-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
In plants, multiple detached tissues are capable of forming a pluripotent cell mass, termed callus, when cultured on media containing appropriate plant hormones. Recent studies demonstrated that callus resembles the root-tip meristem, even if it is derived from aerial organs. This finding improves our understanding of the regeneration process of plant cells; however, the molecular mechanism that guides cells of different tissue types to form a callus still remains elusive. Here, we show that genome-wide reprogramming of histone H3 lysine 27 trimethylation (H3K27me3) is a critical step in the leaf-to-callus transition. The Polycomb Repressive Complex 2 (PRC2) is known to function in establishing H3K27me3. By analyzing callus formation of mutants corresponding to different histone modification pathways, we found that leaf blades and/or cotyledons of the PRC2 mutants curly leaf swinger (clf swn) and embryonic flower2 (emf2) were defective in callus formation. We identified the H3K27me3-covered loci in leaves and calli by a ChIP–chip assay, and we found that in the callus H3K27me3 levels decreased first at certain auxin-pathway genes. The levels were then increased at specific leaf genes but decreased at a number of root-regulatory genes. Changes in H3K27me3 levels were negatively correlated with expression levels of the corresponding genes. One possible role of PRC2-mediated H3K27me3 in the leaf-to-callus transition might relate to elimination of leaf features by silencing leaf-regulatory genes, as most leaf-preferentially expressed regulatory genes could not be silenced in the leaf explants of clf swn. In contrast to the leaf explants, the root explants of both clf swn and emf2 formed calli normally, possibly because the root-to-callus transition bypasses the leaf gene silencing process. Furthermore, our data show that PRC2-mediated H3K27me3 and H3K27 demethylation act in parallel in the reprogramming of H3K27me3 during the leaf-to-callus transition, suggesting a general mechanism for cell fate transition in plants.
Published in the journal: Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues. PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002911
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002911Summary
In plants, multiple detached tissues are capable of forming a pluripotent cell mass, termed callus, when cultured on media containing appropriate plant hormones. Recent studies demonstrated that callus resembles the root-tip meristem, even if it is derived from aerial organs. This finding improves our understanding of the regeneration process of plant cells; however, the molecular mechanism that guides cells of different tissue types to form a callus still remains elusive. Here, we show that genome-wide reprogramming of histone H3 lysine 27 trimethylation (H3K27me3) is a critical step in the leaf-to-callus transition. The Polycomb Repressive Complex 2 (PRC2) is known to function in establishing H3K27me3. By analyzing callus formation of mutants corresponding to different histone modification pathways, we found that leaf blades and/or cotyledons of the PRC2 mutants curly leaf swinger (clf swn) and embryonic flower2 (emf2) were defective in callus formation. We identified the H3K27me3-covered loci in leaves and calli by a ChIP–chip assay, and we found that in the callus H3K27me3 levels decreased first at certain auxin-pathway genes. The levels were then increased at specific leaf genes but decreased at a number of root-regulatory genes. Changes in H3K27me3 levels were negatively correlated with expression levels of the corresponding genes. One possible role of PRC2-mediated H3K27me3 in the leaf-to-callus transition might relate to elimination of leaf features by silencing leaf-regulatory genes, as most leaf-preferentially expressed regulatory genes could not be silenced in the leaf explants of clf swn. In contrast to the leaf explants, the root explants of both clf swn and emf2 formed calli normally, possibly because the root-to-callus transition bypasses the leaf gene silencing process. Furthermore, our data show that PRC2-mediated H3K27me3 and H3K27 demethylation act in parallel in the reprogramming of H3K27me3 during the leaf-to-callus transition, suggesting a general mechanism for cell fate transition in plants.
Introduction
Unlike most animal tissues, a wide variety of plant tissues can easily acquire pluripotency when properly cultured in vitro with two plant hormones, auxin and cytokinin [1]. On a callus-inducing medium (CIM), explants usually first form a pluripotent cell mass, called callus, from which shoots and roots regenerate on the corresponding shoot - or root-inducing medium [2]. Recent studies revealed that callus formation is via a lateral root development pathway involving a process where cells evolve from pericycle-like to form root meristem-like cells [3]–[5]. This novel finding greatly improves our understanding of the plant cell regeneration process, but also raises a new question: what is the underlying mechanism that guides cell fate transition from diverse differentiated plant tissues to callus?
Callus formation from explants requires dramatic changes both in cell identities and cell growth patterns, and such a cell fate transition has been shown to be accompanied by changes in expression of numerous genes [3], [5]–[7]. It seems unlikely that the genome-wide changes in gene expression only involve spatially and temporally regulated transcription factors. Plant epigenetic pathways, which are known to influence genome-wide gene expression [8], may also participate in the large-scale gene regulations occurring during callus formation [7], [9].
Chromatin, which is composed of repeating units termed nucleosomes, is the template of epigenetic information, and changes in chromatin structure could lead to simultaneous expression changes of numerous genes [8], [10]. Changes of histone modifications often affect epigenetic regulation [11]. In Arabidopsis thaliana, silenced euchromatin is usually marked with histone H3 lysine 27 trimethylation (H3K27me3) [12]–[14], while the active gene transcription phase is usually associated with H3K4me3 and H3K36me3 [14]–[16]. We show in this work that reprogramming of H3K27me3 is critical in genome-wide regulation of key genes, leading to cell fate transition from leaf blade to callus.
Reprogramming of H3K27me3 involves the Polycomb group (PcG)-mediated H3K27 trimethylation and H3K27 demethylation. PcG proteins form at least two multiprotein complexes, namely Polycomb Repressive Complex 1 (PRC1) and PRC2. While PRC2 shows histone methyltransferase activity, targeting lysine 27 on the N-tail of histone H3, PRC1 recognizes the trimethylation marker for subsequent chromatin compression [17], [18]. The Arabidopsis proteins CURLY LEAF (CLF), SWINGER (SWN) and MEDEA [19]–[21] were proposed to be the core components of PRC2, and the H3K27 methyltransferase activity of CLF was shown in a biochemical assay [22]. In addition to the core components, PRC2 also contains other components, including EMBRYONIC FLOWER2 (EMF2) in Arabidopsis [21], [23]. On the other hand, RELATIVE OF EARLY FLOWERING 6 (REF6) is an H3K27me3 demethylase identified in Arabidopsis, which efficiently removes the methyl group from H3K27me3 both in vitro and in planta [24]. However, since the ref6 mutation results in H3K27me3 hypermethylation only on a part of the PRC2-targeted loci [24], it is possible that other unidentified H3K27me3 demethylase(s) or additional demethylation mechanism(s) exist.
