-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
The somatic genome of the ciliated protist Tetrahymena undergoes DNA elimination of defined sequences called internal eliminated sequences (IESs), which account for ∼30% of the germline genome. During DNA elimination, IES regions are heterochromatinized and assembled into heterochromatin bodies in the developing somatic nucleus. The domesticated piggyBac transposase Tpb2p is essential for the formation of heterochromatin bodies and DNA elimination. In this study, we demonstrate that the activities of Tpb2p involved in forming heterochromatin bodies and executing DNA elimination are genetically separable. The cysteine-rich domain of Tpb2p, which interacts with the heterochromatin-specific histone modifications, is necessary for both heterochromatin body formation and DNA elimination, whereas the endonuclease activity of Tpb2p is only necessary for DNA elimination. Furthermore, we demonstrate that the endonuclease activity of Tpb2p in vitro and the endonuclease activity that executes DNA elimination in vivo have similar substrate sequence preferences. These results strongly indicate that Tpb2p is the endonuclease that directly catalyzes the excision of IESs and that the boundaries of IESs are at least partially determined by the combination of Tpb2p-heterochromatin interaction and relaxed sequence preference of the endonuclease activity of Tpb2p.
Published in the journal: A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in. PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004032
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004032Summary
The somatic genome of the ciliated protist Tetrahymena undergoes DNA elimination of defined sequences called internal eliminated sequences (IESs), which account for ∼30% of the germline genome. During DNA elimination, IES regions are heterochromatinized and assembled into heterochromatin bodies in the developing somatic nucleus. The domesticated piggyBac transposase Tpb2p is essential for the formation of heterochromatin bodies and DNA elimination. In this study, we demonstrate that the activities of Tpb2p involved in forming heterochromatin bodies and executing DNA elimination are genetically separable. The cysteine-rich domain of Tpb2p, which interacts with the heterochromatin-specific histone modifications, is necessary for both heterochromatin body formation and DNA elimination, whereas the endonuclease activity of Tpb2p is only necessary for DNA elimination. Furthermore, we demonstrate that the endonuclease activity of Tpb2p in vitro and the endonuclease activity that executes DNA elimination in vivo have similar substrate sequence preferences. These results strongly indicate that Tpb2p is the endonuclease that directly catalyzes the excision of IESs and that the boundaries of IESs are at least partially determined by the combination of Tpb2p-heterochromatin interaction and relaxed sequence preference of the endonuclease activity of Tpb2p.
Introduction
Transposons represent harmful genetic elements because they potentially rearrange their host's genome, and their integration into important coding or regulatory regions can have deleterious effects. Transposons are therefore considered “junk” DNAs [1], and hosts have evolved genome defense mechanisms to counteract these selfish elements [2]. However, transposons may not be just junk because they potentially contribute to the evolution of the host by genome rearrangements, alternating gene expression networks, or providing new genes from transposons to the host [3]. Therefore, host organisms must evolve by balancing the harmfulness and usefulness of transposons. An evolutional product likely created by such a balance is the programmed DNA elimination in the ciliated protist Tetrahymena, in which the transposon-related sequences are eliminated by a domesticated piggyBac transposase [4].
Most ciliates display nuclear dimorphism [5]. The germline micronucleus (Mic) is transcriptionally inert during vegetative growth, whereas the somatic macronucleus (Mac) provides the cell with most if not all RNA. When nutrients are scarce, Tetrahymena undergoes sexual reproduction (conjugation), in which two mating partners form a pair (Fig. 1A) and their Mics undergo meiosis (Fig. 1B). Three of the meiotic products are degraded, and the remaining product divides mitotically (Fig. 1C). One of the products is exchanged with the mating partner and afterwards, the two pronuclei fuse to form the zygote (Fig. 1D). The zygotic nucleus divides twice mitotically (Fig. 1E); of the four mitotic products, two become the new Mics, and the other two develop to become the new Macs (Fig. 1F). The parental Mac is degraded at the end of this process, and the progeny resume vegetative growth when nutrients are supplied (Fig. 1G).
Fig. 1. Conjugation of Tetrahymena. 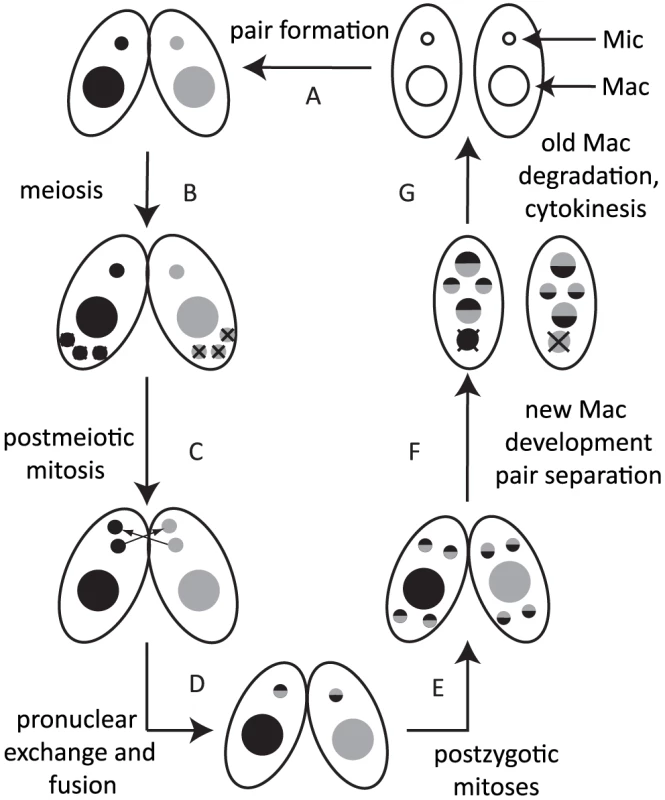
Under starvation conditions, two Tetrahymena cells form a pair (A) and initiate sexual reproduction (conjugation). The Mic undergoes meiosis, and three of the four meiotic products are degraded (B), whereas the remaining product divides mitotically to form two pronuclei in each cell (C). One of the two pronuclei is exchanged with the mating partner, and fusion leads to formation of the zygotic nucleus (D). The zygotic nucleus divides mitotically twice (E); two nuclei become Mics, and the other two differentiate into Macs (F). The parental Mac is degraded. After cell division, the nuclei are distributed to the daughter cells (G). Two major types of programmed genome rearrangements occur in the developing new Mac of Tetrahymena. The first type is chromosome breakage, which leads to the fragmentation of germline chromosomes. The chromosome breakage occurs at conserved 15-nt sequences called chromosome breakage sequences (CBSs). It has been estimated that there are ∼250 CBSs in the Mic genome [6], [7]. The sites of chromosome breakages are healed by de novo telomere formation [8]. The second type of genome rearrangement in Tetrahymena is DNA elimination of internal eliminated sequences (IESs), followed by ligation of their flanking sequences [9] by the non-homologous end joining (NHEJ) pathway [10]. The indispensability of the NHEJ pathway for DNA elimination was also demonstrated for another ciliate, Paramecium [11]. It has been estimated that there are over 8,000 different IESs, which represent ∼30% of the Mic genome [12], [13]. Because many IESs contain transposon-related sequences, it is assumed that DNA elimination is a process that removes potentially harmful transposons from the transcriptionally active somatic genome [14]. Moreover, because some IESs are in regulatory regions and exons of genes, DNA elimination is necessary to create the streamlined functional somatic genome [15]. Despite the fact that different IESs do not share any detectable common sequences within themselves and in their flanking regions, invariable sets of IESs are eliminated from the Mac, and the majority of their boundaries occur within a few to several base pairs.
The identities of IESs are most likely determined epigenetically by an RNAi-related mechanism [16], [17]. While the Mic is transcriptionally inert during the vegetative growth, non-coding RNA transcription occurs in the Mic during the early stages of conjugation. The ∼28–29-nt siRNAs produced from the non-coding RNAs are selected for IES specificity by selective degradation of siRNAs complementary to the parental Mac genome [12]. The selected IES-specific siRNAs eventually induce the establishment of heterochromatin specifically on IESs in the developing new Mac. This heterochromatin comprises tri-methylated histone H3 at lysine 9 and lysine 27 (H3K9me3, H3K27me3) [18], [19] and the chromodomain protein Pdd1p [20], which binds to the histone H3 modifications. Although both H3K9me3 and H3K27me3 have been shown to play an important role in DNA elimination [18], [19], the functional distinction between H3K9me3 and H3K27me3 in the DNA elimination process, if any, is not clear. The heterochromatinized IESs are assembled into heterochromatin bodies located at the nuclear periphery [21], and each IES is eventually excised as one linear or circular piece of DNA [22]–[24]. Artificially tethering Pdd1p to DNA is sufficient to induce DNA elimination [25], indicating that heterochromatin but not the RNAi-related mechanism is the immediate signal inducing DNA elimination.
Previous studies have shown that, in some IES elements, the deletion boundaries are determined by flanking cis-acting sequences located 40–50 bp away, and the precise nature of the deletion is dependent on the sequences at the boundaries [26]–[28]. However, because no sequence homology has been observed across different elements, it is unclear how the boundaries of IESs are determined. In addition it is not known whether and how heterochromatin is involved in the boundary determination. We previously reported that the domesticated piggyBac transposase-like protein Tpb2p (Tetrahymena piggyBac-like transposase 2) localizes to the heterochromatin bodies and is essential for DNA elimination [4]. Furthermore, we reported that Tpb2p has the ability to produce DNA double-strand breaks at a boundary sequence of an IES in vitro [4]. Therefore, we hypothesized that Tpb2p is recruited to the IESs by directly interacting with a heterochromatin component and then inducing a DNA double-strand break at its preferential DNA sequence near the heterochromatin to execute DNA elimination. To validate this hypothesis, we analyze the roles of the individual domains of Tpb2p genetically and biochemically to understand 1) how Tpb2p interacts with heterochromatin; 2) whether the Tpb2p-heterochromatin interaction is necessary for DNA elimination; 3) what is the sequence preference of the endonuclease activity of Tpb2p; and 4) whether the sequence preference contributes to the choice of boundary sequence of IESs. Based on the results, we discuss how Tpb2p is involved in reproducible DNA elimination and how a domesticated transposase has evolved to catalyze the DNA elimination of transposons.
