-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Key Role for Chd1 in Histone H3 Dynamics at the 3′ Ends of Long Genes in Yeast
Chd proteins are ATP–dependent chromatin remodeling enzymes implicated in biological functions from transcriptional elongation to control of pluripotency. Previous studies of the Chd1 subclass of these proteins have implicated them in diverse roles in gene expression including functions during initiation, elongation, and termination. Furthermore, some evidence has suggested a role for Chd1 in replication-independent histone exchange or assembly. Here, we examine roles of Chd1 in replication-independent dynamics of histone H3 in both Drosophila and yeast. We find evidence of a role for Chd1 in H3 dynamics in both organisms. Using genome-wide ChIP-on-chip analysis, we find that Chd1 influences histone turnover at the 5′ and 3′ ends of genes, accelerating H3 replacement at the 5′ ends of genes while protecting the 3′ ends of genes from excessive H3 turnover. Although consistent with a direct role for Chd1 in exchange, these results may indicate that Chd1 stabilizes nucleosomes perturbed by transcription. Curiously, we observe a strong effect of gene length on Chd1's effects on H3 turnover. Finally, we show that Chd1 also affects histone modification patterns over genes, likely as a consequence of its effects on histone replacement. Taken together, our results emphasize a role for Chd1 in histone replacement in both budding yeast and Drosophila melanogaster, and surprisingly they show that the major effects of Chd1 on turnover occur at the 3′ ends of genes.
Published in the journal: A Key Role for Chd1 in Histone H3 Dynamics at the 3′ Ends of Long Genes in Yeast. PLoS Genet 8(7): e32767. doi:10.1371/journal.pgen.1002811
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002811Summary
Chd proteins are ATP–dependent chromatin remodeling enzymes implicated in biological functions from transcriptional elongation to control of pluripotency. Previous studies of the Chd1 subclass of these proteins have implicated them in diverse roles in gene expression including functions during initiation, elongation, and termination. Furthermore, some evidence has suggested a role for Chd1 in replication-independent histone exchange or assembly. Here, we examine roles of Chd1 in replication-independent dynamics of histone H3 in both Drosophila and yeast. We find evidence of a role for Chd1 in H3 dynamics in both organisms. Using genome-wide ChIP-on-chip analysis, we find that Chd1 influences histone turnover at the 5′ and 3′ ends of genes, accelerating H3 replacement at the 5′ ends of genes while protecting the 3′ ends of genes from excessive H3 turnover. Although consistent with a direct role for Chd1 in exchange, these results may indicate that Chd1 stabilizes nucleosomes perturbed by transcription. Curiously, we observe a strong effect of gene length on Chd1's effects on H3 turnover. Finally, we show that Chd1 also affects histone modification patterns over genes, likely as a consequence of its effects on histone replacement. Taken together, our results emphasize a role for Chd1 in histone replacement in both budding yeast and Drosophila melanogaster, and surprisingly they show that the major effects of Chd1 on turnover occur at the 3′ ends of genes.
Introduction
Eukaryotic genomes are packaged as chromatin, whose fundamental repeating subunit, the nucleosome, is composed of 147 bp of DNA wrapped 1.7 times around an octameric histone core. Nucleosomes may interact with each other to form higher-order levels of chromatin packaging necessary to compact an entire genome within a nucleus. This genome packaging strategy leads to a dominant theme in eukaryotic gene regulation: nucleosomes tend to repress gene expression, and a large array of gene regulatory mechanisms in eukaryotes operate by strengthening or weakening the repressive effects of nucleosomes on gene expression [1].
Genome-wide nucleosome mapping studies indicate that although the majority of a eukaryotic genome is typically covered with regularly spaced nucleosomes, nucleosome depleted or nucleosome free regions are frequently found over promoters and at the 3′ ends of genes (reviewed in [2]). Although these studies give a fixed snapshot of chromatin organization, other analyses indicate that chromatin is dynamic. Studies in which histones were pulse-labeled with radioisotopes or tagged with GFP demonstrated that histones can be actively exchanged on chromatin, even in the absence of DNA replication [3], [4]. More recent work has utilized induction of epitope-tagged alleles of histones in G1-arrested yeast cells followed by chromatin immunoprecipitation to examine histone H3 dynamics genome-wide [5], [6]. These studies show that histone H3 exchanges at a high rate on promoters and in other intergenic regions such as downstream of the 3′ ends of genes. With the exception of highly-transcribed genes, the bodies of genes, even those that are transcribed at moderate rates, exhibit much lower H3 exchange rates.
Although nucleosomes over transcribed genes appear to be relatively stable in vivo, nucleosomes form a strong barrier to elongating RNA polymerase II (RNA Pol2) in vitro [7]. Thus, it is likely that accessory factors assist in transcription elongation to alleviate this barrier. These factors may promote the temporary disassembly or displacement of nucleosomes permitting the passage of elongating RNA Pol2, and furthermore, they may assist in nucleosome (re)assembly after polymerases have passed. A wide variety of factors have been implicated in the dynamics and maintenance of chromatin structure over transcribed sequences. These include ATP-dependent chromatin remodeling enzymes, enzymes that post-translationally modify histones, histone chaperones and transcription elongation factors [8]. Interestingly, mutations affecting a number of these factors cause a cryptic transcription initiation phenotype, in which disruption of chromatin in the body of genes leads to activation of internal, normally quiescent promoters [9].
One factor implicated in the regulation of transcribed chromatin is the ATP-dependent chromatin remodeling enzyme Chd1. Chd1 is the founding member of a family of highly conserved chromatin remodeling enzymes found throughout eukaryotes [10]. Although budding yeast only express a single Chd1 protein, at least 9 CHD family proteins are expressed in humans. Mammalian CHD family members have been implicated in diverse roles including promotion of normal organismal development, and the maintenance of pluripotency and prevention of heterochromatin formation in mouse embryonic stem cells [10]. In addition, mutations in CHD protein genes are implicated in several human cancers and CHARGE syndrome, which is characterized by a phenotypically heterogeneous set of developmental defects [10], [11].
CHD proteins typically have a pair of N-terminal chromodomains, a central Snf2/Swi2 type helicase domain and a C-terminal domain that mediates DNA or nucleosome binding [10]. The chromodomains of human Chd1 bind histone H3 tails methylated at lysine 4 (H3K4me) suggesting a mechanism for recruitment [12], [13]. However, yeast Chd1 does not bind H3K4-methylated tails [13], and in Drosophila melanogaster, the chromodomains do not play an important role in its localization to chromatin [14]. Recent structural and biochemical studies suggest that rather than mediating chromatin localization, the chromodomains may regulate enzyme activity [15]. In vitro assays show that Chd1 has the ability to assemble, remodel, slide and promote regular spacing of nucleosomes [16]–[18]. Chromatin immunoprecipitation in budding and fission yeast, and immunostaining of Drosophila polytene chromosomes show that Chd1 associates with both promoters and transcribed regions of active genes [19]–[23]. Consistent with its localization on genes, genetic studies in yeast have implicated Chd1 in the regulation of transcription initiation, elongation and termination [22], [24]–[28]. Although Chd1 can be purified as a monomer, its association with several complexes that regulate initiation and elongation, which include mediator, FACT, the Paf1 complex, SAGA and SLIK, provides further support to these conclusions [22], [29]–[33]. Chd1 also associates with histone chaperones Nap1 in fission yeast, and HirA, a histone chaperone for histone H3.3, in fruit flies [19], [34].
