-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
STAT Is an Essential Activator of the Zygotic Genome in the Early
Embryo
In many organisms, transcription of the zygotic genome begins during the
maternal-to-zygotic transition (MZT), which is characterized by a dramatic
increase in global transcriptional activities and coincides with embryonic stem
cell differentiation. In Drosophila, it has been shown that
maternal morphogen gradients and ubiquitously distributed general transcription
factors may cooperate to upregulate zygotic genes that are essential for pattern
formation in the early embryo. Here, we show that Drosophila
STAT (STAT92E) functions as a general transcription factor that, together with
the transcription factor Zelda, induces transcription of a large number of
early-transcribed zygotic genes during the MZT. STAT92E is present in the early
embryo as a maternal product and is active around the MZT. DNA–binding
motifs for STAT and Zelda are highly enriched in promoters of early zygotic
genes but not in housekeeping genes. Loss of Stat92E in the
early embryo, similarly to loss of zelda, preferentially
down-regulates early zygotic genes important for pattern formation. We further
show that STAT92E and Zelda synergistically regulate transcription. We conclude
that STAT92E, in conjunction with Zelda, plays an important role in
transcription of the zygotic genome at the onset of embryonic development.
Published in the journal: STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo. PLoS Genet 7(5): e32767. doi:10.1371/journal.pgen.1002086
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002086Summary
In many organisms, transcription of the zygotic genome begins during the
maternal-to-zygotic transition (MZT), which is characterized by a dramatic
increase in global transcriptional activities and coincides with embryonic stem
cell differentiation. In Drosophila, it has been shown that
maternal morphogen gradients and ubiquitously distributed general transcription
factors may cooperate to upregulate zygotic genes that are essential for pattern
formation in the early embryo. Here, we show that Drosophila
STAT (STAT92E) functions as a general transcription factor that, together with
the transcription factor Zelda, induces transcription of a large number of
early-transcribed zygotic genes during the MZT. STAT92E is present in the early
embryo as a maternal product and is active around the MZT. DNA–binding
motifs for STAT and Zelda are highly enriched in promoters of early zygotic
genes but not in housekeeping genes. Loss of Stat92E in the
early embryo, similarly to loss of zelda, preferentially
down-regulates early zygotic genes important for pattern formation. We further
show that STAT92E and Zelda synergistically regulate transcription. We conclude
that STAT92E, in conjunction with Zelda, plays an important role in
transcription of the zygotic genome at the onset of embryonic development.Introduction
Embryonic pattern formation is a complex and progressive process. In many multicellular organisms, the initial period of embryogenesis relies on gene products inherited from the mother. In Drosophila, maternally derived morphogen proteins form broad gradients along the major body axes to define body polarities [1]–[3]. Zygotic transcription begins during the maternal-to-zygotic transition (MZT), which is characterized by a decline in maternal mRNA levels and a dramatic increase in a large number of zygotic transcripts [4], [5]. Many of the zygotic genes transcribed the earliest, exhibit region-specific patterns. For instance, the “gap genes”, such as zygotic hunchback (hb), Krüppel (Kr), knirps (kni), and tailless (tll) are transcribed zygotically in broad and mostly non-overlapping domains along the anteroposterior (A/P) body axis. The boundaries of these zygotic genes are determined by morphogen gradients that are set up by maternal gene products, such as Bicoid (Bcd) and maternal Hb [2], [3]. Additional zygotic genes, mostly transcription factors, are induced in more refined embryonic regions as a result of cooperation between the maternal morphogens and gap gene products. The combinatorial input of different transcription factors at different positional coordinates results in expression of thousands of zygotic genes in an increasingly refined pattern, leading to cell fate determination and differentiation [1]–[3], [6].
To date, only a few transcription factors have been implicated in transcription of the zygotic genome during the MZT. For example, the maternal morphogens Bcd and Dorsal activate target genes along the anteroposterior (A/P) and dorsoventral (D/V) axis, respectively [7], [8]. The dramatic increase in gene expression that occurs during the MZT raises the possibility that additional unidentified transcription factors are involved in the rapid initiation and maintenance of the heightened levels of zygotic gene transcription that characterize the MZT. It has been proposed that the few known regionally localized transcription factors, such as Bcd and Dorsal, act in conjunction with ubiquitously present factors to induce and maintain expression of a large number of zygotic genes in cell type-specific patterns. This idea is supported by the identification of a ubiquitous factor encoded by zelda (zld; a.k.a. vielfaltig or vlf) [9], and further by the demonstration that combining Dorsal with Zelda - or STAT-binding sites supports transcription in a broad domain in the embryo [10].
To identify additional ubiquitous transcription factors that are important for transcription of the zygotic genome during the MZT, we first conducted in silico analyses, taking advantage of the large amount of information available in public databases on transcriptional regulation of zygotic genes expressed during early embryogenesis in Drosophila. This approach led to the identification of STAT92E, in addition to Zelda, as a plausible transcription factor important for the upregulation of multiple genes during the MZT. Global expression profiling studies indicate that loss of STAT92E, similarly to loss of Zelda, preferentially causes down-regulation of zygotic genes essential for early embryogenesis. We further demonstrate that STAT92E is indeed involved in transcription of the developmentally important genes dpp, tailless (tll), and Kr during early embryogenesis. Our results suggest that STAT92E is essential for upregulation of a multitude of zygotically transcribed genes during the MZT, and thus is important for transition of the early embryo from a totipotent embryonic stem cell state to a state of cellular differentiation.
Results
In silico identification of factors important for transcription of the zygotic genome
To identify general transcription factors that are required for transcription of a large number of zygotic genes at early embryonic stages, or during the MZT, we performed a meta-analysis to search for candidate transcription factors required for activation of multiple zygotic genes. To this end, we first selected a list of developmentally important zygotic genes transcribed during the MZT (referred to as “zygotic genes”), whose expression patterns altogether cover the entire embryo, and whose transcriptional activation has previously been studied. We analyzed a total of 21 early zygotic genes, including the gap genes: hunchback (hb), huckebein (hkb), Giant (Gt), Krüppel (Kr), knirps (kni), and tailless (tll); the pair-rule genes: even skipped (eve), fushi tarazu (ftz), hairy (h), odd paired (opa), paired (prd), sloppy paired 1 (slp1), and runt (run); the segmental polarity and other genes: engrailed (en) and Sex lethal (Sxl), as well as genes expressed along the D/V axis: decapentaplegic (dpp), zerknüllt (zen), rhomboid (rho), short gastrulation (sog), snail (sna), and twist (twi).
As a second step, for each of these genes, we searched Flybase (http://flybase.org) and PubMed (http://www.ncbi.nlm.nih.gov), and compiled a list of all currently known or potential transcriptional activators or signaling pathways involved in their transcriptional induction (Table S1). We used the RedFly database (http://redfly.ccr.buffalo.edu) [11] to obtain a list of experimentally verified transcription factor binding sites for each target gene, and the FlyEnhancer program (http://genomeenhancer.org/fly) [12] to search for the presence of particular transcription factor binding sites in the promoter region (defined as 4 kb upstream of the transcriptional start site) of all the target genes. Based on these search results, we assigned activation scores to the putative or known transcriptional activators to reflect their importance in the expression of a particular zygotic gene (Table S1). These scores were added to obtain a cumulative score for each activator (Figure 1A; Table S2). The connections between activators and their target genes are represented in an activation map (Figure 1B).
