-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Detection of Host-Derived Sphingosine by Is Important for Survival in the Murine Lung
Pseudomonas aeruginosa is a common environmental bacterium that is also a significant opportunistic pathogen, particularly of the human lung. We must understand how P. aeruginosa responds to the lung environment in order to identify the regulatory changes that bacteria use to establish and maintain infections. The P. aeruginosa response to pulmonary surfactant was used as a model to identify transcripts likely induced during lung infection. The most highly induced transcript in pulmonary surfactant, PA5325 (sphA), is regulated by an AraC-family transcription factor, PA5324 (SphR). We found that sphA was specifically induced by sphingosine in an SphR-dependent manner, and also via metabolism of sphingomyelin, ceramide, or sphingoshine-1-phosphate to sphingosine. These sphingolipids not only play a structural role in lipid membranes, but some are also intracellular and intercellular signaling molecules important in normal eukaryotic cell functions as well as orchestrating immune responses. The members of the SphR transcriptome were identified by microarray analyses, and DNA binding assays showed specific interaction of these promoters with SphR, which enabled us to determine the consensus SphR binding site. SphR binding to DNA was modified by sphingosine and we used labeled sphingosine to demonstrate direct binding of sphingosine by SphR. Deletion of sphR resulted in reduced bacterial survival during mouse lung infection. In vitro experiments show that deletion of sphR increases sensitivity to the antimicrobial effects of sphingosine which could, in part, explain the in vivo phenotype. This is the first identification of a sphingosine-responsive transcription factor in bacteria. We predict that SphR transcriptional regulation may be important in response to many sites of infection in eukaryotes and the presence of homologous transcription factors in other pathogens suggests that sphingosine detection is not limited to P. aeruginosa.
Published in the journal: Detection of Host-Derived Sphingosine by Is Important for Survival in the Murine Lung. PLoS Pathog 10(1): e32767. doi:10.1371/journal.ppat.1003889
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003889Summary
Pseudomonas aeruginosa is a common environmental bacterium that is also a significant opportunistic pathogen, particularly of the human lung. We must understand how P. aeruginosa responds to the lung environment in order to identify the regulatory changes that bacteria use to establish and maintain infections. The P. aeruginosa response to pulmonary surfactant was used as a model to identify transcripts likely induced during lung infection. The most highly induced transcript in pulmonary surfactant, PA5325 (sphA), is regulated by an AraC-family transcription factor, PA5324 (SphR). We found that sphA was specifically induced by sphingosine in an SphR-dependent manner, and also via metabolism of sphingomyelin, ceramide, or sphingoshine-1-phosphate to sphingosine. These sphingolipids not only play a structural role in lipid membranes, but some are also intracellular and intercellular signaling molecules important in normal eukaryotic cell functions as well as orchestrating immune responses. The members of the SphR transcriptome were identified by microarray analyses, and DNA binding assays showed specific interaction of these promoters with SphR, which enabled us to determine the consensus SphR binding site. SphR binding to DNA was modified by sphingosine and we used labeled sphingosine to demonstrate direct binding of sphingosine by SphR. Deletion of sphR resulted in reduced bacterial survival during mouse lung infection. In vitro experiments show that deletion of sphR increases sensitivity to the antimicrobial effects of sphingosine which could, in part, explain the in vivo phenotype. This is the first identification of a sphingosine-responsive transcription factor in bacteria. We predict that SphR transcriptional regulation may be important in response to many sites of infection in eukaryotes and the presence of homologous transcription factors in other pathogens suggests that sphingosine detection is not limited to P. aeruginosa.
Introduction
Pseudomonas aeruginosa is a common, Gram negative, environmental bacterium that can cause significant disease as an opportunistic pathogen, particularly in the lung. P. aeruginosa lung infections are prevalent in people with cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD), as well as individuals undergoing mechanical ventilation [1]–[4]. These infections cause significant morbidity and mortality and continue to be a major health care burden [5]–[8]. P. aeruginosa has a large genome by bacterial standards (∼6 Mbp) containing a high proportion of regulatory genes (∼8% are predicted to be transcriptional regulators) [9]. This large regulatory capacity is likely important for P. aeruginosa success as an opportunist, enabling it to rapidly alter gene expression in response to host-derived factors and environmental conditions. Understanding mechanisms by which P. aeruginosa detects and responds to the host could present new avenues to combat these devastating, and often antibiotic resistant [10], opportunistic infections. Our current understanding of P. aeruginosa response to the host come from transcriptional profiling using epithelial cells, mucus, or CF sputum [11], [12]. We were interested in the response of P. aeruginosa to the environment of the distal airway, particularly the response to mammalian pulmonary surfactant, the lipid rich mixture that coats the airway surface liquid of the lungs and participates in both respiratory physiology and host defense (reviewed in [13], [14]). This mixture is rich in phosphatidylcholine (∼75% by mass) but also has a substantial fraction of other phospholipids, cholesterol and its esters, and sphingolipids [15].
Sphingolipids constitute a class of molecules that are critical components of eukaryotic cell membranes. In addition to this structural role in membranes and their biophysical role in pulmonary surfactant, many sphingolipids have been shown to act as signaling molecules that play critical roles in regulation of diverse physiological processes. The broad importance of sphingolipid signaling in eukaryotic hosts has only recently been appreciated, and the rapidly expanding field has many recent reviews [16]–[19]. Sphingosine serves as a backbone component for all sphingolipids, which include the signaling molecules sphingosine-1-phosphate (S1P) and ceramide, as well as the structural lipid sphingomyelin. S1P, in particular, has been intensely studied in the past decade as a potent immune signaling molecule that plays a critical role in diverse immune functions such as lymphocyte trafficking, myeloid cell activation, and epithelial and endothelial barrier function, mediated by five G-protein coupled receptors [20]–[24]. Importantly, S1P is released by endothelial cells and platelets during the acute phase response and therefore plays an important role in the initial response to infection [25]–[28].
A specific transcriptional response to host derived sphingolipids and S1P has never been previously shown in bacteria. Here we have identified P. aeruginosa genes induced in response to mammalian pulmonary surfactant and subsequently characterized a subset of genes that are specifically and directly regulated by sphingosine or via metabolism of S1P, sphingomyelin, or ceramide to sphingosine. This response to sphingosine and its precursors is dependent on an AraC-family transcription factor in response to physiological levels of sphingosine and its precursors. This transcription factor binds sphingosine, which alters its association with DNA. A bacterial system to detect and respond to sphingosine may have broad implications in the modulation of host immune function and aid P. aeruginosa in altering host immune response in the human lung. In support of this prediction, deletion of the sphingosine-responsive transcription factor confers a survival defect during mouse lung infections.
Results
Transcriptional profiling of P. aeruginosa exposed to pulmonary surfactant (Survanta)
Microarray studies were used to identify a group of P. aeruginosa transcripts that were induced when the bacteria were grown in minimal media supplemented with lung surfactant (Survanta). When wild type PAO1 was exposed to minimal media containing lung surfactant compared to minimal medium with pyruvate alone, 125 transcripts (both predicted open reading frames (ORFs) and intergenic regions) were changed more than 3-fold (p<0.05), with 96 being induced (Table S1) and 29 being reduced (Table S2). Of the induced transcripts, 56 were characterized and 40 were predicted or hypothetical, while in the reduced transcript group, 11 were characterized and 18 were predicted or hypothetical. The induced class was dominated by genes from the Anr-regulon (29 genes, 16 of which were recently demonstrated as induced in surfactant [29]) and the choline catabolic pathway (15 genes) [30]–[32]. One observation of note in the induced group is the preponderance of transcripts encoding stress-related proteins including the chaperones hslU, groEL, dnaK, and dnaJ, and the universal stress protein family members sspK, PA1789, PA4352, and PA5027. We were interested in using the response to lung surfactant to identify gene function and novel biology in P. aeruginosa, and thus we have focused on highly induced genes of unknown function.
PA5325 induction by pulmonary surfactant and eukaryotic cells is in response to sphingosine
The PA5325 transcript was induced ∼18 fold in the presence of lung surfactant and was the most highly induced transcript in these experiments (Table S1). The PA5325 gene is divergently transcribed from PA5324, which encodes a probable AraC-family transcription factor that we hypothesized could be the transcriptional regulator of PA5325 (Fig. 1A). The robust induction of PA5325 in the presence of lung surfactant (Fig. 1B) suggested a possible role of this gene in the early stages of lung infection by P. aeruginosa.
