-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Inflammation Fuels Colicin Ib-Dependent Competition of Serovar Typhimurium and in Blooms
The host's immune system plays a key role in modulating growth of pathogens and the intestinal microbiota in the gut. In particular, inflammatory bowel disorders and pathogen infections induce shifts of the resident commensal microbiota which can result in overgrowth of Enterobacteriaceae (“inflammation-inflicted blooms”). Here, we investigated competition of the human pathogenic Salmonella enterica serovar Typhimurium strain SL1344 (S. Tm) and commensal E. coli in inflammation-inflicted blooms. S. Tm produces colicin Ib (ColIb), which is a narrow-spectrum protein toxin active against related Enterobacteriaceae. Production of ColIb conferred a competitive advantage to S. Tm over sensitive E. coli strains in the inflamed gut. In contrast, an avirulent S. Tm mutant strain defective in triggering gut inflammation did not benefit from ColIb. Expression of ColIb (cib) is regulated by iron limitation and the SOS response. CirA, the cognate outer membrane receptor of ColIb on colicin-sensitive E. coli, is induced upon iron limitation. We demonstrate that growth in inflammation-induced blooms favours expression of both S. Tm ColIb and the receptor CirA, thereby fuelling ColIb dependent competition of S. Tm and commensal E. coli in the gut. In conclusion, this study uncovers a so-far unappreciated role of inflammation-inflicted blooms as an environment favouring ColIb-dependent competition of pathogenic and commensal representatives of the Enterobacteriaceae family.
Published in the journal: Inflammation Fuels Colicin Ib-Dependent Competition of Serovar Typhimurium and in Blooms. PLoS Pathog 10(1): e32767. doi:10.1371/journal.ppat.1003844
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003844Summary
The host's immune system plays a key role in modulating growth of pathogens and the intestinal microbiota in the gut. In particular, inflammatory bowel disorders and pathogen infections induce shifts of the resident commensal microbiota which can result in overgrowth of Enterobacteriaceae (“inflammation-inflicted blooms”). Here, we investigated competition of the human pathogenic Salmonella enterica serovar Typhimurium strain SL1344 (S. Tm) and commensal E. coli in inflammation-inflicted blooms. S. Tm produces colicin Ib (ColIb), which is a narrow-spectrum protein toxin active against related Enterobacteriaceae. Production of ColIb conferred a competitive advantage to S. Tm over sensitive E. coli strains in the inflamed gut. In contrast, an avirulent S. Tm mutant strain defective in triggering gut inflammation did not benefit from ColIb. Expression of ColIb (cib) is regulated by iron limitation and the SOS response. CirA, the cognate outer membrane receptor of ColIb on colicin-sensitive E. coli, is induced upon iron limitation. We demonstrate that growth in inflammation-induced blooms favours expression of both S. Tm ColIb and the receptor CirA, thereby fuelling ColIb dependent competition of S. Tm and commensal E. coli in the gut. In conclusion, this study uncovers a so-far unappreciated role of inflammation-inflicted blooms as an environment favouring ColIb-dependent competition of pathogenic and commensal representatives of the Enterobacteriaceae family.
Introduction
Enteric Salmonella enterica serovar Typhimurium (S. Tm) infection is antagonized by a highly complex intestinal microbiota, a condition termed colonization resistance. Disruption of colonization resistance by oral antibiotic therapy, germfree state or an immature microbiota of low complexity results in increased susceptibility to oral infection with pathogens of the Enterobacteriaceae family [1], [2], [3]. In addition to the named conditions, inflammatory changes in the intestine induce dysbiosis and favour Enterobacteriaceae overgrowth [4], [5], [6]. Recently, we have shown that S. Tm-induced gut inflammation mediates parallel blooms of S. Tm and host-intrinsic commensal E. coli [7], [8]. In these blooms, both bacteria can reach high densities (>108 cfu/ml) while the residual microbiota is outgrown [7]. Therefore, under inflammatory conditions, commensal E. coli are one of the main competitors of S. Tm. In the inflamed gut, environmental conditions encountered by bacteria vastly differ from the situation in the absence of inflammation. By resisting antimicrobial defences, utilizing iron acquisition systems and exploiting anaerobic electron acceptors (e.g. NO3−, tetrathionate), Enterobacteriaceae can capitalize on inflammatory conditions [9], [10], [11], [12], [13], [14].
Besides exploitative competition for resources, bacteria can directly antagonize one another by producing antimicrobials, such as bacteriocins. Bacteriocins produced by Enterobacteriaceae (E. coli, Salmonella and relatives) are termed colicins. They have a narrow spectrum of activity and act only against phylogenetically close relatives. Colicins kill by one of three general mechanisms: pore formation in the inner membrane, nuclease activity or interference with cell wall synthesis [15]. We have shown that growth of S. Tm in inflammation-induced blooms promotes horizontal transfer of the conjugative pCol1B9-plasmid (in the following termed P2) to commensal E. coli strains [7]. P2 encodes the locus for colicin Ib (ColIb) production (cib) and immunity (imm). In blooms, ColIb was shown to increase the fitness of S. Tm in competition with commensal, colicin-sensitive E. coli. However, it has remained unclear if ColIb only affords a benefit for S. Tm in inflammation-inflicted blooms or also in the absence of gut inflammation.
ColIb belongs to the group of pore-forming colicins [16]. ColIb is closely related to ColIa and most of its structural/functional properties can be inferred from ColIa [17]. Free ColIa/b binds to the outer-membrane receptor CirA, a catecholate siderophore receptor, and traverses the outer membrane in a TonB-dependent way [18], [19], [20]. The exact mode of outer-membrane translocation is still unclear but a recent study suggests that two molecules of CirA are required, one for ColIa/ColIb binding, the second one for its translocation [18], [21]. ColIa/ColIb forms a pore in the inner membrane of sensitive bacteria which leads to disruption of the electrochemical membrane gradient and consequent bacterial death [22]. ColIa/ColIb producers protect themselves by expression of the immunity protein (imm) which interferes with ColIa/ColIb action in the inner membrane [23]. In addition, resistance to ColIb can be gained by alterations of the cell-surface receptor CirA or mutations in the TonB-dependent import pathway [24].
ColIa/ColIb expression is regulated in a Fur - and LexA-dependent way [25]. The Fur protein binds FeII and thereby measures the intracellular FeII pool [26]. In the iron-bound state, Fur acts as a transcriptional repressor of iron-regulated promoters which is released under iron-limiting conditions. In addition, the ColIb gene cib is also repressed by the LexA protein, the regulator of the SOS response. The SOS response is launched when bacteria sense DNA damage (e.g. DNA double strands breaks) [27]. As a consequence, the RecA protease is activated and cleaves the LexA repressor protein which in turn activates an array of genes involved in DNA repair, survival, prophages and also colicins [15], [28]. Interestingly, ColIa and ColIb are the only reported colicins which are controlled by both, Fur and LexA. This suggests that maximal expression of ColIa/b requires iron limitation and stress conditions. Interestingly, the majority of other colicins are only regulated by LexA and usually contain two LexA-binding sites in their promoter to ensure tight repression.
Theoretical and experimental studies suggested that colicin producers have a competitive advantage over non-producers when colonizing the same ecological niche [29]. When directly tested in competition experiments with sensitive strains in animal models, colicin production only conferred a competitive advantage after weeks [30], [31]. In some cases, colicin-production even conferred no detectable benefit despite colicin-dependent killing could be demonstrated under in vitro conditions [32], [33], [34]. The underlying reasons were attributed to colicin inactivation by proteases [33] or reduced colicin activity under anaerobic conditions [35], [36].
In the S. Tm mouse colitis model ColIb conferred an overt fitness benefit for S. Tm in competition against a colicin-sensitive E. coli strain [7]. This was somewhat surprising, considering the lack of an apparent competitive advantage of colicin producers in the reports mentioned above. Although different experimental setups were used in previous studies, a key difference to our study was the lack of concomitant gut inflammation. Therefore we reasoned that the inflammatory response may somehow promote ColIb dependent competition of S. Tm and E. coli. We tested this idea and analyzed ColIb-dependent S. Tm and E. coli competition under normal and inflammatory conditions in the mouse colitis model. Our experiments revealed that the inflammatory response creates conditions that potentiate the effects of colicins, both by increasing their production and by mediating susceptibility of the competitor. This finding has implications for colicin ecology and points out the importance of colicins as fitness factor for bacterial competition in Enterobacterial blooms in the inflamed gut.
Results
ColIb affords S. Tm a growth advantage over colicin-sensitive E. coli strains in the inflamed, but not in the normal gut
ColIb confers a fitness benefit to Salmonella Typhimurium (S. Tm) over colicin-sensitive E. coli strains [7]. In vitro, the E. coli Nissle (EcNissle) strain shows intermediate susceptibility to ColIb (turbid inhibition zone), while the K-12 strain E. coli MG1655 (EcMG1655) is highly sensitive (clear inhibition zone; Figure S1A). For this reason we selected EcMG1655 for our initial experiments. First we tested, if the growth benefit of S. Tm over EcMG1655 is ColIb-dependent. We performed the co-infection experiments with S. Tm strains and EcMG1655 in the streptomycin Salmonella mouse colitis model [2]. Here, we used gnotobiotic mice colonized with a low-complexity microbiota (LCM) lacking any kind of Enterobacteriaceae which may interfere with the experiment (i.e. by the production of other colicins). Further, since the S. Tm P2-plasmid is highly mobile and rapidly transferred to co-colonizing E. coli strains in the gut [7], all S. Tm strains carried an additional mutation in the origin of transfer of P2 (ΔoriT) to prevent conjugation (Table 1). LCM mice pre-treated with streptomycin were co-infected with 1∶1 mixtures of EcMG1655 and either ColIb-producing (S. TmΔoriT) or ColIb-deficient strains (S. TmΔoriT Δcib). EcMG1655 was efficiently outcompeted by S. TmΔoriT but not by the ColIb-deficient mutant (Figure 1A,B). Both S. Tm strains induced similar degrees of gut inflammation by day 4 p.i. (Figure 1CD). This confirms that the competitive advantage of S. Tm over EcMG1655 in the inflamed intestine is for the most part ColIb-dependent. Next, we tested if an avirulent strain (S. TmΔoriT avir) defective in triggering an inflammatory response due to the absence of functional type three secretion systems [37] would also benefit from ColIb. Interestingly, in the absence of gut inflammation, S. TmΔoriT avir did not out-compete EcMG1655 (Figure 1).
