-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene
Bacterial leaf streak of rice, caused by Xanthomonas oryzae pv. oryzicola (Xoc) is an increasingly important yield constraint in this staple crop. A mesophyll colonizer, Xoc differs from X. oryzae pv. oryzae (Xoo), which invades xylem to cause bacterial blight of rice. Both produce multiple distinct TAL effectors, type III-delivered proteins that transactivate effector-specific host genes. A TAL effector finds its target(s) via a partially degenerate code whereby the modular effector amino acid sequence identifies nucleotide sequences to which the protein binds. Virulence contributions of some Xoo TAL effectors have been shown, and their relevant targets, susceptibility (S) genes, identified, but the role of TAL effectors in leaf streak is uncharacterized. We used host transcript profiling to compare leaf streak to blight and to probe functions of Xoc TAL effectors. We found that Xoc and Xoo induce almost completely different host transcriptional changes. Roughly one in three genes upregulated by the pathogens is preceded by a candidate TAL effector binding element. Experimental analysis of the 44 such genes predicted to be Xoc TAL effector targets verified nearly half, and identified most others as false predictions. None of the Xoc targets is a known bacterial blight S gene. Mutational analysis revealed that Tal2g, which activates two genes, contributes to lesion expansion and bacterial exudation. Use of designer TAL effectors discriminated a sulfate transporter gene as the S gene. Across all targets, basal expression tended to be higher than genome-average, and induction moderate. Finally, machine learning applied to real vs. falsely predicted targets yielded a classifier that recalled 92% of the real targets with 88% precision, providing a tool for better target prediction in the future. Our study expands the number of known TAL effector targets, identifies a new class of S gene, and improves our ability to predict functional targeting.
Published in the journal: Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene. PLoS Pathog 10(2): e32767. doi:10.1371/journal.ppat.1003972
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003972Summary
Bacterial leaf streak of rice, caused by Xanthomonas oryzae pv. oryzicola (Xoc) is an increasingly important yield constraint in this staple crop. A mesophyll colonizer, Xoc differs from X. oryzae pv. oryzae (Xoo), which invades xylem to cause bacterial blight of rice. Both produce multiple distinct TAL effectors, type III-delivered proteins that transactivate effector-specific host genes. A TAL effector finds its target(s) via a partially degenerate code whereby the modular effector amino acid sequence identifies nucleotide sequences to which the protein binds. Virulence contributions of some Xoo TAL effectors have been shown, and their relevant targets, susceptibility (S) genes, identified, but the role of TAL effectors in leaf streak is uncharacterized. We used host transcript profiling to compare leaf streak to blight and to probe functions of Xoc TAL effectors. We found that Xoc and Xoo induce almost completely different host transcriptional changes. Roughly one in three genes upregulated by the pathogens is preceded by a candidate TAL effector binding element. Experimental analysis of the 44 such genes predicted to be Xoc TAL effector targets verified nearly half, and identified most others as false predictions. None of the Xoc targets is a known bacterial blight S gene. Mutational analysis revealed that Tal2g, which activates two genes, contributes to lesion expansion and bacterial exudation. Use of designer TAL effectors discriminated a sulfate transporter gene as the S gene. Across all targets, basal expression tended to be higher than genome-average, and induction moderate. Finally, machine learning applied to real vs. falsely predicted targets yielded a classifier that recalled 92% of the real targets with 88% precision, providing a tool for better target prediction in the future. Our study expands the number of known TAL effector targets, identifies a new class of S gene, and improves our ability to predict functional targeting.
Introduction
Bacterial leaf streak of rice (Oryza sativa), caused by Xanthomonas oryzae pv. oryzicola (Xoc), and bacterial blight of rice, caused by the closely related Xanthomonas oryzae pv. oryzae (Xoo) are important constraints to production of this staple crop in many parts of the world. Yield losses as high as 50% for blight and 30% for leaf streak have been documented [1]. Leaf steak in particular appears to be growing in importance, as high-yielding but susceptible hybrid varieties of rice are increasingly adopted (C. Vera-Cruz and G. Laha, personal communications). Xoc enters through leaf stomata or wounds and interacts with mesophyll parenchyma cells to colonize the mesophyll apoplast, causing interveinal, watersoaked lesions that develop into necrotic streaks. Quantitative trait loci for resistance to leaf streak have been characterized [2], but native major gene resistance has yet to be identified. In contrast, Xoo typically enters through hydathodes or wounds and travels through the xylem, interacting with xylem parenchyma cells through the pit membranes, and typically resulting in wide necrotic lesions along the leaf margins or following veins down the center of the leaf. Only in later stages of disease development does Xoo colonize the mesophyll. Also in contrast to leaf streak, roughly 30 independent genes for resistance (R) to blight have been identified and seven molecularly characterized [3], [4]. The basis for the distinct tissue specificities of Xoc and Xoo and the disparity in known host resistance, despite the genetic similarity of the two pathogens, is not known.
Virulence of Xoo, and of Xanthomonas that infect citrus, cotton, or pepper, is influenced by transcription activator-like (TAL) effectors [5]–[15]. Widespread in Xanthomonas, TAL effectors are proteins delivered into the plant cell via type III secretion (T3S) that transactivate effector-specific host genes [16], [17]. If activation is important for disease, the target is considered a susceptibility (S) gene [9]. Individual Xoo strains harbor multiple, distinct TAL effector (tal) genes [8], and several bacterial blight S genes have been identified. The first of these were Os8N3 (a sugar transporter gene family member also and hereafter referred to as OsSWEET11), the bZIP transcription factor OsTFXI, and the transcription initiation factor TFIIAγI, upregulated respectively by TAL effectors PthXo1, PthXo6, and PthXo7 of Xoo strain PXO99A [9], [10]. More recently, the closely related OsSWEET11 paralog OsSWEET14 (also Os11N3) was discovered to be an S gene targeted by several distinct TAL effectors from other strains [11], [18], [19]. A third close paralog upregulated during infection by some strains, OsSWEET12, also functions as an S gene, though a TAL effector that upregulates it has not yet been reported [19], [20]. The recessive blight R genes xa13 and xa25 are promoter variant alleles of OsSWEET11 and OsSWEET12, respectively, that are not activated by the corresponding TAL effector (or presumed TAL effector in the case of OsSWEET12) [9], [20]. Some TAL effectors induce host resistance by transcriptionally activating a type of dominant R gene that triggers local cell death when expressed, for example the archetypal TAL effector AvrBs3 from the pepper pathogen X. euvesicatoria [21], which activates the pepper Bs3 gene for resistance to bacterial spot [17], and the Xoo effector AvrXa27, from strain PXO99A, which induces the rice R gene Xa27 [22]. Like Xoo, Xoc strains harbor multiple tal genes [8], [23]. However, though the T3S system through which TAL effectors travel is required for leaf streak [24], the role of Xoc TAL effectors in the disease is uncharacterized, and no leaf streak S genes have been identified.
TAL effectors find their targets via a structurally modular mechanism that allows prediction of DNA specificity and customization to target nucleotide sequences of choice [25]–[29]. The modules are tandem repeats of a 33–35 amino acid sequence, exhibiting polymorphism at residues 12 and 13, together called the repeat variable diresidue (RVD). Different RVDs were shown computationally and experimentally, and later structurally to each specify a single nucleotide through direct interaction with (or exclusion of other bases by) the residue 13 side chain, such that the string of RVDs presented by the repeats “encodes” the sequence of the so-called TAL effector binding element (EBE) on the DNA [25], [26], [30], [31]. The RVD nucleotide associations observed in nature are not strictly one to one, however [26]. Indeed, all known natural EBEs contain one or more mismatches to the corresponding TAL effector RVD sequence, a mismatch being a base different from the one most commonly associated with the RVD. Furthermore, some RVDs have dual or even entirely lax specificity. So, the TAL effector-DNA binding code is partially degenerate, rendering target prediction probabilistic [26], [32]. Finally, EBEs in nature are almost all directly preceded by a 5′ thymine (T) that has been shown, in the few studied cases, to be important for TAL effector-driven gene activation as well as full affinity DNA binding [33]–[35]. The single known exception, EBETalC in the promoter of OsSWEET14, displays a cytosine (C). Although the effect of substituting a T was not tested directly, a perfect match EBE for TalC, with a T at base 0 and corrected mismatches at two other locations, indeed showed higher activity [13]
In this study, we sought to better understand bacterial leaf streak in relation to bacterial blight, particularly with an eye toward identifying determinants of tissue specificity, and to examine the roles of Xoc TAL effectors in disease. We began by comparing transcription profiles in Xoc-, Xoo-, and mock-inoculated plants by microarray analysis. We then combined the transcriptomic data with computational identification of candidate EBEs to predict TAL effector targets, and carried out experiments to differentiate real from falsely predicted ones. Screening a TAL effector mutant library of Xoc, we next identified a TAL effector that plays a major role in virulence, and we discriminated from among its two targets the first known S gene for leaf streak, in part by using designer TAL effectors to independently activate the genes. Using our complete list of newly discovered targets as well as the previously identified Xoo targets represented in our dataset, we next examined general characteristics of TAL effector driven gene expression. Finally, in an attempt to better discriminate real targets from falsely predicted ones in the future, prior to experimentation, we used machine learning to train a classifier on primary and contextual features of EBEs in the respective groups. Our results provide new insight into bacterial leaf streak, increase the number of known natural TAL effector combinations by 20, identify a new class of S gene, and advance our understanding of and ability to predict functional targeting by TAL effectors.
Results
X. oryzae pv. oryzicola BLS256 and X. oryzae pv. oryzae PXO99A induce largely different gene expression changes in rice leaves
We initially set out to determine whether there are differences in host genome-wide expression patterns during bacterial leaf streak vs. bacterial blight that might help to explain the different tissue specificity of Xoc and Xoo. Using a vacuum infiltration approach developed from a dipping method we showed previously to be effective for both pathovars [36], we inoculated rice (cv. Nipponbare) plants en masse with Xoc strain BLS256 (hereafter Xoc refers to this strain unless otherwise specified), Xoo strain PXO99A (likewise), or a mock inoculum, harvested leaves at 2, 4, 8, 24, and 96 hours thereafter, and quantified transcript levels in these leaves for the roughly 56,000 annotated rice genes in parallel using the Affymetrix GeneChip Rice Genome Array [37]. We focused our analysis on patterns of expression across the time course rather than expression levels at a particular time point and examined three pairwise comparisons, Xoc vs. mock, Xoo vs. mock, and Xoc vs. Xoo (see Materials and Methods).
A total of 505 genes showed significantly different expression profile patterns (q≤0.3; see Materials and Methods) in one or more of the pairwise comparisons (Figure 1). Eighty and 94 genes were differentially expressed uniquely in response to Xoc or Xoo, respectively (Figure 1; Table S1 and Table S2). Only five genes were differentially expressed both in response to Xoc and Xoo relative to mock: three similarly between Xoc - and Xoo - and two with different patterns in Xoc - vs. Xoo-inoculated plants (Figure 1; Table S3). Strikingly, all of the statistically significantly differentially expressed genes showed patterns of upregulation in response to Xoc or Xoo. Expression patterns of the ten or fewer most significantly differentially expressed genes in response to Xoc, Xoo, or both are shown in Figure 2.
Fig. 1. Rice transcriptional responses to Xanthomonas oryzae pv. oryzicola BLS256 (Xoc) or X. oryzae pv. oryzae PXO99A (Xoo). 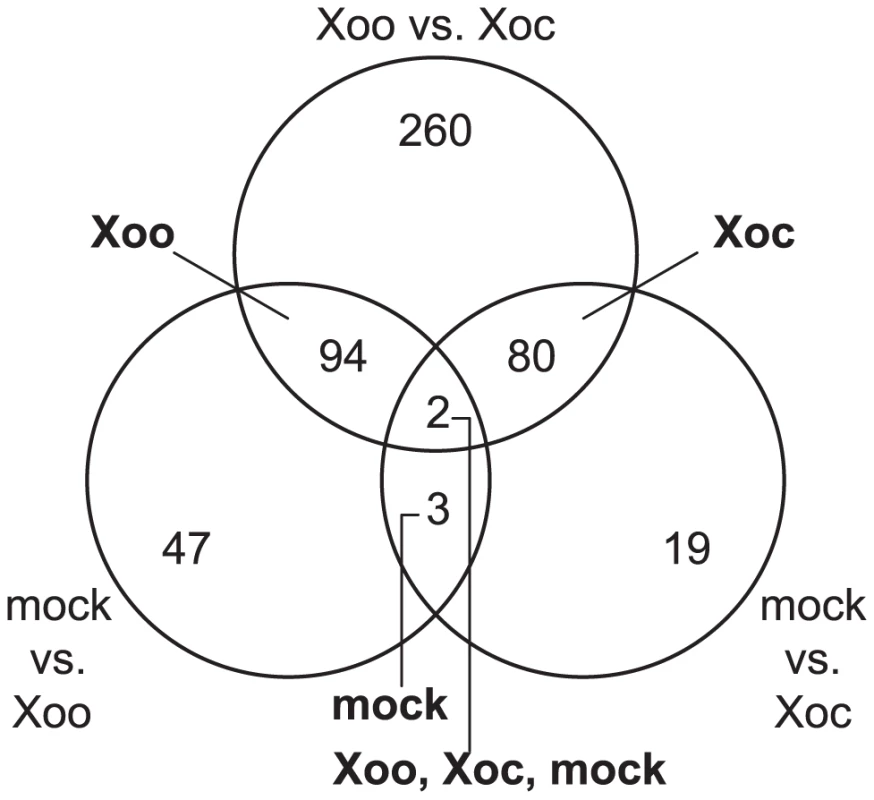
Distribution of genes differentially expressed over a 96(see Materials and Methods) in response to either strain relative to a mock inoculation is shown. Each circle of the Venn diagram represents a different pairwise comparison of treatments, as indicated in non-bold text. Results are based on mixed linear model analysis using four biological replicates for each time point of the study and an estimated false discovery rate of 0.3. The intersections represent the genes differentially expressed uniquely in response to the different treatments, indicated in bold text. Note that differentially expressed uniquely in response to mock means differentially expressed similarly in Xoc and Xoo relative to mock, and differentially expressed uniquely in response to all three treatments means differentially expressed both in Xoc and Xoo relative to mock, but also differentially between Xoc and Xoo. Also, since differential expression in a given pairwise comparison is determined using a statistical cutoff, transitive predictions, i.e., A = B and B = C, therefore A = C, may not hold. Fig. 2. Expression patterns of the most significantly differentially expressed rice genes. 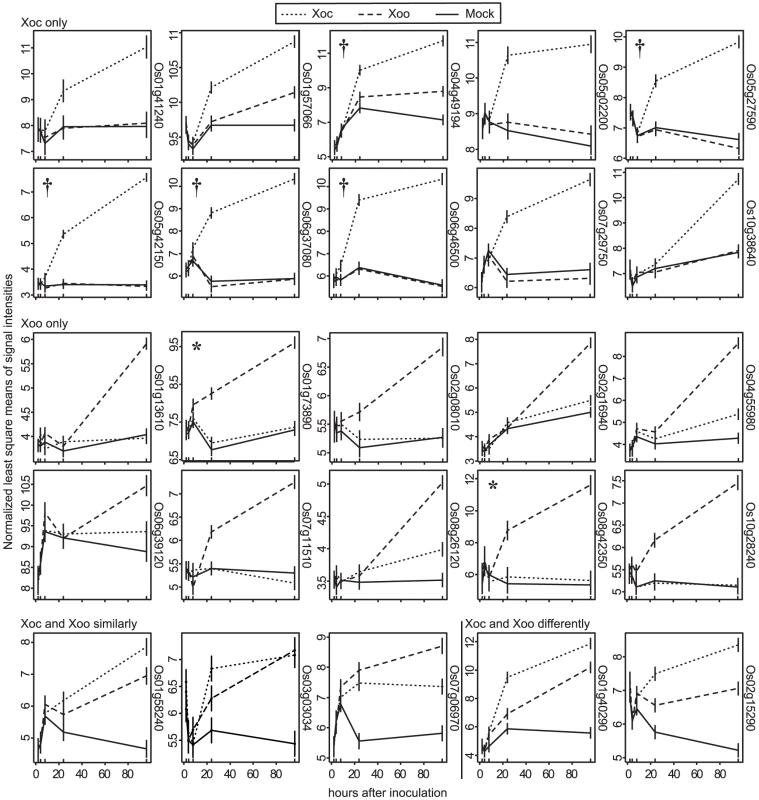
Normalized least square means of signal intensities (y-axis) at 2, 4, 8, 24, and 96 h after inoculation (x-axis) with X. oryzae pv. oryzicola BLS256 (Xoc), X. oryzae pv. oryzae strain PXO99A (Xoo) or mock control are plotted for the genes most significantly differentially expressed relative to mock uniquely in response to Xoc (Xoc only), uniquely in response to Xoo (Xoo only), similarly in response to Xoc and Xoo (Xoc and Xoo similarly), and differently in response to Xoc and Xoo (Xoc and Xoo differently). Where two probe sets correspond to the same gene, the one with the lower q-value was selected for display. Locus IDs are given at right, omitting the prefix “LOC_Os”. Results were derived from a mixed linear model analysis with four replicates. Vertical bars represent standard error. Asterisks mark previously identified targets of Xoo TAL effectors, TFIIaγ1(Os01g73890) and OsSWEET11 (Os08g42350), activated by PthXo7 and PthXo1, respectively. Daggers flag Xoc TAL effector targets discovered in this study. Singular enrichment analysis [38] of gene ontology (GO) for all Xoc - and Xoo-upregulated genes revealed broad differences in the major categories represented (Table S4 and Table S5). Six significant GO terms were identified for Xoc-induced genes. Four of these are categorized under biological processes and include coenzyme metabolic, cofactor metabolic, sulfur metabolic and, cellular amino acid derivative metabolic processes. The other two, catalytic and oxidoreductase activities, are grouped under molecular function (Table S4). For Xoo-induced genes, the significant terms all fall within the cellular component category, including membrane-bounded vesicle, vesicle, cytoplasmic membrane-bounded vesicle, and cytoplasmic vesicle (Table S5). The most abundant ontology category for genes induced by Xoc was catalytic activity, and included several glutathione S-transferase and oxidase genes (Table S4). These were part of a large group of Xoc-induced genes, distributed among several categories, with annotations that suggest roles in reactive oxygen species detoxification and redox status control (assembled together in Table S6).