Several studies suggest mechanisms for how Chd1's biochemical activity may relate to these biological functions. Chd1 can promote transcription and catalyze activator dependent, promoter specific nucleosome remodeling in vitro [35], [36]. Furthermore, in Schizosaccharomyces pombe, Chd1 (Hrp1) acts at a subset of promoters to disassemble nucleosomes close to the transcription initiation site [19]. In Drosophila, following fertilization of an egg, sperm chromatin is decondensed, protamines are removed and replaced with nucleosomes whose only form of histone H3 is the replication-independent variant H3.3 [37]. Interestingly, in chd1 mutants, H3.3 levels in decondensing sperm chromatin are greatly reduced and unevenly distributed, suggesting a role for Chd1 in the replication-independent assembly or distribution of H3.3 nucleosomes [34], [38].
A recent high-resolution genome-wide nucleosome mapping study in budding yeast points to an in vivo role for Chd1's nucleosome remodeling activity. Nucleosomes are typically regularly positioned over genes in wild type yeast cells [39]. However, in a chd1Δ mutant, this positioning is largely lost over gene bodies [40]. Specifically, nucleosome free regions at the 5′ and 3′ ends of genes and the first (+1) nucleosome over the transcribed region were minimally affected by loss of Chd1, but downstream nucleosomes (particularly those starting at the +3 position) were dramatically delocalized in chd1Δ yeast cells. Curiously, micrococcal nuclease digestion patterns of bulk chromatin are not affected in a chd1 mutant, suggesting that Chd1 affects the positioning of nucleosome arrays primarily over the transcribed body of genes, rather that the precise spacing between any given pair of nucleosomes [40], [41]. Although chd1 mutations have modest effects on gene expression in yeast, and are virtually indistinguishable from wild type strains in phenotypic assays, they do cause a cryptic initiation phenotype, consistent with the loss of nucleosome organization over the body of genes [9], [28], [42], [43].
Although these data clearly demonstrate a role for Chd1 in nucleosome positioning in vivo, the mechanism underlying its in vivo function and its relationship to transcription remains unclear. In this study, we examine the role(s) of Chd1 in governing the replication-independent exchange of newly-expressed histone H3 onto chromatin in budding yeast and Drosophila using genome-wide methodologies. Chd1 mutants have dramatic defects in the localization of the replication-independent histone variant H3.3 in flies, while in Saccharomyces cerevisiae, chd1Δ mutants exhibit dramatic defects in H3 turnover in coding regions. Surprisingly, Chd1 predominantly affects histone H3 exchange at the 3′ ends of coding regions, and this effect on turnover depends on gene length – H3 turnover at 3′ ends is fairly concordant between wild type and chd1Δ strains for genes 1 kb and shorter, whereas Chd1 appears to specifically stabilize nucleosomes over the 3′ ends of longer genes. Finally, we show that loss of Chd1 globally alters histone modification patterns related to active transcription, with H3K36me3 in particular shifting in concert with the changed patterns of H3 replacement. Together, our results show that Chd1 plays a key role in histone H3 dynamics, and surprisingly, that yeast Chd1's influence on H3 dynamics is most apparent at the 3′ ends of genes.
Results
Chd1 Influences Replication-Independent Assembly of H3.3 in Drosophila
Previously, Fyodorov and colleagues examined the distribution of epitope-tagged, full length H3.3 in the Drosophila syncytial blastoderm and only observed a modest defect in H3.3 distribution in chd1 null mutants [34]. Because the H3.3 N-terminal tail, which is required for replication-dependent assembly of H3.3 [44], was intact in this experiment, we reasoned that any defect in replication-independent assembly of the tagged H3.3 might have been obscured.
To reassess Chd1's role in replication-independent deposition of H3.3, we imaged GFP-tagged histone H3.3core in live salivary glands from chd1 mutant larvae. We utilized a transgenic fly expressing an AB1-GAL4 driver and a Gal inducible histone H3.3core-GFP [44]. Because the H3.3core protein encoded by the transgene lacks the N-terminal tail, it is only incorporated into chromatin via the replication-independent pathway [44]. In an otherwise wild type background, H3.3core-GFP was deposited into the polytene chromosome arms of salivary glands (Figure 1A). In some cases, we also observed a nucleoplasmic GFP signal in which the entire nucleus, including non-chromosomal territories, exhibited a strong GFP signal, although a chromosomal banding pattern was still evident. In flies that were heterozygous or homozygous for chd15, a null allele of Chd1 [21], we observed salivary gland nuclei with GFP signals similar to those of wild type, i.e. chromosomal or broad nucleoplasmic GFP fluorescence. However, we also observed nuclei with a novel, “non-chromosomal” phenotype where the polytene arms appear almost devoid of GFP signal and a substantial nuclear, non-chromosomal H3.3core-GFP signal was still apparent (Figure 1B, 1C). We determined the relative frequencies of these phenotypes in wild type and mutant flies by blind scoring, and observed that the predominant chromosomal fluorescence pattern observed in wild type cells declined dramatically in chd1 mutants, whereas the nucleoplasmic and non-chromosomal patterns increased in frequency (Figure 1D). We observed similar phenotypes when we repeated these experiments with independently derived chd15 flies using a different balancer chromosome (data not shown). These results do not appear to be due to any peculiarity of the AB1-GAL4 driver as we observed similar fluorescence patterns when we used sgsGAL4 and eyelessGAL4 drivers (data not shown). Furthermore, we did not observe obvious differences in the strength of H3.3core-GFP signals between flies with the three analyzed genotypes (wild type, +/chd15 heterozygous and chd15/chd15 homozygous), nor between nuclei with the three observed staining patterns (chromosomal, non-chromosomal and nucleoplasmic) (Figure S1), suggesting that the observed localization patterns were not due to differences in H3.3core-GFP expression. Rather, we favor the idea that the variability observed here reflects perdurance of maternally contributed Chd1, which has been observed previously [21]. Immunostaining of fixed polytene chromosomes similarly revealed a reduction of H3.3core-GFP on chromosomes derived from chd15 mutant larvae, while levels of full length H3.3-GFP were not affected by loss of Chd1 (Figure S2), consistent with the ability of full length H3.3 to incorporate through both replication-dependent and –independent pathways. Consistent with our observations in the chd15 mutants, we observed decreased association of H3.3core-GFP with polytene chromosomes when we knocked down Chd1 levels with either of two RNAi constructs (Figure S2 and data not shown). Overall, these data are consistent with the possibility that Chd1 may contribute to replication-independent assembly of H3.3 containing nucleosomes.
