-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
Non-coding RNAs are much more common than previously thought. However, for the vast majority of non-coding RNAs, the cellular function remains enigmatic. The two long non-coding RNA (lncRNA) genes DLEU1 and DLEU2 map to a critical region at chromosomal band 13q14.3 that is recurrently deleted in solid tumors and hematopoietic malignancies like chronic lymphocytic leukemia (CLL). While no point mutations have been found in the protein coding candidate genes at 13q14.3, they are deregulated in malignant cells, suggesting an epigenetic tumor suppressor mechanism. We therefore characterized the epigenetic makeup of 13q14.3 in CLL cells and found histone modifications by chromatin-immunoprecipitation (ChIP) that are associated with activated transcription and significant DNA-demethylation at the transcriptional start sites of DLEU1 and DLEU2 using 5 different semi-quantitative and quantitative methods (aPRIMES, BioCOBRA, MCIp, MassARRAY, and bisulfite sequencing). These epigenetic aberrations were correlated with transcriptional deregulation of the neighboring candidate tumor suppressor genes, suggesting a coregulation in cis of this gene cluster. We found that the 13q14.3 genes in addition to their previously known functions regulate NF-kB activity, which we could show after overexpression, siRNA–mediated knockdown, and dominant-negative mutant genes by using Western blots with previously undescribed antibodies, by a customized ELISA as well as by reporter assays. In addition, we performed an unbiased screen of 810 human miRNAs and identified the miR-15/16 family of genes at 13q14.3 as the strongest inducers of NF-kB activity. In summary, the tumor suppressor mechanism at 13q14.3 is a cluster of genes controlled by two lncRNA genes that are regulated by DNA-methylation and histone modifications and whose members all regulate NF-kB. Therefore, the tumor suppressor mechanism in 13q14.3 underlines the role both of epigenetic aberrations and of lncRNA genes in human tumorigenesis and is an example of colocalization of a functionally related gene cluster.
Published in the journal: Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB. PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003373
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003373Summary
Non-coding RNAs are much more common than previously thought. However, for the vast majority of non-coding RNAs, the cellular function remains enigmatic. The two long non-coding RNA (lncRNA) genes DLEU1 and DLEU2 map to a critical region at chromosomal band 13q14.3 that is recurrently deleted in solid tumors and hematopoietic malignancies like chronic lymphocytic leukemia (CLL). While no point mutations have been found in the protein coding candidate genes at 13q14.3, they are deregulated in malignant cells, suggesting an epigenetic tumor suppressor mechanism. We therefore characterized the epigenetic makeup of 13q14.3 in CLL cells and found histone modifications by chromatin-immunoprecipitation (ChIP) that are associated with activated transcription and significant DNA-demethylation at the transcriptional start sites of DLEU1 and DLEU2 using 5 different semi-quantitative and quantitative methods (aPRIMES, BioCOBRA, MCIp, MassARRAY, and bisulfite sequencing). These epigenetic aberrations were correlated with transcriptional deregulation of the neighboring candidate tumor suppressor genes, suggesting a coregulation in cis of this gene cluster. We found that the 13q14.3 genes in addition to their previously known functions regulate NF-kB activity, which we could show after overexpression, siRNA–mediated knockdown, and dominant-negative mutant genes by using Western blots with previously undescribed antibodies, by a customized ELISA as well as by reporter assays. In addition, we performed an unbiased screen of 810 human miRNAs and identified the miR-15/16 family of genes at 13q14.3 as the strongest inducers of NF-kB activity. In summary, the tumor suppressor mechanism at 13q14.3 is a cluster of genes controlled by two lncRNA genes that are regulated by DNA-methylation and histone modifications and whose members all regulate NF-kB. Therefore, the tumor suppressor mechanism in 13q14.3 underlines the role both of epigenetic aberrations and of lncRNA genes in human tumorigenesis and is an example of colocalization of a functionally related gene cluster.
Introduction
Non-coding RNAs (ncRNA) are emerging as an important factor for the aberrant gene expression associated with cancer [1]. NcRNA genes are mostly involved in the regulation of target gene function [2]. Their mode of action varies from posttranscriptional regulation (i.e. miRNA genes) [3] to modulation of transcription in cis or in trans, either via competition or blockage mechanisms [4], by acting as chromatin organizers that target chromatin modifying factors (e.g. HOTAIR, KCNLQT1 and XIST) [5]. NcRNA genes can even act as enhancers themselves [6]. However, for the vast majority of ncRNAs, the specific cellular function remains enigmatic.
Two long ncRNA (lncRNA) genes DLEU1 (Gene ID: 10301) and DLEU2 (Gene ID: 8847) map to a critical region at chromosomal band 13q14.3 that is recurrently deleted in hematopoietic and solid tumors (Figure 1) [7]–[10]. DLEU2 splicing variants have been suggested to represent the primary transcripts (pri-miR) of miR-15a (Gene ID: 406948) and miR-16-1 (Gene ID: 406950) because of their localization and coregulation [11]. MiR-15/16 are among the most strongly and ubiquitously expressed miRNA genes in human cells [12] and appear to exert a crucial role in tumorigenesis [13]. In chronic lymphocytic leukemia (CLL), more than 50% of cases harbor a deletion of the critical region at 13q14.3 [7], [14]. Loss of 13q14.3 is also the most common aberration in the CLL precursor monoclonal B-cell lymphocytosis (MBL) [15]. The tumor suppressor mechanism at 13q14.3 is multifactorial and is likely to involve other genetic elements than miR-15a/16-1, since (i) knocking out miR-15a and miR-16-1 in mice leads to a lymphoproliferative disease [16], but rare cases of CLL have been described where the deletion at 13q14.3 does not encompass the miRNA genes [10], [17], [18]. (ii) Deletion of a larger region at 13q14.3 including adjacent regions in addition to miR-15a/16-1 leads to more aggressive disease in mice and humans that more frequently resembles a CLL phenotype [16], [18]–[20]. (iii) Familial CLL can be associated with deletion of DLEU7 (Gene ID: 220107) localized more proximal in 13q14.3 than with miR-15a/miR-16 [21].
Fig. 1. The critical region at 13q14.3 displays relaxed chromatin at the transcriptional start site (TSS) of the long non-coding RNA genes DLEU1 and DLEU2 in CLL cells. 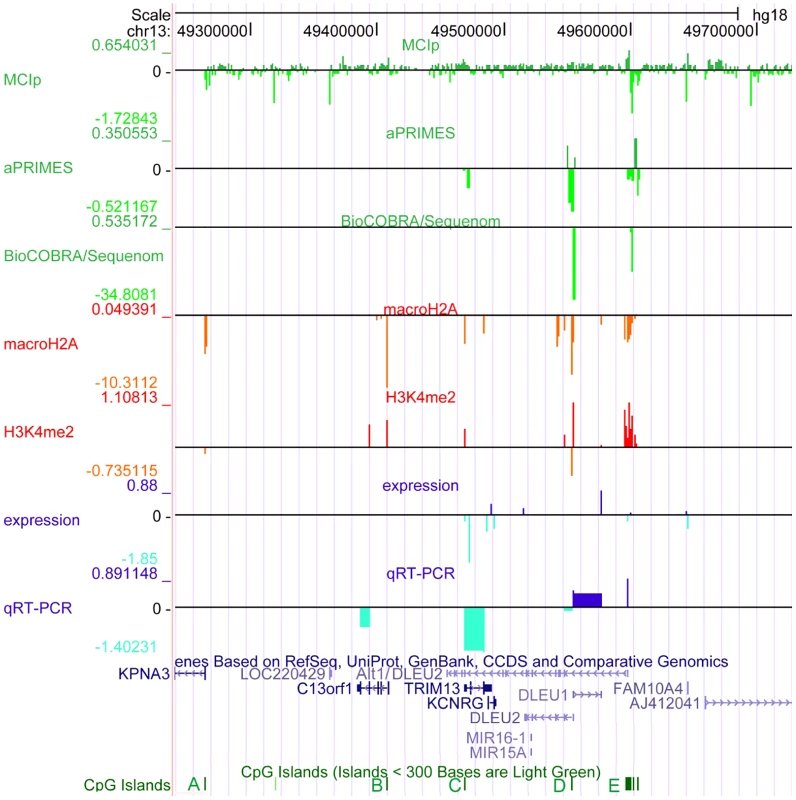
The critical region on 13q14.3 was analyzed for DNA-methylation (lanes 1–3), distribution of histone modifications (lanes 4, 5) and gene expression (lanes 6, 7). DNA-hypomethylation was detected in CLL cells at the transcriptional start sites of the lncRNA genes DLEU1 and DLEU2 variant Alt1. This finding coincides with enrichment of H3K4me2 and depletion of macroH2A, corroborating relaxation of 13q14.3 in CLL. While expression of the protein coding genes and DLEU2 is decreased in CLL cells, lncRNA genes DLEU1 and DLEU2/Alt1 show upregulation, suggesting a direct regulation by DNA-methylation. Base pair positions on top refer to genome GRCh37 build hg18. The conserved CpG islands (A–E) are shown in green, less stringent CpG islands shown in light green. Lane 1: Methyl-CpG Immunoprecipitation (MCIp) allowed semi-quantitative DNA-methylation analysis in non-malignant B-cells sorted from healthy donors (n = 7) and CLL samples (n = 25). Precipitated DNA was hybridized onto a custom tiling microarray covering the 13q14.3 critical region. Depicted is the median log2 fold enrichment of CLL samples from which the median of log2 fold enrichment of healthy donor B-cell samples has been subtracted. Lane 2: Restriction digest-based analysis of DNA-methylation (aPRIMES) was performed in CpG islands C–E at 13q14.3 at 1 kbp resolution. Shown is the median log2 signal intensity of CLL patients from which the median of non-malignant B cell samples has been subtracted. Lane 3: Hypomethylation at D6 and E6 was validated by BioCOBRA and MassARRAY/Sequenome analyses (for details see Figure 2). Shown is the difference of the median percentage of methylation in CLL cells and non-malignant B cells. Lanes 4,5: ChIP was performed for macroH2A (lane 4) and H3K4me2 (lane 5). Precipitated DNA was quantified using qPCR. Enrichment was normalized to non-specific IgG and two control promoters (CDH12, HK2) that displayed similar enrichment for the two histone marks in CLL samples (n = 7) and peripheral blood mononuclear cells (PBMC) from healthy probands (n = 5). Enrichment was calculated as median log2 fold enrichment in precipitate vs. input for CLL samples after subtraction of enrichment from non-malignant B-cells. Lane 6: Gene expression profiling (GEP) was performed using bead chip arrays (Illumina) in CLL patients (n = 25) and sorted B-cells from healthy donors (n = 9). Plotted is the difference of log fold changes of CLL samples and non-malignant B cells. Lane 7: QRT-PCR for gene expression analysis of 13q14.3 candidate genes (for details see Figure 3). It remains unclear how the miRNAs and the other candidate tumor suppressor genes are functionally inactivated in CLL. Sequence mutations in the miRNA genes that lead to aberrant processing from primary transcripts occur only very rarely in CLL [18], [22]–[24]. In addition, the miRNA genes may be more commonly affected by a processing defect (Allegra et al., manuscript submitted). No point mutations have been found in the other candidate genes at 13q14.3 [25]. However, in support of their role as tumor suppressors, the two miRNA genes and the other candidate tumor suppressor genes in the region are downregulated in CLL cells compared to non-malignant B-cells [10], [13], [26], [27].
Thus, epigenetic aberrations play a major role in the pathomechanism of CLL [28]–[30] and not only the genes but also regulatory sequences (e.g. CpG islands) are conserved in the mouse [31]. Accordingly, we have investigated the epigenetic features of the critical region at 13q14.3 in detail to dissect the underlying regulatory network.
Interestingly, two genes in the vicinity of the critical 13q14.3 region are imprinted (RB1, Gene ID: 5925 and HTR2A, Gene ID: 3356) [32], [33]. Parental imprinting is a mechanism where epigenetically regulated lncRNA genes control the expression of genes in cis. Similar to an imprinting mechanism, we recently found a complex epigenetic regulatory mechanism that involves asynchronous replication timing and monoallelic expression in non-malignant B-cells isolated from healthy donors [34], [35]. In addition, the copy of an epigenetic aberration to the homologous chromosome could account in CLL for the observed high incidence of loss of heterozygosity at 13q14.3 without loss of genetic material or the occurrence of mutations [10], [36]. Incomplete inactivation by epigenetic markers could also explain the frequent occurrence of genetic loss of the second copy of 13q14.3 in clonal evolution [34], [37]. In summary, these findings together with the transcriptional deregulation in CLL cells made it very likely that the function of the tumor suppressor mechanism at 13q14.3 is lost through epigenetic aberrations.
We therefore characterized the epigenetic makeup of 13q14.3 in a thoroughly selected cohort of CLL patients (n = 143, see Table S1 for patient characteristics), and found significant DNA-demethylation of two specific sequences within conserved CpG islands at the transcriptional start sites (TSS) of DLEU1 and DLEU2/Alt1 (ENST00000425586). This epigenetic aberration was correlated with transcriptional deregulation of the neighboring candidate tumor suppressor genes. Such a coregulation in cis of several tumor suppressor genes points to a functionally related gene cluster that is involved in the same cellular pathway. In support of this view we found that the 13q14.3 candidate tumor suppressor genes KPNA3 (Gene ID: 3839), RFP2 (Gene ID: 10206) and C13ORF1 (Gene ID: 57213) are positive regulators of NF-kB activity. In addition, we performed an unbiased screen of 810 human miRNAs and showed the miR-15/16 family of genes to be the strongest inducers of NF-kB activity. As one major function of NF-kB in CLL has been shown to be prevention of apoptosis [38]–[40], our findings contrast with the supposed role of the 13q14 genes as tumor suppressor genes. Based on these results it will be tempting to dissect the exact molecular link between 13q14.3 and NF-kB in CLL. In summary, the tumor suppressor mechanism at 13q14.3 is orchestrated by two epigenetically controlled lncRNA genes regulating a cluster of genes that impact on NF-kB.