Among the complete list of Xoo-induced genes are each of the bacterial blight S genes previously reported to be induced by PXO99A TAL effectors, OsSWEET11 (Os08g42350), OsTFXI (Os09g29820), and TFIIAγI (Os01g73890) (Table S2 and Table S7). Notably, none of these three genes nor any of the OsSWEET11 paralogs reported to function as bacterial blight S genes [11], [19], [20] was activated following inoculation with Xoc.
Thus, host genome wide expression patterns during bacterial leaf streak vs. bacterial blight are almost completely different.
The most significant gene expression changes depend on bacterial type III secretion
The TAL effector inventories in Xoc and Xoo are entirely distinct. Xoc harbors 26 unique, intact TAL effector genes and Xoo 14, with no shared predicted EBEs based on RVD sequences [23], [39]. The inventories of predicted non-TAL type III effectors in Xoc and Xoo are similar, but six effector genes present in Xoc are absent from or pseudogenized in Xoo and several minor polymorphisms exist among the shared genes [23]. As a first step to determine the extent to which differences in TAL or other type III effector content might account for the differences in rice global transcription patterns we observed, we asked whether T3S is required for induction of the top ten rice genes most significantly induced uniquely following inoculation with Xoc, the top ten induced by Xoo, and all five induced in common by both strains. We compared, by RT-PCR, transcript accumulation after inoculation with the wild-type strains or with T3S-deficient derivatives BLS256hrcC− [24] and PXO99AME7 [9]. Induction of each gene required bacterial T3S (Figure 3 and [9], [10]).
Fig. 3. Type III secretion system dependence of the most significant rice gene expression changes. 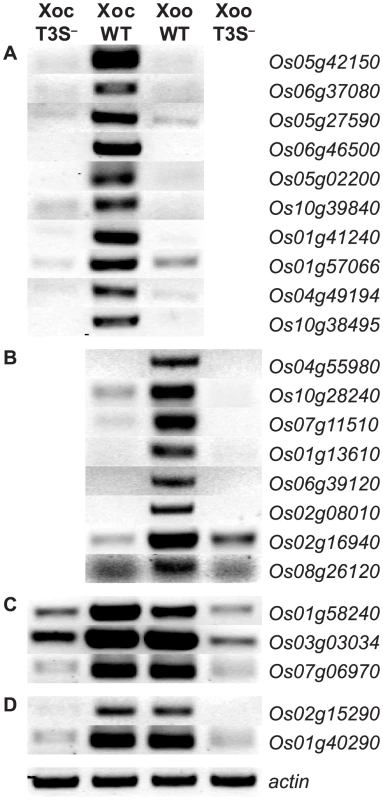
RT-PCR results reflecting transcript abundance are shown for rice genes identified by GeneChip expression analysis as the ten (or fewer) most significantly differentially expressed in response to (A) X. oryzae pv. oryzicola BLS256 (Xoc), (B) X. oryzae pv. oryzae strain PXO99A (Xoo), (C) Xoc and Xoo similarly, or (D) Xoc and Xoo to different extents. Leaf samples were harvested at 36 hours after inoculation with wild-type strains or with the type III secretion (T3S−) deficient derivatives. RT-PCR results for previously reported Xoo-induced genes, OsSWEET11 and TFIIAγ1 [9], [10], are omitted. An actin gene (Os04g57210) that is not differentially expressed was used as a reference for relative transcript abundance across samples. The experiment was repeated twice and yielded the same results. Among the top ten Xoo-induced genes are the TAL effector targets OsSWEET11 (Os08g42350) and TFIIaγ1 (Os01g73890). The patterns of induction of each of the top Xoc - or Xoo-induced genes revealed by the genome-wide expression analysis described in the previous section vary, but some are similar to that of OsSWEET11 and TFIIaγ1 (Figure 2). This similarity and the T3S-dependence of expression suggested that some of these and perhaps others in the complete lists of induced genes are targets of TAL effectors.
Many upregulated genes are predicted targets of TAL effectors
To identify TAL effector targets, we first used the scoring function we developed previously based on observed RVD-nucleotide association frequencies [26], [32] to scan in silico all annotated rice gene promoters (the promoterome) [32] for candidate EBEs for any of the 40 total TAL effectors present in Xoc and Xoo [23], [39]. Some of these TAL effectors have new RVDs whose specificities have not been characterized. The scoring function by default treats new RVDs as wild cards, equally likely to specify any base. However, since structural studies revealed that the second residue of each RVD makes the base-specific contacts while the first stabilizes the inter-helical loop that projects that second residue into the major groove of the DNA [30], [31], we used the specificities of common RVDs for any new RVDs that share the same second position residue. These were limited to two RVDs found in Xoc TAL effector Tal2g, ‘SN’ for which we substituted nucleotide association frequencies of ‘NN’, and ‘YG’ for which we substituted those of ‘NG’. Candidate EBEs were required to be directly preceded by a T at the 5′ end and, for each TAL effector, to score below a cutoff calculated based on the distribution of scores for that effector (see Materials and Methods). This list was then cross-referenced to the GeneChip expression data, and genes with one or more candidate EBEs in the promoter that were also induced following inoculation with the corresponding strain were retained as predicted targets (Table S7).
Thirty-five of these are genes induced by Xoc (three of the 35 are also induced by Xoo), and they collectively contain candidate EBEs for 19 out of the 26 Xoc TAL effectors. Twenty-nine are genes induced by Xoo (five are also induced by Xoc), and they together contain putative EBEs for all 14 of the unique Xoo TAL effectors (Tal7a and 7b are identical to Tal8a and 8b, respectively). The latter include each of the three previously demonstrated targets of Xoo (i.e., PXO99A) TAL effectors in Nipponbare, OsSWEET11 targeted by PthXo1, OsTFXI targeted by PthXo6, and TFIIAγI targeted by PthXo7 [9], [10], [40] (the AvrXa27-activated allele of Xa27 is not present in Nipponbare). Among the five genes induced in common by Xoc and by Xoo, two were predicted to be targeted by a TAL effector from Xoo but not by one from Xoc (Os01g58240 by Tal4 and Os01g40290 by Tal7b/8b of Xoo). In the other three, sequence distinct, candidate EBEs for one or more TAL effectors from each strain were found in the promoters (EBEs for Tal2c and Tal3b of Xoc and AvrXa27 and Tal9b of Xoo in Os03g03034, for Tal1c and Tal3a of Xoc and Tal9a of Xoo in Os07g06970, and for Tal5a and Tal11a of Xoc and Tal9e of Xoo in Os02g15290).
Of the 35 total genes induced by Xoc that harbor a candidate EBE for an Xoc TAL effector, eight harbor EBEs for more than one. Likewise, of the 29 Xoo-induced genes that match an Xoo TAL effector, four genes contain EBEs for multiple Xoo TAL effectors. These results suggest for both pathovars a partial redundancy among effectors for some targets. The Xoc-induced gene Os06g14750 and the Xoo-induced gene Os07g11510 contain overlapping candidate EBEs for three TAL effectors each from those strains, Tal2a, Tal1c, and Tal11b, and PthXo6, Tal2a, and Tal5a, respectively.
The number of predicted targets for individual TAL effectors varies. In the case of Xoc, we identified five predicted targets each for Tal3b and Tal6, and one of the predicted Tal6 targets, Os12g42970, harbors two candidate Tal6 EBEs. Five Xoc TAL effectors, Tal2c, Tal5a, Tal8, Tal9b and Tal11b, have only one predicted target each. For Xoo, we predicted seven targets for PthXo6 and one target each for PthXo1, PthXo7, Tal6a, Tal7a/8a, Tal9d, and Tal9e. AvrXa27 had five predicted targets, two of which, Os06g03080 and Os06g03120, are paralogs nearly identical in their coding sequences and both represented by a single probeset. The promoters of these genes share the same AvrXa27 EBE (one of two AvrXa27 EBEs in Os06g03120), but are otherwise distinct.
In sum, all but a few of the TAL effectors of Xoc and Xoo have candidate binding sites in a gene upregulated by that strain; a total of 61 out of 179, or roughly one-third, of the genes induced following inoculation with Xoc, Xoo, or either strain are predicted targets of those TAL effectors; and within these predictions multiple targets per TAL effector as well as multiple TAL effectors per target were observed.
Experimentation verifies 19 targets for X. oryzae pv. oryzicola BLS256 TAL effectors
The next step was to determine which predicted TAL effector targets are real targets. Because several S genes for bacterial blight of rice have been characterized and all are TAL effector targets, while no S genes have yet been identified for bacterial leaf streak and the roles of TAL effectors in this disease have not been explored, we focused on the 44 TAL effector-target pairs predicted for Xoc (Table 1, taking Tal6 and Os12g42970, with its two Tal6 EBEs, as one pair). To identify real targets, we used both TAL effector loss of function and gain of function assays to test TAL effector dependence of expression. First we generated a library of Xoc TAL effector mutant strains by marker exchange mutagenesis. By mapping the mutation in several strains, we identified loss of function derivatives for all but one (Tal2a) of the TAL effectors for which we had predicted a target. And, we cloned each of the TAL effectors into a broad host range plasmid for complementation and heterologous expression (gain of function). Then we assessed by RT-PCR whether any TAL effector mutant strain failed to activate any of the corresponding predicted targets of that TAL effector, and for any that did, whether the cloned effector specifically complemented the mutation to restore activation. In parallel, we expressed each TAL effector in strain EB08 of the soybean pathogen X. axonopodis pv. glycines (Xag) [41], which neither causes symptoms nor elicits a hypersensitive reaction when inoculated to rice (cv. Nipponbare), and we determined whether the transformants specifically activated corresponding targets.
Tab. 1. Predicted X. oryzae pv. oryzicola BL256 TAL effector targets in rice (cv. Nipponbare) induced during infection and results of verification experiments.a 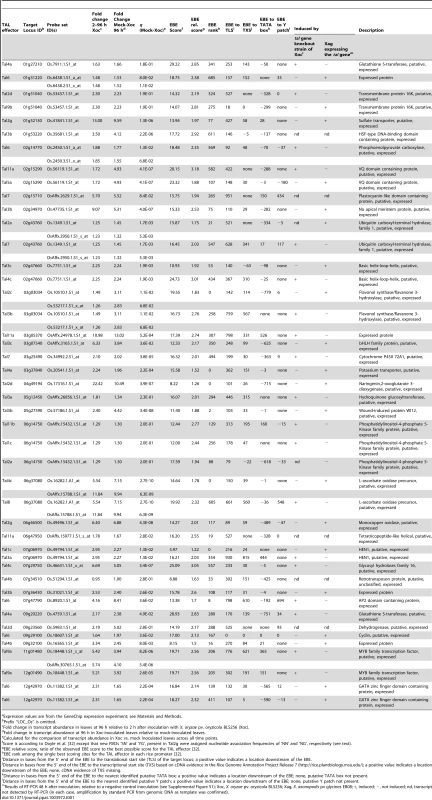
Expression values are from the GeneChip expression experiment; see Materials and Methods. The results verified 19 of the 44 predicted Xoc TAL-effector targets as real (Table 1 and Figure S1; the Tal2a target was verified only by the gain of function experiment). Another 20 were shown not to be activated by the corresponding TAL effector and are hereafter referred to as falsely predicted targets. The remaining five could not be tested because transcript was not detected by RT-PCR, despite induction according to the GeneChip expression data. Interestingly, multiple predicted targets were verified for some TAL effectors, however, for each of the eight genes predicted to be targeted by multiple TAL effectors, only activation by one of those TAL effectors was verified.
Most X. oryzae pv. oryzicola BLS256 TAL effectors have no significant role in virulence
Having identified 19 targets of Xoc TAL effectors, the next challenge was to ascertain whether any are S genes for bacterial leaf streak. Barring redundancy, i.e., targeting of the same S gene by multiple TAL effectors, which our verification experiments excluded for each target tested, loss of a TAL effector that activates an important S gene should by definition result in a reduction of virulence. We therefore first quantified the virulence of each of several mutant strains of Xoc to identify such TAL effectors, using a lesion length assay (Figure 4). Collectively, the mutants account for all 26 Xoc TAL effectors except Tal2a, for which a mutant was not isolated. Assayed on rice cv. Nipponbare plants, only mutations that map on at least one side to the 3′ end of the tal2 cluster, i.e., involving tal2f or tal2g, or that map to the tal11 cluster, which contains tal11a and tal11b, were associated with significantly reduced virulence, 49–64% and 64–79%, respectively. Thus, most of the Xoc TAL effectors, in the context of the Nipponbare host genotype, appear not to make any non-redundant, major contributions to virulence. Interestingly, this includes the TAL effectors that activate genes induced in common by Xoc and Xoo, Tal1c, Tal2c, and Tal5a (Table 1, Table S3, and Table S7).