Fig. 1. Expression of sphA (PA5325) is induced by sphingosine. 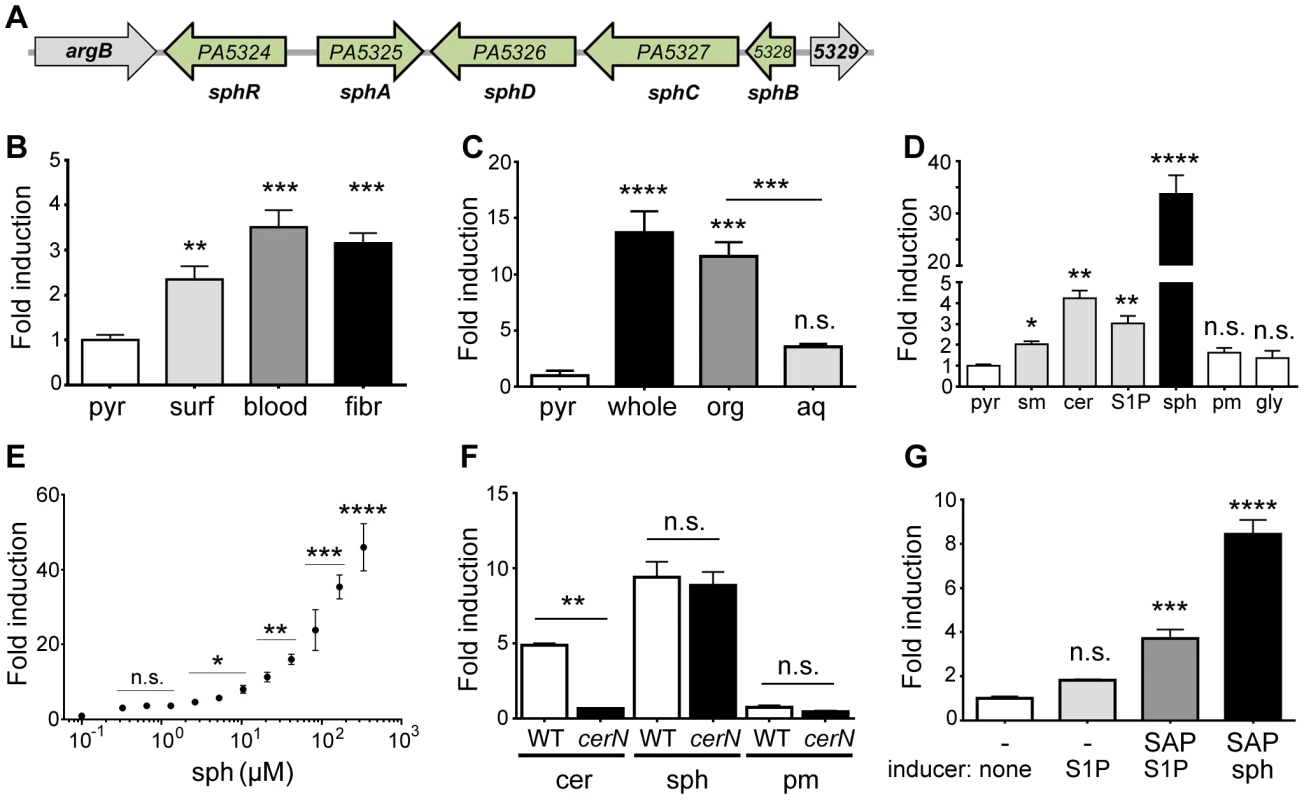
(A) Arrangement of the sphA genomic region in P. aeruginosa PAO1. The genes in green are those discussed further in this study. Data from panels B through F all use a plasmid-borne sphA-lacZ reporter (pAL5) to assess regulation with pyruvate (pyr) used as the non-inducing control condition. Fold induction of sphA-lacZ is calculated compared to its pyruvate control. (B) sphA is induced in the presence of pulmonary surfactant (surf), sheep red blood cells (blood), and mouse fibroblasts (fibr). (C) The primary sphA-inducing component of fibroblasts is present in the organic fraction (org), compared to the aqueous fraction (aq) after the mouse fibroblasts (whole) were extracted with chloroform:methanol. (D) The sphingolipids sphingomyelin (sm), ceramide (cer), sphingosine-1-phosphate (S1P), and sphingosine (sph) induce sphA, while likely catabolic products palmitate (pm) and glycine (gly) do not cause induction. Sphingosine causes the highest level of induction. (E) Induction of sphA by sphingosine is dose dependent and occurs at physiologically relevant concentrations of sphingosine. Lines for statistical significance denote groups of samples with the same magnitude of significance, not grouped comparisons. (F) Induction of sphA by ceramide requires the neutral ceramidase gene, cerN, while sphingosine induction is independent of cerN. (G) Induction of sphA by S1P in a heterologous E. coli sphR-sphA-lacZ reporter system (pAL5) requires phosphatase treatment of S1P. Statistical significance determined using one way ANOVA with Dunnett's post-test for B–E & G with the uninduced condition being the comparator for all other data. In panel F, the wild type and ΔcerN data were compared by student t-test within each treatment condition. p-value summaries: n.s. = not significant; * for p<0.05; ** for p<0.01; *** for p<0.001; **** for p<0.0001. All experiments were performed at least three times and data shown is representative of both the scale and statistical significance levels of all experiments. For the following studies, we generated two reporter plasmids; pAL5 contained both sphR and the PA5325-lacZYA reporter and pAL4 contained only the PA5325-lacZYA reporter. Unless specified, the reporter used was pAL5 as it resulted in more robust induction. In addition to verifying the microarray results with surfactant, the PA5325-lacZ reporter was also induced in response to mouse fibroblasts (L-cells) and defibrinated sheep's blood (Fig. 1B). To determine which component of these eukaryotic-derived mixtures was a specific inducer of PA5325, we extracted mouse fibroblasts into aqueous and organic fractions and tested induction of PA5325-lacZ. The organic fraction contained the inducing activity, suggesting a lipid or other hydrophobic compound (Fig. 1C). Mouse fibroblasts and sheep's blood both contain high percentages of sphingomyelin [33], [34], and sphingomyelin makes up ∼4% of lung surfactant. Therefore, we tested induction of PA5325 by sphingomyelin and related sphingolipids including ceramide, S1P, and sphingosine. PA5325 was induced by sphingomyelin, ceramide, S1P, and sphingosine, but not the likely degradation products of sphingosine: palmitate and glycine (Fig. 1D). Other common lipid components of surfactant such as phosphatidylcholine and cholesterol did not induce transcription of PA5325-lacZ, and neither did unsaturated fatty acids (Supplemental Fig. S1). The strong induction by sphingosine compared to the other sphingolipids led us to hypothesize that the specific inducer of PA5325 is sphingosine. We tested the sensitivity of our PA5325-lacZ reporter to sphingosine (Fig. 1E), which demonstrated PA5325-lacZ induction in a dose-dependent manner and showed response to physiological levels of sphingosine, which range from 200 nM (as S1P) in the serum and lymph up to 2–13 mM of free sphingosine in the skin and some epithelial surfaces [35], [36]. We did not reach saturation in this assay due to a combination of sphingosine insolubility, plastic binding, and bactericidal effects (discussed below).
We hypothesized that the reduced induction of PA5325-lacZ in response to sphingomyelin, S1P, and ceramide compared to sphingosine was due to a processing step required by P. aeruginosa to yield sphingosine. To test this hypothesis we generated a clean deletion in the neutral ceramidase encoded by PA0845 and measured enzyme activity from the PA5325-lacZ reporter construct pAL4. Ceramide fails to induce PA5325-lacZ in the absence of the neutral ceramidase, whereas the response to sphingosine was unaffected (Fig. 1F). This finding strongly supports our hypothesis that induction of PA5325 occurs in response to sphingosine. In addition, PA5325-lacZ is induced in response to S1P in P. aeruginosa (Fig. 1D), but not significantly induced by S1P in E. coli (Fig. 1G), although the reporter in E. coli could still be induced in the presence of sphingosine (Fig. 1G). This suggested that P. aeruginosa may be processing S1P and that E. coli does not possess an orthologous activity under these conditions. When S1P was pretreated with shrimp alkaline phosphatase, induction of PA5325-lacZ was partially restored in E. coli (Fig. 1G). Given the transcriptional control of PA5325 in response to sphingosine, we have renamed it sphingosine regulated gene A, sphA.
sphA transcription is dependent on PA5324 (sphR)
PA5324 encodes a predicted AraC-family transcription factor divergently transcribed from sphA (Fig. 1A). This arrangement led us to suspect that PA5324 was the transcriptional regulator of sphA. To confirm the requirement of PA5324 for induction of sphA we generated an in-frame deletion of PA5324. The PA5324 deletion strain carrying our sphA-lacZ reporter construct (pAL4) showed no induction in the presence of sphingosine (Fig. 2A). Insertion of PA5324 onto the chromosome at the attTn7 site restored induction in the deletion strain (Fig. 2A). Furthermore, PA5324 was necessary to induce sphA-lacZ in a heterologous E. coli system in response to sphingosine (Fig. 2B). Our data suggest that the sphingosine responsiveness via sphA transcription is dependent on PA5324, and PA5324 was sufficient to confer sphingosine responsiveness in an E. coli system, therefore we have renamed PA5324 as the Sphingosine-responsive Regulator, SphR.
Fig. 2. The transcription regulator SphR (PA5324) controls sphA induction in response to sphingosine. 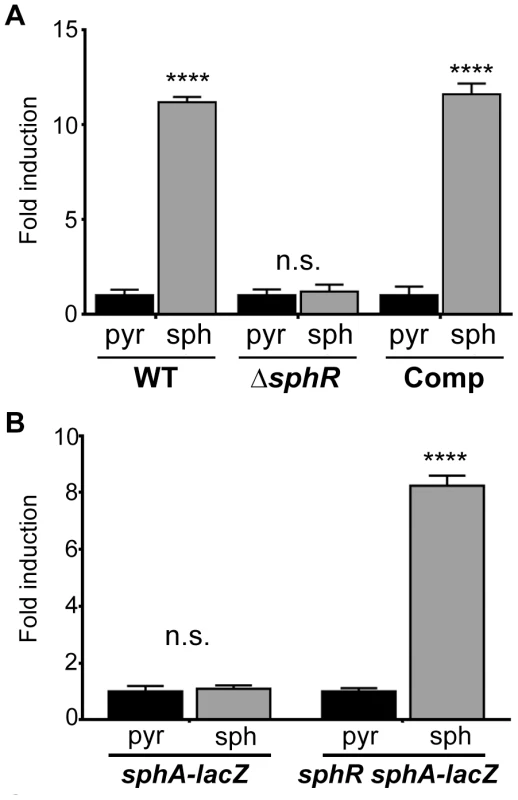
(A) Sphingosine (sph) induces sphA-lacZ expression (pAL4) compared to the pyruvate (pyr) control in wild-type cells (wild type) but not in sphR mutant cells (ΔsphR). This regulation is restored by complementation (Comp) of sphR at the attTn7 locus. (B) In a heterologous E. coli system, the sphA-lacZ reporter (pAL4) is not responsive to sphingosine (second bar) unless the sphR gene is included in the system (pAL5) (fourth bar). Data for these panels were compared by student t-test comparing treatment conditions within each strain. p-value summaries: n.s. = not significant; **** for p<0.0001. All experiments were performed at least three times and data shown is representative of both the scale and statistical significance levels of all experiments. ΔsphR mutants have a survival defect during mouse lung infection
Our a priori prediction was that SphR, controlling expression of a strongly induced gene by pulmonary surfactant, would be important for colonization and/or survival in the mammalian lung. To test this hypothesis, we examined bacterial survival 24 hours after infection in the mouse lung. The sphR deletion strain had significantly lower survival than wild type (7.7-fold decrease, Dunnett's multiple comparisons p<0.001) and the survival defect was complemented by addition of sphR at the attTn7 site (Fig. 3). In this comparison, wild type and ΔsphR both contained the empty attTn7 insertion cassette on the chromosome. The contribution of sphR to survival in the mouse lung led us to a more in-depth study of SphR and its target genes.
Fig. 3. Deletion of sphR reduces P. aeruginosa survival in the mouse lung. 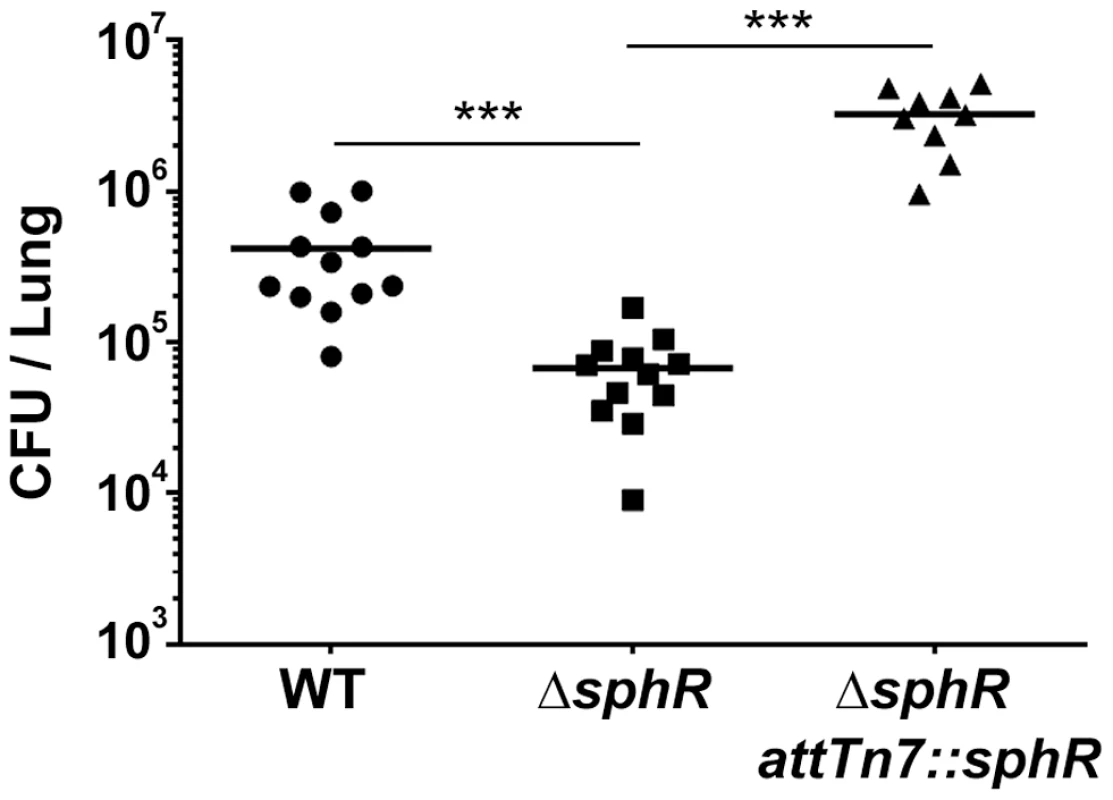
Male C57Bl/6J mice were infected with 2×107 CFU/mouse of each strain via oropharyngeal aspiration. Mice were euthanized and lungs harvested 24 hours after infection. Bacterial counts were determined by serial dilution onto Pseudomonas Isolation Agar (PIA). Deletion of sphR (ΔsphR) reduced P. aeruginosa survival 7.7-fold (p<0.001), an effect that was complemented by addition of sphR at the attTn7 site. Wild-type (WT) and ΔsphR cells carried the empty attTn7 insertion cassette as described in the methods section. Statistical significance determined using one way ANOVA with Tukey's post-test comparing all groups to each other. p-value summaries: n.s. = not significant; *** for p<0.001. Data shown is combined from two experiments. The same effect sizes and variance have been seen in at least one additional experiment for each strain, resulting in each strain having been compared to wild type in at least three independent experiments (described further in the Methods section). Determination of the SphR regulon
Deletion of sphR resulted in reduced P. aeruginosa survival in the mouse lung (Fig. 3), leading us to hypothesize that one or more of the genes in the SphR regulon were likely candidates for this phenotype. To identify SphR-regulated genes in addition to sphA, we conducted microarray transcriptome analyses to compare wild type and the sphR deletion mutant in the presence and absence of pulmonary surfactant. Using a two-fold change cutoff and a p-value <0.05, there are six genes that differ between wild type and ΔsphR in the presence of surfactant (Table 1). Transcripts that are induced in wild type but not in the sphR deletion mutant include sphA, the neutral ceramidase (PA0845), and a three gene operon convergently transcribed toward sphA, PA5328-PA5326. The argB gene (PA5323) was induced more strongly in the sphR deletion than in wild type, which we think is likely due to a cis effect of the sphR (PA5324) deletion, as these genes are convergently transcribed (Fig. 1A). To denote their placement in the SphR regulon, we have renamed the genes in the predicted PA5328-PA5326 operon as sphBCD. The sphB gene encodes a predicted periplasmic cytochrome and sphC and sphD encode a predicted flavin-dependent oxidoreductase and a predicted pyridoxalphosphate-containing threonine aldolase-like enzyme, respectively. The predicted functions of SphC and SphD suggest a potential two-step pathway for sphingosine degradation to glycine and a long chain aldehyde by oxidation to an aldol and subsequent cleavage by the aldolase, a prediction we are currently exploring. The neutral ceramidase (PA0845) was previously designated PaCD [37], which does not conform to standard bacterial nomenclature. We propose that PA0845 be renamed cerN for ceramidase, neutral. The induction of sphA by surfactant in wild type (17.8-fold) versus the difference of sphA induction between wild type and ΔsphR (5.9-fold) suggested altered regulation of sphA in the absence of sphR (Table 1). The relative induction of sphA in the sphR mutant compared to wt under pyruvate (non-inducing) conditions supports a de-repression of sphA transcription in the absence of sphR at baseline. The remaining genes in the operon appear solely regulated by SphR under these conditions, as their induction levels in wild type compared to the difference between wild type and ΔsphR are not different.
