Increase in RNASEL gene expression by miR-29-3p inhibitors in HEK293T cells
Authors:
A. Maleki 1; M. Ravanshad 1; F. Kouhkan 2
Authors‘ workplace:
Department of Virology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
1; Department of Molecular Biology and Genetic Engineering, Stem Cell Technology Research Center, Tehran, Iran
2
Published in:
Epidemiol. Mikrobiol. Imunol. 69, 2020, č. 3, s. 116-120
Category:
Original Papers
Overview
Background: RNase L is known as a terminal component of antiviral and Interferon (IFN) pathways in mammalian cells. On the other hand, the human miR-29 family of microRNAs (miRs) has three mature members, miR-29a, miR-29b, and miR-29c. miR-29 is encoded by two gene clusters and the family members have multifunctional roles in various biological processes.
Objectives: To determine the potential role of miR-29 in the regulation of RNASEL gene expression by designing inhibitors against its targeting miRNA, miR-29a-3p and evaluate the RNase L expression.
Material and Methods: After selecting miR-29a-3p as a main regulating miRNA for RNASEL in silico, two inhibitors were designed against it and synthesized. Synthesized strands were made double-stranded DNA oligos, treated with T4 polynucleotide kinase (PNK), cloned into the pCDH-CMV-MCS-EF1-cGFP-T2A-Puro vector and transformed into DH5α. Colony PCR and sequencing was done for affirmation. Then the miR-29a-3p inhibitors were transfected into HEK-293T cell line and RNASEL gene expression was analyzed.
Results: The miR-29a-3p inhibitors decreased miR-29a-3p expression in vitro. In addition, miR-29a-3p expression reduction resulted in an increase of RNASEL gene expression.
Conclusions: miR-29a-3p inhibitors could increase in RNASEL gene expression which potentially affects the antiviral/IFN pathway. The inhibitors could be considered as drug candidates in different diseases especially viral infections.
Keywords:
Anti-mir – mir-29a-3p – real-Time pcr – rnase l
INTRODUCTION
microRNAs (miRs) are an important class of small, endogenous, non-coding RNA 20-22 nucleotides that mediate post-transcriptional gene silencing [1]. miRNAs bind to 3ˈUTR, of their mRNA targets and prevent their translation through preventing interaction 3ˈUTR and 5ˈUTR ends of mRNA essential for initiation of translation and/or may bind to their target mRNA and recruit of the miRNA induced silencing complex (miRISC). However, miRNAs are not restricted to attachment only to 3ˈ and 5ˈ UTRs of mRNAs as they may also interact with the open reading frame, ORF [1–3]. In mammalian cells, miRNA biogenesis and processing pathway typically have several steps that convert primary miRNA transcripts (pri-miRNAs) into the biologically-active and mature forms of miRNA. Like mRNAs, pri-miRNAs have been transcribed by RNA polymerase II enzyme in the nucleus and they are recognized and cropped by Drosha complex, a ribonuclease enzyme, into new precursor miRNA (pre-miRNA) which has a hairpin structure [4]. Pre-miRNA will be translocated by exportin-5 protein and Ran-GTP into the cytoplasm and now needs to proceed by second ribonuclease enzyme, Dicer RNase III, into a ~22 nt long miRNA duplex [2]. However, some miRNAs like miR-29b mostly return to the nucleus of the cell and in this case, its distinctive hexanucleotide terminal motif acts as a nuclear localization element (NLE) [5]. The miR-29 family is one of the most important miRNA families in human that has a multifunctional role in a wide variety of molecular pathways in mammalian cells. The miR-29 family consists of three members located in two genomic loci in clusters: miR-29-a/b1 on chromosome 7 in human and chromosome 6 in mouse, miR-29-c/b2 on chromosome 1 in human and mouse [6]. The miR-29 family members differ only in two or three bases [7].