Fig. 4. Virulence of X. oryzae pv. oryzicola BLS256 tal gene knockout strains. 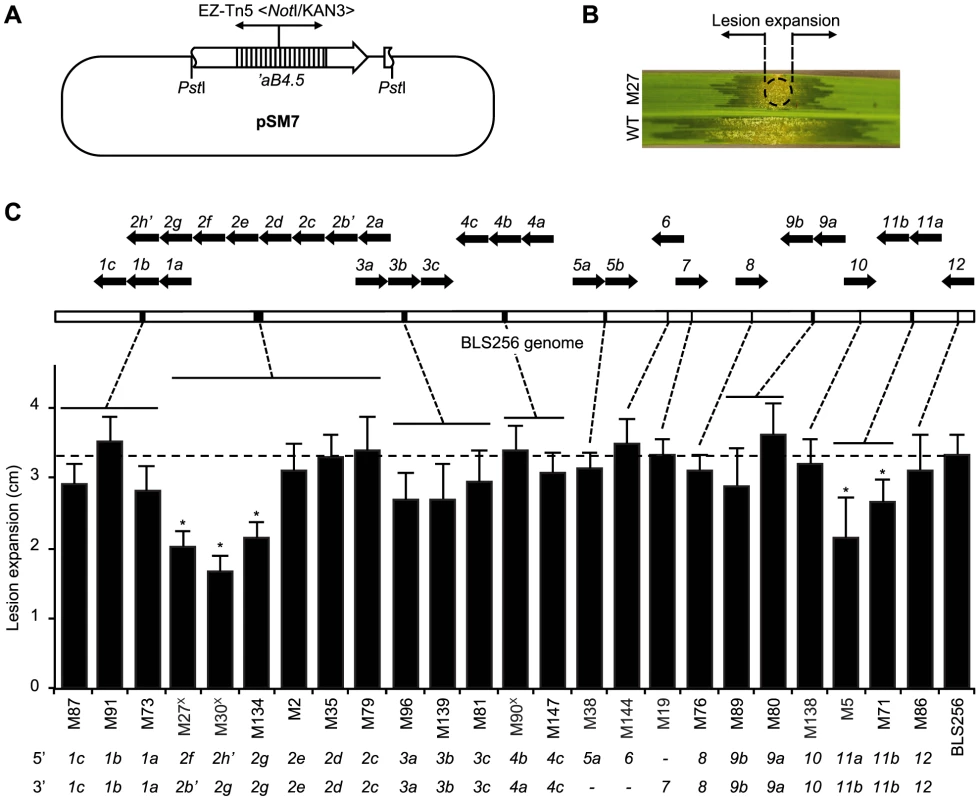
(A) Suicide plasmid pSM7 (Table S8) used for tal gene knockouts by homologous recombination in BLS256. pSM7 harbors a 4.5-kb PstI fragment containing all but the first 80 bp of the ORF of tal gene aB4.5 [12] with an insertion of the EZ-Tn5 <NotI/KAN-3> transposon (Epicentre) in repeat 9, in pBluescript II KS(+) (Agilent), which does not replicate in Xanthomonas. The transposon provides kanamycin resistance for selection. Because the tal ORF is truncated at the 5′ end, either a single or double recombination that retains the transposon results in a tal gene knockout. Double recombination can knock out clustered tal genes. The 4.5 kb PstI fragment also includes the first 85 bp of the avrXa10 tal gene downstream of ab4.5, which might increase the likelihood of complex recombination. (B) Virulence assay used to characterize knockout strains. Suspensions of mutant and wild-type cells are inoculated side by side via leaf infiltration of 4-week old plants using a needless syringe, and expansion of lesions from the inoculation site (circle), as shown for mutant M27 in this example, is measured after 7 days. (C) Virulence of knockout strains and mapped endpoints of integrations. Only strains with single integrations as determined by Southern blot (not shown) were further characterized. Integration endpoints were mapped by PCR amplification of flanking DNA, using transposon-specific and tal gene conserved end specific primers, and sequencing. BLS256 tal gene polymorphisms in most cases enabled unambiguous mapping. Virulence results are plotted left to right in the histogram by integration location, indicated by dashed lines pointing to a linearized representation of the genome, above, with individual tal gene clusters indicated by black bars and magnified at top to show gene content and orientation using block arrows. An apostrophe denotes a pseudogene. At bottom, integration endpoints for each mutant strain are given, by tal gene. A dash means the endpoint could not be unambiguously determined. A superscript “X” after the mutant strain designation denotes an apparent complex recombination, suggested by the 5′ endpoint mapping downstream of the 3′ endpoint. In the histogram, an asterisk indicates significantly reduced virulence (p<0.01, N = 10) relative to wild type. Assays were repeated at least three times with consistent results. Tal2g is a major virulence factor of Xanthomonas oryzae pv. oryzicola BLS256
Of the few Xoc TAL effectors pinpointed by the mutational analysis as possible virulence factors that might lead us to one or more S genes (Tal2f, Tal2g, Tal11a, and Tal11b), we had verified targets only for Tal2g (Table 1). From the code - and GeneChip expression-based analysis, Tal2f had no predicted targets, and two of the three predicted targets of Tal11a and the sole predicted target of Tal11b were shown not to be actual targets by the loss - and gain-of-function RT-PCR experiments (Table 1). So, we focused on Tal2g. Of the three mutant strains in which the mutation endpoints map within or flanking Tal2g (Figure 4: M27, M30, and M134), we chose mutant M27 for further characterization. In M27, the marker exchange endpoints suggest a complex recombination, with a disrupted tal2f on the 5′ end and a disrupted tal2b′, a pseudogene that resides 5′ of tal2f in the native chromosome, on the 3′ end. Because the apparent complex recombination might have affected several genes in the cluster, we assayed each tal2 gene (tal2a, -c, -d, -e, -f, and -g), individually on a plasmid for the ability to complement M27. Only tal2g restored virulence to M27 in the lesion length assay, and it did so fully, confirming Tal2g as the sole virulence factor among the TAL effectors whose expression is disrupted in this mutant (Figure 5A). The marker exchange endpoints in M27 could be explained by a double crossover between tal2b′ and tal2g, concurrent with the marker exchange crossovers, that positioned tal2b′ sequences at the 3′ endpoint of the exchange, with the 5′ end in tal2f, disrupting tal2g but not affecting tal2c, tal2d, or tal2e. Consistent with this, the verified targets of Tal2c and Tal2d (Os03g03034 and Os04g49194) are induced by M27 (Figure S2).
Fig. 5. Virulence contribution of X. oryzae pv. oryzicola BLS256 TAL effector Tal2g. 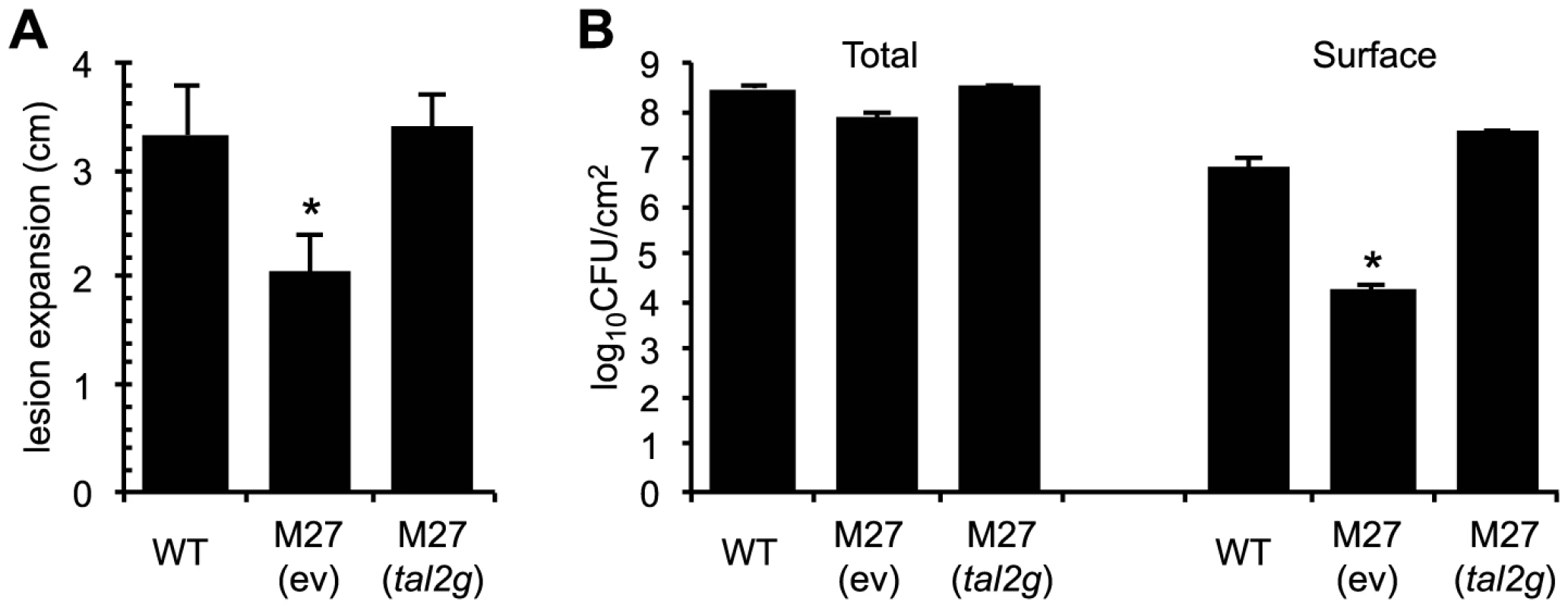
(A) Lengths of lesions caused by X. oryzae pv. oryzicola BLS256 (WT), the tal2g knockout derivative M27 carrying an empty plasmid vector (ev), and M27 carrying the vector with the cloned tal2g gene, measured as in Figure 4, but at 10 days after infiltration. The asterisk indicates a significant difference relative to WT (p<0.01). Error bars represent standard deviation (N≥10). (B) Total and surface (exudate) bacterial populations of leaves seven days after inoculation with the strains in panel A. The asterisk indicates a significant difference relative to WT (p<0.01). Error bars represent standard deviation (N≥6). Experiments were repeated three times with consistent results. Curiously, the total population of M27 isolated from leaf homogenates at seven days after inoculation was not significantly different from that of the wild type (Figure 5B). However, we observed less bacterial exudate on the surface of M27-inoculated leaves than on leaves inoculated with wild type (see Figure 4B). When surface bacteria were isolated and quantified (see Materials and Methods), M27 indeed showed nearly a 400-fold reduction relative to the wild type, and Tal2g on a plasmid fully restored wild-type levels of exudate (Figure 5B). Thus, Tal2g is a major virulence factor in bacterial leaf streak that functions both in lesion expansion and exudation of bacteria to the leaf surface.
A sulfate transporter gene targeted by Tal2g is a major susceptibility gene for bacterial leaf streak
The two verified targets of Tal2g, Os06g46500, encoding a predicted monocopper oxidase, and Os01g52130, encoding a predicted sulfate transporter, OsSULTR3;6 [42], are among the most significantly induced genes in the GeneChip expression dataset (Table S1). To test whether either is a biologically relevant target, i.e., an S gene, we engineered designer TAL effectors (dTALEs) to specifically activate each target individually, and we tested the ability of these dTALEs to restore virulence to M27 (Figure 6). Assayed by RT-PCR, in syringe infiltrated leaves dTALE dT434 expressed in M27 specifically induced the monocopper oxidase gene, and dTALEs dT436 or dT437 induced OsSULTR3;6, each similarly to wild type and to M27 expressing Tal2g (Figure 6B). In the lesion length assay, dT436 and dT437 each restored full virulence to M27, whereas dT434 made no significant difference (Figure 6C). When surface bacterial populations were quantified over time at the inoculation site, and spread of bacteria over time was measured by quantifying total populations in contiguous leaf segments at and extending from the inoculation site, M27 expressing dT437 and M27 expressing Tal2g behaved the same as the wild type, whereas M27 expressing dT434 showed a reduction in surface population and slowed population spread equivalent to M27 carrying the empty vector (Figure 6D and Figure 6E). Scanning the rice promoterome for candidate EBEs as in our original search for potential Xoc and Xoo TAL effector targets, we found no overlap between candidate off-targets of dT436 and dT437, or between off-targets of either with genes harboring a potential Tal2g EBE. Together, the data therefore indicate that OsSULTR3;6 is the relevant Tal2g target and a major S gene for bacterial leaf streak.
Fig. 6. Determination of Os01g52130 as the relevant target of Tal2g using designer TAL effectors. 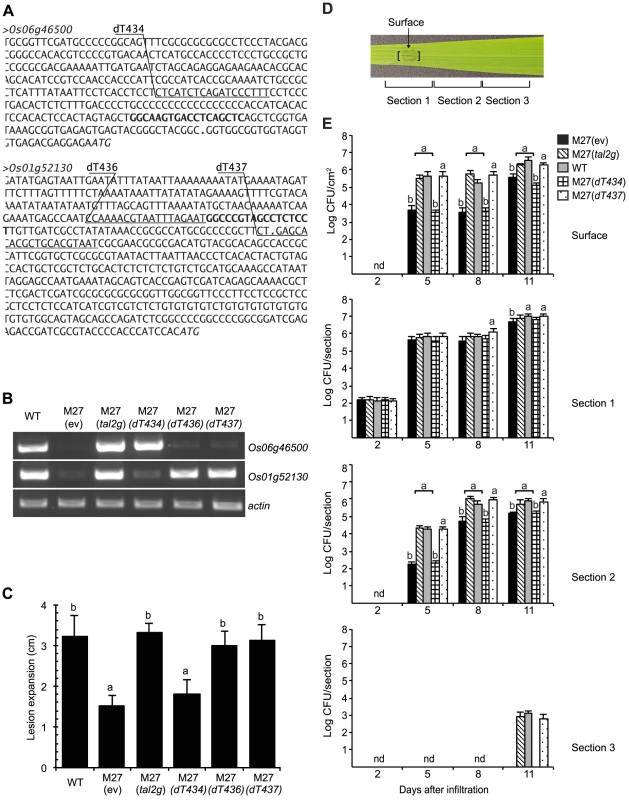
(A) DNA sequence of the promoter regions of Tal2g induced genes Os06g46500 and Os01g52130 in rice cv. Nipponbare. The effector binding elements (EBEs) for Tal2g are in bold. The EBEs for designer TAL effectors dT434 targeting Os06g46500 and dT436 and dT437 targeting Os01g52130 are underlined and labeled above. Periods indicate transcriptional start sites and italics indicate translational start sites, per the Rice Genome Annotation Project (Release 7, http://rice.plantbiology.msu.edu). (B) Activation of Os06g46500 and Os1g52130 by Tal2g, and specific activation respectively of Os06g46500 and Os01g52130 by dT434, and dT436 or dT437. Shown are the results of RT-PCR amplification from leaf RNA isolated 48 h after inoculation by infiltration with X. oryzae pv. oryzicola BLS256 (WT), the tal2g knockout derivative M27 carrying an empty plasmid vector (ev), M27 carrying the vector with the cloned tal2g gene, or M27 carrying the vector with coding sequences for dT436, dT436, or dT437 as indicated. The actin gene Os04g57210 was used as a reference for relative transcript abundance across samples. (C) Rescue of the virulence defect of M27 by dT436 or dT437 but not dT434 in the lesion length assay. Lesion lengths were measured as in Figure 4, 10 days after inoculation with the indicated strains. Values labeled with the same letter are not significantly different and those labeled with different letters are (Student's t-test, p<0.01). Error bars represent standard deviation (N≥10). Experiments were repeated twice with consistent results. (D) A rice (cv. Nipponbare) leaf showing bacterial leaf streak symptoms two days after inoculation with a suspension of WT cells at an OD600 of 0.5 (approximately 1×108 CFU/ml) by infiltration using a needleless syringe over a 4 mm diameter leaf area, and labeled to indicate the site of inoculation, at which surface bacterial populations were quantified, and the three 12 mm long leaf sections in which total bacterial populations were quantified, as presented in panel E. (E) Restoration of the surface population and the total population spread of M27 to wild-type levels by dTAL437 but not dTAL434. Populations were quantified at 2, 5, 8 and 11 days after inoculation. Results are the means and standard deviations of samples from three leaves; nd, not detected. At each time point (not across time points), values labeled with the same letter are not significantly different, and those labeled with different letters are (Student's t-test p<0.0001). Functional characterization of Tal2g EBEs and similarly scored sequences supports presumed specificities of new RVDs ‘SN’ and ‘YG’
As described above, in our search for TAL effector targets, we used specificity values of ‘NN’ and ‘NG’ for the ‘SN’ and ‘YG’ RVDs that are found in Tal2g. As might be expected, the list of candidate Tal2g EBEs generated using these values differed from a second list we generated in parallel using the default, wild card values. Specifically, in the list generated using the default values for ‘SN’ and ‘YG’, hereafter referred to as the default scoring list, the verified Tal2g target Os06g46500 did not make the cutoff (Materials and Methods) to be considered a candidate (indeed no sequence from any Xoc-induced gene beside OsSULTR3;6 scored well enough in this list to be considered a candidate), indicating that substituting the RVD specificity values allowed us to capture an otherwise false negative.