Tab. 1. The SphR (PA5324) regulon in P. aeruginosa. 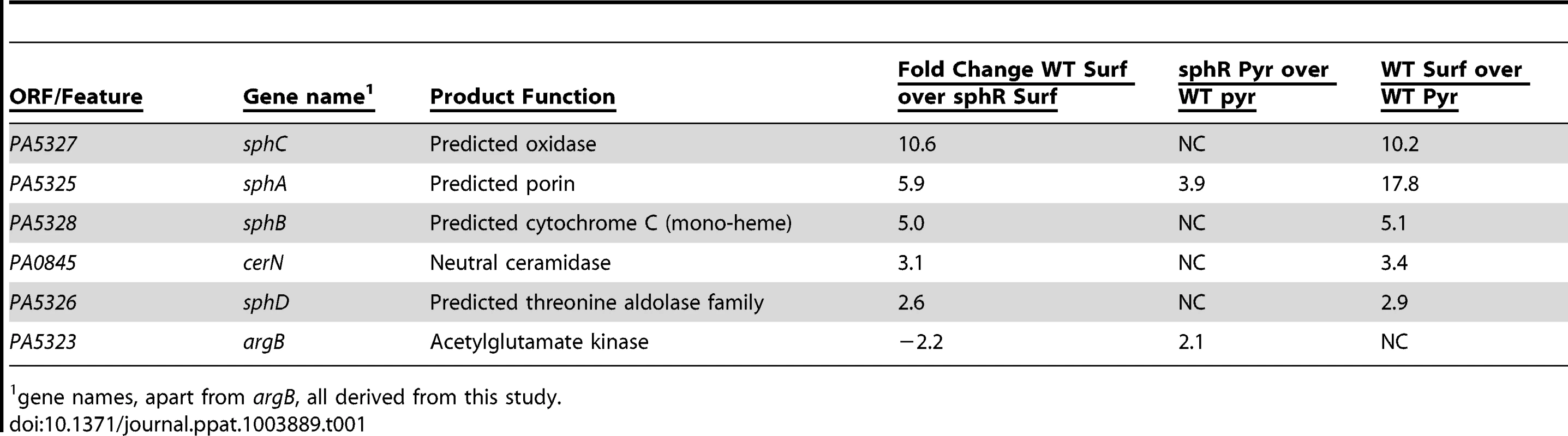
1 gene names, apart from argB, all derived from this study. Mapping of the sphA, sphBCD, and cerN promoters
We used promoter mapping to identify the promoter proximal regions of the sphA, sphBCD, and cerN promoters that were important for sphingosine and sphR-dependent regulation. Using lacZ reporter fusions to each upstream region, we identified a portion of each promoter-proximal region required for responsiveness to sphingosine (Fig. 4A). The regions required for sphingosine responsiveness were aligned using KALIGN [38], which produced an alignment that highlights the general format of an AraC-family binding site (Fig. 4B). The MEME consensus for a single half-site is shown below the alignment (Fig. 4B). Bioinformatic search of the P. aeruginosa genome (DNA Motif Search [39]) turned up only one additional predicted binding site (two direct repeats of the consensus (TGNCCSNNRNNSNCC) separated by 6–8 bp) in the genome apart from those present in the three identified promoters. The additional binding site is in the intergenic region between PA0428 and PA0429, upstream of the PA0428 gene. We did not detect any change in the PA0428 transcript for wild type or ΔsphR in the presence of surfactant or in either strain in the absence of surfactant. Therefore, based on our microarray data and bioinformatic analysis, we predict that sphA, sphBCD, and cerN likely comprise the core SphR regulon. The upstream sequences for the SphR regulon members showing the predicted SphR binding sites, promoter elements, and ribosome binding sites are shown in Supplemental Figure S2.
Fig. 4. Determination of the probable SphR binding site from SphR-regulated promoters. 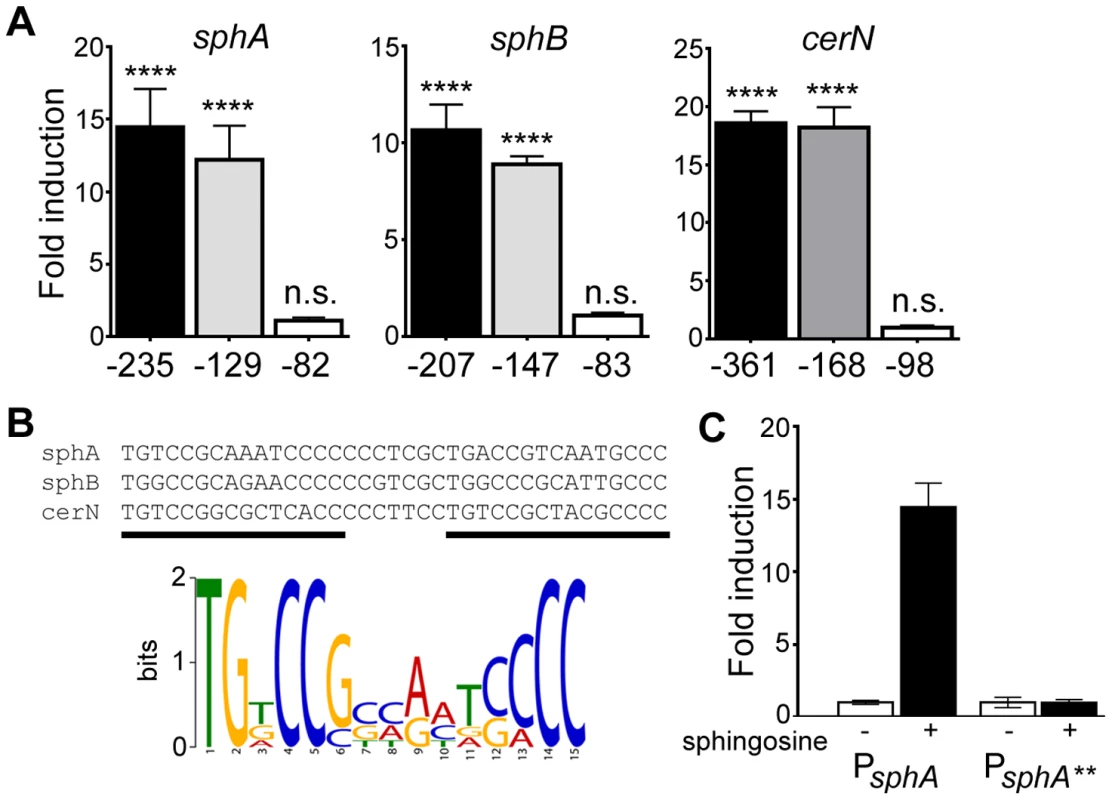
(A) Fold-induction of ß-galactosidase activity from each reporter and the truncations compared to a pyruvate non-induced control (not shown). Promoter deletion mapping demonstrated a minimal region required for each SphR-controlled transcript. The negative numbers below each panel refer to the position relative to the translational start of each gene. (B) KALIGN showing the nucleotide alignment of the conserved region from the minimal regulatory regions defined in (A). The black bars denote the two direct repeat half-sites typical of AraC-family transcription factors. Below the alignments is the MEME-generated logo showing the strength of conservation based on the six half-sites from these three promoters. (C) ß-galactosidase assay for wt sphA-lacZ and the sphA-lacZ** promoter mutant (TG at the beginning of the first half-site was changed to AA) to demonstrate the importance of the conserved region for induction. Statistical significance determined using one way ANOVA with Dunnett's post-test with the uninduced pyruvate condition being the comparator for all other data. p-value summaries: n.s. = not significant; **** for p<0.0001. All experiments were performed at least three times and data shown is representative of both the scale and statistical significance levels of all experiments. To test both specificity and the importance of conserved consensus sequences we mutated the first two residues in the consensus sequence TG to AA in half-site 1 (sphA**) (Fig. 4B), and tested the ability of the mutant sequence to permit induction of the reporter gene in response to sphingosine. The mutant reporter was unable to support reporter induction in response to sphingosine (Fig. 4C), demonstrating the importance of these conserved binding site residues.
SphR directly binds the sphA, sphBCD, and cerN promoter proximal regions
We conducted electrophoretic mobility shift assays (EMSAs) with purified MBP-SphR fusion protein to test if SphR directly bound the sphA, sphBCD, and cerN promoters. The binding of MBP-SphR to the sphA promoter probe was greatly enhanced by the addition of sphingosine to the binding reaction in a concentration-dependent manner, providing evidence that sphingosine was a direct ligand of SphR (Fig. 5A). In the presence of sphingosine, MBP-SphR specifically shifted the sphA, sphBCD, and cerN promoters in a protein concentration-dependent manner and the binding could be competed with unlabeled sphA promoter probe, which gives a sense of the relative affinities for each binding site (Fig. 5B). MBP-SphR did not shift the non-specific plcH probe (Fig. 5B). The plcH probe is a useful negative control and demonstrates the specificity of SphR binding, as it has a known binding site for the AraC-family transcription factor GbdR in P. aeruginosa and its regulation is well described [31], [40]–[43].
