-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Distinct Antiviral Responses in Pluripotent versus Differentiated Cells
article has not abstract
Published in the journal: Distinct Antiviral Responses in Pluripotent versus Differentiated Cells. PLoS Pathog 10(2): e32767. doi:10.1371/journal.ppat.1003865
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003865Summary
article has not abstract
Introduction
There is a mystery unfolding, the solution of which has implications for the understanding of both stem cell biology and the evolution of the vertebrate pathogen defense response. At the heart of this puzzle lies the observation of substantially different antiviral responses in mammalian cells with high potency (e.g., embryonic stem, oocytes, induced pluripotent, teratocarcinoma, and embryonic carcinoma cells) versus differentiated somatic cells (i.e., epithelial, fibroblast, lymphocyte). While differentiated cells are proficient in the interferon (IFN)-associated protein-based response [1]–[3], pluripotent cells have an attenuated IFN response [4]–[8]. Conversely, pluripotent cells can utilize RNA interference (RNAi) to combat viruses [9], [10], while this response is attenuated in differentiated cells [11]. Here we provide an overview of this developing area of virology.
Early Observations of Altered Antiviral Responses in Pluripotent versus Differentiated Cells
Somatic mammalian cells have the capacity to detect double-stranded RNA (dsRNA), a common byproduct of viral replication, and respond by inducing the expression of interferon (reviewed in [12]). Interferon acts in a paracrine and autocrine fashion to induce the expression of hundreds of antiviral interferon-stimulated genes (ISGs), forming the basis of the protein-based antiviral response in mammals [12]. Studies from the 1970s provided the first evidence that pluripotent cells can have altered susceptibility to virus infection. Teratocarcinoma cells were shown to be refractory to infection by murine polyomavirus, while differentiated derivatives of these cells were susceptible [13]. This work inspired further inquiry into infection of undifferentiated cells. Subsequently, Burke et al. demonstrated that pluripotent cells do not produce type I IFN in response to viral infection or treatment with poly I:C, a mimic of double-stranded RNA [4]. In the nearly 40 years since, numerous reports have reiterated these inherent differences [5]–[8]; however, the mechanism is only now being revealed [14]–[16].
Multiple Components of the Protein-Based Antiviral Response Are Attenuated in Pluripotent Cells
Understanding the basic biology of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) is an area of intense research. Recent reports establish that these cell types are deficient in numerous components of the IFN and associated protein-based antiviral innate response. Human ESCs display reduced expression of genes involved in the dsRNA response pathways, including pathogen recognition receptors (PRRs) that lead to IFN induction such as OAS1, PKR, MDA5, TLR3, and others [14]. Similar decreases in TLR3 and MDA5 were observed in mouse ESCs [15], demonstrating that this attenuated antiviral response is at least partially conserved among diverse mammals. In addition to reduced dsRNA-mediated induction of IFN, enhanced expression of SOCS1 (an inhibitor of the IFN-activated transcription factor STAT1) contributes to an attenuated response to IFN stimulation in pluripotent cells [16]. Thus, multiple levels of the innate protein-based immune response, both upstream and downstream of IFN production, are attenuated in pluripotent cells.
About 20 years after the reports that the IFN response of embryonic cells is deficient, the discovery of RNA interference (RNAi) led to these observations being revisited. Several studies demonstrated sequence-specific repression of gene expression following the introduction of long dsRNA into C. elegans, Drosophila, and trypanosomes [17]–[19]. We now know that ∼22 nt, double-stranded RNA duplexes (small interfering RNAs or siRNAs) function as the RNA effectors of RNAi, and that the cytoplasmic exonuclease Dicer can process long dsRNA into mature siRNAs. Argonaute proteins bind the siRNA, forming the RNA-induced silencing complex (RISC), which represses translation and/or directs cleavage of complementary mRNAs. Long dsRNA was effective in eliciting RNAi-mediated silencing in these early experiments, as the invertebrate organisms lack an IFN-based immune response to the presence of dsRNA. However, this approach presented a considerable obstacle for studying RNAi in mammalian cells, which respond to long dsRNA in the cytoplasm by globally inhibiting translation and inducing the interferon response [12]. Because pluripotent cells are deficient in this dsRNA response, several groups were able to overcome this obstacle [5]–[7]. They demonstrated that mammalian cells retained a functional RNAi pathway and affirmed that undifferentiated cells have an attenuated IFN response to long dsRNA.
