-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
HIV-1 subtypes distribution and resistance to ART in HIV-infected persons in Slovakia (2019–2021)
Distribúcia HIV-1 subtypov a rezistencie na ART u HIV-infikovaných osôb na Slovensku (2019–2021)
Cieľ: Cieľom štúdie bolo popísať výskyt HIV-1 subtypov a HIV-1 kmeňov rezistentných na antiretrovírusovú liečbu (ART) u HIV pozitívnych osôb novo diagnostikovaných na Slovensku v rokoch 2019–2021.
Materiál a metódy: Študijnú skupinu tvorilo 184 HIV pozitívnych naivných pacientov novo diagnostikovaných na Slovensku v rokoch 2019–2021. Vírusová HIV-1 RNA bola izolovaná z plazmy pomocou QIAamp Viral RNA Mini Kit (QIAGEN, Nemecko). Pre RT-PCR a sekvenovanie oblasti HIV pol sme použili interné postupy podľa protokolu ANRS AC11 pre RT (reverzná transkriptáza), PRO (proteáza) a IN (integráza) (ANRS AC11 Resistance Study Group, 2015). Analýzu sekvencií sme uskutočnili pomocou softvéru Sequencing Analysis Software v5.3 (Applied Biosystems®). HIV sekvencie boli manuálne upravené pomocou BioEdit (verzia 7.2.5), porovnané s konsenzuálnymi HIV-1 sekvenciami v Los Alamos Sequence Database (URL 2), zarovnané pomocou CLUSTAL W (Labarga et al., 2007) a softvérových balíkov BioEdit (verzia 7.2 .5) (Hall, 1999). Na vyhodnotenie sekvencie sme použili algoritmus HIVDB (verzia 9.0) Stanfordskej databázy HIV liekovej rezistencie (URL 1.). Na analýzu HIV-1 subtypov sme použili nástroj REGA HIV-1 Subtyping Tool (De Oliviera et al., 2005) a fylogenetickú analýzu sme vypracovali pomocou programu MEGA X (Kumar et al., 2018).
Výsledky: Fylogenetickú analýzu sme vykonali zo vzoriek 184 osôb, kde sme odhalili najrozšírenejší subtyp B (129/184, 70,11 %) prevládajúci v populácii u mužov, ktorí majú sex s mužmi (MSM) (96/129 74,42 %). Čo sa týka non-B subtypov (55/184, 29,89 %), najrozšírenejší bol subtyp A (48/184, 26,09 %) v porovnaní so subtypom F (F1) (3; 1,63 %), C (1; 0,54 %) a cirkulujúcimi rekombinantnými formami CRF02_AG (2; 1,09 %), CRF01_AE (1; 0,54 %). U 9,24 % (17/184) vzoriek sme zistili prítomnosť 25 mutácií asociovaných s HIV rezistenciou na ART, z toho 7,07 % (13/184) na inhibítory reverznej transkriptázy, 1,66 % (3/181) na inhibítory proteázy a 1,32 % (2/151) na inhibítory integrázy. Okrem toho u 1,63 % (3/184) pacientov bola prítomná viactriedna rezistencia. Mutácie asociované s rezistenciou HIV na ART sa našli u 9,30 % osôb infikovaných podtypom B.
Záver: Naša štúdia potvrdila pretrvávajúci najvyšší výskyt subtypu B s mierne klesajúcou tendenciou v porovnaní s minulými rokmi. Detekcia mutácií vytvárajúcich rezistenciu HIV na ART podčiarkuje potrebu testovania rezistencie u naivných pacientov ešte pred začatím ART na Slovensku.
Authors: A. Kovářová 1,2; D. Valkovičová Staneková 2
; M. Hábeková 2
; M. Takáčová 2
Authors place of work: Department of Laboratory Investigation Methods in Healthcare, Faculty of Health Sciences and Social Work, The University of Trnava, Trnava, Slovak Republica 1; National Reference Centre for HIV/AIDS Prevention, Slovak Medical University, Bratislava, Slovak Republic 2
Published in the journal: Epidemiol. Mikrobiol. Imunol. 72, 2023, č. 4, s. 203-212
Category: Původní práce
Summary
Aim: The aim of the study was to describe the prevalence of HIV-1 subtypes and HIV-1 strains resistant to antiretroviral therapy (ART) in HIV-positive persons newly diagnosed in Slovakia in 2019–2021.
Materials and Methods: The study group consisted of 184 HIV-positive naïve patients newly diagnosed in Slovakia from 2019 to 2021. The viral HIV-1 RNA was isolated from plasma by the QIAamp Viral RNA Mini Kit (QIAGEN, Germany). For RT-PCR and sequencing of the HIV pol region, in-house procedures were used according to the ANRS AC11 protocol for RT (reverse transcriptase), PRO (protease), and IN (integrase) [ANRS AC11 Resistance Study Group, 2015]. Analysis of sequences was performed using Sequencing Analysis Software v5.3 (Applied Biosystems®). HIV sequences were manually edited using BioEdit (version 7.2.5), compared with consensus HIV-1 sequences in the Los Alamos Sequence Database (URL 2), aligned using CLUSTAL W [Labarga et al., 2007] and BioEdit software packages (version 7.2 .5) [Hall, 1999]. HIVDB Algorithm (version 9.0) of the Stanford HIV Drug resistance database (URL 1.) was used for sequence evaluation. For HIV-1 subtype analysis, the REGA HIV-1 Subtyping Tool [De Oliviera et al., 2005] and phylogenetic analysis MEGA X [Kumar et al., 2018] were used.
Results: Phylogenetic analyses performed in samples of 184 persons revealed the most prevalent subtype B (129/184, 70.11%), detected to the greatest extent in the population of men who have sex with men (MSM) (96/129 74.42%). Concerning non-B subtypes (55/184, 29.89%), subtype A was found with the highest prevalence (48/184, 26.09%) compared to subtype F (F1) (3; 1.63%), C (1; 0.54%) and circulating recombinant forms CRF02_AG (2; 1.09%), CRF01_AE (1; 0.54%). In 9.24% (17/184) of samples, 25 mutations clinically relevant and associated with HIV resistance ART were detected, of which 7.07% (13/184) to reverse transcriptase inhibitors, 1.66% (3/181) to protease inhibitors and 1.32% (2/151) to integrase inhibitors. In addition, multiclass resistance was present in 1.63% (3/184) of patients. Mutations associated with HIV resistance to ART were found in 9.30 % of persons infected with subtype B. Conclusion: Our study confirmed ongoing highest prevalence of subtype B with a slightly decreasing trend compared to last years. Detection of mutations causing HIV resistance to ART underlines the need for resistance testing in naïve patients even before the initiation of ART in Slovakia.