Several studies have shown the important effect of miRNA regulation of the immune response especially of the Interferon (IFN) pathway [8]. From Toll-Like Receptors (TLRs) to Interferon receptor (IFNAR1/2) cascade mediators like Signal Transducer and activator of transcription 2 (STAT1/2), Janus Kinase 1 (JAK1), Interferon Regulatory Factor 9 (IRF9), Interleukin-1 Receptor Associated Kinases (IRAKs) and IFN-Regulated Genes (IRGs) are targeted and regulated by miRNAs [9, 10]. Importantly, miR-29a is one of the miRNA families involved in IFN pathway regulation and miRNA-29a has been shown to decrease IFNAR1 expression on murine thymic epithelial cells. Besides, it has been shown that gene knockout mice lacking miR-29a have increased expression of IFNAR1 and also hypersensitivity to poly I:C treatment [11].
The mammalian innate immune system recognizes double-stranded RNA(dsRNA) as a sign of cell damage and potentially viral infection. Activation and degrade of intracellular RNA is mediated by RNase L which mechanism is considered as a protective process [12]. IFNs induce the expression of oligoadenylate synthetases, OAS, which synthesize 2ˈ-5ˈ oligoadenylates (2-5A) in response to especially viral infection. 2-5A triggers dimerization and conversion of latent RNase L to its activated form. RNase L is a 741 amino acid latent endoribonuclease expressed in mammalian cells [13]. RNase L has a strong antiviral activity and it implements its activity via viral genome/mRNA degradation, cellular mRNA/rRNA degradation and amplification of IFN production [14, 15].
Objectives: The current study aimed to evaluate miR-29a-3p and RNASEL expression in presence of miR-29a-3p inhibitors in the HEK-293T cell line. The studies suggest an important role for miR-9a-3p regulation of RNASEL expression.

MATERIALS AND METHODS
In Silico Analysis and miR Inhibitor Design
The miRNAs that targeted RNASEL were identified from miRNA/target databases, TargetScanHuman and miRTarBase online tools [22, 23]. First of all, miRNAs, which were predicted in both databases with the highest score for targeting RNASEL, were listed and thus miR-29a-3p was selected.
A short or small hairpin RNA (shRNA) is an artificial RNA molecule with a tight hairpin turn that can be used to silence target gene expression via RNA interference (RNAi) [24]. The two strands of the shRNA were designed by a unique manual method. The sequences of the shRNA were engineered from the miR-29a-3p sequence in miRBase [25]. The reverse of the miR-29a-3p sequence was formulated and then was modified for what was predicted to be the best inhibitory effect on miR-29a-3p. A second series of shRNA sequences were designed for comparison of efficacy. The secondary structure of the designs were analyzed by RNAfold [26]. The first and second design of anti-miR-29a-3p were named AD-1 and AD-2, respectively. All of the sequences were synthesized and tested (Table 1).
Cloning of Double-Stranded Anti-miR into Plasmid pCDH and Transformation
For cloning into an expression vector, the sense and antisense were attached together. Briefly, 15µl of each sense and antisense strands (from 100 µM stock solution) was mixed with 15µl of Tris/NaCl buffer (pH = 8) and 30µl H2O. Regarding the secondary structure in sequences, it was necessary to reduce the annealing temperature gradually. Specifically, for the first round, the temperature was set to 110 °C and immediately reduced to 90 °C for 1 min then was gradually decreased to 37 °C as once per 0.5 °C. Finally, the reaction was incubated at 37 °C for 1 h. In order to prepare the sample for ligation, phosphorylation of newborn dsDNA was essential. Therefore, dsDNA was treated with T4 PNK (T4 Poly Nucleotide Kinase) (ThermoFisher, USA) according to the manufacturer’s protocol [27].
The pCDH-CMV-MCS-EF1-cGFP-T2A-Puro vector (Stem Cell Technology, Iran) was selected as an expression vector and the double-stranded DNA was cloned into the EcoRI and XbaI cleavage sites of pCDH. Briefly, at the first pCDH plasmid backbone was cleaved by EcoRI and XbaI restriction enzyme (ThermoFisher, USA) and then, was cleaned up from rest of restriction enzymes and digested segments via plasmid clean-up kit (MN, Germany). In order to assess the digestion, samples were electrophoresed at 1% agarose gel at the side of undigested plasmid and 1Kb gene marker. Therefore, the digested pCDH backbone was incubated with an insert segment of double-stranded Anti-miR in the presence of T4 DNA Ligase and its buffer (ThermoFisher, USA). Ligated plasmids were transformed into DH5α strain of Escherichia Coli bacterium through 0.1M CaCl2 and heat-shock assay.
