-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The duration of SARS-CoV-2 shedding in patients recovering from COVID-19
Délka vylučování SARS-CoV-2 u pacientů uzdravujících se z COVID-19
Znalosti o vylučování SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) jsou významné pro diagnózu, terapii a sledování pacientů s COVID-19 (coronavirus disease 2019). Vzorky získané opakovaným hlubokým nazofaryngeálním výtěrem u kohorty 100 pacientů s COVID-19 byly testovány na přítomnost SARS-CoV-2 RNA pomocí RT-PCR (real-time polymerase chain reaction). Medián doby detekovatelnosti virového genomu byl 15 dnů. Autoři dále testovali hypotézu o vztahu mezi závažností průběhu COVID-19 a délkou období, po které je virový genom detekovatelný. Nenalezli statisticky signifikantní rozdíl v trvání virové clearance mezi osobami s mírnou či asymptomatickou infekcí a pacienty se závažným onemocněním.
Klíčová slova:
COVID-19 – SARS-CoV-2 – RT-PCR – virová clearance – vylučování viru
Authors: Š. Cimrman 1; L. Macková 1; V. Král 2; H. Bartoš 1; I. Stiborová 2; P. Dlouhý 1
Authors place of work: Department of Infectious Diseases, Masaryk Hospital in Ústí nad Labem, Czech Republic 1; Centre of Immunology and Microbiology, Public Health Institute based in Ústí nad Labem, Czech Republic 2
Published in the journal: Epidemiol. Mikrobiol. Imunol. 69, 2020, č. 3, s. 148-151
Category: Krátké sdělení
Summary
The knowledge of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) shedding is highly relevant to the diagnosis, treatment and follow-up of patients with COVID-19 (coronavirus disease 2019). Deep nasopharyngeal swabs repeatedly collected from a cohort of one hundred patients with COVID-19 were tested for SARS-CoV-2 RNA using RT-PCR (real-time polymerase chain reaction). The median period of viral genome detectability was 15 days. Furthermore, the authors tested the hypothesis on the relationship between the severity of COVID-19 and the period in which the viral genome is detectable. They did not find any statistically significant difference in the duration of viral clearance between patients with asymptomatic to mild disease or severe disease.
Keywords:
SARS-CoV-2 – COVID-19 – RT-PCR – viral clearance – viral shedding
SARS-COV-2 AND ITS EXCRETION DYNAMICS
COVID-19 (Coronavirus Disease 2019) is an infectious disease caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) that has affected over 8.000 people in the Czech Republic and approximately 4.5 million people worldwide since December 2019. The infection is transmitted through respiratory secretions during close contact or can be transferred from contaminated hands to mucosal surfaces. The incubation period is estimated to be between two and 14 days. The disease most commonly manifests as febrile respiratory infection with cough and shortness of breath. A considerable number of cases remain completely asymptomatic [1].
The diagnosis is based on the detection of viral genome (E, N and RdRp genes) by RT-PCR (real-time polymerase chain reaction) in deep nasopharyngeal swabs. The viral genome can also be detected in blood, but not all patients with COVID-19 have detectable viral RNA in their blood samples. Tracheal aspiration from the lower respiratory tract is the preferred method for obtaining samples from patients in ICU (intensive care units).
There is a growing body of evidence supporting the detectability of viral RNA in other body fluids and secretions, such as stool, urine and ejaculate. However, it was not detected in maternal milk. The detection of the viral genome in a sample alone does not necessarily prove the presence of infectious viral particles or the possibility for the transmission of the infection [2].
Serological detection methods are limited of diagnostic value in most of respiratory infectious diseases and due to a delayed and unreliable production of antibodies, are neither suitable for an early diagnosis of COVID-19 nor for screening for asymptomatic carriers. The population screening for IgA, IgM and IgG class antibodies can be used in order to estimate the population immunity rate if a proper methodology and result interpretation is utilized. Serological methods can also find use in the treatment of patients with COVID-19 in a critical condition: before use of therapeutic plasma from a recovered patient in the treatment of COVID-19, evidence of virus neutralizing antibodies is required.
The dynamics of SARS-CoV-2 excretion have already been examined by numerous studies [3, 4, 6]. The peak viral load was found in the first week of the disease, followed by a decline in the second week. This is in accord with a considerable decrease of virus transmissibility in time. The rate at which SARS-CoV-2 replicates is dependent on its virulence and the host immune response; it is highest in elderly patients [4]. No evidence was found to support a possible connection between the number of viral particles in nasopharyngeal swab and the severity of COVID-19. Even asymptomatic individuals infected with SARS-CoV-2 can present with a high a viral load [5].