RNAi Is Attenuated in Differentiated Cells Undergoing Antiviral Signaling
The ability of the RNAi pathway to combat virus infection is shared among many metazoans including plants and invertebrates. However, it is still debated whether RNAi is an antiviral response in mammals where the complex IFN-based response exists [20]–[23]. Unlike in invertebrates, strong biochemical evidence of natural antiviral siRNAs produced during infection is lacking in differentiated cells. Furthermore, unlike in plants and insects, genetic experiments have failed to demonstrate that the growth of viruses with mutant suppressors of RNAi is rescued in differentiated cells defective for RNAi. Additionally, recent reports from our lab and others have shown that human Argonaute2 (Ago2), a key component of RISC, is inhibited during both stress [24] and the pathogen response [11], [25]. Therefore, if RNAi is to have antiviral function in somatic cells, the coordinated inhibition of Ago2 creates a significant paradox. Thus, converse to the protein-based response to dsRNA in pluripotent cells, at least two components of the RNAi pathway are impaired in differentiated cells: 1) production and/or stability of siRNA; and 2) the activity of Ago2. In contrast to this, recent reports show that RNAi can act as an antiviral response in pluripotent mammalian cells [9], [10]. These findings suggest an intriguing dichotomy whereby differentiated, somatic cells rely on the protein-based IFN response, while undifferentiated cells can utilize RNAi (Figure 1).
Fig. 1. The dichotomy of antiviral responses in differentiated versus undifferentiated cells. 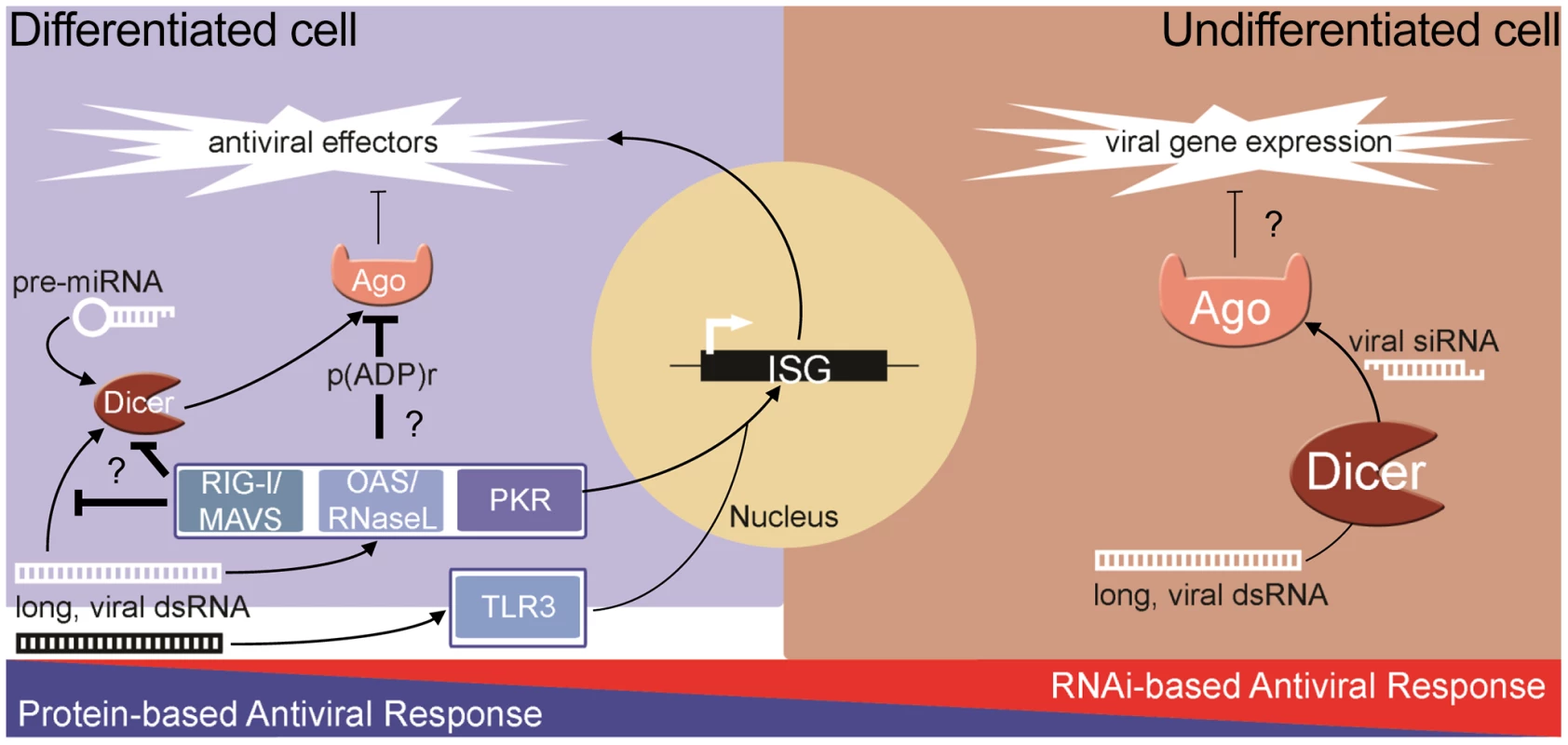
