-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
It has been suggested that imprinted genes are important in the regulation of sleep. However, the fundamental question of whether genomic imprinting has a role in sleep has remained elusive up to now. In this work we show that REM and NREM sleep states are differentially modulated by the maternally expressed imprinted gene Gnas. In particular, in mice with loss of imprinting of Gnas, NREM and complex cognitive processes are enhanced while REM and REM–linked behaviors are inhibited. This is the first demonstration that a specific overexpression of an imprinted gene affects sleep states and related complex behavioral traits. Furthermore, in parallel to the Gnas overexpression, we have observed an overexpression of Ucp1 in interscapular brown adipose tissue (BAT) and a significant increase in thermoregulation that may account for the REM/NREM sleep phenotypes. We conclude that there must be significant evolutionary advantages in the monoallelic expression of Gnas for REM sleep and for the consolidation of REM–dependent memories. Conversely, biallelic expression of Gnas reinforces slow wave activity in NREM sleep, and this results in a reduction of uncertainty in temporal decision-making processes.
Published in the journal: Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice. PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002706
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002706Summary
It has been suggested that imprinted genes are important in the regulation of sleep. However, the fundamental question of whether genomic imprinting has a role in sleep has remained elusive up to now. In this work we show that REM and NREM sleep states are differentially modulated by the maternally expressed imprinted gene Gnas. In particular, in mice with loss of imprinting of Gnas, NREM and complex cognitive processes are enhanced while REM and REM–linked behaviors are inhibited. This is the first demonstration that a specific overexpression of an imprinted gene affects sleep states and related complex behavioral traits. Furthermore, in parallel to the Gnas overexpression, we have observed an overexpression of Ucp1 in interscapular brown adipose tissue (BAT) and a significant increase in thermoregulation that may account for the REM/NREM sleep phenotypes. We conclude that there must be significant evolutionary advantages in the monoallelic expression of Gnas for REM sleep and for the consolidation of REM–dependent memories. Conversely, biallelic expression of Gnas reinforces slow wave activity in NREM sleep, and this results in a reduction of uncertainty in temporal decision-making processes.
Introduction
Mammalian evolution from the reptile lineage involved important changes in gene regulation and sleep. Many genetic mechanisms play important roles in various electrophysiological and behavioral traits that set the three major states of mammalian life: Wakefulness, Rapid Eye Movement (REM) and Non-REM (NREM) sleep [1], [2]. However, here we focus on a particular gene regulation, namely genomic imprinting. Genomic imprinting is an epigenetic mechanism that results in allele-specific expression of some genes according to parental origin and, in vertebrates, is unique to mammals.
Clinical observations of neurodevelopmental disorders of sleep suggest a role of genomic imprinting on various measures of sleep [3], [4]. For example, Prader-Willi syndrome (PWS) and Angelman syndrome (AS), both neurodevelopmental syndromes, exhibit opposing imprinting profiles and opposing sleep phenotypes. PWS is associated with maternal duplications/paternal deletions of alleles on chromosome 15q11–13 and is characterized by temperature control abnormalities and excessive sleepiness as well as REM sleep abnormalities [5]–[8]. Conversely, AS is associated with paternal duplications/maternal deletions on chromosome 15q11–13 and is characterized by severe mental retardation and reductions in sleep. The UBE3A gene, that resides in the PWS/AS imprinting region, has been associated to sleep abnormalities. Ube3a deficient mice are characterized by reduced NREM sleep, deteriorated REM sleep, and an increased frequency of waking during the dark–light transition [9].
Interestingly, the serotonin (5-HT) 2A receptors, which mediate aminergic inhibition of REM-on cells in the parabrachialis lateralis region [10], are primarily expressed from maternal alleles [11], [12]. The above reviewed findings support the idea that epigenetic regulatory mechanisms, such as genomic imprinting, influence sleep-associated mechanisms.
REM and NREM sleep underlie important metabolic, physiological and cognitive processes. REM sleep influences early postnatal developmental behaviors; it facilitates the acquisition of resources from the mother (i.e. by means of suckling behavior) and it promotes the release of hormones such as prolactin and oxytocin which are pivotal in the development of attachment behavior [4]. From an evolutionary and behavioral perspective, REM sleep has been associated with adult reproductive success [4]. NREM sleep is associated with more stable metabolic and autonomic responses compared to REM sleep. Furthermore, the presence of REM-like and NREM-like states has been associated with nutritive behaviors in offspring and mother, respectively [4].
Recent advances in functional studies of sleep strongly suggest that memory consolidation benefits from a slow (<1 Hz) highly synchronized cortical activity in NREM and from subcortical theta (5–9 Hz) rhythms in REM sleep [13]. Despite these advances in functional understanding of REM and NREM sleep states, genetic and epigenetic mechanisms of sleep-dependent plasticity and memory processing are not currently well understood.
Here we report, for the first time, experimental evidence for a role of an imprinted gene, Gnas, in the modulation of REM/NREM sleep physiology. Gnas encodes the stimulatory G-protein subunit Gsα, which is involved in the generation of intracellular cyclic AMP and plays a crucial role in energy expenditure and metabolism by mediating sympathetic effects on many tissues [14]. It is biallelically expressed in most tissues including adult white adipose tissue [15] but it is predominantly maternally expressed and paternally repressed in a subset of tissues such as neonatal brown adipose tissue (BAT) [16], although it is not known if it shows imprinted expression in adult BAT. BAT produces heat by fatty acid oxidation and serves a pivotal thermoregulatory function within the organism [17]. Thanks to the rich presence of mitochondria and by means of the uncoupling protein-1 (UCP1), BAT is implicated in non-shivering thermogenesis [15], [18].