To further probe the validity of substituting the values, we tested the function of two candidate EBEs from the default scoring list that each scored better (lower; see Materials and Methods) than the (default-scored) EBEs in the two verified targets, but that displayed a mismatch to one or each of the two new RVDs in Tal2g based on the presumed specificities of those RVDs (Figure 7A). Though not induced by Xoc, both of the corresponding genes, Os06g13880 and Os12g36920, are induced by Xoo (Table S2), indicating that they are euchromatic. Also, the default-scored candidate EBEs, at 139 bp and 86 bp upstream of the respective annotated transcriptional start sites, are each within the range of locations displayed by the EBEs in all the targets verified in this study (152 bp or less; Table 1), so failure to be induced by Xoc likely does not relate to suboptimal EBE localization. We also chose to test a third sequence with a mismatch to one of the new RVDs, that scored just above the cutoff in the default scoring list (Figure 7A) and was therefore not considered a candidate, but was nonetheless in the promoter of an Xoc-induced gene Os05g10650 (Table S1), and therefore a potential false negative in that list. To test the function of the three sequences, we used a transient, Agrobacterium-mediated, TAL effector-driven reporter gene expression assay in Nicotiana benthamiana [40]. None of the sequences, inserted into a 343 bp fragment of the pepper Bs3 promoter just upstream of the native EBE for the cognate TAL effector AvrBs3 [25], rendered the reporter responsive to Tal2g (Figure 7B). In contrast, the EBEs from the verified targets of Tal2g resulted in strong and specific induction of the reporter by Tal2g similar to induction of the unamended reporter by AvrBs3 (Figure 7C).
Fig. 7. Functional characterization of selected rice promoter sequences similar to the verified Tal2g EBEs. 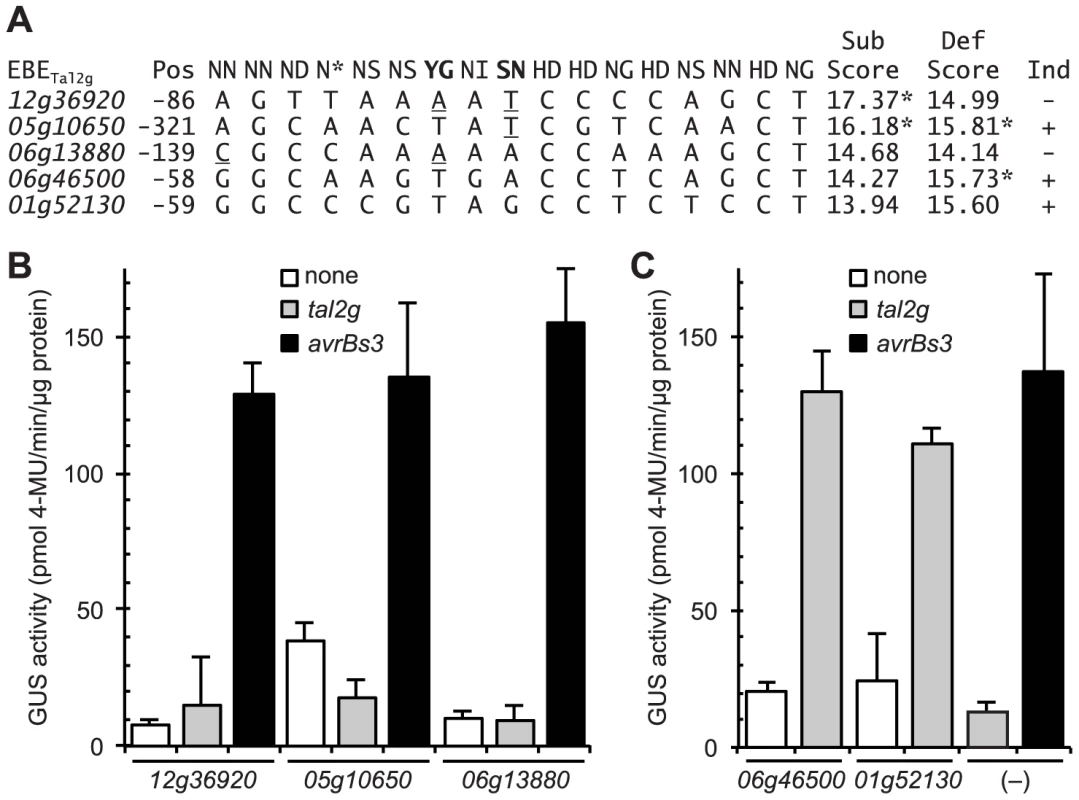
(A) Alignment of selected rice promoter sequences (from loci Os06g13880, Os12g36920, and Os05g10650; see text) and EBEs from the verified Tal2g targets Os01g52130 (OsSULTR3;6) and Os06g46500 with the corresponding sequence of repeat variable diresidues (RVD) of Tal2g. Position (Pos) is that of the 5′ end relative to the annotated transcriptional start site. Rare RVDs ‘YG’ and ‘SN’ of Tal2g are in bold. Scores were calculated according to [32], either substituting the nucleotide association frequencies of common RVDs ‘NN’ and ‘NG’ for the new RVDs ‘SN’ and ‘YG’ (“Sub Scores”) or using the default wild card specificity values for the new RVDs (“Def Scores”). An asterisk indicates that the score is outside the cutoff to be considered a candidate EBE for Tal2g, calculated independently for each scoring method. Nucleotide mismatches to the new RVDs using the substituted specificities are underlined, as is a (5′) mismatch in the 06g13880 sequence to the first RVD (‘NN’) of Tal2g. Whether a gene is induced (Ind) upon infection by Xanthomonas oryzae pv. oryzicola BLS256 is indicated by a plus or minus sign at right. (B) Activity of the selected sequences in an Agrobacterium-mediated transient transformation based reporter assay in Nicotiana benthamiana leaves [40]. In this assay, a TAL effector gene (none, tal2g, or avrBs3) driven by the 35S promoter is introduced together with the GUS gene under the control of a minimal promoter from the pepper Bs3 gene, with the test sequence inserted slightly upstream of the native EBE for AvrBs3 (AvrBs3 is the TAL effector from the pepper pathogen X. euvesicatoria that activates Bs3 upon infection). The inserted sequences are indicated by locus ID on the X axis; “(–)” indicates the minimal Bs3 promoter with only the AvrBs3 EBE and no added sequence. Error bars represent standard deviation (N = 3). Experiments were repeated twice with consistent results. (C) Activity and specificity of the EBEs from the two verified targets of Tal2g, as in panel B. Thus, in addition to capturing the verified target Os06g46500 as a candidate, the substituted scoring correctly classifies the Os12g36920 and Os05g10650 sequences as non-candidates (scored above the cutoff). The substituted scoring scores the Os06g13880 sequence as worse than the EBEs of the two verified targets, consistent with its lack of activity, but still calls it a candidate. This incongruity might be explained by the observation that the Os06g13880 sequence displays a mismatch to the first RVD of Tal2g (Figure 7A), and mismatches at the 5′ end and especially at the first position have been shown to more strongly negatively affect activity than mismatches elsewhere [43] a phenomenon not accounted for by the scoring function. Taken together, the observations overall support the assignment of the common RVD specificities for those of the new cognate RVDs, in agreement with the inference from published structural data discussed earlier.
Target upregulation by TAL effectors on average is moderate
Returning to our list of 19 new, verified TAL effector-target pairs, we next sought to determine whether the expression patterns of the targets might reveal general characteristics of TAL effector-driven gene expression. Using the normalized (log2 transformed) GeneChip expression data, we began by comparing the average transcript levels of the targets at two hours after inoculation in mock - or Xoc-inoculated plants to expression levels of 1) the 20 falsely predicted targets, 2) all genes differentially expressed (DE) in the Xoc vs. mock comparison, and 3) all genes represented on the array (Figure 8). This average basal expression level of the targets was nearly identical in mock - and Xoc-inoculated plants, similar to that of the falsely predicted targets, slightly higher than that of all genes DE in the mock vs. Xoc comparison, and markedly higher than the average expression level for all genes under either condition at any time point (4.4). Indeed, the majority (14 of 19) of the targets showed basal levels (two hours after inoculation with Xoc) higher than that average (Table S7; for the analyses presented here and throughout this section, genes represented by two probesets in any table were assigned the average values of those probesets). The target with the highest normalized basal expression level was Os03g37840 targeted by Tal4a, at 7.6, approximately 1.7 times the genome-wide average at that time for either Xoc - or mock-inoculated plants.
Fig. 8. Expression levels of probesets associated with X. oryzae pv. oryzicola BLS256 (Xoc) TAL effector targets relative to other probesets. 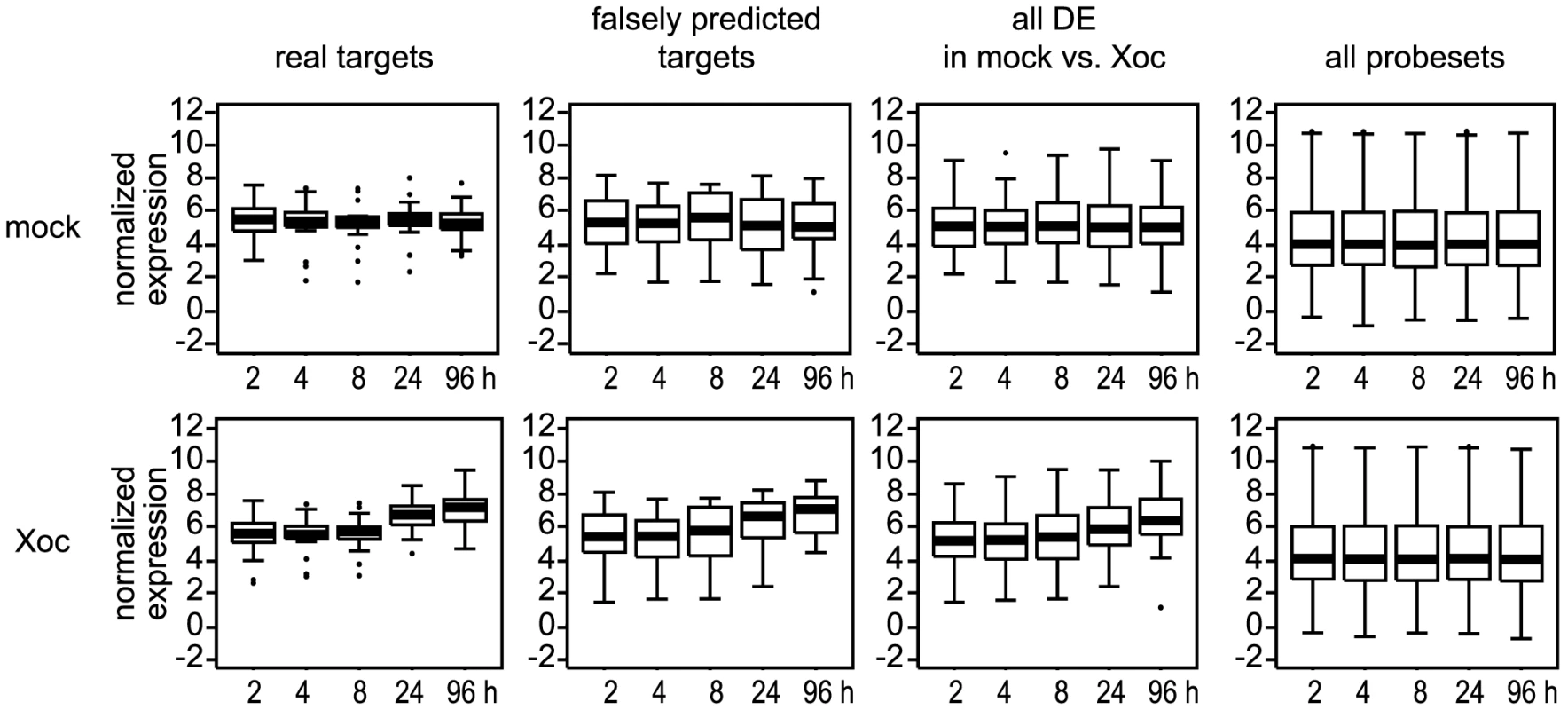
Individual box plots show average normalized expression values over time for probesets associated with verified (real) Xoc TAL effector targets, probesets associated with genes predicted but shown not to be targeted by an Xoc TAL effector (falsely predicted targets), all probesets differentially expressed (DE) in the mock vs. Xoc comparison at q≤0.3, or all probesets on the chip. The top row of plots shows data from mock-inoculated plants and the bottom row data from plants inoculated with Xoc. For each plot, the central bar indicates the median value and the top and bottom of the box indicate the 75th percentile and the 25th percentile, respectively. Whiskers indicate the most extreme data points above and below the median that are not outliers, calculated as ≤1.5*(75th percentile – 25th percentile) above the 75th percentile or below the 25th percentile. Outliers are plotted individually. Boxplots were made using the ‘boxplot()’ function of the statistical software package R (www.r-project.org). We next examined expression at two hours after Xoc inoculation relative to expression at 96 hours after that treatment. The average fold induction (Table 1 and Table S7) of the targets (calculated as 2average(X-Y), where fold induction of a gene is defined as 2(X-Y) for the difference between normalized expression values X and Y; see Materials and Methods) was 3.3, compared to an average of 2.7 for the falsely predicted targets (Table 1 and Table S7) and 2.6 for all the genes DE in the Xoc vs. mock comparison (Table S1 and Table S3). Compared to the average for all 19 targets, induction of 11 of the 14 targets with higher than average basal expression levels was moderate, from the overall minimum of 1.2-fold, exhibited by the Tal2a target Os02g43760, to 3.3-fold, whereas the five targets basally expressed at or below the average for the genome were induced 1.6 - to 9.1-fold. The remaining three targets, which were expressed at higher than average basal levels, varied in their induction from 6.4 - to the overall high of 22.4-fold exhibited by the Tal2d target Os04g49194. This latter value was second only to the 34.2-fold induction of Os01g40290 (Table S1), a gene not predicted to be an Xoc TAL effector target. The normalized expression value for the Tal2d target Os04g49194 at 96 hours after Xoc inoculation, 9.4, was also near the maximum across the genome for that time point and treatment, 10.7 (Os11g47970, probeset Os.11573.2.A2_a_at). Right behind was the sulfate transporter S gene Os01g52130 targeted by Tal2g, exhibiting induction of 13.0-fold to an expression level of 9.1. Overall, though there was not a perfect inverse correlation between basal expression level and fold-induction, expression levels of all targets at 96 hours after Xoc inoculation varied relatively little, averaging 6.9 (standard deviation, SD,1.3), suggesting that regardless of initial target expression level, TAL effectors may generally induce genes to a similar final level.