In most differentiated cells, PRRs (e.g., RIG-I, OAS-1/RNaseL, PKR) recognize pathogen-associated molecular patterns and stimulate the protein-based interferon response. The RNAi pathway functions, through miRNAs, to temper the expression of cytotoxic transcripts, but is inhibited in response to stress and antiviral signaling pathway activation. In undifferentiated/pluripotent cells, the interferon response is attenuated through reduced expression and activity of numerous components. In these cells, the RNAi pathway can function directly as an antiviral defense, using longer, virally derived dsRNA to generate siRNAs that target and silence viral transcripts. The Advantage of Being Different
MicroRNAs (miRNAs) are small RNAs, derived from endogenous precursors, and, similar to siRNAs, mediate the silencing of mRNAs via RISC. Multiple components of the mammalian pathogen defense response, including some subclasses of ISGs, are regulated by miRNAs. We have proposed that with the evolution of a protein-based antiviral response in most differentiated cells, components of RNAi (no longer essential as protectors against virus) became repurposed to control the toxic effectors of the pathogen defense [11]. Consistent with this, the pathogen response can lead to inhibition of RISC and a subsequent increase in translation of antiviral and/or inflammatory transcripts [11], [25]. This model is in line with the established role of miRNAs as important regulators of homeostasis [26], [27].
Key Questions Remaining
Despite such recent progress in the field, important questions regarding the different antiviral responses of pluripotent and differentiated cells remain unresolved.
