-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
Hirschsprung disease (HSCR) is a congenital disorder characterized by aganglionosis of the distal intestine. To assess the contribution of copy number variants (CNVs) to HSCR, we analysed the data generated from our previous genome-wide association study on HSCR patients, whereby we identified NRG1 as a new HSCR susceptibility locus. Analysis of 129 Chinese patients and 331 ethnically matched controls showed that HSCR patients have a greater burden of rare CNVs (p = 1.50×10−5), particularly for those encompassing genes (p = 5.00×10−6). Our study identified 246 rare-genic CNVs exclusive to patients. Among those, we detected a NRG3 deletion (p = 1.64×10−3). Subsequent follow-up (96 additional patients and 220 controls) on NRG3 revealed 9 deletions (combined p = 3.36×10−5) and 2 de novo duplications among patients and two deletions among controls. Importantly, NRG3 is a paralog of NRG1. Stratification of patients by presence/absence of HSCR–associated syndromes showed that while syndromic–HSCR patients carried significantly longer CNVs than the non-syndromic or controls (p = 1.50×10−5), non-syndromic patients were enriched in CNV number when compared to controls (p = 4.00×10−6) or the syndromic counterpart. Our results suggest a role for NRG3 in HSCR etiology and provide insights into the relative contribution of structural variants in both syndromic and non-syndromic HSCR. This would be the first genome-wide catalog of copy number variants identified in HSCR.
Published in the journal: Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:. PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002687
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002687Summary
Hirschsprung disease (HSCR) is a congenital disorder characterized by aganglionosis of the distal intestine. To assess the contribution of copy number variants (CNVs) to HSCR, we analysed the data generated from our previous genome-wide association study on HSCR patients, whereby we identified NRG1 as a new HSCR susceptibility locus. Analysis of 129 Chinese patients and 331 ethnically matched controls showed that HSCR patients have a greater burden of rare CNVs (p = 1.50×10−5), particularly for those encompassing genes (p = 5.00×10−6). Our study identified 246 rare-genic CNVs exclusive to patients. Among those, we detected a NRG3 deletion (p = 1.64×10−3). Subsequent follow-up (96 additional patients and 220 controls) on NRG3 revealed 9 deletions (combined p = 3.36×10−5) and 2 de novo duplications among patients and two deletions among controls. Importantly, NRG3 is a paralog of NRG1. Stratification of patients by presence/absence of HSCR–associated syndromes showed that while syndromic–HSCR patients carried significantly longer CNVs than the non-syndromic or controls (p = 1.50×10−5), non-syndromic patients were enriched in CNV number when compared to controls (p = 4.00×10−6) or the syndromic counterpart. Our results suggest a role for NRG3 in HSCR etiology and provide insights into the relative contribution of structural variants in both syndromic and non-syndromic HSCR. This would be the first genome-wide catalog of copy number variants identified in HSCR.
Introduction
Hirschsprung disease (HSCR, aganglionic megacolon) is a rare, congenital disorder characterized by the absence of enteric ganglia along a variable length of the intestine. It can be classified according to the length of aganglionosis into short segment (S-HSCR; 80% of the cases), long-segment (L-HSCR; 15%) and total colonic aganglionosis (TCA; 5%). The incidence of Hirschsprung disease varies by gender and ethnicity, and is highest among Asians (2.8/10,000 newborns) [1]. The male∶female ratio is ≈4∶1 among S-HSCR patients and ≈1∶1 among L-HSCR patients. The majority of HSCR patients are isolated (non-syndromic and sporadic) S-HSCR whose modes of inheritance are primarily multifactorial.
Since the discovery of the major HSCR gene, receptor tyrosine kinase (RET; 10q11), a number of rare mutations have been reported in genes (EDNRB; 13q22, GDNF; 5p13, PHOX2B; 4p13, SOX10; 22q13, etc) mostly involved in the two interrelated pathways: RET and endothelin receptor B (EDNRB) signaling cascade [1]. However, together these mutations are of incomplete penetrance and account for only 50% of the familial (mostly L-HSCR, TCA) and up to 20% of the sporadic cases (mostly S-HSCR), contributing to only a small proportion of the heritability [2]. On the other hand, common variants in RET and NRG1 (8p12) were found associated with all sub-phenotypes and explained a considerably larger variance [3], [4]. Still, in spite of the vast coding sequence (CDS) mutation screening and the genome-wide association mapping on HSCR patients, a substantial genetic contribution remained elusive.
Copy number variations (CNVs), which represent an important portion of missing heritability, have recently been highlighted as significant genetic risk factors in disease pathogenesis, such as schizophrenia, autism and early-onset obesity [5]–[7]. Through these genome-wide CNV analyses, a number of disease-susceptibility genes have been suggested (e.g. SH2B1 in obesity and NRXN1 in autism and schizophrenia). In fact, CNV discovery has been essential for uncovering genes/risk factors for a wide range of diseases, including Hirschsprung disease. The two major HSCR genes—RET and EDNRB—are indeed the classical examples of how structural variations assist in mapping the disease-predisposing genes. It is estimated that about 12% of Hirschsprung patients have structural abnormalities [1]. Among these, trisomy 21 (Down's syndrome) is the commonest anomalies, involving 2–10% of the patients [8]. Given this early impact of CNVs on gene discovery and the non-random association of HSCR with syndromes, it is highly probable that structural variations underlain HSCR. Thus far, several studies have attempted to survey CNVs in the targeted HSCR genes (RET, GDNF, etc) [9]–[11], albeit the extent to which CNVs contribute to HSCR is still largely unknown.
To systematically explore the global contribution of CNVs to the disease, we performed a genome-wide copy number analysis based on our previously published SNP genotyping data [3]. By performing a comprehensive association analysis on the identified structural variants, we aimed to uncover novel genes conferring risk to HSCR.
Results
After extensive pre - and post - calling quality control (QC), we obtained a stringent dataset of 866 CNVs with a median size of 34.39 kb in 129 HSCR cases (excluding chromosome 21 for 8 Down's syndrome patients) and 1515 CNVs with a median size of 57.90 kb in 331 ethnically matched controls. Apart from the 1.5 fold increase in average CNV count for cases, a higher proportion of deletions, presumably of larger functional impact, were also observed in cases (59.36%) over controls (51.68%), which allowed us to hypothesize that CNVs significantly contribute to the pathogenesis of HSCR.