Results
Conditional knockout of TPB2 phenocopies TPB2 loss via RNAi knockdown
Recombinantly expressed Tpb2p has been demonstrated to have the ability to produce DNA double-strand breaks in vitro [4]. However, it is unclear whether the endonuclease activity of Tpb2p is necessary for DNA elimination in vivo because Tpb2p is also necessary for the formation of the heterochromatin bodies, which is believed to be a prerequisite of DNA elimination. To test whether the endonuclease activity of Tpb2p is needed for DNA elimination in vivo, we attempted to express a catalytically inactive Tpb2p mutant in a TPB2-null background. RNAi knockdown, which has been previously used to study the function of TPB2 [4], only partially down-regulates TPB2 expression and is difficult to use for genetic rescue experiments. Therefore, we attempted to obtain knockout (KO) strains of TPB2 but without success. This lack of success might be because TPB2 is a haplo-insufficient gene, and heterozygous TPB2 KO strains are not viable. This is consistent with the fact that RNAi knockdown of TPB2 caused nearly complete lethality of sexual progeny [4]. To overcome this problem, we created TPB2 conditional knockout (cKO) strains.
To make TPB2 expression conditional, we produced a TPB2 cKO construct in which the endogenous TPB2 promoter was replaced with the cadmium-inducible MTT1 promoter (Fig. 2A). The TPB2 cKO construct was first introduced into the TPB2 locus in the Mic by homologous recombination to produce heterozygous TPB2 cKO strains, and then, two heterozygous TPB2 cKO strains were mated to obtain homozygous TPB2 cKO strains (hereafter referred to as TPB2 cKO strains) (Fig. 2B). In these genetic crosses, TPB2 expression was continuously induced during conjugation to obtain viable progeny.
Fig. 2. Construction and analyses of TPB2 conditional KO (cKO) cells. 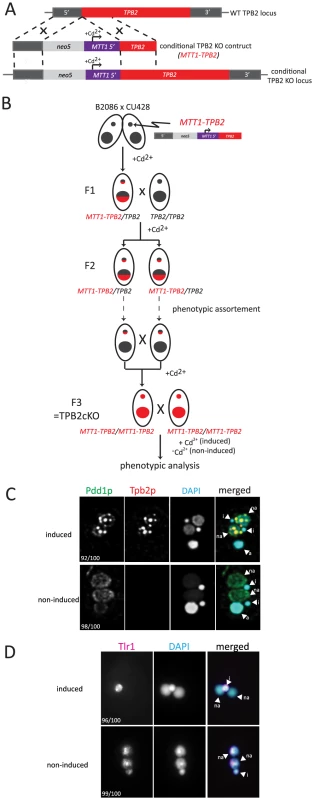
(A) The TPB2 conditional KO construct (MTT1-TPB2) was introduced into the endogenous TPB2 locus (WT TPB2 locus) by homologous recombination to create the TPB2 conditional KO locus, in which TPB2 expression is under the control of the cadmium-inducible MTT1 promoter. (B) Germline transformation with the MTT1-TPB2 construct resulted in one heterozygous TPB2 cKO clone (F1), which was then crossed with WT to obtain the heterozygous TPB2 cKO clones with different mating types (F2). The heterozygous TPB2 cKO clones were phenotypically assorted (dotted arrows) until they lost Mac copies of MTT1-TPB2 and afterwards crossed with each other to obtain homozygous TPB2 cKO strains (F3). (C) Mating TPB2 cKO cells were incubated with (induced) or without (non-induced) cadmium, and exconjugants (progeny) were fixed at 14 hr post-mixing for immunofluorescence staining. The cells were triple stained with a guinea pig anti-Pdd1p antibody (green), which marks heterochromatin, a rabbit anti-Tpb2p antibody (red) and DAPI (blue), which stains DNA. The number of cells displaying the phenotype represented by the pictures in each culture condition are shown. (D) Mating TPB2 cKO cells were incubated with (induced) or without (non-induced) cadmium, and exconjugants (progeny) were fixed at 36 hr post-mixing for DNA FISH against the Tlr1 IES to assess DNA elimination. DNA was counterstained with DAPI. The number of cells displaying the phenotype represented by the pictures in each culture condition are shown. i = micronucleus; na = new macronucleus. It is known that the MTT1 promoter is leaky in the standard culture conditions [29]. Therefore, we used a metal-depleted medium (see Materials and Methods for details) to minimize the basal level activity of the MTT1 promoter. Western blot analysis and immunofluorescent staining using an anti-Tpb2p antibody demonstrated that in our culture conditions, Tpb2p was undetectable in the absence of cadmium during conjugation (see Fig. 3B −Cd2+ lanes) of the TPB2 cKO strains. In contrast, when cadmium was added, Tpb2p expression was clearly detected (see Fig. 3B +Cd2+ lanes). Therefore, Tpb2p expression can be conditionally knocked out in the TPB2 cKO strains.
Fig. 3. Functional analyses of the endonuclease and cysteine-rich domains of TPB2 in vivo. 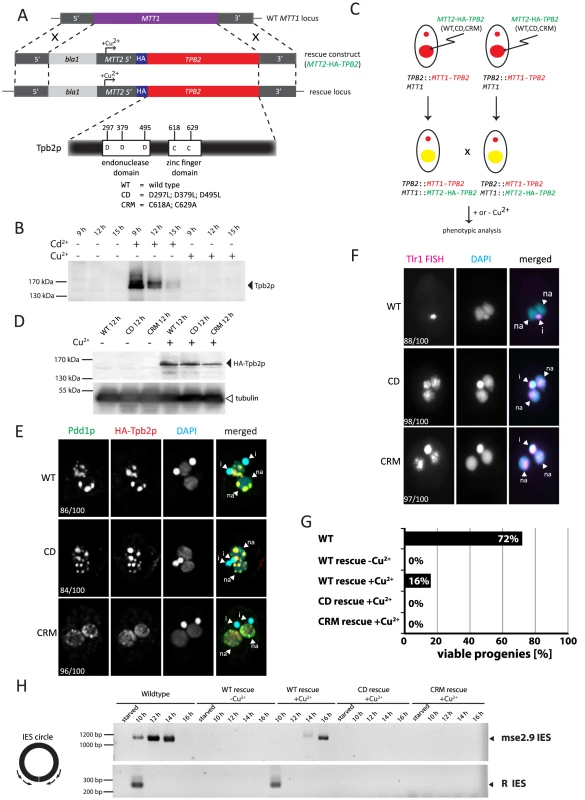
(A) The TPB2 rescue construct (MTT2-TPB2) was introduced into the endogenous MTT1 locus (WT MTT1 locus) by homologous recombination to create the TPB2 rescue locus, in which TPB2 expression is under the control of the copper-inducible MTT2 promoter. Three different TPB2 rescue constructs encoding for wild-type Tpb2p (WT), catalytic-dead Tpb2p (CD) and a cysteine-rich mutant Tpb2p (CRM) were used. (B) TPB2 cKO strains were mated and either left uninduced or cadmium or copper were added during conjugation. Tpb2p expression from the MTT1 promoter at the TPB2 cKO locus at 9, 12 and 15 h post-mixing was detected by western blot using an anti-Tpb2p antiserum. (C) Scheme of production of the TPB2 rescue strains. A TPB2 rescue construct was introduced into the MTT1 locus of the Mac of TPB2 cKO cells. (D) TPB2 rescue strains expressing WT, CD or CRM TPB2 were mated, and Tpb2p expression from the MTT2 promoter at the rescue locus was compared between in the absence (−) or presence (+) of CuSO4 by western blot (top). As a loading control, expression of alpha-tubulin was analyzed (bottom) (E) Mating TPB2-rescued cells with wild-type (WT), catalytic-dead (CD) or cysteine-rich mutant (CRM) construct were incubated with copper, and exconjugants (progeny) were fixed at 14 hr post-mixing for immunofluorescence staining. The cells were triple stained with a guinea pig anti-Pdd1p antibody (green), which marks heterochromatin, a rabbit anti-Tpb2p antibody (red), and DAPI (blue), which stains DNA. The numbers of cells displaying the phenotype represented by the pictures in each culture condition are shown. (F) Mating TPB2 rescued cells were incubated with CuSO4, and exconjugants (progeny) were fixed at 36 hr post-mixing for DNA FISH against the Tlr1 IES to assess DNA elimination. DNA was counterstained with DAPI. The number of cells displaying the phenotype represented by the pictures in each culture condition are shown. i = micronucleus; na = new macronucleus. (G) Viability of sexual progeny of the wild-type (WT)or the TPB2 rescue strains without (−Cu2+) or with (+Cu2+) induction of the wild-type (WT rescue), catalytic-dead (CD rescue) or cysteine-rich mutant (CRM rescue) TPB2 were analyzed. 192 single mating pairs were placed into drops of medium and incubated for ∼60 h at 30°C. Completion of conjugation was confirmed by testing for negative expression of the marker specific for the parental Macs. (H) Genomic DNA was extracted from starved or mating (10, 12, 14 and 16 h post-mixing) cells of the wild-type or the TPB2 rescue strains without (−Cu2+) or with (+Cu2+) induction of the wild-type (WT), catalytic-dead (CD) or cysteine-rich mutant (CRM) rescue constructs, and circularized mse2.9- and R-IES elements were detected by nested PCR as shown on the left. It has been reported that the formation of heterochromatin bodies and DNA elimination can be inhibited by the RNAi knockdown of TPB2 [4]. To determine if the cKO of TPB2 phenocopies the RNAi knockdown of TPB2, the formation of heterochromatin bodies in the TPB2 cKO cells was observed by immunofluorescent staining of the heterochromatin component Pdd1p. When TPB2 expression was induced in the presence of cadmium, the TPB2 cKO strains formed Pdd1p-containing heterochromatin bodies in the new Macs (Fig. 2C, “induced”). In contrast, in the absence of TPB2 induction, Pdd1p-stained heterochromatin did not form large bodies but remained as dispersed small foci in the new Mac (Fig. 2C, “non-induced”).