Fig. 5. SphR directly binds to its target regulatory regions and binding is stimulated by sphingosine. 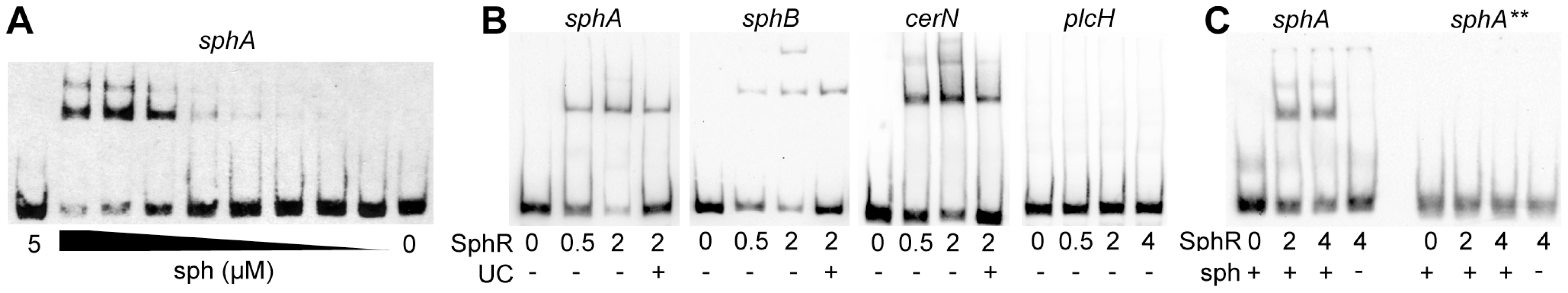
The binding of MBP-SphR to target DNA was measured using electrophoretic mobility shift assays (EMSAs). (A) Sphingosine (sph) is required for robust MBP-SphR binding to target DNA and sphingosine stimulates SphR DNA binding in a dose-dependent manner. Based on this data all subsequent EMSAs were conducted with 10 µM sphingosine. (B) MBP-SphR (SphR) binds in a dose-dependent manner to sphA, sphB, and cerN upstream regulatory regions, but not to the sphR-independent plcH regulatory region. The MBP-SphR concentration (µM) is shown below the lanes. Specificity is shown by lack of MBP-SphR binding to the plcH promoter and by the ability to compete the bulk of the shift with unlabelled competitor DNA (UC) denoted by the + sign under the lanes. (C) MBP-SphR binds a 59-bp oligonucleotide probe containing the predicted SphR binding site upstream of sphA. The extra shifts seen in panels A & B are absent from the EMSAs with these minimal probes and likely are due to additional interaction sites. Mutation of two conserved residues in half-site #1 (TG to AA) (sphA**) results in substantial reduction in MBP-SphR binding. All experiments were performed at least three times and data shown is representative of all experiments. To test the predicted SphR binding site, 59-mer oligonucleotides containing the proposed SphR binding site from the sphA promoter were annealed and the resultant probe was used in binding reactions. MBP-SphR was able to shift the 59-bp sphA probe (Fig. 5C, left), but only in the presence of sphingosine. Based on the inability of the mutated consensus sequence (sphA**) to support sphingosine-dependent reporter expression (Fig. 4C), we predicted that an oligonucleotide carrying these mutations would also be unable to bind SphR. As shown in the right side of Figure 5C, MBP-SphR was unable to bind this mutated probe. Together with the reporter fusions, these data support both the specificity of SphR binding and the importance of the conserved residues in the consensus.
SphR binds sphingosine
Based on the enhancement of SphR DNA binding in the presence of sphingosine and our genetic evidence, we predicted that SphR would directly bind sphingosine. We used 3H-sphingosine to test the ability of SphR to bind sphingosine (Fig. 6). The binding assay conditions were similar to those used for EMSA studies with MBP-SphR in the presence of 3H-sphingosine. Amylose resin beads were used to pull down the MBP-SphR, and bead-associated sphingosine was assayed by liquid scintillation counting. 3H-sphingosine was substantially enriched in the fraction containing amylose-bound MBP-SphR, while relatively little remained associated with the amylose beads alone, or beads bound to a non-specific MBP-tagged P. aeruginosa AraC-family transcription factor, CdhR (MBP-CdhR) [44]. These data, in combination with the EMSAs (Fig. 5), demonstrate direct interaction between sphingosine and SphR.
Fig. 6. SphR directly binds to sphingosine. 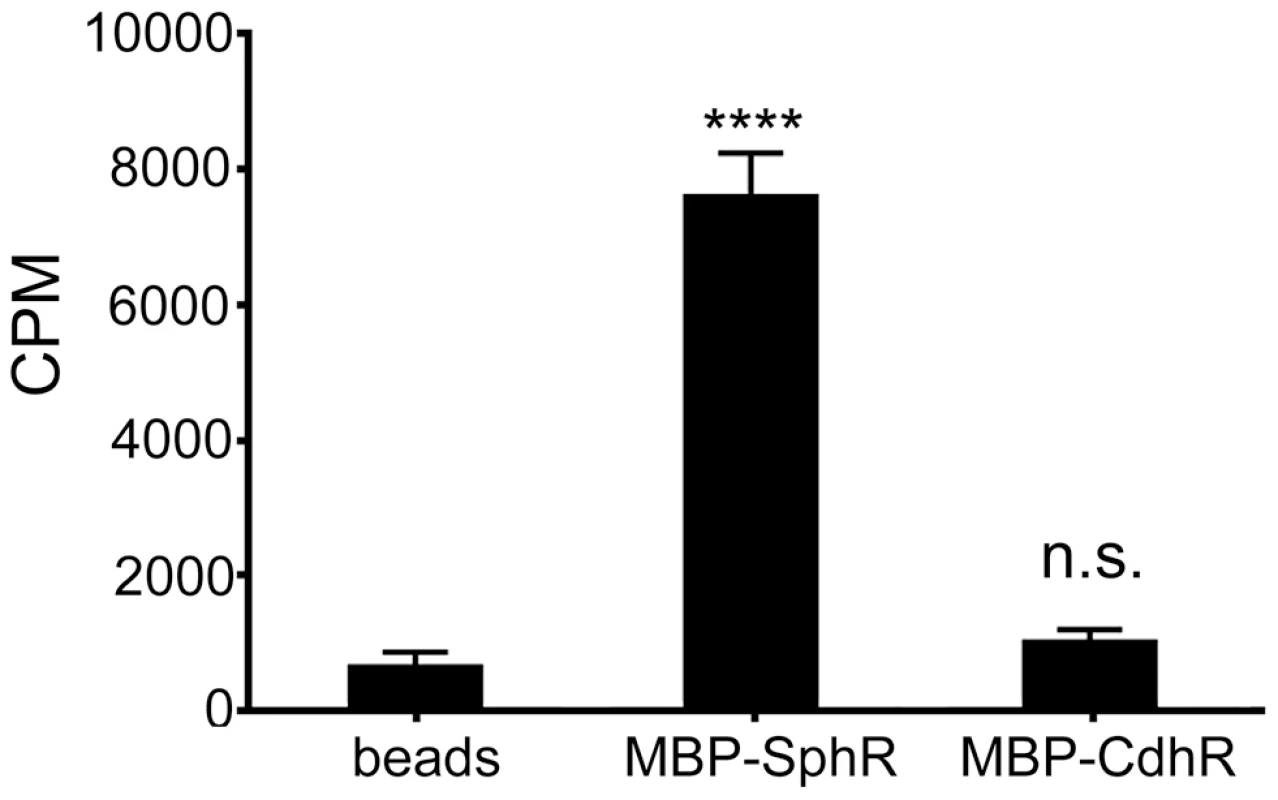
The association of MBP-SphR was determined by measuring binding of 3H-sphingosine and reporting counts per minute (CPM). Sphingosine minimally associates with amylose beads alone (beads) or an un-related AraC-family transcription factor (MBP-CdhR), but approximately 11-fold more 3H-sphingosine binds to MBP-SphR (p<0.0001). Statistical significance determined using one way ANOVA with Dunnett's post-test with the beads alone condition being the comparator for all other data. p-value summaries: n.s. = not significant; **** for p<0.0001. This experiment was performed at least three times and data shown is representative of both the scale and statistical significance levels of all experiments. Deletion of sphA phenocopies the mouse survival phenotype of the sphR deletion
Because deletion of sphR led to reduced survival in the mouse lung, we were interested in determining which of the SphR regulon members contributed to survival in the lung. We generated deletions in cerN, sphA, and sphC and compared to wild type in our 24 hour lung infection model. Deletion of sphA led to a significant reduction in bacterial survival in the mouse lung (9-fold decrease, Dunnett's multiple comparisons p<0.001), while deletion of cerN or sphC had no impact on bacterial survival in vivo (Fig. 7). The sphA mutant phenotype could be complemented by supplying the sphA under its native promoter control at the attTn7 site (Supplemental Fig. S3). These data suggest an important role for sphA in survival during infection. We did not test deletions of sphB and sphD in the animal model, given their predicted coordinate role with sphC in sphingosine metabolism and their similar phenotype to an sphC deletion during in vitro sphingosine killing (Fig. 8 and Figure S4).