The top seven activators identified, in descending order of cumulative interaction score, were Zelda (Zld), Bicoid (Bcd), STAT92E, Torso, Caudal (Cad), Dorsal, and Twist (Twi) (Figure 1A; Table S2). Zelda has previously been shown to be a key transcription activator of the early zygotic genome [9], validating our bioinformatic approach. Both Bcd and Cad are maternal-effect gene products that form gradients along the A/P axis in the early embryo [7], [13], [14]; Torso signaling is activated only at the anterior and posterior poles, and the specific transcriptional activators that it regulates remain unidentified [15]–[17]; Dorsal and Twi are active only in the ventral region of the embryo [18]. On the other hand, STAT92E is ubiquitously distributed in the early embryo as a maternal product [19] and is activated early [20], and thus has the potential to act more universally. STAT92E is the transcriptional activator mediating the JAK/STAT (Hop/STAT92E) pathway [19], [21], [22], and also participates in Torso signaling [23]–[25]. Thus, we decided to investigate whether STAT92E acts as a general transcriptional regulator during early embryogenesis, similar to Zelda.
STAT - and Zelda-binding sites are enriched in promoter regions of early zygotic genes
To test whether STAT92E is important for transcription of early “zygotic genes”, we first assessed the occurrence of consensus STAT92E binding sites (TTCnnnGAA) in the promoter region, defined as 4 kb genomic sequence upstream of the transcription start site, of the 21 zygotic genes in this study. The Drosophila genome is slightly AT-rich, with 57.4% AT and 42.6% GC base pairs [12]. Thus the probability for A or T to occur at any position is 0.287, and for G or C is 0.213, and the probability (p) for random occurrence of one STAT binding site (with 6 fixed nucleotides) at any position is 3.08x10−4 (0.2874x0.2132), and its frequency of occurrence within the 4 kb upstream regulatory regions of 21 genes (n = 84,000 bp) at random is 25.9 (np; expected value). However, when we searched for STAT binding sites within the 4 kb upstream region of the 21 zygotic genes, we found 43 in total (observed value) (Figure 1C). Assuming the actual occurrence of STAT-binding sites exhibits Binomial distribution with a probability of 3.08x10−4, the standard deviation (σ) should be 5.1. The difference between the observed (43) and expected (25.9) values is 17.1, which is beyond three standard deviations (Z = 3.29; p = 0.001).
In contrast, when we searched for STAT-binding sites within a 4 kb window upstream of the transcription start site of 21 housekeeping genes (defined as ubiquitously expressed, both maternally and zygotically, with generally cellular metabolic or structural functions), including rp49, GAPDH, Actin5C, and those encoding ribosomal proteins and RNA polymerases, we found a total of 13 STAT-binding sites (Figure 1D), which is significantly lower than the expected 25.9 sites (Z = 2.48; p = 0.013). (A total of 78 housekeeping genes and the numbers of STAT-binding sites in their upstream regions are listed in Table S3.) Moreover, many of the STAT-binding sites in the upstream regions of the 21 zygotic genes are clustered (defined by two sites occurring within 500 bp), which is characteristic of functional transcription factor binding sequences [12], [19], [25], [26] (Figure 1C), whereas in the promoter regions of the 21 housekeeping genes, the STAT-binding sites occur as single sites (Figure 1D; Table S3).
It has been shown that Zelda-binding sites (the TAGteam motif) are enriched in the promoter regions of “zygotic genes” [9], [27]. We examined the distribution of Zelda-binding sites in the promoter regions of the 21 zygotic and housekeeping genes, respectively. Consistent with the previous report [9], [27] and similar to STAT-binding sites, we found that Zelda-binding sites are similarly enriched in the promoters of the zygotic and very infrequently in the housekeeping genes (Figure 1C, 1D). Since the enhancers for many of the early zygotic genes are not localized in the upstream promoter regions, we also searched for STAT and Zelda-binding sites in the promoter-distal enhancers for these 21 zygotic genes, and found that promoter-distal enhancers are not enriched for STAT-binding sites (Z = 0.63; p = 0.736), but are significantly enriched for Zelda-binding sites (Z = 3.13; p = 0.0017) (Figure S1). Such a result suggests that STAT92E might differ from Zelda and might not be important for regulating promoter-distal enhancers, which usually control spatial expression patterns. Nonetheless, our studies indicate that DNA-binding sites for both STAT and Zelda are enriched in the upstream promoter regions of the 21 zygotic genes that are highly transcribed during the MZT, but are underrepresented in the housekeeping genes that are ubiquitously transcribed. This observation is consistent with the finding that Zelda is required specifically for expression of “zygotic genes” at the MZT [9], raising the possibility that STAT may play a similar role.
Similar to Zelda, STAT92E is required for transcription of the zygotic genome during the MZT
To determine whether STAT92E functions as a general transcriptional activator of the zygotically expressed genes in the early embryo, we determined the expression profiles of early stage embryos (corresponding to nuclear division cycle 8–14, a time window for the MZT) of wild-type control and of those lacking the maternal Stat92E gene products (referred to as Stat92Emat–; see Methods) at the same stage.
We found that in Stat92Emat– embryos, 657 genes were down regulated and 558 genes up-regulated by at least 1.5 fold, compared with wild-type control (Figure 2A). In Stat92Emat– embryos, genes exhibiting >1.5 fold change in expression constituted 8.9% of all genes (n = 13,615) on the Gene Chip, while the majority (91.1%) of the genes exhibited no significant changes (Figure S2). Consistent with the idea that STAT92E is preferentially required for expression of “zygotic genes”, the vast majority (78.2%) of the down-regulated genes in Stat92Emat– embryos were “zygotic genes” (Figure 2B, left; Table S4). In contrast, the up-regulated genes contained more maternally expressed than zygotically expressed genes (Figure 2B, right; Table S5). This observation is reminiscent of gene expression profiles of zld mutant embryos at the same stage, in which more “zygotic genes” than maternal genes are down-regulated [9]. By comparing the two sets of genes, we found that >50% of the “zygotic genes” that were down-regulated in zldmat– embryos (67/120) were also down-regulated in Stat92Emat–embryos, suggesting that these genes might be co-regulated by STAT and Zelda (Table S4).
Fig. 2. Expression profiles of embryos lacking maternal STAT92E. 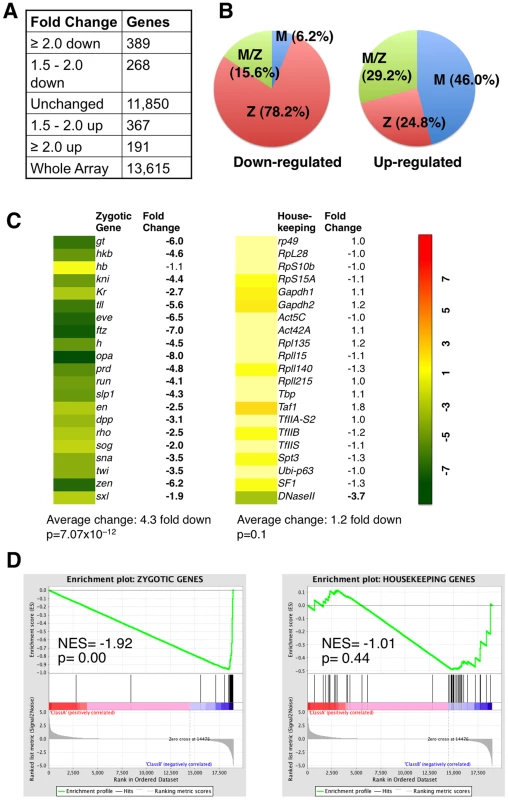
Consistent with the observed difference in the abundance of STAT-binding sites present in their promoter regions, the 21 zygotic genes (except for hb) were all significantly down-regulated, with a 4.3 fold down-regulation on average, whereas the 21 housekeeping genes showed no significant changes in expression, with the exception of DNase II (Figure 2C), in Stat92Emat– embryos. Similar to Stat92Emat– embryos, in zldmat– embryos, many of these 21 zygotic genes were also significantly down-regulated, whereas the housekeeping genes were not significantly changed [9], suggesting that STAT92E and Zelda may both be important for transcription of early zygotic genes. Expression profiling experiments indicate that STAT92E and Zelda do not transcriptionally regulate each other (Liang et al., 2008; this study). We further performed qRT-PCR experiments and found that Zelda mRNA levels were indeed not significantly changed in Stat92E loss-of-function or hop gain-of-function mutants (Figure S3), suggesting that STAT92E does not indirectly control zygotic gene activation by affecting Zelda levels.