Next, DNA elimination in TPB2 cKO strains was observed by DNA fluorescence in situ hybridization (FISH) against Tlr1 IESs, which are moderately repeated (∼30 copies) in the Mic genome [30]. DNA elimination in wild-type cells is normally completed by ∼16-hr post-mixing [31]. In the presence of cadmium, Tlr1 IESs were undetectable in the new Macs at 36-hr post-mixing and detected only in the Mic (Fig. 2D, “induced”, na = new Mac, i = Mic), indicating that these IESs were removed completely from the new Macs. In contrast, the Tlr1 IESs remained in the new Mac even at 36-hr post-mixing when TPB2 expression was not induced (Fig. 2D, “non-induced”). These results indicate that the TPB2 cKO strains exhibit defects in heterochromatin body formation and DNA elimination in the absence of the induction of TPB2, as it was previously reported for the TPB2 RNAi knockdown strains.
Establishment of a genetic rescue system to study the function of Tpb2p domains
Next, we attempted to establish a genetic rescue system in which the non-essential MTT1 locus [29] of the parental Mac in the TPB2 cKO strains was replaced with a MTT2 cassette expressing a gene of interest under the control of the copper-inducible MTT2 promoter [32] (Fig. 3A). Before starting the rescue experiments, we first tested whether the TPB2 cKO locus could be kept silent in the presence of copper. We incubated the conjugating TPB2 cKO cells either with cadmium or copper, and Tpb2p expression was analyzed by western blot using an anti-Tpb2p antibody. Although Tpb2p expression was induced in the presence of cadmium, it was undetectable from the cells incubated with copper (Fig. 3B). These results indicate that the expression of TPB2 in the TPB2 cKO locus, which is under control of the cadmium-inducible MTT1 promoter, is not induced by the addition of copper.
Next, the MTT2 cassette containing the wild-type TPB2 tagged with HA epitope (Fig. 3A, MTT2-HA-TPB2) was introduced into the TPB2 cKO strains (Fig. 3C), and the cells were incubated with or without copper. We analyzed HA-Tpb2p expression by western blot using an anti-HA antibody and observed that HA-Tpb2p was detected only when copper was added to the medium (Fig. 3D, compare WT +/ − Cu2+). Therefore, the expression of a gene in the MTT2 cassette is induced in the presence of copper. When the MTT2 cassette containing the wild-type TPB2 was introduced into the TPB2 cKO strains, heterochromatin body formation (Fig. 3E, WT) and DNA elimination (Fig. 3F, WT) were restored in the presence of copper. Also, expression of the wild-type TPB2 partially restored the progeny viability of the TPB2 cKO strains (Fig. 3G, WT-rescue +Cu2+). These results indicate that the wild-type TPB2 expressed from the MTT2 cassette in the parental Mac is sufficient for all essential steps of DNA elimination, although it might not be enough to restore some non-essential steps of DNA elimination. Therefore, the rescue system using the TPB2 cKO strains and MTT2 cassette can be used to assay functionalities of Tpb2p mutants.
DNA elimination process was also analyzed by observing circularized excised IESs by PCR (see Fig. 3H left). DNA elimination events in the wild-type cells release IESs in two different forms: the major linear form and the minor circular form [23], [24]. The circular form of two different IESs, mse2.9 and R elements, were detected when the wild-type TPB2 was expressed in the TPB2 cKO background (Fig. 3H, WT rescue +Cu2+). However, the appearance of the excised mse2.9 IES was delayed in the conditional TPB2 KO cells expressing the wild-type TPB2 compare to the wild-type cells (Fig. 3H, see Wild-type and WT rescue +Cu2+), possibly because the ectopic expression of TPB2 from the MTT2 promoter in the parental Mac could not fully restore the function of endogenous TPB2. This may explain why the progeny viability was much lower in the conditional TPB2 KO cells expressing the wild-type TPB2 than in the wild-type cells (Fig. 3G).
The endonuclease activity of Tpb2p is essential for DNA elimination but not for heterochromatin body maturation
Tpb2p has the endonuclease catalytic domain that contains three aspartic acids that form the DDD catalytic triad (Fig. 3A). We previously reported that the replacement of these three aspartic acids with leucines compromises the endonuclease activity of Tpb2p in vitro [4]. To understand the role of the endonuclease activity in vivo, the MTT2 cassette expressing the TPB2-CD mutant, in which the catalytic triad of Tpb2p was replaced with leucines (D297L; D379L; D495L, “CD” in Fig. 3A), was introduced into the TPB2 cKO strains. The amount of Tpb2p-CD expressed from the MTT2 cassette after induction with copper was comparable to that of the wild-type Tpb2p from the cassette (Fig. 3D, compare +Cu2+ lanes of WT and CD), indicating that the mutations do not significantly affect the stability of Tpb2p. The expression of the TPB2-CD mutant did restore heterochromatin body maturation, based on the localization of the heterochromatin component Pdd1p (Fig. 3E, “CD”), but did not support the elimination of Tlr1 IESs from the new Macs, as evaluated by the fact that FISH using the probes complementary to Tlr1 IESs stains the new Mac (Fig. 3F, “CD”). The circular form of two different IESs, mse2.9 and R elements, could not be detected by PCR when TPB2-CD expression was induced (Fig. 3H, CD rescue +Cu2+), indicating that no detectable IES excision was induced by the TPB2-CD expression. Consistent with the fact that DNA elimination is essential for the production of viable sexual progeny, expression of TPB2-CD mutant could not restore the progeny viability of the TPB2 cKO strains (Fig. 3G). Altogether, we conclude that the endonuclease activity of Tpb2p is necessary for DNA elimination but dispensable for the heterochromatin body formation.
The cysteine-rich domain of Tpb2p is essential for heterochromatin body formation and DNA elimination
The results above clearly indicate that the necessity of Tpb2p for heterochromatin body formation (Fig. 2C) should be attributed to an activity of Tpb2p other than its endonuclease activity. The endonuclease domain of piggyBac transposases is followed by a PHD finger-like domain [33], [34] (Supplemental Fig. S1). Tetrahymena Tpb2p and its Paramecium homolog Pgm [35] also have a cysteine-rich domain downstream of their endonuclease domains (Fig. 3A, Supplemental Fig. S1). Although this cysteine-rich domain displays similarity to the PHD finger domain, it lacks a potential metal-binding residue in one of the two intermingled zinc-fingers (Supplemental Fig. S1). Therefore, it is unclear whether the cysteine-rich domain of Tpb2p has any biological role or if it is only a non-functional remnant of the PHD finger domain of the ancestral piggyBac transposase.
To determine if the cysteine-rich domain of Tpb2p has any role in DNA elimination, a MTT2 cassette containing the TPB2-CRM mutant, in which two of the seven potential metal-binding core cysteine/histidine residues of the cysteine-rich domain were replaced with alanines (C618A; C629A, “CRM” in Fig. 3, Supplemental Fig. S1), was introduced into the TPB2 cKO strains. The amount of Tpb2p-CRM expressed after induction with copper was comparable to that of the wild-type Tpb2p from the cassette (Fig. 3D, compare +Cu2+ lanes of WT and CRM), indicating that the mutations did not significantly affect the stability of Tpb2p. We observed that the expression of TPB2-CRM did not restore heterochromatin body formation (Fig. 3E, CRM), the elimination of Tlr1 IESs from the new Macs (Fig. 3F, CRM), the formation of circularized excised IESs (Fig. 3H, CRM rescue +Cu2+), and the progeny viability (Fig. 3G, CRM rescue +Cu2+). Therefore, we concluded that the cysteine-rich domain of Tpb2p is essential for both heterochromatin body formation and the following DNA elimination.
Heterochromatin-specific histone H3 methylations enhance the interaction between the cysteine-rich domain of Tpb2p and the N-terminal tail of histone H3
The cysteine-rich domain of Tpb2p resembles the PHD-finger domain (Supplemental Fig. S1), and some PHD finger-containing proteins bind to the N-terminal tail of histone H3 with methylated lysine residues [36]. Because histone H3 on the heterochromatinized IESs is specifically tri-methylated at lysine 9 and lysine 27 (H3K9me3/K27me3), we reasoned that Tpb2p might interact with heterochromatin through the interaction between its cysteine-rich domain and H3K9me3/K27me3. This hypothesis is consistent with the fact that although the wild-type Tpb2p and Tpb2p-CD tightly co-localized with the heterochromatin component Pdd1p (Fig. 3E WT, CD), the localization of Tpb2p-CRM did not completely overlap with that of Pdd1p (Fig. 3E, CRM). Therefore, we aimed to investigate if the cysteine-rich domain of Tpb2p interacts with H3K9me/K27me.