Some studies have claimed that there is a connection between the severity of COVID-19 and the time period in which the viral genome is detectable in a patient’s samples [6]. We tested this claim on our cohort of patients.
CHARACTERISTICS OF THE COHORT
Our cohort consisted of 100 individuals with COVID-19, inpatients or outpatients, treated at the Department of Infectious Diseases of the Masaryk Hospital in Ústí nad Labem, Czech Republic from 19 February 2020 to 12 May 2020. Only patients diagnosed with COVID-19 by RT-PCR of nasopharyngeal swabs or blood samples, who had two negative follow-up RT-PCR tests during recovery, were included.
We used a Rotor-Gene Q thermocycler, Qiagen, Germany, QuickExtractTM RNA Extraction Kit, Lucigen, USA, and Novel Coronavirus (2019-nCoV) Real Time Multiplex RT PCR Kit (Detection for 3 Genes), Liferiver, China. Statistical analysis was performed using the TIBCO Statistica software.
We examined the duration of viral RNA excretion, also referred to as viral shedding and viral clearance, defined as the interval between the first positive RT-PCR test result and the first negative result, assuming that the negative result was then followed by a second confirmatory negative test result. Two negative PCR tests in succession are required as the criterion for COVID-19 recovery. The time interval between sample (swab, blood) collections was dependent on the course of the disease, logistical conditions and the epidemiologist’s opinion.
The median age in our cohort was 49 years; age range 11 to 93 years.
The hypothesis that patients with a more severe course of the disease shed viral RNA longer than those with a milder disease was tested on a subgroup of 74 cohort patients with COVID-19 whose course of illness was well known and documented in detail and who could therefore be stratified into four groups based on the disease severity:
Group1: asymptomatic (n = 16),
Group 2: mild (n = 34) – needed no or short hospital admission, have neither pneumonia nor shortness of breath, were stable and required no oxygen support,
Group 3: moderate (n = 16) – admitted to the hospital and presenting with shortness of breath, low hemoglobin saturation (SpO2 < 93%) and/or pneumonia,
