-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
How Do Microbial Pathogens Make s?
article has not abstract
Published in the journal: How Do Microbial Pathogens Make s?. PLoS Pathog 8(2): e32767. doi:10.1371/journal.ppat.1002463
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1002463Summary
article has not abstract
What Is the Link between Drug Resistance and Aneuploidy in Microbial Pathogens?
In a variety of pathogens, drug resistance and aneuploidy are intimately associated. Anueploidy is believed to alter the dosage of certain genes that can impart drug resistance. Generation of a new chromosome by duplication of chromosome segments followed by telomere addition in a pathogenic yeast Candida glabrata [1] and isochromosome formation by breakage of chromosome 5 at the centromere followed by joining two identical arms of chromosome 5 in an opportunistic yeast Candida albicans [2] have been shown to occur frequently in drug-resistant isolates. Fluconazole-resistant strains of another pathogenic fungus, Cryptococcus neoformans, have been shown to be disomic for certain chromosomes [3]. Experimental evidence suggests that acquisition of chromosomes by the plant pathogenic fungus Fusarium can convert a non-pathogenic strain to a pathogenic one [4]. Anueploidy has been shown to be the cause of drug resistance in the protozoan parasite Leishmania as well [5].
Improper chromosome segregation is one route to aneuploidy. The centromere–kinetochore complex facilitates interaction between a chromosome and the spindle microtubules to ensure equal segregation of chromosomes from a mother to daughter cells. In addition to serving as sites of protein assembly to form kinetochores, centromeres also hold two sister chromatids together until the onset of anaphase by balancing opposing forces acting on them—a pole-ward (outward) force generated by spindle microtubules depolymerizing toward opposite poles, and a cohesive (inward) force between two sister chromatids. Paradoxically, in spite of performing the conserved function of chromosome segregation, centromere (CEN) DNA sequence and organization of CEN DNA elements vary widely among eukaryotes. In contrast, several kinetochore proteins are evolutionarily conserved, although sometimes this conservation is restricted to a group of organisms. For example, the Dam1 complex, an outer kinetochore protein complex, is fungus specific and is essential for viability in C. albicans [6], [7]. Thus, this complex may be a suitable target for development of anti-fungal drugs. However, a CEN-specific histone H3 variant of the CENP-A/Cse4 family is found to be universally associated with the formation of specialized and unique chromatin at all functional CENs despite seemingly diverse CEN DNA sequences.
What Are the Different Types of CENs in Eukaryotic Pathogens?
Some organisms have holocentric chromosomes where the CEN is diffused and CEN elements are distributed throughout a chromosome. However, most organisms have monocentric chromosomes where a CEN is localized to a single region on a chromosome. Monocentric CENs are of three types: a) “point” CENs in which centromere function is retained in a short (<400 bp) stretch of DNA with highly conserved protein binding motifs and often a single CENP-A-containing nucleosome, b) “small regional” CENs with CENP-A-containing chromatin formed on a stretch of DNA (<40 kb) that is longer than the point centromere but often lacks any protein binding sequence motifs or conserved pericentric repeats, and c) “large regional” CENs that span a long region (>40 kb–a few Mbs) usually rich in repeats and often CENP-A-containing nucleosomes are interspersed with H3-containing nucleosomes. Formation of a kinetochore on most point CENs initiates with binding of a DNA sequence-specific kinetochore protein [8]. In contrast, several lines of evidence suggest that epigenetic factors, not the DNA sequence alone, also contribute to centromere identity in organisms carrying small or large regional centromeres [8]–[10].
How Are Centromeres Organized in Fungal Pathogens?