Imprinted expression of Gnas is controlled by a cis-acting differentially methylated region (DMR): the Exon1A-DMR [16]. Paternal transmission of a deletion of the Exon1A-DMR (Gnastm1Jop, hereafter called Ex1a, see Figure 1a) causes derepression of the normally repressed paternal Gnas allele in imprinted tissues resulting in biallelic Gnas expression and loss of imprinting [16]. We show here, for the first time, that loss of imprinting of Gnas results in specific abnormalities in sleep, cognition and thermoregulation in adult mice.
Fig. 1. Exon 1A deletion: Gnas expression and body temperature. 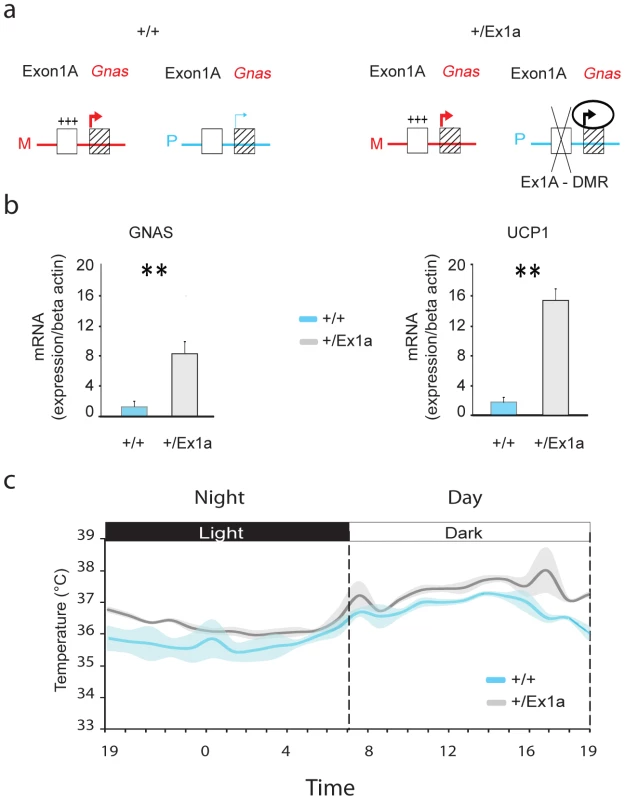
(a) Representation of Gnas expression when the Exon1A-DMR (Ex1A-DMR) is methylated on the maternal (M) allele in +/+ and +/Ex1a or deleted on the paternal (P) allele in +/Ex1a mice. (b) Expression levels, by q-PCR measurements, in BAT tissue of Gnas (left panel) and Ucp1 (right panel) mRNA in +/+ versus mutant +/Ex1a mice. (c) Grand-averages and shadowed ± s.e.m. of body temperature over a 24-hour period for +/+ and +/Ex1a mice. Statistics are reported as following: t-test; ** = P<0.01. Results
On paternal inheritance of the deletion, we have observed that in +/Ex1a mice the level of Gnas mRNA expression is increased, compared to littermate controls, in adult BAT (Figure 1b) indicating a derepression on the paternal allele due to a loss of Gnas imprinting, as previously reported in newborn mice BAT [16]. Moreover, we show here that Ucp1 mRNA in BAT is significantly higher in +/Ex1a mice compared to littermate controls (Figure 1b). As expected from the observation of increased Ucp1 levels a significantly higher body temperature was found in mutant animals (Figure 1c). The major increase of temperature in +/Ex1a mice compared to littermate controls occurs at the end of the subjective day and at the beginning of the subjective night, when there is a strong urge to sleep (Figure 1c).
In order to investigate the sleep-wake profile and the behavioral performance in these mice, we subjected adult +/Ex1a mice and +/+ littermate controls to behavioral and electrophysiological investigation in the home-cage environment. Interestingly, the total REM sleep was significantly reduced in +/Ex1a mice compared to littermate controls (Figure 2a) while the total NREM was unaffected between the two groups (Figure 2b). However, at the electrophysiological level, the contribution (power density) of delta (1–4 Hz) frequencies, the main synchronized rhythm in NREM sleep, was higher in the +/Ex1a mice compared to +/+ mice (Figure 3).