To delineate the global impact of CNVs on disease susceptibility, we compared the overall CNV burden in cases relative to controls, in terms of the estimated CNV size, number of CNVs per individual (rate of CNV) and number of genes overlapped by CNVs (gene count).
Greater burden of rare CNVs in HSCR patients
Rare CNVs (present in <1% of the general population) were found significantly overrepresented in HSCR cases with a ratio of 1.97 (p = 1.50×10−5; conditional permutation p = 4.97×10−3). Such difference was not observed for common CNVs, in accordance with their weak global contribution to diseases [12]. As shown in Table 1, the rate of both rare deletions and duplications were significantly higher in HSCR patients; furthermore, these CNVs intersected with more genes. The association was stronger for deletions with a 2.31 fold increase in rate (p = 9.20×10−5; conditional permutation p = 0.017) and with 7.61 times more genes overlapped when compared to controls (p = 5.00×10−6; conditional permutation p = 2.60×10−5). In particular, long deletions (>100 kb) were 14 times enriched with genes in cases when compared to controls (p = 4.25×10−4; conditional permutation p = 2.61×10−4). This could be partly explained by the increase in number of CNVs per patient (rate) (p = 0.014; conditional permutation p = 0.051) as well as by the larger size of CNVs (p = 0.047; conditional permutation p = 0.015) when compared to controls. Consistently, singleton (single occurrence) genic deletions were found more abundant in patients.
Tab. 1. Global CNV burden in HSCR patients. 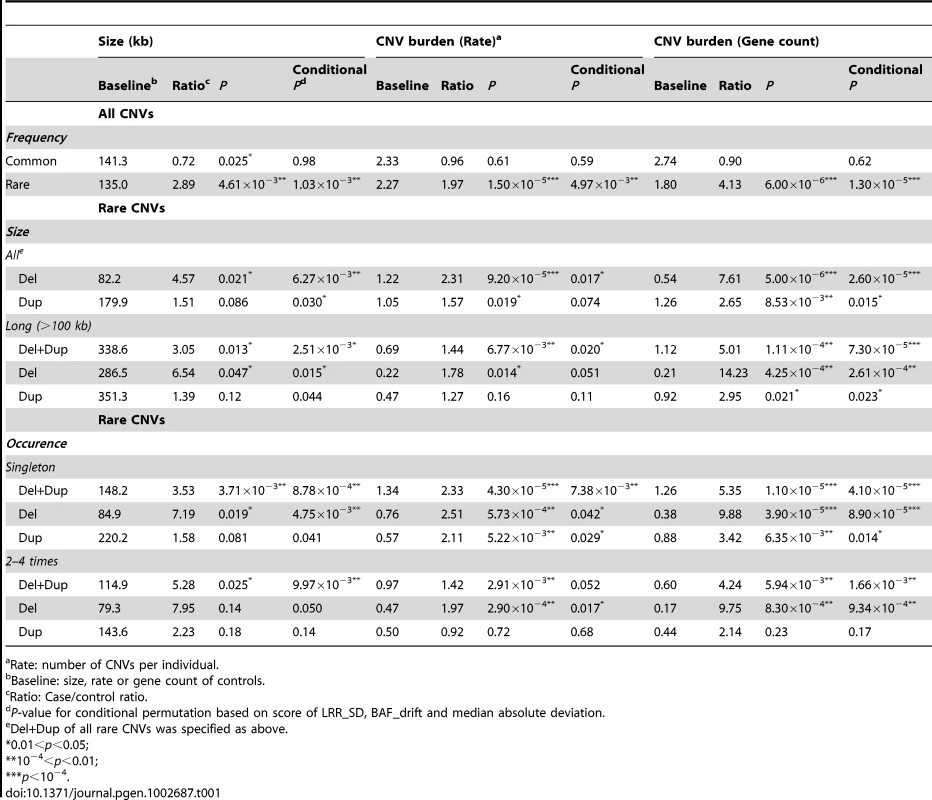
Rate: number of CNVs per individual. Recognizing that CNV analysis is more sensitive to outliers and batch effects, we evaluated if any of these potential factors might account for the observed CNV burden (Text S1, Table S3, Figures S2, S3 and S4). For both CNV rate and gene count, the distinctive overall CNV distribution in HSCR cases (Figure S6) together with the insignificant association with Affymetrix plates confirmed that our findings were not attributed to experimental artifacts. Moreover, the similar level of significance for overrepresentation achieved by conditional permutation on data quality, as illustrated by conditional p-value in Table 1, further demonstrated the robustness of our findings.
Summarizing, HSCR patients have more rare-CNVs and more genes intersected by rare CNVs when compared to controls. The overrepresentation of rare genic CNVs in HSCR patients implies that some CNVs could be pathogenic, regardless of the size and occurrence.
Rare CNVs distribution in syndromic HSCR and isolated (non-syndromic) HSCR patients
We have previously demonstrated that the genetic susceptibility to HSCR varies across sub-phenotypes such as familiality and segment length [2]. To assess if such genetic heterogeneity also occurs at the structural variation level, we further examined the CNV burden for 29 syndromic and 100 non-syndromic (isolated) HSCR separately.
As illustrated in Figure 1, syndromic HSCR cases, on average, harbored longer CNVs than non-syndromic HSCR cases or controls (p = 1.50×10−5 vs. controls) even when the 8 HSCR patients with Down syndrome were excluded from the analysis. Had these patients been taken into account, the significance of the association would have been much lower (p<1×10−8). On the other hand, non-syndromic HSCR cases were enriched with rare CNVs compared to syndromic patients or controls (p = 4.00×10−6 vs. controls). Whilst the involvement of CNVs is somewhat expected in syndromic patients, the excess of rare CNVs in isolated HSCR, irrespective of the size, suggested that copy number variants also contribute to the manifestation of isolated Hirschsprung disease.