INTRODUCTION
Human immunodeficiency virus (HIV) continues to be a major global public health issue, having claimed 40.1 million lives so far [WHO, 2022]. 84.2 million people have become infected with HIV since the start of the epidemic [UNAIDS, 2022]. An estimated 38.4 million people were living with HIV at the end of 2021, two-thirds of whom (25.6 million) are in the WHO African Region [WHO, 2022]. In 2021, 1.5 million people became newly infected with HIV, and 650 000 people died from AIDS-related illnesses [UNAIDS, 2022]. There is no cure for HIV infection. However, with increasing access to effective HIV prevention, diagnosis, treatment, and care, including for opportunistic infections, HIV infection has become a manageable chronic health condition, enabling people living with HIV to lead long and healthy lives [WHO, 2022].
HIV type 1 (HIV-1) shows high genetic diversity between and within human hosts [Cuevas et al., 2015; Abram et al., 2010]. While within-host viral diversity is initially limited by the transmission bottleneck, HIV genomes quickly diversify as a result of a high viral replication rate and high error rates during replication [Lee et al., 2008]. Such high genetic diversity allows the virus to avoid the host’s immune response and can lead to the development of drug resistance during treatment [Cuevas et al., 2015]. HIV-1 consists of four major phylogenetic groups: M (major), N (new), O (outlier), and P, representing independent cross-species transition from SIV in chimpanzees and gorillas to HIV in humans [Sharp and Hahn, 2011]. HIV-1 group M is responsible for the current pandemic and comprises genetically distinct subtypes (A, B, C, D, F, G, H, J, K, and, more recently, L), 101 circulating recombinant forms (CRFs) to date, and numerous unique recombinant forms (URFs) [Hemelaar et al., 2019; Yamaguchi et al., 2019]. HIV-1 subtypes are unequally distributed globally, which has been explained by different founder effects followed by local spread driven by socioeconomic and behavioral factors and circulation within and between specific risk groups [Hemelaar, 2012; Murillo et al., 2013; Junqueira and de Matos Almeida, 2016]. Subtype C is the most abundant strain worldwide and is prevalent in South and Eastern Africa and Southeast Asia [Hemelaar et al., 2019]. Subtype B is predominant in North America, Western Europe, and Australia, while subtype A predominates in Eastern Europe and Central Asia, including Russia [Hemelaar et al., 2019; Magiorkinis et al., 2016]. CRFs and URFs are widely distributed in central Africa and in countries where different subtypes co-circulate [Hemelaar et al., 2019; Alaeus, 2000; Konstantinos et al., 2015]. The wide variety of subtypes, which are distributed mainly in different populations, may be used for molecular epidemiological analysis to track, and understand the dynamics and patterns of HIV-1 transmission and for developing strategic prevention programs [Paraskevis et al., 2016].
Subtype B was determined as the most prevalent genetic form in Slovakia [Hábeková et al., 2010; Chabadová et al., 2014; Čereš et al., 2018; Hábeková et al., 2023]. Slovak Republic belongs for a long time to EU states with the lowest annual incidence of HIV infection. In the last decade, we have observed an upward trend in the occurrence of new cases and the number of people living with HIV infection is also increasing. The majority of HIV infections (70.00%) in the Slovak Republic were recorded in the group of men who have sex with men (MSM). Overall, 20.00 % of infections were acquired through unprotected heterosexual intercourse (HTS), 1.20% through injecting drug use, 0.05% through blood transfusion (one man who got infected abroad in 1986), and 8.75% the route of transmission is still unknown. The highest HIV cumulative incidence has been for a long time recorded in the capital of Slovakia, Bratislava. More than 90.00% of people diagnosed with HIV infection in the Slovak Republic are on antiretroviral therapy (ART). More than 90.00% of people on ART are in viral suppression, so they should not be able to transmit to other people thanks to continuous treatment [Public Health Authority of the Slovak Republic, 2022].
This study aimed to describe the prevalence of HIV-1 subtypes and HIV strains resistant to ART in HIV-positive persons newly diagnosed in 2019–2021 in Slovakia in relationship to socio-demographical factors.
Fig. 1. RT region-based phylogenetic tree of HIV-1 non-B subtype isolates 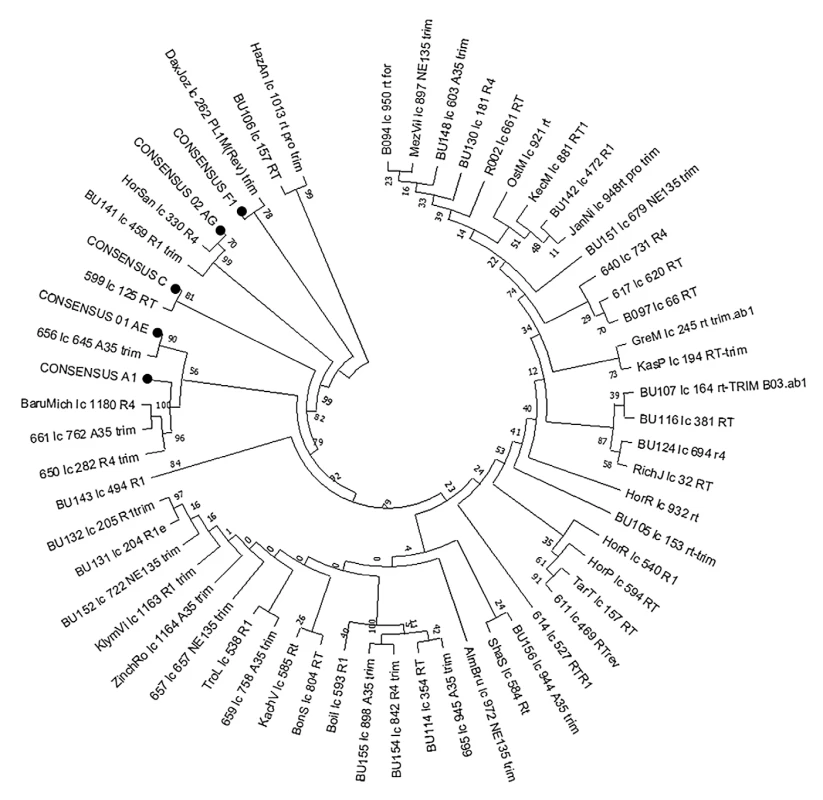
The phylogenetic tree was constructed using the Neighbor-Joining method. Evolutionary distances were calculated using the Kimura 2-parameter method in MEGA X. Consensus sequences A, F, C, CRF01_AE, and CRF02_AG were included. MATERIALS AND METHODS
In our study, 184 HIV-positive patients (naive to ART) newly diagnosed in Slovakia between 2019 and 2021 were included. In addition, personal data of patients regarding the route of transmission and country of origin were routinely obtained for epidemiological purposes during post-test counseling.