Fig. 2. Sleep architecture in Exon 1A mice. 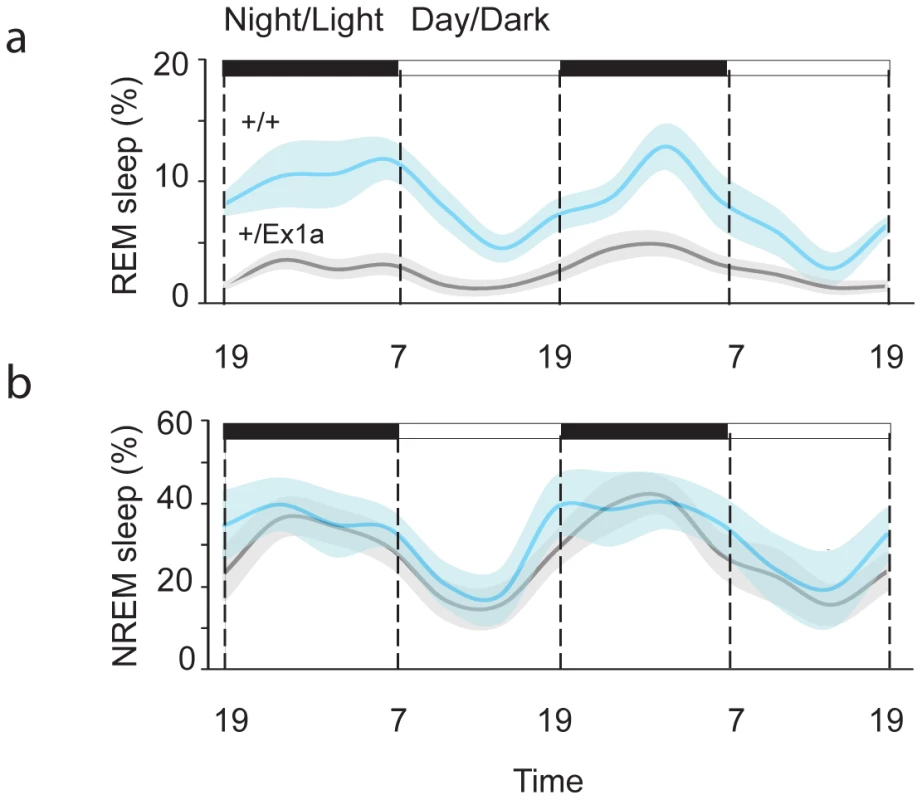
Four-hour bins percentage and ± s.e.m., shadowed, of REM (a) and NREM (b) sleep for a 48-hour period. Fig. 3. Representation of EEG frequencies in REM and NREM sleep. 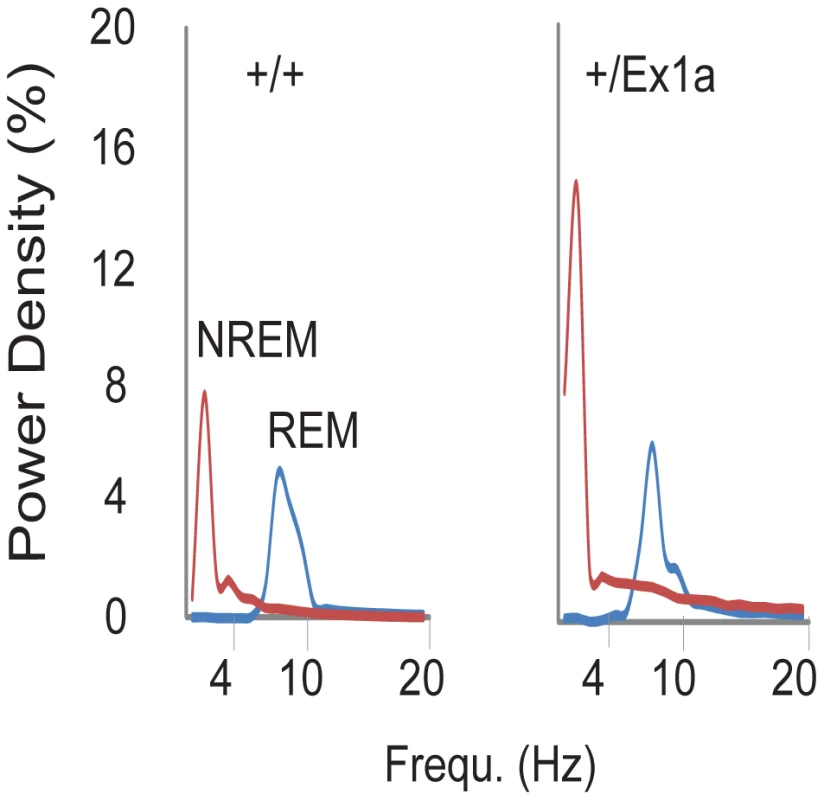
Here is represented the power density (see Methods) for all frequencies in all 4-seconds REM (blue line) and NREM (red line) sleep epochs during a 48-hour baseline. A 6-hour sleep deprivation protocol triggered, immediately after deprivation, a significantly increase (rebound) of REM in +/Ex1a mice, which testifies the need for a homeostatic recovery of REM, but not for NREM sleep, in mutants (Figure S1 and Figure S2). No differences in body temperature occurred during sleep deprivation and during the following recovery period between the two groups (Figure S2).
Thus, we extended our investigation into specific REM/NREM-dependent behavioral functions. We subjected mice to a classical memory task, the fear conditioning (FC) test, which affects REM sleep homeostasis [19]. After the first (conditioning) day of the FC protocol, we have observed an increase of REM sleep but not of NREM sleep in +/+ mice (Figure S1a–S1b). The presence of REM and its theta density were significantly higher in +/+ compared to +/Ex1a mice (Figure S1b). We suggest that this lack of REM increase in mutants is the causal mechanism explaining the reduced freezing behavior in +/Ex1a mice, compared to controls, that occurred when the animals were exposed to the same context (Figure 4) the following day. Indeed, the consolidation of fear responses has been previously associated with REM mechanisms that occur during sleep [19]. Because no difference between the two groups was observed in the cue condition (day 3) we reason that the deficit in in +/Ex1a mice is restricted to context-dependent mechanism, which has been previously associated with REM/fear memory consolidation [20], [21].