The pathogenic fungi show wide diversity in CEN structure (Figure 1; reviewed in [11]). C. glabrata, a human pathogenic ascomycetous yeast, has short point CENs. CENs in C. glabrata, like the well-studied CENs of baker's yeast Saccharomyces cerevisiae, have three CEN DNA elements (CDEs). CDEI (8 bp) and CDEIII (18 bp) are well conserved in all the chromosomes, while CDEII (77–79 bp) is not conserved in sequence but is highly AT-rich (83%–93%) [12]. A circular mini-chromosome containing a short CEN sequence (<160 bp total, inclusive of CDEs) and a replication origin/autonomously replicating sequence (ARS) can stably propagate through many generations. Both CDEI and CDEIII are required for proper function of a centromere. CDEIII has the crucial CCG sequence and certain mutations in CDEIII result in a complete loss of centromere function. Candida maltosa, a hemiascomycetous yeast that is phylogenetically closely related to other human pathogenic Candida species of the CTG clade such as C. albicans, is believed to be non-pathogenic to humans but has been reported to be virulent occasionally when tested in the mouse model [13]. Although a 325-bp C. maltosa CEN region with a conserved CDEI and an AT-rich CDEII sequence can provide mitotic stability to an otherwise unstable ARS plasmid, the conserved CDEIII sequence, frequently found in point centromeres, is absent [14], [15].
Fig. 1. Structural organization of centromeres in various microbial pathogens. 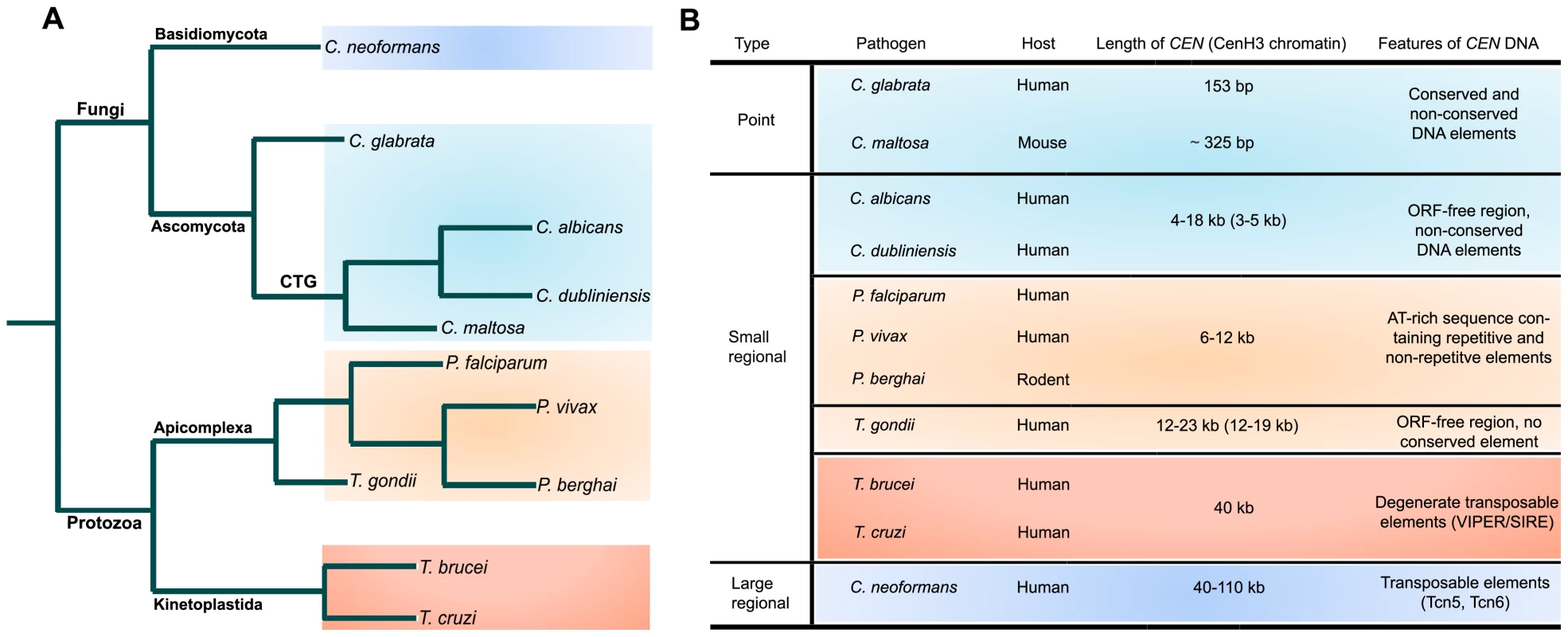
(A) A schematic showing relatedness of various microbial pathogens. (B) A table summarizing essential features of different types of centromeres identified in microbial pathogens. C. albicans, the most common fungal pathogen isolated from patients, and Candida dubliniensis, a less frequently observed but closely related species, have small regional CENs. C. albicans CENs were identified as binding sites of Cse4, a CENP-A homolog [16]. Each of the eight CENs has a 3–5-kb CENP-A-rich sequence present in a 4–18-kb ORF-free region [17]. Only chromosome 5 has long inverted pericentric repeats. The AT-richness at the CEN region is similar to the overall AT-content of the genome. CEN formation in this organism requires a pre-existing epigenetic memory, and thus an exogeneously introduced CEN/ARS plasmid is mitotically unstable in C. albicans [18]. Strikingly, when a native centromere is replaced by a transcribed, selectable gene, the acentric chromosome often is stabilized by forming a neocentromere very efficiently elsewhere on the chromosome on a region with no sequence homology to the native centromere [19]. CENs in C. dublineinsis have been identified by synteny analysis with C. albicans and are found to be rich in CENP-A binding [20]. Although all the features of CdCENs are very similar to those of CaCENs, CEN DNA sequences in these two organisms were found to be rapidly evolving. The CEN sequence of another pathogenic yeast, Candida lusitaniae, has been identified by bioinformatic analysis but not experimentally verified [21]. A GC-poor trough approximately 4 kb in length found in every chromosome is the putative centromere in this organism.