Results
CLL cells display epigenetic marks at 13q14.3 associated with relaxed chromatin and transcriptional activation
For a comprehensive characterization of the epigenetic make-up of the critical region at 13q14.3 in CLL cells, DNA-methylation of the whole region (from ITM2B Gene ID: 9445 to DLEU7) was quantified in primary patient and healthy proband samples (Table S1 for patient and Table S2 for healthy proband characteristics). In addition, the CpG islands of the candidate genes were analyzed for changes in histone modifications. Applying five different techniques for detection and quantification of DNA-methylation [41]–[45], we found that two regions displayed significantly different DNA-methylation patterns in CLL cells compared to non-malignant B-cells (Figure 1, lanes 1–3; Figures S1, S2). The differentially methylated regions are localized within the CpG islands D and E at the transcriptional start sites of the lncRNA genes DLEU1 (region “D6”) and the DLEU2 variant Alt1, respectively (region “E6”; Figure 1, Figure S1 for validation and S2A, S2B for detail). In a region of chromosomal band 3q25.33 that shows a genetic makeup similar to 13q14.3, no aberrant DNA-methylation could be detected (Figure S2C, S2D). Also no differential DNA-methylation was found in CLL at the retinoblastoma tumor suppressor gene RB1 at 13q14.3 that has been implicated in the pathomechanism of the disease [16], [18], or the DLEU7 gene (Figure S2D, S2E) [46]. To corroborate the finding of a relaxed chromatin conformation in CLL, the CpG islands C, D and E were analyzed for the presence of histone modifications that correlate with open chromatin and active transcription (dimethylation of H3K4, “H3K4me2”) [47] or with epigenetic mechanisms leading to transcriptional inactivation (macroH2A) [48]. In line with a more relaxed chromatin in CLL cells as compared to non-malignant cells, H3K4me2 showed significantly more enrichment, while less chromatin was precipitated that carried the macroH2A modification (Figure 1, lanes 4 and 5). Therefore unexpectedly, active chromatin marks were detected in CLL cells at 13q14.3 instead of repressive epigenetic marks that are characteristic for tumor suppressor inactivation.
Epigenetic aberrations at 13q14.3 are independent of clinical and genetic characteristics and affect 95% of all CLL patients
In order to test whether aberrant DNA-methylation is independent of prognostic and cytogenetic characteristics and thus a unifying feature of CLL, we analyzed a larger cohort of CLL patients (Figure 2, Table S1). DNA-hypomethylation was independent of 13q14.3 gene dosage and was also not a result of the advanced age of the patients (Figure 2B and 2C, compare age-matched controls; characteristics are listed in Table S2; for Mann-Whitney Rank Sum Test see Table S6). Interestingly, DNA-methylation was significantly retained in CLL cells with a deletion of 11q22-q23 covering the ATM (Gene ID: 472) gene, and the most pronounced loss of DNA-methylation was found in patients with a deletion of TP53 (Gene ID: 7157; Figure 2B and 2C). It can be speculated that the DNA-damage repair function of the ATM kinase could be involved in aberrant DNA-demethylation [49] or that a defect in 11q could be epistatic to loss of function of 13q14.3 [50], but this needs to be shown in further analyses. Finally, levels of DNA-methylation were not significantly correlated with mutation status of the immunglobulin heavy chain variable segment genes (IGHV), an important prognostic marker in CLL (Figure S3A) [51], or with overall survival (Figure S3B and S3C), implying that DNA-hypomethylation is present in CLL patients from all prognostic subgroups. In order to complement the single time point analyses of DNA-methylation with assessment of the dynamic changes over time, we analyzed peripheral blood mononuclear cell (PBMC) samples collected from patients at different time points during the course of the disease (Figure S3D). Intriguingly, 4/10 CLL PBMC samples (P7-P10) displayed more DNA-hypomethylation at 13q14.3 than would be expected from the content of CLL cells within the PBMC sample. These findings suggest that DNA-demethylation at 13q14.3 could be an ongoing process in CLL and should also be studied as a marker for imminent disease progression.
Fig. 2. Hypomethylation at the TSS of the large ncRNAs affects the majority of CLL patients independent of 13q gene dosage and IGHV mutation status. 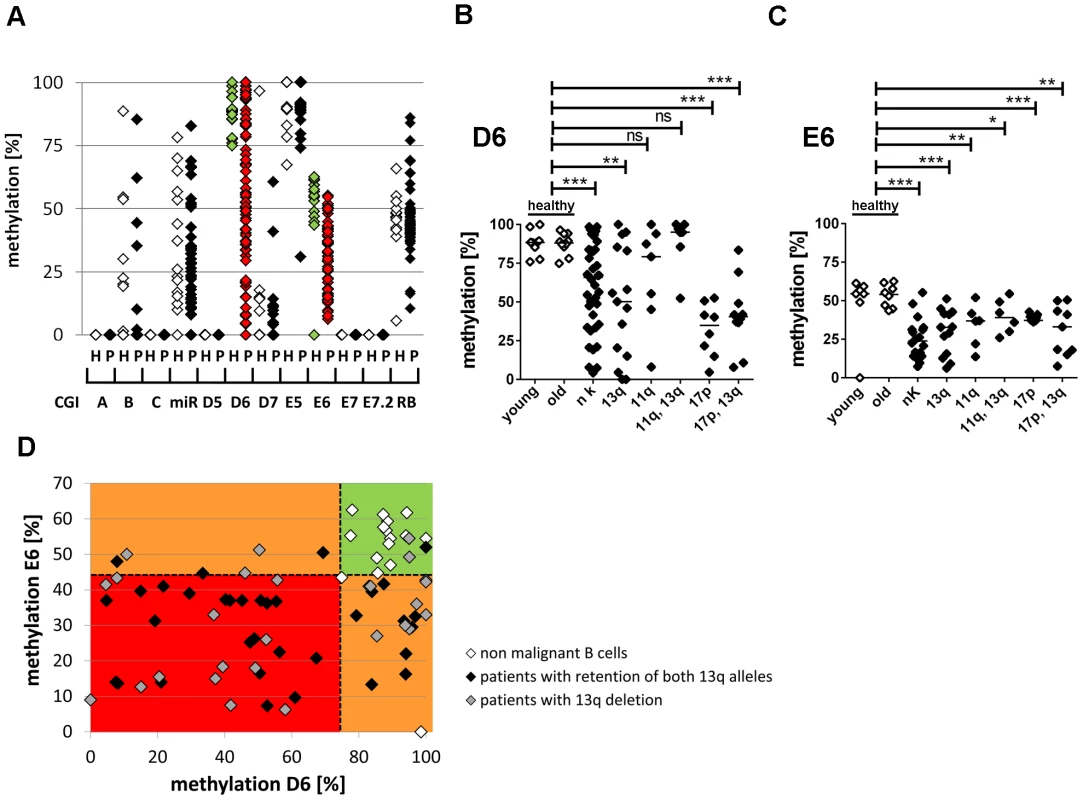
(A) For validation of DNA-hypomethylation at different CpG islands in 13q14.3 (x-axis; for localization see figure 1), quantitative measurement with BioCOBRA and MassARRAY (“Rb”) technology was performed in B-cells of healthy donors (“h”, n = 15–19, white diamonds) and PBMCs of CLL patients (“p”, n = 47–82, black diamonds; “miR” = 308 bp fragment spanning miR-15a; “Rb” = intragenic CpG island). (B, C) CLL patients with different genomic aberrations show different degree of hypomethylation that is distinct from healthy probands younger (“young”) or older (“old”) than 45 years. DNA-methylation was quantified with BioCOBRA (D6) and MassARRAY (E6). Analyzed were patients with normal karyotype (“nk”), deletion of 13q14.3 (“13q”), deletion of 11q22-q23 (“11q”), deletion of 17p (“17p”) or combinations thereof. Statistics were performed using Wilcoxon rank sum test (*** p<0.001, ** p 0.001 to 0.01, * 0.01 to 0.05 and ns not significant p>0.05). (D) DNA-methylation of D6 or E6 is aberrant in 58/61 CLL patients. Black diamonds: Patient samples with two copies of the critical region in 13q, grey diamonds: samples with deletion of one copy of 13q14.3, white diamonds: healthy donor samples. Dashed lines represent lowest levels of methylation of non-malignant B-cells. In summary, DNA-methylation at 13q14.3 was aberrantly lower in 58 of 61 patients (95%) compared to non-malignant B-cells (Figure 2D), proposing that DNA-hypomethylation at 13q14.3 seems to be a universal feature of CLL.
13q14.3 genes are a coregulated gene cluster whose expression correlates with DNA-methylation in CLL cells
Next we investigated the functional impact of the epigenetic aberrations in 13q14.3. As reported previously, the protein-coding and the miRNA candidate tumor suppressor genes (including their host gene DLEU2; [11]) in the critical region are downregulated in CLL cells (Figure 3A and 3B) [26]. In contrast, the lncRNA genes DLEU1 and variant DLEU2/Alt1 that display DNA-hypomethylation at their 5′ ends are significantly upregulated in CLL cells (Figure 3C). To exclude bias caused by the influence of more complex interrelations e.g. by deletion of the critical region, we focused on samples with retention of both copies of 13q14.3. We found a significant inverse correlation of gene expression of the lncRNA genes DLEU1 and the DLEU2 variant Alt1 [52] with DNA-methylation levels in regions D6 and E6 that are localized at their transcriptional start sites. The Pearson correlation coefficient for DLEU2/Alt1b with D6 was R = −0.33 (p = 0.022) and for DLEU1 with E6 the coefficient R = −0.28 (p = 0.044; see Figure 3E, for correlation coefficients see panels F, G). This suggests the direct regulation of DLEU1 and DLEU2/Alt1 by DNA-methylation. In contrast, expression of the protein-coding genes in the region and the miR-15a/-16-1 host gene DLEU2 were positively correlated with DNA-methylation levels (Figure 3D; correlation coefficients F, G), suggesting an indirect regulation by DNA-demethylation e.g. via the lncRNA genes. Levels of mature miR-15a and miR-16 showed no significant correlation with DNA-methylation levels, probably because they are subject to additional posttranscriptional deregulation (Allegra et al., manuscript submitted). Differences in DNA-methylation supposedly reflects differential binding of transcription factors, and we comparatively analysed the sequences at D6 and E6 for binding motifs of transcription factors by comparing it to the TRANSFAC database using PATCH (PatchTM public 1.0, http://www.gene-regulation.com/cgi-bin/pub/programs/patch/bin/patch.cgi). Intriguingly, a number of transcription factor binding motifs are present both in the D6 and E6 sequence, further suggesting that these sequences might be regulated by similar pathways (Table S3).