Group 4: severe (n = 8) – required intensive care due to signs of cardiopulmonary instability and/or non-invasive or invasive ventilation support, had bilateral pneumonia, acute respiratory distress syndrome (ARDS), sepsis and septic shock or multiple organ dysfunction syndrome (MODS).
The remaining 26 cohort patients were only consulted over the phone because of incomplete records regarding their presenting symptoms, and their samples were collected by medical personnel outside of the hospital. For some of these 26 patients, nasopharyngeal swab test results were only available.
RESULTS
The cohort analysis results are shown in Figure 1. The shortest amount of time a COVID-19 positive patient in our cohort required to achieve a negative PCR result for SARS-CoV-2 RNA in nasopharyngeal swab was three days and the longest was 31 days. The cohort (n = 100) median was 15 days.
Fig. 1. Time span of viral shedding in all patients in the cohort (n = 100) using RT-PCR of nasopharyngeal swabs Horizontal axis = number of days of documented COVID-19 positivity, vertical axis = number of patients. The median is 15 days. 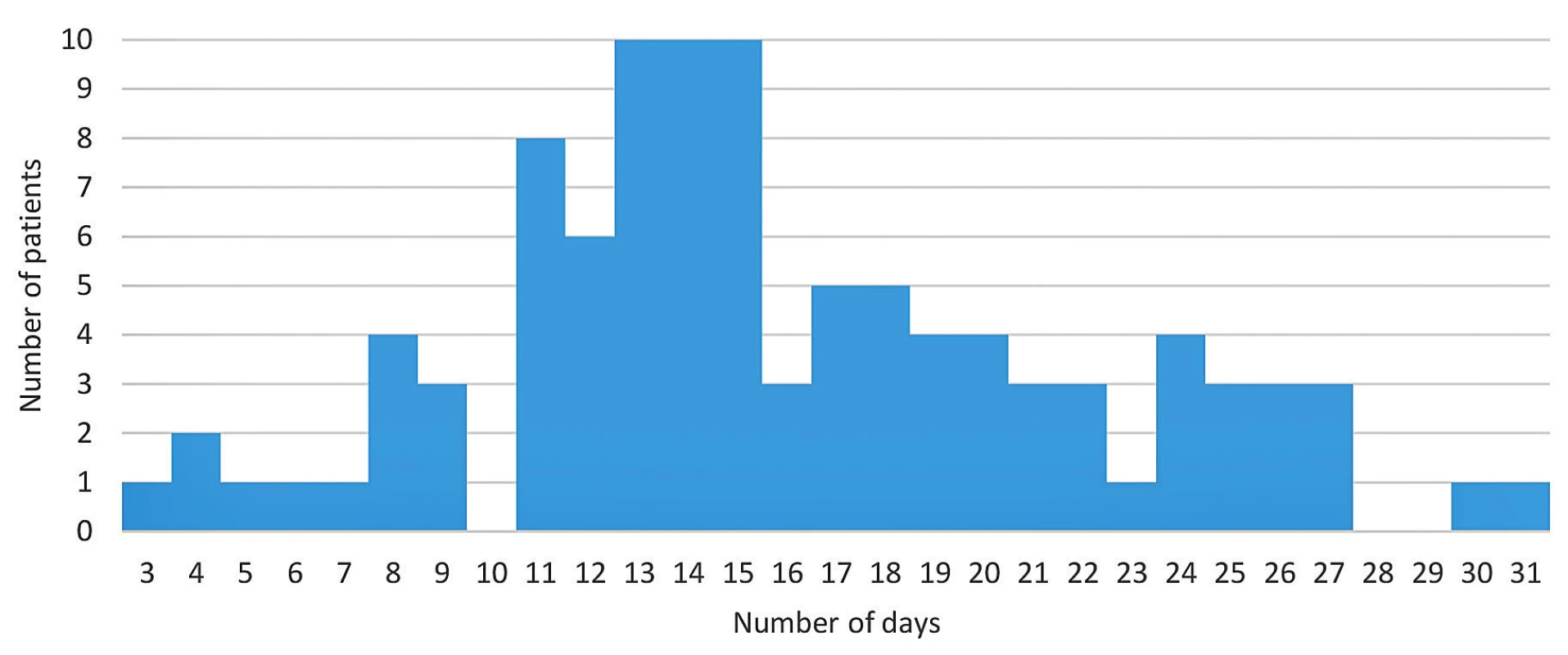
The median period between the first positive viral RNA PCR test result and the first negative test result (followed by another negative result as established earlier) in asymptomatic patients was 16 days, with a range of three to 30 days. The median of viral RNA clearance in patients with mild disease was 14 days; range five to 31 days. The group of patients with moderate disease showed a median clearance of 17.5 days, ranging between four and 27 days. The median viral clearance in patients with severe disease was 14 days; range six to 21 days.
Patients with asymptomatic to mild disease (n = 50, expected value = 15.4, median = 14, standard deviation = 6.17) were compared to those with moderate to severe disease (n = 24, expected value = 75.79, median = 17, standard deviation = 6.83) utilizing survival analysis and t-test, see Figure 2. Neither of the tests demonstrated a statistically significant difference between the two groups (Mantel-Cox test: p = 0.234; Wilcoxon test: p = 0.134; Tarone-Ware: p = 0.137; t-test: p = 0.136). Therefore, we did not confirm the hypothesis that persons with a more severe course of the disease shed viral RNA significantly longer than those with a mild or asymptomatic disease.