Fig. 1. Colicin-dependent competition of S. Tm and E. coli in the gut in inflammation-induced “blooms” in gnotobiotic LCM mice. 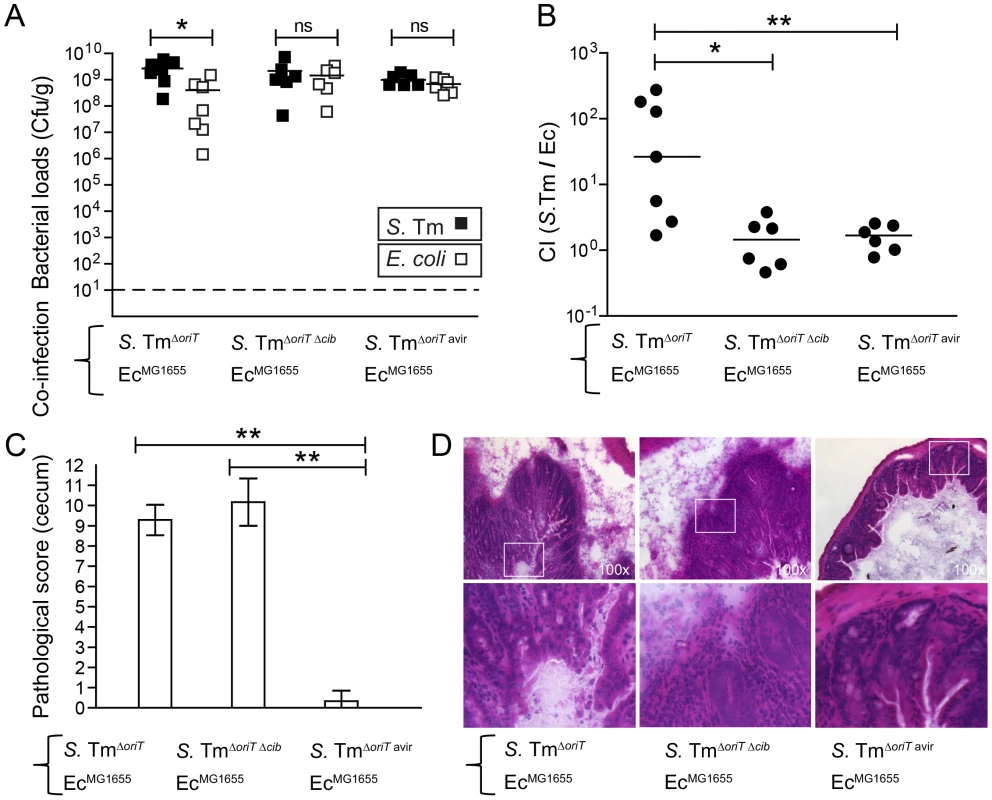
Streptomycin-treated LCM-mice were co-infected with 1∶1 mixtures of S. TmΔoriT and EcMG1655, S. TmoriT Δcib and EcMG1655 or S. TmΔoriT avir and EcMG1655. (A) S. Tm (black) and E. coli (white) colonization densities were determined at day 4 p.i. in the cecum content (cfu/g). (B) Competitive indices (CI; ratio of S. Tm/E. coli) were determined for individual mice shown in (A). Bars show the median. (C) Histopathological analysis of cecal tissue of the infected mice shown in (A). Cecal tissue sections of the mice were stained with hematoxylin/eosin (H&E) and the degree of submucosal edema, neutrophil infiltration, epithelial damage and loss of goblet cells was scored (Materials and Methods). 1–3: no pathological changes; 4–7: moderate inflammation; above 8: severe inflammation. shown mean and StD. (D) Representative H&E–stained cecal sections of mice shown in (A–C). Magnification 100-fold. Enlarged sections (squares) are shown in the lower panels. Dotted line: detection limit (1 cfu/g). Tab. 1. Bacteria and plasmids used in this study. 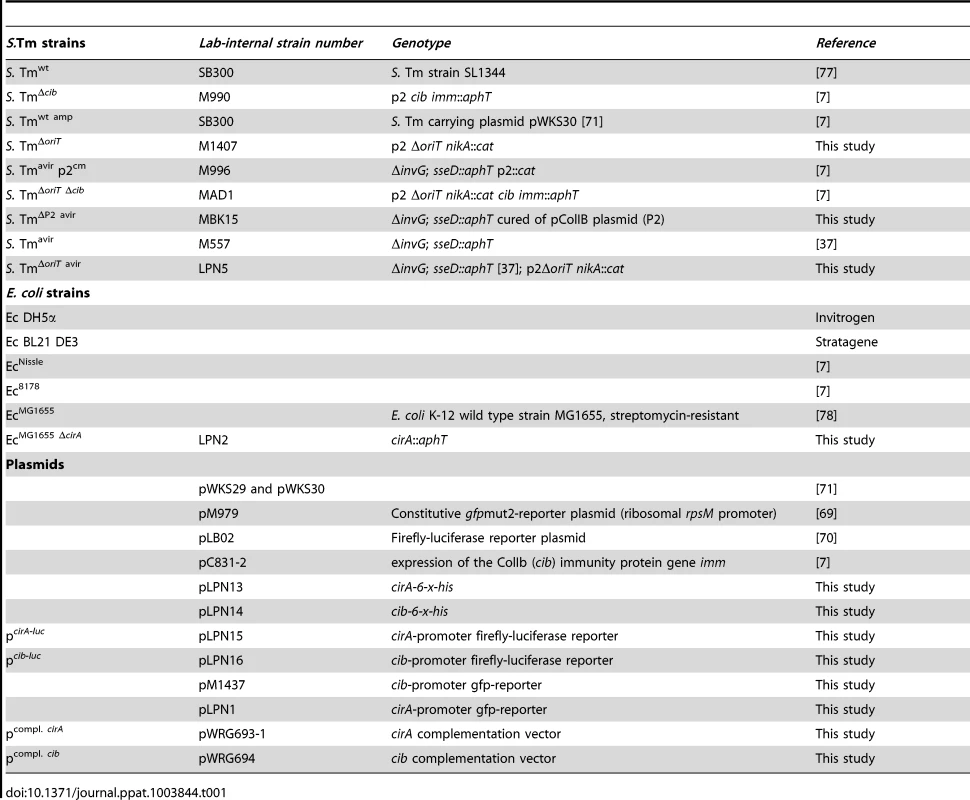
Previous studies on colicin-dependent bacterial competition were using conventional mice. To this end we aimed to verify that our key findings in gnotobiotic mice were also reproducible in a more “natural” mouse model. To this end, we used the streptomycin-treated mice with a conventional complex microbiota. We used EcNissle for the co-infection experiments as in contrast to LCM mice, EcMG1655 only poorly colonized conventional streptomycin-treated, S. TmΔoriT infected mice (not shown). We set up four different experimental groups of streptomycin-treated mice (Figure 2). One group was co-infected with a virulent colicin-producing, the other with a virulent colicin–deficient S. Tm strain (S. TmΔoriT or S. TmΔoriT Δcib, respectively) and EcNissle. Both groups developed strong Salmonella-induced gut inflammation upon infection by day 4 p.i. (Figure 2E,F). The other two groups were either co-infected with avirulent ColIb-producing S. Tm (S. TmΔoriT avir) and EcNissle, or avirulent ColIb-deficient S. Tm (S. TmΔP2 avir) and EcNissle. The latter two groups did not develop gut inflammation within 4 days of infection (Figure 2E,F). In the presence of inflammation, virulent ColIb-producing S. TmΔoriT grew up to similar numbers as EcNissle (to ∼108 cfu/g) while ColIb-deficient S. TmΔoriT Δcib was outcompeted by EcNissle (Figure 2B). This difference is also reflected in the competitive index (Figure 2D). In the absence of inflammation, both co-infecting strains (ColIb-producing S. TmΔoriT avir and EcNissle) and (ColIb-deficient S. TmΔP2 avir and EcNissle) colonized well at day 1 post infection (Figure 2AC) but were out-competed (to ∼107 cfu/g) by the complex conventional microbiota, which recovers by day 5 after streptomycin-treatment (Figure 2B). No benefit of ColIb-production was observed for the avirulent Salmonella strain. The absolute ratio of Salmonella/EcNissle is different when compared to Salmonella/EcMG1655 in LCM mice (Figure 1). We reason that this is due to strain-specific differences between EcMG1655 and EcNissle as well as due to differences between the gnotobiotic and complex gut microbiota. This data verified that S. Tm/EcNissle competition is only ColIb-dependent in the presence of gut inflammation.
Fig. 2. Colicin-dependent competition of S. Tm and E. coli in the gut in inflammation-induced “blooms” in conventional mice. 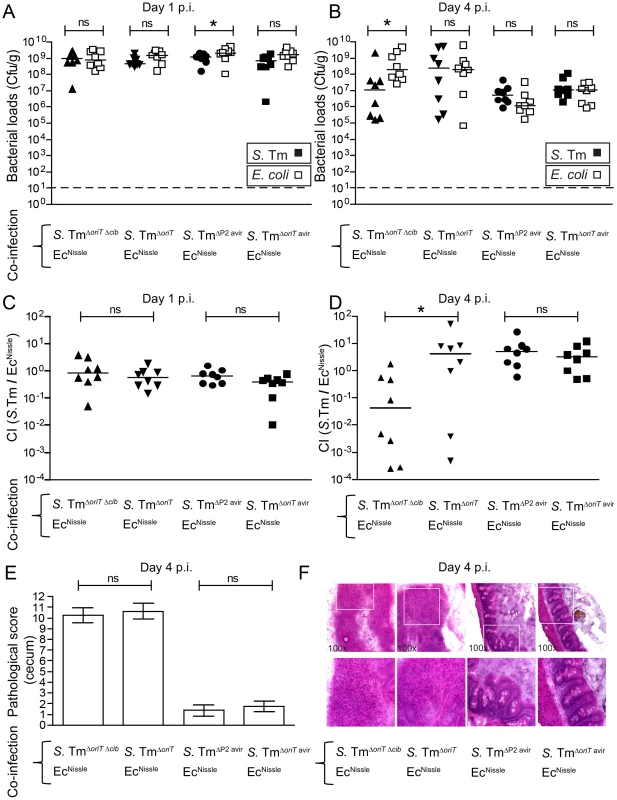
Streptomycin-treated conventional mice were co-infected with 1∶1 mixtures of S. TmΔoriT Δcib and EcNissle, S. TmΔoriT and EcNissle, S. TmΔP2 avir and EcNissle or S. TmΔoriT avir and EcNissle. S. Tm (black) and E. coli (white) colonization densities (Cfu/g) were determined at day 1 (A) and day 4 p.i. (B) in the feces and cecum content respectively. (C, D) Competitive indices (CI; ratio of S. Tm/E. coli) were determined for individual mice shown in (A) and (B). Bars show the median. (E) Histopathological analysis of cecal tissue of the infected mice. Cecal tissue sections of the mice were stained with hematoxylin/eosin (H&E) and the degree of submucosal edema, neutrophil infiltration, epithelial damage and loss of goblet cells was scored (Materials and Methods). 1–3: no pathological changes; 4–7: moderate inflammation; above 8: severe inflammation. Bars show mean and StD. (F) Representative H&E–stained cecal sections of mice shown in (A–E). Magnification 100-fold. Enlarged sections (squares) are shown in the lower panels. Dotted line: detection limit (1 cfu/g). In conclusion, these experiments prompted us to hypothesize that, in the normal non-inflamed gut, either ColIb expression by S. Tm was down-regulated or susceptibility of the Ec strains to ColIb was decreased. To further address the mechanism of colicin-dependent competition in the inflammation-induced blooms we investigated the regulation of cib expression in S. Tm as well as the susceptibility of Ec to ColIb in detail.
Regulation of S. Tm ColIb is induced by iron limitation and the SOS response in vitro
The cib promoter region contains binding sites for the transcriptional repressors Fur and LexA (Figure 3A; Figure S2). We generated a cib promoter firefly-luciferase (luc)-reporter construct as well as an affinity-purified polyclonal rabbit-α-ColIb antiserum to analyze the regulation of ColIb expression. ColIb expression was strongly up-regulated upon induction of the SOS response by the antibiotic mitomycin C (0.25 µg/ml) as confirmed by luc-reporter assays and immunoblot (Figure 3B–D). Depletion of Fe(III) from culture media by chelation with 100 µM diethylenetriaminepentaacetic acid (DTPA) [38] had a comparable inductive effect on ColIb production. Supplementation of both, mitomycin C and DTPA lead to maximal induction of ColIb production and secretion. This result confirmed that ColIb is de-repressed in response to SOS signals and iron starvation.
