IPTG; 200 µg ml−1 for induction of fluorescence in E. coli) were used where appropriate.
Gene deletion and complementation in Bdellovibrio
Chromosomal deletions of the dgcA, dgcB, dgcD and cdgA reading frames (Bd0367, Bd0742, Bd3766 and Bd3125) were created using a modified version of previously described methods [27], [46]. Deletion of the dgcC (Bd1434) ORF and replacement with a kanamycin resistance cassette was initially performed using a modified version of that described by Lambert and co-workers [29] and later followed up, to verify the cell size phenotype with the same dgcC chromosomal deletion using the silent deletion method as used for the four other genes above. Complementation analyses for dgcA, dgcB, dgcD and cdgA deletion strains used single recombination of either full-length mcherry tagged (for dgcA) or wild type dgc or cdg genes (for the other strains) into the Bdellovibrio chromosome from suicide plasmid pK18mobsacB. For strains dgcB and dgcC only, GGAAF expressing (rather than wild type GGDEF or GGEEF) genes were used in single recombination experiments to test the function of the GGDEF domain. (Wild type CdgA does not have a conventional GGDEF domain so was excluded). Construction of each of the mutant and complemented strains is described fully in Text S1.
Electron Microscopy (EM)
Bdellovibrio deletion strains were assayed for morphological changes by EM, after growth (both predatory and axenic) under standard conditions. Cells were stained with 1% phosphotungstic acid (PTA; pH 7.0) and imaged with a JEOL 1200Ex electron microscope at 80 kV. Replicate cultures were grown independently on different days and flagellar patterns and cell morphologies were determined from 45–50 cells observed each time.
Assays for predatory and axenic growth capabilities
Deletion mutants of dgcA, dgcB and cdgA could only be isolated using axenic (HI) growth. HI deletion strains were added in excess to prey in Ca/HEPES buffer and monitored by microscopy for signs of prey entry and replication at several time-points during prolonged incubation over multiple days (up to 10 days for ΔdgcB; whilst wild-type HI strains [HID2, HID13 and HID26 [47], chosen for morphological similarities, see important considerations in Text S1] completed predation when checked after 24 hours of incubation). Both the ΔdgcA and ΔcdgA mutants were seen to enter prey within the first 24–48 hours of incubation in these liquid conditions and these were further analysed by time-lapse microscopy (described below). Cells from prolonged incubation of ΔdgcB mutant strains with prey were re-confirmed to be ΔdgcB by PCR analysis. Deletion mutants of both dgcC and dgcD could be readily obtained from screening predatory cultures, to check whether these strains could grow axenically they were “turned HI” by filtration of attack-phase cells through a 0.45 µm filter to remove any remaining prey, and plating onto rich Peptone-Yeast Extract media [45]. The number of attack-phase cells of the ΔdgcC mutant had to be multiplied 1000 –fold (to 4.5×1011) to obtain a single colony on the PY media. Complemented strains were tested by the same methods.
C-di-GMP activity measurements
The diguanylyl cyclase assays in vitro were performed by measuring the rate of GTP conversion into c-di-GMP by MBP-Dgc fusion proteins. GTP was added to the purified enzyme solution following the protocol by Ryjenkov et al. [48]. Each dignuanylyl cyclase reaction contained 5 micromoles of the MBP-Dgc fusion proteins. The equilibrium dialysis measurements of c-di-GMP binding to CdgA were performed using 10 micromole of MBP-CdgA and varying concentrations of c-di-GMP as described elsewhere [26]. Nucleotide separation and quantitation was accomplished by HPLC as described by Ryjenkov et al. [48].
Extractable c-di-GMP determinations for Bdellovibrio cells
The method of Bobrov [31] was used to determine the extractable levels of c-di-GMP in axenically grown Bdellovibrio cells. with analysis using liquid chromatography tandem mass spectrometry carried out by Lijun Chen and Bev Chamberlin at the Mass Spectrometry Core of RTSF (Research Technology Support Facility) at Michigan State University USA Axenically growing HI Bdellovibrio strains (mutant and wild type HI) were grown in 50ml of PY broth from a starting 0.2 OD600nm until they reached a final 0.6 OD600nm. Cells were then pelleted by centrifugation, the wet weights were determined and matched; they were frozen in liquid nitrogen and then processed using Bobrov's method and extraction buffer (40% methanol 40% acetonitrile in 0.1N formic acid) which was later neutralised with NH4HCO3. The levels of extractable c-di-GMP in the cell extracts were compared to known added standards of pure c-di-GMP.