We prepared C-terminally biotinylated peptides representing the amino acids 1–19 or 16–35 of Tetrahymena histone H3 (Supplemental Table S2). The peptides were either unmodified or tri-methylated at lysine 4, 9 or 27. In addition, unmodified peptides with scrambled amino acid orders were prepared. The peptides were bound to avidin-coated beads and incubated with a recombinant cysteine-rich domain (aa 566–aa 657) of Tpb2p fused to a maltose-binding protein (MBP)-tag (MBP-Tpb2p-CRD), and the proteins co-precipitated with the peptides were analyzed by western blotting using an anti-MBP antibody.
We observed that more MBP-Tpb2p-CRD was precipitated with the unmodified peptides than with the beads only (compare lanes 2 and 4, lanes 2 and 7 or lanes 10 and 12 of Fig. 4, top), whereas the amount of MBP-Tpb2p-CRD precipitated with these unmodified peptides was comparable to the amount precipitated with the corresponding unmodified peptides with scrambled amino acid orders (compare lanes 3 and 4, lanes 6 and 7 or lanes 11 and 12 of Fig. 4, top). Therefore, some physical property of the peptides, such as charge, likely mediates the co-precipitation of MBP-Tpb2p-CRD with the unmodified peptides. Importantly, significantly more MBP-Tpb2p-CRD was co-precipitated with the peptides having tri-methylated lysines 9 or 27 than with the corresponding unmodified peptides (compare lanes 4 and 5 or lanes 7 and 8 of Fig. 4, top). Therefore, the presence of tri-methylations at lysine 9 or lysine 27 enhances the interaction between MBP-Tpb2p-CRD and the peptides. In contrast, tri-methylations at lysine 4 did not enhance co-precipitation of MBP-Tpb2p-CRD with the peptide (compare lanes 12 and 13 of Fig. 4). The mutations (C618A, C629A) at the putative metal-binding core of the cysteine-rich domain, which abolish the heterochromatin body formation in vivo (Fig. 3E), inhibited the co-precipitation of MBP-Tpb2p-CRD with any of the peptides in vitro (Fig. 4, bottom). We conclude that Tpb2 can interact with the histone H3 tail through its cysteine-rich domain, and this interaction is significantly enhanced by the presence of tri-methylated lysines 9 or 27.
Fig. 4. In vitro histone peptide-binding assay. 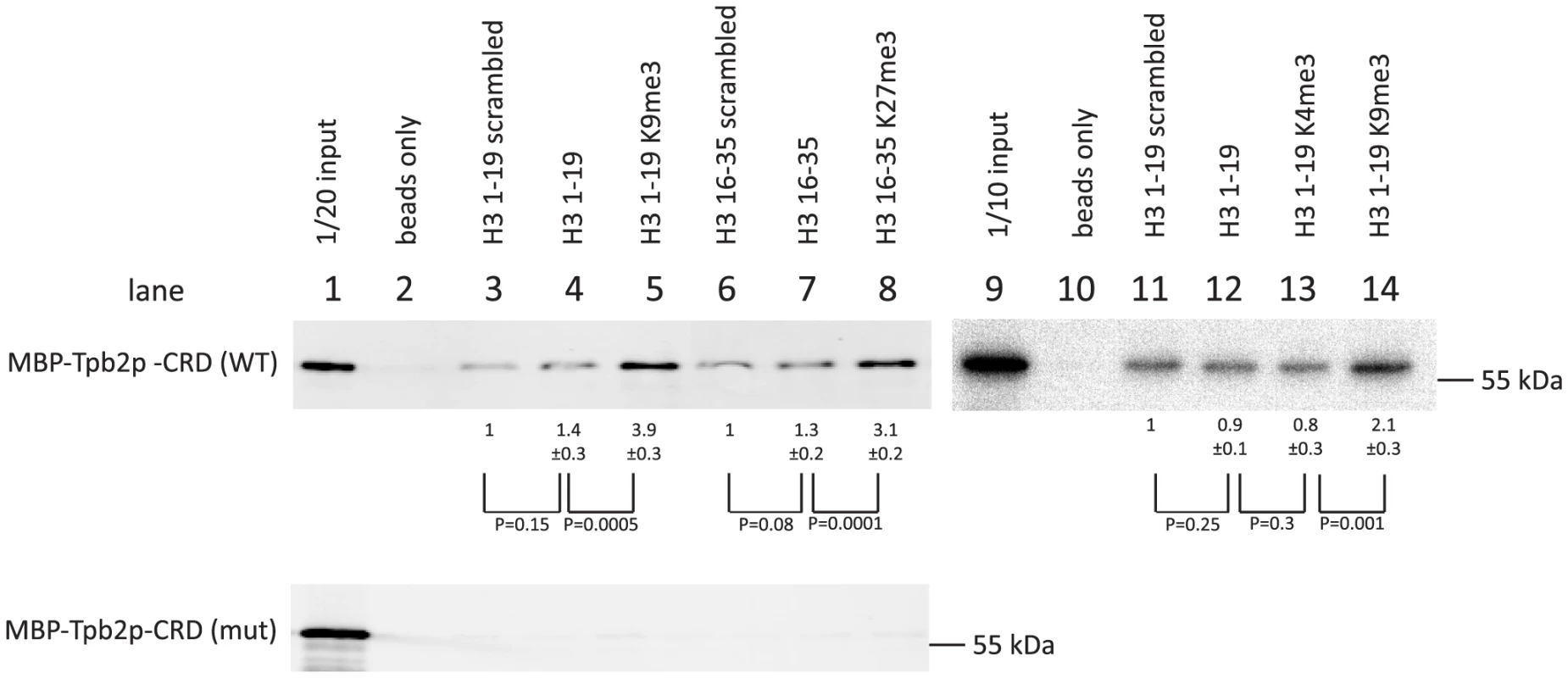
Indicated biotin-tagged histone peptides were immobilized on streptavidin Dynabeads and incubated with the recombinant expressed MBP-tagged cysteine-rich domain (566 aa–657 aa) of Tpb2p (MBP-Tpb2p-CRD (WT)) or peptides having mutations (C618A, C629A) at the putative metal-binding cysteines (MBP-Tpb2p-CRD (mut)). Proteins co-precipitated with the histone peptides were detected by western blot using anti-MBP antiserum. The average intensities of bands from three independent experiments were normalized to the value of “scrambled” peptides. Standard deviations are shown after the average intensity values. P-values from a student's T-test are shown. The integrity of the left R-IES boundary sequence is important for precise cleavage by Tpb2p in vitro
We have previously shown that recombinant Tpb2p, which is expressed from E. coli, produces DNA double-strand breaks (DSB) possessing 4-base 5′ overhangs at the left boundary sequence of Tetrahymena R-IES (5′-AGTGAT-3′) in vitro [4] (see also Fig. 5B). However, it is unclear what sequence feature, if any, is recognized by Tpb2p. To better understand what sequence feature of an IES boundary is recognized by Tpb2p, the left R-IES boundary sequence (5′-AGTGAT-3′) was placed in the middle of otherwise artificial sequence, and every position of the boundary was substituted with every other possible nucleotide (Fig. 5B). The radiolabeled oligo DNA duplexes were incubated with recombinant Tpb2p and analyzed as described above. As previously observed, when Tpb2p was incubated with an oligo DNA duplex having the wild-type R-IES boundary, the major 50-nt product, which is produced by the endonucleolytic cleavage between the first A and first G of the boundary (see Fig. 5A), was detected (Fig. 5B, the leftmost lane). Base substitutions at positions −1, +1, +4 and +5 did not significantly affect the choice of cleavage position by Tpb2p in this in vitro assay (Fig. 5B). In contrast, when position +2 was substituted, the major 50-nt product was greatly reduced, and instead, a few nucleotide longer products were detected (Fig. 5B). Similarly, when position +3 was substituted, a few nucleotide longer products were detected, although the 50-nt product was not significantly reduced (Fig. 5B). Importantly, no obvious cleavage products were detected when the substrates were incubated with the catalytically inactive Tpb2p (Fig. 5C), indicating that the observed products were produced by Tpb2p, but not by any contaminated bacterial endonucleases. All together, these results suggest that most nucleotides of the left R-IES boundary sequence are replaceable without disturbing the boundary recognition by Tpb2p, whereas the 5′-TG-3′ sequence at positions +2 and +3 are important for Tpb2p to execute precise cleavage of the left R-IES boundary in vitro.