Fig. 4. Fear conditioning performance. 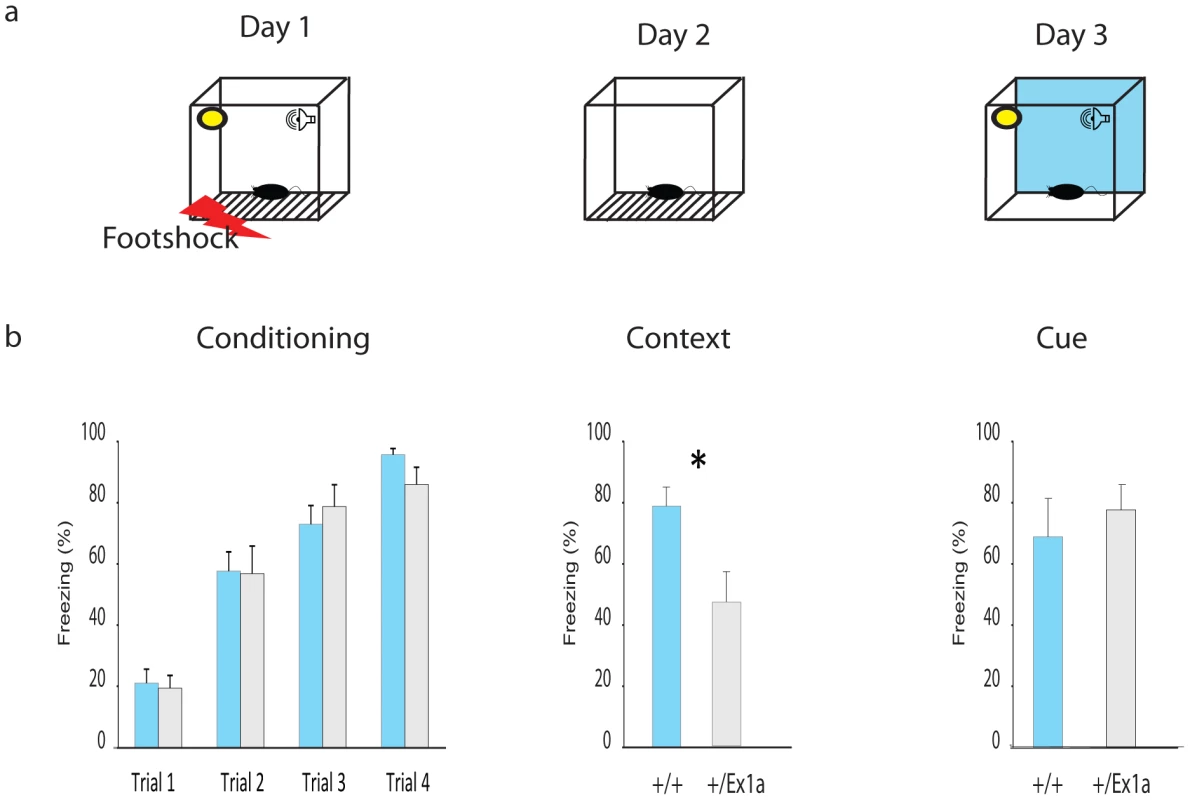
(a) Representation of the FC protocol over three days and three different experimental phases (Conditioning, Context and Cue). (b) Mean percentage values of freezing behavior ± s.e.m. are plotted for each experimental phase. Statistics are reported as following: t-test; * = P<0.05. A different subset of mice was tested in their home-cage with a cortico-striatal cognitive task, the “Switch-test” [22]. The test requires the animal to decide to nose-poke in different hoppers within the home-cage in response to a short - versus a long-light signal to obtain a reward (Figure 5a). Optimal performance in this task implies that the animal has learnt to distinguish between a short-signal associated with the reward coming at the hopper in one of the two locations and a long-signal associated with the reward coming at the hopper in the other location. This test of cognitive performance assesses whether the animal has an accurate representation of both endogenous/implicit (the subjective estimation of the signal duration) and exogenous/explicit (the ratio between short and long signals) temporal variables. In one condition (the “Switch” condition) all trials resulted in a reward if the animal responded correctly. In a second condition (the “Probes” condition) a percentage of trials was never rewarded regardless of the response of the animal. This latter condition was to measure the animal's uncertainty and its cognitive performance during a temporal decision making process [23]. Interestingly, +/Ex1a mice performed better in each phase of the experiment showing higher accuracy and time precision compared to controls (Figure 5b–5d). As expected, this 24-hour home-cage cognitive effort triggered a significant NREM increase and a higher delta power in +/Ex1a mice compared to +/+ mice in both “Switch” and “Probes” conditions (Figure S1a–S1c). Notably, the “Probes” condition, which added a degree of uncertainty to the expectation of obtaining a reward, resulted in an more severe augmentation of NREM sleep respect to the “Switch” condition. REM did not change significantly following both conditions neither in +/+ or +/Ex1a mice (Figure S1a–S1c).