Fig. 1. Chd1 affects H3.3core-GFP localization on Chd1 in Drosophila. 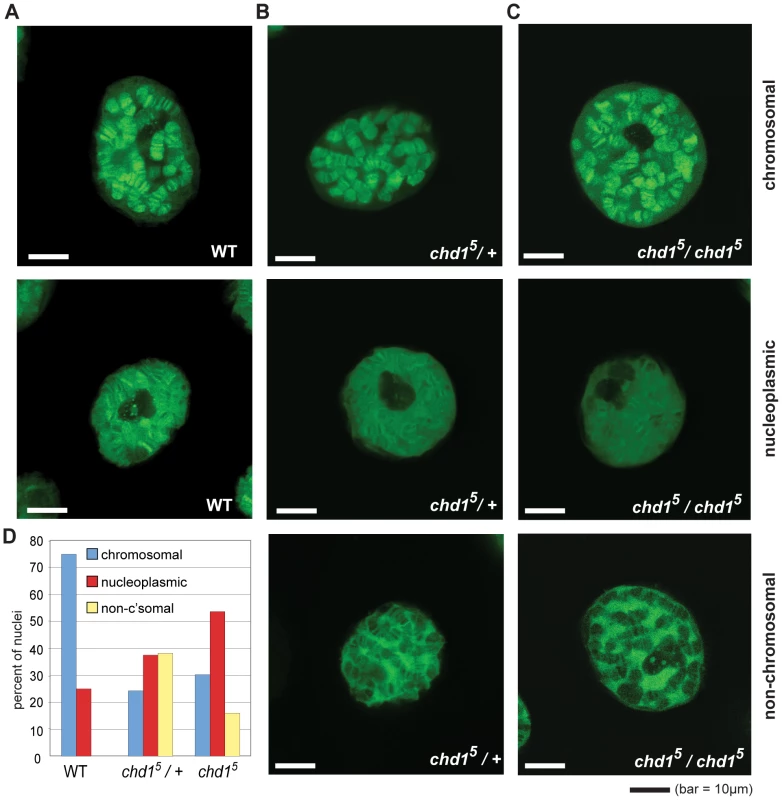
(A) Representative sections from confocal imaging of H3.3core-GFP in nuclei from salivary glands of wild type larvae, (B) chd15 heterozygotes and (C) chd15 homozygotes. The GFP signal is pseudo green. In all cases, H3.3core-GFP was expressed from P[UHS-H3.3core-GFP] and driven by P{GawB} AB1-Gal4. (D) Quantitation of banding patterns observed in nuclei from flies with the indicated genotypes. A total of 44 wild type, 144 heterozygote, and 162 homozygous null nuclei were scored, all blind to genotype. The H3 Tail and Chd1 Function Redundantly in Yeast
To further examine roles of Chd1 in nucleosome dynamics in vivo, we turned to budding yeast. To test the idea that Chd1 may modulate replication-independent nucleosome assembly or dynamics, we took advantage of the observation that the yeast H3 N-terminal tail is important for normal chromatin structure [45]. Reasoning that the H3 N-terminal tail deletion mutation likely interferes with replication-dependent assembly of H3, as is the case in Drosophila and Physarum polycephalum, [44], [46], we predicted that loss of this function would sensitize cells to defects in other chromatin assembly or maintenance pathways, we used a plasmid shuffle strategy to create CHD1+ and chd1Δ yeast strains expressing either wild type histone H3 (H3WT) or a histone H3 N-terminal deletion mutation, H3Δ4-30. Consistent with prior observations, the chd1Δ H3WT strain grew indistinguishably from wild type cells, and the CHD1 H3Δ4-30 strain exhibited a moderate growth defect (Figure 2). Interestingly, the chd1Δ H3Δ4-30 double mutant grew much more poorly than the CHD1 H3Δ4-30 single mutant, indicating that Chd1 and the N-terminal tail of H3 share a redundant function.
Fig. 2. The H3 N-terminal tail functions redundantly with Chd1 and an H3.3-like surface of histone H3 in budding yeast. 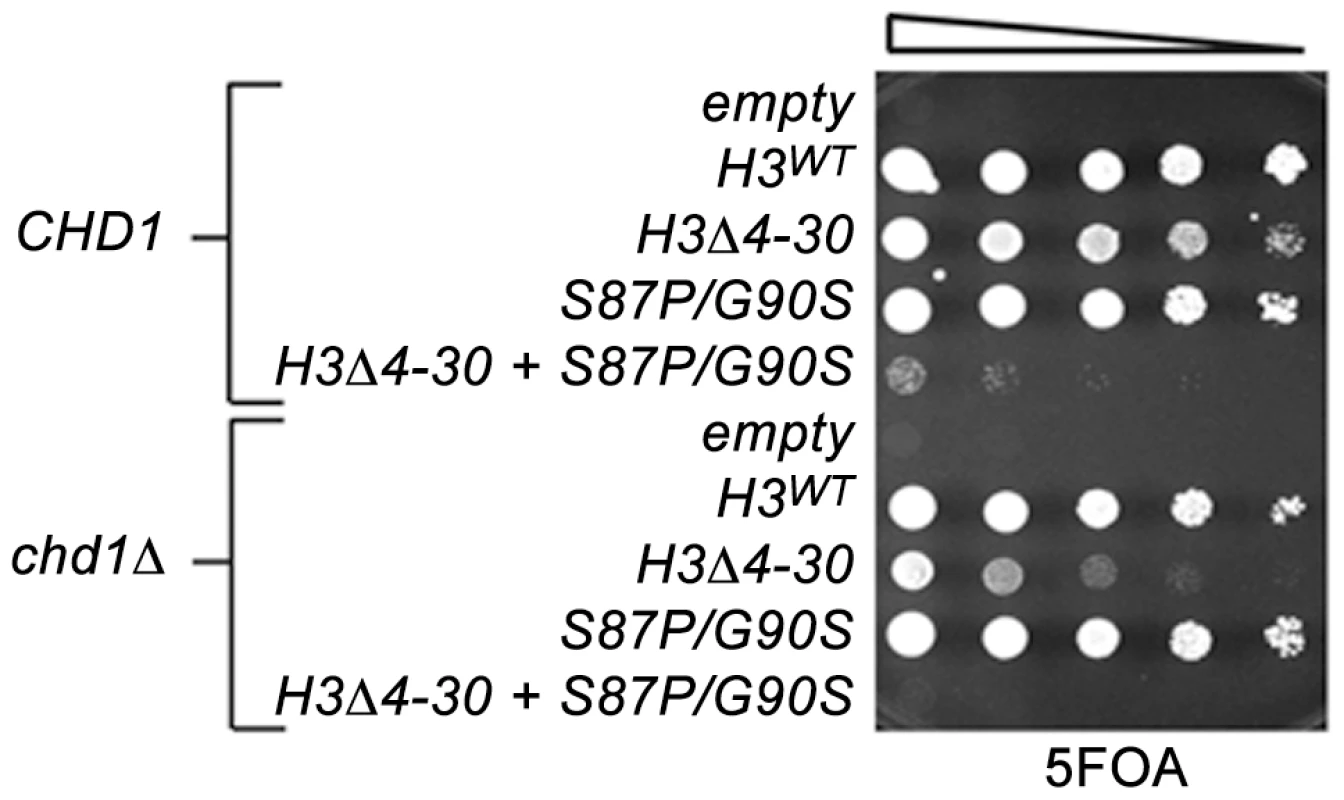
The indicated histone H3 plasmids (which also carried histone H4) were transformed into wild type CHD1 or chd1 null strains that lack both chromosomal copies of the histone H3/H4 genes and contained a URA3 H3/H4 plasmid. Cultures were adjusted to 1×107 cells per ml and five-fold serial dilutions were spotted directly onto 5FOA media, selecting for cells that had lost the URA3 H3/H4 plasmid, and incubated for 2 days at 30°C. In contrast to other model organisms, the budding yeast genome expresses only a single non-centromeric form of histone H3. However, the major histone H3/H4 chaperones, including the H3.3 chaperone HirA, are conserved, suggesting that yeast retain distinctive replication dependent and independent chromatin assembly pathways [47]. We have obtained data consistent with this idea in a screen for genetic suppressors of a cold-sensitive allele of transcription elongation factor SPT5. Among these suppressors were mutations in CHD1, mutations in the H3K4 and H3K36 histone methyltransferases SET1 and SET2, histone H3K4 and H3K36 substitutions, and mutations in members of the RPD3S histone deacetylase complex. Further characterization of these suppressors led us to propose that they act by lowering the chromatin barrier to efficient transcription elongation [28].