Fig. 3. Expression of S. Tm ColIb is induced by iron limitation and the SOS response. 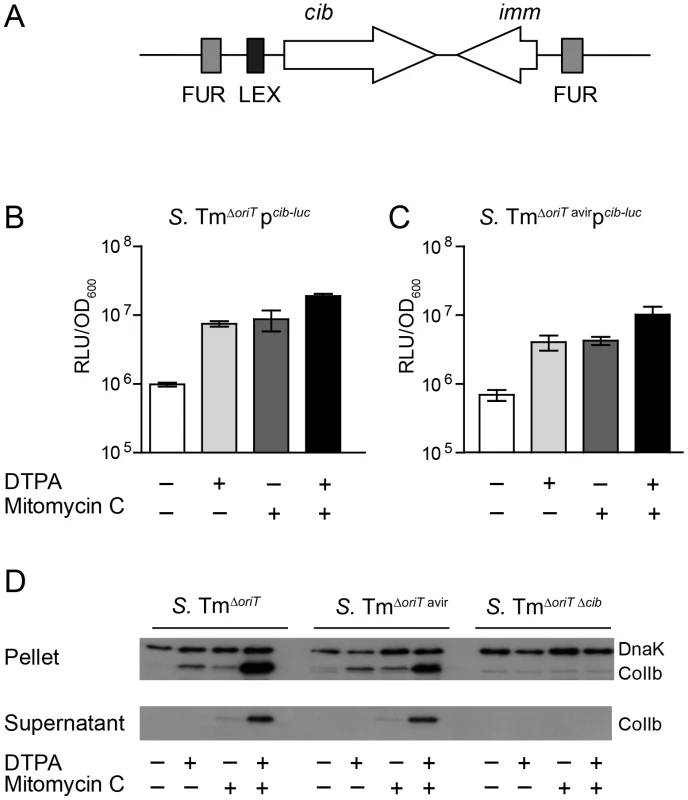
(A) Organization of the ColIb locus showing location of the Fur and LexA repressor binding sites in the ColIb (cib) promoter and the immunity protein gene (imm). Overnight cultures of S. TmΔoriT (B) and S. TmΔoriT avir (C) carrying the reporter plasmid pcib-luc were re-inoculated 1∶20 in fresh LB with indicated supplements (0.25 µg/ml mitomycin C; 100 mM DTPA) and grown under aeration for 4 h. Cultures were normalized to OD600, bacteria were harvested and luciferase-activity was measured in bacterial lysates. Relative luminescence units (RLU) per unit OD600 are indicated. (D) Overnight cultures of indicated S. Tm strains were re-inoculated 1∶20 in fresh LB with indicated supplements (0.25 µg/ml mitomycin C; 100 mM DTPA) and grown under aeration for 4 h. OD600 of the cultures was normalized, bacteria were harvested and ColIb was detected in bacterial lysates as well as in the culture supernatant by immunoblot using an affinity-purified rabbit-α-ColIb antiserum. S. Tm DnaK was detected as loading control. Upregulation of EcMG1655 cirA under iron limitation correlates with increased ColIb susceptibility
The outer membrane protein CirA is the receptor ColIa and ColIb [18], [19], [20]. Expression of cirA is under negative control of Fur [39] (Figure S3). To confirm FeIII-dependent regulation of the cirA expression in EcMG1655, we generated a cirA promoter-luc-reporter as well as a polyclonal rabbit-α-CirA antiserum. CirA expression was strongly upregulated in LB media upon addition of 100 µM DTPA but not by mitomycin C as confirmed by luciferase assay and immunoblot (Figure 4A, B). This confirmed that EcMG1655 cirA was de-repressed in response to FeIII-starvation. Next, we tested if cirA-expression correlated with sensitivity to ColIb-mediated killing. As expected, EcMG1655 ΔcirA was resistant to ColIb (Figure 5A). This phenotype was complemented by expressing cirA on a plasmid in EcMG1655 ΔcirA (Figure S1C). We further investigated, whether successive iron depletion would increase ColIb sensitivity of EcMG1655. To this end, we performed ColIb killing assays of EcMG1655 in M9 minimal media with different concentrations of FeCl3 using the same amounts of purified recombinant ColIb. Indeed, EcMG1655 was most sensitive to ColIb in M9 without FeCl3 addition and became less susceptible upon FeCl3 supplementation (Figure 5A). This correlated with increased cirA expression under this condition, as determined by immunoblot (Figure 5B). Thus, sensitivity of EcMG1655 to ColIb increases with elevated cirA expression.
Fig. 4. E. coli cirA expression is upregulated in response to iron limitation. 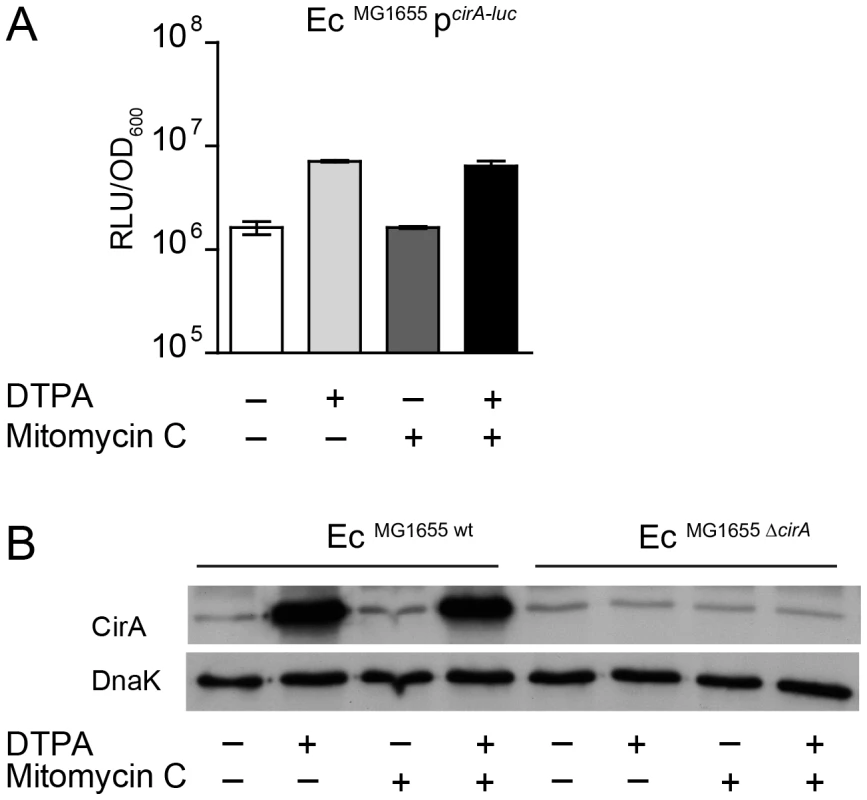
(A) An overnight culture of EcMG1655 carrying the reporter plasmid pcirA-luc was re-inoculated 1∶20 in fresh LB with indicated supplements (0.25 µg/ml mitomycin C; 100 mM DTPA) and grown under aeration for 4 h. OD600 of the cultures was normalized, bacteria were harvested and luciferase-activity was measured in bacterial lysates. Relative luminescence units (RLU) per unit OD600 are indicated. (B) Overnight cultures of EcMG1655 as well as EcMG1655 ΔcirA were re-inoculated 1∶20 in fresh LB with indicated supplements (0.25 µg/ml mitomycin C; 100 mM DTPA) and grown under aeration for 4 h. Cultures were normalized to OD600, bacteria were harvested and CirA was detected in bacterial lysates by immunoblot using a rabbit-α-CirA antiserum. E. coli cytoplasmic protein DnaK was detected as loading control. Fig. 5. Induction of cirA expression increases sensitivity to ColIb of E. coli. 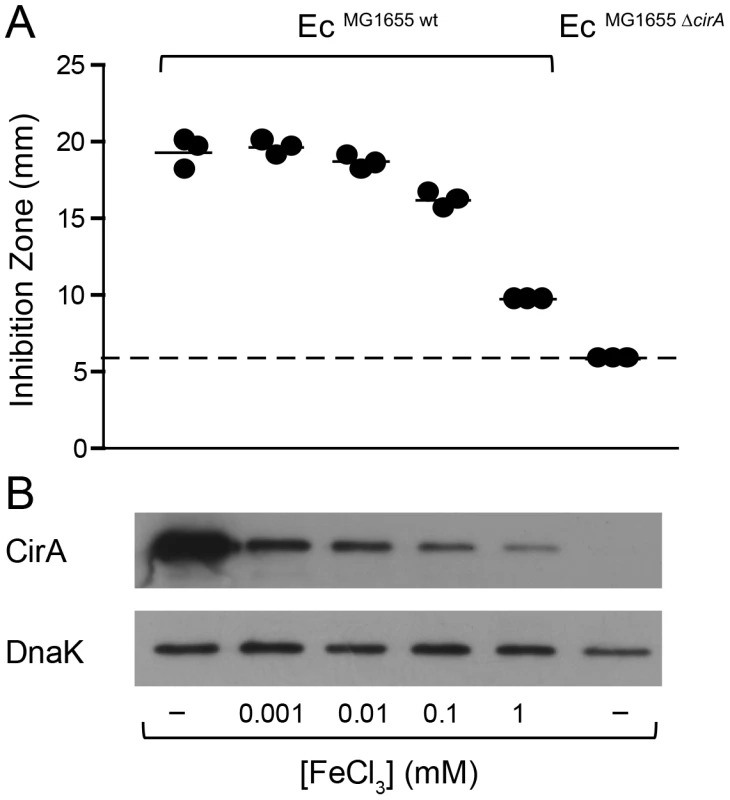
(A) EcMG1655 and EcMG1655 ΔcirA were cultivated in M9 medium o.n., mixed with soft agar and indicated concentrations of FeCl3 and overlaid on M9 agar plates. Paper discs with recombinant ColIb were placed on the agar plate and the diameter of the inhibition zone (halo) was measured after 24 h. The detection limit (dotted line) is the size of the paper-disc (6 mm). (B) cirA expression of E. coli grown in vitro in M9 medium supplemented with different concentrations of FeCl3. Overnight cultures of EcMG1655 and EcMG1655 ΔcirA grown in M9 medium for 12 h were used for inoculation of 2 ml M9 medium supplemented with 1 µM, 10 µM, 0.1 mM or 1 mM FeCl3 and subcultured for 7 h. From each subculture, 250 µl (for an OD600 of 1) was taken, spun down at 4°C, 10 min, 10, 000 rpm. CirA was detected by immunoblot in bacterial lysates using affinity-purified rabbit-α-CirA-antiserum. E. coli DnaK was detected as loading control. ColIb-dependent competition of S. Tm and EcMG1655 in vitro is boosted by iron starvation and SOS-stress
To underscore the importance of environmental conditions for ColIb-dependent competition of S. TmΔoriT and EcMG1655, we performed in vitro co-culture experiments. We followed growth of S. TmΔoriT and EcMG1655 and the respective mutants (S. TmΔoriT Δcib and EcMG1655 ΔcirA) in co-cultures under different conditions (Figure 6). In the absence of supplements, S. TmΔoriT and EcMG1655 grew at similar rate. S. TmΔoriT outcompeted EcMG1655 by ∼7-fold after 8 h (mean titer S. TmΔoriT: 1.7×109 cfu/ml and EcMG1655: 2.3×108 cfu/ml; Figure 6A). In contrast, EcMG1655 was outcompeted by several orders of magnitude after 8 h when either DTPA (7×107-fold), mitomycin C (1×104-fold) or both supplements (6×106-fold) were added to the co-culture (Figure 6D,G,J). S. Tm overgrowth in vitro was indeed ColIb-dependent, as no killing was observed in the absence of ColIb (S. TmΔoriT Δcib) or CirA (EcMG1655 ΔcirA) (Figure 5B,C,E–L). Of note, addition of 100 µM DTPA led to more pronounced killing of EcMG1655 than mitomycin C with comparable amounts of ColIb (Figure 3D), suggesting that iron depletion has a greater impact on ColIb-dependent competition (i.e. by enhancing EcMG1655 cirA expression and thereby its susceptibility to ColIb). The mutant phenotypes of the S. Tm cib mutant as well as the EcMG1655 cirA mutant were complemented using a plasmid-based complementation approach (Figure S1, Figure S4 and Figure S5).
