-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Transcription of a -acting, Noncoding, Small RNA Is Required for Pilin Antigenic Variation in
The strict human pathogen Neisseria gonorrhoeae can utilize homologous recombination to generate antigenic variability in targets of immune surveillance. To evade the host immune response, N. gonorrhoeae promotes high frequency gene conversion events between many silent pilin copies and the expressed pilin locus (pilE), resulting in the production of variant pilin proteins. Previously, we identified a guanine quartet (G4) structure localized near pilE that is required for the homologous recombination reactions leading to pilin antigenic variation (Av). In this work, we demonstrate that inactivating the promoter of a small non-coding RNA (sRNA) that initiates within the G4 forming sequence blocks pilin Av. The sRNA promoter is conserved in all sequenced gonococcal strains, and mutations in the predicted transcript downstream of the G4 forming sequence do not alter pilin Av. A mutation that produces a stronger promoter or substitution of the pilE G4-associated sRNA promoter with a phage promoter (when the phage polymerase was expressed) produced wild-type levels of pilin Av. Altering the direction and orientation of the pilE G4-associated sRNA disrupted pilin Av. In addition, expression of the sRNA at a distal site on the gonococcal chromosome in the context of a promoter mutant did not support pilin Av. We conclude that the DNA containing the G-rich sequence can only form the G4 structure during transcription of this sRNA, thus providing a unique molecular step for the initiation of programmed recombination events.
Published in the journal: Transcription of a -acting, Noncoding, Small RNA Is Required for Pilin Antigenic Variation in. PLoS Pathog 9(1): e32767. doi:10.1371/journal.ppat.1003074
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003074Summary
The strict human pathogen Neisseria gonorrhoeae can utilize homologous recombination to generate antigenic variability in targets of immune surveillance. To evade the host immune response, N. gonorrhoeae promotes high frequency gene conversion events between many silent pilin copies and the expressed pilin locus (pilE), resulting in the production of variant pilin proteins. Previously, we identified a guanine quartet (G4) structure localized near pilE that is required for the homologous recombination reactions leading to pilin antigenic variation (Av). In this work, we demonstrate that inactivating the promoter of a small non-coding RNA (sRNA) that initiates within the G4 forming sequence blocks pilin Av. The sRNA promoter is conserved in all sequenced gonococcal strains, and mutations in the predicted transcript downstream of the G4 forming sequence do not alter pilin Av. A mutation that produces a stronger promoter or substitution of the pilE G4-associated sRNA promoter with a phage promoter (when the phage polymerase was expressed) produced wild-type levels of pilin Av. Altering the direction and orientation of the pilE G4-associated sRNA disrupted pilin Av. In addition, expression of the sRNA at a distal site on the gonococcal chromosome in the context of a promoter mutant did not support pilin Av. We conclude that the DNA containing the G-rich sequence can only form the G4 structure during transcription of this sRNA, thus providing a unique molecular step for the initiation of programmed recombination events.
Introduction
Neisseria gonorrhoeae is an obligate human pathogen and the causative agent of the sexually transmitted infection gonorrhea. Gonococci generally infect the urogenital tract and the infection typically presents as urethritis in men and cervicitis in women, but many women can be asymptomatic carriers [1]. The N. gonorrhoeae type IV pilus is essential for establishing infection [2]. Pili assist in epithelial adherence, gonococcal cell aggregation, and also mediate twitching motility [3]–[5]. Protective immunity never develops, partially because the bacterium can evade host immune selection by varying the expression of surface antigens including lipooligosaccharides, the opacity family of outer membrane proteins, and pili [6]–[10].
Pilin antigenic variation (Av) is a high frequency diversification system that operates via a specialized recombination pathway, and utilizes enzymes that participate in general recombination and repair pathways as well as enzymes that do not participate in either pathway [11]. N. gonorrhoeae possesses one pilin expression locus (pilE) and up to 19 silent pilin loci (pilS) residing in up to 6 discrete locations in the genome [12]. Pilin Av occurs as a result of non-reciprocal DNA recombination between any pilS copy and pilE, leading to the expression of a new variant protein [7]. These new variants can be fully functional, poorly expressed or not expressed, and when the pilus is not expressed or poorly expressed there is an obvious change in colony morphology on solid growth medium [13]. The difference between piliated and nonpiliated colony morphologies thus allows the ability of a strain to undergo pilin (Av) to be easily assayed [14].
A transposon-based genetic screen previously identified insertions in a DNA region upstream of pilE that blocked pilin Av, but did not alter pilin expression [15], [16]. A subsequent targeted genetic screen in this region identified 11 GC base pairs that when individually mutated completely blocked pilin Av, and a 12th GC base pair that when mutated retained residual pilin Av activity [11], [17]. Mutation of an adjacent 13th GC base pair had no effect on pilin Av but mutation of this GC base pair in addition to the 12th resulted in a complete loss of pilin Av, suggesting that the 13th GC base pair could partially substitute for the 12th [11], [17]. The organization of these 12 GC base pairs conforms to a guanine quadruplex or guanine quartet (G4) forming sequence [17]. Biophysical studies demonstrated that this G-rich sequence formed a G4 structure, and mutations that block pilin Av also inhibited structure formation [17]. Growth of N. gonorrhoeae on N-methyl mesoporphyrin IX, a compound that specifically binds G4 structures but not double or single stranded DNA [18], decreased the frequency of pilin Av [17]. Furthermore, point mutations in N. gonorrhoeae that block pilin Av and G4 structure formation prevented single stranded nicks from being detected in both the G4 forming sequence and complement strand [17]. The results from these studies suggested that for pilin Av to occur, the pilE G4 sequence must form a G4 structure [17]. In order for a G4 structure to form, duplex DNA must first be converted into single stranded DNA. In this study, we have now established that pilin Av absolutely requires transcription of a non-coding sRNA which originates within the pilE G4 sequence, thus providing a mechanism for the initiation of this conversion event.
Results
Analysis of the pilin expression locus (pilE) guanine quartet (G4)-associated small RNA (sRNA) and the effects of mutations on pilin antigenic variation (Av)
The region upstream of pilE that contains the pilE G4 is devoid of predicted genes and open reading frames. However, we identified a putative promoter sequence adjacent to the pilE G4 [19] (Fig. 1A & B). The most conserved bases of the −10 element [20], all the bases in the −35 element, and the relative location of this putative promoter to the pilE G4 are conserved in all sequenced N. gonorrhoeae strains and Neisseria meningitidis strains, except N. meningitidis strain FAM18, which expresses a different pilin class that cannot antigenically vary and does not have a pilE G4 sequence [21], [22].