Fig. 7. Deletion of the SphR-regulon member, sphA, reduces P. aeruginosa survival in the mouse lung. 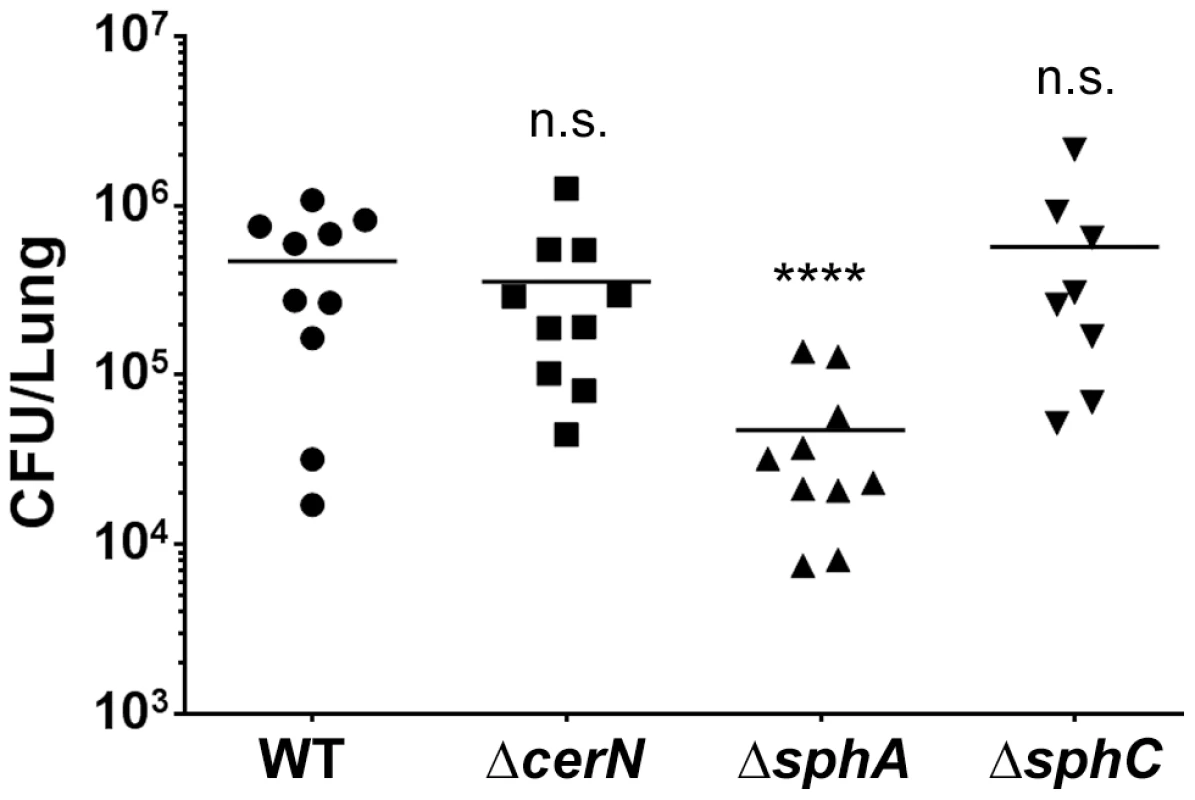
Male C57Bl/6J mice were infected with 2×107 CFU/mouse of each strain via oropharyngeal aspiration. Mice were euthanized and lungs harvested 24 hours after infection. Bacterial counts were determined by serial dilution onto PIA. Deletion of sphA (ΔsphA) reduced P. aeruginosa survival 9-fold (p<0.001). Deletion of the other SphR-regulon members cerN and sphC (as an sphBCD operon representative) resulted in no survival defect. Statistical analysis determined using one way ANOVA with Dunnett's post-test with wild type as the comparator. Data shown is combined from two experiments. The same effect sizes and variance have been seen in at least one additional experiment for each strain, resulting in each strain having been compared to wild type in at least three independent experiments (described further in the Methods section). Fig. 8. Deletion of sphA or sphR renders P. aeruginosa susceptible to killing by sphingosine. 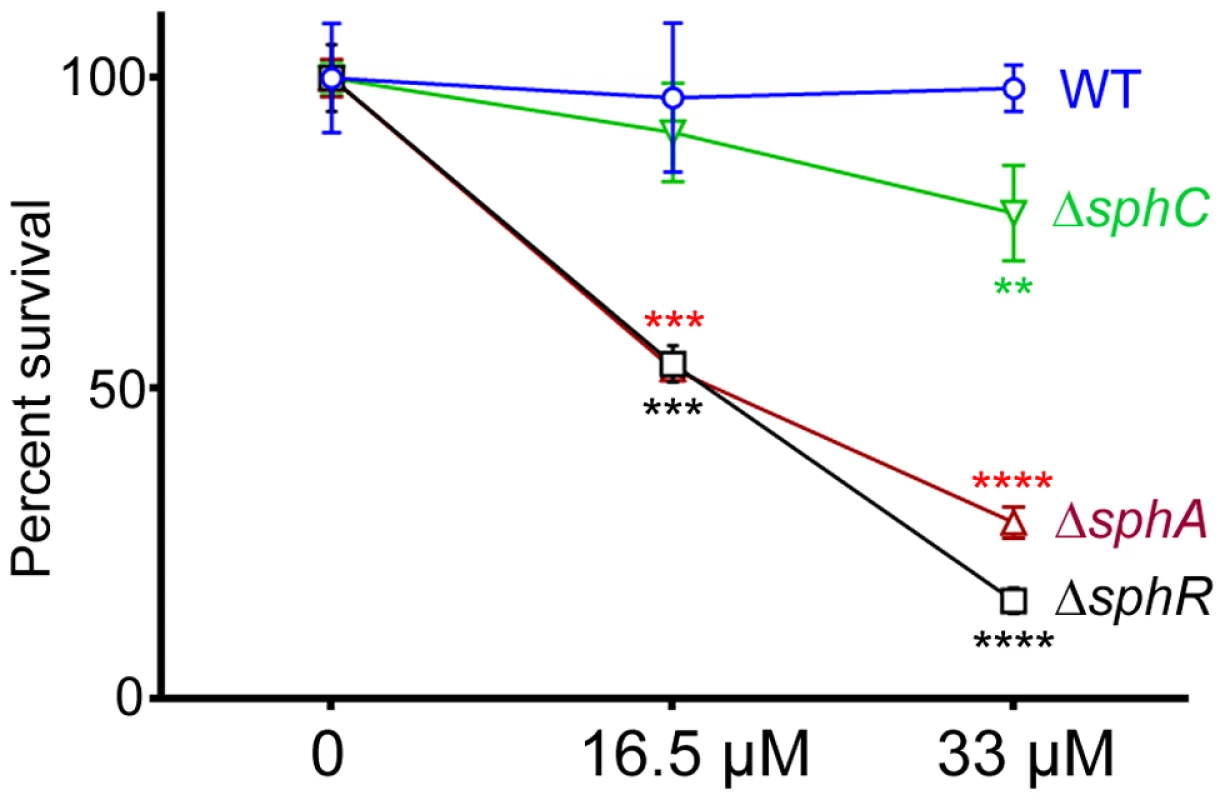
Strains with deletions in sphA (red), sphR (black), and sphC (green) were compared to wild type (blue) using a sphingosine killing assay in neopeptone as described in the Methods section. Cells were exposed to vehicle alone (0) or one of two concentrations of sphingosine for 30 minutes followed by serial dilution and plating. The mean of the vehicle treatment samples for each strain was set as 100% survival for that strain. Statistical significance determined using one way ANOVA with Dunnett's post-test with wild type at each concentration being the comparator for the mutant strain data at the same concentration. p-value summaries: n.s. = not significant; ** for p<0.01; *** for p<0.001; **** for p<0.0001. This experiment was performed more than three times and data shown is representative of both the scale and statistical significance levels of all experiments. SphR & SphA are important for resistance to sphingosine killing
Sphingosine has previously been shown to have antimicrobial properties and is able to inhibit growth and kill many Gram positive and Gram negative bacteria [19]. Previous studies suggest that P. aeruginosa is not sensitive to killing by sphingosine [45]. We hypothesized that SphR might play a role in the response of P. aeruginosa to sphingosine and could regulate sphingosine resistance. Using a modified sphingosine killing assay, we show that the ΔsphR deletion strain is more sensitive to sphingosine compared to wild type (Fig. 8), an effect that could be complemented by sphR on a plasmid (Supplemental Fig. S4). Most of the sensitivity of the ΔsphR strain appears to be due to loss of sphA induction, as the ΔsphA strain is also more sensitive to sphingosine than wild type and is nearly as sensitive as ΔsphR (Fig. 8). The deletion phenotype of sphA could be complemented by sphA on a plasmid (Supplemental Fig. S4). Deletion of sphC and transposon insertions into sphD and sphB also led to small but reproducible decreases in survival on sphingosine, suggesting a minor role for this operon in the response to sphingosine (Fig. 8 and Supplemental Fig. S4). Deletion of cerN, befitting its known function as an extracellular ceramidase, had no effect on survival in sphingosine (data not shown).
Discussion
The induction of ceramidase activity in response to sphingosine has been demonstrated in a few bacteria [37], but the mechanism of sphingosine detection and conversion into a response had not previously been elucidated. In this study we show that sphingosine is directly detected by the AraC-family transcription factor SphR (PA5324) leading to the induction of sphA, sphBCD, and cerN transcripts. Deletion of sphR or sphA resulted in survival defects in a mouse model of acute pneumonia, suggesting that the ability to detect and respond to host-derived sphingolipids is important for survival in the lung. Sphingolipids are abundant in mammals, plants, and fungi, constituting a diverse family of molecules that serve as essential structural components of eukaryotic cell membranes and as dynamic signaling molecules that mediate diverse cellular functions [16]–[19]. In particular, S1P has been implicated as a critical component of mammalian innate and adaptive immune function, particularly in the acute phase response to pathogens [25]–[28]. Interestingly, orthologs of SphR and some of the SphR-regulon members are present in other opportunistic pathogens including Acinetobacter haemolyticus and Burkholderia pseudomallei, as well as the professional pathogen Mycobacterium tuberculosis.
Sphingolipids play important roles in host-pathogen interactions, particularly S1P and ceramide signaling [46]–[48]. In addition to host modulation of sphingolipid pathways to combat infection, pathogens can modulate host sphingolipids. M. tuberculosis alters sphingolipid signaling in macrophages by undetermined mechanisms [49], and S1P levels in the lungs of patients infected with M. tuberculosis are significantly decreased [50]. Interestingly, M. tuberculosis has an AraC-family transcription factor that is 47% similar along the whole length to SphR (RV1395) that was identified though signature-tagged mutagenesis where the RV1395 transposon mutant strain had an ∼1.5 log reduced survival in a mouse lung infection model [51]. Similarity between RV1395 and SphR is not restricted to the helix-turn-helix DNA-binding domain, as the two proteins are 44% similar when the DNA-binding domain is removed from the alignment analysis. RV1395 was characterized and found to be an activator of a divergently transcribed cytochrome gene, however the signals that govern RV1395 activation and its direct contribution to virulence have yet to be determined [52]. Based on the similarity of RV1395 to SphR we predict that a sphingolipid, perhaps sphingosine, may be the inducing ligand of RV1395.
The AraC-family transcription regulators are one of the largest groups of regulatory proteins in bacteria, and are often involved in the regulation of catabolism, stress response, and virulence [53]. Many members of the AraC family have been shown to respond to host-derived chemical signals present at the site of infection, but relatively few inducing ligands have been demonstrated to bind directly to their cognate regulator [54]. We found that addition of sphingosine altered the binding of SphR to the sphA promoter in EMSA studies and observed a dose response curve of SphR DNA binding at physiologically relevant concentrations of sphingosine. Bioinformatic analysis suggest similarity of SphR to ToxT (44% similarity and 20% identity), which directly regulates the major virulence factors in Vibrio cholerae. ToxT activation is inhibited by unsaturated fatty acids found in bile [55]. Subsequently, the crystal structure of ToxT was solved revealing a bound 16-carbon fatty acid that alters the structure of ToxT to prevent DNA binding in the presence of these bile associated fatty acids [56]. The similar size and hydrophobic nature of the regulatory ligands (palmitate vs. sphingosine) coupled with the sequence similarity allows us to speculate that SphR may bind sphingosine in a manner analogous to ToxT binding of palmitate.
Ito et al. identified a neutral ceramidase encoded by PA0845 (renamed cerN in this study) that was induced in the presence of sphingomyelin, ceramide and sphingosine, however the regulatory mechanism was not reported [37]. The discovery of SphR control of neutral ceramidase allows us to expand a model of bacterial utilization of sphingomyelin by linking it to our previous work on regulation of the phospholipase C/sphingomyelinase PlcH. We previously characterized the AraC-family regulator GbdR that is integral to a positive feedback loop controlling PlcH expression in response to a metabolite of the choline headgroup of sphingomyelin [31]. Sphingomyelin hydrolysis by PlcH yields ceramide [57], which P. aeruginosa can further metabolize through the action of ceramidases [54]. Here we show that CerN is produced as part of an SphR-dependent positive feedback loop in response to the ceramide metabolite sphingosine, in a manner analogous to GbdR control of PlcH. Both of these positive feedback loops link induction of secreted catabolic enzymes not to the availability of the substrate itself, but to metabolic products derived from the substrate. In each case, this ensures that the positive feedback loop will robustly operate only if the substrate is being metabolized at sufficient rates.
Sphingolipids such as sphingosine have long been known to have antimicrobial properties and sphingosine is found in high concentration in the skin where it is thought to be part of the barrier function against microbial infections [58]–[61]. A variety of Gram positive and Gram negative bacteria are sensitive to sphingosine, including Staphylococcus aureus and Escherichia coli [62]. The precise bactericidal mechanism of sphingosine remains unknown. However, recent evidence suggests that sphingosine may directly damage bacterial membranes [63]. P. aeruginosa has recently been reported to be resistant to the bactericidal effects of sphingosine [45]. While none of the deletion strains generated in this study showed growth defects under normal conditions, we found that both the sphR and sphA deletion strains were susceptible to the antimicrobial effects of sphingosine compared to wild type in vitro. Strains with deletions in sphR and sphA were also shown to have reduced survival in the mouse lung. We hypothesize that the sensitivity of sphA and sphR mutants to sphingosine contributes to their observed reduced survival in vivo. It is interesting to note that the double deletion ΔcerNΔsphA strain did not survive better or worse than ΔsphA, minimally suggesting that if the defect is due to sphingosine sensitivity, it is not sphingosine derived from P. aeruginosa hydrolysis of host-derived ceramide; in other words, they are not causing their own death by sphingosine derived from sphingomyelin and ceramide hydrolysis. Therefore, while the in vitro sphingosine killing correlates well with the in vivo phenotypes, we currently do not know the mechanism governing reduced survival of the sphR and sphA mutants in the lung.