Pluripotent Cells
What advantage do pluripotent cells gain from lacking multiple arms of the protein-based IFN-associated response? Pluripotent cells undergo rapid cell division, and may mute the IFN response to avoid its antiproliferative effects [28]. Interferon has been shown to stimulate differentiation [28], suggesting that pluripotent cells may inhibit its expression as a means of maintaining potency. An alternative, non–mutually exclusive model predicts that RNAi serves as a more efficient defense against transposons than the IFN response [14]. An extension of this model is that the fitness cost of transposon activity in pluripotent cells is higher than infection by an exogenous virus. Consistent with this proposed role for the RNAi machinery in pluripotent cells, mammalian oocytes have been shown to maintain slicer-dependent RISC activity despite having decreased microRNA-mediated, slicer-independent silencing [29]. Yet another model suggests that relatively “harmless” triggers of the IFN response (i.e., cytoplasmic dsRNA) are readily produced in pluripotent cells and the suppression of the IFN response prevents terminal sacrifice of the lineage. The lack of a protein-based antiviral response in pluripotent cells raises the question of whether the antiviral response is even important in these cells. It is unknown what fraction of pluripotent cells actually come into contact with exogenous virus, and it has even been speculated that such infections would be so detrimental to daughter cells that these cells may purposely not mount a defense to ensure their destruction [14]. Testing these models and understanding the motivation behind attenuating the IFN response gets to the heart of the biology of pluripotency.
Differentiated Cells
It is unknown what accounts for the lack of abundant detectable siRNAs in somatic cells. Is Dicer or one of its cofactors differentially active and/or are the dsRNA-binding proteins that are components of the IFN response sequestering long dsRNA away from Dicer? Alternatively, are unknown factors altering the stability of the derivative siRNAs or impairing their association with RISC?
Downstream of Dicer, exciting questions remain for understanding antiviral-signaling-mediated inactivation of RISC. For example, what is the full repertoire of PRRs, signaling pathways, effectors, and biochemical changes involved in inhibition of RISC? What are the key infection-relevant miRNAs and associated ISG targets that are altered by RISC inhibition? Finally, how prevalent are these phenomena in other animals? We predict that somatic cells of other vertebrates (positive for IFN response) should also have an attenuated RNAi response when undergoing antiviral signaling. It will be interesting to determine if similar phenomena are observed in invertebrates (IFN negative), which often have separate Agos that specialize in either miRNA-mediated or siRNA-mediated regulation. One exciting possibility is that only miRNA-associated RISCs will be inhibited during cytotoxic stress in these organisms.
Conclusion
The last few years have shed much light on virus infection of pluripotent cells. It can be expected that this pace of discovery will continue in the near term. This improved grasp of the antiviral response in pluripotent versus differentiated cells will lead to new inroads in RNAi technology, stem cell biology, and understanding the evolution of vertebrates.
Zdroje
1. LampsonGP, TytellAA, FieldAK, NemesMM, HillemanMR (1967) Inducers of interferon and host resistance. I. Double-stranded RNA from extracts of Penicillium funiculosum. Proc Natl Acad Sci U S A 58 : 782–789.
2. FieldAK, TytellAA, LampsonGP, HillemanMR (1967) Inducers of interferon and host resistance. II. Multistranded synthetic polynucleotide complexes. Proc Natl Acad Sci U S A 58 : 1004–1010.
3. LeeSB, EstebanM (1994) The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 199 : 491–496.
4. BurkeDC, GrahamCF, LehmanJM (1978) Appearance of interferon inducibility and sensitivity during differentiation of murine teratocarcinoma cells in vitro. Cell 13 : 243–248.
5. SvobodaP, SteinP, HayashiH, SchultzRM (2000) Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127 : 4147–4156.
6. YangS, TuttonS, PierceE, YoonK (2001) Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol 21 : 7807–7816.
7. PaddisonPJ, CaudyAA, HannonGJ (2002) Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci U S A 99 : 1443–1448.
8. SteinP, ZengF, PanH, SchultzRM (2005) Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes. Dev Biol 286 : 464–471.
9. LiY, LuJ, HanY, FanX, DingSW (2013) RNA interference functions as an antiviral immunity mechanism in mammals. Science 342 : 231–234.
10. MaillardPV, CiaudoC, MarchaisA, LiY, JayF, et al. (2013) Antiviral RNA interference in mammalian cells. Science 342 : 235–238.