Fig. 6. ColIb dependent competition of S. Tm and E. coli in vitro. 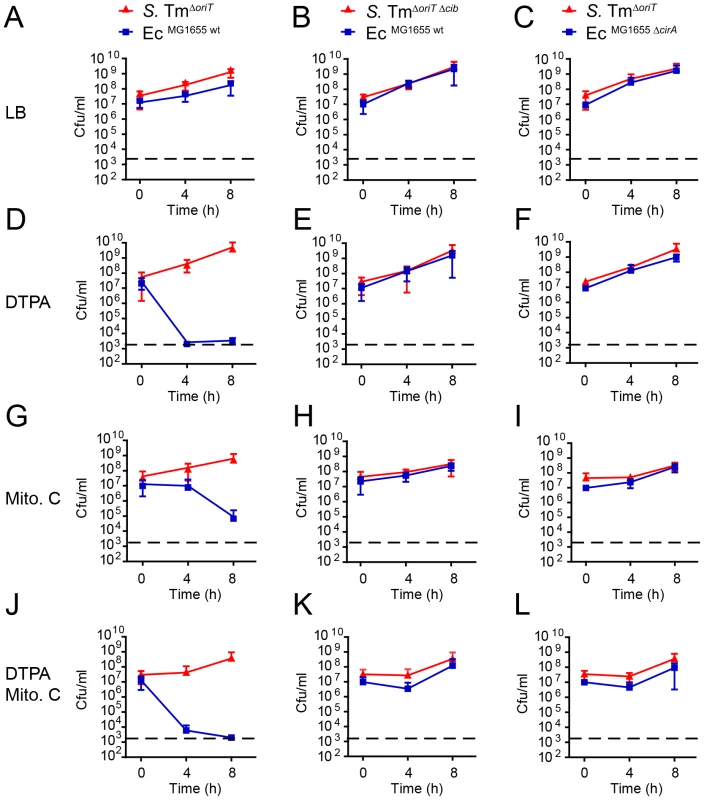
Overnight cultures of S. TmΔoriT and EcMG1655 (A, D, G, J), S. TmΔoriT Δcib and EcMG1655 (B, E, H, K) or S. TmΔoriT and EcMG1655 ΔcirA (C, F, I, L) were diluted and normalized to an OD600 of 0.05 for each strain in fresh LB with indicated supplements (0.25 µg/ml mitomycin C (Mito. C); 100 mM DTPA). Cfu/ml of both strains were determined at 0 h, 4 h, and 8 h after the start of the subculture. Red lines: S. Tm strains, blue lines: E. coli strains. Dotted line: detection limit (2000 cfu/ml). Inflammation-induced Enterobacterial blooms foster cib and cirA expression in vivo
The in vitro co-culture experiments of S. Tm and EcMG1655 showed that under iron excess conditions and in the absence of triggers of the SOS response, ColIb confers little to no detectable benefit to S. Tm. In contrast, under FeIII-limitation or in the presence of stressors, S. Tm outcompetes E. coli, which is dependent on ColIb production by S. Tm and cirA expression by E. coli. Based on these data, we reasoned that ColIb-dependent competition of S. Tm and EcMG1655 in the inflamed gut could indeed be due to increased production of ColIb by S. Tm, or up-regulation of the colicin receptor CirA by E. coli or both. To address this, we analyzed expression of EcMG1655 cirA and S. Tm ColIb (cib) in the streptomycin mouse colitis model using firefly-luciferase reporter-constructs. To generate inflammatory and non-inflammatory conditions in the intestine, LCM mice were infected either with virulent (S. TmΔoriT or S. Tmwt; +inflammation) or avirulent (S. TmΔoriT avir or S. Tmavir; -inflammation) Salmonella strains, respectively. To investigate regulation of EcMG1655 cirA expression, mice were co-infected with EcMG1655 carrying the pcirA-luc-reporter plasmid (Figure 7A,C). To investigate regulation of S. Tm cib expression, mice were co-infected with S. TmΔoriT avir carrying the pcib-luc-reporter plasmid (Figure 7B,D). The experiments showed that luciferase levels for both the pcirA-luc - and the pcib-luc-reporters were significantly increased in the inflamed intestine (Figure 7A–D). Therefore, these data are in agreement with our initial hypothesis and suggest that in response to gut inflammation, ColIb production by S. Tm and susceptibility of EcMG1655 to ColIb are increased. This explains how inflammation fuels ColIb dependent competition of S. Tm and commensal E. coli (Figure 8).
Fig. 7. S. Tm ColIb and the E. coli ColIb receptor CirA are induced in the inflamed gut. 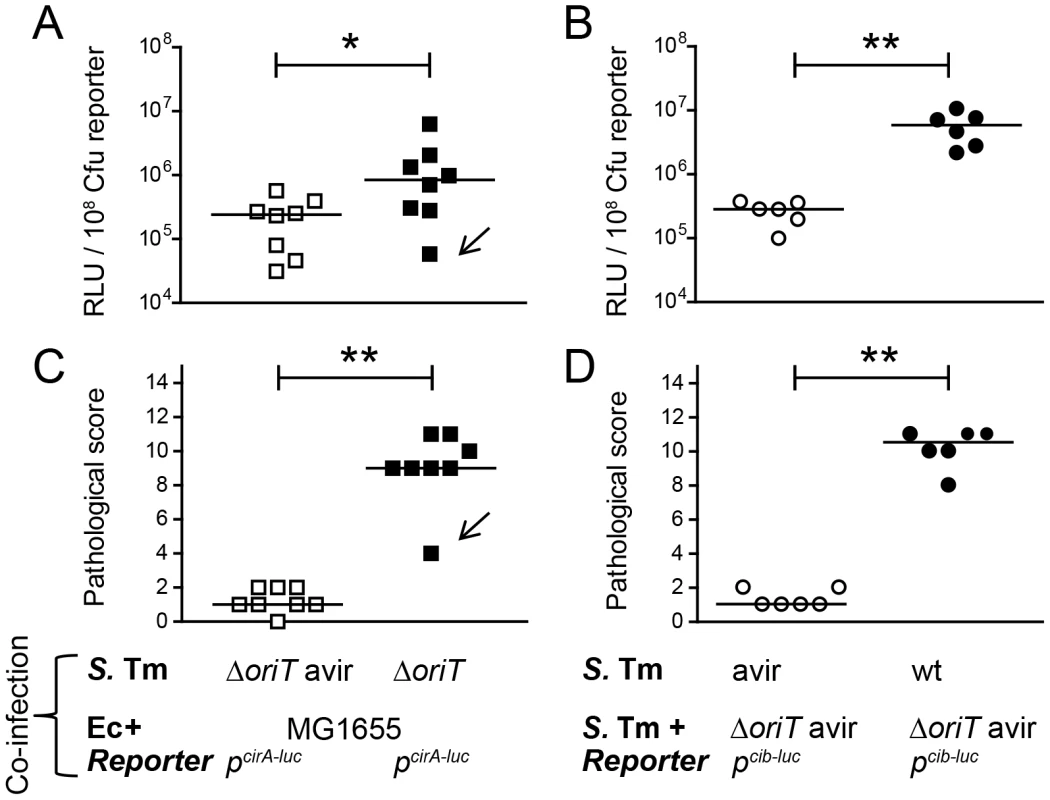
To measure in vivo regulation of E. coli cirA expression, streptomycin-treated LCM-mice were co-infected with 1∶1 mixtures of the luciferase-reporter strain EcMG1655 pcirA-luc and S. TmΔoriT avir (avirulent) or S. TmΔoriT (wildtype), respectively (A, C). To measure in vivo regulation of S. Tm cib expression, streptomycin-treated LCM-mice were co-infected with the luciferase-reporter strain S. TmΔoriT pcib-luc and S. Tmavir (avirulent) or with S. Tmwt (wildtype), respectively (B, D). Bacteria were harvested from cecal content and luciferase-activity was measured in cecum content (A, B). Relative luminescence units (RLU) per 108 cfu of the reporter strain are indicated (C, D). Gut inflammation as determined by pathological score of cecal tissue sections of the infected mice. 1–3: no pathological changes; 4–7: moderate inflammation; above 8: severe inflammation. Bars show the median. Arrow in (A) and (C) point at one animal with atypically mild inflammatory symptoms. Fig. 8. Model for the role of colicins for bacterial competition in inflammation-induced blooms. 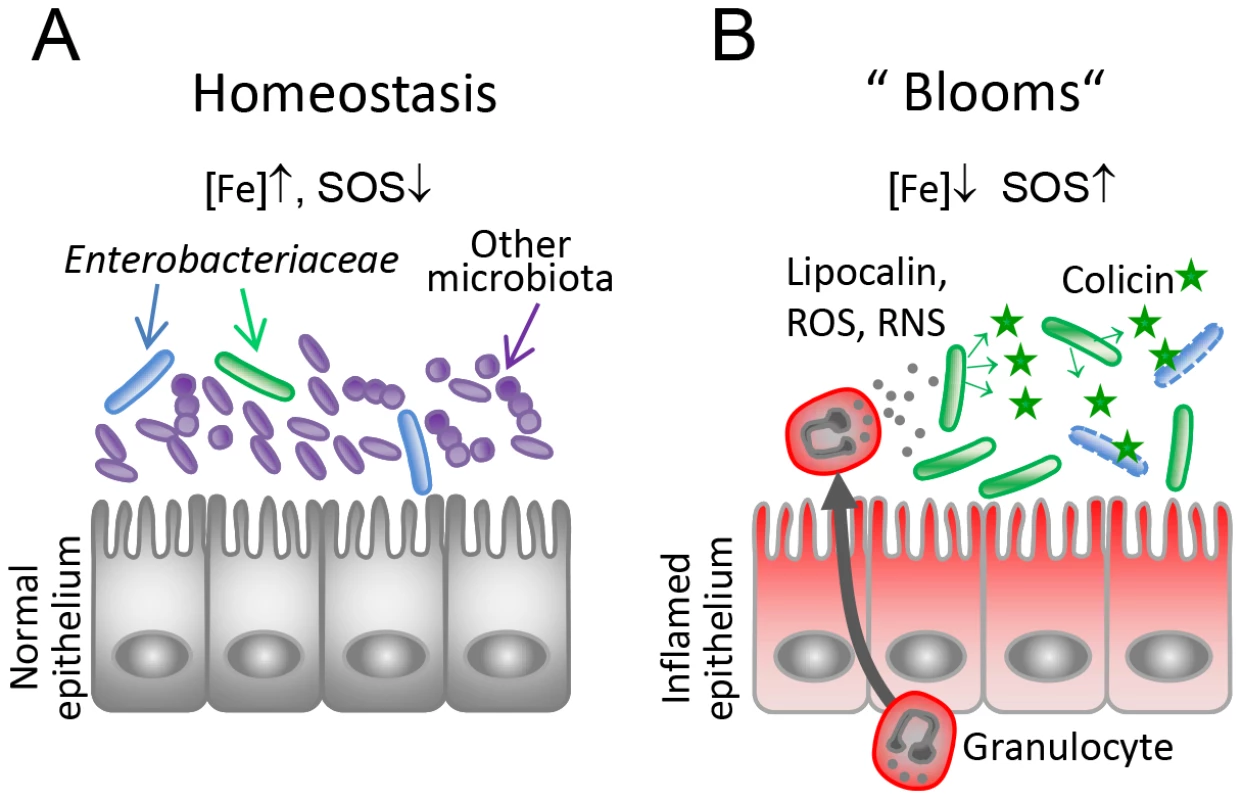
(A) Under homeostatic conditions, Enterobacteriaceae (blue, green) are reduced in numbers as they are kept in check by the obligate anaerobic microbiota (violet-blue). Under this condition, colicin expression as well as expression of the colicin surface receptors are relatively repressed (high iron, no triggers of the SOS response). (B) Upon induction of an inflammatory response, gut microbial ecology is altered leading to Enterobacterial blooms. Neutrophils transmigrate into the gut lumen and produce iron-depleting agents (lipocalin, lactoferrin) and reactive oxygen and nitrogen species (ROS, RNS). This triggers SOS-and Fur-dependent transcriptional responses in Enterobacteriaceae and colicin- and colicin-receptor expression is induced. Thereby, the inflammatory response drives colicin-dependent enterobacterial competition. Discussion
Enterobacterial blooms provide environmental cues for colicin production and susceptibility
Inflammatory conditions in the gut shape gut microbial community structure and are characterized by an increased prevalence of facultative anaerobic bacteria (“blooms”), in particular members of the Enterobacteriaceae family [4], [8], [40]. Commensal E. coli strains can hitchhike gut inflammation induced by inflammatory bowel diseases or enteric pathogens (S. Tm or Citrobacter rodentium) [5], [7], [12]. Commensal and pathogenic representatives of the Enterobacteriaceae efficiently dwell in blooms as they can exploit resources with increased abundance in the inflamed gut (ethanolamine, the anaerobic electron-acceptors tetrathionate and nitrate) [11], [12], [41]. Up to 15 different E. coli strains can be detected in one individual human gut ecosystem [42]. Thus, successful competition for resources should be of major importance for bacteria in order to come out on top and eventually benefit from an episode of gut inflammation [8]. Here, we propose that colicins such as ColIb are effective means to fight off competing bacteria, particularly in inflammation-induced enterobacterial blooms.