Isolation of viral HIV-1 RNA was performed from plasma samples using The QIAamp Viral RNA Mini Kit (QIAGEN, Germany). The nested reverse transcriptase polymerase chain reaction (RT-PCR) and sequencing of HIV pol region were carried out with the use of in-house procedures according to ANRS AC11 protocol for reverse transcriptase (RT), protease (PRO) and integrase (IN) [ANRS AC11 Resistance Study Group, 2015].
Sequence analysis of RT, PRO, and IN was performed using Sequencing Analysis Software v5.3 (Applied Biosystems®). The HIV sequences were edited manually in the BioEdit program (version 7.2.5). We compared the HIV-1 pol sequences with the consensus HIV-1 sequences available in the Los Alamos Sequence Database (URL 4), aligned using CLUSTAL W software [Labarga et al., 2007] and BioEdit software packages (version 7.2.5) [Hall, 1999]. RT sequences included codons 14–248, PRO sequences 1–99 and IN sequences 1–280. To evaluate the sequences and determine the degree of resistance, the HIVDB Algorithm (version 9.0) was used, where based on the database (Stanford HIV Drug Resistance Database, URL 1.) determined the degree of resistance and the occurrence of major and minor mutations. For phylogenetic analysis of HIV-1 subtypes, edited HIV-1 pol sequences were analysed using the REGA HIV-1 Subtyping Tool program [De Oliveira et al., 2005]. The evolutionary history was inferred using the Neighbor-Joining method [Saitou and Nei, 1987]. The percentage of replicate trees in which associated taxa clustered in a bootstrap test (100 replicates) are shown next to branches [Felsenstein, 1985]. Evolutionary distances were calculated using Kimura’s 2-parameter method [Kimura, 1980] and are in units of the number of base substitutions per site. Variation in velocity between sites was modelled using a gamma distribution (shape parameter = 1). This analysis included 60 nucleotide sequences. Codon positions 1st + 2nd + 3rd + non-coding, all were included. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 499 positions in the final data set. Phylogenetic analyses were performed in MEGA X [Kumar et al., 2018]. In addition, the distribution of HIV-1 and CRF subtypes in the relationship to socio-demographical indicators was investigated.
RESULTS AND DISCUSSION
Study group
Gender and Demography
Out of 184 HIV-1 positive persons 169 (91.85%) were men and 15 (8.15 %) were women (Table 1). The mean age of the patients was 38 (20–75) years, for men 38 (20–72) years, and for women 43 (23–75) years. Both, the youngest (20 years old men), as well as the oldest (75 years old men) individuals in the study group, were Slovaks.
Table 1. Distribution of HIV-1 subtypes by gender, country, and route of transmission in 2019 – 2021
Subtype B No. (%)
Non-B subtypes No. (%)
Total No. (%)
Gender
Male
124 (96.12)
45 (81.82)
169 (91.85)
Female
5 (3.88)
10 (18.18)
15 (8.15)
Total
129
55
184
Country of Origin
Slovakia
124 (96.12)
41 (74.55)
165 (89.67)
Other country
5 (3.88)
14 (25.45)
19 (10.33)
Total
129
55
184
Route of transmission
MSM Male
96 (74.42)
29 (52.73)
125 (67.93)
HTS Male
19 (14.73)
11 (20.0)
30 (16.30)
HTS Female
5 (3.87)
9 (16.36)
14 (7.61)
Unknown Male
9 (6.98)
5 (9.09)
14 (7.61)
Unknown Female
0 (0.00)
1 (1.82)
1 (0.55)
Total
129
55
184
MSM – men having sex with men, HTS – heterosexual contact, No. – number of people
Out of 184 participants, there were 165 (89.67%) Slovaks and 19 foreigners (10.33%): from them 14 (7.61%) Ukrainians, 2 (1.09%) Polish, 1 (0.54%) Czech and 2 (1.09%) persons of unknown nationality. Out of 169 men, 158 (85.87%) were Slovaks and 11 (5.98%) were foreigners while out of 15 women, 7 (3.80%) were Slovaks and 8 (4.35%) were Ukrainians.
Routes of HIV transmission
Most of the patients were infected by sexual contact between men (125 : 67.93%), followed by patients infected by unprotected heterosexual contact (44 : 23.91%) and by unknown routes of transmission (15 : 8.15%). Similar results were obtained in Slovakia in 2004–2008 and 2009–2012 when more than half of HIV-1 infected patients were MSM. Similarly, the Czech Republic, reported long-term highest incidence of newly diagnosed HIV infections among MSM (79.00%) [Mravčík et al., 2017]. Proportion of men vs. of women remains also like the years 2004–2012 among newly diagnosed HIV cases in Slovakia [Chabadová et al., 2014; Hábeková et al., 2010]. However, as the number of foreigners (Ukrainian emigrants) increased during 2019 – 2021, the ratio among HIV-infected women decreased [Public Health Authority of the Slovak Republic, 2022].
HIV-1 B subtype
The findings of this study, which describe the distribution of HIV-1 subtypes by gender, country, and route of transmission, as well as the epidemiological data of people infected with HIV-1 non-B viruses diagnosed between 2019 and 2021, are summarized in Tables 1 and 2.