Fig. 3. Downregulation of 13q14.3 candidate tumor suppressor genes and upregulation of lncRNA genes DLEU1 and DLEU2/Alt1 correlates with DNA-methylation. 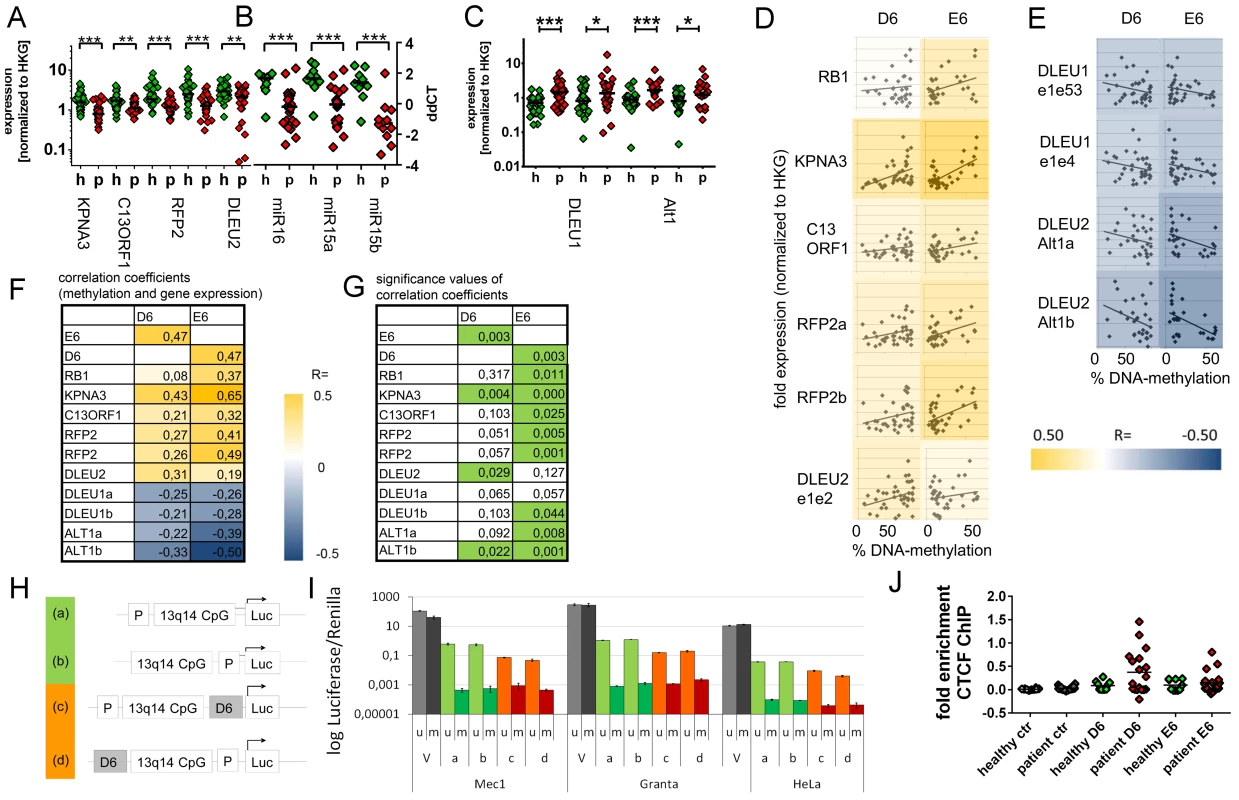
(A–C) Expression of the protein-coding genes KPNA3, C13ORF1 and RFP2, the miRNA genes miR-15a and miR-16-1 and the lncRNA transcripts from genes DLEU1, DLEU2 and alternative transcript Alt1 were quantified with qRT-PCR in CLL cells from patients with retention of both copies of 13q14.3 (“p”, n = 34; see Table S1) and compared to B-cells from healthy donors (“h”, n = 20). Mean expression is indicated by a black horizontal line. “HKG” = housekeeping genes, average of PGK, LMNB1 and PPIA. (D, E) DNA-methylation levels correlate with transcript levels of genes in 13q14.3: While expression (y-axis) of candidate tumor suppressor genes is directly correlated to DNA-methylation of regions D6 and E6 (x-axis; left panel), lncRNA genes are anti-correlated (right panel). Pearson correlation coefficients are color-coded, blue = negative, yellow = positive correlation. (F) Pearson coefficients of correlation of gene expression (rows) and DNA-methylation at D6 and E6 (columns), colour coded (see legend). DLEU1a = exon1 to exon4; DLEU1b = exon 1 to exon 2; ALT1a/b = exon1, two different amplicons (G) Significance values of Pearson correlation coefficients (t-distribution), values p<0.05 are coded green. (H) The bidirectional promoter of DLEU1 and DLEU2 and the flanking CpG island was cloned into pCpGL luciferase vector, either including D6 (orange) or excluding D6 (green). (I) Constructs depicted in (H) were either methylated in-vitro using SSsI methylase (m, dark bars) or left unmethylated (u, light bars) and subsequently transfected into Mec1, Granta519 and HeLa cells and luciferase activity quantified. Grey: Luciferase CMV expression vector (V) not containing CpGs was used as negative control, green: excluding D6 region, orange: including D6 region. Error bars signify standard deviation of 2 independent experiments, each performed in duplicate. No changes were detected for E6 (Figure S4G–S4J). (J) Binding of the DNA-methylation sensitive chromatin reader CTCF was enriched in CLL samples both in the D6 and E6 regions compared to non-malignant B-cells. CTCF-bound chromatin was immunoprecipitated and quantitated with qPCR. Specific binding was shown by quantification of a sequence not bound by CTCF 2 kb upstream of the DM1 insulator (“ctr”). Statistics were performed using Wilcoxon rank sum test (*** p<0.001, ** p 0.001 to 0.01, * 0.01 to 0.05 and ns not significant p>0.05). DNA-demethylating agent 5-aza-2′-deoxycytidine upregulates 13q14.3 genes in cultured cells
In order to understand how DNA-demethylation of D6 and E6 would impact on transcriptional deregulation of 13q14.3, we first tested 16 cell lines for presence of DNA-methylation at 13q14.3 (Figure S4A, S4B) and whether DNA-demethylation results in transcriptional deregulation of 13q14.3 candidate genes. Only Jurkat cells showed DNA-methylation in both loci and could be DNA-demethylated both at D6 or E6 (Figure S4A–S4F). Downregulation of the protein-coding candidate genes detected in CLL cells could not be reproduced in Jurkat cells in-vitro, probably because either the cellular system (T-cells) or the treatment does not faithfully reproduce the complex in-vivo situation (Figure S4F). Interestingly, the levels of mature miR-15a and miR-16 also remained unchanged, which is in line with a recent report where incubation of CLL cells with a inhibitor of histone deacetylases (HDACi) led to upregulation of miR-15a and miR-16-1 in only 35% of patient samples [53]. These findings suggest that the miRNA genes are regulated at the post-transcriptional level in the majority of CLL patients (Allegra, manuscript submitted). However, as expected we could show that both lncRNA genes DLEU1 and DLEU2/Alt1 were upregulated in Jurkat cells upon DNA-demethylation in-vitro (Figure S4F), underlining that their transcriptional activity depends on levels of DNA-methylation.
DNA-demethylation regulates lncRNA expression in vitro and correlates with CTCF binding in a subset of CLL patients
In order to test the functional relevance of the hypomethylated DNA-sequences, their impact on the expression of luciferase reporter constructs was quantified. The two sequence elements D6 and E6 completely lost their capacity to activate transcription when they were in-vitro DNA-methylated (Figure 3H and 3I, Figure S4G–S4J), which is in line with an upregulation of the lncRNA genes in CLL cells upon DNA-demethylation. In addition, inclusion of the non-methylated D6 sequence led to a transcriptional inhibition of the reporter construct in all three cell lines analyzed, suggesting that transcriptional inhibitors might bind to the sequence element (Figure 3H and 3I). In contrast, no changes in transcription were found when the E6 element was included (Figure S4G–S4J), suggesting that either no transcription factors would bind to that sequence or that the reporter system did not faithfully reproduce the in-vivo situation. Both could be the case if E6 would represent an element of higher order chromatin e.g. a boundary element. Such an element could be bound by CTCF protein (Gene ID: 10664), which insulates active chromatin from heterochromatic gene deserts [30] reminiscent of the region distal to DLEU1 that is gene-poor. In addition, CTCF has a central role in transcriptional control exerted by ncRNA genes in cis, probably by segregating regulatory elements like enhancers and promoters [54], and its binding to DNA is sensitive to DNA-methylation [55]. In addition, CTCF binding sites were predicted to be localized close to or within D6 and E6 (Figure S4K). We therefore tested CTCF binding at 13q14.3 using ChIP-qPCR, and in fact CTCF binds to E6 and D6 in a subset of CLL cells but not in sorted B-cells from healthy donors (Figure 3J). Therefore, CTCF is a candidate for modulating transcription at 13q14.3 in cis in a subset of CLL cells.
13q14.3 lncRNA genes do not bind to chromatin
In order to further delineate the regulatory mechanism of the lncRNA genes DLEU1 and DLEU2/Alt1, we tested whether they exert their function by binding to chromatin. As expression levels of DLEU1 and DLEU2/Alt1 were too low for direct visualization of the lncRNA transcripts using RNA-FISH, we used RNA-seq of RNA bound to chromatin [56] that was isolated from murine embryonic stem cells, HeLa and U2OS cells. However, compared to the other genes localized in the critical region, no significant enrichment of DLEU1 or DLEU2 transcripts was found to be bound to chromatin (Figure S5). It is therefore unlikely that DLEU1 or DLEU2 exert their function by binding to chromatin, but rather regulate the neighboring cluster of candidate tumor suppressor genes by divergent transcription (see Discussion). This coregulation of the 13q14.3 genes implies that they are also functionally related, e.g. that the respective gene products are involved in similar cellular processes. To understand which common pathway is targeted by the 13q14.3 candidate genes, we analyzed their gene function.
The epigenetically coregulated 13q14.3 genes form a functional cluster of interacting genes that modulate NF-kB signalling
For most of the 13q14.3 candidate genes, the associated molecular function remains unclear. Examples are miR-15a and miR-16-1, for which a role in regulation of the cell cycle has been shown [16], [57]–[59]. Interestingly, for these miRNA genes and for several additional gene products at 13q14.3, an involvement in the NF-kB pathway has been postulated: miR-15a and miR-16-1 (inducing NF-kB) [60] and DLEU7 (repressing NF-kB) [61] modulate this central signalling pathway. For KPNA3, whose loss leads to an expansion of hemocytes in Drosophila [62], binding of the NF-kB DNA-binding subunit p65/RELA (Gene ID: 5970) has been reported, suggesting a NF-kB inductive role [63]. Because of this suggestive functional link of the 13q14.3 gene cluster, our further experiments focused on their involvement on NF-kB signalling.
MiR-15/miR-16 gene family is the strongest inducer of NF-kB in the miRNome
First we tested whether miR-15a and miR-16 modulate NF-kB with an unbiased whole genome miRNA (miRNome) screen and measured NF-kB activity with a luciferase reporter assay [64]. Of 810 miR-mimics transduced into HEK293 cells, the miR-15a/miR-16 family (miR-15a, miR-15b, miR-16, miR195, miR424, miR497) showed the strongest induction of NF-kB of all tested miRNA families (Figure 4A). Compared to a non-specific control miRNA, transfection of miR-15a and miR-16 miRmics into HEK293 cells significantly enhanced the induction of NF-kB by TNFalpha (Figure 4B). In line with this finding, NF-kB target genes like IL6, IL8, CXCL1 and TNFalpha were induced in three different cell lines derived from embryonic kidney and breast cancer, albeit with different induction patterns (Figure 4C–4E), suggesting that the modulation of NF-kB by the miR-15/16 miRNA family can occur in different tissues. Thus, in addition to their previously reported role in regulation of cell-cycle associated genes [16], [58], [65], the miR-15/-16 family of genes is capable of inducing NF-kB. As activation of NF-kB has been shown in CLL cells to prevent apoptosis [38]–[40], an inducive effect of miR-15/-16 of this pathway is difficult to reconcile with their tumorsuppressive role at least in the tissue analysed here. Therefore to validate an involvement of miR-15/-16 in NF-kB signalling we sought to identify target genes that modulate NF-kB in addition to the previously reported target genes that are associated with cell cycle progression.
Fig. 4. MiR-15/miR-16 family is the strongest inducer of NF-kB. 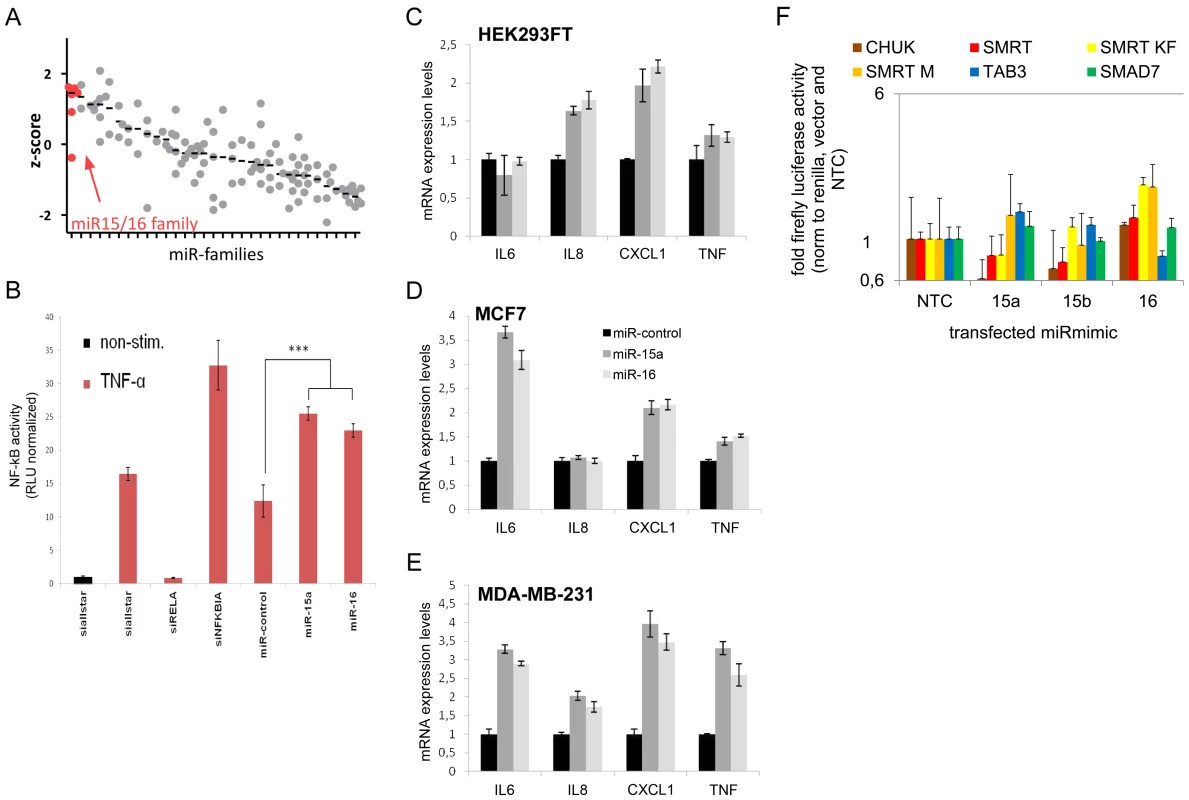
(A) NF-kB induction by miRNA families (see Table S5 for complete list) was measured using NF-kB luciferase reporter assay. Median z-scores are indicated by black horizontal lines. Each dot represents one miRNA-mimic. (B) Transfection of miRNA-mimics miR-15a and miR-16 induces NF-kB activity as compared to unspecific control siRNA. Error bars represent standard deviation of triplicate measurements. (C–E) Upon transfection of miRNA mimic miR-15a and miR-16, expression of NF-kB target genes measured by qRT-PCR is induced in HEK293FT, MCF7 and MDA-MB-231 cells. (F) 3′UTRs of genes involved in the modulation of NF-kB activity and shown to be miR-15a and/or miR-16 targets were cloned into luciferase reporter plasmids. After co-transfection with miR-mimics into HEK cells, firefly luciferase activity was measured and normalized to renilla luciferase activity and empty luciferase plasmid as transfection and background control, respectively, and to no-template control (“NTC”). Data is depicted on a logarithmic scale to balance lower and higher activity compared to control, error bars show standard error of the mean of three independent experiments. miR-15/miR-16 family represses genes that modulate NF-kB activity
The miR-15/miR-16 family of miRNAs has been reported to target several genes involved in NF-kB signalling: IKKa/CHUK, the NF-kB activating kinase itself (Gene ID: 1142) [66], TAB3 (Gene ID:257397), an adaptor protein connecting TRAF6 with the NF-kB activating kinase TAK1 [60], and the transcriptional coregulator NCOR2/SMRT (Gene ID: 9612) [67]. As a control we included SMAD7 (Gene ID: 4092) that is a predicted target of miR-15a (TargetScan6.2 algorithm), and a negative modulator of NF-kB activity [68] but has not been validated as a target so far. In order to delineate the molecular mode of induction of NF-kB activity by miR-15a, miR-15b and miR-16, the respective miR-mimics were cotransfected with luciferase reporter constructs containing 3′UTRs or parts of the 3′UTRs of the candidate target genes into HEK293T cells. While constructs containing 3′UTRs of genes previously reported to be targets of miR-15a and/or miR-16 (CHUK/IKKa, SMRT and TAB3) showed lower luciferase activity after miRmimics-15a/-16 transfection, luciferase activity from the control reporter SMAD7 selected using in-silico prediction remained constant (Figure 4F). Thus we reproduced previously reported findings on gene targets of the miR-15/miR-16 family that modulate NF-kB transcription factor activity either directly (NCOR2/SMRT) or via upstream kinases (IKKa/CHUK) or upstream adaptor proteins (TAB3). The strong induction of NF-kB by the miR-15/miR-16 family in our screen however suggests that additional genes are targeted by these miRNAs that are part of the NF-kB circuitry.