11. SeoGJ, KincaidRP, PhanaksriT, BurkeJM, PareJM, et al. (2013) Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14 : 435–445 doi:10.1016/j.chom.2013.09.002
12. TakeuchiO, AkiraS (2009) Innate immunity to virus infection. Immunol Rev 227 : 75–86.
13. SwartzendruberDE, LehmanJM (1975) Neoplastic differentiation: interaction of simian virus 40 and polyoma virus with murine teratocarcinoma cells in vitro. J Cell Physiol 85 : 179–187.
14. ChenLL, YangL, CarmichaelGG (2010) Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle 9 : 3552–3564.
15. WangR, WangJ, PaulAM, AcharyaD, BaiF, et al. (2013) Mouse embryonic stem cells are deficient in type I interferon expression in response to viral infections and double-stranded RNA. J Biol Chem 288 : 15926–15936.
16. HongXX, CarmichaelGG (2013) Innate immunity in pluripotent human cells: attenuated response to interferon-β. J Biol Chem 288 : 16196–16205.
17. FireA, XuS, MontgomeryMK, KostasSA, DriverSE, et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 : 806–811.
18. KennerdellJR, CarthewRW (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95 : 1017–1026.
19. NgôH, TschudiC, GullK, UlluE (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci U S A 95 : 14687–14692.
20. UmbachJL, CullenBR (2009) The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23 : 1151–1164.
21. CullenBR (2006) Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol 7 : 563–567.
22. JeangKT (2012) RNAi in the regulation of mammalian viral infections. BMC Biol 10 : 58 doi:10.1186/1741-7007-10-58
23. de VriesW, BerkhoutB (2008) RNAi suppressors encoded by pathogenic human viruses. Int J Biochem Cell Biol 40 : 2007–2012.
24. LeungAK, VyasS, RoodJE, BhutkarA, SharpPA, et al. (2011) Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42 : 489–499.
25. MazumderA, BoseM, ChakrabortyA, ChakrabartiS, BhattacharyyaSN (2013) A transient reversal of miRNA-mediated repression controls macrophage activation. EMBO Rep 14 : 1008–1016 doi:10.1038/embor.2013.149
26. HeL, HannonGJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5 : 522–531.
27. BartelDP, ChenCZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5 : 396–400.
28. HertzogPJ, HwangSY, KolaI (1994) Role of interferons in the regulation of cell proliferation, differentiation, and development. Mol Reprod Dev 39 : 226–232.
29. MaJ, FlemrM, SteinP, BerningerP, MalikR, et al. (2010) MicroRNA activity is suppressed in mouse oocytes. Curr Biol 20 : 265–270 doi:10.1016/j.cub.2009.12.042
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic HelixČlánek Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron MobilizationČlánek AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for ImmunityČlánek Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden HamstersČlánek Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 2- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Viral Enhancer Mimicry of Host Innate-Immune Promoters
- The Epstein-Barr Virus-Encoded MicroRNA MiR-BART9 Promotes Tumor Metastasis by Targeting E-Cadherin in Nasopharyngeal Carcinoma
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- The Consequences of Reconfiguring the Ambisense S Genome Segment of Rift Valley Fever Virus on Viral Replication in Mammalian and Mosquito Cells and for Genome Packaging
- Substrate-Induced Unfolding of Protein Disulfide Isomerase Displaces the Cholera Toxin A1 Subunit from Its Holotoxin
- Male-Killing Induces Sex-Specific Cell Death via Host Apoptotic Pathway
- Highly Active Antiretroviral Therapies Are Effective against HIV-1 Cell-to-Cell Transmission
- The microRNAs in an Ancient Protist Repress the Variant-Specific Surface Protein Expression by Targeting the Entire Coding Sequence
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Type III Secretion Protein MxiI Is Recognized by Naip2 to Induce Nlrc4 Inflammasome Activation Independently of Pkcδ
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
- Induction of Type I Interferon Signaling Determines the Relative Pathogenicity of Strains
- Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic Helix