In this study, we show that genome-wide reprogramming of H3K27me3 is critically required for the leaf-to-callus transition. The PcG pathway is responsible for repression of the leaf-regulatory genes in leaf blade explants, and acts in parallel with the Arabidopsis H3K27 demethylation pathway, which derepresses the auxin-pathway and root-regulatory genes to enable the leaf-to-callus transition.
Results
Mutations in PRC2 genes block callus formation from leaf blades
Because the gene expression profiles in calli differ dramatically from those in their original tissues [3], [5]–[7], we hypothesized that one or more epigenetic pathways, which function in genome-wide regulation of gene expression, may participate in this cell fate switching process. To test this hypothesis, we first analyzed callus formation using leaf blades from mutants with reduced levels of H3K4me3, H3K36me3, or H3K27me3, which in Arabidopsis are important for gene regulation in the euchromatin regions [11], [14]. For convenience, explants of leaf blade are referred to as leaf explants hereafter. The mutants used corresponded to the methyltransferases of ARABIDOPSIS THALIANA TRITHORAX1 (ATX1) and SET DOMAIN GROUP2 (SDG2) for H3K4me3 [25]–[27], SDG8 for H3K36me3 [28], [29], and CLF and SWN for H3K27me3 [12]. Compared with the wild type (Figure 1A, 1E), atx1-2, sdg2-3, and sdg8-2 leaf explants formed calli on CIM (Figure 1B–1D), whereas no callus was seen from leaf explants of the clf-50 swn-1 double mutant (Figure 1F). It should be mentioned that swn-1 is a weak allele, and the null clf swn double mutant displays distinct plant phenotypes [21]. Because both CLF and SWN are core components of PRC2 and are functionally redundant, these results suggest that the PRC2-mediated H3K27me3 is required for the leaf-to-callus transition.
Fig. 1. Leaf explants of the PcG double mutant clf-50 swn-1 lose the ability to form a callus. 
Sterilized seeds were grown on plain MS media. The third and fourth rosette leaves of 20-day-old seedlings were cut at the junction between the blade and the petiole and only the blade parts were cultured on CIM for another 20 days. (A–F) Wild-type Col-0 (A), atx1-2 (B), sdg2-3 (C), sdg8-2 (D), wild-type Ws (E), and clf-50 swn-1 (F). Note that only leaf explants, which were from the clf-50 swn-1 rosette leaves, failed to form callus. For each genotype, more than 30 rosette leaves were used in one experiment, and they all exhibited the consistent phenotype. Shown are three randomly picked leaves out of the 30 tested leaves for each genotype. Bars = 2 mm in (A–E) and 400 µm in (F). Genome-wide changes in H3K27me3 distribution in the leaf-to-callus transition
Since H3K27me3 is required for callus formation from leaves, we next analyzed the genome-wide H3K27me3 profile by a ChIP-chip assay to identify genes with the altered H3K27me3 modification between the leaf and callus (Figure 2A; Table S1). A total of 3856 and 3991 genes harboring high levels of H3K27me3 were identified in the leaf and callus, respectively, with many of these included among the 4979 genes previously identified in seedlings [13] (72.7% and 75.6%, respectively) (Figure 2B). Additionally, 2306 genes were common among the leaves, calli, and seedlings (Figure 2B). Furthermore, 434 and 186 genes showed considerable and significant decrease and increase in H3K27me3 levels, respectively, in the callus as compared to the leaf (Figure 2C, Table S1).
Fig. 2. ChIP–chip analysis to identify genes with changes in the H3K27me3 levels between leaf and callus. 
(A) Leaf explants (left) and calli (right) were used for the ChIP-chip experiment. For preparation of experimental materials, leaf explants that were just cut from leaves of 20-day-old wild-type Col-0 seedlings and calli that were on the margins of leaf explants cultured on CIM for 20 days were harvested for nuclei extraction. (B) A comparison of H3K27me3 covered loci taken from three independent genome-wide analyses (seedlings from Zhang et al. [13], and leaves and calli in this work). (C) Classifications by annotated functions of the identified H3K27me3-covered genes with either decreased (left) or increased (right) levels. (D) Expression profiles of the genes identified by the ChIP-chip experiment with the decreased (left) or increased (right) H3K27me3 levels. The original expression data were downloaded from the online program Genevestigator [30]. Genes bearing the altered H3K27me3 levels encode proteins with different functions (Figure 2C), including phytohormone pathway proteins and putative transcription factors. It is possible that some of these putative regulatory genes may have important roles during callus formation, and we thus analyzed expression profiles of these putative regulatory genes in four different tissues: seedlings, inflorescences, rosette leaves, and roots, based on the data from published databases [30]. Compared with leaves, calli contained the increased number of genes which are normally expressed in the root of a plant, and all these genes belonged to the category with decreased H3K27me3 levels (Figure 2D, left). Conversely, among the H3K27me3 level increased category, the number of genes normally expressed in the root of a plant was reduced, whereas the number of genes expressed in the leaf was increased (Figure 2D, right). Since the H3K27me3 modification is generally thought to negatively affect gene expression [12], [13], these results support the previous notion that the callus cells are the root-featured cells [5].
We also performed a microarray experiment to analyze whether and to what degree the changed H3K27me3 levels of these genes affect their expression levels. Our data showed that about 40% genes with decreased H3K27me3 levels were upregulated, and about 46% genes with increased levels of H3K27me3 were downregulated (Table S2). This result indicates that the H3K27me3 modification and gene expression regulation are correlated for a relatively large group of the H3K27me3-covered genes during the leaf-to-callus transition.