Given the observations described above, we recently screened a randomly mutagenized plasmid library for histone H3 mutations that suppress spt5Cs - (to be described in detail elsewhere). Among the suppressor mutations obtained in that screen, we isolated a mutation, H3-S87P/G90S, which simultaneously alters two of the four residues that distinguish histone H3.1 from H3.3 in other eukaryotes. Yeast expressing the S87P/G90S form of histone H3 from the normal HHT2 locus are viable, indicating that this mutation is unlikely to strongly perturb replication coupled chromatin assembly. As with several other of the mutations that suppress spt5Cs - (e.g., H3K36R, set2, mutations affecting Rpd3s), the H3-S87P/G90S mutant caused cryptic initiation of transcription (Figure S3).
We therefore examined genetic interactions between the H3-S87P/G90P, chd1Δ and the H3Δ4-30 mutations using the plasmid shuffle assay described above (Figure 2). Interestingly, the chd1Δ H3-S87P/G90S double mutant exhibited no new mutant phenotypes, whereas combining H3-S87P/G90S with the H3Δ4-30 deletion resulted in a very poor growth phenotype and the chd1Δ H3-S87P/G90S H3Δ4-30 triple mutation showed an even more severe growth defect. Thus, like Chd1, residues 87 and 90 of histone H3 function redundantly with the H3 N-terminal tail. It is tempting to argue that these data indicate that Chd1 interacts with histone H3 via a surface defined by residues 87 and 90. However, the fact that the phenotype of the chd1Δ H3-S87P/G90S H3Δ4-30 triple mutant is more severe than that of the chd1Δ H3Δ4-30 double mutant suggests that H3 residues S87 and G90 may retain functions that are redundant with the H3 tail, even when Chd1 is absent.
Chd1 Affects Histone H3 Dynamics
The data presented above suggest that Chd1 affects replication independent dynamics of histone H3. To test this idea directly in budding yeast, we used a yeast strain carrying galactose-inducible Flag-tagged H3, coupled with chromatin immunoprecipitation and tiling microarray (ChIP on chip) analysis, to follow the incorporation of newly-synthesized H3 genome-wide in cells arrested in the cell cycle [6]. Briefly, wild type or chd1Δ yeast strains are arrested in G1 phase using alpha factor, then Flag-H3 is induced with galactose, and after 60 minutes Flag-H3 and total H3-associated DNA are subject to ChIP enrichment and competitively hybridized on ∼250 bp resolution tiling microarrays. Resulting Flag/total H3 ratios provide locus-specific estimates of H3 turnover rates.
Figure 3A shows a “metagene” analysis of H3 turnover in 3 biological replicate samples for wild type (blue) and chd1Δ (red) strains. The wild type profile recapitulates previous results from multiple labs [5], [6], [48] – H3 replacement is highest over promoters and at the 5′ ends of genes, with coding regions being remarkably protected from H3 replacement, and modest levels of turnover being seen at the 3′ ends of genes.
Fig. 3. Chd1 plays a key role in H3 replacement dynamics at gene ends. 
(A) H3 replacement was measured in G1-arrested cells by induction of Flag-H3 for 60 minutes, followed by ChIP enrichment of both Flag-containing and total H3-associated DNA and subsequent competitive hybridization to tiling microarrays. H3 turnover is represented as log2 of Flag-H3 ChIP enrichment over total-H3 ChIP (y axis). Here, data for all yeast genes is shown in a “metagene” view, with 10 bins of 50 bp each, upstream of the +1 nucleosome, followed by 20 bins representing 5% increments along gene coding regions, scaling for gene length. Three independent replicate experiments are shown along with the averaged profile, as indicated. (B) Average Flag-H3/total-H3 for various classes of genomic element, defined as in [6], [54]. Briefly, 5′ and 3′ CDS refer to probes within the first and last 500 bp of coding regions, with mid-CDS encompassing any remaining probes. TSS indicated probes up to 500 bp upstream of the ATG, and Promoter includes all remaining upstream probes. ARS includes all probes within 200 bp of an ARS. Null indicates all remaining probes, predominantly those that fall between convergently-transcribed genes. Conversely, chd1Δ mutants exhibit H3 turnover patterns in which genes appear to effectively reverse polarity. Turnover is still lowest over coding regions, but the trough of minimal turnover has shifted 5′ along coding regions. Promoter and 5′ turnover are slower in chd1Δ cells, whereas maximal H3 replacement is instead observed at the 3′ ends of genes. This behavior is highly unusual, as several published [5], [6], [49], [50] and a large number of unpublished (OJR, unpublished data) mutants exhibit quite distinct turnover defects. We confirmed the increased 3′ H3 replacement at two model genes (Figure S4) using an entirely independent assay for histone replacement based on Cre-mediated recombination of C-terminal H3 epitope tags [51]–[53].
As a separate visualization, Figure 3B shows the average H3 turnover for various classes of genomic elements [6], [54]. Even though a previous microarray analysis showed only a very modest effect of chd1Δ on transcription [18], we considered the possibility that the altered H3 turnover in chd1 cells could be due to a large shift in cellular transcription. However, we observed strong concordance of ChIP on chip of RNA Pol2 signals for wild type and chd1Δ cells (Figure S5). Moreover, as noted below, Chd1's effects on H3 replacement are strongly gene length-dependent, but we find no correlation between mRNA abundance changes and gene length or transcription frequency (Figure S6). Thus, Chd1's effects on turnover are not secondary effects of altered transcription.
Chd1 Protects Long Genes from 3′ H3 Replacement
We sought to understand what factors might contribute to Chd1 recruitment or function at gene ends. To this end, we first examined the genes with the greatest changes in H3 replacement at their 3′ ends in chd1Δ mutants. Notably, we observed that the genes with the greatest changes in 3′ end H3 turnover were among the longest (>3 kb) genes in budding yeast. We therefore systematically analyzed the effects of gene length on Chd1's role in H3 replacement.
Figure 4 shows H3 turnover levels for wild type and chd1Δ yeast cells at gene ends (the first and last 500 bp of coding regions) as a function of gene length. At both gene ends there is strong length dependence for H3 turnover in wild type yeast cells, with turnover decreasing as a function of gene length. Notably, for both 5′ end and 3′ end H3 turnover, Chd1's effect on H3 turnover was greatest at unusually long genes. In addition, we found that Chd1's effect on 3′ turnover was greater at highly transcribed genes (Figure S7).
Fig. 4. Length dependence of Chd1 effects on H3 replacement. 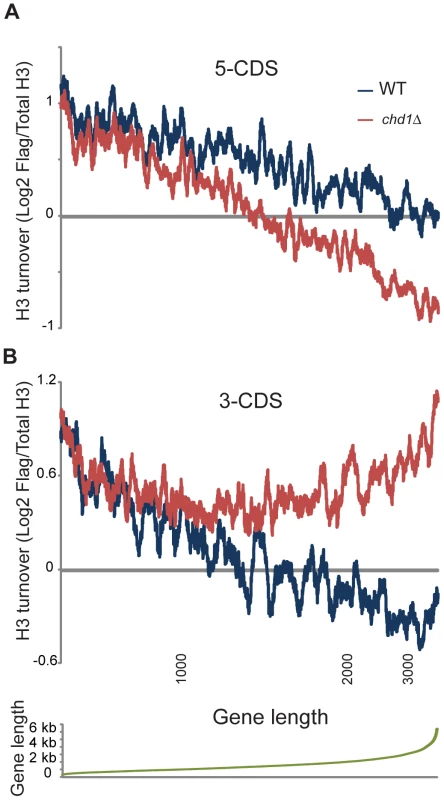
H3 replacement was averaged for the 500 bp at the 5′ ends of genes (A), or the 3′ ends of genes (B). Genes were ordered by length, and an 80 gene window average is shown for wild type and chd1Δ turnover data as indicated. Bottom panel plots gene lengths, and locations for 1, 2, and 3 kb are indicated below panel (B). The length dependence for 3′ end H3 replacement (Figure 4B) is particularly remarkable – H3 turnover is nearly identical in wild type and chd1Δ strains for genes of up to roughly 1 kb in length, at which point 3′ end turnover continues to decrease with gene length in wild type cells but stays essentially constant in chd1Δ cells. In other words, the role of Chd1 in wild type cells seems to be to help stabilize nucleosomes at the 3′ ends of genes over 1 kb in length.