Fig. 5. In vitro Tpb2p endonuclease assay. 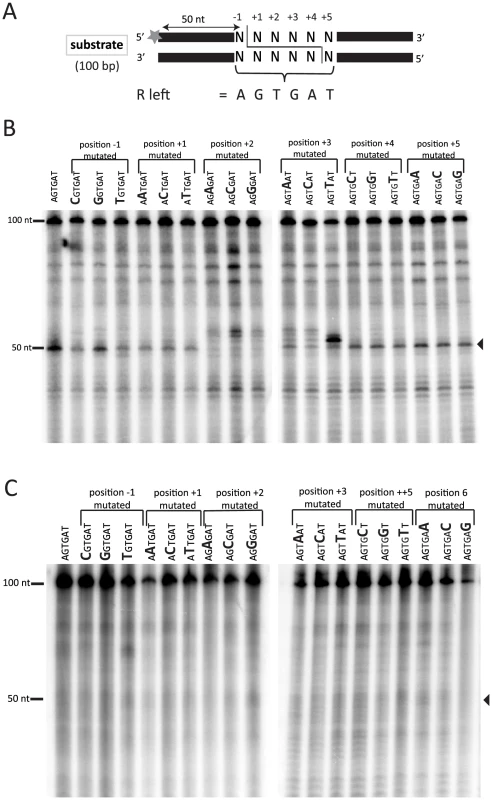
(A) Schematic drawing of the oligo DNA substrates. The substrates were 100 bp long, and 6 bp boundary sequences from the R-IES (5′-AGTGAT-3′) or those having mutated versions of the boundary, in which every position was substituted to every other possible nucleotide, were placed at the 50th to 55th position, which were designated positions −1 to +5. The top strands of the substrates were 32P-labeled at their 5′-ends. The expected cleavage site, where a 4-base 5′-overhang DSB is expected to occur during DNA elimination in vivo, is after the 50th nucleotide. (B, C) The substrates were incubated with the wild-type Tpb2p (B) or the catalytic dead Tpb2p (Tpb2p-CD) (C) expressed recombinantly in E. coli. The products were separated in a denaturing polyacrylamide gel and visualized by autoradiography. The positions of the 50 and 100 nt markers separated in the same gel are shown on the left. Because not all boundary sequences of IESs share 5′-TG-3′ at their +2 and +3 positions [22], [37], [38], this dinucleotide sequence cannot be the sole sequence feature recognized by Tpb2p. Tpb2p may recognize multiple different sequence features, including 5′-TG-3′. Alternatively, Tpb2p may recognize some complex combinatorial sequence feature, which is shared in all boundary sequences of IESs and was not completely disrupted by the single-base substitutions in this study, and 5′-TG-3′ may be a part of this combinatorial sequence feature. From this study, we can conclude that the endonuclease activity of Tpb2p has a relaxed but not completely identified sequence preference for its substrates.
The in vivo effect of base replacement of the left R-IES boundary sequence strongly indicates that Tpb2p is the excisase
Although our genetic analyses indicated that the endonuclease activity of Tpb2p is required for DNA elimination (Fig. 3), none of the results directly demonstrated that Tpb2p is the “excisase,” the enzyme that cuts out IESs in vivo. Above, we observed that the base substitution of +2 or +3 positions of the left R-IES boundary force Tpb2p to cleave a few bases downstream in vitro (Fig. 5B). If the same base substitutions at the left R-IES boundary forced a shift in the position of DSB downward in vivo, it would support the assignation of the excisase function to Tpb2p.
A previously established, in vivo IES elimination assay was used to test this possibility. It has been demonstrated that IESs introduced into a non-coding region of the ribosomal DNA (rDNA) are removed precisely as their endogenous counterparts, albeit with lower efficiency, when the rDNA construct is introduced into the developing new Mac [26]. We prepared three different rDNA constructs (Fig. 6A, right top) having the R-IES and its flanking regions with 1) no base substitution (WT construct); 2) T to G substitution at position +2 of the left boundary (T+2G construct); and 3) G to T substitution at position +3 of the left boundary (G+3T construct). These constructs were introduced into the new Mac of conjugating wild-type cells by electroporation (Fig. 6A). Twenty-four sexual progeny possessing the transgenic rDNA were pooled for each construct, and their genomic DNA was analyzed by PCR to observe elimination of the R-IES of the introduced rDNA (Fig. 6A, bottom).
Fig. 6. In vivo elimination assay using mutated R-IES boundaries. 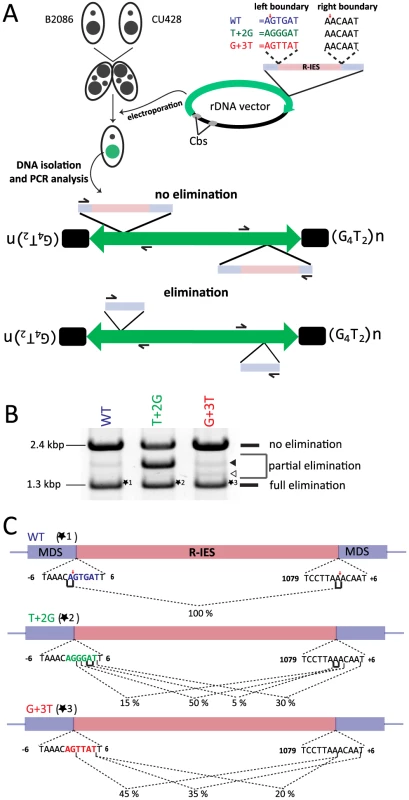
(A) Two wild-type strains (B2086 and CU428) were mated, and the rDNA vector containing the R-IES and its flanking regions was introduced into the new Mac of their progeny. The left boundary of the R-IES was WT, the position +2 T was mutated to G (T+2G) or the position +3 G was mutated to T (G+3T). The introduced circular rDNA vector was rearranged into an rDNA “minichromosome” in which two copies of rDNA are joined in inverted orientations and telomeres are formed at the ends. The R-IES inserted in the 3′ non-coding region of the rDNA is subjected to DNA elimination similar to the endogenous R-IES [26]. The reported cleavage positions at the endogenous R-IES locus are indicated by red arrows. (B) Twenty-four progeny from each construct were pooled, and their genomic DNA was analyzed by PCR using the primer set shown in (A) to observe the elimination of R-IES from the rDNA. The PCR products were separated by agarose gel electrophoresis. The quickly migrating products (∼1.3 kbp, marked with *1, *2 or *3) correspond with the rDNA locus where the full (or nearly full) length of R-IES was eliminated (full elimination). The most slowly migrating product (∼2.4 kbp) bands correspond with the rDNA locus where no R-IES elimination occurred (no elimination). Some products (open and closed arrowheads) migrating between the two products correspond with the rDNA locus where the R-IES were partially eliminated (partial elimination) (C) The ∼1.3-kbp PCR products, marked with *1, *2 or *3 in (B), were cloned, and sequences of 48 clones from each rDNA construct were analyzed to assign cleavage sites. The lines under the boundary sequences indicate the positions where the cleavage sites were assigned at exact positions. The brackets indicate the positions where the cleavage site could not be mapped at exact positions due to sequence redundancies (direct repeats). The fractions of the cleavage sites among the 48 observed clones are shown. The reported cleavage positions at the endogenous R-IES locus are indicated by red arrows. First, the PCR products were analyzed by gel electrophoresis. Two major PCR products were detected from the cells transformed with the WT construct (Fig. 6B, WT). The shorter (1.3 kbp) and longer (2.4 kbp) major products correspond with the R-IES locus on rDNA with or without IES elimination, respectively. A minor PCR product (1.8 kbp), in which a part of R-IES was eliminated (data not shown), was also detected (Fig. 6B, WT, closed arrowhead). Similar PCR products were also detected from cells transformed with the T+2G construct (Fig. 6B, T+2G) and the G+3T construct (Fig. 6B, G+3T). In the cells transformed with the T+2G construct, 1.8-kbp product(s) became as prominent as the other two products (Fig. 6B, T+2G, closed arrowhead). In the cells transformed with the G+3T construct, an extra 1.5-kbp product was detected (Fig. 6B G+2T, open arrowhead). Sequencing analysis revealed that this product had a short deletion in the IES (data not shown). These results indicate that the substitutions at position +2 or +3 of the left boundary change frequencies of occurrence of alternative boundaries in vivo.
Next, the 1.3-kbp products from cells transformed with the different constructs (*1, *2 and *3 in Fig. 6B) were extracted from gel and cloned, and DNA sequences of 20 clones each were analyzed. All of the sequenced 1.3-kbp products from the cells transformed with the WT construct had the same elimination boundary (Fig. 6C, WT), which exactly corresponded with the reported boundary of endogenous R-IES [39] (Fig. 6C, red arrows). In contrast, in all of the sequenced 1.3-kbp products from the cells transformed with the T+2G and G+3T constructs, the left elimination boundaries shifted one to several bases downstream (Fig. 6C, T+2G, G+3T). Interestingly, in the T+2G and G+3T constructs, the choice of the right boundary, which had no base substitutions, was also affected (Fig. 6C, T+2G, G+3T). These results may suggest that there is crosstalk between the ends of an IES during DNA elimination. This crosstalk could be established before DNA cleavage, as would be expected if Tpb2p acts as a typical DNA transposase [40], or during the repair of the excision site as suggested by Saveliev and Cox [22]. Importantly, the observed shifts of the left boundary by the T+2G and G+3T substitutions in the in vivo assay (Fig. 6C, T+2G, G+3T) correlate well with, although not identical to, the patterns of shifts of the cleavage position by the corresponding base substitutions in the in vitro Tpb2p endonuclease assay (Fig. 5B). These results, together with the facts that Tpb2p has an endonuclease activity with wide substrate specificity and is necessary for DNA elimination, strongly suggest that Tpb2p is the excisase, the enzyme that cuts IES boundaries in vivo.
Discussion
In this study, we dissected the roles of the Tetrahymena piggyBac transposase-like protein Tpb2p in DNA elimination. We observed that Tpb2p has two genetically separable functions: the heterochromatin body forming activity, which resides in its cysteine-rich domain, and the DNA excision activity, which requires the endonuclease domain of Tpb2p. Furthermore, our in vitro biochemical studies indicated that the cysteine-rich domain of Tpb2p interacts with heterochromatin-specific histone modifications, and the endonuclease activity of Tpb2p has relaxed sequence specificity with its substrates. Here, we discuss how these biochemical features of Tpb2p potentially determine the reproducible occurrence of IES boundaries in vivo and how these features have been formed during the course of the evolution of DNA elimination in ciliates.
How does Tpb2p execute the reproducible DNA elimination?