We speculate that SphR responds to sphingosine to induce transcripts encoding proteins that protect P. aeruginosa from the bactericidal effects of sphingosine by induction of membrane stabilizing factors and/or catabolism of sphingosine to non-bactericidal metabolites. Here we show that SphR binds to sphingosine to initiate transcription of sphA, sphBCD and cerN. sphA encodes a hypothetical protein with some homology to proteins involved in meta-pathway phenol degradation. Protein localization predictions for SphA using the structure similarity-based prediction of Phrye2 [64] suggests that SphA is an outer membrane porin. Perhaps P. aeruginosa responds to sphingosine by providing a porin for sphingosine import and subsequent degradation that could aid in protecting the outer membrane from the damaging effects of free sphingosine. Okino and Ito demonstrated sphingosine utilization by P. aeruginosa by measuring removal of sphingosine from the culture supernatants and cell fractions [54]. Based on bioinformatic predictions, SphB, SphC and SphD are most likely involved in the metabolism of sphingosine. The sphB gene encodes a predicted cytochrome, while sphC encodes an FMN-linked oxidoreductase, and sphD encodes a pyridoxalphosphate serine-threonine aldolase. The latter two activities could work in concert to oxidize carbon 1, generating an aldol, which SphD could hypothetically act upon, rendering a long chain aldehyde and glycine. Transposon insertion into the sphC coding sequence (PA5327) resulted in reduced bacterial survival in a chronic rat lung infection model [65], suggesting that while our sphC deletion strain did not show a phenotype in the acute mouse lung infection (Fig. 7), it nonetheless impacts survival in the mammalian lung.
The microarray data comparing wild type in the presence and absence of pulmonary surfactant suggests some interesting biology in the presence of surfactant. The first observation has been covered by Jackson et al., who recently analyzed the changes in transcript levels of P. aeruginosa exposed to pulmonary surfactant, and compared wild type to both plcH and gbdR mutants [29], but did not publish results of these strains in the absence of pulmonary surfactant. They noted a reduction in transcript levels for Anr-controlled genes in both the gbdR and plcH mutants grown in surfactant, as do we (Table S1). Given the high levels of phosphatidylcholine and sphingomyelin in pulmonary surfactant, it was not surprising that the transcripts encoding proteins from the choline catabolic pathway were also highly induced in the presence of surfactant (Table S1). In addition to the high proportion of transcripts encoding stress-related proteins (mentioned in the Results section), there are also a high proportion (∼8%) of transcriptional regulators: NalC, BetI, NirG, PsrA, NarL, CgrA, PA3458, and PA4596. It is possible that the effects of induction of these transcription factors is contained in our regulation data, however our transcriptome analyses were a snapshot of transcripts at four hours post-induction and effects from changes in these transcription factors may not have sufficiently accumulated in the transcriptome. Of the reduced transcripts (Table S2), we note that three of the pyrroquinoline quinine biosynthesis genes are down, suggesting a change in requirement for this cofactor between surfactant and pyruvate conditions.
The demonstration of sphingosine detection by P. aeruginosa also opens up the possibility that this bacterium, and others with similar detection systems, could alter sphingosine and related sphingolipid signals, including S1P in the host. We have not yet examined the contribution of host immune signaling effected by the SphR regulon, but the impact of altering such an important and tightly controlled signaling network by bacterial factors has not been elucidated and may be an important contributing factor to the survival of P. aeruginosa in vivo.
Materials and Methods
Ethics statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol for animal infection was approved by the University of Vermont Institutional Animal Care and Use Committee (Permit number A3301-01). All procedures were performed under pentobarbital anesthesia and all efforts were made to minimize animal suffering.
Strains, growth conditions, and chemicals
P. aeruginosa PAO1, isogenic mutant strains, and E. coli (Table 2) were maintained in LB-Lennox (LB) medium. Morpholinepropanesulfonic acid (MOPS) medium [66] supplemented with 25 mM sodium pyruvate, 5 mM glucose and 50 µg/ml gentamicin (for P. aeruginosa) or MOPS with 10% LB (v/v), 5 mM glucose, and 10 µg/ml gentamicin (for E. coli) was used to grow strains prior to transcriptional induction studies. See LaBauve and Wargo (2012) for further details on P. aeruginosa growth methods [67]. For bactericidal assays, 1% neopeptone was supplemented with varying sphingosine concentrations in ethanol to reach a final concentration of 6.25% (w/v) ethanol in the assay. All lipids were purchased from Avanti Polar Lipids and other chemicals were purchased from Sigma-Aldrich or Fisher.
Tab. 2. Bacterial strains and plasmids used in this study. 
Mouse lung infection model
We used the oropharyngeal route of mouse lung infection previously described [68], [69]. Briefly, P. aeruginosa PAO1 and isogenic strains were streaked onto LB plates from −80°C stocks. Colonies from the first plate were restreaked onto a new LB plate after 24 hours and incubated at 37°C for 24 hours. Cells from the second plate were used to start 3 ml cultures in LB that were grown for 16–18 hours at 37°C on a roller drum. From these overnight cultures, cells were collected by centrifugation, washed in Dulbecco's PBS (DPBS), and resuspended to give ∼1×107 viable P. aeruginosa in 40 µL, with actual inoculum determined by serial dilution and plate counting. Eight to twelve week old male C57Bl/6J mice (Jackson Labs) were inoculated with 40 µL of the bacterial suspension via oropharyngeal aspiration. Anesthesia, surgery, bronchoalveolar lavage fluid (BALF) collection, organ harvest, and organ homogenization were done as previously described [68], [69] at 24 hours post-infection. Viable bacterial counts in organs were determined by serial dilution plating onto Pseudomonas Isolation Agar (PIA) (BD-Difco) followed by incubation at 37°C for 24 hours.
Mouse experiments (Fig. 3 and 7 and Supplemental Fig. S3) show CFU counts from all animals from duplicate experiments with each replicate having 4–6 animals per experimental group. All informative comparisons: mutants versus wild type (both Figures) and mutant versus complementation strain (Fig. 3 and Supplemental Fig. S3) were conducted in at least one additional experiment, included with comparator strains from other studies. Therefore, all informative comparisons were assessed three times. All experiments met the same statistical criteria, i.e. all replicates were consistent with regards to effect size and significance of changes. Inoculation order and harvest order alternated between experiments to eliminate potential issues related to the difference between the duration of inoculation (∼20–30 min) and the duration of harvest (∼1.5 h). For group comparisons, data (log10 transformed CFU counts) were analyzed by ANOVA followed by Tukey's (Fig. 3 and Supplemental Fig. S3) or Dunnett's (Fig. 7) Multiple Comparisons tests. All calculations were done using GraphPad Prism.
RNA extraction and microarray methodology
P. aeruginosa PAO1 wild type and ΔsphR were grown overnight in MOPS media supplemented with 20 mM pyruvate and 5 mM glucose. Overnight cultures were collected by centrifugation and resuspended in either MOPS supplemented with 20 mM pyruvate alone or 20 mM pyruvate and a 1∶50 dilution of the bovine surfactant preparation Survanta (Abbott) and induced for 4 hours at 37°C. Bacteria were collected by centrifugation, resuspended in MOPS and RNA Protect Bacterial Reagent (Qiagen), and the resultant pellets stored overnight at −20°C. RNA was extracted using an RNeasy kit (Qiagen), and eluted samples were treated with DNase I followed by a second round of RNeasy purification including an on-column DNase I treatment. Purified RNA samples were checked for DNA contamination by PCR and RNA integrity scores based on Agilent Bioanalyzer analysis were indicative of little to no DNA contamination.
Microarray analysis was performed on a Pseudomonas aeruginosa PAO1 gene chip using raw oligonucleotide probes generated from each condition using the NuGen Pico system. Each condition was analyzed in duplicate (N = 2), and summarized in one probe intensity by the Vermont Genetics Network Microarray Facility using Affymetrix GCOS software. Information from multiple probes was combined to obtain a single measure of expression for each probe set and sample. Probe-level intensities were background-corrected, normalized, and summarized, and Robust Multichip Average (RMA) statistics were calculated for each probe set and sample as is implemented in Partek Genomic Suites, version 6.6 (Copyright 2009, Partek Inc., St. Louis, MO, USA). Sample quality was assessed based on relative log expression (RLE), and normalized unscaled standard error (NUSE). To identify differentially expressed genes, linear modeling of sample groups was performed using ANOVA as implemented in Partek Genomic Suites. The magnitude of the response (fold change calculated using the least square mean) and the p-value associated with each probe set and binary comparison were calculated. The data have been submitted to NCBI GEO with accession number GSE48982.
Construction of deletion strains and complementation
Deletion mutants were generated using the pMQ30 plasmid [70] carrying the flanking regions of each of the four genes, sphR, sphA, sphC, and cerN, using conjugation-mediated deletion as described previously [30], [69]. Primers for these constructs are listed in Table S3. Single cross-over mutants were selected on PIA with gentamicin and selection of double crossover deletion mutants were carried out on LB 5% sucrose plates prepared without NaCl. Unmarked deletion mutants were verified using PCR. Complementation was done by integration of the sphR or sphA coding sequence under control of their native promoter at the attTn7 locus using the pUC18-miniTn7T-Gm vector as we described previously [68], [69] using the method of Choi and Schweizer [71]. This allowed stable complementation in the absence of antibiotic. For complementation where reporter plasmids were used, the gentamicin resistance cassette was excised by FLP-mediated recombination [71]. All sphR::attTn7 and sphA::attTn7 complementation strains were compared with wild type or mutant strains carrying the empty attTn7 integration region from the pUC18-miniTn7T-Gm vector.
Reporter assay to measure sphA transcriptional induction
Two reporter constructs were generated in this study using yeast homologous recombination [70] to generate translational fusions to lacZYA. A target lacZYA-containing vector suitable for yeast cloning (pMW42) was generated by excising the lacZYA region from pMW5 [31] with HindIII and EcoRI and cloning into the similarly cut pMQ80 backbone [70], which removes egfp-mut3. Either the sphA promoter (pAL4), or the entire sphR gene and the sphA promoter (pAL5) were recombined with pMW42 linearized with KpnI and HindIII. P. aeruginosa strains were electrotransformed with the reporter constructs and grown overnight in MOPS media supplemented with 20 mM pyruvate, 5 mM glucose, and 50 µg/ml gentamicin prior to induction. Inductions were carried out in MOPS media supplemented with 20 mM pyruvate and the inducing compound and incubated at 37°C for 6 hours. β-galactosidase assays were done as previously described [31], [72], using the method of Miller [73]. Studies of heterologous sphA induction in E. coli were carried by transforming pAL4 and pAL5 into E. coli NEB5α. Resulting E. coli strains were grown overnight in MOPS media supplemented with 10% LB (v/v), 5 mM glucose and 10 µg/ml gentamicin. For induction assays with S1P in E. coli, 2.4 µg of S1P or sphingosine were pre-treated with or without 5 U shrimp alkaline phosphatase (SAP) in 100 µL of water with 1× SAP buffer (USB), and incubated at 37°C for 60 minutes. Induction assays were carried out in MOPS supplemented with 10% LB (v/v), treated inducing compounds, and 10 µg/ml gentamicin. All E. coli strains were induced for 8 hours prior to ß-galactosidase assays.
Promoter mapping of sphA, sphB, and cerN
Full-length reporter constructs and truncations of sphA, sphB, and cerN promoters were cloned into pMW5 [31]. The resultant lacZYA reporter constructs were transformed into wild type P. aeruginosa and used to identify the region required for response to sphingosine. Inductions were carried out in MOPS media supplemented with 20 mM pyruvate and 150 µM sphingosine and incubated at 37°C for 6 hours followed by ß-galactosidase assays.