Fig. 5. Cognitive performance in Exon 1A mice and littermate controls. 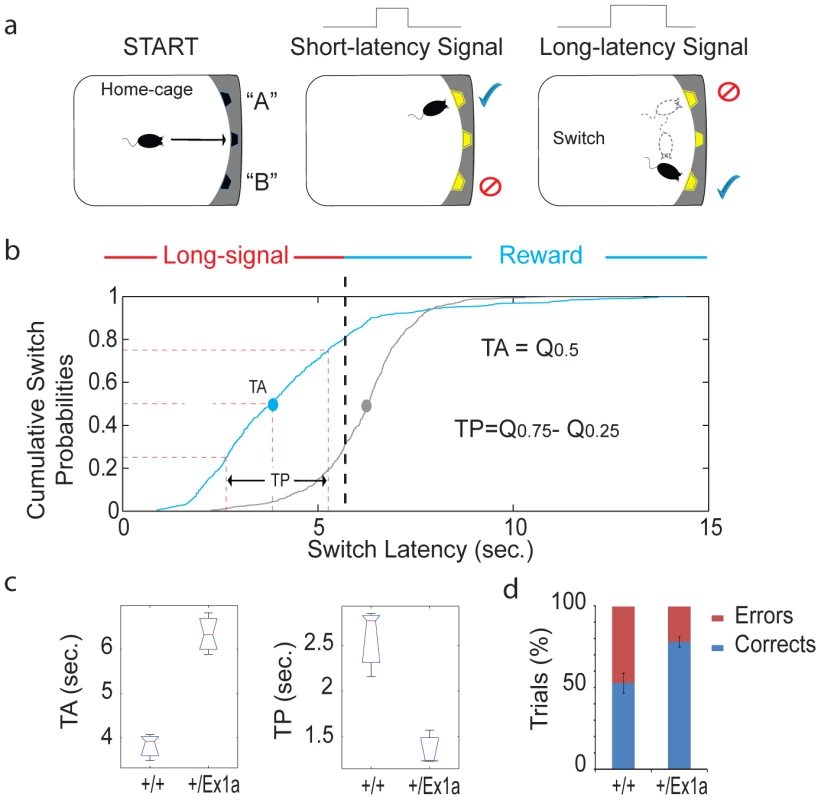
(a) A schematic representation of the “switch task” in which a mouse self-initiates the trial (“START”) and receives a reward according to the duration of the signal (“short-latency” versus “long-latency”) and the location (“A” or “B”). (b) The cumulative distribution of all switch latencies (the time at which the mouse stops nose poking in the short-location and moves to the long location) for all long-signal trials are reported for the +/Ex1a mice and their littermate controls. (c) From the above cumulative distribution of the switch latencies we have derived the Time Accuracy (TA), as the median (Q0.5), and the Time Precision (TP), as the difference of the third and first inter-quartiles (Q0.75−Q0.25), similar to what is described in [22]. Box-plots of TAs (the median values for each subject) and TPs (the inter-quartiles) are shown as well as (d) the percentage of correct versus error trials for the whole experiment. The correct/error trials graph refers only to the “Switch” condition, however, the same trend was observed in the “Probes” condition. Discussion
We have shown here clear evidence for an involvement of the imprinted Gnas transcript in the modulation of sleep and sleep-dependent behaviors.
Our study demonstrated a close link between sleep and thermoregulation. Temperature control is a well-known mechanism that modulates the expression of REM/NREM sleep in humans and mice [24]. When the thermoregulatory demand increases, REM sleep diminishes in rodents [24]–[26]. Hence, the increase of body temperature in +/Ex1a accounts for the significant reduction of REM sleep that we have observed in these mutants. Moreover, several studies have shown that an increase in body temperature is associated with an increase in NREM sleep propensity [24], [27]–[29]. Thus, the hyperthermic phenotype is responsible for the REM defect-NREM improvement that we observed in +/Ex1a mice, indicating a link between imprinted Gnas, thermoregulation and REM/NREM sleep [24]. The molecular pathway joining Gnas and thermoregulation in BAT is well understood. Gsα is considered an important mediator of the activity of the sympathetic nervous system (SNS) on many functions of BAT, included thermogenesis and energy expenditure [30]. SNS stimulation of BAT leads to thermogenesis. Gsα is a constituent of transmembrane G-protein-coupled receptors that translate adrenergic SNS stimulation, in BAT, to the activation of UCP1 via specific intracellular changes and a signaling cascade that involves the production of cAMP and activation of protein kinase A (PKA). Our results showing increased Gnas - Ucp1 expression are in line with a specific SNS-molecularly mediated cAMP-PKA role in non-shivering thermogenesis and therefore sleep. Indeed, the intracellular activity of cAMP-PKA is inversely related to the urge to sleep. In particular, an increased PKA affects sleep according to the specific cells in which it is expressed [31]–[33]. A reduced REM sleep would, presumably, be associated with an elevated cAMP-PKA activity.
We have shown that functional monoallelic expression of Gnas, due to imprinting of this gene, is important, in mice, for the physiology of REM sleep and for the consolidation of fear conditioning contextual memories. Many experimental observations support the idea that hippocampus is crucial for the development of contextual fear conditioning [20], [34]. However, fear conditioning responses are mediated by a complex interaction within limbic and prelimbic areas and this is also modulated by circadian genes [35]. As Chen and colleagues [36], have shown that Gnas is not imprinted in the hippocampus, our study implies that this specific cognitive phenotype must be due to imprinting of Gnas in brain regions other than hippocampus. Within this same study it was shown that Gnas is imprinted in the paraventricular nucleus (PVN) of the hypothalamus, a brain region that subserves important metabolic functions [36]. The PVN receives important projections from neurons located within the suprachiasmatic nucleus (SCN) of the hypothalamus, the master clock for circadian rhythms [37]. The interaction between PVN and SCN is important in orchestrating circadian rhythms and to set proper neuroendocrine responses to stressors [37]. Thus loss of Gnas imprinting, within these structures, can be envisaged as playing a role in both sleep and behavior. The results of our study confirm the idea that REM sleep is fundamental in the consolidation of fear responses [19], likely, involving a complex network of brain activity.