Finally, we tested expanded sets of zygotic and housekeeping genes to include >40 genes in each set (Table S6) using the Gene Set Enrichment Analysis (GSEA) software (http://www.broadinstitute.org/gsea/index.jsp), which is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states (e.g., mutant versus wild-type) [28]. Indeed, by subjecting our microarray data to GSEA analysis, we found that the “zygotic genes” were highly significantly down regulated (p = 0.00), whereas the housekeeping genes were insignificantly changed (p = 0.44), in Stat92Emat– embryos when compared with wild-type control (Figure 2D). Thus, similar to Zelda, STAT92E is preferentially required for transcription of “zygotic genes”.
STAT92E and Zelda co-regulate multiple early “zygotic genes”
To validate our gene profiling results from the microarray studies, we investigated the effects of over-activation and loss of STAT92E on transcript levels of a number of early “zygotic genes”. We chose to examine expression levels of dpp, Kr, tll, and eve, four early zygotic genes whose promoter regions contain STAT-binding sites and whose expression domains span broad and distinct regions of the early embryo (see below).
We first examined mRNA levels of dpp, Kr, tll, and eve in the early embryo (1–2 h after egg laying) using semi-quantitative reverse-transcription polymerase chain reaction (RT-PCR) in Stat92E gain - or loss-of-function genetic backgrounds. We found that in hopGOF embryos, in which STAT92E is overactivated [29]–[31], mRNA of these four genes were all expressed at significantly higher levels relative to wild-type; whereas in Stat92Emat– embryos, these four genes were expressed at approximately 50% of the wild-type levels (Figure 3A, 3B). Moreover, reducing the dosage of zelda by half in Stat92Emat– embryos caused further reductions in the transcript levels of dpp, Kr, tll, and eve (zelda+/–; Stat92Emat– in Figure 3A, 3B). We examined zelda+/–; Stat92Emat– embryos only, because it was technically not possible to examine embryos lacking both Zelda and Stat92E. We further confirmed the expression results by quantitative real-time PCR (Figure 3C). These results were consistent with the microarray data, which suggested that Stat92E and Zelda may co-regulate transcription of many “zygotic genes”.
We next investigated whether STAT92E binds to the putative STAT-binding sites in the respective promoter regions of dpp, Kr, and tll using chromatin immunoprecipitation (ChIP) experiments with early embryo extracts using anti-STAT92E antisera. Binding of STAT92E to the eve enhancer and of Zelda to the TAGteam sequences enriched in “zygotic genes” have been previously shown [9], [19], [21]. Using primers flanking the putative STAT-binding sites in these promoter regions, we detected STAT92E binding to the promoter regions dpp, Kr, and tll (Figure 3D). The results from RT-PCR and ChIP studies were consistent with the bioinformatic and gene profiling studies shown above, suggesting that STAT92E, likely together with Zelda, regulates the transcription of early “zygotic genes” in vivo.
STAT and Zelda cooperate to regulate dpp transcriptional regulation
Having shown that STAT92E regulates expression levels of early “zygotic genes”, and that STAT92E binds to the consensus STAT-binding sites present in the promoter regions of dpp, Kr, and tll, we next investigated whether these consensus STAT-binding sites are indeed essential for mediating STAT92E transcriptional activation, and whether STAT92E and Zelda cooperate to regulate “zygotic genes”, as it has previously been shown that Zelda is essential for expression of dpp, Kr, tll, and eve, among others, in the early embryo [9]. We carried out reporter gene assays in Drosophila S2 cells (Figure 4A).
We first tested whether activated STAT92E binds to the promoter regions of dpp, Kr, tll, and eve in S2 cells as it does in early embryos (see Figure 3C). We transfected a V5-tagged STAT92E into S2 cells and performed ChIP assays. STAT92E activation in S2 cells was achieved by co-expressing Hop, which phosphorylates and activates STAT92E when over-expressed (Figure 4B). By immunoprecipitation with anti-V5 antibody, we found that co-transfection with Hop leads to an enrichment of STAT92E binding to the endogenous dpp promoter (Figure 4C, lane 3). Activation of JAK/STAT signaling thus induces a stronger association of STAT92E with the dpp promoter, consistent with the idea that STAT92E directly regulates dpp expression. However, the same ChIP experiments failed to detect association of STAT92E with the Kr, tll, or eve promoter in S2 cells, in contrast to the ChIP results in early embryos (see Figure 3C), suggesting that the epigenetic states of these promoter sequences may be different in S2 cells than in early embryos. We thus focused on the dpp promoter for reporter gene analysis. To this end, we isolated a 1.3 Kb dpp promoter fragment (Figure 4A; Figure S4), which contains the two clustered STAT92E binding sites we had tested in ChIP experiments (see Figure 3C, Figure 4C).
To test whether the STAT-binding sites in the dpp promoter are important for JAK/STAT-induced dpp expression, we made reporter genes by fusing a wild-type dpp promoter fragment (WT), or a mutant version with both STAT-binding sites mutated (DM), with an enhanced yellow fluorescent protein (EYFP), and transfected S2 cells (Figure 4A). In order to activate reporter gene expression, we first treated the cells with H2O2/vanadate (pervanadate), which causes rapid and efficient STAT92E phosphorylation [32], [33] (Figure S5A) and is more efficient than transient transfection of hop in activating STAT. We found that, indeed, EYFP was expressed 1.5 hours after pervanadate treatment in S2 cells transfected with the wild-type (WT), but not the double mutant (DM) construct (Figure S5B), indicating that these STAT92E-binding sites are important for phosphorylated STAT92E-induced reporter gene expression.
To more accurately quantify transcription from the dpp promoter with or without the two STAT-binding sites, we replaced EYFP with luciferase in the reporter constructs to obtain dppWT-luc and dppDM-luc, respectively. In addition, we used Hop and STAT92E co-transfection, instead of pervanadate, to ensure specific activation of STAT92E. In the presence of co-transfected Hop and STAT92E, we detected an increase in luciferase activity in S2 cells tranfected with dppWT-luc to more than 20 fold when measured 72 hours after transgene expression, and this increase was abolished when dppDM-luc was used in the assay, which showed much less pronounced increase (Figure 4D). These results further substantiate our finding that STAT92E-mediated activation of dpp requires the two STAT92E binding sites.
It has previously been shown that transcription of dpp is significantly down-regulated in the absence of Zelda [9], and that Zelda-binding sites are present in the dpp promoter region (Figure 1C; Figure 4A; also see [9]). To test whether Zelda binds to the putative site in the dpp reporter gene, we carried out ChIP assays in S2 cells after transfecting a Zelda-Flag plasmid. Indeed, we detected Zelda binding to the dpp promoter region using an anti-Flag antibody and ChIP assay (Figure 4E).