Fig. 1. CNV burden for syndromic (Syn) and non-syndromic (Non-syn) HSCR patients relative to controls (Ctrl). 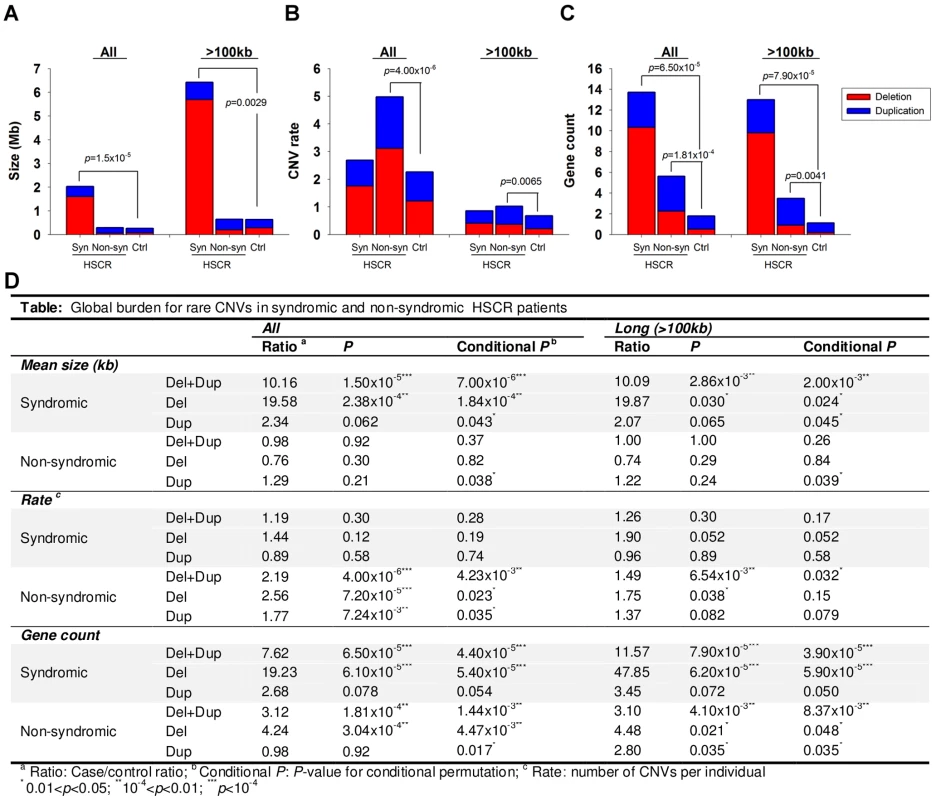
The burden was measured with reference to the (A) size, (B) rate and (C) gene count of CNVs. Red and blue bars denote the mean value of the corresponding test for deletion and duplication respectively. Summary statistics as well as conditional permutation p-value was shown in (D). It is tempting to speculate that the CNVs present in non-syndromic HSCR patients may contribute to the phenotype by affecting the regulation or gene-dosage of genes members of those biological pathways involved in the development of the enteric nervous system. Such variations could add up to the phenotypic expression of HSCR. On the other hand, the presence of longer CNVs in syndromic HSCR implies a larger number of disrupted genes, and consequently, a larger number of systems may be affected during developmental stages. Given that the increase in CNV rate was not evident in syndromic HSCR, long CNVs disrupting multiple genes are likely to have a more deleterious effect. One of these genes is likely to implicate in the development of enteric nervous system, particularly the etiology of Hirschsprung disease.
No differences in CNV rate or type were observed when patients were stratified according to the length of the aganglionic segment.
CNV analysis of syndromic HSCR patients reveals putative candidate genes
Among the 7 large CNVs (>1 Mb) discovered in HSCR patients (Table 2), four were found in unrelated syndromic HSCR patients with mental disabilities (Table 2, in bold). It is interesting to note that three CNVs—a 29 Mb deletion in 11q14.2-q23.2, a 16.68 Mb deletion in 13q21.31-q22.3 and an 11 Mb duplication in 16p11.2-p12.3—coincide with the recently identified candidate regions for intellectual disability (ID) [6], [13]–[15]. In addition, the patient carrying the 16p duplication had also been diagnosed with epilepsy and, incidentally, this 16p11.2 region had also been reported as an epilepsy-implicating loci [16], [17]. While intellectual disability and Hirschsprung disease are frequently associated, it is highly probable that these overlapping regions encompass genes that contribute to the etiology and hence explain the comorbidity of both disorders.
Tab. 2. Large (>1 Mb), rare CNVs identified in HSCR patients. 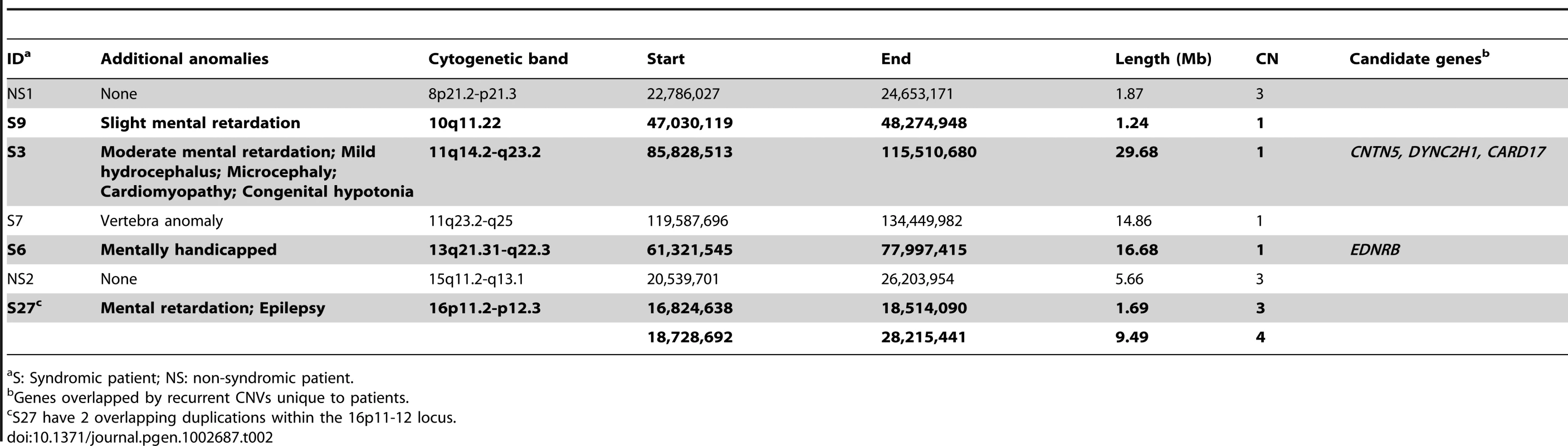
S: Syndromic patient; NS: non-syndromic patient. We subsequently examined these regions on other non-syndromic patients, which further revealed 4 smaller deletions encompassed by the 11q14.2-q23.2 CNV. Altogether, several genes were recurrently and uniquely covered by CNVs in patients, including dynein, cytoplasmic 2, heavy chain 1 (DYNC2H1) and contactin 5 (CNTN5). DYNC1H1 encodes a cytoplasmic dynein implicated in axonal transport and retrograde trafficking. Mutations in DYNC2H1 were reported to be associated not only with ID [18] but also with abnormal skeletogenesis which is occasionally found in HSCR [19], [20]. Recent studies in mice also showed that Dync2h1 mutations disrupted sonic hedgehog (Shh) dependent neural patterning and, most importantly, Shh is essential in gastrointestinal development, plausibly by regulating enteric neural crest cells migration [21]–[24]. Contactin 5, also known as NB-2, is a paralog of DSCAM and L1CAM both previously implicated in HSCR [25]–[28]. All three genes encode neural cell adhesion molecules belonging to the same immunoglobin superfamily. CNTN5, together with its paralogs, is involved in the nervous system. It mediates cell surface interactions and the formation of axon connections [29], [30].