Phenotypic analyses of leaf explants reveal two distinct stages of callus formation
To better understand callus development, we analyzed phenotypes of leaf explants during culturing using scanning electron microscopy (SEM). Shapes of leaf explants on CIM were unchanged 2 days after culturing (DAC) (Figure 3A), compared with explants prior to culturing (i.e., time 0) (Figure S1). Additionally, cell proliferation was not observed in the margin of 2 DAC explants (Figure 3A, 3B). In 4 DAC explants, cells started to proliferate, initially in the middle of the midvein (Figure 3C, 3D). Six days after culturing, cell proliferation occurred in the margin of leaf explants and vigorous cell division surrounding the midvein in the proximal part of the explants was observed (Figure 3E, 3F). Cell proliferation was clearly accelerated in 8 DAC and 10 DAC explants (Figure 3G, 3H), and the newly formed calli from the midvein and margins covered most parts of the 10 DAC explants (Figure 3H).
Fig. 3. The leaf-to-callus transition process could be divided into pre-2 DAC and post-2 DAC stages based on the status of cell proliferation. 
(A–H) SEM analysis of morphology of leaf explants from 2 to 10 DAC, respectively. Green arrowheads indicate the midvein. Red arrowheads indicate leaf epidermal pavement cells. Yellow arrowheads show the dividing callus cells. Images in (B), (D), and (F) are close-ups of the boxed regions in (A), (C), and (E), respectively. (I–L) GUS staining of leaf explants from the CYCB1;1:GUS transgenic line from 2 to 8 DAC. Note that the 2 DAC explants show no GUS staining. Bars = 100 µm in (B, D, F), and 1 mm in (A, C, E, G, H) and 2 mm in (I–L). The CYCB1;1 gene is strictly regulated by the cell cycle, and thus is widely used as a cell division marker [31]. By analyzing a CYCB1;1:GUS transgenic line, we demonstrated that the CYCB1;1 expression pattern was consistent with the results from SEM. The 2 DAC leaf explants did not show any CYCB1;1:GUS staining (Figure 3I), whereas GUS staining was observed in the midvein and other fine veins in the proximal part of 4 DAC explants (Figure 3J). The strong CYCB1;1:GUS staining was observed along the midvein and vein branches, and in the margins of the 6 DAC and 8 DAC explants (Figure 3K, 3L). Based on the time of cell proliferation initiation, callus formation from leaf explants could be mainly divided into two stages: the pre - and post-2 DAC stages, reflecting prearrangement and performance of cell fate transition, respectively.
H3K27me3 hypomethylation was first detected in the auxin response genes
Auxin is a plant hormone required for callus induction [2], and our ChIP-chip data revealed that H3K27me3 levels of a group of auxin-pathway genes were dramatically reduced in the callus compared with the leaf. These included several auxin-inducible genes, such as the GH3 genes, GH3.1, 2, 3, 6, and 17, which are involved in auxin homeostatic control, and the AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) genes, IAA1, 2, 14, 19, 20, and 24 (Table S1), which participate in auxin signaling [32], [33]. Our qRT-PCR results revealed that these genes were induced before or on 2 DAC. For example, compared with the basal expression levels in time 0 explants, GH3.2 expression levels were dramatically increased in 2 DAC explants and showed the highest levels in the 4 DAC explants (Figure 4A). The highest level of IAA2 expression also appeared on 2 DAC (Figure 4A). We also analyzed the time course of the H3K27me3 level changes at the GH3.2 and IAA2 loci by ChIP (Figure 4B), and results were normalized against those at AGAMOUS (AG) because the H3K27me3 levels at the AG locus kept unchanged between the leaf and callus (Figure S2). Our results showed that increased expression levels of these auxin-pathway genes were accompanied by a sharp decrease in H3K27me3 levels at their loci starting from 2 DAC (Figure 4B). This suggested a dramatically altered auxin-related action in the pre-2 DAC explants.
Fig. 4. H3K27me3 hypomethylations at the auxin-pathway genes in the pre-2 DAC stage. 
(A) qRT-PCR analysis of GH3.2 and IAA2. (B) ChIP analysis of GH3.2 and IAA2. Bars show s.e. (C–G) GUS staining of leaf explants of the DR5pro:GUS transgenic line from time 0 to 8 DAC on CIM. (H) ChIP-chip results showed decreased H3K27me3 levels at IAA2, GH3.2, NIT2, YUC4, IAMT1 and PIN1 loci in calli, compared with those in leaves. Bars = 2 mm in (C–G). The expression patterns of GH3.2 and IAA2 were consistent with those of the DR5pro:GUS reporter line [34], in which GUS staining was hardly detected at time 0 (Figure 4C), but was evident in the 2 DAC explants (Figure 4D). The strongest GUS staining was observed in 4 DAC explants (Figure 4E). Compared with the time 0 explants, which only showed very faint GUS signals surrounding the midvein (Figure 4C), deep GUS staining of the 2 DAC DR5pro:GUS explants was detected in the margins and midveins (Figure 4D). As cells were rapidly proliferating in 6 DAC and 8 DAC explants, all freshly formed calli were deeply stained (Figure 4F, 4G), indicating that a strong auxin signal transduction was occurring in these tissues.
In addition to the GH3.2 and IAA2 genes, other genes related to auxin biosynthesis, metabolism, and transport showed a significant loss in H3K27me3 (Figure 4H, Table S1). These genes included YUCCA4 (YUC4) [35], NITRILASE2 (NIT2) [36], IAA CARBOXYL METHYLTRANSFERASE1 (IAMT1) [37], and PIN-FORMED1 (PIN1) [38]. These results suggest that the entire network of auxin regulation is activated during the leaf-to-callus process.
Changes in the H3K27me3 level in post-2 DAC explants may initiate cell fate transition
To understand the molecular basis during the leaf-to-callus transition, we analyzed expression patterns of all putative transcription factor genes that showed altered H3K27me3 levels in the ChIP-chip assay, as these genes may include important contributors to cell fate transition. The analysis was first performed using Genevestigator, which is a program available online (https://www.genevestigator.com/) [30]. Interestingly, many of these transcription factor genes with decreased H3K27me3 levels were those that were either silenced or exhibited low-level expression in the leaf, but were highly expressed in the roots (Figure 5A). Conversely, most genes that showed increased H3K27me3 levels were those that were originally predominantly expressed in the leaf, but were silenced or weakly expressed in the roots (Figure 5A). These results indicate that callus formation from leaf explants experiences a tissue identity transition from leaf to root, and thus support the recent proposal that the callus contains cells resembling the root meristematic pluripotent cells [5]. Our data also suggest that reprogramming of H3K27me3 is important for changes in expression of regulatory genes that are originally preferentially expressed in specific tissues.