- Foxp3 Regulatory T Cells Delay Expulsion of Intestinal Nematodes by Suppression of IL-9-Driven Mast Cell Activation in BALB/c but Not in C57BL/6 Mice
- Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization
- MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Identification of Host-Targeted Small Molecules That Restrict Intracellular Growth
- A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection
- Engagement of NKG2D on Bystander Memory CD8 T Cells Promotes Increased Immunopathology following Infection
- Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress Expression
- CIB1 Synergizes with EphrinA2 to Regulate Kaposi's Sarcoma-Associated Herpesvirus Macropinocytic Entry in Human Microvascular Dermal Endothelial Cells
- A Gammaherpesvirus Bcl-2 Ortholog Blocks B Cell Receptor-Mediated Apoptosis and Promotes the Survival of Developing B Cells
- Metabolic Reprogramming during Purine Stress in the Protozoan Pathogen
- The Post-transcriptional Regulator / Activates T3SS by Stabilizing the 5′ UTR of , the Master Regulator of Genes, in
- Tailored Immune Responses: Novel Effector Helper T Cell Subsets in Protective Immunity
- AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity
- Epstein-Barr Virus Large Tegument Protein BPLF1 Contributes to Innate Immune Evasion through Interference with Toll-Like Receptor Signaling
- The Major Cellular Sterol Regulatory Pathway Is Required for Andes Virus Infection
- Insights into the Initiation of JC Virus DNA Replication Derived from the Crystal Structure of the T-Antigen Origin Binding Domain
- Domain Shuffling in a Sensor Protein Contributed to the Evolution of Insect Pathogenicity in Plant-Beneficial
- Lectin-Like Bacteriocins from spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor
- A Compositional Look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects
- Exploits Asparagine to Assimilate Nitrogen and Resist Acid Stress during Infection
- Interleukin-33 Increases Antibacterial Defense by Activation of Inducible Nitric Oxide Synthase in Skin
- Protective Vaccination against Papillomavirus-Induced Skin Tumors under Immunocompetent and Immunosuppressive Conditions: A Preclinical Study Using a Natural Outbred Animal Model
- Gem-Induced Cytoskeleton Remodeling Increases Cellular Migration of HTLV-1-Infected Cells, Formation of Infected-to-Target T-Cell Conjugates and Viral Transmission
- Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden Hamsters
- Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
- Inflammatory Monocytes Orchestrate Innate Antifungal Immunity in the Lung
- Quantitative and Qualitative Deficits in Neonatal Lung-Migratory Dendritic Cells Impact the Generation of the CD8+ T Cell Response
- Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response
- The Master Regulator of the Cellular Stress Response (HSF1) Is Critical for Orthopoxvirus Infection
- Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene
- Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri ORF57 to the Mammalian Export Adaptor ALYREF
- The Type III Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including TepP, a Tyrosine-phosphorylated Protein Required for the Recruitment of CrkI-II to Nascent Inclusions and Innate Immune Signaling
- Yeasts: How Many Species Infect Humans and Animals?
- Clustering of Pattern Recognition Receptors for Fungal Detection
- Distinct Antiviral Responses in Pluripotent versus Differentiated Cells
- Igniting the Fire: Virulence Factors in the Pathogenesis of Sepsis
- Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane
- Inducible Deletion of CD28 Prior to Secondary Infection Impairs Worm Expulsion and Recall of Protective Memory CD4 T Cell Responses
- Clonal Expansion during Infection Dynamics Reveals the Effect of Antibiotic Intervention
- The Secreted Triose Phosphate Isomerase of Is Required to Sustain Microfilaria Production
- Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2
- ‘Death and Axes’: Unexpected Ca Entry Phenologs Predict New Anti-schistosomal Agents
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