Fig. 1. Analysis of the pilE G4-associated sRNA and effect on pilin Av. 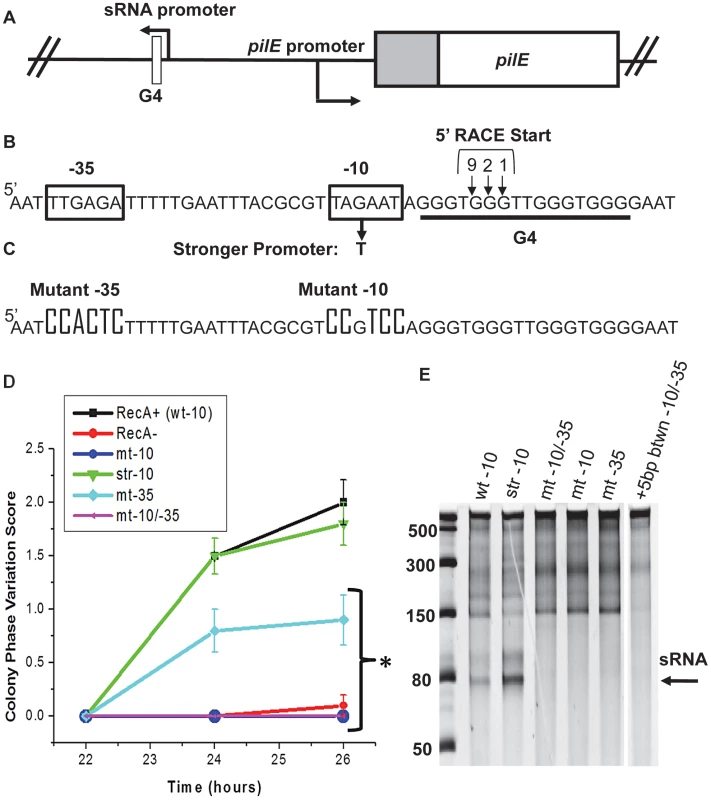
A. Chromosomal map of the pilE G4-associated sRNA locus. In gonococci, the G4-associated sRNA promoter is located upstream of pilE in the opposite orientation of transcription and is adjacent to the pilE G4. B. Predicted sRNA promoter sequence. Shown are the predicted sRNA −10 and −35 promoter sequences (boxed), the G to T mutation that makes a stronger promoter, and the predicted start sites detected by 5′RACE located within the pilE G4 forming sequence (underlined). Bases numbered and arrowed indicate the number of 5′RACE transformants showing the arrowed base as a start site. C. pilE G4 sRNA promoter mutagenesis. Knock-out mutation of the −10 and −35 promoter elements are shown D. Kinetic pilus-dependent colony phase variation assay. This standard assay measures the average number of visible pilus-dependent colony morphology changes occurring over time and is a surrogate measure for pilin Av [16]. Mutation of the −10 (mt-10) and/or −10 & −35 (mt-10/-35) promoter elements causes a loss in phase variation (these lines overlap), whereas mutation of the −35 promoter element alone results in an intermediate phenotype. P<0.05 as determined by two-tailed Student's T-test indicated by an asterisk. Bacteria having a stronger −10 promoter (str −10) element show the same levels of phase variation as the parental strain. Error bars represent the standard error of the mean for 10 colonies. A representative assay of n = 4 is shown. E. In vitro transcription products initated from sRNA promoter sequences. Shown is a 14% denaturing polyacrylamide gel containing in vitro transcribed RNA products using Escherichia coli RNA polymerase. Transcription was initiated from the wild-type promoter (wt-10), str-10, mt −10/−35, mt-10, mt −35, and a promoter that contains an insertion of 5 base pairs (bps) between the −10 and −35 elements (+5 bp btwn −10/−35). The RNA ladder is labeled in bases. The location of the sRNA transcript is shown by the arrow. The higher molecular weight bands represent irrelevant transcription products. To test whether the putative pilE G4-associated promoter was functional in N. gonorrhoeae, 5′ Rapid Amplification of cDNA Ends (5′RACE) adapted for bacterial small RNAs, was used to identify transcripts associated with the pilE G4 [23]. 5′RACE analysis detected a transcript initiating within the pilE G4 forming sequence and three adjacent start sites were identified (Fig. 1B). The existence of this RNA was confirmed by RNAseq analysis of N. meningitidis strain alpha14 (N. Heidrich and J. Vogel, personal communication). Moreover, this RNAseq analysis showed that levels of the pilE G4-associated sRNA in N. meningitidis are 125× lower than the corresponding pilE mRNA transcript suggesting that this sRNA is non-abundant (N. Heidrich and J. Vogel, personal communication). We have been unable to reliably detect this sRNA molecule by Northern Blot using conditions that allow other sRNA molecules to be detected (data not shown).
To determine whether the pilE G4-associated sRNA is required for pilin Av, promoter loss-of-function mutants were constructed and analyzed by the kinetic pilus-dependent colony morphology phase variation assay, which is a surrogate measure for pilin Av [16] (Fig. 1C & D). Mutation of the −10 sequence or both the −10 and −35 promoter elements caused a complete block of pilin Av, suggesting that transcription of the pilE G4-associated sRNA is required for pilin Av (Fig. 1D). Point mutations in the −10 element, a 5 bp insertion between the −10 and −35 elements, or mutation of the −35 element showed decreased pilin Av, suggesting that the level of transcription correlates with the amount of pilin Av (Fig. 1D and Fig. S1A & B). We also tested whether pilin Av was enhanced by substitution of the −10 element with a stronger promoter that was closer to the consensus sequence, but there was no change in the levels of pilin Av [20] (Fig. 1B & D). 5′-RACE analysis of the −10/−35 promoter mutant resulted in a loss of the 5′RACE product (data not shown). Taken together, these results are consistent with a model where pilin Av requires transcription of a sRNA which originates in the pilE G4 sequence, but that transcriptional initiation is not a rate limiting step in this process. However, we cannot ascertain how much more the stronger promoter contributes to transcription in N. gonorrhoeae.
Not surprisingly, a 5 bp insertion upstream of the −35 element, which is not part of the sRNA promoter, had no effect on pilin Av. However 5, 10, and 46 bp insertions downstream of the G4 forming sequence, but within the predicted sRNA transcript, also had no effect on pilin Av (Fig. 2, and Fig. S1B, C & E). Furthermore, deletion of 5, 9, 10 and 32 bps [15], which are also potentially part of the sRNA transcript, had no effect on pilin Av (Fig. 2 and Fig. S1C, D & F). Since these insertions and deletions downstream of the pilE G4 have no effect on pilin Av, and there is no ribosome binding site or open reading frame within the sRNA sequence, we conclude that the active form of this sRNA is non-coding.