Extending the analysis to the four known Xoo TAL effector-target pairs represented in our data (Table S7), we found that the average basal expression (i.e., two hours following Xoo infection) was 5.4 (SD 0.6), nearly identical to the average basal expression of Xoc TAL effector targets (5.2 with SD 1.3). One of the Xoo TAL effector targets (Os07g06970 targeted by Tal9a, also targeted by Tal1c of Xoc) was expressed basally at near genome-average levels. It was moderately induced, 5.0-fold, by 96 hours after Xoo inoculation. The other three, like the majority of the Xoc TAL effector targets, were each basally expressed at higher than average levels. Two of these, Os01g73890 (TFIIAγ1) and Os09g29820 (OsTFX1), targeted by PthXo7 and PthXo6, respectively, also showed relatively low fold induction (3.2 - and 2.2-fold, respectively). The overall average fold induction, 4.9, was higher than that of the Xoc TAL effector targets, but this number is skewed somewhat by the large change, 17.1-fold, in expression of the third target initially expressed at higher than average levels, Os08g42350 (OsSWEET11) targeted by PthXo1. Despite the small sample size, and with the PthXo1 target as a notable exception, the pattern of expression and fold-induction of the Xoo TAL effector targets overall was similar to that observed for Xoc TAL effector targets, tending toward higher than average initial levels and relatively moderate induction.
EBE features are predictive of real targets
Finally, to better understand targeting and to improve prediction, we asked whether there are features of EBEs in the real targets we identified that distinguish them from those in our falsely predicted targets. Indeed, inspection of the features listed in Table 1 revealed some that appear to be characteristic of EBEs in real targets (we included both Tal6 EBEs in Os12g42970 in this analysis, for a total of 20 EBEs in real targets). First, on average, EBEs in real targets had lower relative scores. The relative score is the ratio of the actual score for a TAL effector-target alignment to the hypothetical score of that TAL effector aligned with its perfect match target; it allows comparison across TAL effectors, which is otherwise not possible because repeat number and RVD composition affect actual score [32]. The average relative score for EBEs in real targets was 1.98 (range 1.22–2.81), while for falsely predicted targets it was 2.47 (range 1.70–3.18). Second, EBEs in real targets generally ranked more highly in the collection of scores for the TAL effector across all rice promoters than the EBEs in the falsely predicted targets did: 16 of the 20 in real targets ranked in the top 200, with an average rank of 137 across all 20, while 17 of the 20 in falsely predicted targets ranked lower than 200, with an average rank of 347 for all 20. Finally, the maximum distance of an EBE in a real target from the annotated transcriptional start site was 152 bp upstream, with an average of 47 bp upstream (based on 19 that have an annotated TXS, out of the 20 total; range, 152 bp upstream to 63 bp downstream), whereas for the falsely predicted targets, the EBEs were anywhere from 22 bp downstream to 815 bp upstream, with an average distance of 293 bp upstream (based on the 18 with an annotated TXS). Proximity to a TATA box did not appear to correlate independently: nine of the EBEs in real and six of the EBEs in falsely predicted targets are within 100 bp of a TATA box.
To test whether the apparent differences in EBE features could be used to computationally discriminate between real and falsely predicted TAL effector targets and thereby improve future prediction, we took a machine learning approach and trained several Naive Bayes and logistic regression classifiers using combinations of relative score, rank, distance to TXS, and proximity to a TATA box, as well as actual score, distance to translational start site (TLS), and distance to a Y patch, a core promoter motif commonly found in plants [44]. For this analysis, we included also the known Xoo (PXO99A) TAL effector targets in Nipponbare, each of which, as noted above, was among our predictions (Table S7). Classifiers were generated using leave-one-out cross validation, a method that determines model parameters using all but one of the EBEs as the training set and then asks whether the resulting classifier correctly calls the remaining EBE. This is repeated with each EBE in turn to optimize the model. Recall, precision, and other metrics are computed based on the number of EBEs classified correctly using this procedure. A Naive Bayes classifier trained on all features achieved the highest recall, capturing 92% of the real targets (Table 2). The precision (percent of positives called that are true positives) of the classifier was 88% (Table 2), and no other classifier had a significantly better area under the receiver operating characteristic curve (AUC; Figure S3), a measure of the tradeoff between recall and precision. Notably, a logistic regression classifier using the distance to transcriptional start site alone achieved a recall almost as high as that achieved using all features, and had a similar AUC (Table 2 and Figure S3).
Tab. 2. Performance of a Naive Bayes classifier trained on all EBE features and of a logistic regression classifier trained on distance to transcriptional start site (TXS) using leave-one-out cross validation.a 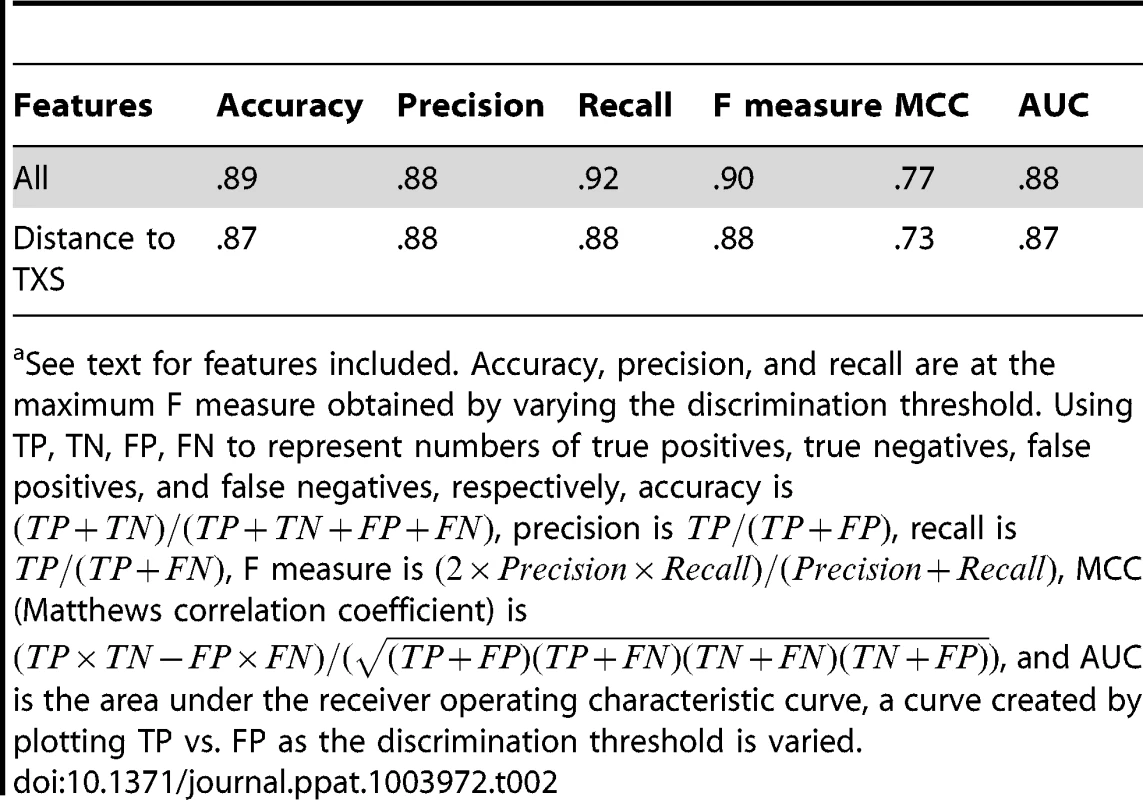
See text for features included. Accuracy, precision, and recall are at the maximum F measure obtained by varying the discrimination threshold. Using TP, TN, FP, FN to represent numbers of true positives, true negatives, false positives, and false negatives, respectively, accuracy is , precision is , recall is , F measure is , MCC (Matthews correlation coefficient) is , and AUC is the area under the receiver operating characteristic curve, a curve created by plotting TP vs. FP as the discrimination threshold is varied. Discussion
In this study we integrated genome-wide expression profiling, computational prediction using the TAL effector-DNA binding code, and functional analyses, and identified a TAL effector target in rice, OsSULTR3;6, that plays a major role in susceptibility of this staple crop species to a disease of increasing global importance, bacterial leaf streak of rice. Key to identifying the S gene was targeted gene activation using designer TAL effectors. Encoding a predicted sulfate transporter, the gene represents a new class of TAL effector-induced S gene, distinct from the handful that has been identified for bacterial blight of rice. Indeed, we discovered that overall, pathogen-induced host transcriptional changes in leaf streak are almost entirely different from those that take place during blight. We found that the T3S-translocated TAL effectors of the leaf streak pathogen are responsible, at a minimum, for nearly a quarter (19/85 genes) of the differential host gene expression during infection that we detected. We identified Tal2g as the major Xoc virulence factor that upregulates OsSULTR3;6, and demonstrated that the upregulation of OsSULTR3;6 contributes specifically to lesion expansion and bacterial exudation. We learned that, on average, TAL effector targets are expressed basally at higher than genome average levels and induced to a moderate extent, though OsSULTR3;6 and the blight S gene OsSWEET11 were exceptions, as two of the most highly induced genes in our dataset. Finally, the targets we identified and predictions we verified to be false allowed us to generate a Naive Bayes classifier that can be used in the future to identify the strongest candidate TAL effector targets prior to verification experiments, and that may also help optimize targeting with dTALEs. These advances leave the key question about tissue specificity unanswered, and raise other questions, but they open promising new avenues of inquiry. Also, they highlight gaps in our understanding of gene activation by TAL effectors, and point to challenges that remain in code-assisted discovery of TAL effector targets, but they demonstrate nonetheless the power of the approach we used to rapidly dissect interactions between TAL effector-wielding pathogens and their hosts.
Tissue specificity and the role of TAL effectors
Regarding the basis for the tissue specificity of Xoc relative to Xoo, the markedly distinct patterns of host global gene expression during bacterial leaf streak compared to bacterial blight suggest a role for host gene manipulation by the pathogens. The results of the gene ontology enrichment analysis we carried out on the differentially expressed genes raise the intriguing possibility that Xoc is uniquely able to control redox status, preventing or dampening the defense-associated oxidative burst and or affecting redox-dependent signaling pathways in the mesophyll. In a preliminary experiment to test this possibility, we observed that Xoc-inoculated leaves do show reduced overall H2O2 content at 96 h after inoculation relative to Xoo - and mock-inoculated plants (Figure S4). The reduction could relate to reduced photosynthesis, as leaves are beginning to exhibit watersoaking by this time, but it could be the direct consequence of Xoc-dependent changes in transcript levels of the redox-modulating genes, as Xoo-infected plants also exhibit watersoaking by 96 hrs, yet are unaltered in their H202 content relative to mock inoculated plants.
In contrast, the abundance of membrane associated and vesicle associated terms unique to the Xoo-induced genes is consistent with an ability to manipulate trafficking pathways that might result in nutrient export from xylem parenchyma cells into the nutrient-poor xylem, an ability Xoc may lack. This possibility aligns with the presumed role of the blight S gene OsSWEET11 as a sucrose exporter.
The extent to which TAL effectors account for the genome-wide differences in gene expression is uncertain. We observed previously that TAL effectors in Xoc and Xoo diversified subsequent to or in concert with divergence of the two pathovars [23], so it is tempting to assume a determinative role for TAL effectors in tissue specificity. However, despite our demonstration that 19 out of the 85 genes induced by Xoc are TAL effector targets, the numbers of identified targets, particularly for Xoo, are still too few to draw any conclusions from ontology enrichment analysis of just the targets. But note that targets of TAL effectors from each pathovar include one or more distinct transcription factor or putative transcription factor (bHLH family) genes: the ontology enrichment results just discussed might reflect a pervasive and determinative role of TAL effectors, through both direct and indirect effects on global gene expression patterns.
Genetic manipulation of host cells tailored to the different conditions in the mesophyll apoplast vs. the xylem is a compelling hypothesis, but one might expect some generic manipulation important for colonization both by Xoc and Xoo as well. Curiously, neither of the two genes targeted by TAL effectors from both pathovars, the OsHen1 RNA methylase gene Os07g06970 or the VQ domain containing protein gene Os02g15290, appears to play an important role in leaf streak, based on the observation that the corresponding TAL effector mutant strains M87 (tal1c) and M38 (tal5a) were not significantly reduced in virulence (Figure 4). Possible roles of these targets in bacterial blight remain to be tested.
Virulence contributions of Xoc TAL effectors
Despite the fact that exactly half of the Xoc TAL effectors were found to activate specific targets, most of the Xoc TAL effectors appear not to play a significant role in virulence, raising the question why the pathogen harbors all 26. We screened over 150 pSM7 integrants of Xoc to find that ones showing significantly reduced lesion lengths when inoculated to rice cv. Nipponbare mapped exclusively to tal clusters 2 or 11 (Figure 4 shows representative mutants). We narrowed this further to those that affect tal2g or the two tal11 genes. We confirmed the virulence contribution of tal2g by complementation, but we did not do the same for the tal11 mutants, leaving open the possibility even that the reduced virulence of the tal11a and tal11b mutants was due to ectopic mutation. The lack of a detectable virulence contribution for most Xoc TAL effectors is not unlike observations for Xoo, in which TAL effectors contribute to virulence to different extents, with typically only one or two out of many per strain playing a major role [8], [12]. Three possible reasons for the phenomenon come to mind, none mutually exclusive. First, the non-contributing TAL effectors may be important in host genotypes other than Nipponbare, in which promoter polymorphisms can influence targeting, or in plants at different growth stages from the one we assayed, in which the physiological context might change the gene activation requirements for the development of leaf streak. Second, having many clusters of tal genes in the genome, even if most are inconsequential to infection, might provide a selective advantage over time by increasing the likelihood of recombination for adaptation to new host genotypes [45]. Finally, the contributions might be redundant, or subtle, similar to those of non-TAL type III effectors [46]. Though predicted, we confirmed no redundant targeting by Xoc TAL effectors. Rather, the functions of distinct targets could themselves be redundant or epistatic to one another, a scenario that would have escaped detection in our study, but again would provide a pathogen advantage in the face of host genotypic variation. Regarding subtle contributions of individual TAL effectors, they might collectively cause an essential perturbation.
The role of Tal2g and its relevant target
The importance of Tal2g and the sulfate transporter gene it upregulates for lesion expansion and bacterial exudation is reminiscent of the phenotype associated with TAL effector Avrb6 of the cotton (Gossypium hirsutum) pathogen X. campestris pv malvacearum. Strains carrying the avrb6 gene cause larger water-soaked symptoms that correlate with more bacterial release to the leaf surface [6]. It has been proposed that bacterial exit and accumulation onto the leaf surface is advantageous as a means of dissemination, particularly for pathogens like Xoc that do not cause systemic infections [14], [47], [48]. AvrBs3 causes cell hypertrophy that may achieve this by reducing the volume of the apoplast and squeezing bacteria out to the surface, by inducing the pepper cell size regulatory gene UPA20 [14], [16]. PthA of X. citri triggers developmental changes that result in canker formation and rupture, releasing bacteria to the leaf surface [49]. Its target has not been reported. We have seen no evidence of hyperplasia or hypertrophy in available micrographs of Xoc infected rice leaves, nor in electron micrographs we have generated, and sulfate transporters are not known to regulate cell growth, but this possibility should be examined more closely in a future study. We hypothesize though that, as suggested by the effect of Avrb6 (the target of which is also yet to be reported), the enhanced watersoaking conferred by Tal2g upregulation of OsSULTR3;6 facilitates bacterial egress.