Chd1 Effects on Histone Modification Patterns
Chd1's effects on H3 turnover are greatest at genomic loci that are enriched in H3K36me3 or H3K4me3 modified nucleosomes [55], and chd1 mutants exhibit synthetic genetic interactions with the H3K4 and H3K36 methyltransferases Set1 and Set2 [56], [57]. We therefore determined if chd1Δ mutants affect histone modification patterns by genome-wide mapping of H3K4me3 and H3K36me3 in wild type and chd1Δ yeast cells. Crosslinked chromatin from these two strains was digested with micrococcal nuclease, immunoprecipitated with H3K4me3 or H3K36me3 antisera and competitively hybridized to microarrays with micrococcal nuclease digested input DNA.
Figure 5 shows average H3K4me3 and H3K36me3 patterns in chd1Δ cells. On average, H3K4me3 patterns were minimally affected by loss of Chd1, although we noticed a subtle increase in H3K4me3 at the 3′ ends of many genes. This may be a consequence of the fact that chd1Δ mutants show increased transcription from “cryptic” internal promoters [9],[28]. Interestingly, the gain in H3K4me3 at the 3′ ends of genes was greatest at longer genes (Figure S8), which also exhibited the greatest defects in H3 turnover.
Fig. 5. Chd1 effects on H3 methylation patterns. 
H3K4me3 and H3K36me3 were mapped genome-wide by ChIP-chip on tiling microarrays. Metagene analysis is shown for wild type and chd1Δ strains, as indicated. More dramatically, H3K36me3 patterns were extensively altered in chd1Δ cells, with loss of H3K36me3 at the 3′ ends of genes and a shift in the H3K36me3 peak towards the 5′ ends of genes. Consistent with the loss of H3K36me3 at the 3′ ends of genes, we previously observed increased H3K9/K14 acetylation at the 3′ ends of several genes in a chd1Δ mutant [28], as would be expected since reduced H3K36me3 results in reduced recruitment or activity of the Rpd3S deacetylase complex [27], [58], [59].
In our prior study, we did not observe any significant change in total levels of H3K4me3 or H3K36me3 in a chd1 mutant [28]. As H3K36me3 typically anticorrelates with H3 turnover [6], we hypothesize that the altered H3K36me3 profile observed here is a consequence of Chd1's effects on H3 turnover – increased H3 turnover at 3′ ends of genes likely results in loss of H3K36me3 at these regions. Consistent with this hypothesis, we found that loss of 3′ H3K36me3 was greatest at longer genes (Figure S8).
Discussion
We present evidence that Chd1 modulates replication-independent turnover of histone H3 in both Drosophila and budding yeast. Chd1's effects on H3 turnover are greatest at genomic loci that normally coincide with peaks of H3K4me3 and H3K36me3 modified nucleosomes. This observation is consistent with prior reports that Chd1 reduces nucleosome density at promoters, can catalyze activator-dependent nucleosome removal and promote transcription in vitro, and that it modulates the efficiency of transcription termination [19], [24], [36].
Chd1's effects on H3 turnover may reflect a direct role in histone eviction or deposition during replication-independent histone exchange, consistent with its ability to catalyze ATP-dependent assembly of nucleosomes in vitro, or it could reflect a role for Chd1 in stabilization of pre-existing nucleosomes. Here, we observed that the predominant effect of Chd1 on H3 turnover in budding yeast was to repress turnover over the 3′ ends of genes. While we do not yet understand the mechanism underlying this observation, we favor the idea that Chd1 acts upon nucleosomes that have been perturbed by elongating RNA polymerase II, restoring them to their normal structures or positions and thereby stabilizing them. Importantly, we do not favor the alternative model, that Chd1's effects on chromatin are secondary to perturbation of transcription; we and others do not observe significant alterations of gene expression in chd1 mutants in yeast (Figure S6 and [21]) and Pol II phospho-Ser2 staining of Drosophila polytene chromosomes is normal in chd1 mutants [21].
Chd1's effects on H3 turnover at the 3′ end of genes depended strongly upon gene length (Figure 4), and was also correlated with transcription rate (Figure S7). Given the model above, it is possible that in the absence of Chd1, perturbation of nucleosome positioning by transcription complexes increases with gene length and nucleosome number. Alternatively, Chd1's function may relate to supercoiling changes driven by transcription. To test this we have preliminarily investigated whether additional deletion of the major topoisomerase Top1 affects the chromatin changes observed in chd1Δ yeast mutants. However, we have not observed any suppression of the chd1Δ turnover phenotype in chd1Δtop1Δ double mutants (not shown). Thus, at present we have no additional evidence that supercoiling per se mediates the length dependence of Chd1 on H3 turnover, although given the ability of other topoisomerases to compensate for loss of Top1 we still consider this an appealing hypothesis.
Previous results show that Chd1 has dramatic effects on nucleosome positioning over coding regions [40]. Our results extend this characterization by showing that Chd1 also has dramatic effects on H3 turnover over coding regions, raising the question of whether these two roles for Chd1 in chromatin structure are related. In other words, does Chd1's effect on H3 replacement follow from its role in establishing wild type nucleosome positions, or vice versa? We have no evidence for either possibility, but note that our prior genetic analyses suggest that chd1Δ mutations lower the nucleosomal barrier to RNA Pol2 elongation [22], [28]. Thus, we speculate that disorganized nucleosomes in chd1Δ mutants could be unusually susceptible to eviction by RNA Pol2. This model is consistent with a recent suggestion that elongating polymerases could cause collisions and eviction of adjacent nucleosomes if they are spaced inappropriately [60]. However, arguing against this are observations that nucleosome ladders are little affected in chd1 mutants [40], [41]. Future studies will be needed to address these mechanistic questions.
Taken together, our results identify an evolutionarily conserved role for Chd1 in histone turnover in yeast and flies. Most surprising is our finding that the major site of Chd1 function appears to be at the 3′ ends of genes, suggesting that this enzyme may be recruited or regulated by 3′ histone marks such as H3K36me3. Finally, we find that Chd1 largely affects H3 turnover over longer coding regions, raising the question of whether resolving superhelical tension could be a key role for Chd1 in maintaining wild type chromatin architecture.
Methods
Drosophila Stocks and Crosses
Flies were raised on cornmeal, agar, yeast, and molasses medium, supplemented with methyl paraben and propionic acid. To drive the P[UHS-H3.3core-GFP] transgene [44], [61] in the salivary gland, flies were crossed to P{GawB} AB1-Gal4 flies (Bloomington Stock Center). Mutant chd15 flies were described previously [21]. All crosses were carried out at 18°C.