Fig. 2. A comparison between a group of patients with less severe illness (n = 50) and more severe illness (n =24) Vertical axis = number of days of documented COVID-19 positivity. 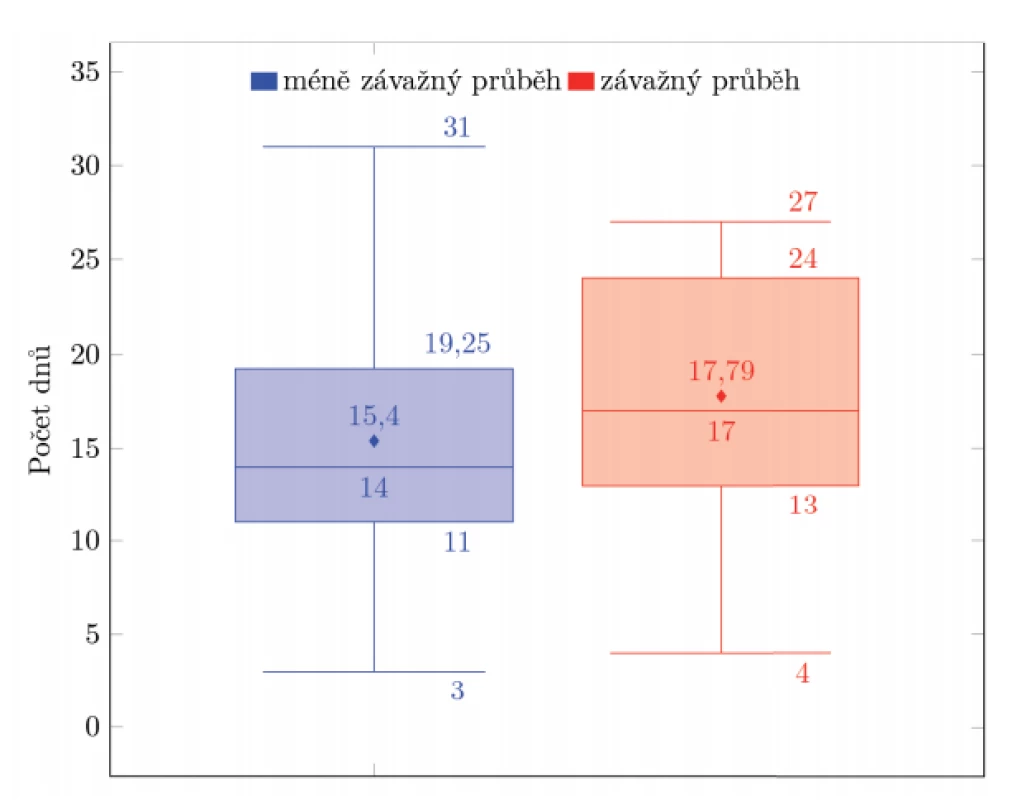
DISCUSSION
The presence of SARS-CoV-2 RNA in the examined material does not necessarily indicate the presence of infectious viral particles. On the other hand, one negative test result does not rule out a longer course of infection. During follow-up, several patients converted from negative to positive and vice versa, some even repeatedly. We therefore agree, that at least two consequent negative PCR test results from samples collected no less than 24 hours apart and at least 12 days after the first positive test result along with the complete absence of symptoms should be the criteria for listing a patient as recovered from COVID-19 and for terminating any isolation measures [7].
Our results show that the process of SARS-CoV-2 viral shedding, or rather the time period in which viral RNA remains detectable in nasopharyngeal swabs, is extensive. The longest interval between the first positive and the first negative test was 31 days. Two patients who continued to be positive at the time of writing were excluded from the cohort, remaining positive for 49 and 37 days, respectively. Although they only exhibit mild symptoms now, both of them had a severe course of the disease at one point.
However, we cannot prove a statistically significant link between the severity of COVID-19 and the length of viral RNA shedding. The distribution of patients who tested positive for extended periods of time was random in all severity groups; the conclusions of other researchers were not confirmed [6]. Our findings are, however, fully in accordance with the known fact that even asymptomatic carriers may shed viral RNA in high concentrations and for extended periods of time [8].
The limitations of this study stem from the impossibility of maintaining shorter and more regular intervals in sample collection. The reasons for that are twofold: limited testing capacity and the inability of many of our patients to commute from their homes in any higher frequency. In some cases, such as in our patient who produced a negative test result for COVID-19 as early as three days after the diagnosis, tests were performed daily to ensure a vital medical procedure would not be further delayed. In most patients, however, samples were collected 12-14 days after the last positive sample, as per official recommendations [7]. Despite these limitations, we believe that the data from such a relatively large cohort can provide important information about the course of viral RNA shedding in patients infected with SARS-CoV-2.
Acknowledgements:
We would like to thank MUDr. K. Hašková for consultation, MUDr. J. Jarý, MUDr. T. Kovačič, T. Čukanová and K. Barková for their contribution to data management. We thank RNDr. A. Fialová, Ph.D. for her contribution in data analysis. We declare no competing interests.
Do redakce došlo dne 21. 5. 2020.