Interestingly, the 13q deletion was found to encompass the second major HSCR gene—EDNRB. Screening among non-syndromic patients revealed an additional 44 kb deletion disrupting the first exon and intron of EDNRB.
Apart from EDNRB, none of the other HSCR genes were found to overlap or encompass any copy number changes even if all patients were considered. This result is in line with the negative findings of previous studies on structural variations intersecting selected HSCR-genes [9], [10]. As for other chromosomal regions known to segregate with HSCR (3p21 [31], [32], 19q12, 4q31-q32 [33] and 9q31 [34], [35]), only 2 rare genic CNVs were observed (Table S4). Neither CNV appeared functionally related to the nervous system development nor to the differentiation or migration of neural crest cells. Likewise, no significant overrepresentation of CNVs was found on chromosome 21 [8]. We also investigated chromosomal regions reported altered in HSCR patients as described by the HSCR consortium, including trisomy 21, 10q11 and 13q22 deletions [1]. We found 11 HSCR-specific CNVs intersecting those implicated regions and in particular, 2 CNVs mapped within 10q11 and 3 within 13q22 where RET and EDNRB reside respectively (Table S5). However, none of the 10q11 deletions encompassed RET. Further investigation of the 6 remaining genic CNVs gene(s) is warranted as it may lead to the discovery of new HSCR-susceptibility genes.
Other genic CNVs
Taken together, a total of 237 non-redundant, rare genic copy number variable regions (CNVRs) were exclusively observed in HSCR patients, corresponding to 246 unique structural variants (see Text S1). To confirm their uniqueness, we compared our HSCR-specific CNVs with the recently published CNV profile on Asian populations [36]. Only two CNVRs were observed in other Asians (Korean or Japanese) and, most importantly, none were observed in the Chinese population. A catalog of these HSCR-specific CNVRs together with the overlapping genes is provided in Table S5. Specially, additional paralogs of CNTN5, including CNTN4, SDK1, DSCAML1, ROBO3 and ROBO4 were disrupted by HSCR-specific CNVs, highlighting the potential relevance of this immunoglobin family in the development of the disease.
NRG3 deletion
We further explored if any particular CNV might be disease causative by comparing the relative frequency in cases to controls. A significant association was found for a CNVR mapping to intron 1 of neuregulin 3 (NRG3) located on 10q23.1 (HSCR-CNVR129.1, chr10 : 84,034,612–84,048,907; hg18; p = 1.64×10−3). Five hemizygous deletions (3.88%), with estimated length ranging from 8 to 14 kb, were observed in patients (2 syndromic and 3 isolated HSCR patients) while none of the controls had such deletion (Figure 2A). Despite its absence in the controls, it is not a novel CNV. It overlaps with 2 deletions (ID:2882 & 48644) reported for normal population according to the Database of Genomic Variants (DGV, Figure 2B). The former one, with similar boundaries as our cases, has been observed in one HapMap Han Chinese from Beijing (CHB) [37]. A much lower frequency (0.1%) was observed for the latter CNV which extends further upstream [38]. Even though the NRG3 deletion may not be deleterious, its ten-fold increase in rate for HSCR patients is highly suggestive of pathogenicity. Intriguingly, the deletion encompasses region marked by strong enhancer chromatin signature (H3K4me1) and DNaseI hypersensitivity, raising the possibility that it might be directly functional (Figure S7) [39].
Fig. 2. NRG3 deletions identified in HSCR patients. 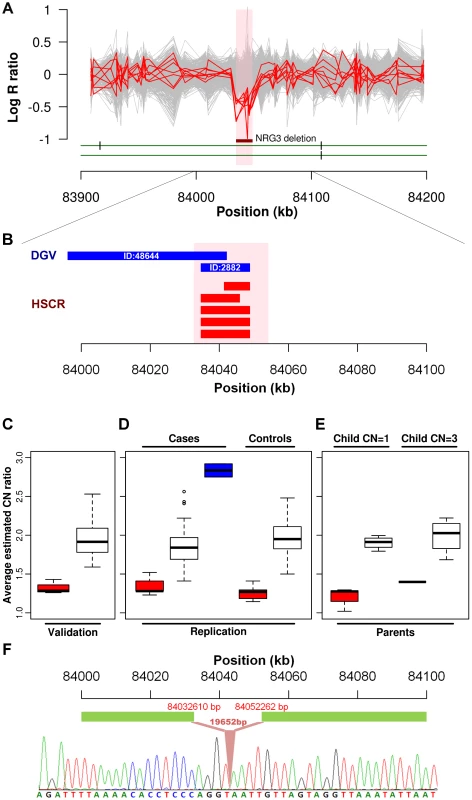
(A) Intensity signals of 5 HSCR patients (CN = 1; red) with NRG3 deletions together with other samples of normal copy number (CN = 2; grey). Deleted regions are shown by the dark red bar and are highlighted in pink. (B) Consensus CNV segments of the 5 NRG3 deletions (red) and the overlapping DGV segments (blue; with DGV ID). (C, D and E) Box plot of NRG3 copy number estimates by real-time PCR. Samples were grouped according to the called copy number states (CN = 1, red; CN = 2, white; CN = 3, blue); (C) Validation of 5 deletions (CN = 1) and 24 copy-neutral (CN = 2) HSCR patients in the discovery phase; (D) Follow-up analysis on independent case-controls set and (E) Transmission analysis for probands with NRG3 deletions (child CN = 1) or duplications (child CN = 3). (F) Sequence of the NRG3 deletion boundary region showing the breakpoint (upstream boundary chr10: 84032610; downstream boundary chr10: 84052262). Neuregulin 3, again, is a paralog of a HSCR-associated gene—neuregulin 1 (NRG1; 8p12)—previously discovered in our genome-wide association study utilizing the same intensity data. It encodes a ligand which physically interacts with a transmembrane tyrosine kinase receptor, ErbB4. NRG3, through activating ErbB4, influences neuroblast proliferation, migration and differentiation [40]. Its paralog, NRG1, in addition to ErbB4 also binds to ErbB2 and ErbB3. Meanwhile, a 35 kb ErbB4 deletion (HSCR-CNVR32.1, chr2 : 212,872,711–212,889,560) was also observed uniquely in a syndromic HSCR patient.