A basidiomycetous fungus, C. neoformans, which causes fungal meningitis in humans, has been predicted to have long regional centromeres. Transposon (Tcn5, Tcn6)-rich sequences, located within 40–110 ORF-free regions that are present once per chromosome, are the presumptive CENs [22]. Moreover, measurement of meiotic recombination rates by random spore analysis also indicates that URA5 and ADE2 are CEN-linked in agreement with the physical map [23]. These CENs are similar to those of the opportunistic pathogen Aspergillus nidulins, an ascomycete [24].
How Are Centromeres Organized in Ancient Eukaryotic Pathogens, the Protozoan Parasites?
Topoisomerase II (Topo-II) has been implicated in chromosome segregation in a range of organisms from yeast to humans [25], [26], [27]. After DNA replication in S phase, sister chromatids remain attached together partly by strand catenation at centromeres as cells enter mitosis. During sister chromatid separation, Topo-II decatenates by creating double-strand DNA breaks followed by passage of uncut DNA through breaks and ligation to repair the breaks. Topo-II has even been implicated to be an epigenetic marker for kinetochore assembly [28]. Etoposide, a Topo-II inhibitor, blocks the ligation step and results in DNA breaks at Topo-II binding sites [29]. In the malaria parasite Plasmodium falciparum, an apicomplexan, an etoposide-mediated Topo-II cleavage site was mapped to a single 10-kb region in each of the 14 chromosomes [30]. Further analysis shows that, with the exception of chromosome 10, this site corresponds to a 6–12-kb ORF-free region with a highly AT-rich region of 2.3–2.5 kb that contains a repetitive region and a core region. Both the core and the repeat region are important for centromere function. Various repetitive DNA elements present in these regions do not show any interchromosomal conservation, but nucleotide content (AT-richness) and the size of these putative CENs are strictly conserved. Another human malaria parasite, Plasmodium vivax, has AT-rich syntenic regions in three chromosomes [30]. Thus, both these Plasmodium species have centromere properties similar to the small regional centromeres found in C. albicans and C. dubliniensis (Figure 1). Based on the syntenic locations of PfCENs, centromeres were identified in genetically tractable Plasmodium berghei, which causes malaria in rodents [31]. A 1.2-kb highly AT-rich (96%) sequence found in PbCEN5 also contains a non-repetitive core and a repetitive region. Centromere function was formally demonstrated by cloning this region in a plasmid. The PbCEN5 plasmid was evenly segregated and was stably maintained at low copy number in 90% of the parasites 21 days post-transfection in the absence of drug selection for the plasmid. A linear Plamodium artificial chromosome (L-PAC), constructed by adding telomeres to the CEN plasmid, was also stably maintained like a natural chromosome. L-PAC exhibited a 10 - to 100-fold increase in transfection efficiency as compared to the circular CEN plasmid. In another apicomplexan, Toxoplasma gondii, centromeres were identified as binding sites of the centromeric histone CENP-A [32]. One CENP-A binding region per chromosome was identified in 12 of the 14 chromosomes. CENP-A binding regions were restricted to 16±3.4 kb sequences that are present in 17±5.6 kb regions largely devoid of ORFs. Etoposide-mediated cleavage in these regions further confirmed centromere identity. Again, no obvious sequence bias or conserved sequence elements were detected, suggesting Toxoplasma centromeres belong to the class of short regional centromere (Figure 1). Centromeres have been identified in two kinetoplastid