Fig. 5. H3K27me3 reprogramming affects cell fate transition in the post-2 DAC stage. 
(A) The online program Genevestigator [30] was used to analyze expression patterns of the putative transcription factor genes that show changes in H3K27me3 levels during callus formation. Asterisks show significant statistical differences by t-tests (*, p<0.05; **, p<0.01; ***, p<0.001). Among the putative transcription factors with decreased H3K27me3 levels, 21 out of 67 (31.3%) genes have significantly higher expression patterns in the root tissues (left), and among those with increased H3K27me3 levels, 16 out of 19 (84.2%) genes have significantly higher expression patterns in the leaf tissues (right). (B) qRT-PCR analysis of SAW1 and SAW2. (C) ChIP analysis of SAW1 and SAW2. Bars show s.e. (D) ChIP-chip results in calli showed increased H3K27me3 levels at SAW1, SAW2, ATH1, and TCP10. We also analyzed gene expression levels over time by qRT-PCR and monitored H3K27me3 levels by ChIP for several selected marker genes of transcription factors during callus formation. SAWTOOTH1 (SAW1) and SAW2 are genes that control leaf development [39] and were selected as the leaf marker genes. Unlike the analyzed auxin-pathway genes, expression levels of SAW1 and SAW2 were largely unchanged 2 DAC, but were reduced markedly 4 DAC and continued to gradually decrease during the subsequent post-2 DAC stages (Figure 5B). The reduced SAW1 and SAW2 expression levels were accompanied by increased H3K27me3 levels beginning 4 DAC (Figure 5C, 5D). The increased H3K27me3 levels were observed for some other leaf development genes, such as the BELL family gene ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) [40] and the TEOSINTE BRANCHED1-CYCLOIDEA-PCF (TCP) transcription factor gene TCP10 [41], [42] (Figure 5D, Table S1). These results suggest that PRC2-mediated H3K27me3 plays a role in silencing leaf regulatory genes during callus formation.
WUSCHEL-RELATED HOMEOBOX 5 (WOX5) is a root gene specifically expressed in the quiescent center (QC) and SHORT-ROOT (SHR) is an important root development-controlling gene [43], [44]. WOX5 was slightly derepressed in the 4 DAC explants and was highly expressed in 6 DAC and 8 DAC explants. A drastic SHR derepression also occurred 6 DAC (Figure 6A). The increased WOX5 and SHR expression levels were consistent with the decreased H3K27me3 levels starting 4 DAC (Figure 6B), suggesting that H3K27 demethylation is also required for derepression of root genes.
Fig. 6. H3K27me3 hypomethylations occur at several root-regulatory genes. 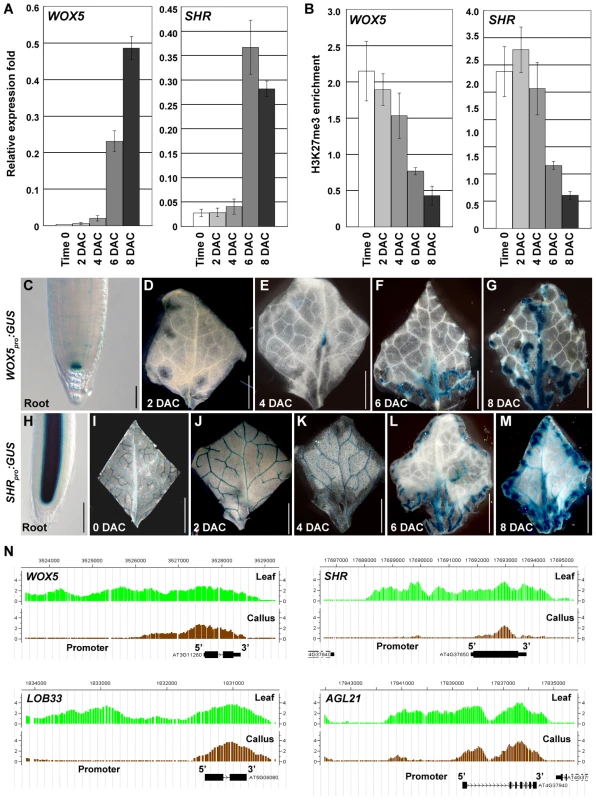
(A) qRT-PCR analyses show increased expression levels of root-regulatory genes WOX5 and SHR. (B) ChIP analysis of WOX5 and SHR. Bars show s.e. (C–G) GUS staining of the WOX5pro:GUS transgenic line in the root (C) and leaf explants cultured for 2 to 8 days on CIM (D–G). (H–M) GUS staining of the SHRpro:GUS transgenic line in the root (H), and in leaf explants at time 0 to 8 DAC (I–M). (N) ChIP-chip results in calli showed the reduced H3K27me3 levels at WOX5, SHR, LOB33 and AGL21. Bars = 100 µm in (C, H) and 2 mm in (D–G, I–M). We then used WOX5pro:GUS and SHRpro:GUS transgenic plants to analyze their expression patterns during callus formation. In the WOX5pro:GUS transgenic line, GUS staining was only detected in the root QC cells (Figure 6C) [43]. WOX5 remained silenced in leaf explants 2 DAC (Figure 6D), but expression in the midvein was observed 4 DAC (Figure 6E). GUS staining in 6 DAC and 8 DAC explants was observed in the midvein and the fine vein ends close to the wounded margins in emerging calli. This staining became more intense with increasing cell proliferation (Figure 6F, 6G). These results indicate that many cells in the newly formed calli from leaf explants possess features of root QC cells.