Due to the biological relevance of NRG3 in HSCR, we used real-time qPCR to experimentally confirm the deletions. A random set of 46 patients called with normal copy number was chosen for validation. We successfully validated the copy number states of all these samples as shown in Figure 2C. Next, we attempted to replicate this finding on an independent set of 96 cases and 220 controls (Figure 2D). In addition to the 5 deletions found in the discovery phase, nine more deletions (9.38%) were detected in cases while 5 were identified in controls (2.27%) (p = 6.92×10−3). Seven of those deletions were found in isolated HSCR patients. In addition, real-time qPCR detected 2 novel duplications (2.08%) among the patients included in the replication phase (one patient affected with isolated HSCR and one with HSCR-Meckel diverticulum). All CNVs detected were verified by an additional TaqMan probe. Dissection of sample origin, i.e., from North or South of China, is detailed in Table S6. We found no evidence of association between sample origin and NRG3 deletion for both cases (p = 0.17) and controls (Fisher exact test p = 1). To assess if the CNVs are de novo, parent-child transmission analysis was performed for those probands (n = 5; 3 with deletion and 2 with duplication) for whom parental DNA was available (Figure 2E). All 3 deletions tested were found to be inherited from normal parents. However, the duplication could not be detected in the parents. Considering deletions alone, we combined association results of both phases, yielding a strong association between HSCR and NRG3 (p = 3.36×10−5; p = 7.76×10−6 when considering all NRG3 deletions and duplications). Such a 7 fold (6.28% in cases and 0.91% in controls) enrichment of deletions together with de novo duplications strongly suggested NRG3 as a candidate HSCR gene.
Given that most NRG3 deletions are inherited, we next attempted to define their nature, i.e. whether they represent a collection of distinct mutations or a low frequency copy number polymorphism instead. In particular, we tried to address two questions, (1) are the deletions on the same haplotype background and if so, (2) do they represent a common ancestral mutation. To achieve this, we performed haplotype analysis on the 206 kb region (chr10 : 83,990,316–84,195,982) where all 5 typed patients with deletion identified through the GWAS shared at least one allele (i.e. identity-by-state of at least 1). Phasing by BEAGLE revealed a 4-SNP common haplotype harboring the deletion (from rs7085458 to rs7897939; chr10 : 83,990,316–84,063,139), which suggested a 73 kb identity-by-descent (IBD) segment (Tables S7 and S8). Indeed, this 4-SNP haplotype is the best tagging genetic variant for the deletion, which had a moderate significance of association in our original GWAS data (p = 0.005). Identification of the CNV breakpoint (Text S1, Figure S8 and Table S9) revealed a deletion of 19,652 bp in length shared by those 5 patients sharing the haplotype. Close inspection of the boundary sequence, revealed a 4 bp homology between the 5′ and 3′ ends of the deletion. Such microhomologies are observed in 70% of the deletion breakpoints and may reflect the mutational mechanisms leading to the formation of a CNV [41]. These analyses therefore suggest that this deletion involving NRG3 is a low frequency copy number polymorphism (Figure 2F).
Discussion
Current data indicates that sporadic HSCR phenotype may result from the interplay and/or accumulation of both common and rare functional DNA variants in genes involved in the enteric nervous system development. These variants may also include structural variations, yet the contribution of CNVs to HSCR had never been investigated at genome-wide level, presumably, because many CNVs are submicroscopic, thus undetectable by conventional karyotyping techniques.
Here we present the first comprehensive survey of copy number variations in HSCR and provide a catalog of rare genic CNVRs possibly implicated in the manifestation of the Hirschsprung disease phenotype. Structural variants identified here are individually rare but collectively common in HSCR patients.
One of the major challenges in CNV discovery is to discriminate between benign and pathological variants. The rarer or longer the CNV, the more likely it is to be pathogenic. Also, the involvement of a gene that lies within a pathway known to contain genes associated with a similar phenotype strengthens the possibility of pathogenicity. Indeed, the CNVs reported here meet the above criteria as we found a plethora of rare CNVs in HSCR patients, in terms of both rate and gene count. In addition, syndromic-HSCR patients were enriched in longer CNV and a number of HSCR-specific CNVs overlapped with paralogs of previously HSCR-implicated genes were observed.
Meanwhile, for rare CNVs, the significant increase in size in syndromic HSCR as well as the overload in number in non-syndromic HSCR suggested a correlation between pathogenesis and genetic heterogeneity at structural level. We attempted to address such correlation across other sub-phenotypes, such as gender and length of aganglionosis. Nonetheless, our study design of random ascertainment limited the sample size of the minor groups and consequently did not permit a detailed investigation of the CNV contribution.