We have also shown that NREM functions are sensitive to Gnas dosage but, in this case, loss of imprinting of Gnas results in a reduction of uncertainty in temporal decision making. This improvement is paralleled by an increased contribution of slow wave activity in NREM sleep. Our result is consistent with the idea that cortical slow oscillations, by modulating synaptic circuits between subcortical and cortical structures, are responsible for the consolidation of daily memories during sleep [13].
In conclusion, in our study, loss of imprinting of Gnas, inhibits REM and primitive REM-linked functions, such as the fear response to a threatening context. Conversely, it enhances NREM physiology and high-level cognitive functions that developed alongside a progressively complex brain.
However, from a behavioral point of view an increased precision in interval timing estimation does not necessarily result in a better performance in other behavioral responses. Indeed, if the subject is less certain about the time of the foot-shock, its fear conditioning response may start earlier and stop later [38], hence resulting in a higher freezing time. Perhaps, evolution has developed a balanced mechanism between temporal uncertainty and fear behavioral responses and then loss of imprinting, in +/Ex1a mice, involves a reduction of freezing because of a higher timing precision. Thus, REM/NREM sleep expression may favor this well-adjusted mechanism.
The results of our study indicate a specific role for the imprinted gene Gnas in thermoregulation, which in turn affects REM/NREM sleep and then, cognitive performance. In addition we also reported a novel effect of specific cognitive mechanisms on sleep, by showing that a specific decision making process, the consolidation of an interval timing task, influences NREM sleep. Furthermore, in our mouse mutant model, the particular NREM physiology, exacerbates the effect of cognition on sleep homeostasis. This study attests the relevance of Gnas in brain functions and that loss of imprinting of Gnas affects cognitive processes.
Another transcript within the Gnas locus, the paternally expressed transcript Gnasxl, is expressed in specific sleep-related brain areas including the locus ceruleus and cholinergic laterodorsal tegmental nuclei [39]. The activity of neurons in the cholinergic laterodorsal tegmental nucleus is particularly important in the regulation of REM sleep [40], [41]. Mice with mutations in paternally derived Gnasxl transcripts show phenotypic deficits associated with growth and development, which is a critical stage for REM sleep across many species [4]. Thus Gnasxl may also play a role in sleep.
Methods
Mouse breeding, genotyping, and procedures
Initial Ex1a stock mice were produced in MRC-Harwell on 129/SvEv background and then transferred to IIT. In IIT mice were bred and maintained, through paternal inheritance, for several generations on C57BL/6J background, as this is a favorite background for behavioral studies. The genotyping of the mice was conducted following the assay as indicated in [42]: Exon1aF 5′cagtcgcgtcggcaccgcggag3′ and Exon1aR 5′gacgcactcacacgcaaagcag3′.
All the behavioral and electrophysiological experiments, in adult +/Ex1a mice and +/+ littermate controls, were conducted in the home-cage environment. In addition, mRNA expression profiles for Gnas and Uncoupling Protein (Ucp)1 were made in naïve adult mice. Each experiment included 8 male mice (10-weeks old) for each genotype and all procedures were done under the guidance issued by the UK authority (Project Licence Numbers 30/2526) and under the Italian Policy (licence issued on 19/06/2009, decreto N°106/2009-B).
Quantitative real-time PCR
Total RNA was extracted from about 0.2 g of snap frozen brown adipose tissue (BAT) and homogenized using Pestel with Trizol Reagent (Invitrogen, Carlsbad, CA) to isolate total RNA according to the manufacturer's procedure. Q-PCR was conducted essentially as previously described [43], [44]. Specific primers were: UCP1:f5′-GTCCCCTGCCATTTACTGTCAG-3′, r5′-TTTATTCGTGGTCTCCCAGCATAG-3′; GNAS: f5′-AGAAGGACAAGCAGGTCTACCG-3′, r5′-GTTAAACCCATTAACATGCAGGA-3′; β-actin, f5′-GGCACCACACCTTCTACAATG-3′, r5′-GGGGTGTTGAAGGTCTCAAAC-3′. Thermal cycling parameters were: denaturation 95°C for 5 min followed by 40 cycles of denaturing - annealing and extending (95°C for 15 sec, 60°C for 30 sec and then 70°C for 1 min). The results were calculated by the comparative Ct method according to the Applied Biosystems ABI-PRISM-7700 User Bulletin#2. Each sample was run in quadruplicate to obtain average Ct values and a ΔCt value for the target gene of the same sample, normalizing each sample to β-actin. The expression relative to β-actin was determined by calculating 2−ΔCt. Mean comparison was performed with unpaired Student's t-test.