The rice cv. Nipponbare genome encodes 14 sulfate transporter genes phylogenetically divided into five groups [42], [50]. OsSULTR3;6 belongs to the less well characterized group 3 that includes five additional members. None of the additional members is induced by Xoc (i.e., they are absent from Table S1). A recent report demonstrated a role for the Arabidopsis group 3 sulfate transporter AtSULTR3;1 in pH-dependent sulfate uptake by chloroplasts [51]. The chloroplast is a main site for sulfate reductive assimilation for the synthesis of cysteine, which together with glutathione maintains the antioxidant capacity of the cytosol [52]–[54]. AtSULTR3;2, AtSULTR3;3 and AtSULTR3;4 also were shown to contribute [51]. In contrast, the last member of the group, AtSULTR3;5, is plasma membrane localized and cooperates in roots with the low affinity transporter AtSULTR2;1 under sulfur deficiency to increase sulfate uptake capacity for root-to-shoot vascular transport [55]. The Tal2g target OsSULTR3;6 is most similar to AtSULTR3;5 (57% identity) yet is expressed, in the absence of Xoc infection, primarily in seeds during later stages of seed development [42]. The physiological consequence of the recruitment of high OsSULTR3;6 expression to mesophyll cells by Tal2g is therefore challenging to predict. Given the M27 phenotype, an attractive hypothesis is that it alters antioxidant capacity, impinging on redox signaling to dampen defense and allow more rapid induction of watersoaking by the pathogen. Another possibility is that it enhances watersoaking more directly, either through a redox-controlled mechanism or simply by altering osmotic equilibrium.
In L. japonica, the group 3 sulfate transporter gene sst1, which is more similar to OsSULTR3;6 (56% identity) than to any other member of the gene family in rice, is essential for normal nodule development and symbiotic nitrogen fixation [56]. Its ortholog in poplar (Populus trichocarpa), PtSultr3;5, is among most highly induced transcripts during the establishment of symbiosis with the fungus Laccaria bicolor [57]. This gene is also strongly induced during both compatible and incompatible interactions with the poplar rust pathogen Melampsora larici-populina [58]. Whether the Tal2g target and these orthologs play analogous roles in such diverse plant-microbe interactions awaits in-depth functional analysis.
That a major S gene for leaf streak is a member of a large gene family recalls the situation in blight, in which five members of the large OsSWEET family can functionally substitute for one another as S genes, three so far have been shown to play that role, and distinct TAL effectors from multiple strains have been identified as the activators of two [11], [13], [19]. Whether any of the five other group 3 paralogs, or of the other 13 total members of the sulfate transporter gene family in rice can substitute for OsSULTR3;6, and whether any are actually targeted by other strains of Xoc is an important question. A tendency for S genes to be members of functionally analogous gene families would make sense from an evolutionary perspective, both for the advantage it would afford the pathogen by providing alternate targets should cis - (e.g. xa13) or trans- (e.g., Bs3) acting types of resistance be encountered, as well as the possibility it would afford the host to adapt through promoter mutation and resist targeting while maintaining essential functions. These processes might indeed drive one another [46]. On the other hand, if OsSULTR3;6 is uniquely capable among its paralogs of serving as an S gene, the likelihood of identifying moderately stable resistance by screening for or engineering promoter variants that retain endogenous function is increased.
General characteristics of TAL effector-driven gene expression
OsSULTR3;6 was one of the most strongly induced and highly expressed genes in Xoc inoculated plants, as OsSWEET11 was in Xoo inoculated plants. These major S genes were exceptional, with the majority of TAL effector targets being induced moderately. The blight S genes OsTFIIAγ1 and OsTFX1, which contribute only moderately, were induced relatively weakly. Whether these differences reflect an evolutionary optimization of transactivation for major S genes, or gene specific differences in induction potential or optima, or chance, is unclear. The general pattern of relatively high basal expression and moderate fold increase across all identified TAL effector targets may be dominated by so-called collateral targeting inconsequential to disease and under no selection, and it suggests that TAL effectors may act as transcriptional enhancers more readily than as activators. However, the low variation in normalized expression levels for all targets at 96 h after inoculation suggests that on average, this enhancement is close to saturating.
We generally did not observe significant expression changes at early time points (i.e. 2 h and 4 h), possibly as a result of a low signal∶noise ratio caused by variation among the replicates, but expression of TAL effector targets generally increased steadily across the later time points. Though some genes were expressed at lower levels in Xoc - or Xoo-inoculated plants than in mock-inoculated plants at 96 h, no significant patterns of downregulation across all time points were observed. We tentatively conclude from these observations that TAL effectors of Xoc and Xoo do not significantly downregulate any genes in their host, despite the potential to do so through indirect effects, or theoretically, through nonfunctional binding that interferes with endogenous expression.
False positives in target prediction
The average number of candidate EBEs in the rice promoterome, per TAL effector across all Xoc and Xoo TAL effectors, was 671. After excluding candidate EBEs in genes not upregulated after inoculation, that average dropped to 1.5, with some TAL effectors having none and some as many as seven. Nearly half of the filtered EBEs that were tested further (i.e., those for Xoc TAL effectors) were real. Thus, combining candidate EBE search results with global gene expression data is a robust and effective approach to identifying TAL effector targets.
Nevertheless, the method still yielded roughly as many false positives as true targets. Though upregulated during infection, false positives might include genes with EBEs that match but are inaccessible or in the wrong context to be functional, or genes with EBEs that score below the cutoff but are not actually sufficiently high affinity sites. In an attempt to decrease the number of falsely predicted targets and improve the efficiency of target identification in the future, we applied machine learning to our set of 24 real (Xoc and Xoo) and 20 falsely predicted (Xoc) targets (Table S7) using several characteristics of their candidate EBEs. The best classifier that resulted calls 22 of the real targets and three of the falsely predicted targets as real, for a recall of 0.92 and a precision of 0.88. Thus, it eliminates 85% (17/20) of the falsely predicted targets at a cost of less than 10% (2/24) of the real ones. The training set was relatively small, so these metrics may not hold strictly when applied to larger numbers of predicted targets, and even if they are relatively stable, if the goal is to comprehensively capture real targets, the classifier clearly can not be used as a strict filter. It is also important to remember that the classifier was trained only on EBEs that passed the score cutoff and were located in up-regulated genes, so performance metrics might not hold if the classifier is used on EBEs that do not meet these requirements. Rather, the probability this classifier provides should be used to prioritize already predicted targets for experimental validation (see Supporting Information for the Weka model file for the Naive Bayes classifier trained on all EBE features). Training on a greater number of targets as they are identified will improve both precision and recall, possibly even uncovering conditional relationships among characteristics of EBEs in real targets that the classifier currently calls incorrectly. With more targets, further comparison of classifiers built on subsets of EBE features might also reveal a smaller set of the most biologically relevant features that are sufficient to effectively discriminate real targets. Even with the small training set used here, the only slightly lower recall of the classifier based just on distance to transcriptional start site strongly suggests a major role for this feature.
As demonstrated by the results of our functional characterization of Tal2g EBEs and candidate EBEs (Figure 7), an important remaining challenge to eliminating false positives is a more nuanced understanding of TAL effector DNA binding. In particular, being able eventually to replace the RVD-nucleotide association frequency-based scoring matrix with one based on biochemically defined contributions of different RVD-nucleotide pairings, weighted to account for effects of position 5′ to 3′, will improve initial candidate EBE calling. Defining specificities for as many rare RVDs as possible will also be important to eliminate false positives and capture real targets for proteins like Tal2g. In this regard, we improved our predictions by substituting values of common RVDs for two rare ones, based on inference from structural data, and supported in the case of ‘SN’ by an experimental study [59].
False negatives in target prediction
Better understanding of TAL effector DNA interactions will also help eliminate false negatives. Without the scoring substitutions for the rare RVDs in Tal2g, one of its targets, the monocopper oxidase gene, would have been overlooked. Another example is suggested by the lack of identified targets for either Tal11a or Tal11b despite the reduced virulence of tal11 mutants (recalling however that complementation analysis was not performed to verify a role for either effector). A very low level of induction may be sufficient for function of some targets, such as an R gene like Xa27 [22] or an S gene that encodes a transcription factor, so false negatives could derive from a failure to detect differential expression in the initial transcript profiling experiment. A false negative could also result for a TAL effector with lax specificity. Xa27 again serves as an example. AvrXa27 contains at several positions an RVD with dual or no specificity; its EBE in Xa27 ranks 5,368th in the rice promoterome, nestled above the low-scoring outlier cutoff [26]. Exclusion of sites preceded by any base other than T, as specified in our search, might also pass over a real EBE. The TalC EBE in OsSWEET14, discussed in the introduction, is a salient if rare example. Two additional, theoretical examples are worth considering. The first is a gene whose expression is activated via read-through transcription by a TAL effector that targets a neighboring gene upstream. The second is a gene for which overall transcript levels do not change detectably, but which yields a unique alternative transcript when driven by the TAL effector due to TAL effector-dependent repositioning of the transcriptional start site [41], [60]. Transcript profiling by next generation sequencing of cDNA (RNAseq) [e.g., 61], in contrast to the GeneChip expression experiment that began this study, should provide the sensitivity to detect weakly expressed or weakly induced genes as well as alternative transcripts, to reduce or eliminate false negatives that might otherwise result. Regarding TAL effectors with lax specificity, EBEs with a non-canonical preceding base, and potential read-through targeting, adjusting EBE search parameters is a simple solution, but will unavoidably increase the number of false positives.
Direct vs. indirect targets
Given the current understanding of TAL effector function and the ability to predict binding sites using the code, we considered each gene that was activated by a TAL effector and that displayed a strong candidate EBE for that effector to be directly activated. Yet even meeting these criteria, it is formally possible that such a gene might be activated indirectly, i.e., in response to expression of another gene directly activated by the TAL effector. In pepper, prior to discovery of the code, direct targets of AvrBs3 were isolated by screening for transcripts whose upregulation by AvrBs3 occurs even in the presence of the eukaryotic translation inhibitor cycloheximide [14], [16]. To address the possibility that some of the Xoc TAL effector targets we identified in rice are indirect targets, using RT-PCR we tested Xoc-triggered transcript accumulation of the targets for sensitivity to cycloheximide, measured at 8, 16, 24, and 36 hr after co-infiltration (Figure S5). At the two earlier time points transcripts of all but one target accumulated similarly in response to Xoc with or without cycloheximide, and most showed identical patterns across all four time points. However, several showed distinct patterns of up and down regulation across the time points in response to cycloheximide treatment alone. Furthermore, cycloheximide treatment strongly and persistently upregulated three pathogenesis-related genes previously observed to be induced by biotic stresses, included as controls, and transiently induced a fourth. Induction of an additional control, Os05g42150, which is the most significantly Xoc-induced gene in our dataset (Table S1) and is not predicted to be a TAL effector target, was unaffected by cycloheximide at the two earlier time points and was slightly repressed at the later ones. The results overall thus reveal differing and confounding epistatic effects of cycloheximide treatment in rice that render conclusive identification of direct and indirect targets by this method impossible.
Regarding the single target showing repression of Xoc-induced transcript accumulation in the presence of cycloheximide, the monocopper oxidase gene Os06g46500, in addition to its being upregulated during infection and harboring an appropriately positioned strong candidate EBE for Tal2g, several other lines of evidence support it being a direct target. First, in the context of the Bs3 promoter, tested in N. benthamiana, that EBE is functional (Figure 7). Second, the pattern of induction of Os06g46500 by Xoc is rapid and robust, virtually identical to that of the SO4 transporter gene targeted by Tal2g (Figure S6) and similar to the patterns displayed by the verified Xoo TAL effector targets (Figure 2). Third, no other Tal2g target that might activate Os06g46500 was predicted other than the SO4 transporter gene, and activation of the SO4 transporter gene by dTALEs was not accompanied by activation of Os06g46500 (Figure 6B). Also, activation of Os06g46500 itself with a dTALE targeting a site in the vicinity of the native EBE (Figure 6) indicates that the promoter is not generally inaccessible to binding by a TAL effector. Finally, assuming that if not all, at least most of the targets in the training set for the classifier we generated by machine learning are direct targets, the leave-one-out validation tests showed that Os06g46500 shares the characteristics of those targets (including the previously confirmed targets of Xoo TAL effectors).
For Os06g46500, and the rest of the targets we identified here, assaying activation following disruption of the endogenous EBEs by site-directed mutagenesis would provide the most direct evidence for or against direct targeting, but such experiments are beyond the scope of the present study. Absent such data, it remains possible that some of the targets we identified are indirect ones. For the reasons detailed in the above two paragraphs, we conclude that this is unlikely, but to the extent that it were true, it would affect the utility of our predictive classifier.
Outlook
Many crops, including rice, wheat, cotton, citrus, tomato, cassava, soybean, and others, suffer losses due to Xanthomonas spp. that deploy TAL effectors. We demonstrated here that TAL effector activity in bacterial leaf streak of rice is directly responsible for nearly a quarter of the gene activation detected during infection. Considering the likely downstream effects, the total proportion is certain to be even greater. Our study provides new insight into bacterial leaf streak of rice in relation to bacterial blight and identifies a major new S gene, but TAL effector target identification in several pathosystems is a critically important ongoing objective. Probing the diversity and functions of TAL effector activated S and R genes will expand our knowledge of disease and defense mechanisms, and our ability to exploit those mechanisms for effective disease control. Patterns of distribution of different S genes in diverse pathosystems might yet reveal causal distinctions between pathogens that colonize the mesophyll and those that invade the xylem. New targets will also refine our understanding of functional TAL effector-DNA interactions, improving our ability to use these proteins [62]. Though improvements can be made, and challenges remain, the overall combined transcriptomic and computational approach we successfully undertook constitutes a moderately high throughput method that can be applied to TAL effector target identification in many Xanthomonas-host interactions, particularly as more, complete pathogen and host genome sequences become available.
Materials and Methods
Plant material and growth conditions
Oryza sativa ssp. japonica cv. Nipponbare plants were grown in LC-1 soil mixture (Sungro, Bellevue, WA) in PGC15 growth chambers (Percival) in trays approximately 60 cm below a combination of fluorescent and incandescent bulbs providing approximately 1,000 µmoles/m2/s measured at 15 cm, under a cycle of 12 h light at 28°C and 12 h dark at 25°C. Fertilizer (Peters Professional, St. Louis, MO) and iron chelate micronutrient (Becker Underwood, Ames, IA) were applied with watering every two days at 0.25 and 4.5 g/l, respectively, until the day before inoculation. Nicotiana benthamiana plants were grown in LC-1 in a PGC15 growth chamber at approximately 90 cm below the lights, under a cycle of 16 h light (fluorescent lighting at 22°C, and 8 h dark at 18°C, and fertilized using a surface amendment of Osmocote granules (ScottsMiracle-Gro, Maryville, OH).
Bacterial strains and plasmids used, culture, and transformation
Bacterial strains and plasmids used for this study are listed in Table S8. E. coli strains were grown in LB medium at 37°C and A. tumefaciens in YEP medium (10 g/l peptone, 10 g/l yeast extract, 5 g/l NaCl, 1.5% agar) at 28°C, and transformed by standard electroporation. X. oryzae strains were cultured in GYE (20 g/l glucose, 10 g/l yeast extract) at 28°C unless otherwise specified, and were transformed by electroporation as described previously [63], except that 1 µl (5 µg) TypeOne Restriction Inhibitor (Epicentre Biotechnologies, Madison, WI) was added prior. Antibiotics were used for selection as follows: ampicillin at 100 µg/ml, gentamycin at 25 µg/ml, kanamycin at 25 µg/ml, spectinomycin at 25 µg/ml, and tetracycline at 10 µg/ml for E. coli or 2 µg/ml for X. oryzae.