Confocal Microscopy
Live analysis of polytene chromosome phenotypes was performed as described previously [62]. To analyze the effect of chd15 on H3.3core-GFP incorporation, chd15 b c sp/BcGla; P[UHS-H3.3core-GFP]/TM6B Tb Hu flies were crossed to chd15 b c sp/BcGla; P{GawB}AB1/TM6B Tb Hu flies at 18°C. Flies with chd15 balanced by CyO Kr-GFP instead of BcGla were also analyzed and yielded similar results. Salivary glands were dissected and imaged from heterozygous and homozygous chd15 third instar larvae. For control nuclei, P{GawB} AB1-Gal4 flies were crossed to P[UHS-H3.3core-GFP]/TM6B Tb Hu flies. H3.3core-GFP expression was quantitated by calculating sum pixel intensity in polytene nuclei using the Volocity software package as described previously [62].
Polytene Chromosome Analysis
Polytene chromosomes were prepared and fixed as described [63] and immunostained using primary antibodies directed against CHD1 ([21], 1∶300 dilution), H5 anti-RNA polymerase II (specific for the Ser 2-phosphorylated form of Pol II CTD, Covance; 1∶50 dilution), and the JL-8 anti-GFP (Clontech, 1∶300 dilution). Secondary antibodies donkey anti-rabbit IgG-Cy3, donkey anti-mouse IgM-Cy2, and donkey anti-mouse IgG Fc2a-DyLight 649 (Jackson ImmunoResearch Laboratories, 1∶200 dilutions) were tested with each individual primary antibody to ensure specificity. Images were examined on an Olympus 1X81 inverted fluorescence microscope and acquired using Image-Pro6.3. Control and mutant chromosomes were photographed using identical exposure times, and images were processed identically in Adobe Photoshop CS3.
Yeast Strains and Media
All S. cerevisiae strains used in this study (see Table S1) were constructed by standard procedures, are isogenic to S288c and are GAL2+ [64]. Yeast media was made as described previously [65].
Plasmids
Plasmids used in this study are described in Table S2. Plasmid pJH18-A06 was obtained by random PCR mutagenesis (GAH, TKQ and Araceli Ortiz unpublished). pJH18-Δ4-30, S87P/G90S was created by site-directed mutagenesis of pJH18-A06. PGAL-H4-FlagH3 contains a KpnI-NotI fragment carrying pGAL-driven Flag-H3 from plasmid MDB61 [50], in pRS416.
Flag-H3 Expression and Chromatin Immunoprecipitation
Strains transformed with pGAL-H4-FlagH3 were grown to ∼1.2×107 cells/ml in SC-Ura media with raffinose as the carbon source. Cells were G1 arrested with alpha factor and Flag-H3/H4WT expression was induced by addition of galactose (2% final concentration). ChIP assays were preformed as described previously [66]. 60 minutes after addition of galactose, cells were crosslinked with 1% formaldehyde for 15 min, disrupted by bead beating and chromatin was sonicated using a Diagenode Bioruptor to obtain an average size of 500 bp. Chromatin was immunoprecipitated using 40 µl (1∶2 slurry) Anti-Flag M2 Affinity gel (A2220; Sigma) or 1 µg of a rabbit polyclonal antibody against the C-terminus of H3 (ab1791; Abcam). Chelex 100 resin (BioRad) was added to the immunoprecipitated material and Input-DNA samples, and the suspensions were placed at 100°C for 10 min to reverse crosslinks. Samples were treated with proteinase K and DNA was recovered.
Initial characterization and confirmatory analyses of ChIP samples were performed by qPCR in a Corbett Life Science Rotor Gene 6000 machine using SYBR Green as the detection dye (qPCR MasterMix Plus for SYBR Green, Eurogentec). The fold difference between immunoprecipitated material (IP) and total Input sample for each qPCR amplified region was calculated as described in [67], following the formula IP/Input = (2InputCt - IPCt). H3 turnover rates were measured as the final ratio between Flag-tagged H3 and total H3 (Flag-H3/Input vs total H3/Input). The sequences of oligonucleotides used in these PCR reactions are listed in Table S3.
The immunoprecipitated DNA was initially PCR amplified using random hexamer primers as described in [68]. The number of cycles used to amplify the samples was adjusted to between 28 and 37 so that there was equal amplification of DNA in the IP vs. Flag-tagged H3 and the IP vs. total H3 samples. Amplified DNA was visualized on a 1% agarose gel and checked for a visible smear of DNA between 500 and 1.2 kB. Amplified DNA from Flag-tagged H3 and total H3 ChIPs samples were labeled and competitively hybridized to tiling microarrays as described below.
Micrococcal Nuclease Digestion and Chromatin Immunoprecipitation of H3K4me3 and H3K36me3 Chromatin
Wt and chd1Δ cells were grown to log phase and fixed with 1% formaldehyde. Cell pellets (from 100 mL cells) were resuspended in 8.8 ml Buffer Z (1 M sorbitol, 50 mM Tris-Cl pH 7.4), with addition 6.5 µl of ß-ME (14.3 M, final conc. 10 mM) and 350 µL of zymolyase solution (10 mg/ml in Buffer Z; Seikagaku America), and the cells were incubated at 30°C shaking at 220 rpm. After spinning at 4000× g, 10 min, 4°C, spheroplast pellets were resuspended in 600 µl NP-S buffer (0.5 mM spermidine, 1 mM ß-ME, 0.075% NP-40, 50 mM NaCl, 10 mM Tris pH 7.4, 5 mM MgCl2, 1 mM CaCl2) per 100 ml cell culture equivalent. 25–40 units (depending on yeast strain and cell density) of micrococcal nuclease (Worthington Biochemical) were added and spheroplasts were incubated at 37°C for 20 minutes. The digestion was halted by shifting the reactions to 4°C and adding 0.5 M EDTA to a final concentration of 10 mM.
All steps were done at 4°C unless otherwise indicated. For each aliquot, Buffer L (50 mM Hepes-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) components were added from concentrated stocks (10–20×) for a total volume of 0.8 ml per aliquot. Each aliquot was rotated for 1 hour with 100 µl 50% Sepharose Protein A Fast-Flow bead slurry (Sigma) previously equilibrated in Buffer L. The beads were pelleted at 3000× g for 30 sec, and approximately 100 µl of the supernatant was set aside for the input sample. With the remainder, antibodies were added to each aliquot (equivalent to 100 ml of cell culture) in the following volumes: 10 µl anti-H3K36me3 (Abcam polyclonal), or 7 µl anti-H3K4me3 (Millipore monoclonal). Immunoprecipitation, washing, protein degradation, and DNA isolation were performed as previously described [69]. The samples were amplified, with a starting amount of up to 75 ng for ChIP samples, using the DNA linear amplification method described previously [54].
Microarray Hybridization of ChIP'ed Material
2.5 µg of aRNA produced from the linear amplification were labeled via the amino-allyl method as described on www.microarrays.org. Labeled probes (a mixture of Cy5 labeled input and Cy3 labeled ChIP'ed material) were hybridized onto an Agilent yeast 4×44 whole genome array. The arrays were scanned at 5 micron resolution with the Agilent array scanner. Image analysis and data normalization were performed as previously described [54].
Microarray Data Availability
Microarray data have been deposited in GEO (Accession #GSE38540).