SHR is also expressed in the leaf vascular system (Figure 6I) in addition to its expression in the root stele cells (Figure 6H) [44]. However, SHR expression in early-stage explants is obviously different from that in the late-stage explants. The SHR expression pattern in the leaf vein did not change from time 0 to 4 DAC (Figure 6I–6K). In the 6 DAC leaf explants, SHR expression was weakened in the original leaf veins but became more intense in the midvein and vein ends near the wounded margin of newly formed calli (Figure 6L, 6M). In addition to WOX5 and SHR, the H3K27me3 levels of some other root genes, such as LATERAL ORGAN BOUNDARY DOMAIN33 (LOB33) [45] and AGAMOUS-LIKE21 (AGL21) [46], were also consistently decreased in the post-2 DAC stage (Figure 6N, Table S1). Changes in expression and H3K27me3 levels of the analyzed leaf and root regulatory genes highlight the process of cell fate transition occurring during callus formation in the post-2 DAC stage, following the auxin-pathway activation in the pre-2 DAC stage.
PcG plays a major role in the elimination of leaf characteristics during callus formation
To determine the role that the PRC2 plays in callus formation, we analyzed the expression of selected regulatory genes in the clf-50 swn-1 mutant. Both GH3.2 and IAA2 showed a similar elevated expression compared to the wild-type counterparts, suggesting that auxin response may not involve PRC2 (Figure 7A, 7B). Expression levels of SAW1 and SAW2 were reduced (Figure 7C, 7D) and those of WOX5 and SHR were elevated (Figure 7E, 7F) in 2 DAC leaf explants of clf-50 swn-1, rather than the expression changes occurring in 4 DAC leaf explants in the wild type. This was indicative of a shortened pre-2 DAC stage in the PRC2 mutant. Interestingly, expression levels of SAW1 and SAW2 in clf-50 swn-1 leaf explants failed to continue to decrease during the post-2 DAC stage (Figure 7C, 7D), as was observed in the wild type (Figure 5B), but instead gradually increased. On the other hand, the H3K27me3 modification at the SAW1 and SAW2 loci consistently retained at a very low level in clf-50 swn-1 (Figure S3). These results indicate that the leaf explants of clf-50 swn-1 consistently retain their leaf identities, although WOX5 and SHR were derepressed during culturing (Figure 7E, 7F).
Fig. 7. PcG is required for silencing leaf-regulatory genes during the leaf-to-callus transition. 
(A–F) Expression level changes of GH3.2 (A), IAA2 (B), SAW1 (C), SAW2 (D), WOX5 (E), and SHR (F) in leaf explants of clf-50 swn-1 from time 0 to 8 DAC, revealed by qRT-PCR analyses. Bars show s.e. Because PcG is required for silencing of the leaf development-controlling genes, but does not affect the derepression of auxin - and root-related genes, it is possible that leaf and root explants of the PRC2 mutant might behave differently during callus formation. To test this hypothesis, we analyzed the ability of the clf-50 swn-1 roots to form calli. Similar to the wild type roots (Figure 8A), the clf-50 swn-1 root explants demonstrated normal callus formation (Figure 8B). A more detailed analysis using SEM showed that the progression of callus formation in wild type (Figure 8C, 8D) and clf-50 swn-1 (Figure 8E, 8F) was also similar, as callus cells were evident on both root explants 6 DAC. These results indicate that PcG is required for the leaf, but not the root, during callus formation.
Fig. 8. The root explants of the PcG mutant can form a callus normally. 
(A, B) Calli of both 20 DAC root explants of wild-type Ws (A) and clf-50 swn-1 (B) are both evident. (C–F) SEM analyses showed that calli on root explants of both Ws (C, D) and clf-50 swn-1 (E, F) 6 DAC are of similar sizes. (D) and (F) are close-ups of the boxed regions in (C) and (E), respectively. Arrows indicate the newly formed calli. Bars = 2 mm in (A, B), 200 µm in (C, E), and 50 µm in (D, F). Discussion
Recent studies have revealed that callus formation occurs through a lateral root development pathway [5]. In this study, we tried to address another layer of questions during callus formation: what is the molecular mechanism that guides different plant tissues to form the pluripotent root meristem-like cells. We show that PcG-mediated epigenetic regulation is an essential step in callus formation, and the PRC2 components CLF and SWN are required for the leaf-to-callus transition. In addition to the rosette leaves, we analyzed cotyledons of clf-50 swn-1, and they were also defective in forming callus (Figure S4). To determine whether PRC2 as a whole is required for cell fate changes, we analyzed another PRC2 component EMF2 [21], [23] by examining the regeneration ability of emf2 cotyledons, because of a lack of leaves in emf2. Similar to those of clf-50 swn-1, cotyledons from two different emf2 alleles, emf2-1 and emf2-11, were both defective in callus formation (Figure S4), indicating the importance of PRC2 complex in the leaf-to-callus transition.
Our ChIP-chip assay revealed that reprogramming of H3K27me3 occurred at several important transcription factor genes, which regulate plant meristematic functions. For example, SAW1, SAW2, and TCP genes are known to control the leaf margin by repression of leaf meristematic features [39], [42], and WOX, KNOX, and several BELL family genes are also known to be involved in root or shoot meristematic activities (Table S1) [47], [48]. It is possible that maintenance of the undifferentiated state of cells requires functions of these genes. In addition, a number of metabolism-related genes showed notable changes in the H3K27me3 level, probably reflecting a secondary effect during changes of cell fates. Interestingly, transposable elements are generally much less regulated by PRC2-mediated H3K27me3 [13], whereas H3K27me3 levels at many transposable elements were elevated during callus formation (Figure 2C; Table S1). It is of interest to test in the future whether reprogramming of H3K27me3 at the loci of these transposable elements contributes to the callus formation.
The PcG pathway plays an important role in the leaf-to-callus transition; however, since PcG regulates many target genes during this process, the exact molecular mechanism still remains elusive. The fact that many leaf genes fail to be repressed in leaf explants of the clf-50 swn-1 double mutant could be one possible reason for the defective callus formation from leaf blades, and several lines of evidence support this hypothesis.