GeneChip expression experiment
Experimental design
The experiment was carried out in four independent replicates, each one week apart. Plants were grown three trays together per replicate, one each for Xoc-, Xoo-, and mock-inoculation. Trays were moved to a new chamber once per week, in order through three chambers total, so that plants for each replicate were incubated in the same chamber (the third one) following inoculation. For the different replicates, position of the trays left to right within the chambers was maintained according to a random assignment specific to each replicate that was also used for the order of inoculation and tissue collection. Tissue was collected at 2, 4, 8, 24 and 96 hours after inoculation, with plants in each tray randomly assigned to time points. Overall, the experiment followed a split-plot design with inoculation as the whole-plot factor and time as the split-plot factor.
Inoculation
Plants were grown four per 5 cm square pot, arranged in trays of 20 pots, one tray for each inoculum. To allow inversion for inoculation (see below), each seed was sown through a short plastic tube (a five ml pipette tip with its tapered end removed) extending from the soil surface through a fiberglass lid (cafeteria tray) that had been perforated with appropriately spaced 1 cm holes and affixed to the top of the tray. Fourteen days after sowing, and 2 h after the beginning of the light period, trays (with seedlings projecting from the tubes) were inverted over a plastic dishpan containing 7 l of inoculum to submerge the seedlings for vacuum infiltration. This was carried out in a custom vacuum chamber by subjecting the submerged plants to 500 mm Hg for two minutes followed by a rapid return to atmospheric pressure, two consecutive times. Xoc and Xoo inoculum was prepared as follows. For each, a single colony from a fresh plate was transferred to 5 ml of liquid medium and incubated for 24 h at 28°C with constant shaking at 250 r.p.m. Then, 2 ml of this culture were transferred to 300 ml of fresh liquid medium and incubated as above for an additional 18 h. Cells were pelleted by centrifugation at 4,000× g for 10 min, washed twice and resuspended in sterile 10 mm MgCl2 to OD600 = 0.05. Tween-20 was added to a final concentration of 0.5%. Seven l of this suspension were used for inoculation. Mock inoculum consisted of 10 mm MgCl2 and Tween-20 only. Following inoculation, plants were returned to a growth chamber.
Tissue collection
Leaves were cut and pooled from plants in four pots (16 plants, approximately 2 g fresh weight) for each timepoint per inoculum per replicate. Harvested tissue was immediately frozen in liquid nitrogen and stored at −80°C until processing.
RNA isolation, probe synthesis, and hybridization
Total RNA was isolated using a hot (60°C) phenol/guanidine thiocyanate method [64]. Probe synthesis and labeling were performed at the Iowa State University GeneChip Core facility (Ames, IA, U.S.A.), using the One Cycle and GeneChip IVT labeling kits. Fifteen µg of fragmented cRNA was used to make each hybridization cocktail containing 10% dimethyl sulfoxide, and 10 µg equivalent was hybridized to the GeneChip Rice Genome Array (Affymetrix, Santa Clara, CA).
Data acquisition and analysis
Stained chips were immediately scanned with the GeneChip Scanner 3000 7G (Affymetrix). Scans were examined for any visible defects and satisfactory image files were analyzed to generate raw data files using the GeneChip Operating Software (GCOS v1.4.0.036; Affymetrix) with default settings. Robust multi-array analysis (RMA) [65], [66] was used to normalize the data. Note that RMA normalization includes a log2 transformation, so fold-change across absolute values for two normalized values X and Y is calculated as 2(X-Y). A mixed linear model was fit separately to RMA-normalized data for each probeset. Each mixed linear model included fixed effects for replicate, treatment, time, and treatment-by-time interaction, as well as random effects for the trays. We used the model for each probeset to test for a difference between its patterns of expression over time in Xoc - and mock-, Xoo - and mock-, and Xoc - and Xoo-inoculated plants. The null hypothesis for each comparison was that the expression difference between inoculations was constant across all five time points. The 55,515 p-values from each of these three inoculation comparisons, representing all probesets, were converted to q-values using the method of Storey and Tibshirani [67]. To better enhance capture of genes that were moderately differentially expressed in this screening experiment, we used a relatively lax q value cutoff of 0.30. This implies that approximately 70% of the genes declared to be differentially expressed are expected to be true positives.
GeneChip expression data access
GeneChip data are available at the PLEXdb gene expression resource (www.plexdb.org) [68] under accession OS3 and at NCBI-GEO under accession GSE16793.
Prediction of TAL effector targets
Promoter sequences, defined as the 1,000 bases upstream of the start codon, for the approximately 56,000 rice genes annotated in the MSU Rice Genome Annotation Project Release 7 (http://rice.plantbiology.msu.edu/) were searched using our previously published TAL effector-target scoring model [32], for the best-scoring site in each promoter for each of the 40 unique Xoo and Xoc TAL effectors. Scoring takes the sum of the negative log probabilities of the RVD-nucleotide pairings at a site, so a lower score is a better score. Sites were required to be directly preceded by a 5′ T. The scoring matrix was used as published and separately with the RVDs ‘SN’ and ‘YG’ assigned nucleotide association frequencies of ‘NN’ and ‘NG’, respectively (see [Results]). The distributions of the approximately 56,000 resulting scores for each TAL effector in each case were then used to calculate a cutoff for outliers, defined as the 25th percentile minus 1.5 times the interquartile range. Promoters were then rescanned to identify all sites scoring below (better than) that cutoff for each TAL effector, and those sites were retained as candidate EBEs. Finally, the list of candidate EBEs for each TAL effector was cross-referenced to the GeneChip expression data. Candidate EBEs in promoters of genes upregulated in Xoc - or Xoo-inoculated plants relative to mock were taken as predicted targets.
Generation of X. oryzae pv. oryzicola BLS256 TAL effector gene knockout mutants
A library of tal gene knockout strains of Xoc was generated by transformation with the suicide (non-replicative) plasmid pSM7 (Figure 4A; Table S8) [69], pSM7 harbors a 4.5-kb PstI fragment containing all but the first 80 bp of the ORF of tal gene aB4.5 [12] with an insertion of the EZ-Tn5 <NotI/KAN-3> transposon (Epicentre) at bp 1,769 of the gene, in repeat number 9 of 17.5 (sequence available on request). The vector is pBluescript II KS(+) (Agilent Technologies, Santa Clara, CA). The transposon provides kanamycin resistance. Selection for this marker yields strains in which the cloned, mutated tal gene has undergone homologous recombination with an endogenous tal gene. Because the tal ORF is truncated at the 5′ end, either a single or double recombination that retains the transposon results in a tal gene knockout. Double recombination can knock out clustered tal genes. The 4.5 kb PstI fragment also includes the first 85 bp of the avrXa10 tal gene downstream of ab4.5, which might increase the likelihood of complex recombination. To determine the number of insertions per strain and to map insertions, genomic DNA was extracted using the GenElute Bacterial Genomic Kit (Sigma-Aldrich, St. Louis, MO). Strains with single insertions were identified by Southern blot using EZ-Tn5 <NotI/KAN-3> as a probe. Insertion endpoints were mapped by amplifying and sequencing the distal ends of 5′ and 3′ fragments flanking the transposon and extending outside the repeat region. Primers used for amplifying 5′ flanking DNA were forward primer p369 (5′-TTCTGfCCCGGACCCCAACCGGATAG), matching a conserved 5′ sequence in Xoc tal genes, and reverse primer p395 (5′-TCCCGTTGAATATGGCTCATAACACCCC), corresponding to the transposon. For the 3′ fragment, forward primer p397 (5′-GTCCACCTACAACAAAGCTCTCATCAACC), corresponding to the transposon, and reverse primer p398 (5′-TCCTCTTCGTTGAATGCC), matching a conserved 3′ sequence of Xoc tal genes, were used. Sequencing of the distal ends of the 5′ and 3′ amplicons (furthest from the repeat region) was done using tal gene plus-strand primer p396 (5′-ACCCCAACCGGATAGG) and p398, respectively. In all but a few cases, insertion endpoints were unambiguously identified by polymorphisms among the 5′ and 3′ sequences of the 26 Xoc tal genes and two tal pseudogenes (Figure 4).
Cloning of TAL effector genes of X. oryzae pv. oryzicola BLS256
Two micrograms of genomic DNA of X. oryzae pv. oryzicola strain BLS256 were digested with BamHI and separated in 1% agarose by electrophoresis. DNA fragments from 2 to 5 kb were gel purified and ligated into pBluescript II SK - (Agilent) previously digested with BamHI and dephosphorylated with alkaline phosphatase (CIP; New England Biolabs, Ipswitch, MA). The ligation reaction was then used to transform E. coli TOP10 cells, and colonies harboring TAL effector clones were identified by colony PCR using oligonucleotides p270 (GCCAAGTCCTGCCCGCG) and p271 (CCTCCAGGGCGCGTGC), which target the conserved 5′ region of Xanthomonas oryzae TAL effector genes. The tal gene fragments in these clones were tentatively identified based on size and 5′ and 3′ sequencing. Next, the corresponding SphI fragment of each tal gene, encoding the repeat region and short flanking sequences, was cloned into the tal1c backbone (i.e., lacking the corresponding SphI fragment) in the entry vector pCS466 [63] and confirmed by 5′ - and 3′ - sequencing with oligos p235 (GGAGGCCTTGCTCACGGATGC) and p236 (GGCCGGTGACAGCACGATCCG), respectively. For tal2g, tal4a, and tal8, BamHI fragments were cloned into pCS466 (cut with BamHI) instead, since those genes are each missing one of the SphI sites. The reconstituted genes in pCS466 were then recombined into the broad host-range destination vector pKEB31 [27], using Gateway LR Clonase (Life Technologies, Carlsbad, CA), for expression in Xanthomonas.
RT-PCR
Xoc strains were cultured for 3–4 days on solid media then resuspended in 10 mM MgCl2 to OD600 = 0.5 (approximately 1×108 CFU/ml) and infiltrated into the abaxial surface of fully expanded leaves of 4-week old rice plants using a needleless syringe. 10 mM MgCl2 alone was infiltrated as the mock. Infiltrated tissue was collected at 48 h and RNA prepared using the RNeasy Mini Kit (Qiagen, Valencia, CA). Before elution, RNA was subjected to in-column digestion with the RNase-Free DNase Set (Qiagen). Two µg of total RNA were used for first-strand cDNA synthesis using SuperScript III reverse transcriptase (Life Technologies) and standard oligo dT20. Reverse transcriptase reactions were diluted 5 times and 1 µl was used as a template for PCR with Phire Hot Start II DNA polymerase (Thermo Scientific, Waltham, MA) together with transcript-specific oligonucleotides for 30 sec at 98°C, followed by 23–25 cycles (depending on transcript abundance) of 10 sec at 98°C, 5 sec at 60°C, and 10 sec at 72°C. The oligonucleotides used are listed in Table S9.
Virulence assays and quantification of bacterial populations
Rice leaves were inoculated by syringe infiltration as described above for RT-PCR. Virulence was quantified at the specified days after inoculation as lesion expansion, in mm, from the inoculation spot (Figure 4B). To measure bacterial populations, duplicate sets of three leaves per treatment per time-point were collected. One set was used to quantify total bacterial populations and the other to quantify surface bacteria. For total bacterial counts, 10 cm leaf sections centered on the infiltration spot or leaf sections as indicated in Figure 6D were cut into small pieces and ground thoroughly in 2 ml of water using a mortar and pestle. For surface bacteria, a leaf section encompassing the watersoaked area was washed with 50 µl of water twice and the wash diluted into 1 ml of water. Samples were thereafter diluted serially in sterile water and spotted on peptone sucrose agar (10 g/l sucrose, 10 g/l peptone, 1 g/l sodium glutamate, 1.5% agar) supplemented with cephalexin at 20 µg/ml. Plates were incubated at 28°C until appearance of single colonies, and colonies at the dilution they were first distinct were counted. For each replicate sample, eight such measurements were made. Results are displayed as the mean and standard deviation of all measurements for all replicates. Experiments were repeated at least three times with consistent results.
Designer TAL effectors
TAL Effector Targeter [32] was used to target designer TAL effectors (dTALEs) to the promoter regions of Os01g52130 and Os06g46500. dTALEs were assembled by golden gate cloning into the entry vector pTAL1 as described [27] and subsequently transferred to the broad host range destination vector pKEB31 [27] by Gateway LR Clonase (Life Technologies). RVD sequences if the dTALEs used in this are provided in Text S1.
GUS reporter gene assay of TAL effector activity
GUS reporter assays were conducted in Nicotiana benthamiana leaves of five-week old plants (from the date of sowing) using the substrate 5-bromo-4-chloro-3-indoyl glucuronide (X-Gluc) as described [70], using three leaf discs from different plants per treatment, collected at 48 hours after infiltration of Agrobacterium. Experiments were repeated twice. Determination of total protein in sample extracts was performed using the Bradford assay kit (Bio-Rad). The vector for T-DNA delivery of avrBs3 under the 35S promoter was pGWB5-avrBs3 [40]. The equivalent construct for tal2g, pGWB5-tal2g, was made by replacing the ∼3.3 kb BamHI fragment of an avrBs3 clone in the entry vector pENTR-D (Life Technologies; gift of T. Lahaye, University of Munich) with the ∼3.2 kb BamHI fragment of tal2g, then moving the reconstituted tal2g equivalent gene to the binary destination vector pGWB5 [71] using Gateway LR Clonase (Life Technologies). The pGWB5 derivatives were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation; transformants were selected with 25 µg/ml each of kanamycin and gentamycin. The reporter constructs were made by first PCR amplifying from a longer Bs3 promoter clone (gift of T. Lahaye) the AvrBs3-responsive 343 bp sequence upstream of the Bs3 start codon, using previously reported primers [70] and inserting it into the Gateway entry vector pCR8/TOPO-TA (Life Technologies). A single base substitution was then introduced by site directed mutagenesis (Agilent) to create an NcoI site 47 bp upstream of the native EBE for AvrBs3. Candidate Tal2g EBEs flanked upstream by 5 bp and downstream by 4 bp matching their native context were synthesized as double stranded oligonucleotides with NcoI overhangs (Text S1) and cloned into the NcoI site of the modified Bs3 promoter. Finally, the modified Bs3 promoter and derivatives were transferred into the binary GUS reporter vector pGWB3 [71] using Gateway LR Clonase (Life Technologies), and the resulting plasmids introduced into A. tumefaciens GV3101 as described above.
Construction and validation of machine learning classifiers
Both Naive Bayes and logistic regression classifiers were implemented using Weka 3.6.9 [72] with default options, which select the discrimination threshold that maximizes F measure. All classifiers were trained on the candidate EBEs in Table S7 that were determined to be either in real or falsely predicted targets (“Yes” or “No” in column P, “Verified”). Classifiers were trained using various subsets of the following features: relative score, actual score, rank, distance to TXS, distance to TLS, proximity to a TATA box, and distance to a Y Patch. If a predicted EBE was located in a promoter without a TATA box or without a Y patch, or with no annotated TXS, the value for that feature was considered missing and replaced with a question mark. All classifiers were evaluated using leave-one-out cross validation. The receiver operating characteristic curve and precision recall curve in Figure S3 were plotted using the ROCR package [73].