Supporting Information
Zdroje
1. KornbergRDLorchY 1999 Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98 285 294
2. Radman-LivajaMRandoOJ 2010 Nucleosome positioning: how is it established, and why does it matter? Dev Biol 339 258 266
3. KimuraHCookPR 2001 Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol 153 1341 1353
4. JacksonVChalkleyR 1985 Histone synthesis and deposition in the G1 and S phases of hepatoma tissue culture cells. Biochemistry 24 6921 6930
5. RufiangeAJacquesPEBhatWRobertFNouraniA 2007 Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 27 393 405
6. DionMFKaplanTKimMBuratowskiSFriedmanN 2007 Dynamics of replication-independent histone turnover in budding yeast. Science 315 1405 1408
7. IzbanMGLuseDS 1991 Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev 5 683 696
8. LiBCareyMWorkmanJL 2007 The role of chromatin during transcription. Cell 128 707 719
9. CheungVChuaGBatadaNNLandryCRMichnickSW 2008 Chromatin - and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6 e277 10.1371/journal.pbio.0060277
10. MarfellaCGImbalzanoAN 2007 The Chd family of chromatin remodelers. Mutat Res 618 30 40
11. HoLCrabtreeGR 2010 Chromatin remodelling during development. Nature 463 474 484
12. FlanaganJFMiLZChruszczMCymborowskiMClinesKL 2005 Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438 1181 1185
13. SimsRJ3rdChenCFSantos-RosaHKouzaridesTPatelSS 2005 Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem 280 41789 41792
14. MorettiniSTribusMZeilnerASebaldJCampo-FernandezB 2011 The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. Nucleic Acids Res 39 3103 3115
15. HaukGMcKnightJNNodelmanIMBowmanGD 2010 The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39 711 723
16. StockdaleCFlausAFerreiraHOwen-HughesT 2006 Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem 281 16279 16288
17. LusserAUrwinDLKadonagaJT 2005 Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12 160 166
18. TranHGStegerDJIyerVRJohnsonAD 2000 The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. Embo J 19 2323 2331
19. WalfridssonJKhorosjutinaOMatikainenPGustafssonCMEkwallK 2007 A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. Embo J 26 2868 2879
20. SrinivasanSArmstrongJADeuringRDahlsveenIKMcNeillH 2005 The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development 132 1623 1635
21. McDanielIELeeJMBergerMSHanagamiCKArmstrongJA 2008 Investigations of CHD1 function in transcription and development of Drosophila melanogaster. Genetics 178 583 587
22. SimicRLindstromDLTranHGRoinickKLCostaPJ 2003 Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. Embo J 22 1846 1856
23. StokesDGTartofKDPerryRP 1996 CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci U S A 93 7137 7142
24. AlenCKentNAJonesHSO'SullivanJArandaA 2002 A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell 10 1441 1452
25. BiswasDDutta-BiswasRStillmanDJ 2007 Chd1 and yFACT act in opposition in regulating transcription. Mol Cell Biol 27 6279 6287
26. ChuYSuttonASternglanzRPrelichG 2006 The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol Cell Biol 26 3029 3038
27. KeoghMCKurdistaniSKMorrisSAAhnSHPodolnyV 2005 Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123 593 605
28. QuanTKHartzogGA 2010 Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4–Spt5 in transcription. Genetics 184 321 334
29. Pray-GrantMGDanielJASchieltzDYatesJR3rdGrantPA 2005 Chd1 chromodomain links histone H3 methylation with SAGA - and SLIK-dependent acetylation. Nature 433 434 438
30. KroganNJKimMAhnSHZhongGKoborMS 2002 RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22 6979 6992
31. SimsRJ3rdMillhouseSChenCFLewisBAErdjument-BromageH 2007 Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28 665 676
32. KelleyDEStokesDGPerryRP 1999 CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108 10 25
33. KhorosjutinaOWanrooijPHWalfridssonJSzilagyiZZhuX 2010 A chromatin-remodeling protein is a component of fission yeast mediator. J Biol Chem 285 29729 29737
34. KonevAYTribusMParkSYPodhraskiVLimCY 2007 CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317 1087 1090
35. LinJJLehmannLWBonoraGSridharanRVashishtAA 2011 Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev 25 2198 2209
36. EhrensbergerAHKornbergRD 2011 Isolation of an activator-dependent, promoter-specific chromatin remodeling factor. Proc Natl Acad Sci U S A 108 10115 10120
37. LoppinBBonnefoyEAnselmeCLaurenconAKarrTL 2005 The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437 1386 1390
38. OrsiGACoublePLoppinB 2009 Epigenetic and replacement roles of histone variant H3.3 in reproduction and development. Int J Dev Biol 53 231 243
39. YuanGCLiuYJDionMFSlackMDWuLF 2005 Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309 626 630
40. GkikopoulosTSchofieldPSinghVPinskayaMMellorJ 2011 A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333 1758 1760
41. XellaBGodingCAgricolaEDi MauroECasertaM 2006 The ISWI and CHD1 chromatin remodelling activities influence ADH2 expression and chromatin organization. Mol Microbiol 59 1531 1541
42. WoodageTBasraiMABaxevanisADHieterPCollinsFS 1997 Characterization of the CHD family of proteins. Proc Natl Acad Sci U S A 94 11472 11477
43. TsukiyamaTPalmerJLandelCCShiloachJWuC 1999 Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev 13 686 697
44. AhmadKHenikoffS 2002 The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9 1191 1200
45. LingXHarknessTASchultzMCFisher-AdamsGGrunsteinM 1996 Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev 10 686 699
46. Ejlassi-LassalletteAMocquardEArnaudMCThirietC 2011 H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol Biol Cell 22 245 255
47. ElsaesserSJGoldbergADAllisCD 2010 New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev 20 110 117
48. JamaiAImoberdorfRMStrubinM 2007 Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell 25 345 355
49. Lopes da RosaJHolikJGreenEMRandoOJKaufmanPD 2011 Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187 9 19
50. KaplanTLiuCLErkmannJAHolikJGrunsteinM 2008 Cell cycle - and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet 4 e1000270 doi:10.1371/journal.pgen.1000270
51. Radman-LivajaMVerzijlbergenKFWeinerAvan WelsemTFriedmanN 2011 Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol 9 e1001075 doi:10.1371/journal.pbio.1001075
52. VerzijlbergenKFMenendez-BenitoVvan WelsemTvan DeventerSJLindstromDL 2010 Recombination-induced tag exchange to track old and new proteins. Proc Natl Acad Sci U S A 107 64 68
53. VerzijlbergenKFvan WelsemTSieDLenstraTLTurnerDJ 2011 A barcode screen for epigenetic regulators reveals a role for the NuB4/HAT-B histone acetyltransferase complex in histone turnover. PLoS Genet 7 e1002284 doi:10.1371/journal.pgen.1002284
54. LiuCLKaplanTKimMBuratowskiSSchreiberSL 2005 Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3 e328 doi:10.1371/journal.pbio.0030328
55. PokholokDKHarbisonCTLevineSColeMHannettNM 2005 Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122 517 527
56. ZhangLSchroederSFongNBentleyDL 2005 Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. Embo J 24 2379 2390
57. BiswasDTakahataSXinHDutta-BiswasRYuY 2008 A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics 178 649 659
58. DrouinSLarameeLJacquesPEForestABergeronM 2010 DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet 6 e1001173 doi:10.1371/journal.pgen.1001173
59. CarrozzaMJLiBFlorensLSuganumaTSwansonSK 2005 Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 581 592
60. EngeholmMde JagerMFlausABrenkRvan NoortJ 2009 Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol 16 151 158
61. SchwartzBEAhmadK 2005 Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 19 804 814
62. SiriacoGDeuringRChiodaMBeckerPBTamkunJW 2009 Drosophila ISWI regulates the association of histone H1 with interphase chromosomes in vivo. Genetics 182 661 669
63. SchwartzBEWernerJKLisJT 2004 Indirect immunofluorescent labeling of Drosophila polytene chromosomes: visualizing protein interactions with chromatin in vivo. Methods Enzymol 376 393 404
64. WinstonFDollardCRicupero-HovasseSL 1995 Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11 53 55
65. RoseMDWinstonFHieterP 1990 Methods in Yeast Genetics: a laboratory course manual Cold Spring Harbor, New York Cold Spring Harbor Laboratory Press
66. ValenzuelaLDhillonNKamakakaRT 2009 Transcription independent insulation at TFIIIC-dependent insulators. Genetics 183 131 148
67. LittMDSimpsonMRecillas-TargaFPrioleauMNFelsenfeldG 2001 Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. Embo J 20 2224 2235
68. LiebJDLiuXBotsteinDBrownPO 2001 Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28 327 334
69. LiuCLSchreiberSLBernsteinBE 2003 Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics 4 19
70. DietzlGChenDSchnorrerFSuKCBarinovaY 2007 A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151 156
71. KimTSLiuCLYassourMHolikJFriedmanN 2010 RNA polymerase mapping during stress responses reveals widespread nonproductive transcription in yeast. Genome Biol 11 R75
Štítky
Genetika Reprodukčná medicína
Článek Allelic Heterogeneity and Trade-Off Shape Natural Variation for Response to Soil MicronutrientČlánek The Chicken Frizzle Feather Is Due to an α-Keratin () Mutation That Causes a Defective RachisČlánek A Trans-Species Missense SNP in Is Associated with Sex Determination in the Tiger Pufferfish, (Fugu)
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2012 Číslo 7- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Functional Evolution of Mammalian Odorant Receptors
- Oocyte Family Trees: Old Branches or New Stems?