First, during callus formation from certain aerial organs of a plant, the differentiated tissues must undergo a common process to eliminate their original characteristics [5]. PcG has long been known to function in switching cell features not only in plants but also in animals. Second, most genes with increased H3K27me3 levels during callus formation are those that are highly expressed in the leaf but weakly in the root, including SAW1, SAW2 and many others (Figure S5). Consistent with SAW1 and SAW2, these genes demonstrated insufficient repression in leaf explants of clf-50 swn-1 (Figure S5). Third, similar to those of wild type, leaf explants of clf-50 swn-1 also show a gradually increased root gene expression, such as the demonstrated WOX5 and SHR genes, and their expression levels between wild-type and clf-50 swn-1 leaf explants are also similar within 4 DAC (Figure 6A, 7E and 7F). Although after 6 DAC, expression levels of these root genes became much higher in wild type than in the double mutant, this dramatic increase of expression levels might result from the newly formed calli but not from the leaf explants, as the clf-50 swn-1 explants failed to form callus. Thus, the clf-50 swn-1 mutant has the same potential to express the root genes but is unable to silence the leaf genes to suspend the leaf program in the leaf explants. The obstacle that must be overcome for callus formation in the leaf explants does not exist in the root explants of clf-50 swn-1 and emf2, providing a possible explanation why clf-50 swn-1 and emf2 root explants possess the ability to form calli (Figure 8B; Figure S4). Although the elimination of leaf characteristics appears to be a necessary step for callus formation from leaf explants, we could not exclude the possibility that this process is controlled by some other not-yet-revealed mechanisms.
To identify key genes that preferentially expressed in the leaf and may contribute to the leaf characteristic, we overexpressed ATH1 and SAW1 and analyzed callus formation of leaf explants from 35Spro:ATH1 and 35Spro:SAW1 transgenic plants. However, overexpression of the genes did not affect the leaf-to-callus transition (Figure S6). It is possible that for maintenance of leaf characteristics, many other leaf-expressed genes may be equally important.
The clf-50 swn-1 double mutant flowers earlier [21], indicating a shortened vegetative growth phase in clf-50 swn-1. This phenotype of clf-50 swn-1 raises a question: whether the clf-50 swn-1 leaves in ages are all equivalent to the very old wild-type ones that may lose the ability to form calli. We thus tested callus formation using wild-type and clf-50 swn-1 tissues at different ages, and found that cotyledons and leaf blades from the 44-day-old Col-0 seedlings were able to form calli, whereas the young 9-day-old leaf blades of the clf-50 swn-1 seedlings were defective in callus formation (Figure S6). These results suggest that the defective PcG function but not the age of tissues in clf-50 swn-1 blocked the callus formation.
During leaf-to-callus transition, the CLF and SWN proteins may act redundantly in regulating downstream genes, as the leaf explants of both clf-50 and the stronger swn-21 single mutants normally form calli (Figure S7). The weak clf-50 swn-1 double mutant usually produces only four rosette leaves. We tested each of these leaves at different leaf developmental stages and found the phenotype of defective regeneration was very consistent (Figure 1F; Figure S7).
Similar to rosette leaves, cotyledons of clf-50 swn-1, emf2-1 and emf2-11 are also defective in callus formation (Figure S4). Although rosette leaves and cotyledons are different plant organs, they may partially share a common program, which must be terminated by PcG for regeneration. Whether the PRC2-mediated H3K27me3 is responsible for initiating or maintaining leaf gene silencing during the leaf-to-callus transition is not yet clear. It has recently been suggested that in both animals and plants, histone modifications might not initiate gene expression regulation [12], [17], [49]. In Arabidopsis, it was found that H3K27me3 alone is not sufficient to initiate the repression of the AG gene [12], [17]. Our data showed that the leaf genes, SAW1 and SAW2, were downregulated in the 2 DAC leaf explants of clf-50 swn-1, whereas such a downregulation could not be maintained at or following 4 DAC. One possible explanation for this is that the PRC2-mediated H3K27me3 is responsible for maintaining rather than initiating the silencing of leaf genes.
A leaf-to-callus transition normally requires both repression of leaf-regulatory genes and activation of auxin-pathway and root-regulatory genes. The PcG-mediated histone modification and the H3K27 demethylation pathways may act in parallel in reprogramming of H3K27me3 during callus formation. How the pre - and post-2 DAC stages are connected is not known. Whether the activated auxin pathway in the pre-2 DAC stage induces a subsequent reprogramming of H3K27me3 at the loci of the leaf - and root-regulatory genes is an interesting question that will be addressed in the future.
Removal of the H3K27me3 marker may depend on an active process involving H3K27 demethyltransferases, such as REF6 [24], or a passive process resulting in the loss of the maintenance of histone methylations during cell division [50]. We propose that in the pre-2 DAC stage, H3K27me3 demethylation of the auxin-pathway genes is mainly via an active process because during this stage, cell division does not occur. However, during the post-2 DAC stage when cells start to proliferate, H3K27 demethylation leading to derepression of the root-regulatory genes may be via both active and passive processes. We tried to address this question by analyzing leaf explants of the ref6-1 mutant, but found that normal ref6-1 calli formed from leaf explants (Figure S7). These results suggest that REF6 may not be the only demethyltransferase in callus formation or may not participate in callus formation at all, and/or passive removal of H3K27 methylation may occur. Elucidation of the molecular mechanism of H3K27me3 demethylation during callus formation is important to fully comprehend plant cell regeneration.
In animals, cell fate determinations also require the PcG function for acquisition of cell pluripotency [51], [52]. In plants, determination of flowering time, floral organ formation, and stem cell restriction are all related to reprogramming of H3K27me3 and require PcG [17], [19], [21], [53]–[55]. Reprogramming of H3K27me3 might represent a general mechanism for cell fate transition in multicellular eukaryotes and the PcG pathway is at least partially involved in the process.
Materials and Methods
Plant materials and callus induction
Seeds of clf-50 swn-1 is in the Ws background, and swn-1 is a weak swn allele [21]. Seeds of sdg2-3 [26], sdg8-2 [28], atx1-2 [56] and ref6-1 [57], swn-21 (GK-783A01), emf2-11 (GK-685A01) [58], emf2-1 [59], clf-29 [60], and the CYCB1;1:GUS [31] and DR5pro:GUS [34] transgenic plants are in the Col-0 background. For callus induction, seeds were first grown on the medium containing Murashige and Skoog (MS) basal salt mixture. Tissues from different organs were prepared from different ages of seedlings and were placed on CIM (Gamborg's B5 medium with 0.5 g/L MES, 2% glucose, 0.2 mmol/L kinetin, and 2.2 mmol/L 2,4-dichlorophenoxyacetic acid and 0.8% agar) [61], followed by incubation at 22°C in the dark.