Colony selection, PCR and Sequencing
Ten bacterial colonies were selected from an LB agar medium dish infected by transformed DH5α. Then each of the colonies was sub-cultured using fresh LB agar medium (Merck, Germany) and were used directly as a template for PCR. A pair of primers from upstream to downstream of multiple cloning sites (MCS) of pCDH were designed. The primers were designed by primer 3 website. PCR products were run on 2% agarose gel electrophoresis. Cytomegalovirus promoter region (CMV-F) primer (AATGGGCGGTAGGCGTGTA) and Elongation Factor1 promoter region (EF1-R) primer (GGACTGTGGGCGATGTGC).
All bacterial colonies contain our shRNA sequence were sequenced (Macrogen, South Korea). Plasmid extraction was done using the manufacture protocol and a Miniprep Kit (Qiagen, Germany). The same primer pairs that are utilized in colony PCR were been used for sequencing.
Transfection of HEK-293T Cells
HEK-293T cells (Stem Cell Technology, Iran) were propagated in DMEM (Dulbecco’s Modified Eagle Medium) (Gibco, USA) supplemented with 5% fetal bovine serum (FBS), 100 U.ml penicillin, 100µg.ml streptomycin and 2% sodium carbonate at 5% CO2 at 37 °C. One day before transfection, HEK-293T cells were seeded around 4.0 × 105 in each well of six-well plate (SPL, Korea). In order to assay that plasmids have an ability to express correctly in eukaryote cells or not, 1µg of each AD-1 and AD-2 plasmids were transfected into HEK-293T cells at 80 to 90% confluency using Lipofectamine 2000 (ThermoFisher, USA) according to manufacturer’s instructions.
RNA Extraction, cDNA Synthesis, and Real-Time PCR
After 48 h, the transfected cells were evaluated using an inverted fluorescence microscope (Nikon TE200) and the total cellular RNA extracted with RNXplus solution (CinnaGen. Iran). The RNA quantity and quality were confirmed via using a UV-Vis spectrophotometer (Biochrom TM WPA Biowave II, UK) and by electrophoresis on 1% agarose gel, respectively. qRT-PCR was performed using 1 µg RNA converted into cDNA using a RevertAid RT reverse transcription kit (ThermoFisher, USA) according to the manufacturer’s instructions. Two types of cDNAs were synthesized for each sample, total and specific, which were synthesized with Random Hexamer (commercial) and specific stem-loop primer for miR-29a-3p (manual design 5-GTCGTATGCAGAGCAGGGTCCGAGGTATTCGCACTGCATACGACTGGGGTA-3), respectively. qRT-PCR has been performed using Evagreen real-time master mix (Solis Biodyne, Estonia) in Step-One plus machine (Applied Biosystem, USA) for 15 min at 95 °C for initial denaturation, 40 cycles including 10 seconds at 95 °C for denaturation, 20 seconds at 56 oC (miR-29a--3p)/60 °C (RNAESL) for annealing and 30 seconds at 72 °C for extension.
Data analysis
For statistical evaluation of the increase in RNASEL expression, data were analyzed for significant differences by Prism 5.0 (GraphPad Company, CA, USA) via using appropriate test, One-Way ANOVA. The probability of 5% was assumed statistically significant (P < 0.05).