Video microscopy of events in the predatory growth cycle and gliding
Time-lapse back-lit microscopy was performed as described fully by Fenton and co-workers [6], with the exception that HI Bdellovibrio, to be tested for predatory capacity, were grown overnight in PY broth and then diluted to a starting OD600nm of 0.75 before addition to prey in the infection culture.
Localisation of fluorescently tagged proteins
Bdellovibrio strains containing GGDEF protein-mCherry fusions, in a wild-type GGDEF gene background, were constructed using a modification of the method described by Fenton and co-workers [49], construction of each tag is described fully in Text S1. Resulting strains were imaged using a Nikon Eclipse E600 epifluorescence microscope with a 100x objective lens and an hcRED filter block (excitation 550 to 600 nm; emission 610 to 665 nm) in conjunction with a Hamamatsu Orca ER camera and the Simple PCI software (version 5.3.1 Hamamatsu). Gliding motility was assayed by timelapse microscopy as in Lambert and co-workers [2].
Genes studied
Bd0367 dgcA GenBank ID: NP_967362.1
Bd0742 dgcB GenBank ID: NP_967706.1
Bd1434 dgcC GenBank ID: NP_968330.1
Bd3125 cdgA GenBank ID: NP_969891.1
Bd3766 dgcD GenBank ID: NP_970474.1
Supporting Information
Zdroje
1. LambertCEvansKJTillRHobleyLCapenessM 2006 Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol Microbiol 60 274 286
2. LambertCFentonAKHobleyLSockettRE 2011 Predatory Bdellovibrio Bacteria Use Gliding Motility to Scout for Prey on Surfaces. J Bacteriol 193 3139 3141
3. DashiffAJunkaRALiberaMKadouriDE 2011 Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol 110 431 444
4. JurkevitchEMinzDRamatiBBarelG 2000 Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl Environ Microbiol 66 2365 2371
5. SockettRELambertC 2004 Bdellovibrio as therapeutic agents: a predatory renaissance? Nat Rev Microbiol 2 669 675
6. FentonAKKannaMWoodsRDAizawaS-ISockettRE 2010 Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol 192 6329 6335
7. HorowitzATKesselMShiloM 1974 Growth cycle of predacious Bdellovibrios in a host-free extract system and some properties of the host extract. J Bacteriol 117 270 282
8. ThomashowLSRittenbergSC 1985 Isolation and composition of sheathed flagella from Bdellovibrio bacteriovorus 109J. J Bacteriol 163 1047 1054
9. BarelGJurkevitchE 2001 Analysis of phenotypic diversity among host-independent mutants of Bdellovibrio bacteriovorus 109J. Arch Microbiol 176 211 216
10. RoschanskiNKlagesSReinhardtRLinscheidMStrauchE 2011 Identification of Genes Essential for Prey-Independent Growth of Bdellovibrio bacteriovorus HD100. J Bacteriol 193 1745 1756
11. HenggeR 2009 Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7 263 273
12. WolfeAJVisickKL 2010 The second messenger cyclic di-GMP. Washington, DC ASM Press
13. TamayoRPrattJTCamilliA 2007 Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol 61 131 148
14. GomelskyM 2011 cAMP, c-di-GMP, c-di-AMP and now cGMP: Bacteria use them all! Mol Microbiol 79 562 565
15. ChanCPaulRSamorayDAmiotNCGieseB 2004 Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA 101 17084 17089
16. AmikamDGalperinMY 2006 PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22 3 6
17. GalperinMY 2004 Bacterial signal transduction network in a genomic perspective. Environ Microbiol 6 552 567
18. GalperinMYHigdonRKolkerE 2010 Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol Biosyst 6 721 728
19. FinnRDMistryJTateJCoggillPHegerA 2010 The Pfam protein families database. Nucleic Acids Res 38 D211 222
20. SchirmerTJenalU 2009 Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7 724 735
21. RendulicSJagtapPRosinusAEppingerMBaarC 2004 A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303 689 692
22. PallenMChaudhuriRKhanA 2002 Bacterial FHA domains: neglected players in the phospho-threonine signalling game? Trends Microbiol 10 556 563
23. KoMParkC 2000 Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol 303 371 382
24. RyjenkovDASimmRRomlingUGomelskyM 2006 The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281 30310 30314
25. DeNPirruccelloMKrastevaPVBaeNRaghavanRV 2008 Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLosONE 6 e67
26. RyjenkovDASimmRRömlingUGomelskyM 2006 The PilZ domain is a receptor for the second messenger c-di-GMP. J Biol Chem 281 30310 30314
27. SteyertSRPineiroSA 2007 Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus. Appl Environ Microbiol 73 4717 4724