- Allelic Heterogeneity and Trade-Off Shape Natural Variation for Response to Soil Micronutrient
- Guidelines for Genome-Wide Association Studies
- GWAS Identifies Novel Susceptibility Loci on 6p21.32 and 21q21.3 for Hepatocellular Carcinoma in Chronic Hepatitis B Virus Carriers
- DNA Methyltransferases Are Required to Induce Heterochromatic Re-Replication in Arabidopsis
- Genomic Data Reveal a Complex Making of Humans
- Let-7b/c Enhance the Stability of a Tissue-Specific mRNA during Mammalian Organogenesis as Part of a Feedback Loop Involving KSRP
- The Secreted Immunoglobulin Domain Proteins ZIG-5 and ZIG-8 Cooperate with L1CAM/SAX-7 to Maintain Nervous System Integrity
- RsfA (YbeB) Proteins Are Conserved Ribosomal Silencing Factors
- Gene Conversion Occurs within the Mating-Type Locus of during Sexual Reproduction
- The Chicken Frizzle Feather Is Due to an α-Keratin () Mutation That Causes a Defective Rachis
- Meta-Analysis of Genome-Wide Scans for Total Body BMD in Children and Adults Reveals Allelic Heterogeneity and Age-Specific Effects at the Locus
- Balancing Selection at the Tomato Guardee Gene Family Maintains Variation in Strength of Pathogen Defense
- Large-Scale Introgression Shapes the Evolution of the Mating-Type Chromosomes of the Filamentous Ascomycete
- OSD1 Promotes Meiotic Progression via APC/C Inhibition and Forms a Regulatory Network with TDM and CYCA1;2/TAM
- Intact p53-Dependent Responses in miR-34–Deficient Mice
- FANCJ/BACH1 Acetylation at Lysine 1249 Regulates the DNA Damage Response
- CED-10/Rac1 Regulates Endocytic Recycling through the RAB-5 GAP TBC-2
- Histone H2A Mono-Ubiquitination Is a Crucial Step to Mediate PRC1-Dependent Repression of Developmental Genes to Maintain ES Cell Identity
- F-Box Protein Specificity for G1 Cyclins Is Dictated by Subcellular Localization
- The Gene Encodes a Nuclear Protein That Affects Alternative Splicing
- A Key Role for Chd1 in Histone H3 Dynamics at the 3′ Ends of Long Genes in Yeast
- Genome-Wide Association Analysis in Asthma Subjects Identifies as a Novel Bronchodilator Response Gene
- GRHL3/GET1 and Trithorax Group Members Collaborate to Activate the Epidermal Progenitor Differentiation Program
- Brain-Specific Rescue of Reveals System-Driven Transcriptional Rhythms in Peripheral Tissue
- Recent Loss of Self-Incompatibility by Degradation of the Male Component in Allotetraploid
- Pregnancy-Induced Noncoding RNA () Associates with Polycomb Repressive Complex 2 and Regulates Mammary Epithelial Differentiation
- The HEI10 Is a New ZMM Protein Related to Zip3
- The SCF Ubiquitin E3 Ligase Ubiquitylates Sir4 and Functions in Transcriptional Silencing
- Induction of Cytoprotective Pathways Is Central to the Extension of Lifespan Conferred by Multiple Longevity Pathways
- Role of Architecture in the Function and Specificity of Two Notch-Regulated Transcriptional Enhancer Modules
- Loss of ATRX, Genome Instability, and an Altered DNA Damage Response Are Hallmarks of the Alternative Lengthening of Telomeres Pathway
- A Regulatory Loop Involving PAX6, MITF, and WNT Signaling Controls Retinal Pigment Epithelium Development
- The Three Faces of Riboviral Spontaneous Mutation: Spectrum, Mode of Genome Replication, and Mutation Rate
- Unmet Expectations: miR-34 Plays No Role in p53-Mediated Tumor Suppression In Vivo
- A Genome-Wide Association Meta-Analysis of Circulating Sex Hormone–Binding Globulin Reveals Multiple Loci Implicated in Sex Steroid Hormone Regulation
- The Role of Rice HEI10 in the Formation of Meiotic Crossovers
- A Trans-Species Missense SNP in Is Associated with Sex Determination in the Tiger Pufferfish, (Fugu)
- Influences Bone Mineral Density, Cortical Bone Thickness, Bone Strength, and Osteoporotic Fracture Risk
- Evidence of Inbreeding Depression on Human Height
- Comparative Genomics of Plant-Associated spp.: Insights into Diversity and Inheritance of Traits Involved in Multitrophic Interactions
- Detecting Individual Sites Subject to Episodic Diversifying Selection
- Regulates Rhodopsin-1 Metabolism and Is Required for Photoreceptor Neuron Survival
- Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Dosage Compensation
- Three Dopamine Pathways Induce Aversive Odor Memories with Different Stability
- TDP-1/TDP-43 Regulates Stress Signaling and Age-Dependent Proteotoxicity in
- Rapid Turnover of Long Noncoding RNAs and the Evolution of Gene Expression
- The Yeast Rab GTPase Ypt1 Modulates Unfolded Protein Response Dynamics by Regulating the Stability of RNA
- Histone H2B Monoubiquitination Facilitates the Rapid Modulation of Gene Expression during Arabidopsis Photomorphogenesis
- Cellular Variability of RpoS Expression Underlies Subpopulation Activation of an Integrative and Conjugative Element
- Genetic Variants in , , and Influence Male Recombination in Cattle
- Differential Impact of the HEN1 Homolog HENN-1 on 21U and 26G RNAs in the Germline of
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Guidelines for Genome-Wide Association Studies
- The Role of Rice HEI10 in the Formation of Meiotic Crossovers
- Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Dosage Compensation
- GWAS Identifies Novel Susceptibility Loci on 6p21.32 and 21q21.3 for Hepatocellular Carcinoma in Chronic Hepatitis B Virus Carriers
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