Fig. 2. Genetic analysis of pilE G4-associated sRNA. 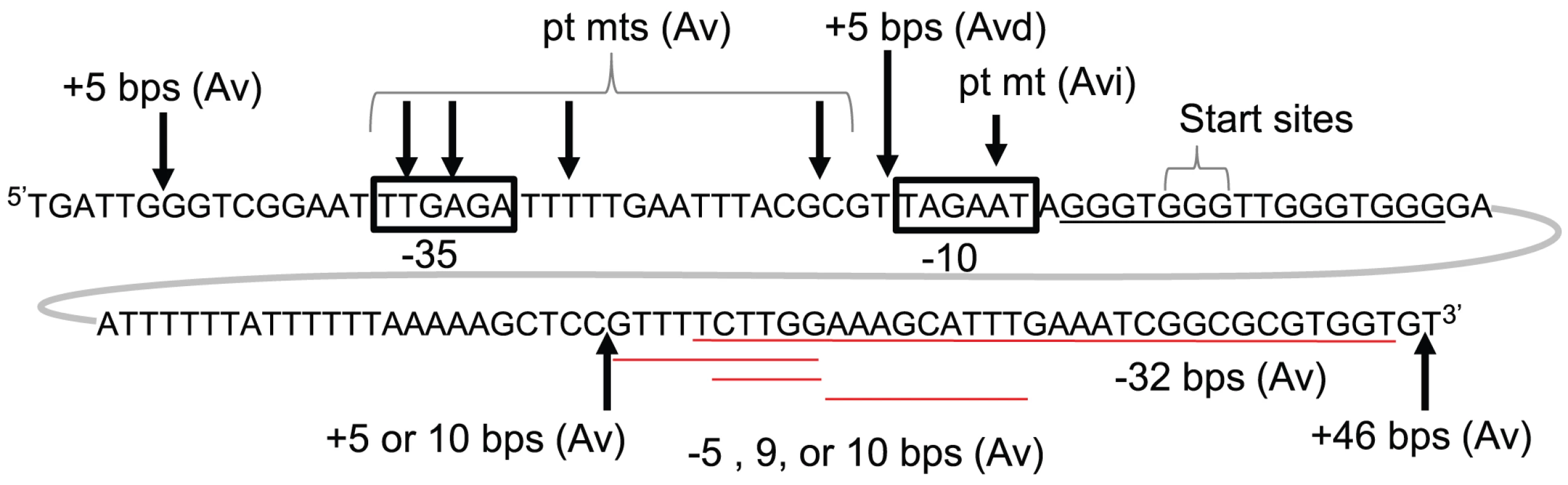
The pilE G4-associated sRNA promoter (boxed), start site (+1) and surrounding sequence are shown. Arrows indicate the location of insertions (+) and point mutations (pt mts) while deletions (−) are underlined in red. Whether these mutations resulted in wild-type levels of pilin Av (Av), an Av deficient (Avd), or an Av intermediate (Avi) phenotype is also indicated. These Av phenotypes were determined by a pilus-dependent colony phase variation kinetic assay [16] which is shown in Fig. S1. The pilE G4-associated sRNA promoter and it's in vitro transcription profile
We further characterized the sRNA promoter using in vitro transcription with Escherichia coli sigma70 RNA polymerase (Fig. 1E). A 74 bp run-off transcript was produced from a 180 bp double stranded DNA template having the wild-type and stronger promoters, where the stronger promoter showed increased transcript levels (Fig. 1E). These results are consistent with a lack of a transcriptional stop sequence within the fragment. No transcript was observed when the −10 element and/or the −35 element were mutated or when 5 bps of heterologous sequence was inserted between the −10 and −35 sequences (Fig. 1E). Primer extension analysis of cDNA synthesized from in vitro transcribed RNA confirmed the products initiated from the wild-type and stronger promoters, but did not detect a product initiated from the mutated promoter (Fig. S2). These transcription profiles are consistent with the pilin Av defects observed in gonococci containing these mutations.
Transcription of the pilE G4-associated sRNA acts upstream of recombination factors involved in pilin Av
RecA, RecG, and RuvB are recombination factors required for pilin Av [16], [24], [25]. RecA is a DNA recombinase, RecG is a 3′ to 5′ helicase involved in branch migration of Holliday junctions and RuvB along with RuvA is a 5′ to 3′ helicase that is also involved in branch migration of Holliday junctions. Previously, the Holliday junction processing double mutant recG/ruvB was found to be synthetically lethal upon recA induction in gonococci [25]. This lethal phenotype was rescued by a pilE G4 mutation that does not allow formation of the G4 structure which demonstrated that formation of the G4 structure was upstream of RecG, RuvB, and RecA during pilin Av [17].
To determine whether transcription of the pilE G4-associated sRNA occurs before the action of RecG, RuvB, and RecA, a −10/−35 G4-associated sRNA promoter mutation was introduced into the recG/ruvB double mutant, recA inducible strain. Gonococcal colonies in the absence of RecA induction do not show a growth defect because the bacteria cannot undergo pilin Av (Fig. S3A). Upon RecA induction, the −10/−35 promoter mutation rescues the synthetically lethal phenotype of the recG/ruvB double mutant showing that transcription of the pilE G4-associated sRNA, like formation of the G4 structure, acts before RecG, RuvB and RecA during pilin Av (Fig. S3B).
A heterologous promoter can substitute for the pilE G4-associated sRNA promoter
To confirm that the pilin Av defects observed in the pilE G4-associated sRNA promoter mutants were a direct consequence of transcriptional defects at the pilE G4 locus and to test whether RNA polymerase was playing an active role in pilin Av, we expressed the pilE G4-associated sRNA with a T7 promoter and T7 RNA polymerase. First, the sRNA −10 promoter element was replaced with a minimal T7 promoter and tested for the ability to drive transcription using T7 RNA polymerase in vitro (Fig. 3A). Next, T7 RNA polymerase was inserted downstream of a lac promoter at the neisserial intergenic complementation site (NICS) [26] in a gonococcal strain where the pilE G4-associated sRNA −10 promoter element was replaced with a minimal T7 promoter (Fig. 3B). Colony morphologies examined after 46 hours of growth demonstrated that expression of T7 RNA polymerase alone in the parental strain (T7 Pol.) does not affect pilin Av, whereas replacement of the sRNA −10 promoter element with a minimal T7 promoter (T7 Prom.) produced an Av deficient (Avd) phenotype because colonies showed a stable piliated morphology after many generations (Fig. 3C). This Avd phenotype was restored to wild-type levels of pilin Av by induction of T7 RNA polymerase expression with IPTG (T7 Prom./NICS::T7 Pol.) (Fig. 3C). These observations were confirmed by the pilus-dependent kinetic colony phase variation assay (Fig. 3D). Since only T7 RNA polymerase can restore pilin Av in the sRNA minimal T7 promoter mutant, we conclude that expression of the pilE G4-associated non-coding sRNA is essential for pilin Av, but that the identity of the polymerase providing the transcription is not important.