28. IidaYHobleyLLambertCFentonAKSockettRE 2009 Roles of multiple flagellins in flagellar formation and flagellar growth post bdelloplast lysis in Bdellovibrio bacteriovorus. J Mol Biol 394 1011 1021
29. LambertCSmithMCSockettRE 2003 A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ Microbiol 5 127 132
30. CotterTWThomashowMF 1992 Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J Bacteriol 174 6018 6024
31. BobrovAGKirillinaORyjenkovDAWatersCMPricePA 2011 Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol Microbiol 79 533 551
32. HickmanJWTifreaDFHarwoodCS 2005 A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102 14422 14427
33. HickmanJWHarwoodCS 2008 Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69 376 389
34. KadouriDO'TooleGA 2005 Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol 71 4044 4051
35. WurtzelODori-BachashMPietrokovskiSJurkevitchESorekR 2010 Mutation detection with next-generation resequencing through a mediator genome. PLosONE 5 e15628
36. GalperinMY 2005 A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol 5 35
37. SolanoCGarciaBLatasaCToledo-AranaAZorraquinoV 2009 Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci U S A 106 7997 8002
38. GomelskyM 2009 The core pathway: diguanylate cyclases, phosphodiesterases, and c-di-GMP-binding proteins. WolfeAVisickK The second messenger cyclic di-GMP Washington, DC ASM Press 37 56
39. DuerigAAbelSFolcherMNicollierMSchwedeT 2009 Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23 93 104
40. GuvenerZTHarwoodCS 2007 Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66 1459 1473
41. MerrittJHHaDGCowlesKNLuWMoralesDK 2010 Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. MBio 19 e00183 10
42. KuchmaSLBallokAEMerrittJHHammondJHLuW 2010 Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J Bacteriol 192 2950 2964
43. SimmRLuschAKaderAAnderssonMRomlingU 2007 Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J Bacteriol 189 3613 3623
44. MorehouseKAHobleyLCapenessMSockettRE 2011 Three motAB Stator Gene Products in Bdellovibrio bacteriovorus Contribute to Motility of a Single Flagellum during Predatory and Prey-Independent Growth. J Bacteriol 193 932 943
45. EvansKJLambertCSockettRE 2007 Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J Bacteriol 189 4850 4859
46. RoschanskiNKlagesSReinhardtRLinscheidMStrauchE 2011 Identification of genes essential for prey independent growth of Bdellovibrio bacteriovorus HD100. J Bacteriol 193 1745 1756
47. LambertCChangCYCapenessMJSockettRE 2010 The first bite–profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5 e8599
48. RyjenkovDATarutinaMMoskvinOVGomelskyM 2005 Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into the biochemistry of the GGDEF protein domain. J Bacteriol 187 1792 1798
49. FentonAKLambertCWagstaffPCSockettRE 2010 Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J Bacteriol 192 1299 1311
50. DeNNavarroMVRaghavanRVSondermannH 2009 Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J Mol Biol 393 619 633
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2012 Číslo 2- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Mechanisms of Pathogenesis, Infective Dose and Virulence in Human Parasites
- Structural and Functional Insights into the Pilotin-Secretin Complex of the Type II Secretion System
- Characterising the Mucosal and Systemic Immune Responses to Experimental Human Hookworm Infection
- Population Genetic Analyses Reveal the African Origin and Strain Variation of var.
- How Do Microbial Pathogens Make s?
- Substance P Causes Seizures in Neurocysticercosis
- Phagosomal Rupture by Results in Toxicity and Host Cell Death
- Absence of HIV-1 Evolution in the Gut-Associated Lymphoid Tissue from Patients on Combination Antiviral Therapy Initiated during Primary Infection
- Five Questions about Viral Trafficking in Neurons
- Selecting an Invertebrate Model Host for the Study of Fungal Pathogenesis
- A Co-Opted DEAD-Box RNA Helicase Enhances Tombusvirus Plus-Strand Synthesis
- Biochemical Properties of Highly Neuroinvasive Prion Strains
- ClpP1 and ClpP2 Function Together in Protein Degradation and Are Required for Viability and During Infection
- Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
- Characterising the Mucosal and Systemic Immune Responses to Experimental Human Hookworm Infection
- How Do Microbial Pathogens Make s?
- Substance P Causes Seizures in Neurocysticercosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