Which environmental cues promote ColIb-dependent bacterial competition in blooms? As an immediate inflammatory defence reaction, the host deprives potential invading pathogens of nutritionally-required iron. Neutrophils release iron-sequestering lactoferrin upon degranulation [43]. Further, lipocalin 2 (LCN2), an antimicrobial protein binding the bacterial siderophore enterochelin [44] is abundantly expressed by neutrophils and intestinal epithelial cells. LCN2 is produced in response to infection with wildtype but not avirulent S. Tm strains in the streptomycin-colitis model [9]. S. Tm overcomes LCN2-imposed inhibition by production of salmochelin [45], [46], a siderophore resistant to binding by LCN2. Salmochelin is produced in a Fur-dependent manner in response to iron limitation and upregulated by S. Tm thriving in the inflamed gut [10]. Therefore, host-mediated iron restriction in the inflamed gut poses an environmental cue for inducing ColIb production by S. Tm. Other cues for ColIb production are compounds triggering the SOS response. Yet, the exact source and identity of those compounds in the inflamed gut is ill-defined. The SOS response in Enterobacteriaceae can be triggered by antibiotics which directly affect DNA integrity (mitomycin C), DNA replication via the DNA gyrase (quinolones) or induce membrane stress (β-lactams) [28], [47]. Further, oxidative stress induced by reactive oxygen species (ROS), superoxides, H2O2 or free radicals formed by UV-light induce DNA damage and thereby the SOS response [28], [48]. Neutrophils infiltrating the intestinal lumen in response to Salmonella infection as well as activated iNOS-producing epithelial cells likely represent a source of free radicals and potential inducer of the SOS response [40], [49]. Recently, it was demonstrated that E. coli indeed upregulates stress-induced proteins such as GroL, RecA, YggE and the Fe-S cluster repair protein NfuA in the inflamed gut [50].
Not only was the expression of cib increased in the inflamed gut, but also the corresponding surface receptor cirA. CirA is the receptor for monomers, dimers and linear trimers of 2,3-dihydroxybenzoylserine, breakdown products of enterochelin [51]. Further, it was shown, that cirA mutants are attenuated in the uptake of monomeric catechol and its analogues [52]. In order to avoid iron overload and its negative consequences (e.g. the formation of hydroxyl-radicals), iron-uptake systems are tightly controlled at the transcriptional level and only de-repressed under iron-limiting conditions. In vitro, susceptibility to ColIb of E. coli was drastically increased under FeIII-depleted conditions, which correlated with elevated CirA protein levels. In the same way, the in vitro competition experiments suggest that S. Tm benefits most from ColIb production under iron-limiting conditions (7×107-fold). The growth advantage was lower in the presence of mitomycin C (1×104-fold) or under both iron limitation and mitomycin C (6×106-fold). Of note, the amount of free S. Tm ColIb in the medium was even higher in the presence of mitomycin C, than with addition of DTPA only (Figure 3D), supporting the idea, that the expression level of cirA has a higher impact on the competition, compared to that of ColIb (Figure 6D,G). Hitherto, the underlying mechanism of ColIb-release or secretion triggered by the SOS response is not known. In conclusion, we reason that increased cirA expression and consequential high susceptibility of E. coli to ColIb may explain for the most part colicin-dependent competition in blooms (Figure 8).
Interestingly, several other colicins parasitize siderophore outer membrane receptors, which are all under control of the Fur-regulon. ColM binds to the ferrichrome receptor FhuA while ColB and ColD bind to FepA, the receptor for enterobactin [15]. This suggests that increased sensitivity to colicin-mediated killing under iron depletion may also apply for other colicins binding to TonB-dependent outer-membrane transporters. Previously, it was shown that killing of susceptible bacteria by pyocin, a bacteriocin produced by Pseudomomas spp., is increased under iron-limited conditions [53]. Pyocin binds the ferri-pyoverdine receptor FpvA, which is controlled by Fur. In summary, physiological changes of the murine intestine upon Salmonella-induced colitis are likely to provide the environmental cues required for upregulation of both, ColIb and its receptor CirA. Yet, we cannot rule out that other physiological parameters altered in the inflamed gut also contribute to the observed phenotype. So far, it is unclear if expression of other types of colicins as well as other Enterobacteriaceae-derived bacteriocins (microcins, pyocins, klebicins) would be upregulated in the inflamed gut. The majority of these bacteriocins is only under control of the SOS response and repressed by LexA and not regulated in a Fur-dependent fashion. Thus, it remains to be shown if the principle of colicin-colicin-receptor upregulation in the inflamed gut also applies to other bacteriocins and their respective receptors.
Colicin-dependent bacterial competition is enhanced in the inflamed gut
Supposedly, colicins play a major role in mediating bacterial population dynamics [54]. Superior fitness of colicinogenic over sensitive strains in vivo could so far only be demonstrated in few studies performing long-term competitive infection experiments (≥12 weeks) [30], [31]. In contrast, a number of other studies reported that high bacteriocidal activity against closely related, sensitive strains observed in vitro could not be recapitulated in in vivo experiments (see below). Our data presented in this paper might explain this puzzling observation: we suggest that the fitness benefit of colicin production strongly depends on the environmental conditions prevailing in the gut. Under normal conditions, colicin expression and expression of their cognate receptors may not be stimulated enough to induce colicin-dependent inhibition of the sensitive strain. In the absence of gut inflammation, S. Tm did not benefit from ColIb in competition with E. coli. Likewise, competition experiments in germfree mice with a colicin-producing E. coli and a sensitive strain resulted in equal colonization levels of both strains over weeks (no inflammation induced) [32], [33], [34] and similar results were obtained for other strains and colicins [55], [56]. The underlying reasons for the absence of an overt fitness-benefit of colicin production were attributed to colicin inactivation by intestinal proteases [33], acquisition of colicin-resistances [55] or absent colicin activity under anaerobic conditions [35], [36]. Our study suggests that absence of inflammatory conditions might be an additional explanation.
Implications for the evolution of colicin-dependent competition
Colicin production is a common trait in E. coli populations [54]. On average, 30% of natural E. coli populations produce one or more colicins [57]. Many experimental and theoretical studies have addressed the ecological consequences of colicin production in bacterial populations [58], [59]. In general, it is assumed, that colicins afford a competitive advantage to the strain producing it. However, the producer pays a fitness cost due to the higher metabolic load of colicin synthesis as well as lethality of production (e.g. lysis-mediated colicin release). Bacteria have partially overcome this limitation by applying the principle of ‘division of labor’. In a population of colicin producers, only a small fraction of bacteria are induced to produce the colicin ( = phenotypic heterogeneity) [60], [61]. In recA negative strains decreased frequencies of colicin producers were observed, suggesting that the rate of colicin production is regulated by the SOS response [60]. This strategy seems to be evolutionary successful, as colicins released by the subpopulation serve as a “common good” for the whole population and secure propagation of the shared genotype. Nevertheless, colicin expression needs to be tightly controlled to ensure, that the fraction of producers is kept at low rates under conditions, when colicin is not required. Those conditions include the lack of stress, nutrient starvation but also the absence of any direct competitors.
Thus, we assume that colicin production of a bacterial population should be confined to environmental niches which are characterized by high density and diversity of competing E. coli and close relatives. E. coli titers in the mammalian gut lumen are rather low as the intestinal tract is dominated by strictly anaerobic bacteria [62]. E. coli predominantly colonizes the mucus layer of the large intestine where it thrives on mucin-derived sugars [63]. Thus, the intestinal mucus layer is one highly competitive environment for E. coli and colicins may be beneficial for competing for the preferred limiting substrates [64]. In contrast, we identify inflammation-induced blooms as an alternative niche for colicin-dependent Enterobacterial competition (Figure 8). Enterobacterial blooms can contain multiple closely-related species at high concentrations which likely compete for the same resources. Under this highly competitive situation, the chances are increased that colicin-sensitive competitors are present at high numbers. Thus, the bacteria may benefit hugely from colicin production under this environmental condition. Moreover, the population size of the colicin-producer is large enough to tolerate loss of a fraction of the population due to suicidal colicin production. In summary, the results presented here provide evidence that intestinal inflammation drives colicin-dependent competition by bacteria of the Enterobacteriaceae family. These findings shed new light on the role of colicins as important fitness factors providing a competitive advantage for growth in Enterobacterial blooms.