Fig. 3. A phage promoter can substitute for the pilE G4-associated sRNA promoter. 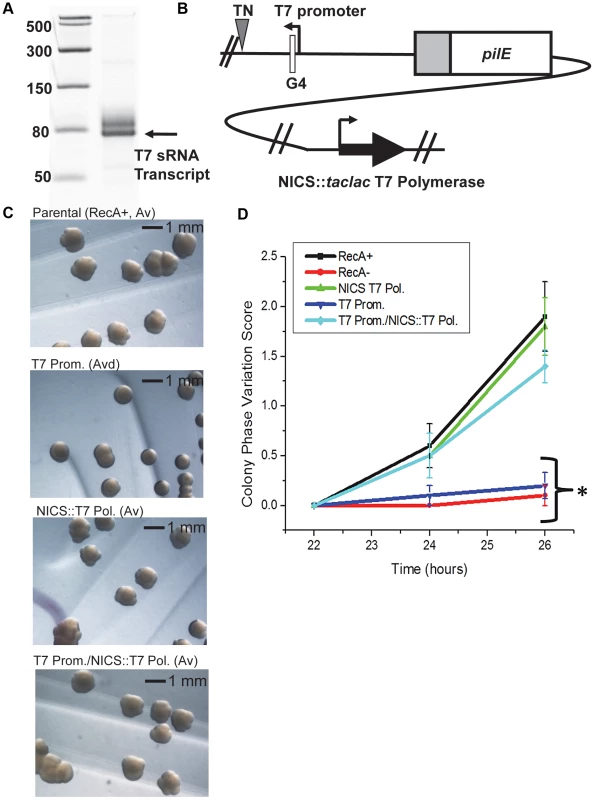
A. In vitro transcription. Shown is a 14% denaturing polyacrylamide gel containing T7 RNA polymerase-transcribed RNA products from a template containing the minimal T7 promoter element. The RNA ladder is labeled in bases. The higher molecular weight band is most likely an alternative start site directed by the T7 polymerase. B. The T7 promoter/T7 RNA polymerase strain. T7 RNA polymerase was cloned downstream of a dual taclac promoter which allows induction with IPTG [26]. The construct was transformed into a strain where the pilE G4 sRNA −10 promoter element was replaced with a minimal T7 promoter and inserted into the neisserial intergenic complementation site (NICS). [A triangle indicates a transposon (TN) encoding kanamycin resistance and was used as a marker for transformation to create the sRNA T7 promoter mutant]. C. Colony morphology of the T7 promoter and/or T7 polymerase strains. Colonies were grown for 46 hours with IPTG. The parental strain shows an Av phenotype, a strain expressing only T7 at the NICS (NICS T7 Pol.) has an Av phenotype, a strain where the G4 sRNA −10 element is replaced by the minimal T7 promoter (T7 Prom.) shows an Av-deficient (Avd) phenotype that is rescued with the expression of T7 polymerase (T7 Prom./NICS T7 Pol.). D. Kinetic pilus-dependent colony phase variation assay. Expression of T7 polymerase at the NICS (NICS T7 Pol.) does not affect phase variation whereas replacement of the G4 sRNA promoter with a minimal T7 promoter (T7 Prom.) causes a decrease in phase variation which is rescued by expression of T7 polymerase at the NICS (T7 Prom./NICS T7 Pol.). P<0.05 as determined by two-tailed Student's T-test (asterisks). Error bars represent the standard error of the mean for 10 colonies. A representative assay of n = 4 is shown. The pilE G4-associated sRNA acts in cis and its direction and orientation are required for function
Since transcription of the pilE G4-associated sRNA allows for melting of duplex DNA containing the pilE G4 sequence which then enables formation of the G4 structure, we reasoned that this sRNA might act in-cis. Therefore, we determined whether expression of the pilE G4-associated sRNA in-trans could complement a promoter mutant at the endogenous locus. The G4 sRNA region with a wild-type or stronger promoter was cloned and inserted at the ectopic NICS locus on the chromosome [26] (Fig. S4A). Kinetic pilus-dependent phase-variation assays demonstrated that expression of the sRNA at the ectopic locus does not complement a promoter mutant at the endogenous locus indicating that the pilE G4-associated sRNA acts in-cis and provides an essential step for the formation of the pilE G4 structure (Fig. S4B).
Because the pilE G4 sRNA acts in-cis, we determined whether the orientation and direction of the sRNA at the endogenous locus was also required for its function. Mutants with an inverted pilE sRNA, a pilE sRNA in reverse, and a pilE sRNA that is both in reversed and inverted were created (Fig. 4A&B). None of the mutants allowed pilin Av, indicating that both correct orientation and direction are required for the sRNA and G4 structure to function (Fig. 4C).