protozoans, Trypanosma cruzi and Trypansoma brucii, the causative agents of Chagas disease in Latin America and of sleeping sickness in humans and nagana in cattle in sub-Saharan Africa, respectively (Figure 1). By telomere-mediated chromosome truncation, a 16-kb GC-rich transcriptional “strand switch” domain was identified as the centromere in T. cruzi [33]. Etoposide cleavage analysis also confirmed that a Topo-II binding site colocalizes to this single locus on T. cruzi chromosome I. This 16-kb region has degenerate retroelements (VIPER/SIRE) and non-LTR transposons but does not contain any satellite repeats. This region is also flanked by transcriptionally quiescent polycistronic satellite units. T. brucii also has a similar 11-kb GC-rich domain in a syntenic region between directionally oriented gene clusters that contain degenerate retroelements (DIRE) and a 5.5-kb AT-rich stretch with 58-bp degenerate repeats [34].
What Are the Factors That Determine CEN Identity?
Since centromeres in certain pathogenic budding yeasts form in a DNA sequence-dependent manner while in most other microbial pathogens centromeres form in a sequence-independent manner, a universal mechanism determining centromere identity seems to be improbable. The best example of DNA sequence-independent assembly is the formation of neocentromeres on non-native loci with no apparent sequence similarity when a native CEN is deleted or inactivated. Centromeres are found to be clustered and occupy a distinct locus at the nuclear periphery in most yeasts, including C. albicans [17] and C. dublineinsis [20], as well as in the protozoan parasite T. gondii [32]. A three-dimensional higher order chromatin structure (clustered centromeres) that occupies a favorable nuclear space (such as the one with high CENP-A concentration) may be the determining factor for CEN identity in these organisms. CENs have been shown to replicate earliest in S phase in C. albicans [35], which indicates that replication timing can be a determinant of CEN identity. Certain post-translational modifications of canonical histone H3 can also support CEN formation [36]. Di - and tri-methylated Lys 9 of histone H3 (H3K9Me2/H3K9Me3), marks associated with pericentric heterochromatin, have been shown to be enriched at centromeric regions of T. gondii [32]. Interestingly, although H3K9Me3 molecules are enriched in transcriptionally silent loci, centromere regions in P. falciparum are largely devoid of such molecules. Non-coding RNA (ncRNA) and the RNAi machinery also play a role in centromere formation in certain eukaryotes [36]. In P. falciparum, short ncRNAs (75–175 nucleotides) transcribed from both strands of CEN regions localize to the nucleus [37] even though Plasmodium species lack components of the RNAi machinery. T. bruceii is the first protozoan parasite in which RNAi has been shown to be functional [38]. Disruption of RNAi causes defects in chromosome segregation in this organism [39]. Strikingly, the related species T. cruzi does not have RNAi machinery. Thus, a variety of factors, often species specific, may contribute to formation of a functional CEN, but elucidation of a specific mechanism that determines CEN identity in most pathogenic eukaryotes is a puzzle to be solved. Understanding centromere-kinetochore structure-function of these pathogens can help us develop specific drugs with fewer side effects to combat microbial infection.