Analysis of HIV-1 strains showed that 129 (70.11%) of all 184 patients, 124 Slovaks, and 5 foreigners were infected with HIV-1 subtype B viruses (see Table 1). Subtype HIV-1 B was discovered to be the most prevalent genetic form in Slovakia during our study as well as in previous years, but with a slight downward trend compared to previous years. The highest incidence of subtype B in Slovakia was observed in 2004–2008 (93.0%), and the lowest was in our observed period (2019–2021, 70.11%). Between those years, the occurrence of subtype B in Slovakia was as follows: in 2009 – 2012 (86.11%), 2015–2016 (80.30%), and in 2017–2018
(82.30%) [Hábeková et al. 2010; Chabadová et al., 2014; Čereš et al., 2018; Hábeková et al., 2023] which is also confirmed by the SPREAD program. SPREAD program analysed a set of samples in newly diagnosed HIV-positive persons in Western, Central, and Eastern Europe and sub-Saharan Africa from 2002–2010, with subtype B being the dominant subtype in these countries (6310/9588, 65.80%) [Hofstra et al., 2016]. This program continued also in the years 2011–2013, including the states of Central, Mediterranean, Northern, and Western Europe, and subtype B prevailed in this period as well [Hofstra et al., 2019]. In Central European countries such as Poland, subtype B remains the most common variant to date, shared by 85.90% of the study population, and is still prevalent among MSM in 2015–2019 [Serwin et al., 2021]. This observation is consistent with a previous report from Poland from 2008–2014, where 86.90% of recorded infections developed from the HIV-1 B lineage [Parczewski et al., 2016]. In 2014–2019 the predominant HIV-1 subtype in Croatians was subtype B (91.30%) [Oroz et al., 2019]. On the contrary, in the study from Greece HIV-1 sequences were found within subtype B only in 50.44% and subtype A1 in up to 49.56% of persons [Kostaki et al., 2022]. In a study from Belgium in 2013–2019 less than half of the patients (47.50%) were infected with the subtype B virus [Mortier et al., 2022]. It appears, a decline in subtype B has been observed in Europe, which may be due to the economic migration and refugee crisis and the increasing circulation of non-B subtypes among the native European population [Beloukas et al., 2016; Neogi et al., 2014; Hauser et al., 2018].
In our study, regarding the data on the route of transmission being infected with a subtype B virus was much more common among MSM than patients in other transmission groups (96/129; 74.42%) – see Table 1. Similar results were observed in Slovakia in studies from 2004–2008 (66.40%) and 2009–2012 (74.20%) [Hábeková et al., 2010; Chabadová et al., 2014]. Other studies also revealed that infection with HIV-1 subtype B is most common in Poland through MSM comparable to the HIV epidemic in the Netherlands [Serwin et al., 2022; Wymant et al., 2022]. A study from Slovenia also confirmed that the HIV-1 epidemic is predominantly affecting MSM infected with subtype B [Lunar et al., 2015]. In Bulgaria 1988–2018 the higher incidence of subtype B was found in MSM (56.70%) compared to heterosexuals (38.60%) [Alexiev et al., 2020]. Mustafa et al. performed phylogenetic and phylodynamic analysis using 21 007 publicly available B sequences from Europe and Asia and found MSM to be the primary highrisk group responsible for subtype B transmission, too [Mustafa et al., 2023].
HIV-1 non-B subtypes
Phylogenetic analysis revealed 55 of 184 HIV-1 non-B subtypes (29.89 %) in 41 Slovaks and 14 foreigners (Figure 1). Subtype A (48; 26.09 %) dominated in Slovakia, while other subtypes and circulating recombinant forms (CRF) were identified as follow: F (F1) (3; 1.63%), C (1; 0.54%), CRF02_AG (2; 1.09%) and CRF01_AE (1; 0.54%) (Table 2). It is obvious that the prevalence of non-B subtypes increased almost twice in Slovakia as compared to the period of 2004–2008, 2009–2012, 2015–2016, and 2017–2018 [Hábeková et al., 2010; Chabadová et al., 2014; Čereš et al., 2018; Hábeková et al., 2023]. We originally assumed that the majority of subtypes A would be transmitted to Slovakia by foreigners originating from Eastern Europe (Ukraine), but up to 71.43% of infected persons were Slovaks (35) and 28.57% were foreigners (13 Ukrainians, 1 unknown). The introduction of subtype A into Slovakia is probably related to the immigration of the population from Eastern Europe, where this variant is most prevalent, as confirmed by the study by Maarten et al., 2022. On the contrary, in southern European countries such as Italy or Spain are non-B subtypes F1 and CRF02_AG dominating [Delgado et al., 2019; Lorenzin et al., 2019], while in Slovakia these non-B subtypes were found in 3 Slovaks, from them, in 1 infected by CRF02_AG in Greece. However, a study from southwestern Greece confirmed the highest incidence of non-B subtype A (51.30%) and lower CRF02_AG (2.70%) [Davanos et al., 2015]. Similarly, Croatia did not record a high incidence of non-B subtypes in 2014–2017 (A1 : 4.20%; C: 1.70%; CRF02_AG: 1.20%; CRF01_AE: 0.50% and CRF06_CPX: 0.20%) [Oroz et al., 2019].
Table 2. Epidemiological data of patients infected with HIV-1 non-B viruses in 2019–2021
Non-B subtypes
A (A1) No. (%)
C No. (%)
F (F1) No. (%)
CRF01_AE No. (%)
CRF02_AG No. (%)
Gender
Male
39 (81.25)
1 (100.00)
2 (66.67)
1 (100.00)
2 (100.00)
Female
9 (18.75)
0 (0.00)
1 (33.33)
0 (0.00)
0 (0.00)
Total
48
1
3
1
2
Route of transmission
MSM Male
25 (52.08)
0 (0.00)
1 (33.33)
1 (100.00)
2 (100.00)
MSM Female
0 (0.00)
0 (0.00)
0 (0.00)
0 (0.00)
0 (0.00)
HTS Male
10 (20.83)
1 (100.0)
0 (0.00)
0 (0.00)
0 (0.00)
HTS Female
8 (16.66)
0 (0.00)
1 (33.33)
0 (0.00)
0 (0.00)
Unknown Male
4 (8.33)
0 (0.00)
1 (33.34)
0 (0.00)
0 (0.00)
Unknown Female
1 (2.10)
0 (0.00)
0 (0.00)
0 (0.00)
0 (0.00)
Total
48
1
3
1
2
Country of Origin
Slovakia
34 (70.83)
1 (100.00)
3 (100.00)
1 (100.00)
2 (100.00)
Ukraine
13 (27.08)
0 (0.00)
0 (0.00)
0 (0.00)
0 (0.00)
Unknown country
1 (2.09)
0 (0.00)
0 (0.00)
0 (0.00)
0 (0.00)
Total
48
1
3
1
2
Region of Slovakia
West
24 (70.58)
1 (100.00)
2 (66.66)
1 (0.00)
1 (50.00)
Central
7 (20.58)
0 (0.00)
1 (33.34)
0 (0.00)
1 (50.00)
East
3 (8.84)
0 (0.00)
0 (0.00)
0 (0.00)
0 (0.00)
Total
34
1
3
1
2
MSM – men having sex with men, HTS – heterosexual contact, No. – Number of people.