Fig. 4. The orientation and direction of the pilE G4 sRNA is required for its function. 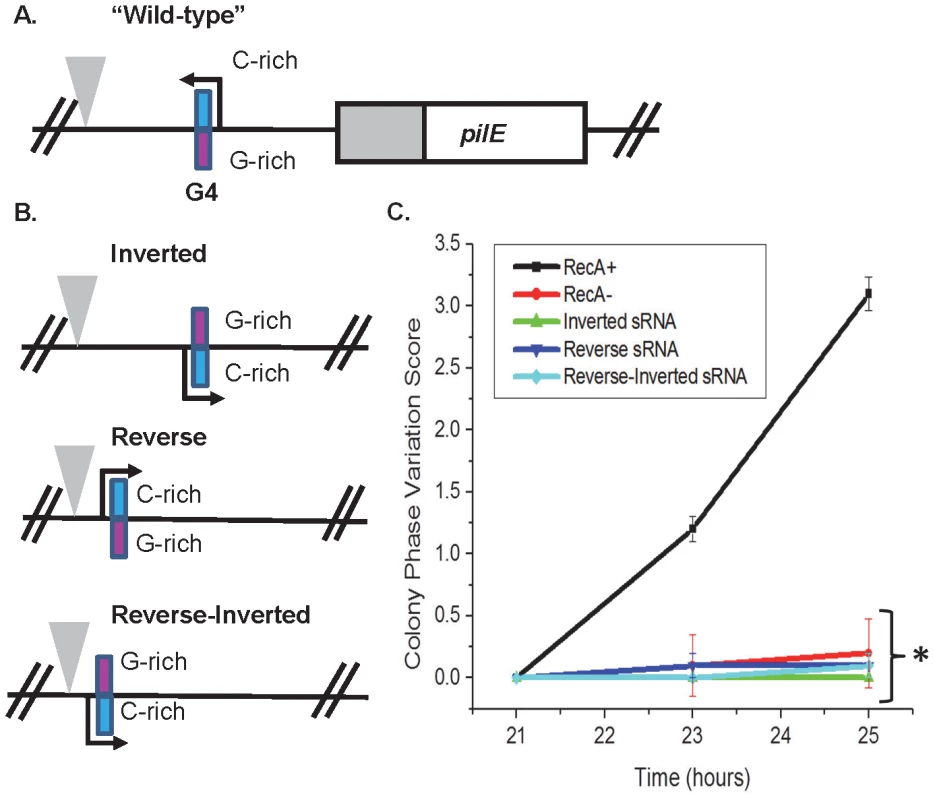
A. The pilE G4 sRNA region. The C-rich strand (blue) and G-rich strand (pink) are designated. A triangle indicates a transposon insertion used for selection of transformation. B. The pilE G4 sRNA endogenous locus mutants. A representation of the inverted, reverse, and reverse-inverted sRNA mutants is shown. A triangle indicates a transposon used as a marker for transformation. C. Kinetic pilus-dependent colony phase variation assay. The pilE G4 sRNA inverted, reverse, and reverse-inverted do not allow pilin Av. P<0.05 as determined by two-tailed Student's T-test (asterisks). Error bars represent the standard error of the mean for 10 colonies. A representative assay of n = 3 is shown. Discussion
We have established a direct link between transcription and G4 structure formation in the process of pilin Av. We show that transcription of the cis-acting pilE G4-associated non-coding sRNA is absolutely required for N. gonorrhoeae pilin Av and is therefore required to initiate the homologous recombination reactions leading to pilin Av. From this work, we can propose a working model for the initiation of pilin Av (Fig. 5). We propose that initiation of pilin Av begins with transcription of the pilE G4-associated sRNA. Pilin Av was not enhanced by substitution of the −10 promoter element with a stronger promoter or a phage promoter, which suggests that pilin Av has a maximal level of efficiency that is not influenced by increased pilE G4 formation potential. However, since we have not been able to detect the sRNA directly, this conclusion is based on the assumption that these promoters function similarly in N. gonorrhoeae as they do in E. coli. It was surprising that that transcription of the sRNA by T7 polymerase in N. gonorrhoeae resulted in wild type levels of phase variation. This result rules out a direct role for RNA polymerase in G4 structure formation and suggests that it is the act of transcription and/or the sRNA product that are critical for G4 structure formation and pilin Av.
Fig. 5. A working model of the initiation of pilin Av in N. gonorrhoeae. 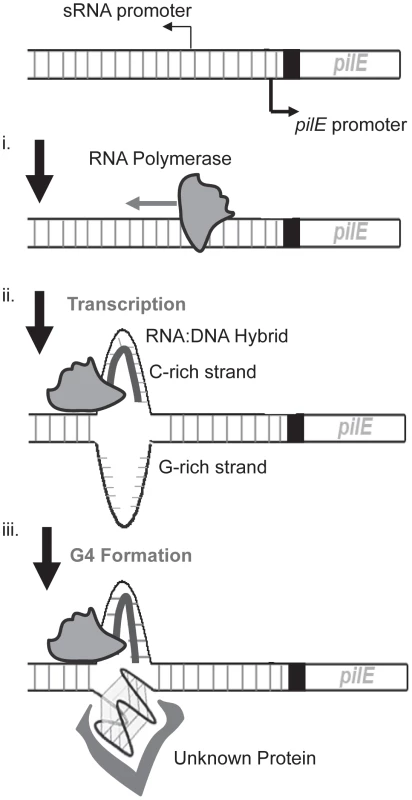
Shown is the pilin expression locus (pilE) and the upstream region containing the pilE G4-associated sRNA promoter. We propose that the initiation of pilin Av begins with transcription of the pilE G4-associated sRNA promoter (i). During transcription, a RNA∶DNA hybrid is made on the C-rich strand of the pilE G4 sequence (ii) facilitating the formation of a G4 structure by the unpaired G-rich strand, which may be mediated by structure specific binding proteins (iii). It is also possible that other processes involved in pilin Av downstream of transcription may be rate-limiting, thus negating any effect of increases in transcription rates. We reason that as transcription proceeds through the pilE G4 sequence, an RNA∶DNA hybrid could form between the sRNA and the C-rich strand leaving the G-rich strand unpaired to aid G4 structure formation, possibly aided by an as yet unknown protein (Fig. 5). How the formation of a G4 structure then initiates gene conversion is presently unknown, but the affinity for the pilE G4 structure by RecA, and the ability of the pilE G4 to stimulate RecA-mediated strand exchange [27], supports a model where the G4 structure acts to recruit recombination factors to the pilE region of the chromosome. In addition, it is likely that the G4 structure would cause a replication fork collapse when the replisome tries to synthesize past the G4 structure [11].
The lack of any observable pilin Av when the orientation and direction of both the sRNA and G4 structure were changed shows that this element does not act like enhancer elements found in eukaryotic cells. We did not observe any growth defect in strains where the G4-forming sequence was moved from the lagging strand onto the leading strand. This result suggests that either the structure doesn't form often enough to produce an observable reduction in viability when the G4 structure interrupts replication, or there are proteins that can remove the G4 structure to prevent a replication stop. The RecQ helicase is a likely candidate for an enzyme that could remove the G4 structure to prevent a replication blockage (Laty A. Cahoon, Kelly A. Manthei, Ella Rotman, James L. Keck and H. Steven Seifert, unpublished).
Regulatory cis and trans-encoded small RNAs have been identified in bacteria, but the pilE G4-associated sRNA does not fit either the cis or trans-encoded sRNA paradigm established for bacteria. Cis-encoded sRNAs are transcribed antisense to the genes they regulate whereas trans-encoded sRNA share limited complementarity. Once base paired with their target mRNA transcript, these regulatory sRNAs can influence the stability of, or modulate translation of, their target [28], [29]. The protein factor Hfq generally binds trans-encoded sRNAs and mediates base-pairing of the sRNAs to their target mRNAs; sRNAs that are not bound to Hfq are extremely labile [30]. Generally, cis-encoded antisense RNAs (asRNA) do not require Hfq [31]. In N. gonorrhoeae, Hfq is an RNA chaperone that functions as a pleiotropic regulator of RNA metabolism [32]. An hfq mutant was found to decrease the amount of pilE protein by an indirect mechanism that has yet to be determined. Since the pilE G4-associated sRNA is not an asRNA or a trans-encoded RNA and does not share complementarity to any known mRNA target, it is unlikely that this sRNA binds Hfq, but this has not been experimentally addressed. Some sRNAs have also been shown to bind and regulate proteins, such as the E. coli 6S RNA that interacts with RNA polymerase [29].