RESULTS
AD-1 and AD-2 Suppress miR-29a-3p Expression
The complementary sequences were hybridized and phosphorylated by T4 PNK treatment. T4 PNK-treated double-stranded DNAs were cloned by T4 DNA ligase into EcoRI and XbaI cloning sites. The bi-directional sequencing results for the dsDNA of anti-miR constructs were determined to be inserted correctly into the pCDH vector (Figure 1). The plasmids then were transfected to HEK293T cell line and transfection efficiency was confirmed through observation of GFP (Green Fluorescent Protein) positive cells using Nikon Eclipse TE2000 fluorescent microscope. After 48 h incubation, total RNA that was extracted, converted to cDNA and used as a template of qRT-PCR were normalized with housekeeping genes HPRT and SNORD47 for RNASEL and miR-29a-3p, respectively. Based on observed data from the above genes, the normalized fold change data (2-ΔΔCt) were analyzed and showed a significant decrease in miR-29a-3p expression level after treatment of both AD-1 and AD-2 Anti-miR-29a-3p plasmids (****P < 0.001). (****P < 0.0001) (Figure 1).
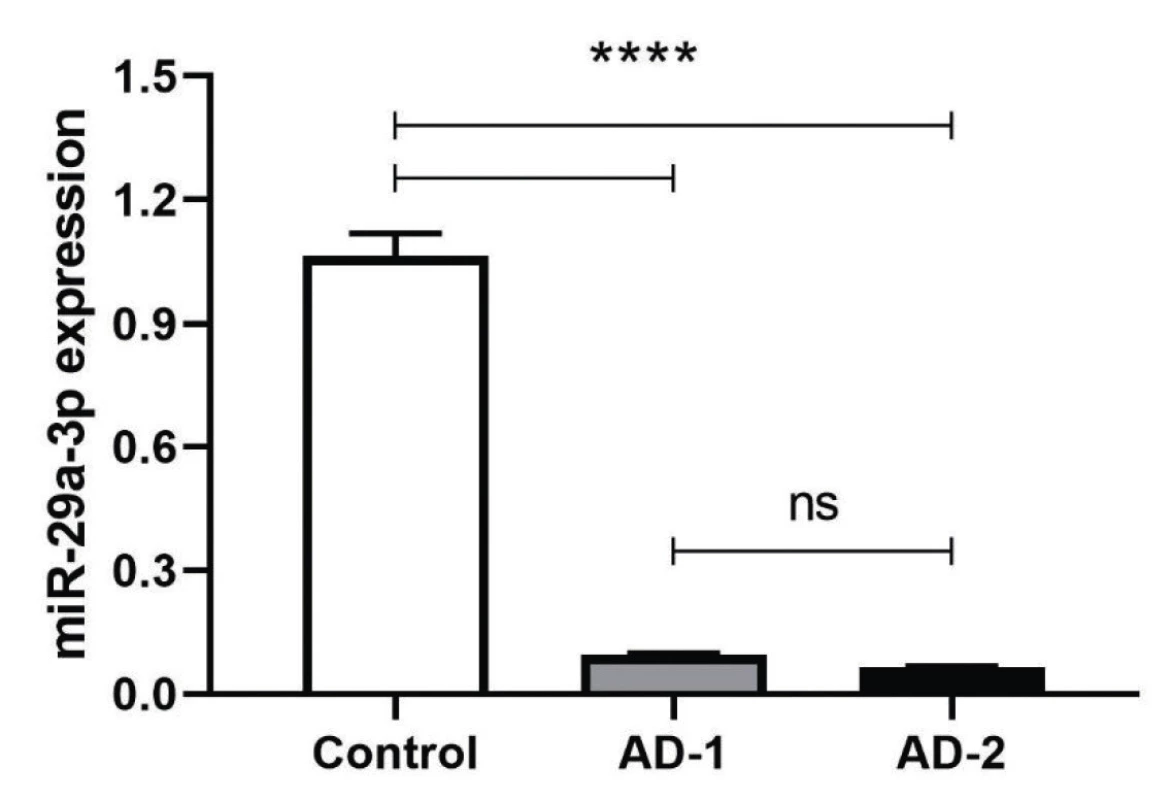
AD-1 and AD-2 Induce Increased RNAESL Expression
It was anticipated the treatment of the HEK-293T cells with AD-1 and AD-2 Anti-miR-29a-3p could increase the expression level of miR-29a-3p’s target, RNASEL. It was tested by Real-Time PCR with primers targeting RNASEL. The efficiency of AD-1 and AD-2 plasmids in the inhibition of miR-29a-3p were approximately the same and were not different, that’s why both of Anti-miR-29a-3p plasmids were pooled and transfected, then RNASEL gene expression was measured and normalized with internal control, HPRT. Treated cells had a significant difference in transcriptional expression of RNASEL in comparison to the scrambled control after 48 h (*P < 0.0155) and after 72 h (**P < 0045) (Figure 2). Although the increased percentage of RNASEL expression was about 10% and 20% after 48 and 72 h, respectively, it seems it could be higher in the next hours if was evaluated.