We demonstrated that the cysteine-rich domain of Tpb2p directly interacts with the N-terminal tail of histone H3, and the interaction is significantly enhanced by the heterochromatin-specific histone modifications H3K9me3 and H3K27me3 in vitro (Fig. 4). Because these histone modifications specifically occur on IESs in the developing new Mac [18], [19], Tpb2p can be recruited to IESs through its direct interaction to H3K9me3 and H3K27me3, and this recruitment may limit the occurrence of Tpb2p-endonuclease cleavage to the near surrounding heterochromatic regions. The interaction between Tpb2p and H3K9/K27me3 may specifically activate the Tpb2p-endonuclease to inhibit Tpb2p to form DNA DSB at non-IES loci. An IES is removed as one piece of DNA [22]–[24]. Therefore, although H3K9me3 and H3K27me3 likely occur throughout an IES [18], [19], the endonucleolytic cleavages of Tpb2p must be restricted to the ends of an IES. Because H3K9me3 and H3K27me3 are also bound by one of the most abundant heterochromatin components, Pdd1p [18], [19], competition of chromatin-binding sites with Pdd1p may exclude Tpb2p from the body of heterochromatin and only allow Tpb2p to bind the edges of heterochromatin regions. Alternatively, Tpb2p may localize throughout the heterochromatin segment, whereas the heterochromatin structure or some heterochromatin proteins may inhibit the action of Tpb2p endonuclease at the body of heterochromatin. Future research should clarify the spatial localization of Tpb2p on chromatin, which will help with understanding how Tpb2p acts only at the ends of IESs.
Regardless of what molecular mechanism limits the nucleolytic action of Tpb2p to the IES ends, the heterochromatin-Tpb2p interaction does not appear to be sufficient to explain the reproducible occurrence of the border of IESs because 1) histone modifications are able to determine a chromatin segment only at the level of a size of a nucleosome, whereas most of the boundaries of IESs occur within a few to several nucleotides, and 2) there are IESs in Tetrahymena that have sizes similar to a single nucleosome [15]. This study demonstrated that the Tpb2p endonuclease has a relaxed sequence preference for its substrate. For the longer IESs, the combination of the heterochromatin localization and the sequence-biased action likely allows Tpb2p to precisely determine the boundaries of IESs at a sub-nucleosomal level. For the shorter IESs, it is possible that the substrate preference of the Tpb2p endonuclease alone is sufficient to determine the precise boundary. Consistent with this idea, many of the shorter IESs have a common 5′-TTAA-3′ sequence at their boundaries [15] on which DNA DSB is efficiently introduced by the endonuclease Tpb2p in vitro [4].
In addition to the heterochromatin-Tpb2p interaction and sequence-biased action of the Tpb2p endonuclease, the cis-acting sequences adjacent to IESs may also play a role in reproducibly determining the border of IESs. Some IESs have cis-acting sequences located 40–50 nucleotides outside of the boundaries of IESs that are necessary in cis for the precise occurrence of DNA elimination boundaries [26], [28], although the mechanism explaining how cis-regulatory elements are involved in DNA elimination is unclear. Because Tpb2p can induce DSB at the IES boundary sequences in oligo DNAs without cis-regulatory elements in vitro (Fig. 5), cis-regulatory elements are not necessary for Tpb2p to recognize the IES boundary sequences, at least on naked DNA. The cis-acting sequences may set nucleosome positioning, and thus, Tpb2p can be recruited to a fixed chromosomal location. Alternatively, the cis-acting sequences may create a nucleosome-free region where DNA is accessible for Tpb2p.
The relationship between the heterochromatin bodies and DNA elimination
The fact that the endonucleolytically inactive Tpb2p still supports heterochromatin body formation (Fig. 3E, “CD”) suggests that the heterochromatin bodies are not a product of DNA elimination but can be formed prior to the initiation of DNA elimination. In contrast, it has been reported that, in the absence of TKU80, the excision of IESs occurs without the formation of the heterochromatin bodies [10]. One fact we should consider to reconcile these seemingly incompatible observations is that the endonucleolytically inactive Tpb2p mutant was expressed in the conditional TPB2 KO background. In the conditional TPB2 KO locus, TPB2 expression was under control of the MTT1 promoter, which can be activated by addition of the cadmium ion. It is known that the MTT1 promoter is leaky [26]. Therefore, although we used a medium containing minimum metals and we could not detect the wild-type Tpb2p by western blotting in the absence of the cadmium ion (Fig. 3B), it is still possible that undetectable amount of Tpb2p from the conditional TPB2 KO locus induces some DNA elimination even without the cadmium ion in the medium. Nonetheless, because no circularized IES products were detected without inducing the wild-type TPB2 expression in the conditional TPB2 KO cells (Fig. 3H, WT rescue −Cu2+), leaky expression of TPB2, if any, causes no or very little IES excision event. Therefore, we conclude that the heterochromatin bodies can be formed without massive DNA elimination. On the other hand, the results reported by Lin et al. (2012) [10] indicate that massive DNA elimination is not sufficient to induce the formation of the heterochromatin bodies in the absence of TKU80. Because TKU80 encodes a KU80 homolog, which binds and senses the end of DNA double-strand breaks, the formation of the heterochromatin bodies may be triggered by a signaling cascade down stream of the KU80-mediated DNA double-strand break sensing. A leaky expression of the wild-type Tpb2p may be enough to activate this signaling cascade, and together with the expression of the endonucleolytically inactive Tpb2p, it may induce the heterochromatin body formation.
The domesticated piggyBac transposase Tpb2p and evolution of DNA elimination
DNA elimination pathways in ciliates are believed to have evolved as a genome defense mechanism against pathogenic invaders, such as transposons [41]. In Tetrahymena, there are several different types of transposons that do not share common boundary sequences, and the previous study demonstrated that Tpb2p is necessary for the elimination of all IESs tested, including the Tlr1 retrotransposon-like element [4]. Therefore, a single molecular mechanism including Tpb2p most likely executes the elimination of all transposons in Tetrahymena.
Tpb2p is evolutionarily related to piggyBac transposases. Tpb2p and piggyBac transposases share a common molecular architecture: the endonuclease domain possessing a DDD catalytic core and the zinc-finger-like cysteine-rich domain that is related to the PHD-finger motif. Although Tpb2p induces DSB at a variety of sequences (Fig. 5), piggyBac transposases specifically cut the 5′-TTAA-3′ sequence [34]. Because insertions of piggyBac transposase in vivo are negatively correlated with the presence of heterochromatin [42], the cysteine-rich domain of piggyBac transposase, in contrast with the domain of Tpb2p, is unlikely to interact with heterochromatin. Therefore, two important changes must have occurred in a piggyBac transposase during the evolution of DNA elimination in ciliates: loss of the strict sequence specificity for its substrate and gaining the ability to interact with the heterochromatin-specific histone H3 modifications.
Although the oligohymenophorean ciliates Paramecium and Tetrahymena use piggyBac transposases for DNA elimination [4], [35], the spirotrich ciliate Oxytricha uses Tc1/mariner class transposases for DNA elimination [43]. Therefore, the involvement of domesticated piggyBac transposases in DNA elimination has most likely evolved in the lineage of oligohymenophorean ciliates. Although IESs in Tetrahymena are not flanked by any common sequence, IESs in Paramecium are flanked by a 5′-TA-3′ [44], [45]. This indicates that the Paramecium piggyBac transposase-like protein Pgm still maintains a certain sequence specificity derived from an ancestral piggyBac transposase that cuts the 5′-TTAA-3′ sequence, whereas the Tetrahymena Tpb2p has evolved to recognize a much greater variety of sequences. Therefore, it appears that the piggyBac transposase has gradually lost its sequence specificity to the substrate during the evolution of oligohymenophorean ciliates.
Relaxation of the substrate specificity of Tpb2p might be compensated for by the heterochromatin-binding ability of the cysteine-rich domain of Tpb2p because heterochromatin is specifically formed on IESs prior to DNA elimination [18], [19]. Heterochromatin formation on IESs is targeted by an RNAi-related mechanism in Tetrahymena [18]. Because transposons in many different eukaryotes are silenced by heterochromatin formation induced by RNAi-related pathways [46], we speculate that heterochromatin formation on IESs in the Tetrahymena lineage evolved from an ancient transposon-silencing mechanism and co-existed in parallel with the DNA elimination, even before the involvement of a piggyBac transposase in the DNA elimination. Such ancient DNA elimination system might be operated by transposases encoded by eliminated transposons, like we see today in Oxytricha [41]. A piggyBac transposon in the Tetrahymena lineage might have first evolved to target heterochromatin, and then, its transposase might have been domesticated to overtake the roles of transposon-encoded transposases in DNA elimination.
Although DNA elimination is widely observed among most ciliates studied, the enzymes used for DNA elimination in different groups of ciliates have distinct properties and even have different transposon origins. Future studies of Tetrahymena Tpb2p and DNA elimination enzymes of other diverse groups of ciliates would help to further understanding of how a transposon has been domesticated for use in regulating a eukaryotic genome.
Materials and Methods
Strains and culture conditions
The wildtype strains B2086 and CU428 were provided by the Tetrahymena Stock Center (http://Tetrahymena.vet.cornell.edu/). Cells were cultured in 1×SPP medium with 2% proteose peptone at 30°C [47]. TPB2 cKO and TPB2 rescue strains were grown in the metal-depleted medium 1×SPPCT to minimize gene expressions from metal-inducible promoters. To make 1×SPPCT, 1×SPP medium was incubated with 5 g/100 ml of Chelex-100 resin (BioRad) with stirring for 2 hr. After filtration to remove the resin, essential trace metals were added (100×stock solution: FeCl3*H2O: 1 mg/ml, Co(NO3)2*6H2O, MnSO4*4H2O: 0.16 mg/ml). Mating was induced by mixing equal numbers of starved cells. Starvation was achieved in 10 mM Tris buffer (pH 7.5) for 12–16 hr at 30°C.