The pilE G4-associated cis-acting sRNA is transcribed in a region devoid of open reading frames and is not complementary to any expressed gene. In addition, the sRNA is most likely non-coding since mutations (5–46 bp insertions or 5–32 bp deletions) just downstream of the G-rich region have no effect on pilin Av (Fig. 2 and Fig. S1). It is possible that this sRNA performs other functions in this bacterium and could be processed or form a structure on its own. While we cannot rule out the possibility that the pilE G4-associated sRNA regulates other molecules, it is likely that the unwinding of the duplex DNA containing the pilE G4 sequence during transcription, and formation of the RNA∶DNA hybrid with the C-rich strand, allows formation of the G4 structure which is required for pilin Av (Fig. S3). It is interesting to note that two other predicted G4 forming sequences in the gonococcal genome also have putative promoters in a similar relative location. Formation of a G4 structure via transcription and occlusion of the C-rich stand has also been previously proposed to act during immunoglobulin class switching [33], [34]. Since both pilin Av and immunoglobulin class switching are high frequency recombination systems, there may exist other diversification systems that use transcriptionally induced alternative DNA structures to initiate programmed recombination reactions, and these may represent a specialized class of transcription-associated recombination systems [35].
While G4 DNA structures have been implicated in many biological processes in eukaryotic cells, less is known about their function in prokaryotic cells. In eukaryotes, G4 structures have been implicated in telomere metabolism [36], the regulation of genes required for normal cell growth and differentiation [37], potentiating genomic instability [38], the regulation of RNA transcription and translation [39], and mediating immunoglobulin gene diversification [33]. Proteins that bind or resolve G4 structures in both eukaryotes and prokaryotes have also been identified [40]–[42]. In prokaryotes, potential G4 forming sequences have been identified by bioinformatics, but functional analysis of these sequences has yet to be reported [43]. Our previous report demonstrating that the pilE G4 is essential for pilin Av in N. gonorrhoeae was the first study to show the requirement of a G4 structure for any biological process in a bacterial cell [17]. This work develops the mechanism of action further by showing that transcription of a cis-acting sRNA is critical for G4 structure formation to direct this specialized diversity generation system.
Materials and Methods
Bacterial growth conditions
E. coli One Shot TOP10 competent cells (Invitrogen) were grown in Luria-Bertani (LB) broth or on solid media containing 15 g/L agar at 37°C and used to propagate plasmids. E. coli selected for plasmids containing kanamycin or erythromycin resistance were selected on media containing 50 or 100 µg/ml of the respective antibiotic. Gonococcal strains were grown on GC Medium Base (Difco) plus Kellogg supplements (GCB) [22.2 mM glucose, 0.68 mM glutamine, 0.45 mM co-carboxylase, 1.23 mM Fe(NO3)3; all from Sigma] at 37°C in 5% CO2, when applicable 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added for induction. Gonococcal transformants were selected on media containing 50 µg/ml kanamycin or 2 µg/ml erythromycin.
Pilin Av assays
Gonococci were revived from frozen stocks, then after 24 hours of growth a single piliated colony was passaged onto solid media to obtain single isolated colonies. Colony variation was scored every 2 hours after 18 hours of growth up to 28 hours by observing the number of non-piliatied (P-) sectors arising over time. 10 colonies per strain were scored per assay. A colony that showed no P - sectors was given a score of 0, one P - sector was given a score of 1 and so forth. Colonies with 4 or more P - secors were given a score of 4.
5′ Rapid Amplification of DNA Ends (5′RACE)
Gonococci swabbed from a confluent lawn grown to 20 generations were resuspended (in RNA protect, Qiagen) and harvested for RNA (RNeasy Kit, Qiagen). 1 µg total RNA was treated with DNase I and enriched for non rRNA (using a Microbe Express Kit, Ambion). Then RNA was ethanol precipitated followed by the 5′RACE Protocol adapted for bacterial small non-messenger RNA [23].
Gonococcal sRNA promoter mutants and kinetic Pilus-dependent colony phase variation
A plasmid containing the region between USS2-pilArev2 from FA1090 (1-81-S2) pilE::mTn#9 recA6 [17] was digested at PacI and Mlu1 sites. Annealed linkers with PacI (TopA/BotA), an overlapping overhang and MluI ends for the −10 promoter mutant (10KOTop/10KOBot), stronger −10 mutant (10strTop/10strBot), T7 promoter mutant (T7G4PTop/T7G4PBot), or PacI and MluI ends for the 5 bp insertion between the −10 and −35 mutant (5in-10-35Top/5in-10-35Bot), were ligated to the PacI/MluI digested plasmid and transformed into TOP10 competent cells (Table S1). The −35 promoter mutant was created by digesting the USS2-pilArev2 plasmid (above) at MluI and EcoRV sites. Then the −35KO linker (35KOTop/35KOBot) was ligated to the digested plasmid. To increase the amount of homologous DNA available for recombination, a plasmid containing the −35 mutation was PCR amplified with GCUUSS2 and a −35 lengthening primer using KOD (Novagen) and cloned into pCRBluntII-Topo (Table S1). To create the −10 and −35 promoter mutant, a plasmid containing the −35 promoter mutation was digested with PacI and MluI. Then, the linker used to create the −10 promoter mutant was ligated to the digested plasmid (Table S1). Then gonococcal strain FA1090 (variant 1-81-S2) recA6 was transformed as described previously [44] and selected for kanamycin resistance. Kinetic phase variation assay was performed as described [16].
Gonococcal promoter region mutants
The USS2-PilArev2 plasmid was digested with PacI and MluI. Annealed linkers with MluI (G4Bot/G4Top), an overlapping overhang and PacI ends for the 10 bp insertion or deletion downstream of the G4 (in10bpDTop/in10bpDBot and del10bpDTop/del10bpDBot, respectively), the 5 bp insertion or deletion downstream of the G4 (in5bpDTop/in5bpDBot and del5bpDTop/del5bpDBot, respectively), were ligated to the PacI/MluI digested plasmid (Table S1). The 5 bp insertion upstream of the sRNA promoter mutant was created by digestion of the USS2-pilArev2 plasmid at unique MluI and EcoRV sites follow by ligation to the linker (in5bpUTop/in5bpUBot) (Table S1). Then gonococcal strain FA1090 (1-81-S2) recA6 was transformed as described [44] and selected for kanamycin resistance. Other mutants (Fig. 2, the 9 bp deletion and 46 bp insertion downstream of the G4) were identified during screens for the desired mutations listed above. Point mutants (Fig. 2) were identified in a previous screen [17] and backcrossed into the parental strain and selected for kanamycin resistance.