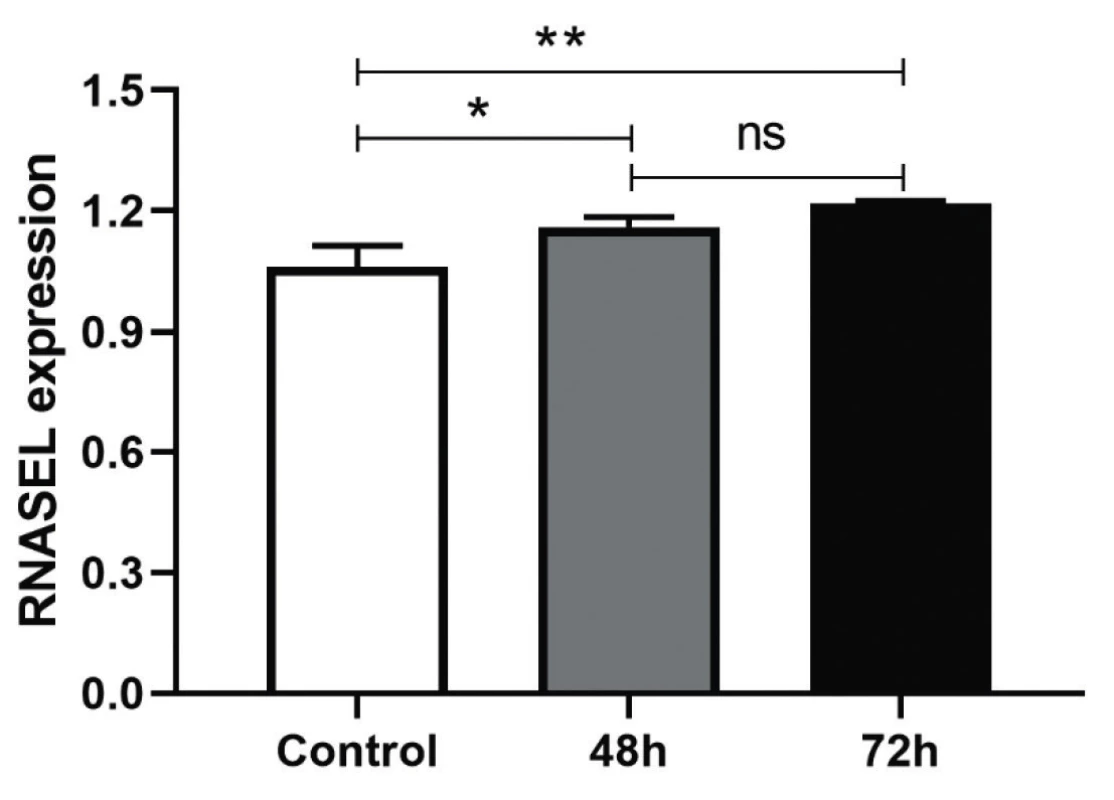
DISCUSSION
The miR-29 family, especially miR-29a-3p, has so important regulating role in different molecular processes and pathways. miR-29 has a direct role in lifespan and reproduction in a gender-dependent manner [16]. Song et al found that miR-29 family members can regulate glucose metabolism in type 2 diabetes mellitus (T2DM) through inhibiting SPARC, secreted protein acidic rich in Cysteine, negatively [17]. As another example, Yu et al observed that the coding region of Human cytochrome P450 2C19 (CYP2C19) has been targeted by miR-29a-3p and miR-23a-3p which has a suppressive effect on CYP2C19 expression in HepaRG cells [18]. There has been shown overexpression of miR-29b leads Hepatic stellate cells (HSCs) to remain in the quiescent state and prevents the expression of Extra Cellular Matrixes (ECMs) like Col1a1 and Col1b1 [19]. On the other hand, Park et al found that miR-29 family members can upregulate the p53 expression and therefore induce apoptosis in a p53-dependent manner [21]. The miR-29 family has also been linked to skin diseases, for instance, miR-29b has been downregulated during skin wound healing that improves angiogenesis and synthesis of collagen [20]. These data approve the important role of miR-29a-3p in diverse biological pathways.