We next investigated the role of Zelda in dpp transcription using dppWT-luc and a mutant promoter fragment with the Zelda-binding site and the two STAT-binding sites mutated (designated as dpp™-luc as it bears triple mutations; Figure 4A). To evaluate whether Zelda and STAT cooperate in regulating dpp transcription, we co-transfected S2 cells with STAT92E (together with Hop to achieve STAT activation) or Zelda, or both STAT92E (with Hop) and Zelda, in the presence of dppWT-luc or dpp™-luc, and carried out luciferase assays. When assayed at 72 h after induction of transgene expression, we found that STAT activation alone induced dppWT-luc transcription by 22 fold, and Zelda alone caused upregulation of dppWT-luc by 48 fold, whereas in the presence of both Zelda and activated STAT, dppWT-luc was up-regulated by 230 fold (Figure 4F). Mutating STAT and Zelda binding sites prevented the dramatic increase in transcription as measured by luciferase activity (Figure 4F). These results suggest that Zelda and STAT have synergistic effects on dppWT-luc transcription. Interestingly, an increase in luciferase activity was observed even when binding sites for STAT or Zelda, or both, were mutated, albeit to a much less pronounced level than with the wild-type promoter (Figure 4D, 4F), suggesting that there might be other cryptic binding sites present in the promoter, or that other molecules were activated by over-expressed JAK or Zelda.
The apparent synergy between STAT92E and Zelda could explain the results from the gene profiling experiments. Microarray results show that embryos without STAT92E (in which Zelda presumably remains active) exhibit a 3.1 fold decrease in dpp expression (Figure 2B), and that Zld mutant embryos (in which presumably STAT92E is still active) have reduced dpp expression by 5.7 fold [9]. These data suggest that in the early embryo either Zelda or STAT activation could induce dpp transcription to a limited extent, whereas the presence of both Zelda and STAT activation synergistically promote dpp transcription.
STAT92E regulates transcription levels, but not spatial domains, of early zygotic genes
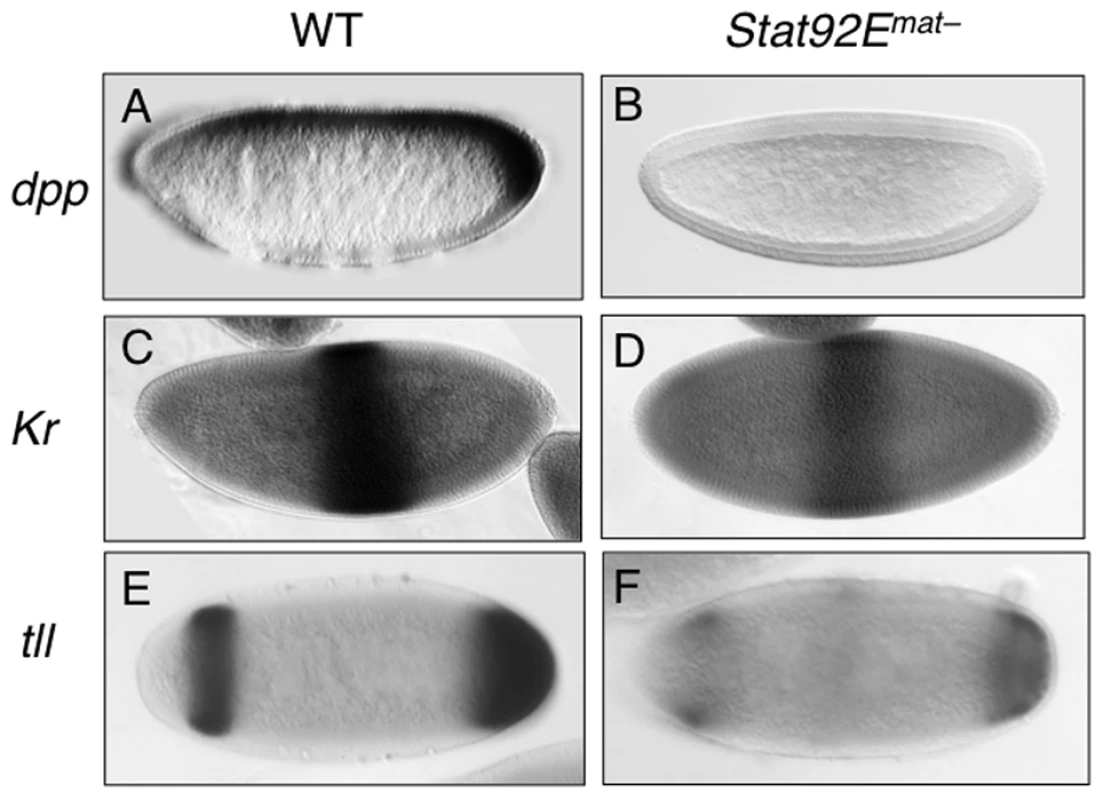
Having shown that STAT92E, possibly acting synergistically with Zelda, is important for expression levels of many early “zygotic genes”, we next investigated whether loss of STAT92E also affects the spatial expression patterns of the early “zygotic genes”. We examined the expression of dpp, Kr, and tll in the early embryo, by in situ hybridization, while the effects of Stat92E mutation on eve expression have previously been documented [19], [21]. These genes are expressed in distinct spatial domains that altogether cover nearly the entire early embryo (see below).
The dpp expression domain spans nearly the entire A/P axis in the dorsal regions of the early embryo [34]-[37] (Figure 5A). It has been shown that dpp transcription in the ventral region is repressed by Dorsal, a Rel family transcription factor [38], and that general transcription factors, such as Zelda and STAT, are responsible for dpp expression in the dorsal region ([9]; this study). By employing in situ hybridization, we found that compared to wild type, the overall level of dpp mRNA is much reduced in Stat92Emat – embryos, especially in the posterior pole region (Figure 5B). Moreover, we found that JAK/STAT signaling also regulates dpp expression during late embryogenesis (Figure S6). These results are consistent with previous findings in other developmental contexts [39], [40] as well as with the above microarray results and mRNA measurements (Figure 2, Figure 3A–3C).
Kr is expressed in the central region of the early embryo [41] (Figure 5C). Other than the maternal morphogens Bcd and Hb, it is not known whether additional factors contribute to Kr transcriptional activation. We found that in Stat92Emat – embryos, although the overall expression pattern of Kr mRNA was little changed, its levels were reduced (Figure 5D), consistent with the microarray and qPCR results.
tll is expressed in two domains along the A/P axis-the anterior and posterior pole regions [42] (Figure 5E). The Torso pathway controls tll expression by antagonizing its repressors [17], [43]; the identity of transcriptional activators of tll remains obscure, although STAT92E has been speculated to contribute to tll expression [25]. We have previously shown that STAT92E is essential for the expansion of tll expression domains caused by Torso, over-activation, but not for the extent of tll spatial expression domains under normal conditions [25]. In addition, we have previously shown that there are two consensus STAT binding sites in the tll promoter region that are particularly important for Torso overactionvation-induced ectopic tll expression [25]. In light of our finding that STAT92E is important for the expression levels of dpp, Kr, and tll, we reexamined the role of STAT92E in endogenous tll expression in Stat92Emat– and wild-type control embryos by in situ hybridization done under identical conditions. We found that, similar to dpp and Kr mRNA, while the spatial patterns of tll expression were not dramatically changed as previously shown [25], the overall levels of tll mRNA were significantly reduced in Stat92Emat– embryos (Figure 5F).