KPNA3 modulates and RFP2 induces NF-kB in HEK293 and primary CLL cells
We confirmed that knockdown of KPNA3 located in 13q14.3 and the family member KPNA4 (Gene ID: 3840) located in 3q25.33 (Figure S2D) leads to a loss of inducibility of NF-kB activity by TNFalpha (Figure 5A). However, double knockdown of both genes did not lead to a full loss of NF-kB induction. In addition we analysed whether C13ORF1, RFP2 and its bicistronic ORF KCNRG (Gene ID: 283518), the protein-coding genes closest to or included in the minimally deleted region, are also involved in NF-kB signalling. To this end we knocked down candidate genes from the minimally deleted region and induced NF-kB with TNFalpha (Figure 5B). Even though TNFalpha activates NF-kB via several pathways, knocking down RFP2 and, depending on the NF-kB recognition sequence used, also KCNRG and C13ORF1 led to a decrease in the activation of NF-kB (Figure 5B, Figure S6A). To further validate the role of RFP2 in NF-kB signalling, we exogenously overexpressed RFP2 in HEK293-T, HEK293, controlled overexpression of RFP2 with a specific antibody we raised in guinea pig against recombinant full length RFP2 and quantified NF-kB activity with a luciferase reporter assay (Figure 5C; Figure S6B). An induction of NF-kB was also observed when RFP2 was overexpressed in primary CLL cells (Figure 5D). As overexpression of recombinant proteins may lead to artificial activation of NF-kB signalling, we separately overexpressed recombinant GFP as a negative control using 50 fold more plasmid than RFP2 expression plasmid and did not observe activation of NF-kB (Figure S6C), underlining that the effect of RFP2 is specific.
Fig. 5. 13q14.3 candidate genes are a functionally related gene cluster that modulates NF-kB signalling. 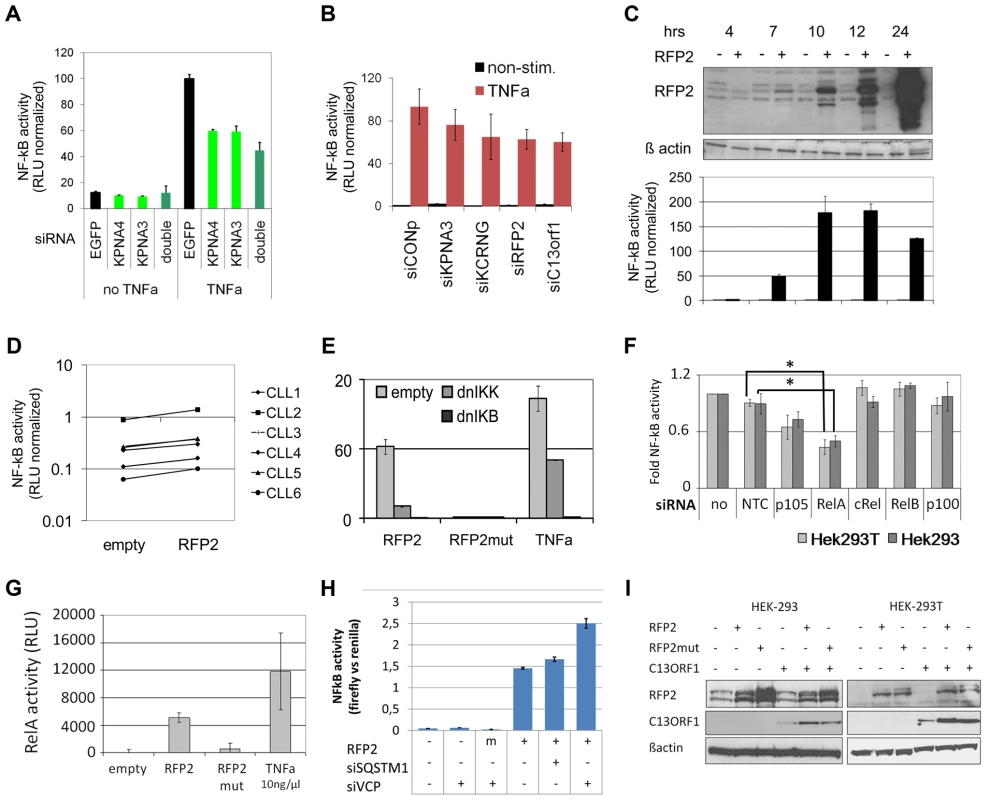
(A) Knockdown of KPNA3 and KPNA4 (x-axis) resulted in reduced stimulation of NF-kB by TNFa in U2OS cells. NF-kB activity was measured after 24 hrs with a reporter driving luciferase expression under the control of 5 canonical NF-kB recognition sites. SiRNA directed against EGFP was used as negative control. Standard deviation of duplicate measurements is shown. (B) KPNA3, KCNRG, RFP2 and C13ORF1 were knocked down in HEK293T cells and activity of NF-kB was measured. As negative control, siRNA without a physiological target was used (siCONP). Error bars signify SEM of 3 independent experiments. (C) RFP2 induces NF-kB activity in HEK293T and HEK293 (Figure S6B) cells. HEK293T cells were transiently transfected with CMV RFP2 expression plasmids or empty vector and NF-kB activity measured after 4, 7, 10, 12 and 24 hrs (bottom panel). The top panel is a representative Western blot of two experiments, error bars in bottom panel represent standard deviation of triplicate measurements. (D) RFP2 induces NF-kB activity in primary CLL cells (n = 6 patients; CLL3 is beneath CLL5). Experimental setup as in (C). (E) Induction of NF-kB by RFP2 can be inhibited with dominant-negative IkB kinase (dnIKK) and dominant-negative IkB (dnIKB). HEK293T cells were transfected with expression vector encoding wt-RFP2 or mutant RFP2 in combination with dnIKK or dnIkB or empty vector, and activity of NF-kB was measured with a luciferase reporter. Inhibition of NF-kB activity by dnIKK and dnIkB was controlled by stimulation with TNFa 6 hrs after transfection and quantification of NF-kB activity after 24 hrs. (F) RFP2 induces NF-kB activity via p65 and p105. p105, RELA (p65), cREL, RELB and p100 were knocked down in HEK293 and HEK293T cells that were transfected with RFP2 expression plasmid. NF-kB activity was measured with luciferase reporter assay. *p<0.05 with students t-test. (G) RFP2 induces DNA-binding of RELA. HEK293T cells were transfected with empty vector (“empty”), expression plasmid containing wildtype RFP2 (“RFP2”), RFP2 with mutated ubiquitin ligase activity (“RFP2mut”) or were stimulated with TNFalpha (“TNFa”). After 24 hrs, DNA-binding capacity of RELA was quantified by co-ELISA. Error bars represent standard deviation of 2 independent experiments. (H) Knockdown of VCP but not SQSTM1 modulates activation of NF-KB by RFP2. SQSTM1 and VCP were knocked down and plasmids for overexpression of RFP2 and luciferase reporters detecting NF-kB activity were transfected after 24 hrs into HEK-293. While only a minor change could be observed after knockdown of SQSTM1, knockdown of VCP led to a substantial increase in the activation of NF-kB after cotransfection of RFP2. (I) Cotransfection of RFP2 leads to stabilization of C13ORF1 protein. HEK-293 and HEK-293T cells were transfected with wildtype RFP2 or mutated RFP2 lacking ubiquitin ligase activity alone or in combination with C13ORF1. Protein levels of RFP2 and C13ORF1 were monitored by Western blot. β-actin served as loading control. Experiment is representative of two independent experiments. RFP2 activates NF-kB subunits RelA and p105 via its ubiquitin ligase activity
These findings raise the question how RFP2 activates NF-kB. NF-kB activation is only induced 7hrs after transfection of the RFP2 expression plasmid, which is the same time point when the exogenous RFP2 protein can first be detected (Figure 5C, Figure S6B). This suggests a direct effect by overexpression of RFP2 and excludes transcriptional induction of other factors, which would require at least an additional 1–2 hrs. The activation of NF-kB by RFP2 could be blocked by dominant negative (dn) IKK and dnIkB [69], suggesting that the effect of RFP2 takes place upstream of these factors (Figure 5E). In addition, loss of the ubiquitin-ligase activity of RFP2 by mutating C13A [70] completely abrogated the activation of NF-kB by RFP2 (Figure 5E). In order to identify the NF-kB component that is targeted by RFP2 activation, all DNA-binding components of the NF-kB signalling pathway were knocked down individually. Downregulation of RELA and to lesser extent of p105 (Gene ID: 4790) reduced the activation of NF-kB by RFP2 (Figure 5F). Corroborating this finding in a custom oligonucleotide-coupled ELISA (co-ELISA) [71], RFP2 specifically induced the activity of RELA (Figure 5G).
Activation of NF-kB by RFP2 is modulated by VCP
It has recently been shown that the RFP2 protein interacts with VCP, ATP2A2/SERCA2 [70] and SQSTM1 [72]. We therefore asked whether these proteins would be involved in the modulation of NF-kB activity by RFP2. However, knockdown of ATP2A2 and SQSTM1 did not result in enhanced activity of NF-kB (Figure 5H; Figure S6D). In contrast, knockdown of VCP substantially increased the activation of NF-kB by RFP2 (Figure 5H and Figure S6D). This finding is intriguing as VCP and SQSTM1 link RFP2 not only to endoplasmatic reticulum associated protein degradation (ERAD) and autophagy [70], [72], but also to regulation of TRAF6 [73], [74] that is involved in signalling pathways such as CD40/CD40L and TLR that are central to the pathogenesis of CLL.
RFP2 overexpression stabilizes C13ORF1 protein
RFP2 is a member of the family of tripartite motif proteins (TRIM) but lacks the SPRY domain (pfam00622) common to other TRIM proteins [75]. Intriguingly, the neighboring C13ORF1 gene has a SPRY domain, suggesting a functional interaction of RFP2 and C13ORF1. While knockdown of C13ORF1 led to a reduction in NF-kB inducibility (Figure 5B), co-expression of RFP2 and C13ORF1 did not lead to a synergistic induction of NF-kB activity (Figure S6D). As RFP2 is an integral membrane protein [70] and requires disruptive RIPA buffer extraction for analysis, physical interaction with C13ORF1 could not be shown by pulldown experiments. However, support for the interaction of RFP2 and C13ORF1 proteins came from the observation that coexpression of RFP2 stabilized expression of C13ORF1 (Figure 5I), even though RFP2 has a destabilizing (auto-)ubiquitin ligase activity [70].
Discussion
The functional gene cluster at 13q14.3 is transcriptionally deregulated by hypomethylation in malignant cells
Several lines of evidence suggest that the tumor suppressor function of 13q14.3 distal to RB1 is multigenic [16], [20] and is not inactivated by mutation [25], but rather by transcriptional deregulation [10], [13], [26], [27]. In line with this notion we found DNA-demethylation at the 5′ends of the lncRNA genes DLEU1 and DLEU2 in more than 95% of CLL patients (Figure 2D, Figure 6A). Hypomethylation in cancer cells usually coincides with chromosomal instability, which we cannot exclude for 13q14.3, or with reexpression of silenced oncogenes. In fact, a genome-wide DNA-demethylation has been observed for CLL cells [76], but the functional consequence is unclear. At 13q14.3, DNA-methylation levels comparable to non-malignant B-cells were observed at all tested loci except for the elements D6 and E6. DNA-demethylation of the regions D6 and E6 in CLL cells is directly correlated with an increase in the expression of DLEU1 and DLEU2 and inversely correlated with the expression of the neighboring candidate tumor suppressing protein-coding genes (Figure 6B). The expression of antisense transcripts is usually lower and is not necessarily coupled to expression of the respective sense transcripts [77], which is what we observed at 13q14.3 for the lncRNA genes DLEU1 and DLEU2 and the protein-coding genes.