In our study, 29 (52.73%) MSM were associated with non-B infections, followed by heterosexuals (20, 36.36 %), and cases of the unknown route of transmission (6, 10.91%) – see Table 2. It appears that in Slovakia male-to-male sexual contact surpassed heterosexual contact as the most common route of HIV transmission in patients infected with the non-B subtype from 2004 to 2012 [Hábeková et al., 2010; Chabadová et al., 2014]. We assume that HIV-1 diversity is increasing in Slovakia due to immigration flows and travel, which causes the presence of new viral subtypes and new recombinant forms with consequences for public health.
Spread of resistant strains of the HIV virus
In our study, 25 mutations were associated with ART resistance in 17 of 184 (9.24%) HIV-infected individuals. In addition, mutations associated with HIV resistance to non-nucleoside/non-nucleotide inhibitors of reverse transcriptase (NNRTIs), nucleoside/nucleotide inhibitors of reverse transcriptase (NRTIs) and NRTIs/NNRTIs were found in 13 (7.07%), 2 (1.09%) and 2 (1.09%) out of 184 patients, respectively. Moreover, mutations associated with resistance to protease inhibitors (PIs) were present in 3 of 181 (1.66%) persons and to integrase inhibitors (INSTIs) in 2 of 151 (1.32%) persons (Table 3).
Table 3. Occurrence of HIV-1 resistance to ART in HIV-positive patients newly diagnosed in the Slovak Republic in 2019–2021
NRTI
NNRTI
PI
INSTI
Patients
S/R
No. (%)
No. (%)
No. (%)
No. (%)
Slovaks
S
166 (99.40)
157 (94.01)
164 (98.20)
136 (98.55)
R
1 (0.60)
10 (5.99)
3 (1.80)
2 (1.45)
total
167
167
167
138
Foreigners
S
16 (94.12)
14 (82.35)
14 (100.00)
13 (100.00)
R
1 (5.88)
3 (17.65)
0 (0.00)
0 (0.00)
total
17
17
14
13
All
S
182 (98.91)
171 (92.93)
178 (98.34)
149 (98.68)
R
2 (1.09)
13 (7.07)
3 (1.66)
2 (1.32)
total
184
184
181
151
S – sensitive, R – resistant, NRTI – nucleoside/nucleotide inhibitor of reverse transcriptase, NNRTI – non-nucleoside/non-nucleotide inhibitor of reverse transcriptase, PI – protease inhibitor, INSTI – integrase inhibitor, No. – Number of patients
Multiclass resistance was observed in 3 (1.63%; 2 Slovaks and 1 foreigner) patients. Mutations associated with NRTIs were as follow: M41l, D67N, T69D, K70R, M184V, T215F, K219Q and T215S, with NNRTIs A98G, K101E, K103N, V108I, V108VI, E138A, V179VD, Y181YCFS, F227L, with PIs: M46I, M4V6L, I84 and with INSTIs:
L74M, S230SGR, D232DN, R263K. The incidence of mutations associated with resistance to ART in HIV-infected naive patients was firstly described in Slovakia in 2015–2016 [Čereš et al., 2018]. The higher prevalence of mutations associated with NNRTIs in Western Europe was confirmed by a study by Mortier et al., 2022. They found that many resistance mutations impacted the sensitivity for NNRTIs (11.40%), followed by NRTIs (6.20%), PIs (2.40%), and INSTIs (0.56%) respectively. Multiclass resistance was observed in 2.40% (88/3708) of persons in Belgium in 2013–2019 [Mortier et al., 2022]. Pingarilho et al. found the highest level of NNRTIs resistance (9.70%) in patients captured in Portugal in 2014–2019 [Pingarilho et al., 2022]. Klundert et al. sequenced the RT and PRO genes from 812 HIV-1 positive persons (from Ukraine: 191 patients, 2019–2020; Georgia: 201 patients, 2013–2017 and Russia: 420 patients, 2019–2020) for ART initiation and confirmed the highest the prevalence of mutations against NNRTIs (106, 12.68%) [Klundert et al., 2022].
Mutations A98G and E138A associated with NNRTIs represented the largest proportion of all detected mutations in our study. A98G mutation occurred in 4 of 184 (2.17%; all Slovaks) patients. A98G is a relatively non-polymorphic accessory NNRTI-resistance mutation. It occurs in 0.10–0.50% of untreated persons depending on subtype. It has been selected primarily in persons receiving Nevirapine (NVP) and Efavirenz (EFV). It reduces NVP, EFV, Rilpivirine (RPV) and Doravirine (DOR) susceptibility about 2-fold [Melikian et al., 2014; Vingerhoets et al., 2010; Westen et al., 2013; Asante-Appiah et al., 2021; Rhee et al., 2022]. The E138A mutation was present in 4 of 184 patients (2.17%; 3 Slovaks and 1 foreigner). E138A is a polymorphic mutation that ranges in prevalence from about 2.00–5.00% in ART-naive persons depending on subtype. It reduces Etravirine (ETR) and RPV susceptibility about 2-fold [Azijn et al., 2010; Melikian et al., 2014; Lai et al., 2014; Tambuyzer et al., 2011, Rizzardini et al., 2020]. Another mutation K101E causing resistance to NNRTIs was observed in 2 of 184 individuals (1.09%; all Slovaks). This mutation was also detected in 3.7% of patients in Croatia in 2014–2017 [Oroz et al., 2019]. K101E is a non-polymorphic mutation and usually occurs in combination with other NNRTI-resistance mutations. Alone it confers 3–10-fold reduced susceptibility to NVP and about 2-fold reduced EFV, ETR, and RPV susceptibility [Azijn et al., 2010; Rimsky et al., 2012; Melikian et al., 2014; Feng et al., 2015; Lai et al., 2014].