Oligo DNAs
The oligo DNAs used in this study are shown in Supplementary Table S1.
Creation of conditional TPB2 KO (TPB2 cKO) strains
A total of 475 bp of the TPB2 5′ flanking region and the first 1132 bp of the TPB2 genomic sequence were amplified from genomic DNA with the primer pairs TN5MT 5′ fw/rv and TN5MT3′ fw/rv, respectively. The neo5 cassette fused to an MTT1 promoter was amplified from a pMNMM3 vector with the primers TN5MTneo fw/rv [48]. After PCR purification, the fragments were combined using overlapping PCR as described previously [49], resulting in the TPB2 cKO construct, which was directly used for germline transformation of mating Tetrahymena UMPS strains at 3 hr post-mixing. The UMPS strains were created by introducing a uridine monophosphate synthase (UMPS) gene from Dictyostelium into the Mac of Tetrahymena. The expression of this gene makes Tetrahymena cells sensitive to 5-fluoroorotic acid (5-FOA). After conjugation, these strains should lose the UMPS gene because it is only in the Mac, and thus, the progeny are 5-FOA resistant. Biolistic transformation was performed as previously described [50]. After transformation of the TPB2 cKO construct into the UMPS strains, 0.1 µg/ml cadmium chloride was added to the cells to induce TPB2 expression from the MTT1 promoter. One paromomycin and 5-FOA resistant clone was obtained, which was confirmed as a heterozygous TPB2 cKO strain. After mating to the WT strain, the heterozygous conditional KO strains were genotyped by PCR and crossed to each other to obtain homozygous conditional KO strains. During all matings, 0.1 µg/ml cadmium chloride was added.
TPB2 rescue system using TPB2 cKO strains
A blasticidin-resistance cassette was amplified by PCR from pBla1 vector using the primers Bra1_OL_5RACErv and Bra1_OL_fw. In parallel, the MTT2 promoter was amplified from the pDET2 vector with the primers MTT2_OL_fw and MTT2_HA_AvrII_rv. The two PCR constructs were combined with overlapping PCR as described previously [49]. The overlapping PCR product was cloned into pMNMM1 with AvrII and SalI restriction enzymes, resulting in pMBM2M. The TPB2 open reading frame was amplified from genomic DNA (strain B2086) using the primers TPB2ORF_AvrII_fw and TPB2ORF_MluI_rv and subsequently cloned with the enzymes MluI and AvrII in pMBM2M, resulting in pMBM2M-TPB2. Via site-directed mutagenesis, a catalytically dead version and cysteine-rich mutant were created from the template pMBM2M-TPB2 using the primers TPB2_D297L_fw/rv, TPB2_D379L_fw/rv and TPB2_D495L_fw/rv or TPB2_C618A_fw/rv or C629A_fw/rv, respectively. These rescue vectors were then transformed into the somatic nucleus of two different mating types of conditional TPB2 KO strains by ballistic transformation as described previously [50]. The transformants were selected with blasticidin and phenotypically assorted (up to 10 mg/ml). To assess the phenotype of the TPB2 rescue strains, they were crossed with each other, and the expression of the respective rescue construct was induced by the addition of copper sulfate in a two-step procedure. Equal amounts of CuSO4 were added at 7 and 8 hr post-mixing, for a final concentration of 100 µM.
Transformation of Tetrahymena thermophila with ribosomal vector
The Mic genomic region containing the R-IES was amplified as two overlapping pieces from the Tetrahymena total genomic DNA using the primers R-leftFW/R-midRV and R-midFW/R-rightRV. The two pieces were combined by overlapping PCR and cloned into the ribosomal vector pD5H8 [51] using the NotI restriction site. pD5H8 containing the R-IES, including the flanking regions, was electroporated into mating wild-type strains. Electroporation was performed as described previously [52] with slight modifications. Mating WT cells in 10 mM Tris (concentration: 7×10∧5 cells/ml) were used for transformation at 8.5 hr post-mixing. The cells were washed in 10 mM HEPES pH 7.5 and resuspended in 120 µl of 10 mM HEPES and then mixed with 120 µl of plasmid DNA in 10 mM HEPES (300 ng/µl) and electroporated (220 V, 50 Ω, 50 µF, exponential pulse) using a BioRad Gene Pulser MXcell. After transformation, the cells were incubated in 1×SPP medium at 30°C overnight without shaking. Transformants were selected in 100 µg/ml paromomycin.
Immunofluorescence analysis
Cells were fixed in 3.7% formaldehyde and 0.5% Triton-X 100 for 30 min at room temperature. The cells were resuspended in 3.7% formaldehyde and 3.4% sucrose and dried on Superfrost Ultra Plus slides (Thermo Fisher). The samples were blocked for 2 hr with 3% BSA, 10% normal goat serum (Invitrogen) and 0.1% Tween 20 in PBS, followed by overnight incubation at 4°C in blocking solution containing a 1∶1000 dilution of anti-HA (Covance), 1∶2000 dilution of rabbit anti-Pdd1p (Abcam), 1∶2000 dilution of guinea pig anti-Pdd1p or 1∶2000 dilution of anti-Tpb2p antiserum. The guinea pig anti-Pdd1p antibody was obtained by immunizing a guinea pig with a peptide (CTAHRSGSRLSQIQSNANQV). The anti-Tpb2p antibody was obtained by immunizing a rabbit with N-terminal half (1 aa to 556 aa) of Tpb2p. After washing, the samples were incubated with a 1∶2000 dilution of secondary antibody against mouse or rabbit conjugated to Alexa 488 or Alexa 568 (Invitrogen). The samples were washed, incubated with 10 ng/ml of DAPI (Sigma) in PBST, and observed by fluorescent microscopy.
DNA elimination assays
The DNA elimination assay using FISH was performed as previously described [53]. The plasmids Tlr1IntB, Tlr1 2 and Tlr1 4C1 [30] were mixed as templates to make probes against the Tlr1 IES. The labeling of the DNA with Cy3 was achieved by nick translation. Cells were fixed at 36 hr post-mixing as described above for immunofluorescence analysis.
The excision of the mse2.9 - and the R-IES elements were examined by detecting circularized IESs by nested PCR using the primers listed in Supplementary Table S1.
Progeny viability assay
Cells were mated at the cell density of 5×105 cells/ml and single pairs were picked into a drop of metal depleted SPP medium (1×SPPCT). Pairs from the WT mating were picked at 7 h post-mixing. Tpb2 expression from the MTT2 cassette was induced in the rescued strains as described earlier and pairs were picked at around 10 h post mixing. As a control, WT rescue strain without addition of copper was used. At 24 h post-mixing 1×SPP medium was added to the drops to recover normal growth speed of the surviving cells. Sexual progeny formation was confirmed either by their 6-methylpurine resistance in the wild-type cells or by their blasticidin sensitivity in the rescue strains.
Production of recombinant proteins
A codon-optimized TPB2 coding region was amplified by PCR from the previously created vectors pGEX-TPB2 or pGEX-TPB2-CD [4] using the primers T2CPfw/T2ECrv. The PCR products were cut with EcoRI and XhoI and cloned into the EcoRI and SalI sites of the pMalC2X vector to obtain pMAL-TPB2 and pMAL-TPB2-CD. To generate pMAL-TPB2-CRM, C618 to A and C629 to A mutations were introduced into pMAL-TPB2 by site-directed mutagenesis using the DNA oligos T2EC C618A fw/rv and T2EC C629A fw/rv. To create the cysteine rich domain or its point mutant fused to the MBP protein, the primers TPB2_CRD_fw/rv were used with pMAL-TPB2 or pMAL-TPB2-CRM.
The plasmids were introduced into the E. coli strain BL21(DE3), which was cultivated to an A600 of ∼0.8 and then incubated with 0.5 mM IPTG for 10 hr at 16°C. The cells were lysed in 500 mM NaCl, 80 mM Tris, pH 8.0, 0.2 mM PMSF and 1×complete proteinase inhibitor cocktail (Roche). The lysate was incubated with Amylose resin (NEB) at 4°C, washed with 500 mM NaCl and 80 mM Tris pH 8.0 and finally eluted with 500 mM NaCl, 80 mM Tris, pH 8.0, and 20 mM maltose, followed by dialysis in 20 mM Tris-HCl, pH 7.5, 100 mM KCl, 4 mM MgCl2, 4 mM MnSO4 and 10% glycerol.
Tpb2p endonuclease assay
The endonuclease assay was performed as previously described [4]. The oligomeric DNA substrates used for the experiments are listed in Supplementary Table S1.
Histone-peptide pull-down assay
Peptides corresponding to the N-terminal tail of histone H3 were synthesized and biotin-tagged at their C terminal with a PEG linker (Supplementary Table S2). In addition, 10 µl (bed volume) of streptavidin-coupled Dynabeads (Invitrogen) were blocked with 2% BSA in interaction buffer (10 mM Tris pH 7.5, 0.1 mM ZnSO4, 0.05% NP40 and 250 mM NaCl) for 1 hr at RT and then incubated with 1 µg of peptide in the blocking solution for 30 min at RT. After washing with interaction buffer and blocking again with 2% BSA 1 µg of MBP-Tpb2p-CRD was added to the beads and incubated overnight at 4°C. After five washing steps with interaction buffer, followed by two washing steps in PBS-T, the beads were resuspended in SDS-PAGE loading buffer, transferred to a fresh tube and boiled for 5 min. The samples were then separated on a 10% SDS-polyacrylamide gel, followed by a western blot. Detection was accomplished using anti-MBP antibody (NEB). The secondary antibody was coupled with an infrared dye, which was visualized with an Odyssey scanner (LI-COR Biosciences).