Neuregulin 3 (NRG3) encodes a protein similar to its paralog NRG1 and both play important roles in the developing nervous system. As seen with other pathologies, including autism and schizophrenia, several members of a given protein family may associate with the same phenotype, individually or together [7]. Thus far the genes involved in HSCR belong to the two major signaling pathways (RET and EDNRB). The current finding on NRG3 and ErbB4 together with our previous study have established the contribution of a new protein family—NRGs—to the disease. Although we have confirmed that both rare and common variants of NRG1 are associated with HSCR [3], [42], the molecular properties and mechanisms leading to the disease remains unclear. As both NRG1 and NRG3 share the same receptor, they may work synergistically or antagonically [43]. Importantly, rare and common variants in NRGs and their receptors have been implicated in schizophrenia [44]–[52]. Furthermore, rare deletions of NRG1, NRG3 and ErbB4 were described in schizophrenic patients as well [44], [53], [54]. The association of these genes with two different disorders in the nervous system not only emphasizes the relevance of NRGs in the nervous system but also strengthens the validity of the findings. Based on the inheritance we observed, we proposed a two-hit hypothesis for the deletion where the “second hit” could be a mutation or copy number variant in NRG or other inter-related pathways, which explained the incomplete penetrance in parents [55].
One of the intriguing observations from this study is the genetic overlap between Hirschsprung disease and schizophrenia. In addition to the pleiotropic effect of neuregulins and ErbB families, the major HSCR gene, RET, was found deleted exclusively in schizophrenia patients in a recent genome-wide CNV analysis [15]. Interestingly, this report, together with Wang et al. (2010), also suggested an association between our candidate genes CNTN5 and schizophrenia [56]. CNTNAP2, which was recurrently and exclusively deleted in HSCR cases, was also associated with multiple neurodevelopmental and neuropsychiatric disorders [57]–[59]. It is thus tempting to speculate that pathogenic alterations affecting common pathway(s) may act in the development of both diseases. Such hypothesis is further supported by the frequently observed association of intestinal dysmotility with psychiatric disorders [60]–[62]. Further investigation into the suggestive genetic link is required. By understanding the pleiotropy and the intersecting pathway(s), one can optimize the search for other causal variants underlying HSCR.
Despite none of the known HSCR genes other than EDNRB nor HSCR-implicated regions was deleted or duplicated in our analysis, the observation did not elude the presence of structural variations affecting these genes. Rather, it suggested that the rare deletions observed previously, like rare mutations for HSCR patients, might not be a global phenomenon but segregate within individual families.
Similarly, we did not find evidence of global contribution of copy number polymorphisms (CNPs) to HSCR in this study. It could be that indeed these common CNVs are not implicated on the manifestation of the phenotype or that our observation results from the limitations posed by the early genotyping platforms. Our data was generated by Affymetrix 500K, well known to suffer from relatively low SNP density and no CNV probes. Regions with CNPs are more likely to violate Hardy-Weinberg equilibrium and could be preferentially excluded from SNP genotyping. Consequently, the power to detect short and/or common CNVs is limited. It should be noted that our stringent quality control to maximize false positive findings in scarify of false negatives also added a further complication to the interpretation of the role of CNPs in HSCR. These limitations applied also to the discovery of the NRG3 deletions. The higher frequency of copy number changes in the replication samples might only reflect the lower power to detect shorter CNVs with high confidence in the discovery phase.
To conclude, our study provides not only a catalog of rare genic HSCR CNVs but also valuable insights into the contribution of rare CNVs in the phenotypic heterogeneity of HSCR. Our finding illuminates the potential of discovering new HSCR genes and provides grounds for further investigation of the role of NRG family in the disease mechanisms.
Materials and Methods
Samples
Discovery phase
We started with 173 HSCR Chinese sporadic probands and 340 controls passing SNP-based quality control (QC) as described previously (see Text S1) [3], [35]. All HSCR patients had been screened for the main HSCR genes, namely RET, NRG1, EDNRB, EDN3 and GDNF. Samples were genotyped using Affymetrix GeneChip 500K array in which ∼500,000 SNP probes were interrogated separately on two chips (Nsp and Sty). Further characteristics of the patients can be found in Table S1 and in Garcia-Barcelo et al. (2009) and Tang et al. (2010) [3], [35]. Only autosomal SNPs were considered in the CNV analysis.
After pre - and post-CNV calling QCs, 129 HSCR cases and 331 controls were left, 29 of whom have additional congenital anomalies in conjunction with Hirschsprung disease. Among these, 8 patients have Down's syndrome and an additional 6 patients with intellectual disability. Details regarding the associated anomalies and known CDS mutations in HSCR genes were listed in Table S2. Out of these 129 patients passing QC, parental DNA was available for 46 probands.
The study was approved by the institutional review board of The University of Hong Kong together with the Hospital Authority (IRB: UW 06-349 T/1374).
Replication phase
To replicate our finding on NRG3 deletion, an independent set of 96 Chinese HSCR cases and 220 controls were subject to the genomic DNA quantification using quantitative real-time PCR. We further determined the inheritance pattern of each NRG3 CNV discovered (n = 5) for which parental DNA was available.
CNV calling and quality control (QC)
The overview of CNV calling as well as quality control was summarized in Figure S1 and was detailed in Text S1. Briefly, pre-calling QCs were carried out to remove samples showing relatively low quality in SNP genotyping and samples prone to bias in CNV calling were excluded [63]. Next, CNVs were called by two programs, PennCNV [64] and Birdsuite [65], and were then filtered for abnormal calls. In order to obtain a high-quality CNV dataset, we restricted our analysis to consensus CNV segments consistently called by both programs (Figure S5). Finally, a total of 866 and 1515 CNVs passing quality controls in 129 cases and 331 controls respectively were used for CNV analysis.
CNV analysis
Global CNV burden analysis
CNVs were defined as rare if their frequencies were <1% in the total sample (cases and controls) and were otherwise considered as common. Tests of CNV burden (1-sided) in terms of size, number of CNV segments and number of genes overlapped were performed using permutation by PLINK [66]. Gene annotation was based on UCSC RefSeq (hg18) and NCBI Build 36 was used throughout the study. We defined genic CNVRs as those with more than 1 bp overlapped with any genic region (from −10 kb upstream of the transcription start site to +10 kb downstream).
CNV analysis on HSCR gene and HSCR–implicated regions
Four HSCR-implicated regions (3p21 [31], [32], 19q12, 4q31-q32 [33], 9q31 [34], [35]) and 12 HSCR genes (RET, GDNF, NRTN, SOX10, EDNRB, EDN3, ECE1, ZFHX1B, PHOX2B, TCF4, KIAA1279 and NRG1) [1] were evaluated for the presence of rare CNVs.