In vitro transcription and primer extension analysis
For E. coli RNA polymerase (holoenzyme, sigma saturated, Epicentre) in vitro transcription reactions, template DNA was PCR generated using KOD, gonococcal DNA having the desired pilE G4 sRNA region mutation, and primers RS1For and pilARevnested2Bot (Table S1). Then using 100 ng of purified template, in vitro transcription reactions were performed as specified by the manufacture. After 1–2 hours at 37°C, reactions were DNase I treated and run on a 14% polyacrylamide denaturing gel (UreaGel System, National Diagnostics). 0.5 µl of the above reactions were used for primer extension analysis, cDNA was synthesized using 3 pmol of 5′FAM-labeled PEx1 primer and Superscript III, EtOH precipitated, and sent for fragment analysis (Northwestern Genomics Core Facility). For T7 RNA polymerase in vitro transcription reactions, template DNA was PCR generated using KOD, gonococcal DNA having the replacement T7 promoter at the pilE G4-associated sRNA locus, and primers RS1For and T7G4invitro (Table S1). Then using 100 ng of purified template, in vitro transcription reactions were performed (Maxiscript Kit, Ambion).
The T7 promoter/T7 polymerase strain
Creation of the pilE G4-associated sRNA T7 promoter replacement mutant is detailed above. To create the IPTG inducible T7 polymerase strain, we used KOD and primers PacIT7F engineered to have a Pac I site and T7R to amplify T7 polymerase from BL21 (DE3) E. coli (Table S1). This PCR product was digested with PacI and cloned into the PacI and PmeI digested pGCC4 vector [26]. This construct was allowed to recombine into strain FA1090 (1-81-S2) at the NICS by liquid transformation and was selected for on media containing erythromycin [26], [45], [46]. Then chromosomal DNA from this strain was transformed into FA1090 (1-81-S2) recA6, and the pilE G4-associated sRNA T7 promoter replacement mutant. These mutants were then assayed for pilus-dependent colony phase variation [16].
Transformation of the Holliday junction processing mutant
The −10 and −35 pilE G4-associated sRNA promoter mutant plasmid detailed above was allowed to recombine into the Holliday junction processing mutant by liquid transformation [25], [46]. After limited exposure to IPTG, colonies were selected for kanamycin resistance, tested for erythromycin resistance associated with the ruvB functional deletion and chloramphenicol associated with the recG functional deletion. Gonococcal transformants containing the −10 and −35 sRNA promoter mutations were verified by sequencing analysis.
The pilE G4 sRNA associated ectopic expression strain
The region from the edge of the transposon insertion in RS1 which does not affect pilin Av [15] to 25 bps upstream of the pilE G4-associated sRNA −35 promoter element were PCR amplified using KOD and primers G4compR1PacI engineered to have a Pac I site and RS1For (Table S1). This PCR product was digested with PacI and cloned into the digested PacI and PmeI pGCC4 vector. This construct was allowed to recombine into the pilE G4-assoicated sRNA −10 promoter mutant strain at the NICS as described previously [44] and selected for erythromycin resistance. Transformants were verified by sequencing both the ectopic and endogenous pilE G4-assoiciated sRNA site. These mutants were then assayed for pilus-dependent colony phase variation [16].
The pilE G4-associates sRNA direction and orientation mutants
The sRNA inverted, reverse, reverse-inverted and respective −10 and −35 promoter element mutants were created by ligating a PacI digested 433 bp synthesized dsDNA fragment (IDT) to the PacI/EcoRV digested USS2-pilArev2 plasmid (Table S1). Then gonococcal strain FA1090 (1-81-S2) recA6 was transformed as described [44] and selected for kanamycin resistance.
Supporting Information
Zdroje
1. EdwardsJL, ApicellaMA (2004) The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin Microbiol Rev 17 : 965–981, table of contents.
2. CohenMS, CannonJG (1999) Human experimentation with Neisseria gonorrhoeae: progress and goals. J Infect Dis 179 Suppl 2: S375–379.
3. ParkHS, WolfgangM, van PuttenJP, DorwardD, HayesSF, et al. (2001) Structural alterations in a type IV pilus subunit protein result in concurrent defects in multicellular behaviour and adherence to host tissue. Mol Microbiol 42 : 293–307.
4. RudelT, van PuttenJP, GibbsCP, HaasR, MeyerTF (1992) Interaction of two variable proteins (PilE and PilC) required for pilus-mediated adherence of Neisseria gonorrhoeae to human epithelial cells. Mol Microbiol 6 : 3439–3450.
5. WolfgangM, LauerP, ParkHS, BrossayL, HebertJ, et al. (1998) PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol 29 : 321–330.
6. DanaherRJ, LevinJC, ArkingD, BurchCL, SandlinR, et al. (1995) Genetic basis of Neisseria gonorrhoeae lipooligosaccharide antigenic variation. J Bacteriol 177 : 7275–7279.
7. HagblomP, SegalE, BillyardE, SoM (1985) Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature 315 : 156–158.
8. JenningsMP, HoodDW, PeakIR, VirjiM, MoxonER (1995) Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol 18 : 729–740.
9. SternA, BrownM, NickelP, MeyerTF (1986) Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell 47 : 61–71.
10. BoslegoJW, TramontEC, ChungRC, McChesneyDG, CiakJ, et al. (1991) Efficacy trial of a parenteral gonococcal pilus vaccine in men. Vaccine 9 : 154–162.
11. CahoonLA, SeifertHS (2011) Focusing homologous recombination: pilin antigenic variation in the pathogenic Neisseria. Mol Microbiol 81 : 1136–1143.
12. HamrickTS, DempseyJA, CohenMS, CannonJG (2001) Antigenic variation of gonococcal pilin expression in vivo: analysis of the strain FA1090 pilin repertoire and identification of the pilS gene copies recombining with pilE during experimental human infection. Microbiology 147 : 839–849.
13. VinkC, RudenkoG, SeifertHS (2011) Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol Rev
14. SwansonJ, BergstromS, BoslegoJ, KoomeyM (1987) Gene conversion accounts for pilin structural changes and for reversible piliation “phase” changes in gonococci. Antonie Van Leeuwenhoek 53 : 441–446.