Taken together, the above results indicate that loss of STAT92E led to much reduced expression levels of dpp, Kr, and tll, without affecting their spatial expression domains. Similarly, it has been shown that loss of STAT92E results in reductions, but not complete loss of, eve stripe 3 and 5, without affecting the overall spatial expression pattern of eve [19], [21]. Thus, STAT92E is likely required for regulating the expression levels of early “zygotic genes”, but not for controlling their spatial patterns.
Loss of STAT results in multiple defects in embryonic pattern formation
Finally, we investigated the biological consequences of reducing expression levels, without altering spatial domains, of multiple zygotically expressed early genes, as with loss of STAT92E. The correct expression of the early zygotic genes during the MZT is essential for formation of different tissues and body parts at the correct positions, i.e., pattern formation [1]–[3]. Pattern formation in Drosophila can be conveniently visualized by examining the exoskeleton (cuticle) morphology of the larva or late embryo [1]–[3].
In the wild-type cuticle (Figure 6A), anteroposterior (A/P) polarity is defined by the head skeleton and three thoracic segments in the anterior, followed by the abdominal segments, and the posterior and terminal structure, consisting of the 8th abdominal segment and the Filzkörper (Figure 6A; Arrow). Dorsoventral (D/V) polarity can easily be seen by the positions of the eight abdominal denticle belts, which form in the ventral region, while bare cuticle marks the dorsal region (Figure 6A). Removal of STAT92E from the early embryo resulted in heterogeneous defects, mostly notably along the A/P axis as seen in the larval cuticles, which were missing part or all of A3, A4, A5, and A8 to various degrees (Figure 6B; also see [19], [25]). Thus, loss of STAT92E, which significantly reduces multiple early “zygotic genes” but does not completely eliminate their expression (see Figure 5), leads to heterogeneous patterning defects, consistent with defects in multiple pathways.
To understand the role of STAT92E in individual signaling pathways important for pattern formation, we investigated whether loss of STAT92E could further compromise pattern formation in sensitized genetic backgrounds. To this end, we examined cuticles of Stat92Emat– embryos that were also heterozygous for tll, Kr, or dpp, and indeed found patterning defects (see below).
The gap gene tll is essential for the development of terminal structures [17], [42], and tll mutant homozygous embryos do not have A8 and the Filzkörper (Figure 6C). tll heterozygous flies, in contrast, are perfectly viable and normal, with cuticles indistinguishable from wild-type controls, according to our own observation. In the absence of STAT92E, however, we found that tll+/– embryos were missing the terminal structures (A8 and Filzkörper) (Figure 6D). This suggests that without STAT92E, a half dose of tll+ is no longer sufficient for development, consistent with the idea that STAT92E is partially required for tll transcriptional output.
Kr is required for development of the thoracic and anterior segments, and these segments are missing in Kr–/– embryos (Figure 6E; also see [44]). Kr+/– embryos are mostly normal but have subtle anterior defects (Figure S7; also see [44]). In the absence of STAT92E, however, we found that Kr+/– embryos were missing a large area of the thoracic and anterior regions (Figure 6F), suggesting a haploinsufficiency in the absence of STAT92E, similar to what we observed for tll.
The dorsally expressed dpp specifies dorsal cell fates and is crucial for the dorsoventral polarity of the embryo, which is reflected in the cuticle by the presence of naked cuticles in the dorsal region and eight abdominal denticle belts in the ventral region (Figure 6A) [37]. Notably, although dpp expression was significantly reduced in Stat92Emat– embryos (Figure 2, Figure 3A–3C, Figure 5B), they did not exhibit gross D/V polarity defects (Figure 6B), suggesting that the residual dpp transcripts present in Stat92Emat– embryos are sufficient for specifying dorsal cell fates, or that the reduction in dpp expression is compensated for by a reduction in a dpp antagonist that is also regulated by STAT92E. Despite the fact that dpp is haploinsufficient for viability, dpp heterozygous embryos exhibit normal D/V polarity, with clearly discernable ventral denticle belts and bare dorsal cuticles (Figure S7), suggesting that a half dose of dpp+ suffices for D/V patterning (also see [37]. Embryos homozygous for dpp, nonetheless, are completely “ventralized,” having denticle belts that extend into the dorsal region to surround the entire D/V axis (Figure 6G; also see [36], [37]). The combination of Stat92Emat– and dpp heterozygosity caused partial ventralization of the embryo; in 13% of Stat92Emat–; dpp+/– embryos (n = 11/86), the posterior-most denticle belt extended significantly dorsally to cover approximately 80% of the circumference (Figure 6H, arrow). Similar ventralization defects were never observed in Stat92Emat– and dpp+/– embryos (n>500). Thus, in the absence of STAT92E, a half dose of dpp is no longer sufficient for dorsoventral patterning, consistent with the notion that STAT92E normally regulates dpp expression levels.
In summary, loss of STAT92E caused heterogeneous patterning defects, as revealed by varying cuticle defects, consistent with an insufficiency of multiple pathways. A further reduction in the dosage of genes in different pathways, such as tll, Kr, and dpp, uncovered the role of STAT92E in regulation of specific early zygotic genes important for pattern formation.
Discussion
We have undertaken a bioinformatics approach to investigating the mechanisms controlling transcription of the zygotic genome that occurs during the MZT, and have identified STAT92E as an important general transcription factor essential for up-regulation of a large number of early “zygotic genes”. We have further investigated the role of STAT92E in controlling transcription of a few representative early zygotic genes, such as dpp, Kr, and tll, that are important for pattern formation and/or cell fate specification in the early embryo. Our studies suggest that STAT92E cooperate with Zelda to control transcription of many “zygotic genes” expressed during the MZT. While STAT mainly regulates transcription levels, but not spatial patterns, of dpp, tll, and Kr, and possibly also other “zygotic genes”, Zelda is essential for both levels and expression patterns of these genes [9].
The transcriptional network that controls the onset of zygotic gene expression during the MZT has remained incompletely understood. It has been proposed that transcription of the zygotic genome depends on the combined input from maternally derived morphogens and general transcription factors. The former are distributed in broad gradients in the early embryo and directly control positional information (e.g., Bicoid, Caudal, and Dorsal), whereas the latter are presumably uniformly distributed regulators that augment the upregulation of a large number of “zygotic genes”. Other than Zelda, which plays a key role as a general regulator of early zygotic expression [9], the identities of these general transcriptional activators have remained largely elusive. It has been shown that combining Dorsal with Zelda - or STAT-binding sites supports transcription in a broad domain in the embryo [10]. The demonstration of STAT92E as another general transcription factor sheds light on the components and mechanisms of the controlling network in the early embryo. Moreover, we have found that STAT92E and Zelda may cooperate to synergistically regulate “zygotic genes”. Our results thus validate the bioinformatics approach as useful in identifying ubiquitously expressed transcription factors that may play redundant roles with other factors and thus might otherwise be difficult to identify.
Our conclusion that STAT92E is important for the levels but not the spatial domains of target gene expression in the early embryo is consistent with several previous reports. It has been shown that in Stat92E or hop mutant embryos, expression of eve stripes 3 and 5 are significantly reduced but not completely abolished [19], [21]. In addition, JAK/STAT activation is required for the maintenance of high levels, but not initiation, of Sxl expression during the MZT [45], [46]. Moreover, it has previously been shown that STAT92E is particularly important for TorsoGOF-induced ectopic tll expression but not essential for the spatial domains of tll expression in wild-type embryos under normal conditions [25]. On the other hand, Zelda may be important for both levels and spatial patterns of gene expression. This idea is consistent with our finding that Zelda-binding sites are enriched in both promoter and promoter-distal enhancers regions, whereas STAT-binding sites are enriched in promoter regions only. It has been reported that pausing of RNA polymerase II is prominently detected at promoters of highly regulated genes, but not in those of housekeeping genes [47]. In light of our results that STAT and Zelda sites are highly enriched in the early zygotic gene promoters, we suggest that these transcription factors might contribute to chromatin remodeling that favors RNA polymerase II pausing at these promoters.