Behavioral experiment
Mice were subjected to a long-term investigation in home-cage environment after the implant of a wireless system (Data Sciences) that enables to record electroencephalography (EEG), electromyography (EMG), locomotor activity and body temperature for off-line sleep-stage analysis (see [19]). Automated sleep scoring followed by visual manual inspection was performed using all sleep criteria for mice [19]. We performed Fast Fourier Transform analysis of the EEG signals with SleepSign software. The contributions of EEG frequencies was expressed as power densities in each frequency bin in all NREM and REM sleep epochs (as described in [45]).
A 2 week post-surgery period of recovery were given to each mouse to ensure a full recovery of normal sleep. At the end of the recovery period, we started recording all the physiological signals uninterruptedly for 48 consecutive hours (sleep baseline). Then, mice went under 6-hour sleep deprivation (SD) and 6-hour recovery period. After an additional 1-week the mice underwent fear conditioning (FC) or timing learning (Switch task) conditions.
The “Switch task.”
Mice were maintained in home-cage (equipped with a novel three-hopper operant wall from TSE) to perform in the switch task [22], The test requires the animal to decide to nose-poke in response to a light-signal to obtain a reward (Figure 5a). All trials were self-initiated by the animals by nose poking in one hopper (located in the middle of the operant wall) in the home-cage. The rewards were delivered through the two lateral hoppers; one hopper was always associated with short light-signals while the other hopper was always associated with long light-signals. On a fraction of the trials, the reward pellets were available in location “A” after a short-signal duration (3-seconds). On the other trials, the pellets were available after a long-signal duration (6-seconds) at the location “B”. If and when they reckoned that the short-latency had elapsed, they decided to leave the location “A” and move (switch) to the location “B”. The rewards were obtained only if the first nose poke of the animal occurred at the correct location. If mice switched too soon or too late for those long-signal trials, they ended up without payoff.
We trained all mice to a steady-state performance, keeping 1∶3 ratio between short - and long - signals (3 vs. 9 sec respectively). Then we reduced the short-/long-signal ratio to 1∶2 (3 vs. 6 sec), which is effectively similar to increased endogenous timing uncertainty (i.e. [23]). The test included two experimental conditions: “Switch” and “Probes”. The “Probes” condition differs from the “Switch” condition as we introduced 20% of no-reward(probe)-trials to independently test the timing uncertainty. We analysed for each condition the number of errors as well as switch latencies, time accuracy and time precision (as described in [22], [38]).
Fear conditioning
Eight mice for each genotype were tested while on their active/dark phase. On day 1 each mouse was placed in a fear conditioning chamber and, after a 2-min baseline, were given four light/white-noise (55-dB) cue-stimuli lasting 30 s and overlapping in the last 2 s with a footshock, (0.5 mA) lasting further 2 s. Each pairing was 2 min apart (inter-trial interval). After a final 30 s post-footshock delay, mice were returned to their home cages. To test contextual memory, we placed the mice back in the original chamber (24-hr post-training, day 2) for 3 minutes with no footshock during which freezing bouts were scored. During day 3 we placed all mice, for 3 minutes, in the testing chamber but with a different context respect to the original one. During this exposure, we subjected all the animals to the light/white-noise cues of day 1 and we measured their freezing behavior.
Supporting Information
Zdroje
1. CirelliC 2009 The genetic and molecular regulation of sleep: from fruit flies to humans. Nat Rev Neurosci 10 549 560
2. SehgalAMignotE 2011 Genetics of sleep and sleep disorders. Cell 146 194 207
3. McNamaraP 2004 Genomic imprinting and neurodevelopmental disorders of sleep. Sleep and Hypnosis 6 82 90
4. McNamaraPDowdallJAuerbachS 2002 REM sleep, early experience, and the development of reproductive strategies. Human Nature 13 405 435
5. Vela-BuenoAKalesASoldatosCRDobladez-BlancoBCampos-CastelloJ 1984 Sleep in the Prader-Willi syndrome. Clinical and polygraphic findings. Arch Neurol 41 294 296
6. HertzGCatalettoMFeinsilverSHAnguloM 1993 Sleep and breathing patterns in patients with Prader Willi syndrome (PWS): effects of age and gender. Sleep 16 366 371
7. VgontzasANBixlerEOKalesACenturioneARoganPK 1996 Daytime sleepiness and REM abnormalities in Prader-Willi syndrome: evidence of generalized hypoarousal. Int J Neurosci 87 127 139
8. VgontzasANKalesASeipJMascariMJBixlerEO 1996 Relationship of sleep abnormalities to patient genotypes in Prader-Willi syndrome. Am J Med Genet 67 478 482
9. ColasDWagstaffJFortPSalvertDSardaN 2005 Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiol Dis 20 471 478