Materials and Methods
Ethics statement
All animal experiments were approved by the Regierung von Oberbayern and the Kantonales Veterinäramt Zürich performed according to national German and Swiss guidelines (Deutsches TschG; Schweizer Kantonale TschV). The permit no. 55.2-1-54-2532-49-11 (Germany) and 201/2007 (Switzerland).
Animal experiments
All mice used in the study were on C57BL/6J background and bred at the Rodent Center, ETH Zürich and the Max-von-Pettenkofer Institute, LMU Munich under SPF conditions in individually ventilated cages. Low-complexity microbiota (LCM) mice were generated by associating germfree mice with members of the Altered Schaedler flora [65] as described previously [66]. Conventional SPF C57BL/6J mice were purchased from Janvier, Le Genest Saint Isle. For infections, conventional and LCM mice were pretreated with streptomycin and infected by gavage with 5×107 cfu S. Tm or mixtures of S. Tm and E. coli as described [7]. For in vivo luciferase-assays, LCM mice were pretreated with ampicillin (20 mg/animal 24 h prior to infection). Live bacterial loads in the cecal content were determined by plating on MacConkey-agar (Roth) with respective supplements (streptomycin 100 µg/ml; kanamycin 30 µg/ml; chloramphenicol 30 µg/ml; ampicillin 100 µg/ml and tetracycline 12,5 µg/ml). Histology of the cecum was done at necropsy. Cecum tissue was embedded in O.C.T. (Sakura, Torrance) and flash frozen. Cryosections (5 µm) of the cecal tissue were H&E-stained and scored as described in detail in [6]. The parameters submucosal edema, PMN infiltration, loss of goblet cells and epithelial damage were scored according to the severity of inflammatory symptoms yielding a total score of 0–13 points. For infections, E. coli and S. Tm strains were grown as described [67]. Briefly, cultures in LB supplemented with 0.3M NaCl were inoculated with 2–3 bacterial colonies from plates. Bacteria were grown over night for 12 h and subcultures (1∶20) for an additional 4 h. Bacteria were mixed (as indicated) washed in PBS and applied to the mice in a total volume of 50 µl by gavage.
Construction of bacterial mutants and plasmids
Bacterial strains and plasmids used in this study are listed in Table 1. EcMG1655 ΔcirA (LPN2) was constructed using the lambda Red recombinase system as described using pKD4 as template for the kanamycin-resistance gene including the FRT-sites [68]. Briefly, EcMG1655 was transformed with the plasmid pKD46. The kanamycin resistance cassette from plasmid pKD4 was amplified by PCR using primers K12ΔcirA_Fwd/K12ΔcirA_Re (Table S1). Correct recombination was verified by PCR using primers cirA-up/cirA-down and cirA-up/cirA-d1 (Table S1). S. Tmavir ΔoriT (LPN5) was generated by P22-transduction of the ΔoriTnikA::cat allele from M1407 into M557 [37]. Correct insertion was verified by PCR using primers ΔoriTnikArev_val, ΔoriTnikA_val. The P2 plasmid was cured from S. Tm ΔinvG; sseD::aphT cured as described previously [7]. For the generation of c-terminal CirA-His-tag fusion, the open reading frame of cirA was amplified from E coli Nissle genomic DNA by PCR, using Fow_cirA_NheI and Re_cirA_XhoI primers (Table S1) and cloned into pET-24c (Novagen) via NheI and XhoI to yield pLPN13. For the generation of c-terminal ColIb-His-tag fusion, the ColIb gene cib was amplified from S. Tmwt genomic DNA by PCR, using primers Fow_colicin_NheI and Re_colicin_XhoI (Table S1) and cloned into pET-24c via NheI and XhoI to yield pLPN14. To generate pLPN1, the cirA promoter was amplified from E. coli Nissle using pcirA-BamHI, pcirA-XbaI (Table S1) and inserted in BamHI and XbaI digested pM979 [69]. For generation of pM1437, the cib promoter from E. coli8178 was amplified using pColIb-XbaI, pColIb-BamHI (Table S1) and inserted in pM968 [69] via restriction with XbaI and BamHI. To generate pLPN15 and pLPN16, the firefly-luciferase gene luc from pLB02 [70] was amplified with Luc-for-BamHI and Luc-rev-HindIII primers (Table S1) and inserted into BamHI/HindIII digested pLPN1 or pM1437, respectively.
Primers pWSK29-Gbs-for and pWSK29-Gbs-rev were used in a PCR with pWSK29 [71] as a template to amplify the low-copy-number plasmid (Table S1). Primers CirA-pWSK29-Gbs-for and CirA-pWSK29-Gbs-rev were used in a PCR with chromosomal DNA of EcMG1655 as a template to amplify cirA including its natural promoter. Primers Cib-Imm-pWSK29-Gbs-for and Cib-Imm-pWSK29-Gbs-rev were used in a PCR with S. Typhimurium strain SL1344 plasmid pCol1B9_SL1344 as a template to amplify the cib/imm locus including both natural promoters. The pWSK29 PCR fragment was combined with the cirA or cib/imm fragment in a Gibson assembly reaction [72]. Four microliters of the Gibson assembly mix were transformed into chemically competent E. coli Mach1 T1 cells (Life Technologies). Constructs were verified using colony PCR, restriction analysis and sequencing.
Identification of regulator binding sites
For annotation of transcription factor binding sites (Fur and LexA regulon), all known transcription factor binding sites of each family one were taken from RegulonDB (version 8.0) [73] and a binding motif was created using MEME [74]. The nucleotide sequences of the cib (S. Tm SL1344; EMBL accession no. FQ312003) and cirA promoter regions (E. coli MG1655 genome accession no. NC_000913.2) were searched for the computed MEME binding site motifs using MAST [74].
Generation and affinity purification of recombinant proteins
For expression of ColIb-His, we used E. coli BL21 transformed with pC831-2 (expression of the ColIb immunity protein gene imm [7]) and pLPN14. For expression of cirA-His we used E. coli BL21 transformed with pLPN13 (Table S1). Over-night cultures of bacteria, grown at 37°C, 180 rpm in Luria-Bertani (LB) medium containing antibiotics were used for inoculation of subcultures (dilution 1∶20). At OD600 between 0.6–0.8 the subcultures was induced with 0.1 mM isopropyl β –D-thiogalactopyranoside (IPTG) and incubated for additional 4 h at 37°C, 180 rpm. Bacteria were harvested (4,500 rpm, 30 min at 4°C), resuspended in 40 ml 1×PBS and spun down at 5,000 rpm, 20 min at 4°C. The pellet was frozen at −20°C. Thereafter, the pellet was thawed and resuspended in 25 ml loading buffer (40 mM Na2HPO4, 0.3M NaCl, 5 mM Imidazol, pH 7.8), supplemented with 2 mM phenylmethylsulfonyl fluoride (PMSF) and benzonase nuclease (Novagen). Bacteria were lysed in the French Press (1,000 PSI) and the lysate was filtered (0.22 µm). Further, the lysate was loaded on a 5 ml HisTrap column (GE Healthcare), and purified using the ÄKTA system (GE Healthcare). ColIb-His was eluted with 5 mM Imidazole. The fractions containing the protein were desalted on a 5 ml HiTrap desalting column (GE Healthcare), using the ÄKTA system and exchange buffer (20 mM Na2HPO4, 100 mM NaCl, pH 7.4). CirA-His was purified as outlined above for ColIb-His, but with following exceptions: loading buffer for the French Press (8M Urea, 40 mM Na2HPO4, 0.3M NaCl, 5 mM Imidazol, pH 7.8, 2 mM PMSF, and Benzonase nuclease): loading buffer for HisTrap column (6M Urea, 40 mM Na2HPO4, 0.3M NaCl, 5 mM Imidazol, pH 7.8); exchange buffer (4M Urea, 20 mM Na2HPO4, 100 mM NaCl, pH 7.4). Rabbit antisera against ColIb-His and CirA-His were raised using standard protocols (Pineda Antikörper-Service, Berlin, Germany). 6 mg/ml ColIb-His (in 20 mM Na2HPO4, 100 mM NaCl, pH 7.4) and 6 mg/ml CirA-His (in 20 mM Na2HPO4, 100 mM NaCl, 4M Urea, pH 7.4) were used for immunization. Control and immune serum were received from bleedings day 61, 90 and 135 post immunization.
Affinity purification of rabbit-antisera
Affinity purification of polyclonal rabbit α-ColIb-His antiserum was done using the Aminolink kit (Thermo Scientific) following the manufacturer's protocol with some minor modifications. For ColIb-His, PBS was used as binding/wash buffer and 1M Glycine, pH 2.7 was used as elution buffer. ColIb-His (stored in 20 mM Na2HPO4, 100 mM NaCl, pH 7.4) was added to the binding buffer at 1∶3 ratio. Desalting of the affinity-purified rabbit-α-ColIb-His antiserum was done using PD-10 desalting columns with PBS as exchange buffer (GE Healthcare). For CirA-His, His-tagged CirA (20 mM Na2HPO4, 100 mM NaCl, 4M Urea, pH 7.4) was dialyzed against PBS using 5 ml Zebra Spin desalting columns (Thermo scientific) shortly before coupling to the column. Coupling was done with CirA-His (in PBS) and coupling buffer supplemented with 4M Urea (MP Biomedicals). Desalting of the affinity-purified rabbit-α-CirA-His was done with Zebra Spin desalting columns with PBS containing 0.05% sodium azide (Merk). Purified antisera were stored at −80°C with addition of sodium azide to 0.01%.
Colicin killing-assay
For measuring colicin production and -sensitivity, the colicin-producing strain was grown o.n. as small spot (ø5 mm) on LB agar containing 0.25 µg/ml mitomycin C (Roth). The plate was overlaid with the tester strain in top-agar (0.75% agar). Growth of the tester strain was analyzed after 24 h. Formation of an inhibition zone (halo) around the producer indicated production of colicin and sensitivity of the tester strain. For determining ColIb sensitivity dependent on the FeIII concentration, the assay was modified accordingly. Starter cultures of E. coli in 3 ml M9 medium (40 mM Na2HPO4×2H2O, 20 mM KH2PO4, 9 mM NaCl, 2 g/L NH4Cl, 1 mM MgSO4, 100 µM CaCl2, 2 g/l D-glucose, 10 mg/l thiamine, 500 mg/l histidine) were grown for 10 h and used for inoculation (1∶20) of 2 ml M9 medium, supplemented with 1 µM, 10 µM, 0.1 mM or 1 mM FeCl3 and grown for 12 h. From each subculture, 50 µl was added to 5 ml 50°C 0.7% M9 top-agar (0.75% agar), which was used to overlay M9 agar plates. Further, antimicrobial susceptibility test discs (Oxoid) were laid on each plate and supplemented with 8 µl 1.3 mg/ml recombinant His-tagged ColIb. The plates were incubated over-night at 37°C.
Growth of bacterial strains for in vitro assays
Bacteria were grown in a starter culture in LB or M9 media and used for inoculation of subcultures (1∶20), except of in vitro co-cultures, where subcultures were inculcated to an OD600 of 0.05 for each strain. Following supplements were used: 0.25 µg/ml mitomycin C (Roth); 100 µM diethylenetriaminepentaacetic acid (DTPA; Sigma), 1 µM, 10 µM, 0.1 mM or 1 mM FeCl3 (Sigma). All cultures were grown at 37°C on a wheel rotor, except of in vitro co-cultures, where subcultures were grown in Erlenmeyer-flasks in a shaker at 200 rpm.