Cloning, expression and purification of SphR
We constructed a maltose binding protein (MBP) fusion to SphR by using the pMALc2 vector system (NEB). The sphR gene was amplified from genomic DNA. The PCR product was gel purified and ligated into the pCR Blunt vector (Invitrogen). The insert was excised with KpnI and HindIII, gel purified, and ligated into a similarly digested pMALc2 vector to generate pAL11. E. coli NEB5α (New England Biolabs) carrying the pAL11 plasmid were grown overnight in LB supplemented with 120 µg/ml carbenicillin. The overnight culture was transferred to two 500 ml flasks containing 100 ml of LB-carbenicillin and shaken at 220 rpm for 5 hours. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cells were induced for 3 hours. Cells were collected by centrifugation, lysed in column buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA) supplemented with 3 mg/ml lysozyme and Halt protease inhibitor 1× cocktail (Thermo Scientific). Lysates were clarified by centrifugation, and the soluble fraction was applied to a column containing amylose resin (NEB). The column was washed with ten volumes of column wash buffer (20 mM Tris-HCl, 150 mM NaCl 1 mM EDTA pH 7.4), followed by elution with column wash buffer supplemented with 10 mM maltose. Elution fractions were run on 10% SDS-PAGE gels and visualized by Coomassie staining. Fractions containing the MBP-SphR were pooled and dialyzed against 20 mM Tris-HCl, pH 7.5 at 4°C in a 20,000 kDa cutoff Slide-A-lyzer cassette (Pierce). The full length MBP-SphR fusion protein was used in electrophoretic mobility shift assays, as the MBP tag did not prevent sequence specific DNA binding (Fig. 5) or binding to sphingosine (Fig. 6).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA DNA probes were generated using PCR (Primers in Table S3) and were spot dialyzed against 2.5 mM Tris-HCl, 0.25 mM EDTA, pH 8.0. Labeled probes, generated using a primer with a covalently linked 5′ biotin tag (IDT), were used at 0.5 fmol/µl, and unlabeled competitor probes were used at a final concentration of 0.5 pmol/µl. EMSA was carried out using a Thermo Scientific Thermoshift kit. The final binding buffer was modified to contain 1× binding buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM dithiothreitol), 0.1 mM glycine betaine, and 2 µg/ml poly-dI-dC. Various concentrations of sphingosine dissolved in ethanol were added to reaction tubes and allowed to dry to eliminate ethanol prior to binding reactions. Binding reactions were carried out at 37°C for 15 minutes and electrophoresed on a 5% non-denaturing polyacrylamide gel then transferred to a BioDyne B membrane (Thermo Scientific). Detection was carried out using streptavidin-linked horseradish peroxidase according to the supplied protocol (Thermo Scientific).
Measurements of sphingosine association with SphR
Sphingosine association with SphR was measured by conducting binding reactions using 3H-D-erytho-sphingosine (Perkin-Elmer). Binding reactions were carried out as described for EMSA except 3H-D-erytho-sphingosine was used at a final concentration of 50 nM. Samples were incubated with and without either 10 µM MBP-SphR or 10 µM MBP-CdhR for 30 minutes then added to amylose resin. The amylose beads were collected by centrifugation and washed 3 times with amylose column wash buffer. After washes, amylose beads were resuspended in 200 µl of amylose wash buffer and transferred to a glass vial containing 10 ml of Biosafe II scintillation cocktail (RPI). Samples were quantified using a Tri-Carb 2910 TR liquid scintillation analyzer (Perkin-Elmer).
Sphingosine killing assay
Killing assays were carried out as previously described [61]. Briefly, overnight P. aeruginosa strains were grown in trypticase soy broth (TSB) and diluted 1∶40. Diluted cultures (100 µl) were added to glass tubes containing 250 µl of 1% neopeptone supplemented with 50 µl of the appropriate sphingosine stock in ethanol or ethanol alone as the vehicle control. The cultures were shaken at 170 rpm for one hour. Survival was determined by serial dilution plating on PIA. Colonies were counted after 24 hour incubation and survival calculated by comparison to vehicle only controls.
Supporting Information
Zdroje
1. LiebermanD, LiebermanD (2003) Pseudomonal infections in patients with COPD: epidemiology and management. Am J Respir Med 2 : 459–468.
2. ChastreJ, FagonJY (2002) Ventilator-associated pneumonia. Am J Respir Crit Care Med 165 : 867–903.
3. Crouch BrewerS, WunderinkRG, JonesCB, LeeperKVJr (1996) Ventilator-associated pneumonia due to Pseudomonas aeruginosa. Chest 109 : 1019–1029.
4. BurnsJL, EmersonJ, StappJR, YimDL, KrzewinskiJ, et al. (1998) Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis 27 : 158–163.
5. BriesacherBA, QuittnerAL, FouayziH, ZhangJ, SwensenA (2011) Nationwide trends in the medical care costs of privately insured patients with cystic fibrosis (CF), 2001–2007. Pediatr Pulmonol 46 : 770–776.
6. BurnsJL, GibsonRL, McNamaraS, YimD, EmersonJ, et al. (2001) Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis 183 : 444–452.
7. RajanS, SaimanL (2002) Pulmonary infections in patients with cystic fibrosis. Semin Respir Infect 17 : 47–56.
8. Butorac-PetanjekB, ParnhamMJ, Popovic-GrleS (2010) Antibiotic therapy for exacerbations of chronic obstructive pulmonary disease (COPD). J Chemother 22 : 291–297.
9. KlockgetherJ, MunderA, NeugebauerJ, DavenportCF, StankeF, et al. (2010) Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol 192 : 1113–1121.
10. FischbachMA, WalshCT (2009) Antibiotics for emerging pathogens. Science 325 : 1089–1093.
11. FriskA, SchurrJR, WangG, BertucciDC, MarreroL, et al. (2004) Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect Immun 72 : 5433–5438.
12. PalmerKL, MashburnLM, SinghPK, WhiteleyM (2005) Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol 187 : 5267–5277.
13. ChroneosZC, Sever-ChroneosZ, ShepherdVL (2010) Pulmonary surfactant: an immunological perspective. Cell Physiol Biochem 25 : 13–26.
14. GlasserJR, MallampalliRK (2012) Surfactant and its role in the pathobiology of pulmonary infection. Microbes Infect 14 : 17–25.
15. CaminitiSP, YoungSL (1991) The pulmonary surfactant system. Hosp Pract (Off Ed) 26 : 87–90, 94–100.
16. HannunYA, ObeidLM (2008) Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9 : 139–150.
17. SpiegelS, MilstienS (2011) The outs and the ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol 11 : 403–415.
18. OhanianJ, OhanianV (2001) Sphingolipids in mammalian cell signalling. Cell Mol Life Sci 58 : 2053–2068.
19. van MeerG, VoelkerDR, FeigensonGW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9 : 112–124.
20. SchwabSR, PereiraJP, MatloubianM, XuY, HuangY, et al. (2005) Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science 309 : 1735–1739.
21. VogelP, DonovielMS, ReadR, HansenGM, HazlewoodJ, et al. (2009) Incomplete inhibition of sphingosine 1-phosphate lyase modulates immune system function yet prevents early lethality and non-lymphoid lesions. PLoS One 4: e4112.
22. WalzerT, ChiossoneL, ChaixJ, CalverA, CarozzoC, et al. (2007) Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol 8 : 1337–1344.
23. WangF, Van BrocklynJR, HobsonJP, MovafaghS, Zukowska-GrojecZ, et al. (1999) Sphingosine 1-phosphate stimulates cell migration through a G(i)-coupled cell surface receptor. Potential involvement in angiogenesis. J Biol Chem 274 : 35343–35350.
24. PostmaFR, JalinkK, HengeveldT, MoolenaarWH (1996) Sphingosine-1-phosphate rapidly induces Rho-dependent neurite retraction: action through a specific cell surface receptor. EMBO J 15 : 2388–2392.
25. RoviezzoF, BrancaleoneV, De GruttolaL, VelleccoV, BucciM, et al. (2011) Sphingosine-1-phosphate modulates vascular permeability and cell recruitment in acute inflammation in vivo. J Pharmacol Exp Ther 337 : 830–837.
26. HammadSM (2011) Blood sphingolipids in homeostasis and pathobiology. Adv Exp Med Biol 721 : 57–66.
27. OskeritzianCA, PriceMM, HaitNC, KapitonovD, FalangaYT, et al. (2010) Essential roles of sphingosine-1-phosphate receptor 2 in human mast cell activation, anaphylaxis, and pulmonary edema. J Exp Med 207 : 465–474.
28. CamererE, RegardJB, CornelissenI, SrinivasanY, DuongDN, et al. (2009) Sphingosine-1-phosphate in the plasma compartment regulates basal and inflammation-induced vascular leak in mice. J Clin Invest 119 : 1871–1879.
29. JacksonAA, GrossMJ, DanielsEF, HamptonTH, HammondJH, et al. (2013) Anr and Its Activation by PlcH Activity in Pseudomonas aeruginosa Host Colonization and Virulence. J Bacteriol 195 : 3093–3104.
30. WargoMJ, SzwergoldBS, HoganDA (2008) Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol 190 : 2690–2699.
31. WargoMJ, HoTC, GrossMJ, WhittakerLA, HoganDA (2009) GbdR regulates Pseudomonas aeruginosa plcH and pchP transcription in response to choline catabolites. Infect Immun 77 : 1103–1111.
32. HampelKJ, LabauveAE, MeadowsJA, FitzsimmonsLF, NockAM, et al. (2013) Characterization of the GbdR regulon in Pseudomonas aeruginosa. J Bacteriol 196 : 7–15.
33. NelsonGJ (1967) Studies on the lipids of sheep red blood cells. I. Lipid composition in low and high potassium red cells. Lipids 2 : 64–71.
34. LengleE, GeyerRP (1972) Comparison of cellular lipids of serum-free strain L mouse fibroblasts. Biochim Biophys Acta 260 : 608–616.
35. YatomiY, IgarashiY, YangL, HisanoN, QiR, et al. (1997) Sphingosine 1-phosphate, a bioactive sphingolipid abundantly stored in platelets, is a normal constituent of human plasma and serum. J Biochem 121 : 969–973.
36. LawSLS, SquierCA, WertzPW (1994) Free Sphingosine in Oral Epithelium. Journal of Dental Research 73 : 108–108.
37. OkinoN, TaniM, ImayamaS, ItoM (1998) Purification and characterization of a novel ceramidase from Pseudomonas aeruginosa. J Biol Chem 273 : 14368–14373.
38. LassmannT, SonnhammerEL (2005) Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6 : 298.
39. WinsorGL, LamDK, FlemingL, LoR, WhitesideMD, et al. (2011) Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39: D596–600.
40. LucchesiGI, LisaTA, DomenechCE (1989) Choline and betaine as inducer agents of Pseudomonas aeruginosa phospholipase C activity in high phosphate medium. FEMS Microbiol Lett 48 : 335–338.
41. SageAE, VasilAI, VasilML (1997) Molecular characterization of mutants affected in the osmoprotectant-dependent induction of phospholipase C in Pseudomonas aeruginosa PAO1. Mol Microbiol 23 : 43–56.
42. SageAE, VasilML (1997) Osmoprotectant-dependent expression of plcH, encoding the hemolytic phospholipase C, is subject to novel catabolite repression control in Pseudomonas aeruginosa PAO1. J Bacteriol 179 : 4874–4881.
43. ShortridgeVD, LazdunskiA, VasilML (1992) Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol Microbiol 6 : 863–871.
44. WargoMJ, HoganDA (2009) Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155 : 2411–2419.
45. FischerCL, DrakeD, DawsonDV, BlanchetteDR, BrogdenKA, et al. (2012) Antibacterial activity of sphingoid bases and fatty acids against gram-positive bacteria and gram-negative bacteria. Antimicrob Agents Chemother 56 : 1157–1161 Epub ahead of print.
46. TeichgraberV, UlrichM, EndlichN, RiethmullerJ, WilkerB, et al. (2008) Ceramide accumulation mediates inflammation, cell death and infection susceptibility in cystic fibrosis. Nat Med 14 : 382–391.