Association analysis
Copy number variable regions (CNVRs) were defined as described in Conrad et al. (2009) [67] and in Text S1. Two-sided Fisher's exact test was used to test for association between NRG3 deletions and Hirschsprung disease, both for the discovery and subsequent replication phases. To combine the association results, meta-analysis was performed by pooling the p-values while weighted by the sample sizes of each phase.
Haplotype analysis on the NRG3 deletion
We phased the genotype calls (1 Mb upstream and downstream) of all 129 HSCR cases and 331 controls using BEAGLE [68], [69]. To increase accuracy (as phasing a small sample set may be somehow inaccurate), we included the unphased genotypes of the 3 carrier parents (from whom NRG3 deletions were inherited) and phased genotypes of 193 Asians (HG00578 was removed due to relatedness) from the 1000 Genomes Project as reference panel [70]. SNPs within the deleted region were recorded as a single bi-allelic marker, with deletion and non-deletion as the two alleles. Association between HSCR and the 4-SNP haplotype encompassing NRG3 deletion was performed in PLINK.
NRG3 deletion validation and replication
Copy number validation and replication was performed by quantitative real-time PCR (ABI Prism 7900 Sequence Detection System; Applied Biosystems) using TaqMan Copy Number Assay. The assay was carried out in quadruplicates with the TaqMan Copy Number Reference Assay according to the manufacturer's protocol. The reference assay targets a copy-number neural region of RNaseP gene, serving as an internal standard. To achieve high confidence, copy number changes for replication samples were detected and verified by 2 NRG3 probes (Hs03732951_cn, chr10 : 84,043,528 & Hs03749105_cn, chr10 : 84,045,098) which fall within the 4 kb minimal overlapping region of the NRG3 deletions (chr10 : 84,041,355–84,045,997). Relative levels of NRG3 to reference probes were determined using comparative CT method. In brief, the mean differences in cycle threshold (CT) ΔCT between the NRG3 and the reference probes for all replicates were computed and were subsequently normalized for copy number prediction.
Supporting Information
Zdroje
1. AmielJSproat-EmisonEGarcia-BarceloMLantieriFBurzynskiG 2008 Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet 45 1 14
2. EmisonESGarcia-BarceloMGriceEALantieriFAmielJ 2010 Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet 87 60 74
3. Garcia-BarceloMMTangCSNganESLuiVCChenY 2009 Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung's disease. Proc Natl Acad Sci U S A 106 2694 2699
4. EmisonESMcCallionASKashukCSBushRTGriceE 2005 A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 434 857 863
5. 2008 Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455 237 241
6. BochukovaEGHuangNKeoghJHenningEPurmannC 2010 Large, rare chromosomal deletions associated with severe early-onset obesity. Nature 463 666 670
7. PintoDPagnamentaATKleiLAnneyRMericoD 2010 Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466 368 372
8. ArnoldSPeletAAmielJBorregoSHofstraR 2009 Interaction between a chromosome 10 RET enhancer and chromosome 21 in the Down syndrome-Hirschsprung disease association. Hum Mutat 30 771 775
9. Nunez-TorresRFernandezRMLopez-AlonsoMAntinoloGBorregoS 2009 A novel study of copy number variations in Hirschsprung disease using the multiple ligation-dependent probe amplification (MLPA) technique. BMC Med Genet 10 119
10. SerraAGorgensHAlhadadKZieglerAFitzeG 2009 Analysis of RET, ZEB2, EDN3 and GDNF genomic rearrangements in 80 patients with Hirschsprung disease (using multiplex ligation-dependent probe amplification). Ann Hum Genet 73 147 151
11. JiangQHoYYHaoLNichols BerriosCChakravartiA 2011 Copy number variants in candidate genes are genetic modifiers of hirschsprung disease. PLoS ONE 6 e21219 doi:10.1371/journal.pone.0021219
12. CraddockNHurlesMECardinNPearsonRDPlagnolV 2010 Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464 713 720
13. NowakowskaBStankiewiczPObersztynEOuZLiJ 2008 Application of metaphase HR-CGH and targeted Chromosomal Microarray Analyses to genomic characterization of 116 patients with mental retardation and dysmorphic features. Am J Med Genet A 146A 2361 2369
14. BallaratiLRossiEBonatiMTGimelliSMaraschioP 2007 13q Deletion and central nervous system anomalies: further insights from karyotype-phenotype analyses of 14 patients. J Med Genet 44 e60
15. BallifBCHornorSAJenkinsEMadan-KhetarpalSSurtiU 2007 Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet 39 1071 1073
16. HeinzenELRadtkeRAUrbanTJCavalleriGLDepondtC 2010 Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet 86 707 718
17. ShinawiMLiuPKangSHShenJBelmontJW 2010 Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet 47 332 341
18. VissersLEde LigtJGilissenCJanssenISteehouwerM 2010 A de novo paradigm for mental retardation. Nat Genet 42 1109 1112
19. MerrillAEMerrimanBFarrington-RockCCamachoNSebaldET 2009 Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am J Hum Genet 84 542 549
20. DagoneauNGouletMGenevieveDSznajerYMartinovicJ 2009 DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am J Hum Genet 84 706 711
21. FuMLuiVCShamMHPachnisVTamPK 2004 Sonic hedgehog regulates the proliferation, differentiation, and migration of enteric neural crest cells in gut. J Cell Biol 166 673 684
22. ReichenbachBDelalandeJMKolmogorovaEPrierANguyenT 2008 Endoderm-derived Sonic hedgehog and mesoderm Hand2 expression are required for enteric nervous system development in zebrafish. Dev Biol 318 52 64
23. OcbinaPJEggenschwilerJTMoskowitzIAndersonKV 2011 Complex interactions between genes controlling trafficking in primary cilia. Nat Genet 43 547 553
24. Ramalho-SantosMMeltonDAMcMahonAP 2000 Hedgehog signals regulate multiple aspects of gastrointestinal development. Development 127 2763 2772
25. KorbelJOTirosh-WagnerTUrbanAEChenXNKasowskiM 2009 The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci U S A 106 12031 12036
26. YamakawaKHuotYKHaendeltMAHubertRChenXN 1998 DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet 7 227 237