Zdroje
1. PolakovaSBlumeCZarateJÃMentelMJorck-RambergD 2009 Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci U S A 106 2688 2693
2. SelmeckiAForcheABermanJ 2006 Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313 367 370
3. SionovELeeHChangYCKwon-ChungKJ 2010 Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog 6 e1000848 doi:10.1371/journal.ppat.1000848
4. MaL-Jvan der DoesHCBorkovichKAColemanJJDaboussiM-J 2010 Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464 367 373
5. UbedaJ-MLegareDRaymondFOuameurABoisvertS 2008 Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol 9 R115
6. ThakurJSanyalK 2011 The essentiality of the fungus-specific Dam1 complex is correlated with a one-kinetochore-one-microtubule interaction present throughout the cell cycle, independent of the nature of a centromere. Eukaryot Cell 10 1295 1305
7. Burrack LSApplen SEBermanJ 2011 The requirement for the Dam1 complex is dependent upon the number of kinetochore proteins and microtubules. Curr Biol 21 889 896
8. ClevelandDWMaoYSullivanKF 2003 Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112 407 421
9. EkwallK 2007 Epigenetic control of centromere behavior. Annu Rev Genet 41 63 81
10. BlackBEClevelandDW 2011 Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 144 471 479
11. RoyBSanyalK 2011 Diversity in requirement of genetic and epigenetic factors for centromere function in fungi. Eukaryot Cell 10 1384 1395
12. KitadaKYamaguchiEHamadaKArisawaM 1997 Structural analysis of a Candida glabrata centromere and its functional homology to the Saccharomyces cerevisiae centromere. Curr Genet 31 122 127
13. YoshidaMHashimotoK 1986 Potential pathogenicity of Candida maltosa IAM 12248. Agric Biol Chem 50 2119 2120
14. OhkumaMKKKawaiSHwangCWOhtaATakagiM 1995 Identification of a centromeric activity in the autonomously replicating TRA region allows improvement of the host-vector system for Candida maltosa. Mol Gen Genet 249 447 455
15. NakazawaTMotoyamaTHoriuchiHOhtaATakagiM 1997 Evidence that part of a centromeric DNA region induces pseudohyphal growth in a dimorphic yeast, Candida maltosa. J Bacteriol 179 5030 5036
16. SanyalKCarbonJ 2002 The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc Natl Acad Sci U S A 99 12969 12974
17. SanyalKBaumMCarbonJ 2004 Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci U S A 101 11374 11379
18. BaumMSanyalKMishraPKThalerNCarbonJ 2006 Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci 103 14877 14882
19. KetelCWangHSWMcClellanMBouchonvilleKSelmeckiA 2009 Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5 e1000400 doi:10.1371/journal.pgen.1000400
20. PadmanabhanSThakurJSiddharthanRSanyalK 2008 Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci U S A 105 19797 19802
21. LynchDBLogueMEButlerGWolfeKH 2010 Chromosomal G+C content evolution in yeasts: systematic interspecies differences, and GC-poor troughs at centromeres. Genome Biol Evol 2 572 583
22. LoftusBJFungERoncagliaPRowleyDAmedeoP 2005 The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307 1321 1324
23. IdnurmA 2010 A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 185 153 163
24. AleksenkoANielsenMLClutterbuckAJ 2001 Genetic and Physical mapping of two centromere-proximal regions of chromosome IV in Aspergillus nidulans. Fungal Genet Biol 32 45 54
25. BachantJAlcasabasABlatYKlecknerNElledgeSJ 2002 The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell 9 1169 1182