Finally, the MZT marks the transition from a totipotent state to that of differentiation of the early embryo. As a general transcription factor at this transition, STAT, together with additional factors (such as Zelda [9]), is important for embryonic stem cell differentiation. Further investigation is required to understand the molecular mechanism by which STAT and Zelda [9] cooperate in controlling zygotic transcription in the early Drosophila embryo. Moreover, it would be interesting to investigate whether STAT plays similar roles in embryonic stem cell differentiation in other animals.
Materials and Methods
Fly stocks and genetics
All crosses were carried out at 25°C on standard cornmeal/agar medium unless otherwise specified. Fly stocks of hopTum-l, Stat92E6346, and dppH46 were from the Bloomington Drosophila Stock Center (Bloomington, IN). To generate Stat92Emat– embryos, hsp70-flp; FRT82B Stat92E6346/TM3 females were crossed to hsp70-Flp; FRT82B [ovoD1, w+]/TM3 males. Their 3rd instar larval progeny were heat-shocked at 37°C for 2 hrs daily for 3–4 days, and resulting adult females of the genotype hsp70-flp; FRT82B Stat92E6346/FRT82B [ovoD1, w+] were used to produce embryos that lack maternal Stat92E gene products, as described in the dominant female-sterile “germline clone” technique [48].
Bioinformatic analyses
The following rules were used for assigning a score to known or putative activators of each of the “zygotic genes”. We placed top importance on genetically demonstrated activation during early embryogenesis, with such an activator receiving an activation score of 10. For instance, Torso was assigned a score of 10 as an activator of tll transcription based on the reports that tll is not expressed in torso loss-of-function mutants and is overexpressed in torso gain-of-function mutants [17], [49]. Activators identified by biochemical/promoter studies in early embryos or by genetic studies at other developmental stages were assigned a score of 5. Lower scores were assigned to other less stringent evidence of interaction, such as unconfirmed genetic screen results (5), in vitro biochemical assays (2), or bioinformatics studies (1) (Table S1).
Databases and programs used in this study:
Flybase (http://flybase.org); PubMed (http://www.ncbi.nlm.nih.gov); RedFly (http://redfly.ccr.buffalo.edu/); FlyEnhancer (http://genomeenhancer.org/fly).
DNA constructs and plasmids
The dpp promoter used in this study was a 1.3 kb genomic DNA fragment including the upstream regulatory sequences and the non-coding exon 1 of the of dpp transcript A (Figure S2). This genomic region has previously been shown to be the core promoter of dpp [38]. Standard cloning was used to generate transcription fusions between the dpp promoter and cDNAs of reporter genes, such as enhanced yellow fluorescent protein (EYFP) and luciferase. Mutagenesis of two STAT92E binding sites within the dpp promoter was done by PCR, and was verified by sequencing. V5-Hop and V5-STAT92E are gifts from S.X. Hou [50].
Examination of embryos
Cuticle preparations were performed according to a standard protocol with minor modifications. Embryos were dechorionated with 50% Clorox, washed extensively with 0.1% Triton, mounted in Hoyer's, and photographed using dark-field optics. In situ hybridization for detecting dpp, Kr, and tll mRNA was performed according to a standard protocol using digoxigenin-incorporated antisense RNA probes made from dpp, Kr, and tll cDNA, respectively, according to the supplier's protocol. A standard protocol was used for antibody staining of embryos, and a biotinylated secondary antibody and the Vectastain ABC kit (Vector Laboratories, Inc.) were used according to the manufacturer's instructions. Stained embryos were mounted in DAPI-containing mounting medium for accurate staging, when necessary. Mounted embryos were photographed using Normaski optics on a Zeiss Axioscope and images were analyzed using Photoshop or ImageJ software.
Microarray, semi-quantitative RT-PCR, and quantitative real-time PCR
Total RNA was isolated from embryos (from flies raised at 25°C) collected at 1–2 h after egg laying (corresponding to nuclear division cycles 8–14) using trizol (Invitrogen) or the RNeasy Kit (QIAGEN) according to the manufacturer's instructions. RNA quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit (Agilent Technologies Inc., Palo Alto, CA).
For RT-PCR analysis, first strand complementary DNA (cDNA) was generated from 5 µg of purified total RNA using Superscript III reverse transcriptase (Invitrogen) and oligo(dT)12–18 in 50 µl total reaction volume. The cDNA (at 1∶100 dilution) was used as template for either semi-quantitative PCR reactions or real time PCR analysis using SYBR green based detection on a BioRad iCycler. Reactions were carried out in triplicate, and melting curves were examined to ensure single products. Results were quantified using the “delta-delta Ct” method to normalize to rp49 transcript levels and to control genotypes. Data shown are averages and standard deviations from at least three independent experiments. The following primer pairs were used.
rp49: TCCTACCAGCTTCAAGATGAC, CACGTTGTGCACCAGGAACT.
dpp: AATCAATCTTCGTGGAGGAGCCGA, TTGGTGTCCAACAGCAGATAGCTC.
eve: TGCACGGATACCGAACCTACAACA, GTTCTGGAACCACACCTTGATCGT.
Kr: CAAGACGCACAAACGCGAACCTTA, TTGACGGTTTGCAGCCAGAAGTTG.
tll: AATACAACAGCGTGCGTCTTTCGC, ACATTGGTTCCTGTGCGTCTTGTC.
For microarray analysis, 200 ng of total RNA was used to prepare biotin-labeled RNA using Ambion MessageAmp Premier RNA Amplification Kit (Applied Biosystems, Foster City, CA). Briefly, the first strand of cDNA was synthesized using ArrayScript reverse transcriptase and an oligo(dT) primer bearing a T7 promoter. Then DNA polymerase I was used (in the presence of E. coli RNase H and DNA ligase) to convert single-stranded cDNA into a double-stranded DNA (dsDNA). The dsDNA was then used as a template for in vitro transcription in a reaction containing biotin-labeled UTP and T7 RNA Polymerase to generate biotin-labeled antisense RNA (aRNA). Twenty µg of labeled aRNA was fragmented and fifteen µg of the fragmented aRNA was hybridized to Affymetrix Drosophila Genome 2.0 Array Chips according to the manufacterer's Manual (Affymetrix, Santa Clara, CA). Array Chips were stained with streptavidin-phycoerythrin, followed by an antibody solution (anti-streptavidin) and a second streptavidin-phycoerythrin solution, performed by a GeneChip Fluidics Station 450.
The Array Chips were then scanned with the Affymetrix GeneChip Scanner 3000. The microarray image data were converted to numerical data with Genespring software (Agilent Technologies Inc., Palo Alto, CA) and normalized using the recommended defaults. The signals from 11 perfect matched oligonucleotides for a specific gene and 11 mis-matched oligonucleotides were used to make comparisons of signals. Genes were identified as present when the present (P) assignment was significant (p<0.05).
The Gene Set Enrichment Analysis (GSEA) online software (http://www.broadinstitute.org/gsea) was used to determine whether the predetermined gene sets (e.g., zygotic versus housekeeping; see Figure S6) show statistically significant, concordant differences between wild-type and Stat92Emat– embryos.