β-glucuronidase (GUS) assay and microscopy
DNA fragments with 4.6 kb and 2.5 kb in length upstream to the translation initiation site of WOX5 and SHR were PCR amplified from wild-type plants, respectively, and were subcloned into the plant transformation vector pBI101 (Clonetech, USA) with the 3′ in-frame fusion to GUS to yield WOX5pro:GUS and SHRpro:GUS, respectively. The constructs were verified by sequencing and were introduced into wild-type plants by Agrobacterium-mediated transformation. Primers used in cloning are shown in Table S3. For GUS staining, plant tissues were incubated in the GUS assay solution (50 mM sodium phosphate buffer pH 7, 5 mM Na2EDTA, 2 mM K3Fe(CN)6, 2 mM K4Fe(CN)6, 0.1% Triton X-100 and 0.04% X-Gluc) at 37°C for 90 min for the DR5pro:GUS or for 3 h for the CYCB1;1:GUS, WOX5pro:GUS, and SHRpro:GUS leaf explants, and the stained tissues were then incubated in 70% alcohol at 37°C for 12 h. GUS staining was observed by using the SMZ1500 and Eclipse 80i microscopes (Nikon, Japan). SEM analysis was performed as previously described [62].
ChIP and qRT–PCR
For the Chromatin immunoprecipitation (ChIP) assay, leaf explants cultured at different time points on CIM or the 20 DAC calli were vacuum-infiltrated with formaldehyde crosslinking solution. ChIP was performed as previously described [29], by using the antibody against H3 trimethyl-Lys 27 (Upstate, USA, Cat. 07-449). Results from real-time PCR represented the relative methylation levels, which were normalized against those produced by the primers for AG (Figure S2), whose values were arbitrarily fixed at 1.0.
For quantitative reverse transcription-polymerase chain reaction (qRT-PCR), RNA extraction and reverse transcription were performed as described previously [62], [63]. Three biological replicates were analyzed and each was tested by three technical replicates. Real-time PCR was performed using gene specific primers (Table S3) and the results represented the relative expression levels, which were normalized against those produced by the primers for ACTIN, whose values were arbitrarily fixed at 1.0.
Microarray, ChIP–chip assay, and data analysis
For microarray analysis, leaves from 20-day-old seedlings of wild type and 20 DAC calli were used for RNA preparation. Microarray was performed using the Affymetrix GeneChip system (Affymetrix, USA, and Gene Tech Biotechnology Company Limited, Shanghai, China). Three independent experiments were performed, and expression of genes was considered significantly perturbed when the change was more than 2.0-fold with a P value<0.05. The microarray data were deposited in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE36479 and the analyzed data were shown in Table S2.
For ChIP-chip, about 100 ng DNA from leaves or calli was enriched, respectively, in the ChIP assay, and the ChIP-chip experiment was carried out by using Affymetrix high density tiling arrays according to the manufacturer's protocol (Cat. 900594, Affymetrix, USA, and Gene Tech Biotechnology Company Limited, Shanghai, China). ChIP-chip data were analyzed using the CisGenome software with standard procedures [64], [65]. Genes with the significantly decreased H3K27me3 levels were defined as the peak cutoff MA (Leaf-Callus)≥3.5 and MA (leaf)≥2.5, and those with the significant increased H3K27me3 levels were defined as MA (Callus-Leaf)≥3.5 and MA (Callus)≥2.5. FDR (<5%) was settled as “Left ta”, which was recommended by the CisGenome software. The peak-gene association was settled from 3,000 bp upstream to the transcription start site (TSS) to the transcription end site (TES). The database TAIR8 was used for annotation of the identified genes. For genes cluster based on the tissue specific expression, an online program Genevestigator (www.genevestigator.com) [30] was used. The ChIP-chip data were deposited in the GEO (http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE34596 and the analyzed data are shown in Table S1.
Supporting Information
Zdroje
1. DuclercqJ, Sangwan-NorreelB, CatterouM, SangwanRS (2011) De novo shoot organogenesis: from art to science. Trends Plant Sci 16 : 597–606.
2. SkoogF, MillerCO (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11 : 118–130.
3. CheP, LallS, HowellSH (2007) Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226 : 1183–1194.
4. AttaR, LaurensL, Boucheron-DubuissonE, Guivarc'hA, CarneroE, et al. (2009) Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J 57 : 626–644.
5. SugimotoK, JiaoY, MeyerowitzEM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18 : 463–471.
6. CheP, LallS, NettletonD, HowellSH (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141 : 620–637.
7. TanurdzicM, VaughnMW, JiangH, LeeTJ, SlotkinRK, et al. (2008) Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 6: e302 doi:10.1371/journal.pbio.0060302.
8. Russo VEA, Martienssen RA, Riggs AD (1996) Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Woodbury.
9. LiW, LiuH, ChengZJ, SuYH, HanHN, et al. (2011) DNA Methylation and Histone Modifications Regulate De Novo Shoot Regeneration in Arabidopsis by Modulating WUSCHEL Expression and Auxin Signaling. PLoS Genet 7: e1002243 doi:10.1371/journal.pgen.1002243.
10. LoidlP (2004) A plant dialect of the histone language. Trends Plant Sci 9 : 84–90.
11. LiuC, LuF, CuiX, CaoX (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61 : 395–420.
12. SchubertD, PrimavesiL, BishoppA, RobertsG, DoonanJ, et al. (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25 : 4638–4649.
13. ZhangX, ClarenzO, CokusS, BernatavichuteYV, PellegriniM, et al. (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129 doi:10.1371/journal.pbio.0050129.
14. RoudierF, AhmedI, BerardC, SarazinA, Mary-HuardT, et al. (2011) Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30 : 1928–1938.
15. OhS, ParkS, van NockerS (2008) Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet 4: e1000077 doi:10.1371/journal.pgen.1000077.
16. ZhangX, BernatavichuteYV, CokusS, PellegriniM, JacobsenSE (2009) Genome-wide analysis of mono-, di - and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10: R62.
17. SchatlowskiN, CreaseyK, GoodrichJ, SchubertD (2008) Keeping plants in shape: polycomb-group genes and histone methylation. Semin Cell Dev Biol 19 : 547–553.