Luciferase assay
Luciferase assays were performed as described [75]. Briefly, overnight cultures (3 ml LB, 100 µg/ml ampicillin) were grown for 12 h and used for 1∶20 inoculation of 3 ml subcultures (LB, 100 µg/ml ampicillin with respective supplements) and grown for 4 h. From each subculture, 250 µl (of an OD600 of 1) was spun down for 5 min, 14,000 rpm, 4°C. The supernatant was removed and the bacterial pellet was frozen at −80°C for 1 h. Further, the pellet was thawn and resuspended in 500 µl lysis buffer (100 mM K2HPO4/KH2PO4 buffer, pH 7.8, 2 mM EDTA, 1% Triton X-100, 5 mg/ml BSA, 1 mM DTT and 5 mg/ml lysozyme) and incubated for 15 min at room temperature while vortexing every 3 minutes. Bacterial lysates (25 µl) were transferred in 96-well plates (white; Thermo scientific) and 50 µl luciferase reagent [1 mM (MgCO3)4Mg(OH)2×5H2O, 20 mM tricine, 0.1M EDTA, 470 µM D(−) luciferin (Sigma), 530 µM Mg-ATP (Sigma), 125 µM glycylglycine (Sigma), 270 µM Li3-coenzym A (Sigma), 33 mM DTT] was added to each well. Luminescence was measured using a FLUOstar Optima plate reader (BMG Labtech).
For luciferase assay from bacteria extracted from cecum content, the cecum content was harvested from infected mice and shortly stored on ice. The cecum content was resuspended in 500 µl PBS (0.1% tergitol) and mixed in a tissue-lyser (Qiagen; 5 min; 50 Hz). Further, the cecum content was filtered through a 40 µm cell-sieve (Milian). Samples were taken to determine the cfu/ml of the reporter strain by plating on MacConkey agar with respective antibiotics. A defined volume (i.e. 900 µl) was pelleted at 4°C, 2 min, 14,000 rpm. The supernatant was removed and the pellet was frozen in dry ice and stored at −80°C. The samples were then thawn and processed as described above. Only values above detection limit (control cecum content) were considered. The relative luminescence units (rlu) per cfu luciferase-reporter strain were calculated.
Generation of samples for immunoblot
Overnight cultures of 3 ml M9 medium, grown for 12 h were used for inoculation (1∶20) of 2 ml M9 medium supplemented with 1 µM, 10 µM, 0.1 mM or 1 mM FeCl3 subculture, grown for 7 h. Starter culture of 3 ml LB, grown for 12 h was used for inoculation of 3 ml LB with supplements grown for 4 h. From each subculture, 250 µl (for an OD600 of 1) was taken, spun down at 4°C, 10 min, 10, 000 rpm. The supernatant was removed and bacterial pellet was frozen in liquid nitrogen and thawn at room temperature for 15 min (repeated three times), resuspended in 100 µl lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.25% nonidet P-40, 1 mg/ml lysozyme) and incubated in thermomixer at 550 rpm at 23°C, for 1 h and thereafter spun down at 4°C, 20 min, 14 000 rpm. Total protein was quantified in the lysate using protein assay reagent (BioRad). Further, bacterial lysate was added to protein loading buffer (50 mM Tris, 100 mM DTT, 2% SDS, 0.1% bromphenolblue, 10% glycerol) and incubated for 10 min at 95°C. For supernatant fractions 500 µl (for an OD600 of 1) of the subculture were spun down twice, supernatant was added to 5× protein loading buffer and incubated for 10 min at 95°C.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot
Proteins were separated by SDS gel electrophoresis [76]. Proteins were transferred onto a nitrocellulose membrane (GE Healthcare) at 300 mA for 2 h. The membrane was blocked in PBS (0.1% tween; 5% milk powder) and probed with antisera (affinity-purified α-CirA-His (1∶50) or affinity-purified α-ColIb-His (1∶500). Goat-α-rabbit-HRP (GE-Healthcare) was used as secondary antibody. For detection of E. coli and S. Tm DnaK, the mouse monoclonal α-E. coli DnaK antibody (mAb 8E2/2; Enzo Life Sciences) and a secondary goat-α-mouse-HRP (Sigma) was used. Blots were developed with ECL detection system (GE Healthcare).
Statistical analysis
Statistical analysis was performed using the exact Mann-Whitney U Test (Graphpad Prism Version 5.01). P-values less than 0.05 (2-tailed) were considered statistically significant.
Supporting Information
Zdroje
1. MushinR, DubosR (1965) Colonization of the mouse intestine with Escherichia coli. J Exp Med 122 : 745–757.
2. BarthelM, HapfelmeierS, Quintanilla-MartinezL, KremerM, RohdeM, et al. (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71 : 2839–2858.
3. StecherB, HardtWD (2010) Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol 14(1): 82–91.
4. SansonettiPJ (2008) Host-bacteria homeostasis in the healthy and inflamed gut. Curr Opin Gastroenterol 24 : 435–439.
5. LuppC, RobertsonML, WickhamME, SekirovI, ChampionOL, et al. (2007) Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of enterobacteriaceae. Cell, host and microbe 2 : 119–129.
6. StecherB, RobbianiR, WalkerAW, WestendorfAM, BarthelM, et al. (2007) Salmonella enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota. PLoS Biol 5: e244.
7. StecherB, DenzlerR, MaierL, BernetF, SandersMJ, et al. (2012) Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A 109 : 1269–1274.
8. StecherB, MaierL, HardtWD (2013) ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol 11 : 277–284.
9. RaffatelluM, GeorgeMD, AkiyamaY, HornsbyMJ, NuccioSP, et al. (2009) Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5 : 476–486.
10. LoetscherY, WieserA, LengefeldJ, KaiserP, SchubertS, et al. (2012) Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS ONE 7: e34812.
11. WinterSE, ThiennimitrP, WinterMG, ButlerBP, HusebyDL, et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467 : 426–429.
12. WinterSE, WinterMG, XavierMN, ThiennimitrP, PoonV, et al. (2013) Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339 : 708–711.
13. LopezCA, WinterSE, Rivera-ChavezF, XavierMN, PoonV, et al. (2012) Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio 3.
14. Rivera-ChavezF, WinterSE, LopezCA, XavierMN, WinterMG, et al. (2013) Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog 9: e1003267.
15. CascalesE, BuchananSK, DucheD, KleanthousC, LloubesR, et al. (2007) Colicin biology. Microbiol Mol Biol Rev 71 : 158–229.
16. ScheinSJ, KaganBL, FinkelsteinA (1978) Colicin K acts by forming voltage-dependent channels in phospholipid bilayer membranes. Nature 276 : 159–163.
17. KoniskyJ (1972) Characterization of colicin Ia and colicin Ib. Chemical studies of protein structure. J Biol Chem 247 : 3750–3755.
18. CardelliJ, KoniskyJ (1974) Isolation and characterization of an Escherichia coli mutant tolerant to colicins Ia and Ib. J Bacteriol 119 : 379–385.
19. LazdunskiCJ, BouveretE, RigalA, JournetL, LloubesR, et al. (1998) Colicin import into Escherichia coli cells. J Bacteriol 180 : 4993–5002.
20. BuchananSK, LukacikP, GrizotS, GhirlandoR, AliMM, et al. (2007) Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J 26 : 2594–2604.
21. JakesKS, FinkelsteinA (2010) The colicin Ia receptor, Cir, is also the translocator for colicin Ia. Mol Microbiol 75 : 567–578.
22. WeaverCA, KaganBL, FinkelsteinA, KoniskyJ (1981) Mode of action of colicin ib: formation of ion-permeable membrane channels. Biochim Biophys Acta 645 : 137–142.
23. MankovichJA, LaiPH, GokulN, KoniskyJ (1984) Organization of the colicin Ib gene. Promoter structure and immunity domain. J Biol Chem 259 : 8764–8768.
24. DaviesJK, ReevesP (1975) Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group B. J Bacteriol 123 : 96–101.
25. MankovichJA, HsuCH, KoniskyJ (1986) DNA and amino acid sequence analysis of structural and immunity genes of colicins Ia and Ib. J Bacteriol 168 : 228–236.
26. EscolarL, Perez-MartinJ, de LorenzoV (1999) Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181 : 6223–6229.
27. ButalaM, Zgur-BertokD, BusbySJ (2009) The bacterial LexA transcriptional repressor. Cell Mol Life Sci 66 : 82–93.
28. KelleyWL (2006) Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol Microbiol 62 : 1228–1238.
29. RileyMA, WertzJE (2002) Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56 : 117–137.
30. GillorO, GiladiI, RileyMA (2009) Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol 9 : 165.
31. KirkupBC, RileyMA (2004) Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428 : 412–414.
32. IkariNS, KentonDM, YoungVM (1969) Interaction in the germfree mouse intestine of colicinogenic and colicin-sensitive microorganisms. Proc Soc Exp Biol Med 130 : 1280–1284.
33. KelstrupJ, GibbonsRJ (1969) Inactivation of bacteriocins in the intestinal canal and oral cavity. J Bacteriol 99 : 888–890.
34. CravenJA, MiniatsOP, BarnumDA (1971) Role of colicins in antagonism between strains of Escherichia coli in dual-infected gnotobiotic pigs. Am J Vet Res 32 : 1775–1779.
35. BraunV, FrenzJ, HantkeK, SchallerK (1980) Penetration of colicin M into cells of Escherichia coli. J Bacteriol 142 : 162–168.
36. de GraafFK (1973) Effects of cloacin DF13 on the functioning of the cytoplasmic membrane. Antonie Van Leeuwenhoek 39 : 109–119.
37. HapfelmeierS, StecherB, BarthelM, KremerM, MüllerA, et al. (2005) The Salmonella Pathogenicity Island (SPI)-1 and SPI-2 Type III Secretion Systems Allow Salmonella Serovar Typhimurium to trigger Colitis via MyD88-Dependent and MyD88-Independent Mechanisms. J Immunol 174 : 1675–1685.
38. TaylorCM, OsmanD, CavetJS (2009) Differential expression from two iron-responsive promoters in Salmonella enterica serovar Typhimurium reveals the presence of iron in macrophage-phagosomes. Microb Pathog 46 : 114–118.
39. GriggsDW, TharpBB, KoniskyJ (1987) Cloning and promoter identification of the iron-regulated cir gene of Escherichia coli. J Bacteriol 169 : 5343–5352.
40. WinterSE, LopezCA, BaumlerAJ (2013) The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 14 : 319–327.
41. ThiennimitrP, WinterSE, WinterMG, XavierMN, TolstikovV, et al. (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108 : 17480–17485.
42. Apperloo-RenkemaHZ, Van der WaaijBD, Van der WaaijD (1990) Determination of colonization resistance of the digestive tract by biotyping of Enterobacteriaceae. Epidemiol Infect 105 : 355–361.
43. WardPP, Uribe-LunaS, ConneelyOM (2002) Lactoferrin and host defense. Biochem Cell Biol 80 : 95–102.
44. BachmanMA, MillerVL, WeiserJN (2009) Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog 5: e1000622.
45. BaumlerAJ, NorrisTL, LascoT, VoightW, ReissbrodtR, et al. (1998) IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol 180 : 1446–1453.