Antibodies and cell culture
Primary antibodies used in this study include mouse anti-V5 (Invitrogen; 1∶500 for Western blots), Rabbit anti-V5 (QED; 1∶200 for immunoprecipitation), goat anti-STAT92E (Santa Cruz; Cat# sc-15708; affinity-purified against the N-terminus of STAT92E; 1∶200), rabbit anti-Kr (1∶5000; a kind gift from C. Rushlow), and anti-phospho-STAT92E (Cell Signaling Technology; 1∶1000). Common secondary antibodies were used in whole-mount immunostaining or Western blots.
Drosophila Schneider L2 (S2) cells were cultured at 25°C in Drosophila Serum-Free Medium (SFM; Invitrogen) supplemented with 10% Fetal Bovine Serum (FBS; Invitrogen) and 0.5x Antibiotic-Antimycotic (Invitrogen). Cells were cultured at 2.5×106/ml prior to transfection. Transfections were performed with FuGene 6 (Roche) according to the manufacturer's instructions. Cu2SO4 (Sigma) was added to the medium at a final concentration of 0.5 mM 16 hours after transfection, and cells were harvested 48 hours after induction. To stimulate JAK/STAT activation in S2 cells, 2 mM H2O2 and 1 mM sodium vanadate (pervanadate) were added to the medium and cells were harvested at desired times after treatment. Treated S2 cells were harvested in cell lysis buffer (from Cell Signaling Tech.) for Western blotting or ChIP experiments.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were carried out according to standard protocols with the following modifications. 200 µl of early embryos (1–2 h AEL) or 1×107 S2 cells were treated with 1% formaldehyde at room temperature for 20 min (embryos) or 10 min (cells) to crosslink protein with chromatin/genomic DNA. Embryos or cells were homogenized and lysed in 300 µl of RIPA lysis buffer with 2 mM EDTA and protease inhibitors on ice. The lysate was sonicated to shear the genomic DNA to lengths between 500 and 1000 bp. An aliquot (50 µl) of sonicated sample was saved as the input control. 5 µg goat anti-STAT92E (Santa Cruz, CA) or rabbit anti-V5 antibodies were added to 200 µl experimental samples in RIPA buffer with 2 mM EDTA and protease inhibitors, and the mixture was incubated overnight at 4°C with rotation. Protein G beads (Sigma), pre-blocked with sonicated salmon sperm DNA (Stratagene), were added to precipitate the antibody-bound chromatin and the precipitate was washed extensively. After reversing the crosslink, DNA was recovered by using a Qiagen PCR purification kit and quantified by PCR. The following forward and reverse primers (flanking two STAT-binding sites in the respective promoter regions) were used for PCR reactions.
dpp: AATTCCGGATAGCGCCTGG, AAAGATGGCACACGCTGGG.
Kr: CATGCGTTTGCATACTGGAG, CTATTCGAATCGCCCTTGTC.
tll: AGTGCTTTGAGGTCGGAATG, AAGAAACCGTGGTGTCCTTG.
Stat92E: TGACTGCCCGCTTTTATACC, CAAACGGCGGTCAATAGTTT.
Supporting Information
Zdroje
1. EphrussiASt JohnstonD
2004
Seeing is believing: the bicoid morphogen gradient
matures.
Cell
116
143
152
2. St JohnstonDNusslein-VolhardC
1992
The origin of pattern and polarity in the Drosophila
embryo.
Cell
68
201
219
3. Nusslein-VolhardC
1991
Determination of the embryonic axes of
Drosophila.
Dev
Suppl 1
1
10
4. WieschausE
1996
Embryonic transcription and the control of developmental
pathways.
Genetics
142
5
10
5. De RenzisSElementoOTavazoieSWieschausEF
2007
Unmasking activation of the zygotic genome using chromosomal
deletions in the Drosophila embryo.
PLoS Biol
5
e117
doi:10.1371/journal.pbio.0050117
6. LawrencePAStruhlG
1996
Morphogens, compartments, and pattern: lessons from
drosophila?
Cell
85
951
961
7. DrieverWNusslein-VolhardC
1988
The bicoid protein determines position in the Drosophila embryo
in a concentration-dependent manner.
Cell
54
95
104
8. RothSSteinDNusslein-VolhardC
1989
A gradient of nuclear localization of the dorsal protein
determines dorsoventral pattern in the Drosophila embryo.
Cell
59
1189
1202
9. LiangHLNienCYLiuHYMetzsteinMMKirovNRushlowC
2008
The zinc-finger protein Zelda is a key activator of the early
zygotic genome in Drosophila.
Nature
10. LibermanLMStathopoulosA
2009
Design flexibility in cis-regulatory control of gene expression:
synthetic and comparative evidence.
Dev Biol
327
578
589
11. GalloSMLiLHuZHalfonMS
2006
REDfly: a Regulatory Element Database for
Drosophila.
Bioinformatics
22
381
383
12. MarksteinMMarksteinPMarksteinVLevineMS
2002
Genome-wide analysis of clustered Dorsal binding sites identifies
putative target genes in the Drosophila embryo.
Proc Natl Acad Sci U S A
99
763
768
13. MacdonaldPMStruhlG
1986
A molecular gradient in early Drosophila embryos and its role in
specifying the body pattern.
Nature
324
537
545
14. MlodzikMGehringWJ
1987
Expression of the caudal gene in the germ line of Drosophila:
formation of an RNA and protein gradient during early
embryogenesis.
Cell
48
465
478
15. SprengerFStevensLMNusslein-VolhardC
1989
The Drosophila gene torso encodes a putative receptor tyrosine
kinase.
Nature
338
478
483
16. CasanovaJStruhlG
1989
Localized surface activity of torso, a receptor tyrosine kinase,
specifies terminal body pattern in Drosophila.
Genes Dev
3
2025
2038
17. LiWX
2005
Functions and mechanisms of receptor tyrosine kinase Torso
signaling: Lessons from Drosophila embryonic terminal
development.
Dev Dyn
232
656
672
18. StathopoulosALevineM
2002
Dorsal gradient networks in the Drosophila
embryo.
Dev Biol
246
57
67
19. HouXSMelnickMBPerrimonN
1996
Marelle acts downstream of the Drosophila HOP/JAK kinase and
encodes a protein similar to the mammalian STATs [published erratum
appears in Cell 1996 Apr 19;85(2):following 290].
Cell
84
411
419
20. LiJLiWCalhounHCXiaFGaoFBLiWX
2003
Patterns and functions of STAT activation during Drosophila
embryogenesis.
Mech Dev
120
1455
1468
21. YanRSmallSDesplanCDearolfCRDarnellJEJr
1996
Identification of a Stat gene that functions in Drosophila
development.
Cell
84
421
430
22. LiWX
2008
Canonical and non-canonical JAK-STAT signaling.
Trends Cell Biol
18
545
551
23. LiJLiWX
2003
Drosophila gain-of-function mutant RTK torso triggers ectopic Dpp
and STAT signaling.
Genetics
164
247
258
24. LiJXiaFLiWX
2003
Coactivation of STAT and Ras is required for germ cell
proliferation and invasive migration in Drosophila.
Dev Cell
5
787
798
25. LiWXAgaisseHMathey-PrevotBPerrimonN
2002
Differential requirement for STAT by gain-of-function and
wild-type receptor tyrosine kinase Torso in Drosophila.
Development
129
4241
4248
26. JinksTMPolydoridesADCalhounGSchedlP
2000
The JAK/STAT signaling pathway is required for the initial choice
of sexual identity in Drosophila melanogaster.
Molecular Cell
5
581
587
27. ten BoschJRBenavidesJAClineTW
2006
The TAGteam DNA motif controls the timing of Drosophila
pre-blastoderm transcription.