Adresa pro korespondenci:
MUDr. Pavel Dlouhý
Infekční oddělení Masarykovi nemocnice
Sociální péče 3316/12 a
400 11 Ústí nad Labem-Severní terasa
e-mail: pavel.dlouhy@kzcr.eu
Zdroje
1. Ioannidis JPA, Axfors C, Contopoulos-Ioannidis DG. Population-level COVID-19 mortality risk for non-elderly individuals overall and for non-elderly individuals without underlying diseases in pandemic epicenters. MedRxiv [online]. May 5, 2020. [cit. 2020-05-20]. DOI: 10.1101/2020.04.05.20054361. Dostupné na www: https://www.medrxiv.org/content/10.1101/2020.04.05.20054361v2q.
2. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA [online]. [cit. 2020-05-20]. DOI: 10.1001/jama.2020.3786. ISSN 0098-7484. Dostupné na www: https://jamanetwork.com/journals/jama/fullarticle/2762997.
3. Chen Y, Li L. SARS-CoV-2: virus dynamics and host response. The Lancet Infectious Diseases [online], 2020;20(5):515–516 [cit. 2020-05-20]. DOI: 10.1016/S1473-3099(20)30235-8. ISSN 14733099. Dostupné na www: https://linkinghub.elsevier.com/retrieve/pii/S1473309920302358.
4. To K, Tsang O, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. The Lancet Infectious Diseases [online], 2020;20(5): 565–657 [cit. 2020-05-20]. DOI: 10.1016/S1473-3099(20)30196-1. ISSN 14733099. Dostupné na www: https://linkinghub.elsevier.com/retrieve/pii/S1473309920301961.
5. WHO. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected. Interim guidance [online], March 13, 2020 [cit. 2020-05-20]. WHO reference number: WHO/2019-nCoV/Clinical/2020.4.
6. Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID-19. The Lancet Infectious Diseases [online], 2020 [cit. 2020-05-20]. DOI: 10.1016/S1473-3099(20)30232-2. ISSN 14733099. Dostupné na www: https://linkinghub.elsevier.com/retrieve/pii/S1473309920302322.
7. Ministerstvo zdravotnictví ČR. Algoritmus testování osob s klinickými příznaky COVID-19 metodou PCR [online]. April 21, 2020 [cit. 2020-05-20]. Dostupné na www: https://www.infekce.cz/Covid2019/AlgTest210420.pdf
8. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. New England Journal of Medicine [online], 2020;382(12):1177–1179 [cit. 2020-05-20]. DOI: 10.1056/NEJMc2001737. ISSN 0028-4793. Dostupné na www: http://www.nejm.org/doi/10.1056/NEJMc2001737.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Mikrobiológia
Článok vyšiel v časopiseEpidemiologie, mikrobiologie, imunologie
Najčítanejšie tento týždeň
2020 Číslo 3- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Epidemiologie vankomycin-rezistentních enterokoků ve Fakultní nemocnici Hradec Králové v roce 2017
- Increase in RNASEL gene expression by miR-29-3p inhibitors in HEK293T cells
- Séroprevalence IgG protilátek proti spalničkám u zaměstnanců Nemocnice Strakonice, a.s.
- Mohou hodnoty biomarkerů, imitující gramnegativní zánětlivou odpověď, negativně ovlivnit iniciální volbu antibiotika u pacientů se sepsí vyvolanou Streptococcus pyogenes?
- Implementace a využití metody sekvenace celého genomu (WGS) v surveillance invazivního pneumokokového onemocnění, Česká republika, 2017–2019
- Koutule skvrnitá – Clogmia albipunctata (Diptera: Psychodidae) – muška s epidemiologickým potenciálem a rizikem myiáz
- The duration of SARS-CoV-2 shedding in patients recovering from COVID-19
- Jubileum profesora Vladimíra Vonky
- Životní jubileum RNDr. Vratislava Němečka, CSc.
- Vzpomínky na MUDr. Evu Jílkovou
- MUDr. Jarmila Kaustová (* 8. 3. 1945 – † 1. 5. 2020)
- Smuteční oznámení: zemřel doc. MUDr. Vlastimil Obdržálek, CSc.
- Epidemiologie, mikrobiologie, imunologie
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Epidemiologie vankomycin-rezistentních enterokoků ve Fakultní nemocnici Hradec Králové v roce 2017
- Koutule skvrnitá – Clogmia albipunctata (Diptera: Psychodidae) – muška s epidemiologickým potenciálem a rizikem myiáz
- The duration of SARS-CoV-2 shedding in patients recovering from COVID-19
- Séroprevalence IgG protilátek proti spalničkám u zaměstnanců Nemocnice Strakonice, a.s.
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