18. MullerJ, VerrijzerP (2009) Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 19 : 150–158.
19. GoodrichJ, PuangsomleeP, MartinM, LongD, MeyerowitzEM, et al. (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386 : 44–51.
20. GrossniklausU, Vielle-CalzadaJP, HoeppnerMA, GaglianoWB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 : 446–450.
21. ChanvivattanaY, BishoppA, SchubertD, StockC, MoonYH, et al. (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131 : 5263–5276.
22. SchmitgesFW, PrustyAB, FatyM, StutzerA, LingarajuGM, et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42 : 330–341.
23. YoshidaN, YanaiY, ChenL, KatoY, HiratsukaJ, et al. (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13 : 2471–2481.
24. LuF, CuiX, ZhangS, JenuweinT, CaoX (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43 : 715–719.
25. Alvarez-VenegasR, PienS, SadderM, WitmerX, GrossniklausU, et al. (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13 : 627–637.
26. BerrA, McCallumEJ, MenardR, MeyerD, FuchsJ, et al. (2010) Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22 : 3232–3248.
27. GuoL, YuY, LawJA, ZhangX (2010) SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc Natl Acad Sci U S A 107 : 18557–18562.
28. ZhaoZ, YuY, MeyerD, WuC, ShenWH (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7 : 1256–1260.
29. XuL, ZhaoZ, DongA, Soubigou-TaconnatL, RenouJP, et al. (2008) Di - and tri - but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28 : 1348–1360.
30. HruzT, LauleO, SzaboG, WessendorpF, BleulerS, et al. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008 : 420747.
31. Colon-CarmonaA, YouR, Haimovitch-GalT, DoernerP (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20 : 503–508.
32. StaswickPE, SerbanB, RoweM, TiryakiI, MaldonadoMT, et al. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17 : 616–627.
33. MockaitisK, EstelleM (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24 : 55–80.
34. UlmasovT, MurfettJ, HagenG, GuilfoyleTJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 : 1963–1971.
35. ChengY, DaiX, ZhaoY (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 : 1790–1799.
36. BartelB, FinkGR (1994) Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA 91 : 6649–6653.
37. QinG, GuH, ZhaoY, MaZ, ShiG, et al. (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17 : 2693–2704.
38. GalweilerL, GuanC, MullerA, WismanE, MendgenK, et al. (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 : 2226–2230.
39. KumarR, KushalappaK, GodtD, PidkowichMS, PastorelliS, et al. (2007) The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19 : 2719–2735.
40. QuaedvliegN, DockxJ, RookF, WeisbeekP, SmeekensS (1995) The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell 7 : 117–129.
41. KoyamaT, FurutaniM, TasakaM, Ohme-TakagiM (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19 : 473–484.
42. KoyamaT, MitsudaN, SekiM, ShinozakiK, Ohme-TakagiM (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22 : 3574–3588.
43. SarkarAK, LuijtenM, MiyashimaS, LenhardM, HashimotoT, et al. (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446 : 811–814.
44. HelariuttaY, FukakiH, Wysocka-DillerJ, NakajimaK, JungJ, et al. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101 : 555–567.
45. BerckmansB, VassilevaV, SchmidSP, MaesS, ParizotB, et al. (2011) Auxin-Dependent Cell Cycle Reactivation through Transcriptional Regulation of Arabidopsis E2Fa by Lateral Organ Boundary Proteins. Plant Cell 23 : 3671–3683.
46. BurgeffC, LiljegrenSJ, Tapia-LopezR, YanofskyMF, Alvarez-BuyllaER (2002) MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214 : 365–372.
47. HakeS, SmithHM, HoltanH, MagnaniE, MeleG, et al. (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20 : 125–151.
48. AichingerE, KornetN, FriedrichT, LauxT (2012) Plant Stem Cell Niches. Annu Rev Plant Biol
49. HenikoffS, ShilatifardA (2011) Histone modification: cause or cog? Trends Genet 27 : 389–396.
50. Radman-LivajaM, LiuCL, FriedmanN, SchreiberSL, RandoOJ (2010) Replication and active demethylation represent partially overlapping mechanisms for erasure of H3K4me3 in budding yeast. PLoS Genet 6: e1000837 doi:10.1371/journal.pgen.1000837.
51. EzhkovaE, PasolliHA, ParkerJS, StokesN, SuIH, et al. (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136 : 1122–1135.
52. PereiraCF, PiccoloFM, TsubouchiT, SauerS, RyanNK, et al. (2010) ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6 : 547–556.
53. KatzA, OlivaM, MosqunaA, HakimO, OhadN (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37 : 707–719.
54. XuL, ShenW-H (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18 : 1966–1971.
55. LafosM, KrollP, HohenstattML, ThorpeFL, ClarenzO, et al. (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040 doi:10.1371/journal.pgen.1002040.
56. PienS, FleuryD, MylneJS, CrevillenP, InzeD, et al. (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20 : 580–588.
57. NohB, LeeSH, KimHJ, YiG, ShinEA, et al. (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16 : 2601–2613.
58. RossoMG, LiY, StrizhovN, ReissB, DekkerK, et al. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 : 247–259.
59. YangCH, ChenLJ, SungZR (1995) Genetic regulation of shoot development in Arabidopsis: role of the EMF genes. Dev Biol 169 : 421–435.
60. BouveretR, SchonrockN, GruissemW, HennigL (2006) Regulation of flowering time by Arabidopsis MSI1. Development 133 : 1693–1702.
61. GamborgOL, MillerRA, OjimaK (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50 : 151–158.
62. XuL, XuY, DongA, SunY, PiL, et al. (2003) Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 : 4097–4107.
63. LiY, PiL, HuangH, XuL (2012) ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture. J Exp Bot 63 : 1423–1433.
64. JiH, JiangH, MaW, JohnsonDS, MyersRM, et al. (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26 : 1293–1300.
65. JiH, JiangH, MaW, WongWH (2011) Using CisGenome to analyze ChIP-chip and ChIP-seq data. Curr Protoc Bioinformatics Chapter 2: Unit2 13.
Štítky
Genetika Reprodukčná medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