Fig. 6. Model of the tumor suppressor mechanism localized in 13q14.3. 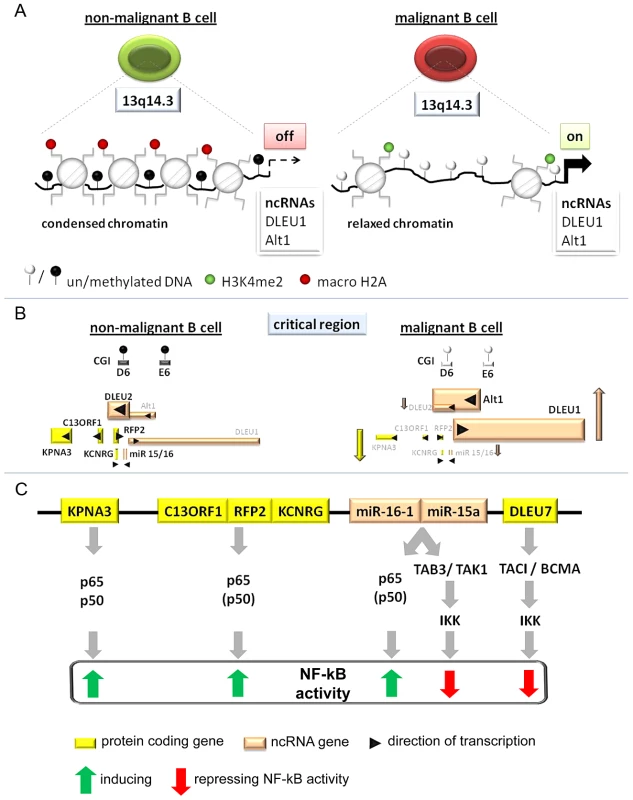
(A) Regions in CpG islands D and E that are DNA-methylated in non-malignant B-cells (left) become demethylated in the vast majority of CLL patients (right). This coincides with relaxed chromatin characterized by absence of macroH2A and enrichment of H3K4me2 at the promoters of the lncRNA genes DLEU1 and DLEU2/Alt1. (B) DNA-hypomethylation is correlated with transcriptional upregulation of splicing variants of the two lncRNA genes DLEU1 and DLEU2 and inversely correlated with the protein-coding genes in 13q14.3. No correlation could be found with levels of mature miR-15a and miR-16, probably because these transcripts are also deregulated by a posttranscriptional processing defect in CLL cells (Allegra, manuscript submitted). (C) Candidate genes localized in the critical region in 13q14.3 are functionally related and all modulate NF-kB signalling, albeit with different impact. Nuclear transporter KPNA3 is binding NF-kB components p65 and p50 and is therefore likely a positive regulator like e.g. RFP2. RFP2 binds to C13ORF1 and induces NF-kB activity via the canonical pathway components p50 and p65, for which its ubiquitin ligase actvity is required. The miRNA genes miR-15a and miR-16-1 were identified together with other members of this miR family to be among the strongest activators of NF-kB activity. Previously they have been shown to both modulate cell cycle regulators and inhibit NF-kB via TAB3/TAK1. DLEU7 has recently also been reported to inhibit NF-kB by binding and inactivating the TACI/BCMA receptors. As for the cis-regulatory mode of action of lncRNA genes, a direct RNA-DNA interaction has been shown to recruit repressors, which leads to changes in chromatin conformation [4], [78]. However, this option seems unlikely as for none of the transcriptional units at 13q14.3 we found substantial enrichment in the chromatin-bound RNA fraction.
A second mode of action could be competition by (i) “divergent transcription” of the ncRNA genes that recruits essential factors away from the candidate tumor suppressor genes [78], (ii) collision of transcription complexes initiated from different promotors (e.g. E6 and the DLEU2 promotor) but transcribing through the same sequences, or (iii) elongation through transcriptional regulators like enhancers/repressors that leads to the deposit of specialized epigenetic marks inhibiting transcription from the opposite strand [79]. Interestingly, D6 seems to inherently harbor transcriptional repressive properties. The described mechanisms are only dependent on the initiation of transcription and can be independent of the resulting (antisense) RNA molecule itself. The dispensable role of the DLEU1 transcript itself is also suggested by the lack of conservation of the DLEU1 gene sequence and its multitude of splicing variants [8]. In fact, lncRNAs involved in regulation in cis are in general poorly conserved, probably because these mechanisms are mostly topological [80] and thus sequence independent. A topological regulation is also suggested by the presence of a homologous region on 3q25.33 (Figure S2D) and the conservation of the orientation of the genes and CpG islands in Mus musculus [31]. However, it should be noted that in the mouse there is no overlap of DLEU2 with RFP2, but the sequence similarity of the first exon of RFP2 and exon 11 of DLEU2 is conserved [9]. This suggests at least for this pair of genes the possibility of a RNAi-like regulatory mechanism. Finally, transcription in the region could be regulated by a central locus control region, organizing the intranuclear localization of 13q14.3 e.g. by binding of chromatin organizing proteins like CTCF. While DNA-methylation dependent aberrant binding of CTCF could be observed at D6 and E6 in a patient subset, more advanced experiments (e.g. 3–6C analyses) are required to assess the functional impact of changed binding properties of chromatin organizing factors such as CTCF.
Interestingly, epigenetic deregulation of lncRNA genes leading to aberrant transcription of neighboring genes occurs also in acute leukemia. The lncRNA HOTAIRM1 for example is expressed exclusively in the myeloid lineage and controls expression of the proximal HOXA gene cluster [81]. Similarly, the ncRNA vault RNA2-1 (vtRNA2-1) in the commonly deleted region of chromosome 5q is monoallelically methylated and expressed in healthy individuals, while it is epigenetically inactivated in AML, leading to activation of NF-kB via RNA-binding protein kinase R (PKR, [82]). Another example resembling the molecular mechanism of the 13q14.3 locus is silencing of the tumor suppressor WT1 by the overlapping WT1-antisense lncRNA WT1-AS, which is monoallelically expressed in non-malignant cells and becomes activated in AML by hypomethylation [83]. Thus, epigenetic deregulation of ncRNA genes seems to be a recurrent disease related phenomenon both in chronic and acute leukemias, leading to aberrant function of tumor suppressor - or oncogenes.
The tumor suppressor genes at 13q14.3 are transcriptionally coregulated and modulate NF-kB signalling by either activating or repressing the pathway
The transcriptional activities of the 13q14.3 candidate genes all correlate with the DNA-methylation levels in the region. This co-regulation suggests that 13q14.3 genes are also functionally related, i.e. are involved in the same cellular pathways. Such clusters of genes seem more common in drosophila than in mammalian cells [84]. In human cells, only a subset of ubiquitously expressed genes and a small set of atypical genes is grouped together into coregulated clusters [85]. A reason for evolutionary conservation of a genetic neighbourhood of functionally connected genes is the coregulation of these genes [85]. Examples in the mammalian system are the globin gene family, groups of olfactory receptors, histone-coding genes, HOX genes, genes of the major histocompatibility complex and imprinted genes. In addition, most long non-coding RNA genes are involved in regulating functionally related gene clusters [2]. One major unifying scheme of these gene clusters seems to be the transcriptional activity from the same chromosomal strand [85], which has been shown for 13q14.3 [35]. Similarly, the topological organization of these gene clusters is highly conserved between mammals, which is also true for 13q14.3 [31], [50] and its homologous cluster at 3q25.33. It is therefore very likely that the 13q14.3 candidate genes are also functionally related, and we and others could show that they activate or repress the NF-kB signalling pathway (Figure 6C).
NF-kB signalling is centrally involved in the homeostasis of the hematopoietic system where it is induced in inflammation and inhibits apoptosis [86]. NF-kB signalling has already been shown to be activated in CLL cells [38]–[40], [87], where it is postulated to help in cellular survival [87]. In CLL, NF-kB is activated by the interaction with the microenvironment [39], which is crucial for the survival of CLL cells [51]. NF-kB is also activated via the B-cell receptor (BCR) that plays an important role in the pathogenesis of CLL [51]. Similarly, NF-kB is activated by interaction of TCL1 (Gene ID: 8115) and ATM in CLL [88], two genes that are coregulated in CLL cells [89] and centrally involved in the pathogenesis of CLL. In contrast, in early developmental stages of a CLL-like disease in transgenic mice, repressive p50/p50 NF-kB dimers (Gene ID: 4790) cause epigenetic lesions that even precede genetic lesions [29], suggesting that at different stages of the disease, NF-kB could play different roles.
Genome - and exome-wide analyses of CLL cells have recently shown that mutations are present in genes involved in NF-kB signalling [90], and intriguingly mutations in a NF-kB-pathway associated gene (MYD88, Gene ID: 4615) seem to be even enriched in del(13q) patients [91]. This is most interesting as MYD88 is required for TLR signalling via TRAF6, a protein that is bound by SQSTM1 and VCP [73], which interact with RFP2 [70], [72]. Further studies will be required to understand the molecular interplay of these proteins in full detail and especially to accommodate the unexpected induction of NF-kB activity by several genes localized at 13q14 with their tumorsuppressive function.
After recent reports have shown 13q14.3 genes to be inhibitors of NF-kB signalling [60], [61], here we demonstrate that the miR-15a/16 cluster, KPNA3 (and KPNA4 from 3q25.33) and RFP2 are positively correlated with NF-kB function (Figure 6C): the miRNA15/16 family of genes were among the strongest inducers of NF-kB in an unbiased screen, KPNA3 is the transporter of p65 and RFP2 induces canonical NF-kB signalling. NF-kB activity is normally associated with an inhibition of apoptosis, and in fact has been shown to be induced in CLL cells by pro-survival microenvironmental stimulants like e.g. CD40L, BAFF; stromal cells or B-cell receptor stimulation [38], [87], [92]–[94]. However, there are a few instances where NF-kB activity can also induce apoptosis. The most relevant example is probably the negative selection of T-cells [95], where strong signalling from the T-cell receptor upon recognition of self-antigens induces apoptosis via activation of NF-kB above a certain threshold. While in B-cells negative selection is somewhat dissimilar, loss of negative selection in CLL cells would make sense considering i) the autoreactivity of CLL cells and ii) the importance of consistent BCR signalling induced by self-antigens in the pathogenesis of the disease [96]. Thus, even though within the same cell, NF-kB activity cannot be at the same time silenced and activated, the activity of NF-kB can change during the leukemogenesis of CLL and the role of the tumor suppressor mechanism in 13q14 could be required only at specific timepoints. The speculative involvement of 13q14 genes in negative B-cell selection could explain how deletion of NF-kB-inducing genes at 13q14 would lead to CLL leukemogenesis at an early timepoint, while malignant B-cells from of terminal stage CLL then exhibit increased levels of NF-kB that prevent apoptosis as has been shown previously (see above). In this respect it should be borne in mind that the functional assays quantifying the impact of 13q14 genes on NF-kB signalling performed both by us and by others depend on in-vitro experiments mostly in cell lines and not in primary cells except for RFP2 (Figure 5D). In addition, overexpression and knockdown of 13q14 genes was performed using recombinant constructs. These experimental settings and their results might therefore not properly reflect the physiological situation, especially when looking at such finely tuned systems like NF-kB signalling. However, NF-kB has been shown to be a promising target for therapeutic intervention in CLL cells [97], [98], and further functional experiments and especially in-vivo analyses should be performed to fully understand the mechanistic link between 13q14 and NF-kB in CLL.
Conclusion and outlook
In summary, we uncovered a cluster of functionally related genes that are coregulated by long non-coding RNA genes in cis and are epigenetically deregulated in malignant cells. We previously speculated that the epigenetic deregulation could explain a stepwise inactivation of the tumor suppressor mechanism [34]. This would complement the findings of clonal evolution and/or extent of 13q14 deletion being associated with a more aggressive form of CLL [18], [37], and the presence of pre-malignant stages of the disease (e.g. MBL) [15]. Further work is required to identify transcription factors binding to the demethylated regions and characterization of their intranuclear localization. It will also be of interest to test whether the observed epigenetic aberrations are present already in premalignant cells of mouse models [23], [99] or whether they constitute the aberrations that have been postulated to be present in hematopoietic stem cells of CLL patients [100].
Materials and Methods
Magnetic depletion and isolation of cells
Mononuclear cells were isolated from peripheral blood by density centrifugation using Ficoll (Biochrom AG) according to the manufacturer's instructions. For positive selection of CD19+ B-cells and CLL cells from peripheral blood, mononuclear cells (PBMCs) were labeled with CD19 MACS magnetic MicroBeads and isolated using MACS LS Column placed in the magnetic field of MACS Separator. The purity of the CD19+ fraction was 95%±3% (± SEM) after purification from PBMCs from healthy probands and 97%±2% for purification from PBMCs of CLL patients as measured by flow cytometry (FACSCalibur, BD Biosciences) using anti-CD19 FITC-labeled antibodies (anti-CD19 MicroBeads, Dako) that specifically binds the CD19 epitope.
Ethics statement
Peripheral blood samples were obtained from patients after informed consent by a procedure approved by the Ethics Committee of Ulm University (approval 96/08), and peripheral blood was drawn from fully anonymised age-matched healthy probands at the german red cross (DRK) in accordance with the Declaration of Helsinki.
QPCR (quantitative PCR) analysis and reverse transcription (RT)
Standard 20 µl qPCR reactions contained 10 µl SYBR Green mixture (Absolute QPCR SYBR Green ROX Mix, Thermo Scientific) and primers at 70 nM final concentration. Thermal cycling conditions were 15 minutes at 95°C, 40 cycles of 15 s at 95°C and 30 s at 60°C, dissociation curve 15 s at 95°C, 15 s at 60°C and heated to 95°C (within 20 minutes), held for 15 s and cooled down to 4°Ç using the 7300 Real-Time PCR system (Applied Biosystems). A standard curve, using template dilutions of HeLa and HEK293 cDNA was measured to determine PCR efficiency and allow exact quantification of template. All primers used for qPCR are listed in the Table S4. Reverse transcription of total RNA was carried out using the AffinityScript QPCR cDNA Synthesis Kit (Agilent), a reaction lacking reverse transcriptase (-RT) was included for each template where primers did not span an intron and amplification of product would have been possible from contaminating genomic DNA. For mRNA detection, Ct-values were normalized using dilution standard curves and three housekeeping genes (PGK2, LMNB1, PPIA) or for the miRNA genes using the ddCT method with RNU6B and SNORA73A as internal normalization controls.
MiRNA detection
10 ng of total RNA was reverse transcribed using the miScript Reverse Transcription Kit (QIAGEN) where reactions were scaled down to 10 µl. The completed RT reactions were diluted to 50 µl with DEPC-treated water and PCR amplification for real-time quantitative analysis was performed using the miScript SYBR Green PCR kit (QIAGEN). Total reaction volume of qPCR was 20 µl, and 2 µl of the diluted RT reactions were used as template. For miRNAs custom forward primers were used to final 0.5 µM (sequences see Table S4) and primers for normalization controls RNU6B and SNORA73A were purchased from QIAGEN. The annealing temperature was 55°C.
Cloning of 13q14.3 constructs into pCpGL vector
To study the effect of DNA-methylation in vitro, the regions D6 and E6 were cloned with their physiological promoter into the pCpGL vector (kind gift from Michael Rehli) [101] to investigate their impact on transcription and whether this is dependent on DNA-methylation. The promoters of the large ncRNAs DLEU1/DLEU2 and DLEU2/Alt1 were cloned in both directions with and without the putative regulatory elements D6 and E6. The construct containing D6 was 2 kb in size and the construct lacking D6 was 1.3 kb in size. These products could be amplified from placenta DNA (SIGMA-Aldrich) using the HotStarTaq Plus PCR system (Qiagen; cycling: 95°C: 5 min; 40 cycles of 95°C: 30 sec, 58°C: 30 sec, 72°C: 1/1.5 min; hold 10°C). Constructs containing or not containing E6 were 4000 and 3500 bp in size and amplified with the Expand High Fidelity PCR System (Roche) using PAC 372-3 from 13q14.3 as template [7]. Cycling was performed 95°C 2 min, 10× [95°C 20 s, 60°C 30 s, 68°C 4 min], 20× [95°C 20 s, 60°C 20 s, 68°C 4 min +20 s in each cycle], 68°C 7 min, hold 4°C. The desired constructs were amplified with primers containing BamH1 and SpeI recognition sites (see Table S4) and cloned into the TOPO TA cloning vector (Invitrogen). Plasmids from positive clones were digested with BamH1 and SpeI (NEB) using 3 µg TOPO-plasmid-insert-DNA or 1 µg pCpGL vector for 1 h at 37°C. The insert was isolated on a 1% agerose gel (50 min, 150 V) and extracted using QIAEX II Kit (Qiagen). For sticky end ligation a 3 times molar excess of insert over pCpGL vector backbone was used in a ligation reaction with 0.1u T4 ligase (Invitrogen) incubating 1 h at 37°C or at 16°C over night.
The ligation reaction was purified by ethanol precipitation, resuspended in 5 µl water and 1 µl was used to transform competent PIR1 E.coli cells (Invitrogen) bacterial cells via electroporation (Gene Pulser II, BIO-RAD). After electroporation in a 2 mm cuvette at 25 µF and 2.5 kV setting the pulser at 200×, transforming 50 ng DNA within a total volume of 400 µl, cells were plated on zeocin containing plates and incubated at 37°C overnight. For transient transfections, plasmids were isolated and purified using the EndoFree Plasmid Kit (Qiagen). In vitro methylation was performed using SssI methylase (NEB) according to manufacturer's instructions but incubating for 4 h at 37°C and adding 1 µl fresh SAM after 2 h.
Cloning of 3′UTRs of miR target genes into pMIR-Report plasmid
To measure the impact of miR-15a, miR-15b and miR-16 on potential target genes, parts of or the whole 3′UTRs of TAB3, CHUK, SMAD7 and SMRT were cloned into the vector pMIR-Report (Applied Biosystems). Sequences containing the miR target sites in the 3′UTRs of TAB3, CHUK, SMRT “KF” and SMAD7 were amplified from HEK293T genomic DNA using the corresponding primers (containing restriction sites for HindIII, SpeI or SacI; see Table S4) and the PRECISOR high-fidelity DNA polymerase (BioCat) according to the manufacturers instructions. Amplified products were purified using the PCR Purification Kit from Qiagen, digested with HindIII, SpeI or SacI (FastDigest Enzymes, Fermentas) and ligated with pMIR-Report (T4 DNA Ligase, Fermentas). Plasmid backbone had been digested with the respective enzymes and purified via agarose gel extraction (Qiagen). Reporters containing just the miR target site and the respective mutated sequence (“SMRT” and “SMRTmut”) were cloned as described previously [67].
Bisulfite conversion
1 µg of genomic DNA was converted using EpiTect 96 Bisulfite Kit or EpiTect Bisulfite Kit (Qiagen) in a GeneAmp PCR System 2700 (Applied Biosystems) with a reaction volume of 100 µl. After desulphonation, converted DNA was eluted 2 times in 20 µl prewarmed (65°C) water. Bisulfite conversion was performed on dilution series (different degree of methylation) of placenta DNA (SIGMA-Aldrich), DNA from CLL patients and from B cells of healthy individuals for quantitative methylation analysis by BioCOBRA or massARRAY as well as bisulfite sequencing.
BioCOBRA
For BioCOBRA analysis (combined bisulfite restriction analysis with the Agilent 2100 Bioanalyzer platform, [102], bisulfite converted DNA was amplified using primers specific for converted template (see Table S4). After purification of the PCR products using Rapid PCR Purification System (Marligen Biosciences), products were digested with BstUI (NEB) over night at 60°C. Fragments were subsequently analysed with DNA 1000 LabChip (Agilent) on the Agilent 2100 Bioanalyzer. For all amplicons a calibration curve was measured with defined mixtures of methylated and unmethylated DNA (Figure S1A). Fully unmethylated DNA was generated by whole genome amplification (REPLI-g Kit, Qiagen), and after purification (QIAamp DNA Mini Kit, Qiagen), half of the amplified and purified DNA was in vitro methylated using SssI methylase (NEB).
MassARRAY
Mass-spectrometric methylation analysis was performed using MassARRAY (Sequenom) analysis according to [42] for the potential regulatory element E6 within 13q14.3, because lack of a BstUI recognition site precluded BioCOBRA. The target gene regions were amplified by PCR (see Table S4) after sodium-bisulfite conversion of template DNA using primers specific for converted template. In this amplification, reverse PCR primers were tagged with the T7 recognition sequence for reverse transcription. Deoxynucleotides in the PCR reaction were dephosphorylated using shrimp alkaline phosphatase (SAP) at 37°C for 20 min followed by 5 min heat inactivation of SAP at 85°C. Making use of the T7 recognition sequence, a single-stranded RNA copy of the template was generated by in vitro transcription. The produced RNA was cleaved specifically at Uracil by RNase A. The cleavage products were analyzed using matrix-assisted laser desorption ionization – time of flight (MALDI-TOF) mass spectrometry in a final elution volume of 27 µl. Cleavage product signals with a 16 Da shift (or a multiple thereof) represent methylation events; signal intensity was correlated with the degree of DNA-methylation.
Bisulfite sequencing
Bisulfite converted DNA (EpiTect Bisulfite Kit, Qiagen) was amplified using primers specific for converted DNA. After PCR purification (Rapid PCR Purification System, Marligen Biosciences) the product was cloned into pCR2.1-TOPO vector and subsequently transformed into One Shot Mach1-T1 competent E. coli cells (Invitrogen). Positive clones were selected by colony PCR using M13 primers; cycling: 95°C: 12 min; 40 cycles of 95°C: 30 sec, 55°C: 30 sec, 72°C: 1 min; 72°C: 7 min; hold 10°C. PCR products of the expected size were purified (Marligen Biosciences) and sequenced (BigDye Terminator v3.1 Cycle Sequencing Kit, Applied Biosystems) using M13 forward primer with the ABI Prism 3100 Genetic Analyzer 3130xl (Applied Biosystems). Cycling: 96°C: 1 min; 25 cycles of 96°C: 10 sec, 52°C: 5 sec, 60°C: 2 min; hold 10°C. The sequencing reactions were purified with the DyeEx 96 Kit or DyeEx 2.0 Spin Kit (Qiagen) to remove non-incorporated nucleotides. Analysis of sequences was performed using MethTools (http://genome.imb-jena.de/methtools/).
Array-based profiling of reference-independent methylation status (aPRIMES)
aPRIMES was performed according to [45] using 500 ng genomic DNA that was digested using 10 U MseI (NEB) for 3 h. The MseI-fragments were then subjected to linker mediated PCR using primer ddMse11 and primer Lib1 at an initial annealing temperature of 65°C that was shifted down to 15°C with a ramp of 18°C/min (MWG, Ebersberg, Germany) and ligation using T4-DNA-Ligase (10 U, Roche) was performed at 15°C overnight. Half of the resulting ligated MseI fragments were digested with the restriction enzyme McrBC (NEB) for 8 h and the other half of was digested with two methylation-sensitive endonucleases, HpaII and BstUI 3 h each. Proteinase K (Invitrogen) was used for digestion before amplification using Expand Long Template system (Boehringer) and Lib1 primer in a MWG thermo cycler; cycling: 72°C: 3 min 20 cycles (94°C: 30 s, 62°C: 30 s, 72°C: 90 s), 72°C: 10 min. The PCR products were recovered by ethanol precipitation and DNA was eluted in 30 µL 0.1× TE, pH 8. In vitro methylated CpG islands from rice were used as positive controls for methylation and 10 pg were spiked in DNA samples used for aPRIMES to control methylation and methylation-sensitive digestion. Mitochondrial CpG island clones that were present in the original library were used as controls for unmethylated and allelically/partially methylated CGIs.
Methyl-CpG-immuno-precipitation (MCIp)
Genomic DNA (2 µg) isolated from CD19 sorted B cells of either CLL patients or healthy individuals was immuno-precipitated using recombinant MBD2–Fc fusion protein [103]. DNA was homogenized through a 22G needle and fragmented to a mean size of 400–500 bp using ultrasonication (2×30pulses, 24 s, 10% amplitude, Bioruptor, Diagenode). 30 µg of MBD-Fc protein was coupled to SIMAG protein-A magnetic beads (Chemicell) 3 h at 4°C in TBS. After completing MBD-Fc protein binding to the magnetic beads, precipitation of the sonicated sample DNA was performed in low salt buffer for 3 h at 4°C. Fractionated elution from the beads was performed using buffers A–F with increasing salt concentrations. In order to ensure complete elution of methylated DNA, elution with buffer F was repeated once. The collected fractions were desalted using the MinElute Kit (Qiagen) and eluates were diluted 1∶10 and analyzed for control genes (SNRPN, ZAP70; primer sequences see Table S4) with qPCR. Samples were subsequently processed for array hybridization.
Chromatin immunoprecipitation (ChIP)
For ChIP 1–5×107 viably frozen CD19-sorted B cells from either CLL patients or healthy individuals were washed once with DMEM medium, taken up in 1 ml PBS and formaldehyde cross link was performed at a final concentration of 1% for 10 min at RT while rotating. Cross-linked samples were sonicated in 300 µl SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris/HCl pH 8.1, 167 mM NaCl, protease inhibitors) 8 times 30 seconds on/off at high amplitude using a Bioruptor (Diagenode). The sonicated material was diluted 1∶10 with dilution buffer (0.01% SDS, 1.2 mM EDTA, 16.7 mM Tris/HCl pH 8.1, 1.1% TritonX100, protease inhibitors), subjected to 1 h preclearing with 30 µl of salmon sperm saturated protein A/G agarose beads (Millipore). Precleared chromatin samples were incubated over night with either 5 µg specific antibody (CTCF, H3K4me2, macroH2a1.2, Millipore), or 5 µg normal IgGs (Santa Cruz) at 4°C. Antibody bound chromatin was precipitated by adding 50 µl of salmon sperm saturated protein A/G agarose beads 4 h at 4°C and unspecifically bound material was removed by washing with low salt buffer, high salt buffer, LiCl buffer and two times with TE buffer. Cross link was reversed over night at 65°C and RNaseA (30 min, 37°C) as well as ProteinaseK digest (2 h, 45°C) was performed before purification of precipitated DNA using GFXTM PCR DNA and Gel Band Purification Kit (GE Healthcare). Precipitation efficiency was analyzed by qPCR for positive and negative control regions (for primers see Table S4) on antibody and control IgG precipitated fractions and expressed as percentage of input DNA using a calibration curve for quantification. Predicted CTCF binding sites were identified at http://www.essex.ac.uk/bs/molonc/binfo/ctcfbind.htm.
Array production and hybridization
Microarrays were either produced by spotting PCR-amplified 1 kbp fragments from the promotors of RFP2, DLEU1 and DLEU2/Alt1 (CpG islands C, D and E) for aPRIMES. For MCIp arrays were custom designed (eArray, Agilent) to tile promotors −3.8 to +1.8 kbp from the transcriptional start sites of the region chr13 : 47702475–49164179 (ITM2B – EBPL) and complete tiling of the region chr13 : 49265143–50317955 (C13ORF1 – DLEU7; GRCh37 hg18). 60 bp oligonucleotides were designed with 30 bp nonoverlapping spacing. The resulting 9863 oligonucleotides were combined with 10 bp linker sequence and had an average melting temperature of 70.43°C. For custom arrays, the 13q14.3 oligonucleotides were complemented with the Agilent normalization group (1262) and replicate group (4626) oligonucleotides. Labeling of ChIP and MCIp samples was performed using the BioPrime Total Genomic Labeling System (Invitrogen). For CTCF ChIP samples the precipitate was labeled using Cy5 and the input was labeled with Cy3. For the MCIp samples only the elutions from the high salt fraction were labeled, the common reference (T cell pool) was labeled with Cy3 and the CLL/healthy donor sample with Cy5. In order to predict labeling efficiency, the samples were measured at the wavelengths A260, A320, A555, A650, A750, and the following equations were used to determine the yield:
Cy3: DNA amount [ µg] (A260–A320)*50*0.04, Dye incorporation (A555–A650)/0.15*40
Cy5: DNA amount [ µg] (A260–A320)*50*0.04, Dye incorporation (A650–A750)/0.24*40
The hybridization of the MCIp samples was performed according to protocol number G4170-90012 for Agilent Microarray Analysis of Methlylated DNA Immunoprecipitation version. 1.0. The hybridization of the ChIP samples was performed as described in the protocol number G4481-90010 for Agilent Mammalian ChIP-on-chip version 10.1 applying the instructions given for the 4× format.
Aza-deoxy-cytidine treatment
Cell lines were seeded at a density of 5×106 in 4 ml of the appropriate medium in 6-well plates. After 24 hrs, they were treated with a final concentration of 1.5 µM 5-Aza-2′-deoxycytidine (Sigma-Aldrich) or the respective amount of DMSO solvent in the control reaction for 6 days with daily medium and drug replacement.
Chromatin fractionation for sequencing of chromatin-bound RNA
The chromatin fraction of RNA was prepared from isolated nuclei after shearing in a Covaris sonicator (Covaris, Inc.). The sample was then centrifuged and the soluble chromatin was loaded on a sucrose gradient as described [104]. Fractions containing DNA fragments > 5000 bp (equivalent to 25 nucleosomes with a 200 bp nucleosome repeat length) were pooled. RNA was phenol/chloroform-extracted after proteinase K and DNase I treatments and RNA-sequencing was performed. After rRNA depletion, RNAs were subjected to metal ion catalyzed cleavage to sizes between 60–200 nucleotides with the Ambion RNA fragmentation reagents. Libraries for Solexa sequencing were generated according to the standard protocol for mRNA (Illumina) that comprised first strand cDNA synthesis, second strand cDNA synthesis, end repair, addition of a single A base and adapter ligation. PCR products were size excised from low melting agarose gels (200–400 bp range) and phenol extracted. Sequencing was performed on the Illumina GAIIx platform at the sequencing core facilities of the EMBL, DKFZ and BioQuant in Heidelberg, Germany. Initial RNA sequence analysis was performed with the Bioconductor (http://www.bioconductor.org) package for the R statistical programming language to assess the read quality and to produce a reads coverage file. The integrative genomics viewer (http://www.broadinstitute.org/igv) was used to visualize the coverage file and the RefSeq genes (NCBI). Reads were aligned on the GRCh37/hg19 (2009) assembly version of the human genome reporting unique hits without mismatches and with and without trimming of the 3′ and 5′ ends. Data is available at ArrayExpress (www.ebi.ac.uk/arrayexpress), Experiment name: lncRNAs at 13q14.3; ArrayExpress accession: E-MTAB-1335 (U2OS) and E-MTAB-582 (HeLa).
Transfection and knockdown
All cell lines were cultured according to DSMZ (www.dsmz.de) recommendations. Adherent cell lines were transiently transfected with indicated constructs using the Nanofectin Kit (PAA). A half confluent flask of cells was transfected according to manufactures instructions using 5 µg of DNA for a 25 cm2 flask and 8 µg for a 75 cm2 culture vessel. Suspension cell lines were transfected according to the Nucleofection protocol of Amaxa (Lonza). For each cell line 2×106 cells were transfected using 5 µg plasmid DNA (2 µg pmax GFP as transfection control) and 100 µl Nucleofector solution. The whole procedure was performed following manufacturer's instructions and preparing the 12-well plates with 1.5 ml prewarmed medium. For Nucleofection the protocol A-023 was used. Gene transcripts were knocked down using either the Universal probe library system (Roche Diagnostics) and validated by q-RT-PCR to be below 30% of siCONP-treated cells. siRNAs were also synthesized using the Silencer siRNA construction kit (Ambion) and modified according to [105], or validated siRNAs were ordered from Applied biosystems. For simultaneous transfection of plasmids and miR-mimics (“miRVANAs”) or miR-inhibitors (both Life Technologies, Darmstadt, Germany) into HEK293T cells, Lipofectamine2000 (Invitrogen, Karslruhe, Germany), Hyperfect (Quiagen) or miRus transit-LT1 (Geneflow) was used. 4×105 cells/well were seeded in 24 well plates and transfected according to the manufacturers instructions after 24 h using 0.5 µg Plasmid and 10 pmol miR-mimics or miR inhibitors, respectively. For single transfection of 10 pmol miR-mimics or miR-inhibitors, the same protocol was used and the cells harvested after 24 h for Western blot or expression analysis. Transfection efficiency was tested by transfection of pmaxGFP (Lonza, Cologne, Germany) or siGlo (Dharmacon, Darmstadt) and subsequent detection by flow cytometry.
Luciferase assay
RFP2, C13ORF1, and D6/E6 elements in pCpGL
4×105 cells were seeded within a 6-well plate in 2 ml medium and co-transfected with 250 ng pRL-CMV-Renilla and 750 ng of the luciferase containing constructs using the appropriate transfection reagent after 24 h. For the constructs containing/not containing the D6/E6 element, equimolar amounts of construct were transfected. After 20–24 h, the cells were stimulated where indicated with 20 ng/ml TNFa for 10 min. The medium of the cells was removed and to each well 250 µL of Passive Lysis buffer (Promega) was added and incubated at RT for 15 min on a shaker. 15 µL of the cell lysates were placed into a 96-well Nunclon White plate while Luciferase buffer and Renilla buffer were brought to RT. The plates were measured with the Glomax Luminometer (Promega) applying the following parameters for both injectors: injection volume: 75 µl; delay between injection and measurement: 0.4 s; integration time: 5 s. The obtained values for Firefly Luciferase activity were normalized to Renilla readings to standardize for transfection efficiency.
miRNA screen
For the NF-kB reporter assay, HEK293FT cells were transfected in 96-well plates with siRNAs or miRNA-mimics from Dharmacon (Lafayette, CO, USA) together with 75 ng of the NF-κB reporter 3×KBL (kind gift from George Mosialos, Aristotle University, Thessaloniki, Greece) and 2,5 ng of pMIR-REPORT β-gal vector (Ambion, Austin, TX, USA). 48 h after transfection cells were stimulated with TNF-α (20 ng/ml), and luciferase activity was measured after another 5 h. β-galactosidase activity was used for normalization, and measured by beta-glo Luminescent Assay Kit (Promega, Madison, WI, USA).
Impact of miR-15a, miR-15b, and miR-16 on the expression of TAB3, IKKa/CHUK, SMRT, and SMAD7
Parts or complete 3′UTRs of TAB3, CHUK, SMRT and SMAD7 cloned into pMIR-Report Luciferase Plasmid were transfected into HEK293T cells. To this end, 4×105 cells were seeded in the wells of 24 well plates with 0.45 µg of pMIR-Report, 0.05 µg TK Renilla and 10 pmol of either miR-15a-3p, miR-15a-5p, miR-15b-5p or miR-16 miR-mimics or miR-inhibitors (Life Technologies, Darmstadt, Germany), respectively. The cells were harvested after 24 h and firefly and renilla luciferase activity measured using the Dual-Luciferase Reporter Assay (Promega) with injection volumes of 50 µl for LARII and Stop & Glo solution. Luminescence was measured using the LB940 Multimode Reader Mithras (Berthold Technologies).
Generation of RFP2 antibodies in guinea pigs
Specific antibodies were raised by immunizing guinea pigs with recombinant RFP2 S154-E264 10×His-tagged purified proteins. Guinea pigs were immunized for the first time at 8 weeks of age with 100 µg of protein per animal diluted 1∶1 with Complete Freud's adjuvant (Sigma). Subsequently animals were immunized monthly with 100 µg of protein per animal diluted 1∶1 with Incomplete Freud's adjuvant (Sigma). During this process, serum was taken periodically by heart-punction. After 24 months animals were bled. Blood was collected in Vacutainer Blood collection tubes (BD) left at RT for 1 hr and blood cells sedimented by spinning at 2000 rpm 1 hr at RT. After sedimentation, serum was collected and aliquoted for storage at −80°C or complemented with 0.02% sodium azide as preservative and kept at 4°C. The polyclonal antibodies were tested against purified recombinant protein in Western blot analysis and against protein over-expressed in mammalian cells by Western blot analysis and immunofluorescence.
Statistical methods
Overall survival curves were estimated by the Kaplan-Meier method. Logrank tests were used for comparing survival distributions between groups. Wilcoxon rank sum tests were used to test for differences in expression or methylation distributions between two groups. For significance of Pearson correlation coefficients, t-distribution was calculated with t = r/sqrt[(1−r∧2)/(N−2)]. For assessment of statistical significance, test results with p-values p<0.05 were considered to be statistically significant.
Supporting Information
Zdroje
1. HuarteM, RinnJL (2010) Large non-coding RNAs: missing links in cancer? Hum Mol Genet 19: R152–161.
2. MercerTR, DingerME, MattickJS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10 : 155–159.
3. BartelDP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 : 281–297.
4. PontingCP, OliverPL, ReikW (2009) Evolution and functions of long noncoding RNAs. Cell 136 : 629–641.
5. NaganoT, FraserP (2011) No-nonsense functions for long noncoding RNAs. Cell 145 : 178–181.
6. OromUA, DerrienT, BeringerM, GumireddyK, GardiniA, et al. (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143 : 46–58.
7. StilgenbauerS, NickolenkoJ, WilhelmJ, WolfS, WeitzS, et al. (1998) Expressed sequences as candidates for a novel tumor suppressor gene at band 13q14 in B-cell chronic lymphocytic leukemia and mantle cell lymphoma. Oncogene 16 : 1891–1897.
8. WolfS, MertensD, SchaffnerC, KorzC, DohnerH, et al. (2001) B-cell neoplasia associated gene with multiple splicing (BCMS): the candidate B-CLL gene on 13q14 comprises more than 560 kb covering all critical regions. Hum Mol Genet 10 : 1275–1285.
9. CorcoranMM, HammarsundM, ZhuC, LernerM, KapanadzeB, et al. (2004) DLEU2 encodes an antisense RNA for the putative bicistronic RFP2/LEU5 gene in humans and mouse. Genes Chromosomes Cancer 40 : 285–297.
10. OuilletteP, ErbaH, KujawskiL, KaminskiM, SheddenK, et al. (2008) Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res 68 : 1012–1021.
11. LernerM, HaradaM, LovenJ, CastroJ, DavisZ, et al. (2009) DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp Cell Res 315 : 2941–2952.
12. Lagos-QuintanaM, RauhutR, LendeckelW, TuschlT (2001) Identification of novel genes coding for small expressed RNAs. Science 294 : 853–858.
13. CalinGA, DumitruCD, ShimizuM, BichiR, ZupoS, et al. (2002) Frequent deletions and down-regulation of micro - RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99 : 15524–15529.
14. LiuY, CorcoranM, RasoolO, IvanovaG, IbbotsonR, et al. (1997) Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene 15 : 2463–2473.
15. RawstronAC, BennettFL, O'ConnorSJ, KwokM, FentonJA, et al. (2008) Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med 359 : 575–583.
16. KleinU, LiaM, CrespoM, SiegelR, ShenQ, et al. (2010) The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17 : 28–40.
17. RondeauG, MoreauI, BezieauS, CadoretE, MoisanJP, et al. (1999) Exclusion of Leu1 and Leu2 genes as tumor suppressor genes in 13q14.3-deleted B-CLL. Leukemia 13 : 1630–1632.
18. OuilletteP, CollinsR, ShakhanS, LiJ, LiC, et al. (2011) The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin Cancer Res 17 : 6778–6790.
19. LichterP (2010) All you need is a Mir-acle: the role of nontranslated RNAs in the suppression of B cell chronic lymphocytic leukemia. Cancer Cell 17 : 3–4.
20. LiaM, CaretteA, TangH, ShenQ, MoT, et al. (2012) Functional dissection of the chromosome 13q14 tumor suppressor locus using transgenic mouse lines. Blood 119 : 2981–2990.
21. BrownJR, LawrenceMS, HannaM, TesarB, StojanovP, et al. (2011) Novel Germline Genetic Variants Associated with Familial Chronic Lymphocytic Leukemia (CLL). ASH Annual Meeting Abstracts 118 : 465-.
22. CalinGA, FerracinM, CimminoA, Di LevaG, ShimizuM, et al. (2005) A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353 : 1793–1801.
23. RavecheES, SalernoE, ScaglioneBJ, ManoharV, AbbasiF, et al. (2007) Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood 109 : 5079–5086.
24. AllegraD, MertensD (2011) In-vivo quantification of primary microRNA processing by Drosha with a luciferase based system. Biochem Biophys Res Commun 406 : 501–505.
25. MigliazzaA, BoschF, KomatsuH, CayanisE, MartinottiS, et al. (2001) Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood 97 : 2098–2104.
26. MertensD, WolfS, SchroeterP, SchaffnerC, DohnerH, et al. (2002) Down-regulation of candidate tumor suppressor genes within chromosome band 13q14.3 is independent of the DNA methylation pattern in B-cell chronic lymphocytic leukemia. Blood 99 : 4116–4121.
27. FulciV, ChiarettiS, GoldoniM, AzzalinG, CarucciN, et al. (2007) Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood 109 : 4944–4951.
28. RavalA, TannerSM, ByrdJC, AngermanEB, PerkoJD, et al. (2007) Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell 129 : 879–890.
29. ChenSS, RavalA, JohnsonAJ, HertleinE, LiuTH, et al. (2009) Epigenetic changes during disease progression in a murine model of human chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 106 : 13433–13438.
30. XieX, MikkelsenTS, GnirkeA, Lindblad-TohK, KellisM, et al. (2007) Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A 104 : 7145–7150.
31. KapanadzeB, MakeevaN, CorcoranM, JareborgN, HammarsundM, et al. (2000) Comparative sequence analysis of a region on human chromosome 13q14, frequently deleted in B-cell chronic lymphocytic leukemia, and its homologous region on mouse chromosome 14. Genomics 70 : 327–334.
32. BunzelR, BlumckeI, CichonS, NormannS, SchrammJ, et al. (1998) Polymorphic imprinting of the serotonin-2A (5-HT2A) receptor gene in human adult brain. Brain Res Mol Brain Res 59 : 90–92.
33. KanberD, BerulavaT, AmmerpohlO, MitterD, RichterJ, et al. (2009) The human retinoblastoma gene is imprinted. PLoS Genet 5: e1000790 doi:10.1371/journal.pgen.1000790.
34. MertensD, PhilippenA, RuppelM, AllegraD, BhattacharyaN, et al. (2009) Chronic lymphocytic leukemia and 13q14: miRs and more. Leuk Lymphoma 50 : 502–505.
35. MertensD, WolfS, TschuchC, MundC, KienleD, et al. (2006) Allelic silencing at the tumor-suppressor locus 13q14.3 suggests an epigenetic tumor-suppressor mechanism. Proc Natl Acad Sci U S A 103 : 7741–7746.
36. LehmannS, OgawaS, RaynaudSD, SanadaM, NannyaY, et al. (2008) Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer 112 : 1296–1305.
37. StilgenbauerS, SanderS, BullingerL, BennerA, LeupoltE, et al. (2007) Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica 92 : 1242–1245.
38. CuniS, Perez-AciegoP, Perez-ChaconG, VargasJA, SanchezA, et al. (2004) A sustained activation of PI3K/NF-kappaB pathway is critical for the survival of chronic lymphocytic leukemia B cells. Leukemia 18 : 1391–1400.
39. HerishanuY, Perez-GalanP, LiuD, BiancottoA, PittalugaS, et al. (2010) The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood 117 : 563–574.
40. VallatLD, ParkY, LiC, GribbenJG (2007) Temporal genetic program following B-cell receptor cross-linking: altered balance between proliferation and death in healthy and malignant B cells. Blood 109 : 3989–3997.
41. FrommerM, McDonaldLE, MillarDS, CollisCM, WattF, et al. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A 89 : 1827–1831.
42. EhrichM, NelsonMR, StanssensP, ZabeauM, LiloglouT, et al. (2005) Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A 102 : 15785–15790.
43. BrenaRM, AuerH, KornackerK, PlassC (2006) Quantification of DNA methylation in electrofluidics chips (Bio-COBRA). Nat Protoc 1 : 52–58.
44. SchillingE, RehliM (2007) Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics 90 : 314–323.
45. PfisterS, SchlaegerC, MendrzykF, WittmannA, BennerA, et al. (2007) Array-based profiling of reference-independent methylation status (aPRIMES) identifies frequent promoter methylation and consecutive downregulation of ZIC2 in pediatric medulloblastoma. Nucleic Acids Res 35: e51.
46. HammarsundM, CorcoranMM, WilsonW, ZhuC, EinhornS, et al. (2004) Characterization of a novel B-CLL candidate gene–DLEU7–located in the 13q14 tumor suppressor locus. FEBS Lett 556 : 75–80.
47. BernsteinBE, HumphreyEL, ErlichRL, SchneiderR, BoumanP, et al. (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A 99 : 8695–8700.
48. GambleMJ, FrizzellKM, YangC, KrishnakumarR, KrausWL (2010) The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev 24 : 21–32.
49. BarretoG, SchaferA, MarholdJ, StachD, SwaminathanSK, et al. (2007) Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445 : 671–675.
50. AuerRL, RiazS, CotterFE (2007) The 13q and 11q B-cell chronic lymphocytic leukaemia-associated regions derive from a common ancestral region in the zebrafish. Br J Haematol 137 : 443–453.
51. ZenzT, MertensD, KuppersR, DohnerH, StilgenbauerS (2010) From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 10 : 37–50.
52. BullrichF, FujiiH, CalinG, MabuchiH, NegriniM, et al. (2001) Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res 61 : 6640–6648.
53. SampathD, LiuC, VasanK, SuldaM, PuduvalliVK, et al. (2012) Histone deacetylases mediate the silencing of miR-15a, miR-16 and miR-29b in chronic lymphocytic leukemia. Blood 119 : 1162–1172.
54. BellAC, WestAG, FelsenfeldG (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98 : 387–396.
55. De La Rosa-VelazquezIA, Rincon-AranoH, Benitez-BribiescaL, Recillas-TargaF (2007) Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res 67 : 2577–2585.
56. MondalT, RasmussenM, PandeyGK, IsakssonA, KanduriC (2010) Characterization of the RNA content of chromatin. Genome Res 20 : 899–907.
57. BonciD, CoppolaV, MusumeciM, AddarioA, GiuffridaR, et al. (2008) The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 14 : 1271–1277.
58. LiuQ, FuH, SunF, ZhangH, TieY, et al. (2008) miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36 : 5391–5404.
59. LinsleyPS, SchelterJ, BurchardJ, KibukawaM, MartinMM, et al. (2007) Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27 : 2240–2252.
60. RoccaroAM, SaccoA, ThompsonB, LeleuX, AzabAK, et al. (2009) MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 113 : 6669–6680.
61. PalamarchukA, EfanovA, NazaryanN, SantanamU, AlderH, et al. (2010) 13q14 deletions in CLL involve cooperating tumor suppressors. Blood 115 : 3916–3922.
62. TorokI, StrandD, SchmittR, TickG, TorokT, et al. (1995) The overgrown hematopoietic organs-31 tumor suppressor gene of Drosophila encodes an Importin-like protein accumulating in the nucleus at the onset of mitosis. J Cell Biol 129 : 1473–1489.
63. FagerlundR, KinnunenL, KohlerM, JulkunenI, MelenK (2005) NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem 280 : 15942–15951.
64. KeklikoglouI, KoernerC, SchmidtC, ZhangJD, HeckmannD, et al. (2012) MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-kappaB and TGF-beta signaling pathways. Oncogene 31 : 4150–4163.
65. BandiN, ZbindenS, GuggerM, ArnoldM, KocherV, et al. (2009) miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res 69 : 5553–5559.
66. LiT, MorganMJ, ChoksiS, ZhangY, KimYS, et al. (2010) MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 11 : 799–805.
67. ZhouR, LiX, HuG, GongAY, DrescherKM, et al. (2012) miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PLoS ONE 7: e30772 doi:10.1371/journal.pone.0030772.
68. HongS, LeeC, KimSJ (2007) Smad7 sensitizes tumor necrosis factor induced apoptosis through the inhibition of antiapoptotic gene expression by suppressing activation of the nuclear factor-kappaB pathway. Cancer Res 67 : 9577–9583.
69. MatsudaA, SuzukiY, HondaG, MuramatsuS, MatsuzakiO, et al. (2003) Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene 22 : 3307–3318.
70. LernerM, CorcoranM, CepedaD, NielsenML, ZubarevR, et al. (2007) The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell 18 : 1670–1682.
71. BhattacharyaN, SarnoA, IdlerIS, FuhrerM, ZenzT, et al. (2010) High-throughput detection of nuclear factor-kappaB activity using a sensitive oligo-based chemiluminescent enzyme-linked immunosorbent assay. Int J Cancer 127 : 404–411.
72. TomarD, SinghR, SinghAK, PandyaCD (2012) TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim Biophys Acta 1823 : 316–326.
73. DaroszewskaA, RalstonSH (2005) Genetics of Paget's disease of bone. Clin Sci (Lond) 109 : 257–263.
74. McManusS, RouxS (2012) The adaptor protein p62/SQSTM1 in osteoclast signaling pathways. J Mol Signal 7 : 1.
75. ReymondA, MeroniG, FantozziA, MerlaG, CairoS, et al. (2001) The tripartite motif family identifies cell compartments. Embo J 20 : 2140–2151.
76. StachD, SchmitzOJ, StilgenbauerS, BennerA, DohnerH, et al. (2003) Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Res 31: E2.
77. HeY, VogelsteinB, VelculescuVE, PapadopoulosN, KinzlerKW (2008) The antisense transcriptomes of human cells. Science 322 : 1855–1857.
78. FlynnRA, ChangHY (2012) Active chromatin and noncoding RNAs: an intimate relationship. Curr Opin Genet Dev 22 : 172–178.
79. FaghihiMA, WahlestedtC (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10 : 637–643.
80. SmitsG, MungallAJ, Griffiths-JonesS, SmithP, BeuryD, et al. (2008) Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet 40 : 971–976.
81. ZhangX, LianZ, PaddenC, GersteinMB, RozowskyJ, et al. (2009) A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113 : 2526–2534.
82. TreppendahlMB, QiuX, SogaardA, YangX, Nandrup-BusC, et al. (2012) Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood 119 : 206–216.
83. DallossoAR, HancockAL, MalikS, SalpekarA, King-UnderwoodL, et al. (2007) Alternately spliced WT1 antisense transcripts interact with WT1 sense RNA and show epigenetic and splicing defects in cancer. RNA 13 : 2287–2299.
84. StolcV, GauharZ, MasonC, HalaszG, van BatenburgMF, et al. (2004) A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306 : 655–660.
85. SproulD, GilbertN, BickmoreWA (2005) The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet 6 : 775–781.
86. SiebenlistU, BrownK, ClaudioE (2005) Control of lymphocyte development by nuclear factor-kappaB. Nat Rev Immunol 5 : 435–445.
87. HewamanaS, AlghazalS, LinTT, ClementM, JenkinsC, et al. (2008) The NF-kappaB subunit Rel A is associated with in vitro survival and clinical disease progression in chronic lymphocytic leukemia and represents a promising therapeutic target. Blood 111 : 4681–4689.
88. GaudioE, SpizzoR, PaduanoF, LuoZ, EfanovA, et al. (2012) Tcl1 interacts with Atm and enhances NF-kappaB activation in hematologic malignancies. Blood 119 : 180–187.
89. GardingA, BhattacharyaN, HaebeS, MuellerF, WeichenhanD, et al. (2013) TCL1A and ATM are coexpressed in chronic lymphocytic leukemia cells without deletion of 11q. Haematologica 98 : 269–273.
90. QuesadaV, CondeL, VillamorN, OrdonezGR, JaresP, et al. (2011) Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 44 : 47–52.
91. WangL, LawrenceMS, WanY, StojanovP, SougnezC, et al. (2011) SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 365 : 2497–2506.
92. SchattnerEJ (2000) CD40 ligand in CLL pathogenesis and therapy. Leuk Lymphoma 37 : 461–472.
93. KernC, CornuelJF, BillardC, TangR, RouillardD, et al. (2004) Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood 103 : 679–688.
94. EdelmannJ, Klein-HitpassL, CarpinteiroA, FuhrerA, SellmannL, et al. (2008) Bone marrow fibroblasts induce expression of PI3K/NF-kappaB pathway genes and a pro-angiogenic phenotype in CLL cells. Leuk Res 32 : 1565–1572.
95. JimiE, StricklandI, VollRE, LongM, GhoshS (2008) Differential role of the transcription factor NF-kappaB in selection and survival of CD4+ and CD8+ thymocytes. Immunity 29 : 523–537.
96. ChiorazziN, RaiKR, FerrariniM (2005) Chronic lymphocytic leukemia. N Engl J Med 352 : 804–815.
97. HewamanaS, LinTT, JenkinsC, BurnettAK, JordanCT, et al. (2008) The novel nuclear factor-kappaB inhibitor LC-1 is equipotent in poor prognostic subsets of chronic lymphocytic leukemia and shows strong synergy with fludarabine. Clin Cancer Res 14 : 8102–8111.
98. Escobar-DiazE, Lopez-MartinEM, Hernandez del CerroM, Puig-KrogerA, Soto-CerratoV, et al. (2005) AT514, a cyclic depsipeptide from Serratia marcescens, induces apoptosis of B-chronic lymphocytic leukemia cells: interference with the Akt/NF-kappaB survival pathway. Leukemia 19 : 572–579.
99. BichiR, ShintonSA, MartinES, KovalA, CalinGA, et al. (2002) Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A 99 : 6955–6960.
100. KikushigeY, IshikawaF, MiyamotoT, ShimaT, UrataS, et al. (2011) Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell 20 : 246–259.
101. KlugM, RehliM (2006) Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 1 : 127–130.
102. BrenaRM, AuerH, KornackerK, HackansonB, RavalA, et al. (2006) Accurate quantification of DNA methylation using combined bisulfite restriction analysis coupled with the Agilent 2100 Bioanalyzer platform. Nucleic Acids Res 34: e17.
103. GebhardC, SchwarzfischerL, PhamTH, SchillingE, KlugM, et al. (2006) Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res 66 : 6118–6128.
104. Caudron-HergerM, Muller-OttK, MallmJP, MarthC, SchmidtU, et al. (2011) Coding RNAs with a non-coding function: maintenance of open chromatin structure. Nucleus 2 : 410–424.
105. KimDH, LongoM, HanY, LundbergP, CantinE, et al. (2004) Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol 22 : 321–325.
Štítky
Genetika Reprodukčná medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 4- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