Supporting Information
Zdroje
1. Mew TW (1993) Xanthomonas oryzae pathovars on rice: cause of bacterial blight and bacterial leaf streak. In: Swings JG, Civerolo EL, editors. Xanthomonas. New York: Chapman and Hall. pp. 30–40.
2. ShengZ-J, ZhenL-Y, JunF-X (2005) Detection of QTL conferring resistance to bacterial leaf streak in rice chromosome 2 (O. sativa L. spp. indica). Scientia Agricultura Sinica 38 : 1923–1925.
3. WhiteFF, YangB (2009) Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol 150 : 1677–1686.
4. Nino-LiuDO, RonaldPC, BogdanoveAJ (2006) Xanthomonas oryzae pathovars∶model pathogens of a model crop. Mol Plant Pathol 7 : 303–324.
5. SwarupS, YangY, KingsleyMT, GabrielDW (1992) A Xanthomonas citri pathogenicity gene pthA pleiotropically encodes gratuitous avirulence on nonhosts. Mol Plant-Microbe Interact 5 : 204–213.
6. YangY, De FeyterR, GabrielDW (1994) Host-specific symptoms and increased release of Xanthomonas citri and X. campestris pv. malvacearum from leaves are determined by the 102-bp tandem repeats of pthA and avrb6, respectively. Mol Plant-Microbe Interact 7 : 345–355.
7. YangY, YuanQ, GabrielDW (1996) Watersoaking function(s) of XcmH1005 are redundantly encoded by members of the Xanthomonas avr/pth gene family. The American Phytopathological Society 9 : 105–113.
8. YangB, WhiteFF (2004) Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol Plant-Microbe Interact 17 : 1192–1200.
9. YangB, SugioA, WhiteFF (2006) Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci U S A 103 : 10503–10508.
10. SugioA, YangB, ZhuT, WhiteFF (2007) Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIA{gamma}1 and OsTFX1 during bacterial blight of rice. Proc Natl Acad Sci U S A 104(25): 10720–5.
11. AntonyG, ZhouJ, HuangS, LiT, LiuB, et al. (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os11N3. Plant Cell 22 : 3864–3876.
12. BaiJ, ChoiSH, PoncianoG, LeungH, LeachJE (2000) Xanthomonas oryzae pv. oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Mol Plant-Microbe Interact 13 : 1322–1329.
13. YuY, StreubelJ, BalzergueS, ChampionA, BochJ, et al. (2011) Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol Plant-Microbe Interact 24 : 1102–1113.
14. MaroisE, Van den AckervekenG, BonasU (2002) The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol Plant-Microbe Interact 15 : 637–646.
15. WichmannG, BergelsonJ (2004) Effector genes of Xanthomonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics 166 : 693–706.
16. KayS, HahnS, MaroisE, HauseG, BonasU (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318 : 648–651.
17. RömerP, HahnS, JordanT, StraussT, BonasU, et al. (2007) Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318 : 645–648.
18. SongCX, ClarkTA, LuXY, KislyukA, DaiQ, et al. (2012) Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods 9 : 75–77.
19. StreubelJ, PesceC, HutinM, KoebnikR, BochJ, et al. (2013) Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol 200(3): 808–19.
20. LiuQ, YuanM, ZhouY, LiX, XiaoJ, et al. (2011) A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant, Cell Environ 34 : 1958–1969.
21. BonasU, StallRE, StaskawiczB (1989) Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol Gen Genet 218 : 127–136.
22. GuK, YangB, TianD, WuL, WangD, et al. (2005) R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435 : 1122–1125.
23. BogdanoveAJ, KoebnikR, LuH, FurutaniA, AngiuoliSV, et al. (2011) Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J Bacteriol 193 : 5450–5464.
24. MakinoS, SugioA, WhiteF, BogdanoveAJ (2006) Inhibition of resistance gene-mediated defense in rice by Xanthomonas oryzae pv. oryzicola. Mol Plant-Microbe Interact 19 : 240–249.
25. BochJ, ScholzeH, SchornackS, LandgrafA, HahnS, et al. (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326 : 1509–1512.
26. MoscouMJ, BogdanoveAJ (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326 : 1501.
27. CermakT, DoyleEL, ChristianM, WangL, ZhangY, et al. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82.
28. ChristianM, CermakT, DoyleEL, SchmidtC, ZhangF, et al. (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186 : 757–761.
29. MorbitzerR, RömerP, BochJ, LahayeT (2010) Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A 1078 : 21617–21622.
30. DengD, YanC, PanX, MahfouzM, WangJ, et al. (2012) Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335 : 720–723.
31. MakAN, BradleyP, CernadasRA, BogdanoveAJ, StoddardBL (2012) The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335 : 716–719.
32. DoyleEL, BooherNJ, StandageDS, VoytasDF, BrendelVP, et al. (2012) TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 40: W117–122.
33. BochJ, ScholzeH, SchornackS, LandgrafA, HahnS, et al. (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326 : 1509–1512.
34. RömerP, RechtS, StraussT, ElsaesserJ, SchornackS, et al. (2010) Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol 187 : 1048–1057.
35. RömerP, StraussT, HahnS, ScholzeH, MorbitzerR, et al. (2009) Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol 150 : 1697–1712.
36. Nino-LiuDO, DarnielleL, BogdanoveAJ (2005) A simple method of mass inoculation of rice effective for both pathovars of Xanthomonas oryzae, and the construction of comparable sets of host cDNA libraries spanning early stages of bacterial leaf blight and bacterial leaf streak. J Phytopathol 153 : 500–504.
37. WiseRP, MoscouMJ, BogdanoveAJ, WhithamSA (2007) Transcript profiling in host-pathogen interactions. Annu Rev Phytopathol 45 : 329–369.
38. DuZ, ZhouX, LingY, ZhangZ, SuZ (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–70.
39. SalzbergSL, SommerDD, SchatzMC, PhillippyAM, RabinowiczPD, et al. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9 : 204.
40. RömerP, RechtS, StraussT, ElsaesserJ, SchornackS, et al. (2010) Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol 187 : 1048–1057.
41. HummelAW, DoyleEL, BogdanoveAJ (2012) Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol 195 : 883–893.
42. KumarS, AsifMH, ChakrabartyD, TripathiRD, TrivediP (2011) Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Functional & Integrative Genomics 11 : 259–273.
43. MecklerJF, BhaktaMS, KimMS, OvadiaR, HabrianCH, et al. (2013) Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res 41 : 4118–4128.
44. YamamotoYY, IchidaH, MatsuiM, ObokataJ, SakuraiT, et al. (2007) Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics 8 : 67.
45. BogdanoveAJ, VoytasDF (2011) TAL effectors: customizable proteins for DNA targeting. Science 333 : 1843–1846.
46. KvitkoBH, ParkDH, VelasquezAC, WeiCF, RussellAB, et al. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Path 5: e1000388.
47. GreenbergJT, YaoN (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6 : 201–211.
48. BruningsAM, GabrielDW (2003) Xanthomonas citri: breaking the surface. Mol Plant Pathol 4 : 141–157.
49. DuanYP, CastanedaA, ZhaoG, ErdosG, GabrielDW (1999) Expression of a single, host-specific, bacterial pathogenicity gene in plant cells elicits division, enlargement, and cell death. Mol Plant-Microbe Interact 12 : 556–582.
50. TakahashiH, BuchnerP, YoshimotoN, HawkesfordMJ, ShiuSH (2011) Evolutionary relationships and functional diversity of plant sulfate transporters. Front Plant Sci 2 : 119.
51. CaoMJ, WangZ, WirtzM, HellR, OliverDJ, et al. (2013) SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J 73 : 607–616.
52. TakahashiH, KoprivaS, GiordanoM, SaitoK, HellR (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62 : 157–184.
53. Lopez-MartinMC, BecanaM, RomeroLC, GotorC (2008) Knocking out cytosolic cysteine synthesis compromises the antioxidant capacity of the cytosol to maintain discrete concentrations of hydrogen peroxide in Arabidopsis. Plant Physiol 147 : 562–572.
54. Lopez-MartinMC, RomeroLC, GotorC (2008) Cytosolic cysteine in redox signaling. Plant Signal Behav 3 : 880–881.
55. KataokaT, HayashiN, YamayaT, TakahashiH (2004) Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136 : 4198–4204.
56. KrusellL, KrauseK, OttT, DesbrossesG, KramerU, et al. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17 : 1625–1636.
57. FeltenJ, KohlerA, MorinE, BhaleraoRP, PalmeK, et al. (2009) The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol 151 : 1991–2005.
58. PetreB, MorinE, TisserantE, HacquardS, Da SilvaC, et al. (2012) RNA-Seq of early-infected poplar leaves by the rust pathogen Melampsora larici-populina uncovers PtSultr3;5, a fungal-induced host sulfate transporter. PLOS ONE 7: e44408.
59. StreubelJ, BlucherC, LandgrafA, BochJ (2012) TAL effector RVD specificities and efficiencies. Nat Biotechnol 30 : 593–595.
60. RömerP, StraussT, HahnS, ScholzeH, MorbitzerR, et al. (2009) Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol 150 : 1697–1712.
61. StraussT, van PoeckeRM, StraussA, RömerP, MinsavageGV, et al. (2012) RNA seq pinpoints a Xanthomonas TAL effector activated resistance gene in a large crop genome. Proc Natl Acad Sci USA 109 : 19480–19485.
62. DoyleEL, StoddardBL, VoytasDF, BogdanoveAJ (2013) TAL effectors: highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol 23 : 390–398.
63. VerdierV, TriplettLR, HummelAW, CorralR, CernadasRA, et al. (2012) Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol 196 : 1197–1207.
64. CaldoRA, NettletonD, WiseRP (2004) Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell 16 : 2514–2528.
65. BolstadBM, IrizarryRA, AstrandM, SpeedTP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 : 185–193.
66. IrizarryRA, BolstadBM, CollinF, CopeLM, HobbsB, et al. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15.
67. StoreyJD, TibshiraniR (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100 : 9440–9445.
68. DashS, Van HemertJ, HongL, WiseRP, DickersonJA (2012) PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res 40: D1194–1201.
69. Makino S (2005) Molecular mechanisms of pathogenesis in pathovars of the rice pathogenic bacterial species Xanthomonas oryzae. [MS thesis]. Ames, IA: Iowa State University.
70. RömerP, RechtS, LahayeT (2009) A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci USA 106 : 20526–20531.
71. NakagawaT, KuroseT, HinoT, TanakaK, KawamukaiM, et al. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104 : 34–41.
72. HallM, FrankE, HolmesG, PfahringerB, ReutemannP, et al. (2009) The WEKA data mining software: an update. SIGKDD Explor Newsl 11 : 10–18.
73. SingT, SanderO, BeerenwinkelN, LengauerT (2005) ROCR: visualizing classifier performance in R. Bioinformatics 21 : 3940–3941.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic HelixČlánek Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron MobilizationČlánek AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for ImmunityČlánek Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden HamstersČlánek Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 2- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Viral Enhancer Mimicry of Host Innate-Immune Promoters
- The Epstein-Barr Virus-Encoded MicroRNA MiR-BART9 Promotes Tumor Metastasis by Targeting E-Cadherin in Nasopharyngeal Carcinoma
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- The Consequences of Reconfiguring the Ambisense S Genome Segment of Rift Valley Fever Virus on Viral Replication in Mammalian and Mosquito Cells and for Genome Packaging
- Substrate-Induced Unfolding of Protein Disulfide Isomerase Displaces the Cholera Toxin A1 Subunit from Its Holotoxin
- Male-Killing Induces Sex-Specific Cell Death via Host Apoptotic Pathway
- Highly Active Antiretroviral Therapies Are Effective against HIV-1 Cell-to-Cell Transmission
- The microRNAs in an Ancient Protist Repress the Variant-Specific Surface Protein Expression by Targeting the Entire Coding Sequence
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Type III Secretion Protein MxiI Is Recognized by Naip2 to Induce Nlrc4 Inflammasome Activation Independently of Pkcδ
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
- Induction of Type I Interferon Signaling Determines the Relative Pathogenicity of Strains
- Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic Helix
- Foxp3 Regulatory T Cells Delay Expulsion of Intestinal Nematodes by Suppression of IL-9-Driven Mast Cell Activation in BALB/c but Not in C57BL/6 Mice
- Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization
- MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Identification of Host-Targeted Small Molecules That Restrict Intracellular Growth
- A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection
- Engagement of NKG2D on Bystander Memory CD8 T Cells Promotes Increased Immunopathology following Infection
- Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress Expression
- CIB1 Synergizes with EphrinA2 to Regulate Kaposi's Sarcoma-Associated Herpesvirus Macropinocytic Entry in Human Microvascular Dermal Endothelial Cells
- A Gammaherpesvirus Bcl-2 Ortholog Blocks B Cell Receptor-Mediated Apoptosis and Promotes the Survival of Developing B Cells
- Metabolic Reprogramming during Purine Stress in the Protozoan Pathogen
- The Post-transcriptional Regulator / Activates T3SS by Stabilizing the 5′ UTR of , the Master Regulator of Genes, in
- Tailored Immune Responses: Novel Effector Helper T Cell Subsets in Protective Immunity
- AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity
- Epstein-Barr Virus Large Tegument Protein BPLF1 Contributes to Innate Immune Evasion through Interference with Toll-Like Receptor Signaling
- The Major Cellular Sterol Regulatory Pathway Is Required for Andes Virus Infection
- Insights into the Initiation of JC Virus DNA Replication Derived from the Crystal Structure of the T-Antigen Origin Binding Domain
- Domain Shuffling in a Sensor Protein Contributed to the Evolution of Insect Pathogenicity in Plant-Beneficial
- Lectin-Like Bacteriocins from spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor
- A Compositional Look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects
- Exploits Asparagine to Assimilate Nitrogen and Resist Acid Stress during Infection
- Interleukin-33 Increases Antibacterial Defense by Activation of Inducible Nitric Oxide Synthase in Skin
- Protective Vaccination against Papillomavirus-Induced Skin Tumors under Immunocompetent and Immunosuppressive Conditions: A Preclinical Study Using a Natural Outbred Animal Model
- Gem-Induced Cytoskeleton Remodeling Increases Cellular Migration of HTLV-1-Infected Cells, Formation of Infected-to-Target T-Cell Conjugates and Viral Transmission
- Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden Hamsters
- Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
- Inflammatory Monocytes Orchestrate Innate Antifungal Immunity in the Lung
- Quantitative and Qualitative Deficits in Neonatal Lung-Migratory Dendritic Cells Impact the Generation of the CD8+ T Cell Response
- Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response
- The Master Regulator of the Cellular Stress Response (HSF1) Is Critical for Orthopoxvirus Infection
- Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene
- Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri ORF57 to the Mammalian Export Adaptor ALYREF
- The Type III Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including TepP, a Tyrosine-phosphorylated Protein Required for the Recruitment of CrkI-II to Nascent Inclusions and Innate Immune Signaling
- Yeasts: How Many Species Infect Humans and Animals?
- Clustering of Pattern Recognition Receptors for Fungal Detection
- Distinct Antiviral Responses in Pluripotent versus Differentiated Cells
- Igniting the Fire: Virulence Factors in the Pathogenesis of Sepsis
- Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane
- Inducible Deletion of CD28 Prior to Secondary Infection Impairs Worm Expulsion and Recall of Protective Memory CD4 T Cell Responses
- Clonal Expansion during Infection Dynamics Reveals the Effect of Antibiotic Intervention
- The Secreted Triose Phosphate Isomerase of Is Required to Sustain Microfilaria Production
- Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2
- ‘Death and Axes’: Unexpected Ca Entry Phenologs Predict New Anti-schistosomal Agents
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