In our study, the T215F/S mutation was detected in 2 of 184 persons (1.09%) from them 0.60% (1/167) was Slovak. This mutation causes resistance to the drug Zidovudine (AZT) (URL 2). Matuzzi et al. reported the occurrence of the T215D/S mutation in 1.50% (10/668) of Italians [Matuzzi et al., 2020] like as in Croatia (T215S: 7.40%; T215D: 1.70 %) in 2014–2017 [Oroz et al., 2019].
M46I/L was the most commonly detected mutation among PIs (2/181; 1,10%) it was found in 1.20% of Slovaks (2/167). M46IL mutation is non-polymorphic creating resistance to Atazanavir (ATV) and Lopinavir (LPV) (URL 3). The study by Klundert et al. also confirmed this mutation as the most common (0.86%) in patients from Eastern Europe and Russia in 2013 – 2020 [Klundert et al., 2022]. In Portugal, and Italy this mutation was described in 0.60% and 0.40% of patients, respectively [Pingarilho et al., 2022; Matuzzi et al., 2020].
Mutation R263K causing resistance to INSTIs was found in 2 of 151 (1.32%) of which 1.45% (2/138) patients were Slovaks. No mutations to any INSTIs were found in patients in Ukraine. Similarly, mutations against PI were not confirmed in a study of patients from Ukraine [Klundert et al., 2022]. R263K is a non-polymorphic mutation and decreases sensitivity to DTG, Bictegravir (BIC), and Cabotegravir (CAB) approximately 2-fold [Tsiang et al., 2016; Rhee et al., 2019; Rhee et al., 2022].
In HIV-1B and HIV-1A infected patients, mutations associated with HIV resistance to ART were present in 12 of 129 (9.30%) and 5 of 48 (10.42%) persons, respectively. The predominant route of transmission of all resistant strains was MSM (12/17; 70.59%) – Table 4.
CONCLUSION
The findings of our study confirmed the long-term highest incidence of subtype B in HIV-1 positive patients newly diagnosed in Slovakia in 2019 – 2021, with a slightly decreasing trend compared to last year. Sexual contact between men remained the main risk factor of HIV-1 infection associated mostly with HIV-1 B subtype infection. Despite increased migration in Europe in recent years, no new subtypes have been detected in Slovakia. Evidence of mutations causing resistance to ART in naïve patients highlights the significance of continuing ART resistance testing in newly diagnosed patients before initiating adequate treatment.
Table 4. Prevalence of most frequently detected mutations in newly diagnosed antiretroviral-naive HIV-infected patients in Slovakia in 2019–2021
No. of patient
NRTI
NNRTI
PI
INSTI
Subtype
Route of transmission
S/R
S/R
S/R
S/R
1. Slovak
S
A98G, V179VD
S
S
B
MSM
2. Slovak
S
E138A
S
S
A(A1)
HTS
3. Slovak
S
V108I
S
S
B
HTS
4. Slovak
S
A98G, K101E
S
R263K, L74M
B
MSM
5. Slovak
M41l, D67N, T69D, K70R, M184V, T215F, K219Q
K103N
S
S
B
MSM
6. Slovak
S
A98G, K101E
S
S
B
unknown
7. Slovak
S
S
S
R263K, S230SGR, D232DN
B
MSM
8. Slovak
S
S
I84V
S
B
MSM
9. Slovak
S
E138A
S
S
B
MSM
10. Slovak
S
A98G
S
S
B
MSM
11. Slovak
S
Y181YCFS
S
S
B
MSM
12. Slovak
S
E138A
S
S
A(A1)
MSM
13. Slovak
S
S
M46I, L89V
S
B
MSM
14. Slovak
S
S
M46L
S
A(A1)
MSM
1. foreigner (Poland)
T215S
F227L
S
S
B
MSM
2. foreigner (Ukraine)
S
V108VI
S
S
A(A1)
HTS
3. foreigner (Ukraine)
S
E138A
S
S
A(A1)
HTS
S – sensitive, R – resistant, NRTI – nucleoside/nucleotide inhibitor of reverse transcriptase, NNRTI – non-nucleoside/non-nucleotide inhibitor of reverse transcriptase, PI – protease inhibitor, INSTI – integrase inhibitor, MSM – men having sex with men, HTS – heterosexual contact, No. – Number of patients.
Do redakce došlo dne 22. 3. 2023.
Adresa pro korespondenci:
Ing. Alexandra Kovářová
Žltá 4 851 07 Bratislava
Slovenská republika
e-mail: alexandra.kovarova@szu.sk
Zdroje
- Abram ME, Ferris AL, Shao W, et al. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol, 2010;84(19):9864–9878.
- Alaeus A. Significance of HIV-1 genetic subtypes. Scand J Infect Dis, 2000;32(5):455–463.
- Alexiev I, Campbell EM, Knyazev S, et al. Molecular Epidemiology of the HIV-1 Subtype B Sub-Epidemic in Bulgaria. Viruses, 2020;12(4):441.
- ANRS AC11 Resistance Study Group (2015): PCR and sequencing procedures: HIV-1 [online]. Version January 2015 [cit. 2023-0206]. Available at www: https://hivfrenchresistance.org/wp-content/uploads/2021/10/ANRS-procedures.pdf
- Asante-Appiah E, Lai J, Wan H, et al. Impact of HIV-1 Resistance-Associated Mutations on Susceptibility to Doravirine: Analysis of Real-World Clinical Isolates. Antimicrob Agents Chemother, 2021; 65(12):e01216–1221.
- Azijn H, Tirry I, Vingerhoets J, et al. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother, 2010;54(2):718–727.
- Beloukas A, Psarris A, Giannelou P, et al. Molecular epidemiology of HIV-1 infection in Europe: An overview. Inf Gen Evol, 2016;46 : 180–189.
- Cuevas JM, Geller R, Garijo R, et al. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS Biol, 2015;13(9):1002251.
- Čereš A, Staneková D, Hábeková M, et al. 2018. HIV genetic diversity and occurrence of the HLA-B*57 : 01 allele in newly diagnosed HIV-positive patients in Slovakia in 2015–2016. Clinical Microbiology Reports. pp. 39, EV 2992/09, Year XVIII., Number SA/2018. ISSN 1338–645X.
- Davanos N, Panos G, Gogos Ch, Mouzaki A. HIV-1 subtype characteristics of infected persons living in southwestern Greece. HIV AIDS (Auckl), 2015;7 : 277–283.