Additionally, 2-5A/RNase L pathway is a host RNase pathway involved in various viral infections. For instance, an elevated level of RNase L and presence of the degraded RNA that is characteristic of RNase L activation were correlated with IFN-mediated inhibition of vaccinia virus [28], reovirus [29] and infection of encephalomyocarditis virus (EMCV) [30]. RNase L also could induce autophagy in response to viral infections. Chakrabarti and his colleagues showed infection of vesicular stomatitis virus (VSV) or encephalomyocarditis virus (EMCV) triggered a higher level of autophagy in wild-type mouse embryonic fibroblast (MEF) than RNase L-null MEF [31]. In addition to the anti-viral feature, RNase L has a role in the regulation of cell cycle [32], apoptosis [33], prevention of translation by cleavage of 18s & 28s rRNA [34].
In this mean, investigation of the role of miR-29 in immune responses and mechanisms of defense of cells needs to be considered more. In this study, we showed the role of miR-29a-3p in the regulation of RNASEL expression which is important in the anti-viral response. Consistent with our results, Lee et al have shown that miR-29 family represses RNASEL expression in some cell lines. They demonstrated miR-29 family has 4 target sites in 3’-UTR region of RNASEL’s mRNA and a novel oncogenic role in CML (chronic myelogenous leukemia). It has been demonstrated that loss of RNase L inhibits proliferation in vitro and tumor growth in a xenograft model as well [35]. Hassel and his colleagues also showed miR-29 family could repress the expression of RNase L protein level across several cells. However, unlike present data in this study, they did not observe any change in RNASEL mRNA level [36]. There are some studies shown RNASEL is targeted by other miRNAs. A previous report revealed that RNASEL is a direct target of miR-146b. Li et al showed RNase L protein level was increased in the presence of miR-146b inhibitors compared with control in H9c2 cell line [37]. It seems augment of the RNase L might be an effective strategy to fight against viral and bacterial infections. It would be roughly said that the inhibition of miR-29a-3p can help to augment RNASEL expression and activity. However, it totally depends on the type of infection and generally miR-29-3p expression level. In the case of increased level of miR-29a-3p expression, inhibition and reduction level of the mentioned miR-29a-3p could increase in RNASEL mRNA significantly. But in the case of decreased level miR-29a-3p expression, miR-29-3p inhibition has too limited effect on RNASEL mRNA level.
CONCLUSION
The significantly decreased transcriptional expression level of RNASEL in this study showed at least miR-29a-3p can suppress RNASEL by targeting and cleaving its mRNA. miR-29a-3p could prevent the translation of RNASEL mRNA as well. Despite the significant but low level of RNASEL augment (about 20%), a further assessment like western blot assay or luciferase assay could determine the exact effectiveness of miR-29a-3p suppression in RNSAEL’s biological role.
Acknowledgment
We are so grateful to Dr. Ralph A. Tripp for the general edition of the manuscript.
Ethics approval: Ethics committee of Tarbiat Modares University approved this study (IR.TMU.REC.1394.277).
Do redakce došlo dne 9. 11. 2019.