27. OkamotoNDel MaestroRValeroRMonrosEPooP 2004 Hydrocephalus and Hirschsprung's disease with a mutation of L1CAM. J Hum Genet 49 334 337
28. Basel-VanagaiteLStraussbergRFriezMJInbarDKorenreichL 2006 Expanding the phenotypic spectrum of L1CAM-associated disease. Clin Genet 69 414 419
29. WalshFSDohertyP 1991 Glycosylphosphatidylinositol anchored recognition molecules that function in axonal fasciculation, growth and guidance in the nervous system. Cell Biol Int Rep 15 1151 1166
30. OgawaJLeeSItohKNagataSMachidaT 2001 Neural recognition molecule NB-2 of the contactin/F3 subgroup in rat: Specificity in neurite outgrowth-promoting activity and restricted expression in the brain regions. J Neurosci Res 65 100 110
31. GabrielSBSalomonRPeletAAngristMAmielJ 2002 Segregation at three loci explains familial and population risk in Hirschsprung disease. Nat Genet 31 89 93
32. Garcia-BarceloMMFongPYTangCSMiaoXPSoMT 2008 Mapping of a Hirschsprung's disease locus in 3p21. Eur J Hum Genet 16 833 840
33. BrooksASLeegwaterPABurzynskiGMWillemsPJde GraafB 2006 A novel susceptibility locus for Hirschsprung's disease maps to 4q31.3-q32.3. J Med Genet 43 e35
34. BolkSPeletAHofstraRMAngristMSalomonR 2000 A human model for multigenic inheritance: phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc Natl Acad Sci U S A 97 268 273
35. TangCSSribudianiYMiaoXPde VriesARBurzynskiG 2010 Fine mapping of the 9q31 Hirschsprung's disease locus. Hum Genet 127 675 683
36. ParkHKimJIJuYSGokcumenOMillsRE 2010 Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet 42 400 405
37. RedonRIshikawaSFitchKRFeukLPerryGH 2006 Global variation in copy number in the human genome. Nature 444 444 454
38. ShaikhTHGaiXPerinJCGlessnerJTXieH 2009 High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res 19 1682 1690
39. RaneyBJClineMSRosenbloomKRDreszerTRLearnedK 2011 ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res 39 D871 875
40. HowardBA 2008 The role of NRG3 in mammary development. J Mammary Gland Biol Neoplasia 13 195 203
41. ConradDFBirdCBlackburneBLindsaySMamanovaL 2010 Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet 42 385 391
42. TangCSNganESTangWKSoMTChengG 2011 Mutations in the NRG1 gene are associated with Hirschsprung disease. Hum Genet
43. BenzelIBansalABrowningBLGalweyNWMaycoxPR 2007 Interactions among genes in the ErbB-Neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav Brain Funct 3 31
44. ChenPLAvramopoulosDLasseterVKMcGrathJAFallinMD 2009 Fine mapping on chromosome 10q22-q23 implicates Neuregulin 3 in schizophrenia. Am J Hum Genet 84 21 34
45. StefanssonHSigurdssonESteinthorsdottirVBjornsdottirSSigmundssonT 2002 Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 71 877 892
46. MunafoMRThiseltonDLClarkTGFlintJ 2006 Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry 11 539 546
47. NicodemusKKLunaAVakkalankaRGoldbergTEganM 2006 Further evidence for association between ErbB4 and schizophrenia and influence on cognitive intermediate phenotypes in healthy controls. Mol Psychiatry 11 1062 1065
48. Walss-BassCLiuWLewDFVillegasRMonteroP 2006 A novel missense mutation in the transmembrane domain of neuregulin 1 is associated with schizophrenia. Biol Psychiatry 60 548 553
49. KaoWTWangYKleinmanJELipskaBKHydeTM 2010 Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci U S A 107 15619 15624
50. MorarBDragovicMWatersFAChandlerDKalaydjievaL 2010 Neuregulin 3 (NRG3) as a susceptibility gene in a schizophrenia subtype with florid delusions and relatively spared cognition. Mol Psychiatry
51. LawAJKleinmanJEWeinbergerDRWeickertCS 2007 Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet 16 129 141
52. LawAJLipskaBKWeickertCSHydeTMStraubRE 2006 Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci U S A 103 6747 6752
53. XuBWoodroffeARodriguez-MurilloLRoosJLvan RensburgEJ 2009 Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci U S A 106 16746 16751
54. WalshTMcClellanJMMcCarthySEAddingtonAMPierceSB 2008 Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320 539 543
55. GirirajanSRosenfeldJACooperGMAntonacciFSiswaraP 2010 A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet 42 203 209
56. WangKSLiuXFAragamN 2010 A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res 124 192 199
57. AlarconMAbrahamsBSStoneJLDuvallJAPerederiyJV 2008 Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet 82 150 159
58. FriedmanJIVrijenhoekTMarkxSJanssenIMvan der VlietWA 2008 CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry 13 261 266
59. ZweierCde JongEKZweierMOrricoAOusagerLB 2009 CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet 85 655 666
60. SonnenbergATsouVTMullerAD 1994 The “institutional colon”: a frequent colonic dysmotility in psychiatric and neurologic disease. Am J Gastroenterol 89 62 66
61. PeupelmannJQuickCBergerSHockeMTancerME 2009 Linear and non-linear measures indicate gastric dysmotility in patients suffering from acute schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry 33 1236 1240
62. Vande VeldeSVan BiervlietSVan GoethemGDe LoozeDVan WinckelM 2010 Colonic transit time in mentally retarded persons. Int J Colorectal Dis 25 867 871
63. PughTJDelaneyADFarnoudNFlibotteSGriffithM 2008 Impact of whole genome amplification on analysis of copy number variants. Nucleic Acids Res 36 e80
64. WangKLiMHadleyDLiuRGlessnerJ 2007 PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17 1665 1674
65. KornJMKuruvillaFGMcCarrollSAWysokerANemeshJ 2008 Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 40 1253 1260
66. PurcellSNealeBTodd-BrownKThomasLFerreiraMA 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
67. ConradDFPintoDRedonRFeukLGokcumenO 2010 Origins and functional impact of copy number variation in the human genome. Nature 464 704 712
68. BrowningBLBrowningSR 2009 A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84 210 223
69. BrowningSRBrowningBL 2007 Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81 1084 1097
70. DurbinRMAbecasisGRAltshulerDLAutonABrooksLD 2010 A map of human genome variation from population-scale sequencing. Nature 467 1061 1073
Štítky
Genetika Reprodukčná medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