Development
133
1967
1977
28. SubramanianATamayoPMoothaVKMukherjeeSEbertBLGilletteMAPaulovichAPomeroySLGolubTRLanderESMesirovJP
2005
Gene set enrichment analysis: a knowledge-based approach for
interpreting genome-wide expression profiles.
Proc Natl Acad Sci U S A
102
15545
15550
29. HarrisonDABinariRNahreiniTSGilmanMPerrimonN
1995
Activation of a Drosophila Janus kinase (JAK) causes
hematopoietic neoplasia and developmental defects.
Embo J
14
2857
2865
30. LuoHHanrattyWPDearolfCR
1995
An amino acid substitution in the Drosophila hopTum-l Jak kinase
causes leukemia-like hematopoietic defects.
Embo J
14
1412
1420
31. ShiSCalhounHCXiaFLiJLeLLiWX
2006
JAK signaling globally counteracts heterochromatic gene
silencing.
Nat Genet
38
1071
1076
32. SweitzerSMCalvoSKrausMHFinbloomDSLarnerAC
1995
Characterization of a Stat-like DNA binding activity in
Drosophila melanogaster.
J Biol Chem
270
16510
16513
33. ShiSLarsonKGuoDLimSJDuttaPYanSJLiWX
2008
Drosophila STAT is required for directly maintaining HP1
localization and heterochromatin stability.
Nat Cell Biol
10
489
496
34. PadgettRWSt JohnstonRDGelbartWM
1987
A transcript from a Drosophila pattern gene predicts a protein
homologous to the transforming growth factor-beta family.
Nature
325
81
84
35. St JohnstonRDGelbartWM
1987
Decapentaplegic transcripts are localized along the
dorsal-ventral axis of the Drosophila embryo.
Embo J
6
2785
2791
36. RayRPAroraKNusslein-VolhardCGelbartWM
1991
The control of cell fate along the dorsal-ventral axis of the
Drosophila embryo.
Development
113
35
54
37. WhartonKARayRPGelbartWM
1993
An activity gradient of decapentaplegic is necessary for the
specification of dorsal pattern elements in the Drosophila
embryo.
Development
117
807
822
38. HuangJDSchwyterDHShirokawaJMCoureyAJ
1993
The interplay between multiple enhancer and silencer elements
defines the pattern of decapentaplegic expression.
Genes Dev
7
694
704
39. Lopez-OnievaLFernandez-MinanAGonzalez-ReyesA
2008
Jak/Stat signalling in niche support cells regulates dpp
transcription to control germline stem cell maintenance in the Drosophila
ovary.
Development
135
533
540
40. WangLLiZCaiY
2008
The JAK/STAT pathway positively regulates DPP signaling in the
Drosophila germline stem cell niche.
J Cell Biol
180
721
728
41. HulskampMPfeifleCTautzD
1990
A morphogenetic gradient of hunchback protein organizes the
expression of the gap genes Kruppel and knirps in the early Drosophila
embryo.
Nature
346
577
580
42. PignoniFBaldarelliRMSteingrimssonEDiazRJPatapoutianAMerriamJRLengyelJA
1990
The Drosophila gene tailless is expressed at the embryonic
termini and is a member of the steroid receptor superfamily.
Cell
62
151
163
43. ParoushZWainwrightSMIsh-HorowiczD
1997
Torso signalling regulates terminal patterning in Drosophila by
antagonising Groucho-mediated repression.
Development
124
3827
3834
44. CarrollSBScottMP
1986
Zygotically active genes that affect the spatial expression of
the fushi tarazu segmentation gene during early Drosophila
embryogenesis.
Cell
45
113
126
45. SeftonLTimmerJRZhangYBerangerFClineTW
2000
An extracellular activator of the Drosophila JAK/STAT pathway is
a sex - determination signal element.
Nature
405
970
973
46. AvilaFWEricksonJW
2007
Drosophila JAK/STAT pathway reveals distinct initiation and
reinforcement steps in early transcription of Sxl.
Curr Biol
17
643
648
47. GilchristDADos SantosGFargoDCXieBGaoYLiLAdelmanK
2010
Pausing of RNA polymerase II disrupts DNA-specified nucleosome
organization to enable precise gene regulation.
Cell
143
540
551
48. ChouTBNollEPerrimonN
1993
Autosomal P[ovoD1] dominant female-sterile insertions
in Drosophila and their use in generating germ-line
chimeras.
Development
119
1359
1369
49. PignoniFSteingrimssonELengyelJA
1992
bicoid and the terminal system activate tailless expression in
the early Drosophila embryo.
Development
115
239
251
50. ChenHWChenXOhSWMarinissenMJGutkindJSHouSX
2002
mom identifies a receptor for the Drosophila JAK/STAT signal
transduction pathway and encodes a protein distantly related to the
mammalian cytokine receptor family.
Genes Dev
16
388
398
51. CapovillaMBrandtMBotasJ
1994
Direct regulation of decapentaplegic by Ultrabithorax and its
role in Drosophila midgut morphogenesis.
Cell
76
461
475
52. ImmergluckKLawrencePABienzM
1990
Induction across germ layers in Drosophila mediated by a genetic
cascade.
Cell
62
261
268
53. PanganibanGEReuterRScottMPHoffmannFM
1990
A Drosophila growth factor homolog, decapentaplegic, regulates
homeotic gene expression within and across germ layers during midgut
morphogenesis.
Development
110
1041
1050
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2011 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Structural and Functional Differences in the Long Non-Coding RNA in Mouse and Human
- Identification, Replication, and Functional Fine-Mapping of Expression Quantitative Trait Loci in Primary Human Liver Tissue
- A −436C>A Polymorphism in the Human Gene Promoter Associated with Severe Childhood Malaria
- A Decline in p38 MAPK Signaling Underlies Immunosenescence in
- The Operon Balances the Requirements for Vegetative Stability and Conjugative Transfer of Plasmid R388
- Novel and Conserved Protein Macoilin Is Required for Diverse Neuronal Functions in
- Ixr1 Is Required for the Expression of the Ribonucleotide Reductase Rnr1 and Maintenance of dNTP Pools
- Genome of Strain SmR1, a Specialized Diazotrophic Endophyte of Tropical Grasses
- A Deficiency of Ceramide Biosynthesis Causes Cerebellar Purkinje Cell Neurodegeneration and Lipofuscin Accumulation
- A Latent Pro-Survival Function for the Mir-290-295 Cluster in Mouse Embryonic Stem Cells
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time
- Prion Formation and Polyglutamine Aggregation Are Controlled by Two Classes of Genes
- Integrated Genome-Scale Prediction of Detrimental Mutations in Transcription Networks
- Post-Embryonic Nerve-Associated Precursors to Adult Pigment Cells: Genetic Requirements and Dynamics of Morphogenesis and Differentiation
- A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
- A Genetic and Structural Study of Genome Rearrangements Mediated by High Copy Repeat Ty1 Elements
- A Missense Mutation in Causes a Major QTL Effect on Ear Size in Pigs
- A Flurry of Folding Problems: An Interview with Susan Lindquist
- Meiotic Recombination Intermediates Are Resolved with Minimal Crossover Formation during Return-to-Growth, an Analogue of the Mitotic Cell Cycle
- A Nervous Origin for Fish Stripes
- The ISWI Chromatin Remodeler Organizes the hsrω ncRNA–Containing Omega Speckle Nuclear Compartments
- The Telomerase Subunit Est3 Binds Telomeres in a Cell Cycle– and Est1–Dependent Manner and Interacts Directly with Est1
- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Characterizing Genetic Risk at Known Prostate Cancer Susceptibility Loci in African Americans
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy