-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step
Entry of human immunodeficiency virus type 1 (HIV-1) commences with binding of the envelope glycoprotein (Env) to the receptor CD4, and one of two coreceptors, CXCR4 or CCR5. Env-mediated signaling through coreceptor results in Gαq-mediated Rac activation and actin cytoskeleton rearrangements necessary for fusion. Guanine nucleotide exchange factors (GEFs) activate Rac and regulate its downstream protein effectors. In this study we show that Env-induced Rac activation is mediated by the Rac GEF Tiam-1, which associates with the adaptor protein IRSp53 to link Rac to the Wave2 complex. Rac and the tyrosine kinase Abl then activate the Wave2 complex and promote Arp2/3-dependent actin polymerization. Env-mediated cell-cell fusion, virus-cell fusion and HIV-1 infection are dependent on Tiam-1, Abl, IRSp53, Wave2, and Arp3 as shown by attenuation of fusion and infection in cells expressing siRNA targeted to these signaling components. HIV-1 Env-dependent cell-cell fusion, virus-cell fusion and infection were also inhibited by Abl kinase inhibitors, imatinib, nilotinib, and dasatinib. Treatment of cells with Abl kinase inhibitors did not affect cell viability or surface expression of CD4 and CCR5. Similar results with inhibitors and siRNAs were obtained when Env-dependent cell-cell fusion, virus-cell fusion or infection was measured, and when cell lines or primary cells were the target. Using membrane curving agents and fluorescence microscopy, we showed that inhibition of Abl kinase activity arrests fusion at the hemifusion (lipid mixing) step, suggesting a role for Abl-mediated actin remodeling in pore formation and expansion. These results suggest a potential utility of Abl kinase inhibitors to treat HIV-1 infected patients.
Published in the journal: Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step. PLoS Pathog 6(6): e32767. doi:10.1371/journal.ppat.1000956
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000956Summary
Entry of human immunodeficiency virus type 1 (HIV-1) commences with binding of the envelope glycoprotein (Env) to the receptor CD4, and one of two coreceptors, CXCR4 or CCR5. Env-mediated signaling through coreceptor results in Gαq-mediated Rac activation and actin cytoskeleton rearrangements necessary for fusion. Guanine nucleotide exchange factors (GEFs) activate Rac and regulate its downstream protein effectors. In this study we show that Env-induced Rac activation is mediated by the Rac GEF Tiam-1, which associates with the adaptor protein IRSp53 to link Rac to the Wave2 complex. Rac and the tyrosine kinase Abl then activate the Wave2 complex and promote Arp2/3-dependent actin polymerization. Env-mediated cell-cell fusion, virus-cell fusion and HIV-1 infection are dependent on Tiam-1, Abl, IRSp53, Wave2, and Arp3 as shown by attenuation of fusion and infection in cells expressing siRNA targeted to these signaling components. HIV-1 Env-dependent cell-cell fusion, virus-cell fusion and infection were also inhibited by Abl kinase inhibitors, imatinib, nilotinib, and dasatinib. Treatment of cells with Abl kinase inhibitors did not affect cell viability or surface expression of CD4 and CCR5. Similar results with inhibitors and siRNAs were obtained when Env-dependent cell-cell fusion, virus-cell fusion or infection was measured, and when cell lines or primary cells were the target. Using membrane curving agents and fluorescence microscopy, we showed that inhibition of Abl kinase activity arrests fusion at the hemifusion (lipid mixing) step, suggesting a role for Abl-mediated actin remodeling in pore formation and expansion. These results suggest a potential utility of Abl kinase inhibitors to treat HIV-1 infected patients.
Introduction
HIV-1 enters cells in a pH-independent manner by fusion at the plasma membrane or from within endosomes [1]–[3]. HIV-1 entry requires multiple conformational changes in the HIV-1 glycoprotein, and rearrangement of the actin cytoskeleton. These events are triggered by binding of the viral envelope (Env) surface subunit gp120 to the primary receptor CD4 and one of two chemokine coreceptors, CCR5 or CXCR4 [1], [4]. This interaction activates signaling events in the cell, similar to those initiated by natural ligands, such as Ca2+ mobilization, activation of RhoGTPases, and phosphorylation of tyrosine kinases, pyk2, Zap70 and p56lck [4]–[6]. Rho family GTPases, which include the Cdc42, Rac, and Rho subfamilies, are responsible for regulating signaling from membrane receptors to the actin cytoskeleton. The Rho sub-family stimulates myosin based contractility, and drives the formation of stress fibers and focal adhesions. The Rac sub-family stimulates lamellipodia and membrane ruffles, and the Cdc42 subfamily stimulates the formation of filopodia [7]–[9]. We showed that HIV-1 Env binding to target cells induces activation of Rac, stimulates membrane ruffles and lamellipodia, and fusion is inhibited by dominant negative Rac [4], [10]. Furthermore, HIV-1 Env-induced Rac activation depends on activation of Gαq, phospholipase C (PLC), Ca2+ mobilization, protein kinase C (PKC), pyk2 and the GTPase Ras [5]. In the current study we identified the fusion-specific effectors of Rac required for actin cytoskeleton rearrangements that mediate membrane fusion and entry.
Guanine nucleotide exchange factors (GEFs) activate GTPases, facilitating the GDP to GTP switch, and regulate their downstream effects by participating in scaffolding protein complexes, thereby linking GTPase activity to specific effectors [7]–[9]. HIV-1 Env-induced Rac activation is mediated by a specific Rac GEF, either Tiam-1 or Trio [10], [11]. There are multiple effectors of Rac, including serine/threonine kinases, lipid kinases, actin-binding proteins, and adaptor/scaffold molecules [7], [12]. PAK is a downstream effector of Rac and Cdc42 that promotes stabilization of actin networks. Another downstream effector of Rac that nucleates actin polymerization is the Arp2/3 complex. The Arp2/3 complex is activated by the Wave2 complex through IRSp53, an adaptor protein that binds Rac and Wave2 [7]. The Wave2 complex includes Rac-associated protein 1, Nck-associated protein, Abl-interacting protein 2, and heat shock protein C300. Wave2 also associates with Abl, and Abl-mediated phosphorylation of Wave2 promotes its activation [13], [14]. In addition to determining which Rac effectors are critical for membrane fusion, we studied the steps in the membrane fusion process affected by these signaling molecules. These data demonstrate that the Wave2 signaling complex and Abl are required for Env-mediated membrane fusion, entry, and infection and that Abl kinase inhibitors arrest the fusion process at hemifusion.
Results
HIV-1 Env-Mediated Fusion Depends on the Wave2 Signaling Complex
To determine whether Abl, Trio, or Tiam-1 were required for HIV-1 Env-mediated cell-cell fusion, expression of these proteins was down regulated by RNA interference (RNAi) in U87.CD4.CCR5 cells. Cells expressing siRNA were then mixed with BSC40 cells expressing different Env subtypes and Env-dependent cell-cell fusion was measured. Transfection of target cells with siRNA to Tiam-1 and Abl decreased levels of Env-mediated cell-cell fusion by an average of 79±5% and 74±5% respectively for both HIV-1 R5 and dual-tropic Env-subtypes (Figure 1A, left). There was no significant fusion observed with CCR5 expressing target cells and X4 Env expressing cells with or without siRNA, as expected. The decrease in the levels of fusion correlated well with the decreased steady-state level of Tiam-1, and Abl as detected by immunoblot (Figure 1C). A siRNA directed against Trio had no effect on Env-induced cell-cell fusion despite a 70% reduction in expression of the Trio protein (Figure 1A and C). To determine whether Tiam-1 and Abl are acting exclusively upstream of Rac, a constitutively active Rac mutant, RacV12 was expressed in siRNA transfected cells. Expression of RacV12 in cells expressing siRNA to Tiam-1 reversed the effects of this siRNA on fusion, suggesting that Tiam-1 is functioning upstream of Rac. In contrast, levels of fusion in cells expressing RacV12 and siRNA to Abl were only 53±1% that of cells expressing RacV12 and control siRNA, suggesting a role for Abl upstream and downstream of Rac (Figure 1A, right).
Fig. 1. Down regulation of Wave2 signaling complex with siRNA reduces HIV-1 Env-mediated cell-cell fusion and virus-cell fusion. 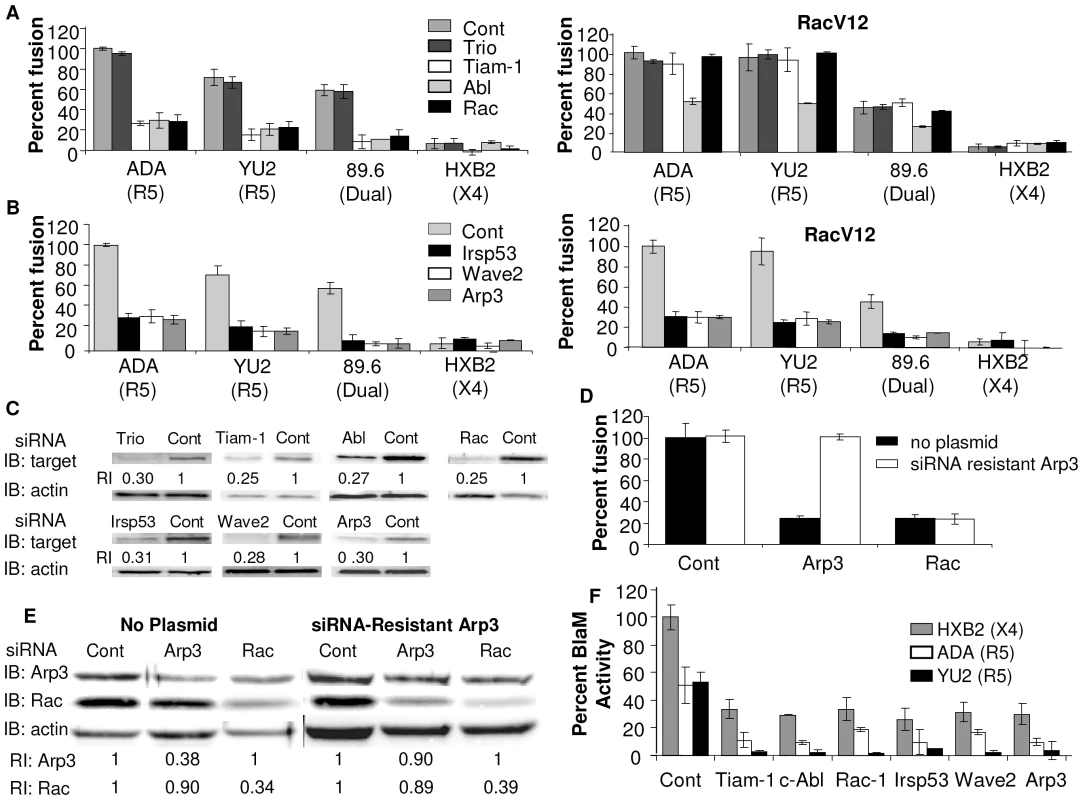
U87.CD4.CCR5 cells were transfected with control siRNA (control) or siRNA targeted against (A) Trio, Tiam-1, Abl, Rac, (B) IRSp53, Wave2, and Arp3. Cells were serum starved 24 h post-transfection (pt), infected with vCB21R alone or with vRacV12 48 h pt, and 72 h pt cells were incubated for 3 h with HIVUNC (subtracted as background), HIVADA, HIVYU2, HIV89.6 or HIVHXB2 Env-expressing cells and β-gal activity was measured (C) Each population of transfected cells was analyzed by Western blot with antibodies to the designated protein or actin. The relative reduction index (RI) is the quotient of the densitometry signal for the target band and that for actin, normalized by the ratio obtained with control siRNA. (D) U87.CD4.CCR5 cells engineered to express a siRNA resistant clone of Arp3 were transfected with control siRNA, or siRNA targeted against Arp3 or Rac. Cell fusion was measured by β-gal activity, and was normalized using control siRNA transfected cells incubated with HIVADA Env as 100%. (E) Western blots were performed on siRNA-resistant U87.CD4.CC5 cells expressing siRNA-resistant Arp3. (F) TZM-BL cells were transfected with 200 nM of targeted siRNA indicated and 48 h pt cells were incubated for 90 min with X4 HIVHXB2 virus R5 HIVADA virus or HIVYU2 virus. Fusion was stopped by adding lysis buffer with BlaM substrate. Cells were then incubated at rt overnight in the dark. OD values for no virus samples was subtracted as background and percent Blam activity was normalized using control siRNA transfected cells incubated with X4 HIVHXB2 virus as 100%. All data are representative of results from three similar experiments performed in triplicate. Tiam-1 binds to the Rac and Cdc42 effector IRSp53, enhancing IRSp53 binding to Rac and activation of the Wave2 scaffolding complex [15]. To determine the role of these Rac effectors in Env-mediated membrane fusion, their expression was down regulated by RNAi in U87.CD4.CCR5 cells. The siRNA expressing cells were mixed with Env-expressing cells and cell-cell fusion was measured. Expression of siRNA to IRSp53, Wave2, and Arp3 decreased fusion by 74±5% 77±4% and 78±4%, respectively. The decrease in fusion with these siRNAs was not overcome by expression of RacV12, suggesting that these proteins are required downstream of Rac (Figure 1B). The decrease in levels of fusion correlated with the decrease in protein expression in cells expressing these siRNAs, as seen by immunoblot (Figure 1C), and each siRNA was specific for its target protein (Figure S1A). Treatment of cells stably expressing siRNA resistant Arp3, with Arp3 targeted siRNA had no effect on Env-mediated cell-cell fusion (Figure 1D, E). In contrast, with untransfected cells, and cells stably expressing siRNA resistant Arp3, treatment with siRNA to Rac decreased fusion by 75±5% and 76±3% respectively (Figure 1D). These results show that the effects of RNAi on fusion were specific to inhibition of their target molecules.
To demonstrate the role of Tiam-1, Abl, Rac, IRSp53, Wave2 and Arp3 in virus-cell fusion, their expression was down regulated by RNAi in TZM-BL cells, a derivative of HeLa cells that express CD4, CCR5, and CXCR4, and these cells were then used in a Vpr-Blam assay [16], [17]. In this assay siRNA expressing cells were mixed for 90 min with HIV-1 strains with cores carrying a β-lactamase (BlaM)-Vpr chimera, and pseudotyped with Env from ADA (R5), YU2 (R5) or HXB2 (X4), and fusion was quantified by measuring the cytosolic activity of viral core-associated BlaM [18]. Expression of siRNA to Tiam-1, Abl, Rac, IRSp53, Wave2, and Arp3 decreased virus-cell fusion by an average of 80±4%, 83±1%, 76±4%, 82±6%, 77±3% and 82±6%, respectively, for HIV-1 R5 and X4 Env subtypes (Figure 1F). These results show that activation of the Wave2 signaling complex is required for Env-dependent cell-cell fusion and virus-cell fusion.
Small Molecule Inhibitors of Abl Kinase Activity Inhibit HIV-1 Entry
Since treatment of cells with Abl targeted siRNA led to a decrease in Env-dependent cell-cell fusion and virus-cell fusion we wanted to determine whether treatment of target cells with commercially available Abl kinase inhibitors, imatinib (IMB), nilotinib (NIL), and dasatinib (DAS), block fusion. IMB is a relatively specific inhibitor of Bcr-Abl, Abl, Arg, and class III receptor tyrosine kinases. NIL is an Abl kinase inhibitor 20–50 fold more potent than IMB at inhibiting Abl. DAS, originally designed as a Src family kinase inhibitor, antagonizes Abl, ephrin and platelet-derived growth factor receptor kinases, and kit. DAS is 300 fold more potent than IMB at inhibiting Abl [19], [20]. To determine the concentrations of these Abl kinase inhibitors that inhibit Abl kinase activity and Env-mediated cell-cell fusion, without non-specific effects, Abl kinase activity, trypan blue analysis, vaccinia virus infection, and T7 polymerase activity were measured in addition to Env-dependent cell-cell fusion (Figure S1B, S2, and data not shown). Treatment of U87.CD4.CCR5 cells with 10 uM IMB, 500 nM NIL, and 300 nM DAS for 1 h prior to and during 3 h incubation with Env-expressing cells decreased Env-mediated cell-cell fusion by an average of 95±2%, 92±5%, and 92±6%, respectively, and Abl kinase activity by 85–87% (Figure 2A and S1B). The CCR5 inhibitor TAK-779, which completely blocks Env-mediated cell-cell fusion and infection of CCR5 expressing cells, was included as a control, and it decreased Env-dependent cell-cell fusion by 99±1% and Env-mediated Abl kinase activation by 98% (Figure 2A and S1B). Similar results were observed with U87.CD4.CXCR4 cells treated with CXCR4 inhibitor AMD3100 and Abl kinase inhibitors and incubated with cells expressing HIV-1 X4 or dual-tropic Env subtypes (Figure S3A). There was no decrease in T7 polymerase activity, or localization of CD4 and CCR5 on the cell surface (Figure S4 and data not shown). Expression of RacV12 in U87.CD4.CCR5 cells treated with IMB, NIL and DAS increased the level of fusion by an average of 3.5-fold (*, P<0.05) compared to treated cells without RacV12, suggesting a role of Abl kinase activity upstream of Rac (Figure 2B).
Fig. 2. Abl kinase is required upstream and downstream of Rac for HIV-1 entry. 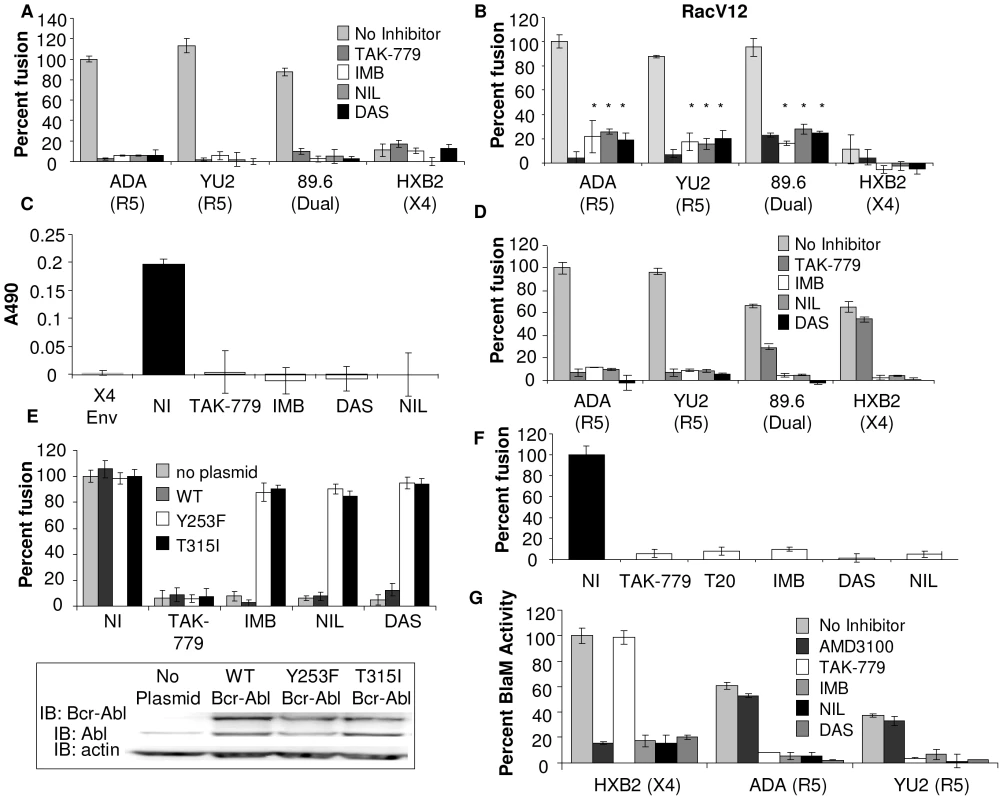
(A) U87.CD4.CCR5 cells were infected with vCB21R alone, or with (B) vRacV12 overnight, then treated with DMSO alone, TAK-779, IMB, NIL, or DAS for 1 h and the inhibitors were also present during 3 h incubation with HIV-1 Env-expressing cells and β-gal activity as measured. (C) U87.CD4.CCR5 cells were treated for 1 h with TAK-779, IMB, NIL, or DAS and during 30 min incubation with BSC40 cells expressing no Env (subtracted as background), HIVADA Env or HIVHXB2 Env. Whole cell lysates were analyzed by Rac specific G-LISA activation assay. Average A490 of triplicate wells ± standard deviation are shown. (D) PBMCs were infected with vCB21R in complete media overnight, treated with DMSO, TAK-779, IMB, NIL, or DAS for 1 h prior to addition of HIV-1 Env-expressing cells and β-gal activity was measured. (E) U87.CD4.CCR5 cells engineered to express indicated clones of Bcr-Abl were treated with Abl inhibitors and HIVUNC or HIVADA Env as described above or analyzed by Western blot with anti-Abl or anti-actin antibody (inset). (F) U87.CD4.CCR5 cells were infected overnight with vCB21R or vPT7-3, then mixed (1∶1) in triplicate wells, treated for 1 h with DMSO, TAK-779, T20, IMB, NIL, DAS, and with 100 ng of HIVYU2 for 3 h at 37°C. β-gal activity was measured and cell fusion was normalized using DMSO treated cells mixed with HIVYU2 as 100%. (G) TZM-BL cells were treated with DMSO, 1 µM TAK-779 or AMD3100, 10 µM IMB, 500 nM NIL, or 150 nM DAS for 1 h prior to 90 min incubation with indicated HIV viruses and BlaM activity was measured. Activity was normalized using DMSO treated cells mixed with HIVHXB2 virus as 100%. All data are representative of results from three similar experiments performed in triplicate. To determine the effect of these Abl kinase inhibitors on Env-induced Rac activation, U87.CD4.CCR5 cells were treated with inhibitors for 1 h prior to mixing with BSC40 cells expressing no HIV-1 Env, HIV-1 X4 Env, or HIV-1 R5 Env for 30 minutes in the presence of inhibitor. The mismatched X4 Env, that does not induce Rac activation in CCR5 expressing cells, and the CCR5 inhibitor TAK-779, which completely blocks Env-mediated Rac activation in CCR5 expressing cells, were included as controls [5]. Env-induced Rac activation was abolished in cells treated with TAK-779, and all three of the Abl kinase inhibitors (Figure 2C). To validate these effects in a relevant HIV-1 target cell, peripheral blood lymphocytes (PBLs), which express CD4, CCR5 and CXCR4, were used as the target cell population in an Env-dependent cell-cell fusion assay. Treatment of PBLs with IMB, NIL, and DAS decreased fusion by an average of 92±1%, 92±3%, and 99.5±1%, respectively, for HIV-1 R5, dual-tropic and X4 Env subtypes (Figure 2D). The CCR5 inhibitor TAK-779, as expected, completely blocked fusion mediated by R5 Env-expressing cells, inhibited fusion mediated by dual-tropic Env by 56±2%, and had no effect on fusion mediated by X4 Env (Figure 2D).
A long term infection assay was also performed where PBLs were infected with 150 ng of the X4 HIVHXB2 virus after 1 h preincubation with no inhibitor, DMSO, 10 µM IMB, 250 nM NIL, or 75 nM DAS. After 3 h, virus and inhibitors were washed off, inhibitors were added back and the plate was incubated at 37° for 21 days with addition of the inhibitors every 24 h. After 21 days the samples were assayed for cell viability and p24 antigen content. Treatment with IMB, NIL, and DAS decreased cell viability of HIVHXB2 infected cells by 17±4%, 8±5%, and 8±3% respectively and decreased infection by 52%, 51% and 94% compared to DMSO treated cells (Figure S5).
To validate the specificity of these effects, we performed an Env-dependent cell-cell fusion assay with cells stably expressing two different drug resistant Bcr-Abl mutants (Y253F and T315I), or expressing wild type (WT) Bcr-Abl [21]. Expression of the drug resistant Bcr-Abl mutants but not WT Bcr-Abl resulted in recovery of fusion (Figure 2E), demonstrating that the effects of these inhibitors on Env-dependent cell-cell fusion are specific to inhibition of Abl.
To confirm these results using virus particles with relevant levels of virus-associated glycoprotein, we used a virus-dependent cell-cell fusion assay based on the ability of virus particles to bridge two cells and allow transfer of cytoplasmic contents, and we also used the Vpr-BlaM assay described above [4], [10]. For the virus-dependent cell-cell fusion assay we used two populations of U87.CD4.CCR5 cells, one expressing the T7 polymerase and the other expressing the β-galactosidase (β-gal) gene under the T7 promoter. Both populations were incubated with inhibitors for 1 h prior to 3 h incubation with R5 virus HIVYU2. In this assay, controls included untreated and inhibitor treated cells that were not incubated with virus, the CCR5 inhibitor TAK-779, and T-20 which blocks entry by inhibiting the conformational change in HIV-1 gp41 required for fusion [17]. R5 Virus-dependent cell-cell fusion was reduced by an average of 94±3% in cells treated with IMB, DAS, and NIL compared to cells treated with DMSO alone, and treatment with TAK-779 and T-20 completely inhibited fusion (Figure 2F). Treatment of U87.CD4.CXCR4 cells incubated with the X4 virus HIVHXB2 with AMD3100 IMB, NIL, and DAS decreased virus-dependent cell-cell fusion by 88±7%, 98.6±1%, 87±5%, and 96±17%, respectively (Figure S3B).
For the Vpr-BlaM assay, TZM-BL cells were treated with 1 µM AMD3100, 1 µM TAK-779, 10 µM IMB, 500 nM NIL and 150 nM DAS for 1 hr prior to and during the 90 min incubation with HIV-1 Vpr-BlaM viruses expressing R5 and X4-tropic Env. AMD3100 treatment decreased X4-Vpr-BlaM activity by 84±1%, but had no effect on R5-Vpr-BlaM activity. TAK-779 treatment decreased R5-Vpr-BlaM activity by an average of 89±2%, but had no effect on X4-Vpr-BlaM activity, as expected. However, treatment of TZM-BL cells with IMB, NIL, and DAS decreased virus-cell fusion by an average of 81±4%, 89±5%, and 90±1%, respectively, for both HIV-1 R5 and X4 Env subtypes (Figure 2G). These results together with the results of the Env-dependent and virus-cell fusion assay demonstrate that Abl kinase is required for HIV-1 entry mediated by CXCR4 and CCR5.
Infection of TZM-BL Cells with HIV-1 Particles, but not Particles Pseudotyped with Amphotropic Murine Leukemia Virus (A-MLV) Env or Vesicular Stomatitis Virus Glycoprotein (VSV-G), Depends on Abl and the Wave2 Signaling Complex
To determine whether the Wave2 signaling complex and Abl are required exclusively for HIV-1 entry, or virus-induced fusion and infection in general, we examined infection with HIV-1 versus A-MLV Env (A-MLV-ENV-HIV-1) or VSV-G pseudotyped HIV-1 (VSV-G-HIV-1) using the TZM-BL assay. HIV-1 Env induces pH independent virus-cell fusion to facilitate entry, whereas viruses pseudotyped with VSV-G or A-MLV Env induce pH-dependent clathrin mediated endocytosis or caveolin-mediated endocytosis, respectively [22]–[25]. TZM-BL cells, a derivative of HeLa cells that express CD4, CCR5, CXCR4, and luciferase (luc) under the control of the HIV-1 LTR, were pretreated with the 10 µM IMB, 500 nM NIL and 150 nM DAS for 1 h prior to incubation with virus for 3 h, and a subsequent 24 h incubation with inhibitor only [16], [17]. The CCR5 inhibitor TAK-779, the CXCR4 inhibitor AMD3100, and ammonium chloride (NH4Cl) which inhibits endosomal acidification required for VSV-G mediated entry, were included as controls [22], [23], [26]. The top two panels of Figure 3A shows that treatment with IMB, NIL, and DAS decreased infection with R5 HIVYU2 virus and X4 HIVHXB2 virus by an average of 91±7%, 88±4%, and 91±5%, respectively, comparable to the reductions observed with Env-dependent cell-cell fusion, virus-dependent cell-cell fusion and virus-cell fusion (Figure 2). The Abl kinase inhibitors had no effect on infection of TZM-BL cells with A-MLV-ENV-HIV-1 or VSV-G-HIV-1, but treatment of cells with NH4Cl blocked infection with VSV-G-HIV-1 as expected (Figure 3A, bottom two panels). These data show that Abl-kinase inhibitors were able to block HIV-1 Env-mediated fusion specifically and had no effect on infection via pH-dependent clathrin-mediated or caveolin-mediated endocytosis, and post-entry steps were not affected by these inhibitors.
Fig. 3. Infection with HIV-1 particles but not particles pseudotyped with MLV Env or VSV-G depends on the Abl and Wave2 signaling complex. 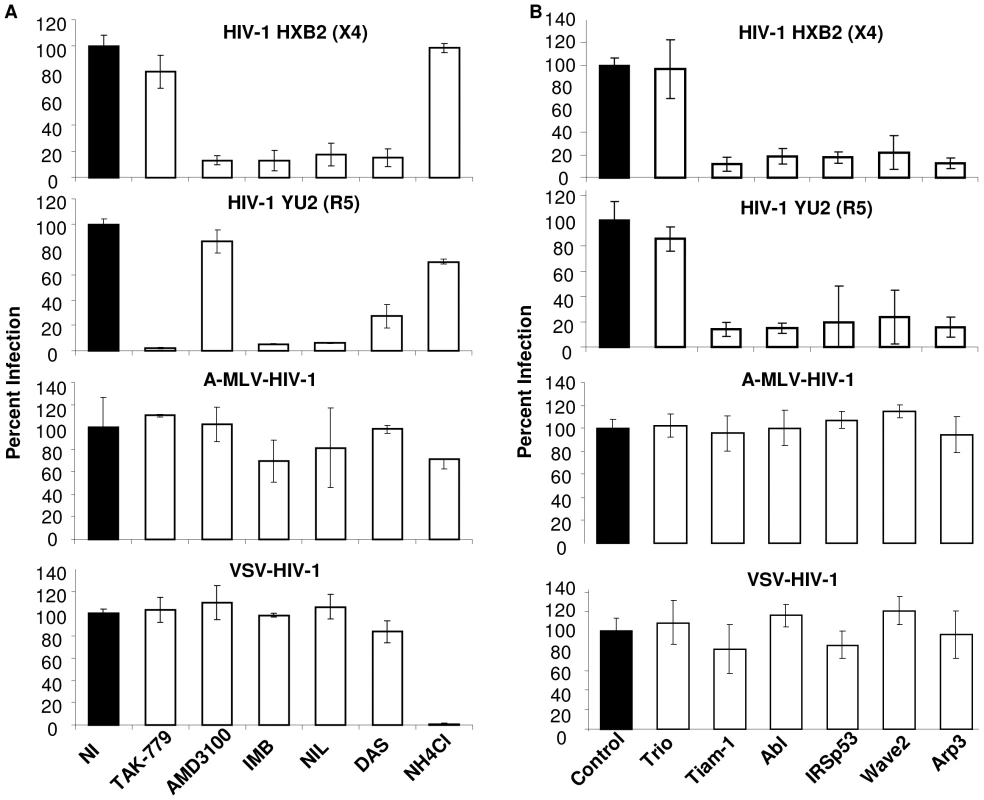
(A) TZM-BL cells were incubated for 1 h with DMSO, TAK-779, AMD3100, NH4Cl, IMB, NIL, or DAS and 150 ng of HIVHXB2, HIVYU2, A-MLV-or VSV-G-HIV-1 per well was added for 3 h, washed, and cells were incubated with inhibitors overnight and luc activity was measured. (B) TZM-BL cells were transfected with control siRNA or siRNA directed against indicated target proteins and 48 h later infected with 150 ng of HIVHXB2, HIVYU2, A-MLV- or VSV-G-HIV-1 for 3 h, cells were washed and incubated overnight, and luc activity measured. Cell infection was normalized using (A) DMSO treated cells or (B) cells transfected with control siRNA as 100%. All data are representative of results from three similar experiments performed in triplicate. To test the effect of Wave2 complex targeted siRNAs on infection, TZM-BL cells were transfected with 200 nM control siRNA or siRNA directed towards Tiam-1, Trio, Abl, IRSp53, Wave2 and Arp3. These cells were incubated with virus for 3 h, and media alone for 24 h. The decreased levels of HIV-1YU2 and HIV-1HXB2 infection of TZM-BL cells expressing siRNA targeted to Tiam-1, Abl, IRSp53, Wave2, and Arp3 were comparable to levels of Env-mediated cell fusion with U87.CD4.CCR5 cells expressing these siRNAs, whereas siRNA to Trio had no effect (Figure 3B, top two panels). Steady state levels of target proteins in cells expressing targeted siRNAs were decreased to similar levels as in U87 cells (Figure 1C and data not shown). Infection of TZM-BL cells with A-MLV-ENV-HIV-1 or VSV-G-HIV-1 was not affected by expression of the targeted siRNAs, suggesting that Tiam-1, Abl, IRSp53, Wave2, and Arp3 are required for HIV-1 Env-mediated entry and are not necessary for post-fusion steps in the virus life cycle (Figure 3B, bottom 2 panels).
The Abl Kinase Inhibitors Arrest Fusion at the Hemifusion Step
HIV-1 Env-induced fusion, and release of the viral capsid into the cytosol is a multistep process. First, gp120 binds to CD4 inducing conformational changes in gp120, and actin cytoskeletal rearrangements in the target membrane that bring the coreceptor CCR5 or CXCR4 into close proximity with CD4. Next, coreceptor binding to gp120 triggers conformational changes in gp41 to produce a prebundle conformation that inserts into the target cell membrane, allowing lipid mixing or hemifusion, and then pore formation. Additional conformational changes induce formation of the gp41 6-helix-bundle which prevents pore closure and facilitates pore enlargement and full fusion [2], [27], [28]. To determine which step(s) in the membrane fusion process are blocked by the Abl kinase inhibitors, we examined the effect on infection of membrane curving agents. Oleic acid (OLA), chlorpromazine (CPZ), and trifluoperazine (TFP) are lipid analogs that insert into the inner leaflet of the cell membrane. OLA induces negative curvature in the membrane that promotes formation of a hemifusion intermediate (i.e. lipid mixing), but cannot induce pore formation if there is a block at hemifusion. CPZ and TFP are membrane-permeable weak bases that partition into inner leaflets of cell membranes, induce positive curvature, and relieve a block at hemifusion [29]–[32].
To determine the effect of inhibitors and lipid analogs on HIV-1 infection, TZM-BL cells were treated with 1 µM AMD3100, 1 µM TAK-779, 10 µM IMB, 500 nM NIL, and 150 nM DAS for 1 h, prior to and during 1 h incubation with no virus, HIVΔENV, R5 HIVYU2, X4 HIVHXB2, A-MLV-ENV-HIV-1, or VSV-G-HIV-1. After 1 h, cells were treated with CPZ or TFP for 1 min or OLA for 5 min, followed by 2 h incubation with inhibitor and virus, and subsequent 24 h incubation with inhibitor only. Addition of CPZ and TFP to cells treated with Abl kinase inhibitors and infected with HIVYU2 or HIVHXB2 resulted in an 8 fold increase in infection compared to inhibitor treated cells infected in the absence of lipid analogs (Figure 4A), The exogenous cone shaped lipid OLA, which induces negative curvature of the membrane resulting in lipid mixing, had no affect on infection (Figure 4A). TAK-779 mediated inhibition of HIVYU2 infection and AMD3100 mediated inhibition of HIVHXB2 infection was not affected by these lipid analogs. No increase in luc activity was observed with lipid analog treatment of cells infected with HIVΔENV versus no virus, indicating that Env is required to observe an increase in infection (Figure S6A). Treatment of A-MLV-ENV-HIV-1 and VSV-G-HIV-1 infected cells with CPZ and TFP decreased overall infection by 2 fold and had no effect on cells treated with Abl kinase inhibitors, indicating that the increase in HIV-1 infection observed with Abl kinase inhibitor treated cells was specific (Figure 4A, lower panels). CPZ also partially reversed the inhibitory effects of nilotinib as measured by the Vpr-BlaM assay (Figure S6B).
Fig. 4. Abl kinase inhibitors and expression of siRNA targeted to the Wave2 complex block HIV-1 Env-mediated infection at a post-hemifusion step. 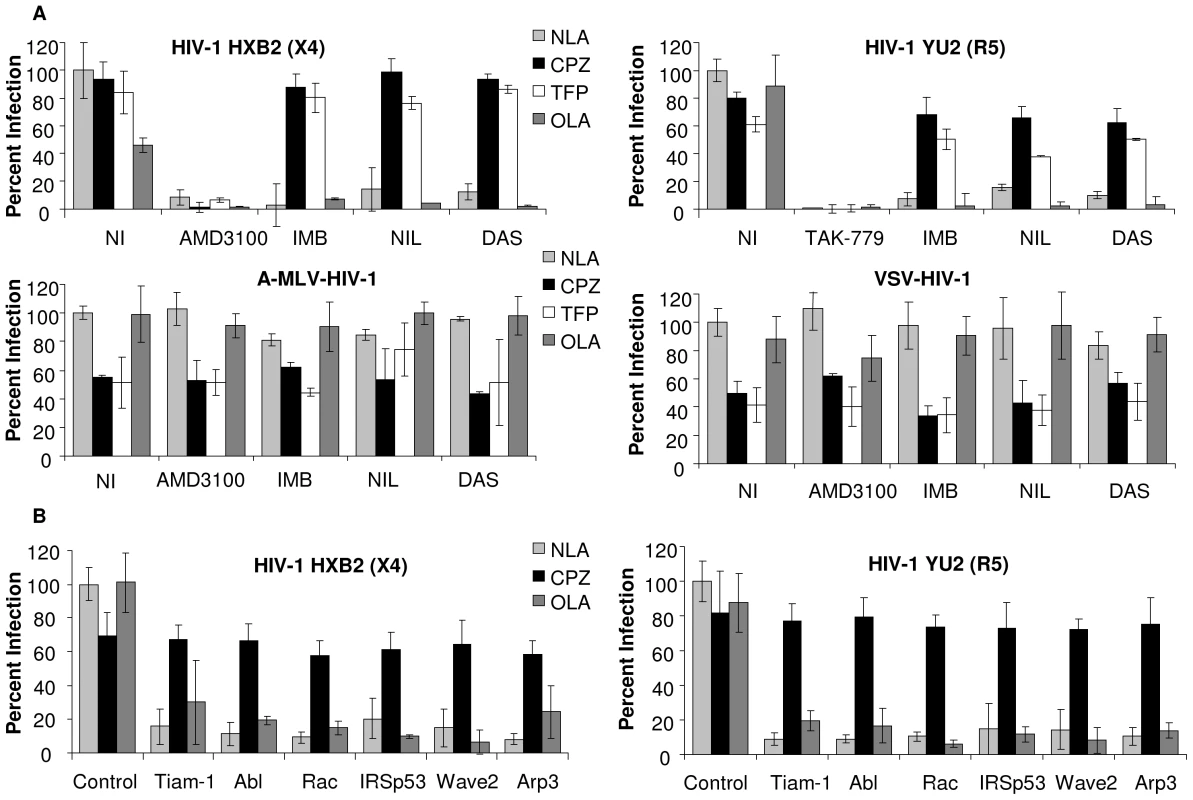
(A) TZM-BL cells were treated with DMSO, TAK-779, AMD3100, IMB, NIL, or DAS for 1 h. HIVYU2 HIVHXB2, A-MLV-Env-HIV-1 or VSV-G-HIV-1 (150 ng) was added for 1 h then cells were then treated with indicated lipid analogs for 1–5 min. Cells were washed and incubated in inhibitor overnight and luc activities were measured. (B) TZM-BL cells were transfected with 200 nM control siRNA or siRNA targeted to Tiam-1, Abl, Rac, IRSp53, Wave2 or Arp3 and 48 h pt cells were incubated with 150 ng of HIVYU2 or HIVHXB2. After 3 h cells were washed and incubated at 37° in complete media overnight and luc activities were measured. Data are representative of results from three similar experiments performed in triplicate. Cell infection was normalized using DMSO treated cells or control siRNA transfected cells infected with HIVYU2 or HIVHXB2 as 100%. Similar increases in virus-dependent cell-cell fusion were observed when U87.CD4.CCR5 cells were treated with inhibitors and lipid analogs and HIVYU2 mediated fusion was measured after 3 h (Figure S6C). Cells were also incubated with the lipid analogs in the absence of HIVYU2 to account for the effects of these agents on the cells and on T7 polymerase activity. Addition of OLA did not increase fusion in cells treated with any of the inhibitors (Figure S6B). To confirm the results obtained with the Abl kinase inhibitors we incubated TZM-BL cells transfected with Tiam-1, Abl, Rac, IRSp53, Wave2, and Arp3 targeted siRNA, for 1 h with no virus, HIVΔENV, R5 HIVYU2, or X4 HIVHXB2. After 1 h cells were treated with CPZ for 1 min or OLA for 5 min, followed by 2 h incubation with virus, and subsequent 24 h incubation with media alone. As with the Abl kinase inhibitors, treatment of siRNA transfected cells with CPZ increased infection by an average 8.4 fold compared to untreated cells, and OLA had no effect (Figure 4B). These results suggest that inhibition of Tiam-1, Abl, Rac, IRSp53, Wave2 or Arp3 arrests fusion at hemifusion, preventing pore formation, pore enlargement and content mixing.
To confirm that Abl kinase inhibitors cause arrest at hemifusion, we used a modification of a fusion assay described previously [33]. CHO-K1 cells that lack expression of the lipid ganglioside GM1, were engineered to express GFP and the HIV-1ADA (R5) Env protein. U87.CD4.CCR5 cells were used as the target cell, and lipid mixing was detected when GM1, detected by a TRITC-conjugated form of cholera toxin β-subunit (CTX), was transferred from the target cell to CHO-K1-GFP cells. Complete fusion is detected when cells express GM1, GFP, and are multinucleated. Quantification was performed for three independent experiments and the percentage of hemifused GFP+, GM1+ cells with single nuclei and the percentage of multinucleated fully fused cells was enumerated for 68 cells from each condition (Figure 5, S7, and Table S1) There were 83.1±10.9% hemifused cells with IMB-treated cells mixed with HIVADA-expressing CHO-K1 cells (Figure 5), compared to DMSO treated cells with 22.3±4.9% hemifused cells and 75.5±6.2% fully fused cells. With no HIV-1 Env or with the addition of TAK-779 there was little or no hemifusion or full fusion (Figure 5).
Fig. 5. Abl kinase inhibitors arrest HIV-1 Env-mediated fusion at the hemifusion step. 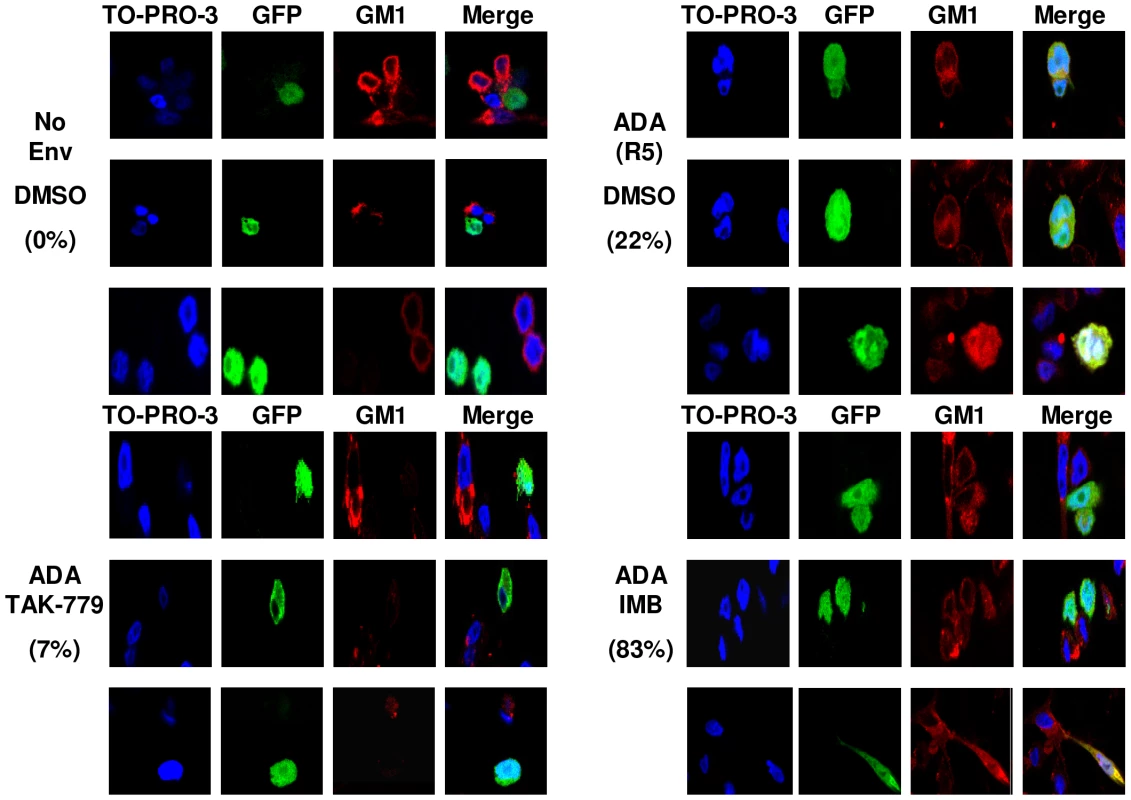
CHO-K1 cells that do not express GM1 were transfected with a GFP expressing plasmid (green), and 24 h later infected with WT vaccinia virus or vaccinia virus expressing HIVADA Env. After another 24 h, CHO-K1 cells were overlaid for 3 h with U87.CD4.CCR5 cells pre-treated for 1 h with DMSO, TAK-779, or IMB. Cells were fixed and stained with CTX-555 (red), and counterstained with TO-PRO 3 (blue). Images were collected using an oil objective (magnification X63). Images were cropped but relative cell size was maintained. The percentage of hemifused cells is listed. To demonstrate the effects of the lipid analog CPZ on HIV-1 Env mediated cell-cell fusion and to observe the effect of CPZ and the Abl kinase inhibitors on A-MLV Env or VSV-G induced cell-cell fusion we treated U87.CD4.CCR5 cells with DMSO, TAK-779, or IMB for 1 hr prior to incubation with CHO-K1 cells expressing no Env, HIVADA, A-MLV Env or VSV-G for 1 hr. After 1 h cells were treated with CPZ for 1 min and OLA for 5 min, then washed and incubated with inhibitor for an additional 2 h prior to fixation and GM1 staining. Incubation of IMB treated cells with HIVADA and CPZ promoted the transition from hemifusion to full fusion as expected (Figure S8). Fusion of A-MLV Env and VSV-G Env expressing cells with U87.CD4.CCR5 cells was unaffected by treatment with IMB or CPZ (Figure S9) and all Env-mediated fusion was unaffected by OLA treatment (data not shown). These results confirm that Abl kinase activity is required at a post-hemifusion step for HIV-1 Env mediated fusion and entry.
Discussion
Dynamic regulation of the actin cytoskeleton is required for fusion of biological membranes. Multiple reports have demonstrated that actin remodeling is required for HIV-1 mediated fusion and entry [4], [5], [10], [11], [34]–[36]. Some studies showed that treatment of target cells expressing physiologically relevant levels of receptor and coreceptor with the actin filament capping drug cytochalasin D prevented the formation of the gp120-CD4-coreceptor complex [35], [37], [38]. Another more recent study, demonstrated a role for CD4 and coreceptor-mediated filamin-A interactions in receptor clustering that is dependent on RhoA and ROCK mediated phosphorylation of ADF/cofilin [34]. Previous work from our lab with the actin filament stabilizing drug jasplakinolide and the actin monomer sequestering drug latrunculin A (LA) suggested a role for actin remodeling at a post binding step in fusion [4]. To further substantiate the role of actin polymerization in HIV-1 entry, we treated cells with 1 µM LA and 5 µM latrunculin B (LB). Both drugs blocked HIV-1 fusion for multiple cell types, as measured by the Env-dependent cell-cell fusion assay, the virus-dependent cell-cell fusion assay, the virus-cell fusion assay, and infection (Figure S10).
Our previous data demonstrated that the GTPase Rac was activated by HIV-1 Env ligation of CCR5, resulting in membrane ruffles and lamellipodia in the target cell membrane. Inhibition of this activation by dominant negative Rac or by a Rac GEF inhibitor completely abolished Env-dependent cell-cell fusion, virus dependent cell-cell fusion and infection [4], [5], [10], [11]. Our lab went on to show that Env-induced Rac activation is mediated by Gαq and its downstream effectors, including Ras. Other studies showed that Ras promotes Rac activation via direct interaction with Tiam-1, or by phosphatidylinositol 3-kinase (PI3K)-mediated activation of Tiam-1 [39]. Env-dependent Rac activation likely occurs through the first mechanism, since treatment of target cells with PI3K inhibitors had no effect on Env-dependent cell-cell fusion [40].
The nonreceptor tyrosine kinase, Abl, modulates actin upstream and downstream of Rac [41], [42]. In the current study, we used siRNAs and specific inhibitors to show that the activity of Abl kinase is required both upstream and downstream of Rac for Env-induced membrane fusion. Upstream of Rac, Abl phosphorylation of the Ras GEF complex promotes the activity of the Rac GEF Tiam-1, which was shown in the current study to be required for HIV-1 fusion. Downstream of Rac, Abl promotes phosphorylation and activation of Wave2 and its interaction with the Arp2/3 complex, events also demonstrated here to be critical for HIV-1 infection, but not VSV-G or A-MLV Env-mediated infection. These results suggest that these signaling mediators are important for HIV-1 Env mediated entry, are not necessary for pH dependent clathrin or caveolin-mediated endocytosis, and are not required at post-entry steps in the virus life cycle.
There is some conflict in the literature as to the location and mechanism of virus cell fusion. A recent report used microscopic imaging to track HIV-1 Env-pseudotyped MLV virus particles and observed virus-membrane fusion in endosomes [3]. This study also showed that virus-cell fusion and infection were inhibited in the presence of the dynamin inhibitor dynasore (DYN) which is known to block both clathrin and caveolin-mediated endocytosis [3]. The results in our current study suggest that fusion is occurring via a mechanism that is distinct from that of VSV (clathrin-mediated endocytosis) or A-MLV (caveolin-mediated endocytosis). In order to address this conundrum, we treated cells with the dynamin inhibitor DYN, and then used these cells for the Env-dependent cell-cell fusion assay, the virus-dependent cell-cell fusion assay, the virus-cell fusion assay and the TZM-BL infection assay. DYN treatment decreased HIV-1 Env-mediated infection and virus-cell fusion by an average of 58±7% and 50±3% respectively (Figure S10 and Figure S11). However treatment with DYN decreased A-MLV-Env-HIV-1 infection and VSV-G-HIV-1 infection by 75±5% and 89±1% respectively, showing that the affect on HIV-1 Env-mediated infection was not as significant (Figure S11). DYN treatment also decreased Env-dependent cell-cell fusion and virus-dependent cell-cell fusion by 53±8% and 50±10%, respectively which was unexpected since these assays both measure cell-cell plasma membrane fusion. Dynamins are a group of large GTPases that are involved in multiple processes in addition to endocytosis, such as vesicle transport, cytokinesis, organelle division, cell movement and cell signaling [43]–[45]. Therefore, the inhibition observed with the dynamin inhibitor DYN could be due to nonspecific effects on cellular processes. In support of this conclusion, a recent study used the Rev-dependent indicator cell line Rev-CEM to study the effects of DYN on HIV-1 replication and VSV-G-HIV-1 infection [44]. Using this assay they observed a dosage dependent decrease in VSV-G-HIV-1 infection with DYN treatment but did not see any decrease in HIV-1 infection [44]. These results as well as the results in Figure 3 show a clear distinction between HIV-1 Env-mediated entry and VSV-G - and A-MLV-mediated entry.
The current study also showed that the block in fusion caused by inhibition of Tiam-1, Abl, Rac, IRSp53, Wave2 and Arp3 occurs after hemifusion and before cytoplasmic mixing. This conclusion was based on the 1) confocal microscopy demonstration that addition of IMB to the fusion reaction allowed membrane but not cytoplasmic mixing, and 2) observation that lipid analogs that overcome a block at hemifusion overcame inhibition of HIV-1 virus dependent cell fusion, virus-cell fusion and infection caused by Abl kinase inhibitors and siRNA expression. These results support a model whereby HIV-1 Env binding to CCR5 stimulates activation of Gαq resulting in activation of Rac and activated Rac interacts with IRSp53. IRSp53 promotes Rac activation of the Wave2 complex, which is also activated by Abl, and activated Wave2 induces subsequent activation of Arp2/3-mediated actin rearrangements which facilitate pore formation, pore enlargement, and entry of HIV-1.
Many microbial pathogens depend on Abl family kinases to mediate efficient infection of their targeted host, including Shigella flexneri, enteropathogenic Escherichia coli, Helicobacter pylori, Anaplasma phagocytophilum, coxsackievirus, poxvirus, and murine AIDS virus. Abl kinases are involved in pathogen entry, intracellular movement, and exit from target cells; proliferation of target cells; and phosphorylation of microbial effectors. Many of these processes involve reorganization of the target cell actin cytoskeleton and depend on the same signaling pathways as HIV-1 [4], [5], [46]. Discovery of these signaling mediators as fundamental components of microbial pathogenesis provides new targets for therapeutic intervention. The clinical application of IMB, NIL, and DAS, which block deregulated Abl kinases in leukemia patients, demonstrate that inhibition in vivo is possible with manageable side effects [19], [20]. In addition IMB has been shown to be an effective inhibitor of anti-apoptotic pathways induced by HIV-1 in macrophages [47]. Most current antiviral therapies target viral proteins and mutation of the virus leads to therapy resistance. Therefore, using inhibitors that target host signaling proteins essential for HIV-1 entry may be an efficient new strategy for treatment of infected patients.
Materials and Methods
Reagents and Cell Lines
U87.CD4.CCR5 cells are astroglioma cells expressing CD4, CCR5-GFP or HA-CCR5. U87.CD4.CXCR4 cells are astroglioma cells expressing CD4 and CXCR4-GFP. CHO-K1 cells (ATCC) were grown in F-12K media with 10% serum and other cells maintained as described [48]. pMSCVneo-WT, Y253F, and T315I Bcr-Abl were gifts from Dr. R. Van Etten [21]. The siRNA resistant mutations were generated in Arp3 based on sequences obtained from Santa Cruz Biotechnology, Inc (SCBT, Santa Cruz, CA,), by PCR-mediated mutagenesis of a sub fragment that was sequenced to confirm the presence of mutations before sub cloning into the corresponding cDNA. WT and mutant cDNAs were cloned in pcDNA3.1+zeo for expression by transduction. IMB, NIL, and DAS were from LC Laboratories and were used at 10 uM, 500 nM, and 300 nM respectively unless indicated; CPZ (0.5 mM), TFP (0.3 mM), OA (100 nM), OLA (50 uM) and NH4Cl (50 mM) were from Sigma; TAK-779 (1 uM), and T-20 (10 ug/ml) were from the AIDS Research and Reference Reagent Program. The control siRNA constructs (non-targeting 20–25 nt siRNA designed as a negative control), the siRNA constructs and antibodies used for Western blots were from SCBT [5]. The siRNA constructs were transfected using GeneEraser siRNA Transfection Reagent or Lipofectamine RNAiMAX Transfection Reagent according to the manufacturer's instructions (Stratagene, La Jolla, CA, Invitrogen, Carlsbad, CA).
Viruses
Wild-type (WT) vaccinia (WR strain) and recombinant vaccinia viruses expressing β-galactosidase (vCB21R), T7 polymerase (vPT7-3), constitutively active Rac GTPase (vRacV12), or HIV-1 Env proteins were described [48]. HIV with R5 YU2 or X4 HXB2 Env in HIVNL4-3 backbone were generated from 293T cells; some were pseudotyped with amphotropic murine leukemia virus (MLV) or vesicular stomatitis virus (VSV) glycoproteins [5]. TZM-BL assays were performed as described [5]. For the BlaM assay pseudoviruses were produced by co-transfecting 293T cells with HIVNL4-3ΔVpr expressing YU2, ADA, or HXB2 Env and BlaM-Vpr expressing pMM310 vector. Transfected 293T cell supernatants were harvested 48 h postlipofection, filtered, and assayed for p24 antigen content by enzyme-linked immunosorbent assay. Viruses were resuspended in culture media, aliquoted and stored at −80°C.
BlaM Assay for Virus-Cell Fusion
TZM-bl cells were serum starved for 24–36 h then plated (4×104 cells/well) in 96-well plates in complete media overnight. Cells were treated with indicated concentrations of inhibitors for 1 hr prior to and during 90 min incubation with DEAE-dextran (20 µg/ml) alone or DEAE-dextran (20 µg/ml) and 150 ng p24 HIVYU2Vpr-BlaM, HIVADAVpr-BlaM, or HIVHXB2Vpr-BlaM. After 90 min virus and media were aspirated off cells and 100 ul 1X Lysis and Detection Solution was added to wells (LyticBlazer-BODIPY FL, Invitrogen). The plate was incubated at room temperature in the dark overnight. The BlaM activity was quantified using TECAN fluorescence plate reader (Tecan, Switzerland). The extent of virus-cell fusion was measured with excitation centered at 485 nm and emission centered at 535 nm. The green signal for samples incubated with no inhibitors or inhibitors and no virus was subtracted as background from their respective virus treated samples.
TZM-BL Assay
TZM-BL cells were serum starved for 12–24 h then plated overnight in complete media in 96 well plate at 2×104 cells per well. Cells were treated for 1 h with indicated concentrations of inhibitors prior to addition of media alone or 150 ng p24 of HIVYU2 HIVHXB2 or VSVG or A-MLV-pseudotyped HIV in the presence of 20 ug/ml DEAE-dextran for 3 h at 37°C. After 3 h cells were washed 3 times with PBS and inhibitors were added in fresh media. Following a 24 h incubation cells were lysed and luciferase (luc) units determined. Infected wells and uninfected wells with inhibitor were compared to wells with no inhibitor. For the TZM-Bl assay with lipid analogs serum starved TZM-BL cells were treated with inhibitors for 1 h, then 150 ng of indicated virus was added for 1 h prior to treatment with CPZ or TFP for 1 min or OLA for 5 min. Cells were washed three times with PBS and virus and inhibitors were added back. After 2 h cells were washed two times with PBS and incubated in inhibitor overnight and luc activities were measured.
Long-Term PBMC Infection and Viability Assay
PBMCs that were isolated and stimulated as previously described [5]. They were plated at 5×105 cells per well in 96 well plate and were treated with 10 µM IMB, 250 nM NIL, or 75 nM DAS for 1 h prior to addition of 150 ng p24 of HIVHXB2 in the presence of 20 ug/ml DEAE-dextran for 3 h at 37°C. After 3 h cells were washed three times with PBS and incubated in inhibitor for 24 h. Inhibitors were added back at the same concentration every 24 h for three weeks. 100 ul of supernatant was collected every fourth day and all samples were assayed for p24 antigen content by enzyme-linked immunosorbent assay. Two separate plates were set up under the exact same conditions and one plate was used for p24 measurement and the other was incubated with 20 ul cell viability substrate per 100 ul of sample (Promega, Madison, WI).
Fusion and Hemifusion Assays
Envelope-mediated and virus-dependent fusion assays were described. Average fusion compared to untreated control reactions were detected by β-galactosidase activity ± standard deviation [5]. To account for any effect of inhibitors on vaccinia virus infection and/or on T7 polymerase function, vCB21R and vPT7-3 co-infected cells were similarly treated with inhibitors. Concentration curves were performed with all of the inhibitors to determine the concentration that resulted in the maximum decrease in fusion without altering vaccinia virus infection or T7 polymerase activity. Hemifusion assays were performed with 2×106 CHO-K1 cells nucleofected with a GFP expression plasmid, and after 24 h infected with vaccinia virus expressing HIVADA Env or no Env. After 16 h, 4×105 U87.CD4.CCR5.HA cells were added for 3 h, fixed with paraformaldehyde, stained with TRITC-conjugated CTX-555 (EMD), and analyzed on a 510 Meta LSM confocal microscope.
Statistical Analysis
Fusion and infectivity results were compared using a two-tailed t-test. All p values, unless indicated, were <0.03.
Supporting Information
Zdroje
1. GreeneWC
PeterlinBM
2002 Charting HIV's remarkable voyage through the cell: Basic science as a passport to future therapy. Nature Med 8 673 680
2. GalloSA
2003 The HIV Env-mediated fusion reaction. Biochim Biophys Acta 1614 36 50
3. MiyauchiK
2009 HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137 1 12
4. PontowS
HeydenN
WeiS
RatnerL
2004 Actin cytoskeletal reorganizations and coreceptor-mediated activation of Rac during human immunodeficiency virus-induced cell fusion. J Virol 78 7138 7147
5. HarmonB
RatnerL
2008 Induction of the G{alpha}q signaling cascade by the human immunodeficiency virus envelope is required for virus entry. J Virol 82 9191 9205
6. StantchevTS
BroderCC
2001 Human immunodeficiency virus type-1 and chemokines: beyond competition for common cellular receptors. Cytokine Growth Factor Rev 12 219 43
7. BurridgeK
WennerbergK
2004 Rho and Rac take center stage. Cell 116 167 79
8. DebreceniB
2004 Mechanisms of guanine nucleotide exchange and Rac-mediated signaling revealed by a dominant negative Trio mutant. J Biol Chem 279 3777 3786
9. Etienne-MannevilleS
HallA
2002 Rho GTPases in cell biology. Nature 420 629 35
10. PontowSE
HarmonB
CampbellN
RatnerL
2007 Antiviral activity of a Rac GEF inhibitor characterized with a sensitive HIV/SIV fusion assay. Virology 368 1 6
11. GaoY
2004 Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA 101 7618 7623
12. SunD
XuD
ZhangB
2006 Rac signaling in tumorigenesis and as target for anticancer drug development. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy 9 274 287
13. SoderlingSH
ScottJD
2006 Wave signaling: from biochemistry to biology. Biochem Soc Trans 34 73 76
14. KonigR
2002 Interactions between MHC molecules and co-receptors of the TCR. Curr Opin Immunol 14 75 83
15. ConnollyBA
RiceJ
FeigLA
BuchsbaumRJ
2005 Tiam1-IRSp53 complex formation directs specificity of Rac-mediated actin cytoskeleton regulation. Mol Cell Biol 25 4602 4614
16. DerdeynCA
2000 Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 Loop of gp120. J Virol 74 8358 8367
17. WeiX
2002 Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46 1896 1905
18. CavroisM
De NoronhaC
GreeneW
2002 A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol 20 1151 1154
19. MeloJV
ChuahC
2008 Novel agents in CML therapy: tyrosine kinase inhibitors and beyond. Hematology 2008 427 435
20. KantarjianHM
GilesF
Quintas-CardamaA
CortesJ
2007 Important therapeutic targets in chronic myelogenous leukemia. Clin Cancer Res 13 1089 1097
21. RoumiantsevS
2002 Clinical resistance to the kinase inhibitor STI-571 in chronic myeloid leukemia by mutation of Tyr-253 in the Abl kinase domain P-loop. Proc Natl Acad Sci USA 99 10700 10705
22. BlumenthalR
1987 pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J Biol Chem 262 13614 13619
23. RocheS
2008 Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited. Cell Mol Life Sci 65 1716 1728
24. JohannsdottirH
2008 Host cell factors and functions involved in Vesicular stomatitis virus entry. J Virol
25. BeerC
AndersenDS
RojekA
PedersenL
2005 Caveola-dependent endocytic entry of amphotropic murine leukemia virus. J Virol 79 10776 10787
26. PakC
PuriA
BlumenthalR
1997 Conformational changes and fusion activity of vesicular stomatitis virus glycoprotein: [125I]iodonaphthyl azide photolabeling studies in biological membranes. Biochemistry 36 8890 8896
27. MelikyanG
2008 Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology 5 111
28. GalloS
2006 Kinetic studies of HIV-1 and HIV-2 envelope glycoprotein-mediated fusion. Retrovirology 3 90
29. MelikyanG
EgelhoferM
von LaerD
2006 Membrane-anchored inhibitory peptides capture human immunodeficiency virus type 1 gp41 conformations that engage the target membrane prior to fusion. J Virol 80 3249 3258
30. ChernomordikL
1998 The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol 140 1369 1382
31. ZavorotinskayaT
QianZ
FranksJ
AlbrittonLM
2004 A point mutation in the binding subunit of a retroviral envelope protein arrests virus entry at hemifusion. J Virol 78 473 481
32. MelikyanGB
BrenerSA
OkDC
CohenFS
1997 Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J Cell Biol 136 995 1005
33. SubramanianRP
GeraghtyRJ
2007 Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc Natl Acad Sci USA 104 2903 2908
34. Jimenez-BarandaS
2007 Filamin-A regulates actin-dependent clustering of HIV receptors. Nat Cell Biol 9 838 846
35. IyengarS
HildrethJ
SchwartzD
1998 Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J Virol 72 5251 5255
36. del RealG
2004 Statins inhibit HIV-1 infection by down-regulating Rho activity. J Exp Med 200 541 547
37. GalloS
PuriA
BlumenthalR
2001 HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 40 12231 12236
38. ViardM
2002 Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells. J Virol 76 11584 11595
39. LambertJM
2002 Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol 4 621 625
40. ViardM
2004 The role of glycosphingolipids in HIV signaling, entry and pathogenesis. Glycoconj J 20 213 222
41. ZandyNL
PendergastAM
2008 Abl tyrosine kinases modulate cadherin-dependent adhesion upstream and downstream of Rho family GTPases. Cell Cycle 7 444 448
42. ZandyNL
PlayfordM
PendergastAM
2007 Abl tyrosine kinases regulate cell-cell adhesion through Rho GTPases. Proc Natl Acad Sci USA 104 17686 17691
43. PraefckeGJK
McMahonHT
2004 The dynamin superfamily: universal membrane tubulation and fission molecules? 5 133 147
44. YuD
2009 The HIV Envelope but Not VSV Glycoprotein Is Capable of Mediating HIV Latent Infection of Resting CD4 T Cells. PLoS Pathog 5 e1000633
45. KruegerEW
OrthJD
CaoH
McNivenMA
2003 A Dynamin-Cortactin-Arp2/3 Complex Mediates Actin Reorganization in Growth Factor-stimulated Cells. Mol Biol Cell 14 1085 1096
46. BackertS
FellerSM
WesslerS
2008 Emerging roles of Abl family tyrosine kinases in microbial pathogenesis. Trends in Biochemical Sciences 33 80 90
47. SwinglerS
2007 Apoptotic killing of HIV-1 infected macrophages is subverted by the viral envelope glycoprotein. PLoS Pathog 3 e134
48. PontowS
RatnerL
2001 Evidence for common structural determinants of human immunodeficiency virus type 1 coreceptor activity provided through functional analysis of CCR5/CXCR4 chimeric coreceptors. J Virol 75 11503 11514
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin NeutralizationČlánek Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane KinasesČlánek Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 6- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Cognitive Dysfunction Is Sustained after Rescue Therapy in Experimental Cerebral Malaria, and Is Reduced by Additive Antioxidant Therapy
- DNA Watermarking of Infectious Agents: Progress and Prospects
- Self-Protection against Gliotoxin—A Component of the Gliotoxin Biosynthetic Cluster, GliT, Completely Protects Against Exogenous Gliotoxin
- Modifies the Tsetse Salivary Composition, Altering the Fly Feeding Behavior That Favors Parasite Transmission
- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- The Enteropathogenic Effector EspF Targets and Disrupts the Nucleolus by a Process Regulated by Mitochondrial Dysfunction
- Epithelial p38α Controls Immune Cell Recruitment in the Colonic Mucosa
- Coexpression of PD-1, 2B4, CD160 and KLRG1 on Exhausted HCV-Specific CD8+ T Cells Is Linked to Antigen Recognition and T Cell Differentiation
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- EspA Acts as a Critical Mediator of ESX1-Dependent Virulence in by Affecting Bacterial Cell Wall Integrity
- An RNA Element at the 5′-End of the Poliovirus Genome Functions as a General Promoter for RNA Synthesis
- Tetherin Restricts Productive HIV-1 Cell-to-Cell Transmission
- Epstein-Barr Virus-Encoded LMP2A Induces an Epithelial–Mesenchymal Transition and Increases the Number of Side Population Stem-like Cancer Cells in Nasopharyngeal Carcinoma
- Protein Kinase A Dependent Phosphorylation of Apical Membrane Antigen 1 Plays an Important Role in Erythrocyte Invasion by the Malaria Parasite
- Requirement for Ergosterol in V-ATPase Function Underlies Antifungal Activity of Azole Drugs
- The SOCS-Box of HIV-1 Vif Interacts with ElonginBC by Induced-Folding to Recruit Its Cul5-Containing Ubiquitin Ligase Complex
- A Crucial Role for Infected-Cell/Antibody Immune Complexes in the Enhancement of Endogenous Antiviral Immunity by Short Passive Immunotherapy
- Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator
- Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step
- Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane Kinases
- Epigenetic Repression of by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP
- Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
- Role of PKR and Type I IFNs in Viral Control during Primary and Secondary Infection
- A Kinome RNAi Screen Identified AMPK as Promoting Poxvirus Entry through the Control of Actin Dynamics
- Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity
- NleG Type 3 Effectors from Enterohaemorrhagic Are U-Box E3 Ubiquitin Ligases
- Human Cytomegalovirus UL29/28 Protein Interacts with Components of the NuRD Complex Which Promote Accumulation of Immediate-Early RNA
- Complement Receptor 1 Is a Sialic Acid-Independent Erythrocyte Receptor of
- A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs
- Immunotoxin Complementation of HAART to Deplete Persisting HIV-Infected Cell Reservoirs
- Protein Expression Redirects Vesicular Stomatitis Virus RNA Synthesis to Cytoplasmic Inclusions
- Entry and Fusion of Emerging Paramyxoviruses
- Paramyxovirus Entry and Targeted Vectors for Cancer Therapy
- Suppressing Glucose Transporter Gene Expression in Schistosomes Impairs Parasite Feeding and Decreases Survival in the Mammalian Host
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Fungal Cell Gigantism during Mammalian Infection
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
- Rotavirus Structural Proteins and dsRNA Are Required for the Human Primary Plasmacytoid Dendritic Cell IFNα Response
- The Epigenetic Landscape of Latent Kaposi Sarcoma-Associated Herpesvirus Genomes
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