- Delgado E, Benito S, Montero V, et al. Diverse Large HIV-1 Non-subtype B Clusters Are Spreading Among Men Who Have Sex With Men in Spain. Front Microbiol, 2019;10.
- De Oliveira T, Deforche K, Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics, 2005;21(19):3797–3800.
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 1985;39(4):783–791.
- Feng M, Wang D, Grobler JA, et al. In vitro resistance selection with doravirine (MK-1439), a novel nonnucleoside reverse transcriptase inhibitor with distinct mutation development pathways. Antimicrob Agents Chemother, 2015;59(1):590–598.
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser, 1999;41 : 95–98.
- Hauser A, Hofmann A, Meixenberger K, et al. Increasing proportions of HIV-1 non-B subtypes and of NNRTI resistance between 2013 and 2016 in Germany: Results from the national molecular surveillance of new HIV-diagnoses. PLoS ONE, 2018;13(11):e0206234.
- Hábeková M, Kovářová A, Takáčová M, Valkovičová Staneková D. Distribution of subtypes HIV1 in Slovakia: update 2017 – 2018. Acta Virol, 2023. ISSN 0001–723X.
- Hábeková M, Takáčová M, Lysy J, et al. Genetic Subtypes of HIV Type 1 Circulating in Slovakia. AIDS Research and Human Retroviruses, 2010;26(10):1103–1107.
- Hemelaar J, Elangovan R, Yun J, et al. Global and regional molecular epidemiology of HIV-1, 1990–2015: A systematic review, global survey, and trend analysis. Lancet Infect Dis, 2019;19 : 143 – 155.
- Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med, 2012;18(3):182–192.
- Hofstra LM, Sauvageot N, Albert J, et al. Ryding, for the SPREAD Program, Transmission of HIV Drug Resistance and the Predicted Effect on Current First-line Regimens in Europe. Clin Infect Dis, 2016;62(5):655–663.
- Hofstra LM, Seguin-Devaux C, Struck D, et al. The prevalence of drug resistance mutations in newly-diagnosed HIV-patients in Europe: interactive monitoring. In: Hofstra M. HIV Drug Resistance. Utrecht: University of Utrecht; 2019. pp. 63–77. ISBN: 978-94-6361-225-8
- Chabadová Z, Habeková M, Truska P, et al. Distribution of HIV-1 subtypes circulating in Slovakia (2009–2012). Acta Virol, 2014;58(4):317–324.
- Junqueira DM, de Matos Almeida SE. HIV-1 subtype B: Traces of a pandemic. Virology, 2016;495 : 173–184.
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol, 1980;16(2):111–120.
- Klundert MAA, Antonova A, Di Teodoro G et al. Molecular Epidemiology of HIV-1 in Eastern Europe and Russia. Viruses, 2022;14(10):2099.
- Konstantinos A, Albert J, Mamais I, et al. Global dispersal pattern of HIV-1 CRF01_AE: A genetic trace of human mobility related to heterosexual activities centralized in South-East Asia J Infect Dis, 2015;211 : 1735–1744.
- Kostaki EG, Limnaios S, Adamis G, et al. Estimation of the determinants for HIV late presentation using the traditional definition and molecular clock-inferred dates: Evidence that older age, heterosexual risk group and more recent diagnosis are prognostic factors. HIV Medicine, 2022;23(11):1143–1152.
- Kumar S, Stecher G, Li M, et al. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol, 2018;35(6):1547–1549.
- Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res, 2007; 35(Web Server issue): W6–W11.
- Lai MT, Feng M, Falgueyret JP, et al. In vitro characterization of MK-1439, a novel HIV-1 nonnucleoside reverse transcriptase inhibitor. Antimicrob Agents Chemother, 2014;58(3):1652–1663.
- Lee HY, Perelson AS, Park SC, Leitner T. Dynamic correlation between intrahost HIV-1 quasispecies evolution and disease progression. PLoS Comput Biol, 2008;4(12):e1000240.
- Lorenzin G, Gargiulo F, Caruso A, et al. Prevalence of Non-B HIV-1 Subtypes in North Italy and Analysis of Transmission Clusters Based on Sequence Data Analysis. Microorganisms, 2020;8(1):36.
- Lunar MM, Vandamme AM, Tomažič J, et al. Bridging epidemiology with population genetics in a low incidence MSM-driven HIV-1 subtype B epidemic in Central Europe. BMC Infect Dis, 2015;15 : 65.
- Magiorkinis G, Angelis K, Mamais I, et al. The global spread of HIV-1 subtype B epidemic. Infect Genet Evol, 2016;46 : 169–179.
- Matuzzi L, Melengu T, Falasca F, et al. Transmitted drug resistance mutations and trends of HIV-1 subtypes in treatment-naïve patients: A single-centre experience. J Glob Antimicrob Resist, 2020;20 : 298–303.
- Melikian GL, Rhee SY, Varghese V, et al. Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. J Antimicrob Chemother, 2014;69(1):12–20.
- Mravčík V, Pitoňák M, Hejzák R, et al. HIV epidemic among men who have sex with men in the Czech Republic, 2016: high time for targeted action. Euro Surveill. 2017;22(48):17–00079.
- Murillo W, Veras NMC, Prosperi MCF, et al. A single early introduction of HIV-1 subtype B into Central America accounts for most current cases. J Virol, 2013;87 : 7463–7470.
- Mustafa A, Akbay B, Davlidova S, et al. Origin and evolution of subtype B variants in the former Soviet Union countries. Infect Genet Evol, 2023;108 : 105402.
- Neogi U, Häggblom A, Santacatterina M, et al. Temporal Trends in the Swedish HIV-1 Epidemic: Increase in Non-B Subtypes and Recombinant Forms over Three Decades. PLoS ONE, 2014;9(6):e99390.
- Oroz M, Begovac J, Planinić A, et al. Analysis of HIV-1 diversity, primary drug resistance and transmission networks in Croatia. Sci Rep, 2019;9(1):17307.
- Paraskevis D, Nikolopoulos GK, Magiorkinis G, et al. The application of HIV molecular epidemiology to public health. Infect Genet Evol, 2016;46 : 159–168.
- Parczewski M, Leszczyszyn-Pynka M, Witak-Jedra M, et al. Distribution and time trends of HIV-1 variants in Poland: Characteristics of non-B clades and recombinant viruses. Infect Genet Evol, 2016;39 : 232–240.