47. ParkK, EliasPM, ShinKO, LeeYM, HupeM, et al. (2013) A novel role of a lipid species, sphingosine-1-phosphate, in epithelial innate immunity. Mol Cell Biol 33 : 752–762.
48. GargSK, VolpeE, PalmieriG, MatteiM, GalatiD, et al. (2004) Sphingosine 1-phosphate induces antimicrobial activity both in vitro and in vivo. J Infect Dis 189 : 2129–2138.
49. MalikZA, ThompsonCR, HashimiS, PorterB, IyerSS, et al. (2003) Cutting edge: Mycobacterium tuberculosis blocks Ca2+ signaling and phagosome maturation in human macrophages via specific inhibition of sphingosine kinase. J Immunol 170 : 2811–2815.
50. GargSK, SantucciMB, PanittiM, PucilloL, BocchinoM, et al. (2006) Does sphingosine 1-phosphate play a protective role in the course of pulmonary tuberculosis? Clin Immunol 121 : 260–264.
51. CamachoLR, EnsergueixD, PerezE, GicquelB, GuilhotC (1999) Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol 34 : 257–267.
52. RecchiC, SclaviB, RauzierJ, GicquelB, ReyratJM (2003) Mycobacterium tuberculosis Rv1395 is a class III transcriptional regulator of the AraC family involved in cytochrome P450 regulation. J Biol Chem 278 : 33763–33773.
53. GallegosMT, SchleifR, BairochA, HofmannK, RamosJL (1997) Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61 : 393–410.
54. OkinoN, ItoM (2007) Ceramidase enhances phospholipase C-induced hemolysis by Pseudomonas aeruginosa. J Biol Chem 282 : 6021–6030.
55. ChatterjeeA, DuttaPK, ChowdhuryR (2007) Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect Immun 75 : 1946–1953.
56. LowdenMJ, SkorupskiK, PellegriniM, ChiorazzoMG, TaylorRK, et al. (2010) Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A 107 : 2860–2865.
57. LubertoC, StonehouseMJ, CollinsEA, MarchesiniN, El-BawabS, et al. (2003) Purification, characterization, and identification of a sphingomyelin synthase from Pseudomonas aeruginosa. PlcH is a multifunctional enzyme. J Biol Chem 278 : 32733–32743.
58. RickettsCRSJ, TopleyE, LillyHA (1951) Human skin lipids with particular reference to the self-sterilising power of the skin . Clinical Science 10 : 89–111.
59. WertzPW, DowningDT (1990) Free sphingosine in human epidermis. J Invest Dermatol 94 : 159–161.
60. BibelDJ, AlyR, ShahS, ShinefieldHR (1993) Sphingosines: antimicrobial barriers of the skin. Acta Derm Venereol 73 : 407–411.
61. BibelDJ, AlyR, ShinefieldHR (1992) Antimicrobial activity of sphingosines. J Invest Dermatol 98 : 269–273.
62. FischerCL, DrakeDR, DawsonDV, BlanchetteDR, BrogdenKA, et al. (2012) Antibacterial activity of sphingoid bases and fatty acids against Gram-positive and Gram-negative bacteria. Antimicrob Agents Chemother 56 : 1157–1161.
63. FischerCL, WaltersKS, DrakeDR, BlanchetteDR, DawsonDV, et al. (2013) Sphingoid Bases Are Taken Up by Escherichia coli and Staphylococcus aureus and Induce Ultrastructural Damage. Skin Pharmacol Physiol 26 : 36–44.
64. KelleyLA, SternbergMJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4 : 363–371.
65. PotvinE, LehouxDE, Kukavica-IbruljI, RichardKL, SanschagrinF, et al. (2003) In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol 5 : 1294–1308.
66. NeidhardtFC, BlochPL, SmithDF (1974) Culture medium for enterobacteria. J Bacteriol 119 : 736–747.
67. LaBauveAE, WargoMJ (2012) Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr Protoc Microbiol Chapter 6: Unit 6E 1.
68. WargoMJ, GrossMJ, RajamaniS, AllardJL, LundbladLK, et al. (2011) Hemolytic Phospholipase C Inhibition Protects Lung Function During Pseudomonas aeruginosa Infection. Am J Respir Crit Care Med 184 : 345–354.
69. WargoMJ (2013) Choline Catabolism to Glycine Betaine Contributes to Pseudomonas aeruginosa Survival during Murine Lung Infection. PLoS One 8: e56850.
70. ShanksRM, CaiazzaNC, HinsaSM, ToutainCM, O'TooleGA (2006) Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72 : 5027–5036.
71. ChoiKH, SchweizerHP (2006) mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1 : 153–161.
72. FitzsimmonsLF, HampelKJ, WargoMJ (2012) Cellular choline and glycine betaine pools impact osmoprotection and phospholipase C production in Pseudomonas aeruginosa. J Bacteriol 194 : 4718–4726.
73. Miller JH (1972) Experiments in molecular genetics. Cold Spring, NY: Cold Spring Harbor Laboratory.
74. WurtzelO, Yoder-HimesDR, HanK, DandekarAA, EdelheitS, et al. (2012) The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8: e1002945.
75. JacobsMA, AlwoodA, ThaipisuttikulI, SpencerD, HaugenE, et al. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100 : 14339–14344.
76. SimonR, PrieferU, PuhlerA (1983) A Broad Host Range Mobilization System for Invivo Genetic-Engineering - Transposon Mutagenesis in Gram-Negative Bacteria. Bio-Technology 1 : 784–791.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite MovementČlánek Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi ScreenČlánek IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed ChildrenČlánek Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive SubjectsČlánek Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 1- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- HIV-1 Accessory Proteins Adapt Cellular Adaptors to Facilitate Immune Evasion
- Ranaviruses: Not Just for Frogs
- Effectors and Effector Delivery in
- Plasmacytoid Dendritic Cell Dynamics Tune Interferon-Alfa Production in SIV-Infected Cynomolgus Macaques
- Lu/BCAM Adhesion Glycoprotein Is a Receptor for Cytotoxic Necrotizing Factor 1 (CNF1)
- A Substrate-Fusion Protein Is Trapped inside the Type III Secretion System Channel in
- Parvovirus-Induced Depletion of Cyclin B1 Prevents Mitotic Entry of Infected Cells
- Red Blood Cell Invasion by : Structural Basis for DBP Engagement of DARC
- NsrR, GadE, and GadX Interplay in Repressing Expression of the O157:H7 LEE Pathogenicity Island in Response to Nitric Oxide
- Loss of Circulating CD4 T Cells with B Cell Helper Function during Chronic HIV Infection
- TREM-1 Deficiency Can Attenuate Disease Severity without Affecting Pathogen Clearance
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- Glutamate Utilization Couples Oxidative Stress Defense and the Tricarboxylic Acid Cycle in Phagosomal Escape
- Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite Movement
- Recovery of an Antiviral Antibody Response following Attrition Caused by Unrelated Infection
- Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi Screen
- Absence of Intestinal PPARγ Aggravates Acute Infectious Colitis in Mice through a Lipocalin-2–Dependent Pathway
- Induction of a Stringent Metabolic Response in Intracellular Stages of Leads to Increased Dependence on Mitochondrial Metabolism
- CTCF and Rad21 Act as Host Cell Restriction Factors for Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Lytic Replication by Modulating Viral Gene Transcription
- Gammaherpesviral Gene Expression and Virion Composition Are Broadly Controlled by Accelerated mRNA Degradation
- The Arabidopsis Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-transcriptional Control of Disease Resistance Genes
- Inflammatory Stimuli Reprogram Macrophage Phagocytosis to Macropinocytosis for the Rapid Elimination of Pathogens
- Alphavirus Mutator Variants Present Host-Specific Defects and Attenuation in Mammalian and Insect Models
- Phosphopyruvate Carboxylase Identified as a Key Enzyme in Erythrocytic Carbon Metabolism
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Electron Tomography of HIV-1 Infection in Gut-Associated Lymphoid Tissue
- Characterisation of a Multi-ligand Binding Chemoreceptor CcmL (Tlp3) of
- Single Cell Stochastic Regulation of Pilus Phase Variation by an Attenuation-like Mechanism
- Cell Tropism Predicts Long-term Nucleotide Substitution Rates of Mammalian RNA Viruses
- Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive Subjects
- RNA-seq Analysis of Host and Viral Gene Expression Highlights Interaction between Varicella Zoster Virus and Keratinocyte Differentiation
- Kaposi's Sarcoma Associated Herpesvirus Tegument Protein ORF75 Is Essential for Viral Lytic Replication and Plays a Critical Role in the Antagonization of ND10-Instituted Intrinsic Immunity
- DAMP Molecule S100A9 Acts as a Molecular Pattern to Enhance Inflammation during Influenza A Virus Infection: Role of DDX21-TRIF-TLR4-MyD88 Pathway
- Variable Suites of Non-effector Genes Are Co-regulated in the Type III Secretion Virulence Regulon across the Phylogeny
- Reengineering Redox Sensitive GFP to Measure Mycothiol Redox Potential of during Infection
- Preservation of Tetherin and CD4 Counter-Activities in Circulating Alleles despite Extensive Sequence Variation within HIV-1 Infected Individuals
- KSHV 2.0: A Comprehensive Annotation of the Kaposi's Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features
- Nutrient Limitation Governs Metabolism and Niche Adaptation in the Human Nose
- Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
- Investigation of Acetylcholine Receptor Diversity in a Nematode Parasite Leads to Characterization of Tribendimidine- and Derquantel-Sensitive nAChRs
- Intranasal Vaccination Promotes Detrimental Th17-Mediated Immunity against Influenza Infection
- -Mediated Inhibition of Iron Export Promotes Parasite Replication in Macrophages
- Variation in RNA Virus Mutation Rates across Host Cells
- A Single Amino Acid in the Stalk Region of the H1N1pdm Influenza Virus HA Protein Affects Viral Fusion, Stability and Infectivity
- Group B Engages an Inhibitory Siglec through Sialic Acid Mimicry to Blunt Innate Immune and Inflammatory Responses
- Synthesis and Biological Properties of Fungal Glucosylceramide
- HIV Protective KIR3DL1/S1-HLA-B Genotypes Influence NK Cell-Mediated Inhibition of HIV Replication in Autologous CD4 Targets
- Recruitment of PfSET2 by RNA Polymerase II to Variant Antigen Encoding Loci Contributes to Antigenic Variation in
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Enhancing Virus-Specific Immunity by Combining Therapeutic Vaccination and PD-L1 Blockade in Chronic Hepadnaviral Infection
- Suppression of Interferon Lambda Signaling by SOCS-1 Results in Their Excessive Production during Influenza Virus Infection
- Inflammation Fuels Colicin Ib-Dependent Competition of Serovar Typhimurium and in Blooms
- Host-Specific Enzyme-Substrate Interactions in SPM-1 Metallo-β-Lactamase Are Modulated by Second Sphere Residues
- STING-Dependent Type I IFN Production Inhibits Cell-Mediated Immunity to
- From Scourge to Cure: Tumour-Selective Viral Pathogenesis as a New Strategy against Cancer
- Lysine Acetyltransferase GCN5b Interacts with AP2 Factors and Is Required for Proliferation
- Narrow Bottlenecks Affect Populations during Vertical Seed Transmission but not during Leaf Colonization
- Targeted Cytotoxic Therapy Kills Persisting HIV Infected Cells During ART
- Murine Gammaherpesvirus M2 Protein Induction of IRF4 via the NFAT Pathway Leads to IL-10 Expression in B Cells
- iNKT Cell Production of GM-CSF Controls
- Malaria-Induced NLRP12/NLRP3-Dependent Caspase-1 Activation Mediates Inflammation and Hypersensitivity to Bacterial Superinfection
- Detection of Host-Derived Sphingosine by Is Important for Survival in the Murine Lung
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