Supporting Information
Zdroje
1. OrgelLE, CrickFH (1980) Selfish DNA: the ultimate parasite. Nature 284 : 604–607.
2. AlmeidaR, AllshireRC (2005) RNA silencing and genome regulation. Trends Cell Biol 15 : 251–258 doi:10.1016/j.tcb.2005.03.006
3. VolffJ-N (2006) Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28 : 913–922 doi:10.1002/bies.20452
4. ChengC-Y, VogtA, MochizukiK, YaoM-C (2010) A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell 21 : 1753–1762 doi:10.1091/mbc.E09-12-1079
5. PrescottDM (1994) The DNA of ciliated protozoa. Microbiol Rev 58 : 233–267.
6. EisenJA, CoyneRS, WuM, WuD, ThiagarajanM, et al. (2006) Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol 4: e286 doi:10.1371/journal.pbio.0040286
7. HamiltonE, BrunsP, LinC, MerriamV, OriasE, et al. (2005) Genome-wide characterization of tetrahymena thermophila chromosome breakage sites. I. Cloning and identification of functional sites. Genetics 170 : 1611–1621 doi:10.1534/genetics.104.031401
8. FanQ, YaoM (1996) New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol Cell Biol 16 : 1267–1274.
9. YaoMC, ChoiJ, YokoyamaS, AusterberryCF, YaoCH (1984) DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell 36 : 433–440.
10. LinI-T, ChaoJ-L, YaoM-C (2012) An essential role for the DNA breakage-repair protein Ku80 in programmed DNA rearrangements in Tetrahymena thermophila. Mol Biol Cell 23 : 2213–2225 doi:10.1091/mbc.E11-11-0952
11. KapustaA, MatsudaA, MarmignonA, KuM, SilveA, et al. (2011) Highly precise and developmentally programmed genome assembly in Paramecium requires ligase IV-dependent end joining. PLoS Genet 7: e1002049 doi:10.1371/journal.pgen.1002049
12. SchoeberlUE, KurthHM, NotoT, MochizukiK (2012) Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev 26 : 1729–1742 doi:10.1101/gad.196493.112
13. CoyneRS, StoverNA, MiaoW (2012) Whole genome studies of Tetrahymena. Methods Cell Biol 109 : 53–81 doi:10.1016/B978-0-12-385967-9.00004-9
14. ChalkerDL, YaoM-C (2011) DNA elimination in ciliates: transposon domestication and genome surveillance. Annu Rev Genet 45 : 227–246 doi:10.1146/annurev-genet-110410-132432
15. FassJN, JoshiNA, CouvillionMT, BowenJ, GorovskyMA, et al. (2011) Genome-Scale Analysis of Programmed DNA Elimination Sites in Tetrahymena thermophila. G3 (Bethesda) 1 : 515–522 doi:10.1534/g3.111.000927
16. SchoeberlUE, MochizukiK (2011) Keeping the soma free of transposons: programmed DNA elimination in ciliates. J Biol Chem 286 : 37045–37052 doi:10.1074/jbc.R111.276964
17. KataokaK, MochizukiK (2011) Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism. Adv Exp Med Biol 722 : 156–173 doi:_10.1007/978-1-4614-0332-6_10
18. LiuY, MochizukiK, GorovskyMA (2004) Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci USA 101 : 1679–1684 doi:10.1073/pnas.0305421101
19. LiuY, TavernaSD, MuratoreTL, ShabanowitzJ, HuntDF, et al. (2007) RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev 21 : 1530–1545 doi:10.1101/gad.1544207
20. MadireddiMT, CoyneRS, SmothersJF, MickeyKM, YaoMC, et al. (1996) Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell 87 : 75–84.
21. SmothersJF, MadireddiMT, WarnerFD, AllisCD (1997) Programmed DNA degradation and nucleolar biogenesis occur in distinct organelles during macronuclear development in Tetrahymena. J Eukaryot Microbiol 44 : 79–88.
22. SavelievSV, CoxMM (2001) Product analysis illuminates the final steps of IES deletion in Tetrahymena thermophila. EMBO J 20 : 3251–3261 doi:10.1093/emboj/20.12.3251
23. SavelievSV, CoxMM (1994) The fate of deleted DNA produced during programmed genomic deletion events in Tetrahymena thermophila. Nucleic Acids Res 22 : 5695–5701.
24. YaoMC, YaoCH (1994) Detection of circular excised DNA deletion elements in Tetrahymena thermophila during development. Nucleic Acids Res 22 : 5702–5708.
25. TavernaSD, CoyneRS, AllisCD (2002) Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110 : 701–711.
26. ChalkerDL, La TerzaA, WilsonA, KroenkeCD, YaoMC (1999) Flanking regulatory sequences of the Tetrahymena R deletion element determine the boundaries of DNA rearrangement. Mol Cell Biol 19 : 5631–5641.
27. GodiskaR, JamesC, YaoMC (1993) A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev 7 : 2357–2365.
28. GodiskaR, YaoMC (1990) A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell 61 : 1237–1246.
29. ShangY, SongX, BowenJ, CorstanjeR, GaoY, et al. (2002) A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci USA 99 : 3734–3739 doi:10.1073/pnas.052016199
30. WellsJM, EllingsonJL, CattDM, BergerPJ, KarrerKM (1994) A small family of elements with long inverted repeats is located near sites of developmentally regulated DNA rearrangement in Tetrahymena thermophila. Mol Cell Biol 14 : 5939–5949.
31. AusterberryCF, AllisCD, YaoMC (1984) Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci USA 81 : 7383–7387.
32. BoldrinF, SantovitoG, FormigariA, BisharyanY, Cassidy-HanleyD, et al. (2008) MTT2, a copper-inducible metallothionein gene from Tetrahymena thermophila. Comp Biochem Physiol C Toxicol Pharmacol 147 : 232–240 doi:10.1016/j.cbpc.2007.10.002
33. KeithJH, SchaeperCA, FraserTS, FraserMJ (2008) Mutational analysis of highly conserved aspartate residues essential to the catalytic core of the piggyBac transposase. BMC Mol Biol 9 : 73 doi:10.1186/1471-2199-9-73
34. MitraR, Fain-ThorntonJ, CraigNL (2008) piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J 27 : 1097–1109 doi:10.1038/emboj.2008.41
35. BaudryC, MalinskyS, RestituitoM, KapustaA, RosaS, et al. (2009) PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev 23 : 2478–2483 doi:10.1101/gad.547309
36. BienzM (2006) The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31 : 35–40 doi:10.1016/j.tibs.2005.11.001
37. KatohM, HironoM, TakemasaT, KimuraM, WatanabeY (1993) A micronucleus-specific sequence exists in the 5′-upstream region of calmodulin gene in Tetrahymena thermophila. Nucleic Acids Res 21 : 2409–2414.
38. AusterberryCF, SnyderRO, YaoMC (1989) Sequence microheterogeneity is generated at junctions of programmed DNA deletions in Tetrahymena thermophila. Nucleic Acids Res 17 : 7263–7272.
39. SavelievSV, CoxMM (1996) Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. EMBO J 15 : 2858–2869.
40. DydaF, ChandlerM, HickmanAB (2012) The emerging diversity of transpososome architectures. Q Rev Biophys 45 : 493–521 doi:10.1017/S0033583512000145
41. KlobutcherLA, HerrickG (1997) Developmental genome reorganization in ciliated protozoa: the transposon link. Prog Nucleic Acid Res Mol Biol 56 : 1–62.
42. WangH, MayhewD, ChenX, JohnstonM, MitraRD (2012) “Calling cards” for DNA-binding proteins in mammalian cells. Genetics 190 : 941–949 doi:10.1534/genetics.111.137315
43. NowackiM, HigginsBP, MaquilanGM, SwartEC, DoakTG, et al. (2009) A functional role for transposases in a large eukaryotic genome. Science 324 : 935–938 doi:10.1126/science.1170023
44. SteeleCJ, Barkocy-GallagherGA, PreerLB, PreerJR (1994) Developmentally excised sequences in micronuclear DNA of Paramecium. Proc Natl Acad Sci USA 91 : 2255–2259.
45. ArnaizO, MathyN, BaudryC, MalinskyS, AuryJ-M, et al. (2012) The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet 8: e1002984 doi:10.1371/journal.pgen.1002984
46. GrewalSI (2010) RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20 : 134–141 doi:10.1016/j.gde.2010.02.003
47. GorovskyMA, YaoMC, KeevertJB, PlegerGL (1975) Isolation of micro - and macronuclei of Tetrahymena pyriformis. Methods Cell Biol 9 : 311–327.
48. BuschCJ-L, VogtA, MochizukiK (2010) Establishment of a Cre/loxP recombination system for N-terminal epitope tagging of genes in Tetrahymena. BMC Microbiol 10 : 191 doi:10.1186/1471-2180-10-191
49. KataokaK, SchoeberlUE, MochizukiK (2010) Modules for C-terminal epitope tagging of Tetrahymena genes. J Microbiol Methods 82 : 342–346 doi:10.1016/j.mimet.2010.07.009
50. BrunsPJ, Cassidy-HanleyD (2000) Biolistic transformation of macro - and micronuclei. Methods Cell Biol 62 : 501–512.
51. YaoMC, YaoCH (1989) Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol Cell Biol 9 : 1092–1099.
52. Sweet MT, Allis CD (2006) Transformation of Tetrahymena thermophila by Electroporation. CSH Protoc 2006. doi:10.1101/pdb.prot4502
53. LoidlJ, ScherthanH (2004) Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci 117 : 5791–5801 doi:10.1242/jcs.01504
Štítky
Genetika Reprodukčná medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