10. AmiciRSanfordLDKearneyKMcInerneyBRossRJ 2004 A serotonergic (5-HT2) receptor mechanism in the laterodorsal tegmental nucleus participates in regulating the pattern of rapid-eye-movement sleep occurrence in the rat. Brain Res 996 9 18
11. KatoMVShimizuTNagayoshiMKanekoASasakiMS 1996 Genomic imprinting of the human serotonin-receptor (HTR2) gene involved in development of retinoblastoma. Am J Hum Genet 59 1084 1090
12. KatoMVIkawaYHayashizakiYShibataH 1998 Paternal imprinting of mouse serotonin receptor 2A gene Htr2 in embryonic eye: a conserved imprinting regulation on the RB/Rb locus. Genomics 47 146 148
13. DiekelmannSBornJ 2010 The memory function of sleep. Nat Rev Neurosci 11 114 126
14. ChenMNemechekNMMemaEWangJWeinsteinLS 2011 Effects of deficiency of the G protein Gsalpha on energy and glucose homeostasis. Eur J Pharmacol 660 119 124
15. ChenMChenHNguyenAGuptaDWangJ 2010 G(s)alpha deficiency in adipose tissue leads to a lean phenotype with divergent effects on cold tolerance and diet-induced thermogenesis. Cell Metab 11 320 330
16. WilliamsonCMBallSTNottinghamWTSkinnerJAPlaggeA 2004 A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat Genet 36 894 899
17. JakusPBSandorAJanakyTFarkasV 2008 Cooperation between BAT and WAT of rats in thermogenesis in response to cold, and the mechanism of glycogen accumulation in BAT during reacclimation. J Lipid Res 49 332 339
18. CannonBNedergaardJ 2004 Brown adipose tissue: function and physiological significance. Physiol Rev 84 277 359
19. SanfordLDYangLWellmanLLLiuXTangX 2010 Differential effects of controllable and uncontrollable footshock stress on sleep in mice. Sleep 33 621 630
20. CaiDJShumanTHarrisonEMSageJRAnagnostarasSG 2009 Sleep deprivation and Pavlovian fear conditioning. Learn Mem 16 595 599
21. CaiDJMednickSAHarrisonEMKanadyJCMednickSC 2009 REM, not incubation, improves creativity by priming associative networks. Proc Natl Acad Sci U S A 106 10130 10134
22. BalciFFreestoneDGallistelCR 2009 Risk assessment in man and mouse. Proc Natl Acad Sci U S A 106 2459 2463
23. BalciFSimenPNiyogiRSaxeAHughesJA 2011 Acquisition of decision making criteria: reward rate ultimately beats accuracy. Atten Percept Psychophys 73 640 657
24. KrauchiKDeboerT 2010 The interrelationship between sleep regulation and thermoregulation. Front Biosci 15 604 625
25. ValatxJLRousselBCureM 1973 [Sleep and cerebral temperature in rat during chronic heat exposure]. Brain Res 55 107 122
26. AmiciRCerriMOcampo-GarcesABaracchiFDenticoD 2008 Cold exposure and sleep in the rat: REM sleep homeostasis and body size. Sleep 31 708 715
27. MorairtySRSzymusiakRThomsonDMcGintyDJ 1993 Selective increases in non-rapid eye movement sleep following whole body heating in rats. Brain Res 617 10 16
28. GaoBOFrankenPToblerIBorbelyAA 1995 Effect of elevated ambient temperature on sleep, EEG spectra, and brain temperature in the rat. Am J Physiol 268 R1365 1373
29. ObalFJrAlfoldiPRubicsekG 1995 Promotion of sleep by heat in young rats. Pflugers Arch 430 729 738
30. PuigserverPSpiegelmanBM 2003 Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev 24 78 90
31. HendricksJCWilliamsJAPanckeriKKirkDTelloM 2001 A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci 4 1108 1115
32. GravesLAHellmanKVeaseySBlendyJAPackAI 2003 Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol 90 1152 1159
33. JoinerWJCrockerAWhiteBHSehgalA 2006 Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441 757 760
34. AnagnostarasSGGaleGDFanselowMS 2001 Hippocampus and contextual fear conditioning: recent controversies and advances. Hippocampus 11 8 17
35. GarciaJAZhangDEstillSJMichnoffCRutterJ 2000 Impaired cued and contextual memory in NPAS2-deficient mice. Science 288 2226 2230
36. ChenMWangJDickersonKEKelleherJXieT 2009 Central nervous system imprinting of the G protein G(s)alpha and its role in metabolic regulation. Cell Metab 9 548 555
37. BuijsRMvan EdenCGGoncharukVDKalsbeekA 2003 The biological clock tunes the organs of the body: timing by hormones and the autonomic nervous system. J Endocrinol 177 17 26
38. BalciFPapachristosEBGallistelCRBrunnerDGibsonJ 2008 Interval timing in genetically modified mice: a simple paradigm. Genes Brain Behav 7 373 384
39. PlaggeAGordonEDeanWBoianiRCintiS 2004 The imprinted signaling protein XL alpha s is required for postnatal adaptation to feeding. Nat Genet 36 818 826
40. BerridgeCWIsaacSOEspanaRA 2003 Additive wake-promoting actions of medial basal forebrain noradrenergic alpha1 - and beta-receptor stimulation. Behav Neurosci 117 350 359
41. BerridgeCW 2008 Noradrenergic modulation of arousal. Brain Res Rev 58 1 17
42. WilliamsonCMTurnerMDBallSTNottinghamWTGlenisterP 2006 Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet 38 350 355
43. WinnVDHaimov-KochmanRPaquetACYangYJMadhusudhanMS 2007 Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 148 1059 1079
44. PossentiRal.E 2011 Characterization of a novel peripheral pro-lipolytic mechanism in mice: role of VGF-derived peptide TLQP-21. Biochem J In Press
45. FrankenPMalafosseATaftiM 1998 Genetic variation in EEG activity during sleep in inbred mice. Am J Physiol 275 R1127 1137
Štítky
Genetika Reprodukčná medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