26. Floridia GZAZuffardiOTyler-SmithC 2000 Mapping of a human centromere onto the DNA by topoisomerase II cleavage. EMBO Rep 1 489 493
27. RattnerJBHendzelMJFurbeeCSMullerMTBazett-JonesDP 1996 Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle - and species-specific manner and is required for proper centromere/kinetochore structure. J Cell Biol 134 1097 1107
28. FukagawaT 2004 Centromere DNA, proteins and kinetochore assembly in vertebrate cells. Chromosome Res 12 557 567
29. ChenGLYangLRoweTCHalliganBDTeweyKM 1984 Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem 259 13560 13566
30. KellyJMMcRobertLBakerDA 2006 Evidence on the chromosomal location of centromeric DNA in Plasmodium falciparum from etoposide-mediated topoisomerase-II cleavage. Proc Natl Acad Sci U S A 103 6706 6711
31. IwanagaSKhanSMKanekoIChristodoulouZNewboldC 2010 Functional identification of the Plasmodium centromere and generation of a Plasmodium artificial chromosome. Cell Host Microbe 7 245 255
32. BrooksCFFranciaMEGissotMCrokenMMKimK 2011 Toxoplasma gondii sequesters centromeres to a specific nuclear region throughout the cell cycle. Proc Natl Acad Sci U S A 108 3767 3772
33. ObadoSBotCNilssonDAnderssonBKellyJ 2007 Repetitive DNA is associated with centromeric domains in Trypanosoma brucei but not Trypanosoma cruzi. Genome Biol 8 R37
34. ObadoSOBotCEcheverryMCBayonaJCAlvarezVE 2010 Centromere-associated topoisomerase activity in bloodstream form Trypanosoma brucei. Nucleic Acids Res 39 1023 1033
35. KorenATsaiH-JTiroshIBurrackLSBarkaiN 2010 Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet 6 e1001068 doi:10.1371/journal.pgen.1001068
36. AllshireRCKarpenGH 2008 Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9 923 937
37. LiFSonbuchnerLKyesSAEppCDeitschKW 2008 Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. J Biol Chem 283 5692 5698
38. NgoHTschudiCGullKUlluE 1998 Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci U S A 95 14687 14692
39. ShiHDjikengATschudiCUlluE 2004 Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol Cell Biol 24 420 427
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2012 Číslo 2- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Mechanisms of Pathogenesis, Infective Dose and Virulence in Human Parasites
- Structural and Functional Insights into the Pilotin-Secretin Complex of the Type II Secretion System
- Characterising the Mucosal and Systemic Immune Responses to Experimental Human Hookworm Infection
- Population Genetic Analyses Reveal the African Origin and Strain Variation of var.
- How Do Microbial Pathogens Make s?
- Substance P Causes Seizures in Neurocysticercosis
- Phagosomal Rupture by Results in Toxicity and Host Cell Death
- Absence of HIV-1 Evolution in the Gut-Associated Lymphoid Tissue from Patients on Combination Antiviral Therapy Initiated during Primary Infection
- Five Questions about Viral Trafficking in Neurons
- Selecting an Invertebrate Model Host for the Study of Fungal Pathogenesis
- A Co-Opted DEAD-Box RNA Helicase Enhances Tombusvirus Plus-Strand Synthesis
- Biochemical Properties of Highly Neuroinvasive Prion Strains
- ClpP1 and ClpP2 Function Together in Protein Degradation and Are Required for Viability and During Infection
- Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
- Characterising the Mucosal and Systemic Immune Responses to Experimental Human Hookworm Infection
- How Do Microbial Pathogens Make s?
- Substance P Causes Seizures in Neurocysticercosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