15. KlineKA, CrissAK, WallaceA, SeifertHS (2007) Transposon mutagenesis identifies sites upstream of the Neisseria gonorrhoeae pilE gene that modulate pilin antigenic variation. J Bacteriol 189 : 3462–3470.
16. SechmanEV, RohrerMS, SeifertHS (2005) A genetic screen identifies genes and sites involved in pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol 57 : 468–483.
17. CahoonLA, SeifertHS (2009) An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 325 : 764–767.
18. RenJ, ChairesJB (1999) Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry 38 : 16067–16075.
19. MunchR, HillerK, GroteA, ScheerM, KleinJ, et al. (2005) Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21 : 4187–4189.
20. ShultzabergerRK, ChenZ, LewisKA, SchneiderTD (2007) Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res 35 : 771–788.
21. AhoEL, BottenJW, HallRJ, LarsonMK, NessJK (1997) Characterization of a class II pilin expression locus from Neisseria meningitidis: evidence for increased diversity among pilin genes in pathogenic Neisseria species. Infect Immun 65 : 2613–2620.
22. HelmRA, SeifertHS (2010) Frequency and rate of pilin antigenic variation of Neisseria meningitidis. J Bacteriol 192 : 3822–3823.
23. ArgamanL, HershbergR, VogelJ, BejeranoG, WagnerEG, et al. (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11 : 941–950.
24. KoomeyM, GotschlichEC, RobbinsK, BergstromS, SwansonJ (1987) Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117 : 391–398.
25. SechmanEV, KlineKA, SeifertHS (2006) Loss of both Holliday junction processing pathways is synthetically lethal in the presence of gonococcal pilin antigenic variation. Mol Microbiol 61 : 185–193.
26. MehrIJ, SeifertHS (1998) Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol 30 : 697–710.
27. Kuryavyi V, Cahoon LA, Seifert HS, Patel DJ, (2012) RecA-Binding pilE G4 Sequence Essential for Pilin Antigenic Variation Forms Monomeric and 5′ End Stacked Dimeric Parallel G-Quadruplexes. Structure In Press.
28. GeorgJ, HessWR (2011) cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75 : 286–300.
29. StorzG, VogelJ, WassarmanKM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43 : 880–891.
30. AndradeJM, PobreV, MatosAM, ArraianoCM (2012) The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18 : 844–855.
31. WatersLS, StorzG (2009) Regulatory RNAs in bacteria. Cell 136 : 615–628.
32. DietrichM, MunkeR, GottschaldM, ZiskaE, BoettcherJP, et al. (2009) The effect of hfq on global gene expression and virulence in Neisseria gonorrhoeae. FEBS J 276 : 5507–5520.
33. MaizelsN (2005) Immunoglobulin gene diversification. Annu Rev Genet 39 : 23–46.
34. DuquetteML, HandaP, VincentJA, TaylorAF, MaizelsN (2004) Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 18 : 1618–1629.
35. AguileraA (2002) The connection between transcription and genomic instability. Embo J 21 : 195–201.
36. LippsHJ, RhodesD (2009) G-quadruplex structures: in vivo evidence and function. Trends Cell Biol 19 : 414–422.
37. GonzalezV, HurleyLH (2010) The c-MYC NHE III(1): function and regulation. Annu Rev Pharmacol Toxicol 50 : 111–129.
38. ZhaoJ, BacollaA, WangG, VasquezKM (2010) Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci 67 : 43–62.
39. MillevoiS, MoineH, VagnerS (2012) G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA
40. KangSG, HendersonE (2002) Identification of non-telomeric G4-DNA binding proteins in human, E. coli, yeast, and Arabidopsis. Mol Cells 14 : 404–410.
41. MatillaI, AlfonsoC, RivasG, BoltEL, de la CruzF, et al. (2010) The conjugative DNA translocase TrwB is a structure-specific DNA-binding protein. J Biol Chem 285 : 17537–17544.
42. WuX, MaizelsN (2001) Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res 29 : 1765–1771.
43. RawalP, KummarasettiVB, RavindranJ, KumarN, HalderK, et al. (2006) Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res 16 : 644–655.
44. StohlEA, SeifertHS (2001) The recX gene potentiates homologous recombination in Neisseria gonorrhoeae. Mol Microbiol 40 : 1301–1310.
45. MehrIJ, LongCD, SerkinCD, SeifertHS (2000) A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154 : 523–532.
46. SerkinCD, SeifertHS (2000) Iron availability regulates DNA recombination in Neisseria gonorrhoeae. Mol Microbiol 37 : 1075–1086.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Biosafety Level-4 Laboratories in Europe: Opportunities for Public Health, Diagnostics, and ResearchČlánek Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis inČlánek The Tomato Prf Complex Is a Molecular Trap for Bacterial Effectors Based on Pto TransphosphorylationČlánek Schmallenberg Virus Pathogenesis, Tropism and Interaction with the Innate Immune System of the HostČlánek The Importance of Prions
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2013 Číslo 1- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Preclinical Therapy of Disseminated HER-2 Ovarian and Breast Carcinomas with a HER-2-Retargeted Oncolytic Herpesvirus
- Biosafety Level-4 Laboratories in Europe: Opportunities for Public Health, Diagnostics, and Research
- Glucose Phosphorylation Is Required for Persistence in Mice
- and Host Determinants of Susceptibility to Invasive Candidiasis
- TNFα-Mediated Liver Destruction by Kupffer Cells and Ly6C Monocytes during Infection
- Transcription of a -acting, Noncoding, Small RNA Is Required for Pilin Antigenic Variation in
- Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in
- Intravital Placenta Imaging Reveals Microcirculatory Dynamics Impact on Sequestration and Phagocytosis of -Infected Erythrocytes
- The Tomato Prf Complex Is a Molecular Trap for Bacterial Effectors Based on Pto Transphosphorylation
- Schmallenberg Virus Pathogenesis, Tropism and Interaction with the Innate Immune System of the Host
- Make It, Take It, or Leave It: Heme Metabolism of Parasites
- Viral and Bacterial Interactions in the Upper Respiratory Tract
- The Importance of Prions
- Loss and Retention of RNA Interference in Fungi and Parasites
- Granzyme A Produced by γδ T Cells Induces Human Macrophages to Inhibit Growth of an Intracellular Pathogen
- Innate Sensing of Chitin and Chitosan
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Biosafety Level-4 Laboratories in Europe: Opportunities for Public Health, Diagnostics, and Research
- Loss and Retention of RNA Interference in Fungi and Parasites
- Make It, Take It, or Leave It: Heme Metabolism of Parasites
- Innate Sensing of Chitin and Chitosan
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