- Pingarilho M, Pimentel V, Miranda MNS, et al. HIV-1-Transmitted Drug Resistance and Transmission Clusters in Newly Diagnosed Patients in Portugal Between 2014 and 2019. Front Microbiol, 2022;13 : 823208.
- Public Health Authority of the Slovak Republic. Incidence of infection in the Slovak Republic until June 30, 2022 [online]. 2022 [cit. 2023-02-01]. Available at www: <https:// www.uvzsr.sk/index.php?option=com_content&view=ar- ticle&id=5463%3Avyskyt-hiv-infekcie-v-slovenskej-repub like-k-306-2022&catid=283%3Ahivaids&Itemid=112>.
- Rhee SY, Grant PM, Tzou PL, et al. A systematic review of the genetic mechanisms of dolutegravir resistance. J Antimicrob Chemother, 2019;74(11):3135–3149.
- Rhee SY, Parkin N, Harrigan PR, et al. Genotypic Correlates of Resistance to the HIV-1 Strand Transfer Integrase Inhibitor Cabotegravir. Review 2022 Sept. 01, PREPRINT (Version 1). Available at www: https://doi.org/10.21203/rs.3.rs-2012078/v1.
- Rhee SY, Schapiro JM, Saladini F, et al. Potential Role of Doravirine for the Treatment of Patients with Transmitted Drug Resistance. AIDS Res Ther, 2023;20 : 8.
- Rimsky L, Vingerhoets J, Van Eygen V, et al. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. J Acquir Immune Defic Syndr, 2012 Jan 1;59(1):39–46.
- Rizzardini G, Overton ET, Orkin C, et al. Long-Acting Injectable Cabotegravir + Rilpivirine for HIV Maintenance Therapy: Week 48 Pooled Analysis of Phase 3 ATLAS and FLAIR Trials. J Acquir Immune Defic Syndr, 2020;85(4):498–506.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 1987;4(4):406 – 425.
- Serwin K, Scheibe K, Horecki M, et al. Detection of Polish cases of highly virulent subtype B of HIV-1 originating in the Netherlands. Journal of Medical Virology, 2022;95(1):28154.
- Serwin K, Urbańska A, Scheibe K, et al. Molecular epidemiology and HIV-1 variant evolution in Poland between 2015 and 2019. Sci Rep, 2021;11 : 16609.
- Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med, 2011;1(1):a006841.
- Tambuyzer L, Nijs S, Daems B, et al. Effect of mutations at position E138 in HIV-1 reverse transcriptase on phenotypic susceptbility and virologic response to etravirine. J Acquir Immune Defic Syndr, 2011;58(1):18–22.
- Tsiang M, Jones GS, Goldsmith J, et al. Antiviral Activity of Bictegravir (GS-9883), a Novel Potent HIV-1 Integrase Strand Transfer Inhibitor with an Improved Resistance Profile. Antimicrob Agents Chemother, 2016;60(12):7086–7097.
- UNAIDS. Global HIV & AIDS statistics – Fact sheet [online]. 2023 [cit. 2023-02-03]. Available at www: <https://www.unaids.org/ en/resources/fact-sheet>.
- URL 1. STANFORD HIVDB PROGRAM. Release notes [online]. 2021 [cit. 2023-02-04]. Available at www: <https://hivdb.stanford.edu/page/release-notes/>.
- URL 2. NRTI Resistance Notes [online]. 2022 [cit. 2023-02-04]. Available at www: <https://hivdb.stanford.edu/dr-summary/ resistance-notes/NRTI/>.
- URL 3. PI Resistance Notes [online]. 2022 [cit. 2023-02-04]. Available at www: <https://hivdb.stanford.edu/dr-summary/resistance-notes/PI/>.
- URL. 4. HIV sequence database [online]. 2019 [cit. 2023-0119]. Available at www: <https://www.hiv.lanl.gov/content/se- quence/LOCATE/locate.html>.
- Vingerhoets J, Tambuyzer L, Azijn H, et al. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. AIDS, 2010;24(4):503–514.
- Mortier V, Debaisieux L, Dessilly G, et al. Prevalence and Evolution of Transmitted Human Immunodeficiency Virus Drug Resistance in Belgium Between 2013 and 2019. Open Forum Infect Dis, 2022;9(7):195.
- Westen GJP, Hendriks A, Wegner JK, et al. Significantly improved HIV inhibitor efficacy prediction employing proteochemometric models generated from antivirogram data. PLoS Comput Biol, 2013. Available at www: https://doi.org/10.1371/journal. pcbi.1002899.
- WHO. HIV [online]. 2022 [cit. 2023-02-04]. Available at www:<https://www.who.int/news-room/fact-sheets/detail/hiv- aids>.
- Wymant CH, Bezemer D, Blanquart F, et al. A highly virulent variant of HIV-1 circulating in the Netherlands. Science, 2022;375(6580):540–545.
- Yamaguchi J, McArthur C, Sthreshley L, et al. Brief Report: Complete genome sequence of CG-0018a-01 establishes HIV-1 subtype L. J Acquir Immune Defic Syndr, 2020;83(3):319–322.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Mikrobiológia
Článek Rejstřík
Článok vyšiel v časopiseEpidemiologie, mikrobiologie, imunologie
Najčítanejšie tento týždeň
2023 Číslo 4- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- HIV-1 subtypes distribution and resistance to ART in HIV-infected persons in Slovakia (2019–2021)
- Risk of Streptococcus pneumoniae-associated haemolytic uraemic syndrome in industrialised nations: a systematic review of the literature
- Patogeneze infekcí vyvolaných virem SARS-CoV-2
- Vzteklina ve světě a program „Zero by 30“
- Analýza očkování vakcínami proti meningokokovým onemocněním u pacientů s invazivním meningokokovým onemocněním, Česká republika, 2006–2022
- Významné životní jubileum RNDr. Pavly Urbáškové, CSc.
- Pokyny pro autory a recenzenty
- Rejstřík
- Epidemiologie, mikrobiologie, imunologie
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Patogeneze infekcí vyvolaných virem SARS-CoV-2
- Vzteklina ve světě a program „Zero by 30“
- HIV-1 subtypes distribution and resistance to ART in HIV-infected persons in Slovakia (2019–2021)
- Risk of Streptococcus pneumoniae-associated haemolytic uraemic syndrome in industrialised nations: a systematic review of the literature
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