46. FischbachMA, LinH, ZhouL, YuY, AbergelRJ, et al. (2006) The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc Natl Acad Sci U S A 103 : 16502–16507.
47. MillerC, ThomsenLE, GaggeroC, MosseriR, IngmerH, et al. (2004) SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305 : 1629–1631.
48. GoerlichO, QuillardetP, HofnungM (1989) Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J Bacteriol 171 : 6141–6147.
49. SegalAW (2005) How neutrophils kill microbes. Annu Rev Immunol 23 : 197–223.
50. SchumannS, AlpertC, EngstW, LohG, BlautM (2012) Dextran sodium sulfate-induced inflammation alters the expression of proteins by intestinal Escherichia coli strains in a gnotobiotic mouse model. Appl Environ Microbiol 78 : 1513–1522.
51. HantkeK (1990) Dihydroxybenzoylserine–a siderophore for E. coli. FEMS Microbiol Lett 55 : 5–8.
52. NikaidoH, RosenbergEY (1990) Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with beta-lactam antibiotics containing catechol and analogous groups. J Bacteriol 172 : 1361–1367.
53. OhkawaI, ShigaS, KageyamaM (1980) Effect of iron concentration in the growth medium on the sensitivity of Pseudomonas aeruginosa to pyocin S2. J Biochem 87 : 323–331.
54. RileyMA, GordonDM (1999) The ecological role of bacteriocins in bacterial competition. Trends Microbiol 7 : 129–133.
55. HardyKG (1975) Colicinogeny and related phenomena. Bacteriol Rev 39 : 464–515.
56. WadolkowskiEA, LauxDC, CohenPS (1988) Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect Immun 56 : 1030–1035.
57. RileyMA, GordonDM (1992) A survey of Col plasmids in natural isolates of Escherichia coli and an investigation into the stability of Col-plasmid lineages. J Gen Microbiol 138 : 1345–1352.
58. ChaoL, LevinBR (1981) Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci U S A 78 : 6324–6328.
59. HibbingME, FuquaC, ParsekMR, PetersonSB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8 : 15–25.
60. MrakP, PodlesekZ, van PuttenJP, Zgur-BertokD (2007) Heterogeneity in expression of the Escherichia coli colicin K activity gene cka is controlled by the SOS system and stochastic factors. Mol Genet Genomics 277 : 391–401.
61. KamensekS, PodlesekZ, GillorO, Zgur-BertokD (2010) Genes regulated by the Escherichia coli SOS repressor LexA exhibit heterogeneous expression. BMC Microbiol 10 : 283.
62. TenaillonO, SkurnikD, PicardB, DenamurE (2010) The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8 : 207–217.
63. ChangDE, SmalleyDJ, TuckerDL, LeathamMP, NorrisWE, et al. (2004) Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 101 : 7427–7432.
64. Conway TC, P.S. (2007) Escherichia coli at the Intestinal Mucosal Surface. American Society for Microbiology Press, Washington, D C 4th Edition Virulence Mechanisms of Bacterial Pathogens: 175–196,.
65. DewhirstFE, ChienCC, PasterBJ, EricsonRL, OrcuttRP, et al. (1999) Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol 65 : 3287–3292.
66. StecherB, ChaffronS, KappeliR, HapfelmeierS, FreedrichS, et al. (2010) Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 6: e1000711.
67. HapfelmeierS, EhrbarK, StecherB, BarthelM, KremerM, et al. (2004) Role of the Salmonella Pathogenicity Island 1 Effector Proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica Subspecies 1 Serovar Typhimurium Colitis in Streptomycin-Pretreated Mice. Infect Immun 72 : 795–809.
68. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 : 6640–6645.
69. StecherB, HapfelmeierS, MullerC, KremerM, StallmachT, et al. (2004) Flagella and Chemotaxis Are Required for Efficient Induction of Salmonella enterica Serovar Typhimurium Colitis in Streptomycin-Pretreated Mice. Infect Immun 72 : 4138–4150.
70. GunnJS, Alpuche-ArandaCM, LoomisWP, BeldenWJ, MillerSI (1995) Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J Bacteriol 177 : 5040–5047.
71. WangRF, KushnerSR (1991) Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100 : 195–199.
72. GibsonDG, YoungL, ChuangRY, VenterJC, HutchisonCA3rd, et al. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6 : 343–345.
73. SalgadoH, Peralta-GilM, Gama-CastroS, Santos-ZavaletaA, Muniz-RascadoL, et al. (2013) RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res 41: D203–213.
74. BaileyTL, GribskovM (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14 : 48–54.
75. GerlachRG, HolzerSU, JackelD, HenselM (2007) Rapid engineering of bacterial reporter gene fusions by using Red recombination. Appl Environ Microbiol 73 : 4234–4242.
76. LaemmliUK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 : 680–685.
77. HoisethSK, StockerBA (1981) Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291 : 238–239.
78. MollerAK, LeathamMP, ConwayT, NuijtenPJ, de HaanLA, et al. (2003) An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect Immun 71 : 2142–2152.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite MovementČlánek Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi ScreenČlánek IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed ChildrenČlánek Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive SubjectsČlánek Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 1- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- HIV-1 Accessory Proteins Adapt Cellular Adaptors to Facilitate Immune Evasion
- Ranaviruses: Not Just for Frogs
- Effectors and Effector Delivery in
- Plasmacytoid Dendritic Cell Dynamics Tune Interferon-Alfa Production in SIV-Infected Cynomolgus Macaques
- Lu/BCAM Adhesion Glycoprotein Is a Receptor for Cytotoxic Necrotizing Factor 1 (CNF1)
- A Substrate-Fusion Protein Is Trapped inside the Type III Secretion System Channel in
- Parvovirus-Induced Depletion of Cyclin B1 Prevents Mitotic Entry of Infected Cells
- Red Blood Cell Invasion by : Structural Basis for DBP Engagement of DARC
- NsrR, GadE, and GadX Interplay in Repressing Expression of the O157:H7 LEE Pathogenicity Island in Response to Nitric Oxide
- Loss of Circulating CD4 T Cells with B Cell Helper Function during Chronic HIV Infection
- TREM-1 Deficiency Can Attenuate Disease Severity without Affecting Pathogen Clearance
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- Glutamate Utilization Couples Oxidative Stress Defense and the Tricarboxylic Acid Cycle in Phagosomal Escape
- Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite Movement
- Recovery of an Antiviral Antibody Response following Attrition Caused by Unrelated Infection
- Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi Screen
- Absence of Intestinal PPARγ Aggravates Acute Infectious Colitis in Mice through a Lipocalin-2–Dependent Pathway
- Induction of a Stringent Metabolic Response in Intracellular Stages of Leads to Increased Dependence on Mitochondrial Metabolism
- CTCF and Rad21 Act as Host Cell Restriction Factors for Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Lytic Replication by Modulating Viral Gene Transcription
- Gammaherpesviral Gene Expression and Virion Composition Are Broadly Controlled by Accelerated mRNA Degradation
- The Arabidopsis Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-transcriptional Control of Disease Resistance Genes
- Inflammatory Stimuli Reprogram Macrophage Phagocytosis to Macropinocytosis for the Rapid Elimination of Pathogens
- Alphavirus Mutator Variants Present Host-Specific Defects and Attenuation in Mammalian and Insect Models
- Phosphopyruvate Carboxylase Identified as a Key Enzyme in Erythrocytic Carbon Metabolism
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Electron Tomography of HIV-1 Infection in Gut-Associated Lymphoid Tissue
- Characterisation of a Multi-ligand Binding Chemoreceptor CcmL (Tlp3) of
- Single Cell Stochastic Regulation of Pilus Phase Variation by an Attenuation-like Mechanism
- Cell Tropism Predicts Long-term Nucleotide Substitution Rates of Mammalian RNA Viruses
- Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive Subjects
- RNA-seq Analysis of Host and Viral Gene Expression Highlights Interaction between Varicella Zoster Virus and Keratinocyte Differentiation
- Kaposi's Sarcoma Associated Herpesvirus Tegument Protein ORF75 Is Essential for Viral Lytic Replication and Plays a Critical Role in the Antagonization of ND10-Instituted Intrinsic Immunity
- DAMP Molecule S100A9 Acts as a Molecular Pattern to Enhance Inflammation during Influenza A Virus Infection: Role of DDX21-TRIF-TLR4-MyD88 Pathway
- Variable Suites of Non-effector Genes Are Co-regulated in the Type III Secretion Virulence Regulon across the Phylogeny
- Reengineering Redox Sensitive GFP to Measure Mycothiol Redox Potential of during Infection
- Preservation of Tetherin and CD4 Counter-Activities in Circulating Alleles despite Extensive Sequence Variation within HIV-1 Infected Individuals
- KSHV 2.0: A Comprehensive Annotation of the Kaposi's Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features
- Nutrient Limitation Governs Metabolism and Niche Adaptation in the Human Nose
- Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
- Investigation of Acetylcholine Receptor Diversity in a Nematode Parasite Leads to Characterization of Tribendimidine- and Derquantel-Sensitive nAChRs
- Intranasal Vaccination Promotes Detrimental Th17-Mediated Immunity against Influenza Infection
- -Mediated Inhibition of Iron Export Promotes Parasite Replication in Macrophages
- Variation in RNA Virus Mutation Rates across Host Cells
- A Single Amino Acid in the Stalk Region of the H1N1pdm Influenza Virus HA Protein Affects Viral Fusion, Stability and Infectivity
- Group B Engages an Inhibitory Siglec through Sialic Acid Mimicry to Blunt Innate Immune and Inflammatory Responses
- Synthesis and Biological Properties of Fungal Glucosylceramide
- HIV Protective KIR3DL1/S1-HLA-B Genotypes Influence NK Cell-Mediated Inhibition of HIV Replication in Autologous CD4 Targets
- Recruitment of PfSET2 by RNA Polymerase II to Variant Antigen Encoding Loci Contributes to Antigenic Variation in
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Enhancing Virus-Specific Immunity by Combining Therapeutic Vaccination and PD-L1 Blockade in Chronic Hepadnaviral Infection
- Suppression of Interferon Lambda Signaling by SOCS-1 Results in Their Excessive Production during Influenza Virus Infection
- Inflammation Fuels Colicin Ib-Dependent Competition of Serovar Typhimurium and in Blooms
- Host-Specific Enzyme-Substrate Interactions in SPM-1 Metallo-β-Lactamase Are Modulated by Second Sphere Residues
- STING-Dependent Type I IFN Production Inhibits Cell-Mediated Immunity to
- From Scourge to Cure: Tumour-Selective Viral Pathogenesis as a New Strategy against Cancer
- Lysine Acetyltransferase GCN5b Interacts with AP2 Factors and Is Required for Proliferation
- Narrow Bottlenecks Affect Populations during Vertical Seed Transmission but not during Leaf Colonization
- Targeted Cytotoxic Therapy Kills Persisting HIV Infected Cells During ART
- Murine Gammaherpesvirus M2 Protein Induction of IRF4 via the NFAT Pathway Leads to IL-10 Expression in B Cells
- iNKT Cell Production of GM-CSF Controls
- Malaria-Induced NLRP12/NLRP3-Dependent Caspase-1 Activation Mediates Inflammation and Hypersensitivity to Bacterial Superinfection
- Detection of Host-Derived Sphingosine by Is Important for Survival in the Murine Lung
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