Corresponding author:
Mehrdad Ravanshad
No7, Jalal Al-Ahmad Street, Chamran Highway
Tarbiat Modares University
Tehran
Iran
P.O.BOX: 14115-331
e-mail: Ravanshad@modares.ac.ir
Sources
1. Lee HJ. Exceptional stories of microRNAs. Experimental Biology and Medicine, 2013;238(4):339–343.
2. Kim VN, Nam JW. Genomics of microRNA. Trends in Genetics, 2006;22(3):165–173.
3. Lin SL, Kim H, Ying SY. Intron-mediated RNA interference and microRNA (miRNA). Front Biosci, 2008;13 : 2216–2230.
4. Suzuki HI, Miyazono K. Emerging complexity of microRNA genera-tion cascades. Journal of biochemistry. 2011;149(1):15–25.
5. Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science, 2007;315(5808):97–100.
6. Ouyang YB, Xu L, Yue S, Liu S, Giffard RG. Neuroprotection by astrocytes in brain ischemia: importance of microRNAs. Neuroscience letters, 2014;565 : 53–58.
7. Noetel A, Kwiecinski M, Elfimova N, Huang J, Odenthal M. MicroRNA are Central Players in Anti - and Profibrotic Gene Regulation during Liver Fibrosis. Front Physiol, 2012;3 : 49.
8. Savan R. Post-transcriptional regulation of interferons and their signaling pathways. Journal of Interferon & Cytokine Research, 2014;34(5):318–329.
9. Forster SC, Tate MD, Hertzog PJ. MicroRNA as type I interferon-regulated transcripts and modulators of the innate immune response. Frontiers in immunology, 2015;6.
10. David M. Interferons and microRNAs. Journal of Interferon & Cytokine Research, 2010;30(11):825–828.
11. Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-alpha receptor. Nat Immunol, 2011;13(2):181–187.
12. Rath S, Donovan J, Whitney G, Chitrakar A, Wang W, Korennykh A. Human RNase L tunes gene expression by selectively destabilizing the microRNA-regulated transcriptome. Proceedings of the National Academy of Sciences, 2015;112(52):15916–15921.
13. Drappier M, Michiels T. Inhibition of the OAS/RNase L pathway by viruses. Current opinion in virology, 2015;15 : 19–26.
14. Malathi K, Dong B, Gale M, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature, 2007;448(7155):816–819.
15. Nilsen TW, Baglioni C. Mechanism for discrimination between viral and host mRNA in interferon-treated cells. Proceedings of the National Academy of Sciences, 1979;76(6):2600–2604.
16. Takeda T, Tanabe H. Lifespan and reproduction in brain-specific miR-29-knockdown mouse. Biochem Biophys Res Commun, 2016;471(4):454–458.
17. Song H, Ding L, Zhang S, Wang W. MiR-29 family members interact with SPARC to regulate glucose metabolism. Biochem Biophys Res Commun, 2018;497(2):667–674.
18. Yu D, Green B, Tolleson WH, Jin Y, Mei N, Guo Y, et al. MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. Biochemical pharmacology, 2015;98(1):215–223.
19. Sekiya Y, Ogawa T, Yoshizato K, Ikeda K, Kawada N. Suppression of hepatic stellate cell activation by microRNA-29b. Biochemical and Biophysical Research Communications, 2011;412(1):74–79.
20. Guo J, Lin Q, Shao Y, Rong L, Zhang D. miR-29b promotes skin wound healing and reduces excessive scar formation by inhibition of the TGF-β1/Smad/CTGF signaling pathway. Canadian Journal of Physiology and Pharmacology, 2016;95(4):437–442.
21. Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol, 2009;16(1):23–29.
22. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. elife, 2015;4:e05005.
23. Chou CH, Shrestha S, Yang C-D, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic acids research, 2017;46(D1):D296–D302.
24. Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: Similarities and differences. Advanced Drug Delivery Reviews, 2009;61(9):746–759.
25. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Research, 2018;47(D1):D155–D62.
26. Lorenz R, Bernhart SH, Zu Siederdissen CH, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms for Molecular Biology, 2011;6(1):26.
27. Berkner KL, Folk WR. Polynucleotide kinase exchange reaction: quantitave assay for restriction endonuclease-generated 5’-phosphoroyl termini in DNA. Journal of Biological Chemistry, 1977;252(10):3176–3184.
28. Díaz-Guerra M, Rivas C, Esteban M. Inducible expression of the 2-5A synthetase/RNase L system results in inhibition of vaccinia virus replication. Virology, 1997;227(1):220–228.
29. Nilsen T, Maroney P, Baglioni C. Synthesis of (2’-5’) oligoadenylate and activation of an endoribonuclease in interferon-treated HeLa cells infected with reovirus. Journal of virology. 1982;42(3):1039–1045.
30. Li XL, Blackford JA, Hassel BA. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. Journal of virology, 1998;72(4):2752–2759.
31. Chakrabarti A, Ghosh PK, Banerjee S, Gaughan C, Silverman RH. RNase L Triggers Autophagy in Response to Viral Infections. Journal of Virology, 2012;86(20):11311–11321.
32. Al-Ahmadi W, Al-Haj L, Al-Mohanna F, Silverman R, Khabar K. RNase L downmodulation of the RNA-binding protein, HuR, and cellular growth. Oncogene, 2009;28(15):1782.
33. Brennan-Laun SE, Li X-L, Ezelle HJ, Venkataraman T, Blackshear PJ, Wilson GM, et al. RNase-L Attenuates Mitogen-stimulated Gene Expression via Transcriptional and Post-transcriptional Mechanisms to Limit the Proliferative Response. Journal of Biological Chemistry, 2014:jbc. M114. 589556.
34. Cooper DA, Jha BK, Silverman RH, Hesselberth JR, Barton DJ. Ribonuclease L and metal-ion–independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic acids research, 2014;42(8):5202–5216.
35. Lee TY, Ezelle HJ, Venkataraman T, Lapidus RG, Scheibner KA, Hassel BA. Regulation of human RNase-L by the miR-29 family reveals a novel oncogenic role in chronic myelogenous leukemia. Journal of Interferon & Cytokine Research, 2013;33(1):34–42.
36. Hassel B, Hsi T, Ezelle H, Scheibner K. P140 MicroRNA-29 regulation of RNase-L reveals a novel function in oncogenesis. Cytokine, 2012;59(3):565.
37. Li JW, He SY, Feng ZZ, Zhao L, Jia WK, Liu P, et al. MicroRNA--146b inhibition augments hypoxia-induced cardiomyocyte apoptosis. Molecular medicine reports, 2015;12(5):6903–6910.
Labels
Hygiene and epidemiology Medical virology Clinical microbiologyArticle was published in
Epidemiology, Microbiology, Immunology

2020 Issue 3
-
All articles in this issue
- Epidemiology of vancomycin-resistant enterococci in the Hradec Králové University Hospital in 2017
- Seroprevalence of IgG antibodies against measles in health care workers of the Strakonice Hospital
- Implementation and use of whole genome sequencing (WGS) in the surveillance of invasive pneumococcal disease, Czech Republic, 2017–2019
- Drain fly – Clogmia albipunctata (Diptera: Psychodidae) – a fly with epidemiological potential and posing risk of myiasis
- Jubileum profesora Vladimíra Vonky
- Životní jubileum RNDr. Vratislava Němečka, CSc.
- Vzpomínky na MUDr. Evu Jílkovou
- MUDr. Jarmila Kaustová (* 8. 3. 1945 – † 1. 5. 2020)
- Smuteční oznámení: zemřel doc. MUDr. Vlastimil Obdržálek, CSc.
- Increase in RNASEL gene expression by miR-29-3p inhibitors in HEK293T cells
- Can gram-negative-like biomarker values in Streptococcus pyogenes sepsis negatively influence right choice of initial antibiotic therapy?
- The duration of SARS-CoV-2 shedding in patients recovering from COVID-19
- Epidemiology, Microbiology, Immunology
- Journal archive
- Current issue
- About the journal
Most read in this issue
- Epidemiology of vancomycin-resistant enterococci in the Hradec Králové University Hospital in 2017
- Drain fly – Clogmia albipunctata (Diptera: Psychodidae) – a fly with epidemiological potential and posing risk of myiasis
- The duration of SARS-CoV-2 shedding in patients recovering from COVID-19
- Seroprevalence of IgG antibodies against measles in health care workers of the Strakonice Hospital
