-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
Mitosis and meiosis are two distinct cell division programs. During mitosis, sister chromatids separate, whereas during the first meiotic division, homologous chromosomes pair and then segregate from each other. In most organisms, germ cells do both programs sequentially, as they first amplify through mitosis, before switching to meiosis to produce haploid gametes. Here, we show that autosomal chromosomes are unpaired at their centromeres in Drosophila germline stem cells, and become paired during the following four mitosis of the differentiating daughter cell. Surprisingly, we further demonstrate that components of the central region of the synaptonemal complex are already expressed in the mitotic region of the ovaries, localize close to centromeres, and promote de novo association of centromeres. Our results thus show that meiotic proteins and meiotic organization of centromeres, which are key features to ensure reductional segregation, are laid out in amplifying germ cells, before meiosis has started.
Published in the journal: Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells. PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004012
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004012Summary
Mitosis and meiosis are two distinct cell division programs. During mitosis, sister chromatids separate, whereas during the first meiotic division, homologous chromosomes pair and then segregate from each other. In most organisms, germ cells do both programs sequentially, as they first amplify through mitosis, before switching to meiosis to produce haploid gametes. Here, we show that autosomal chromosomes are unpaired at their centromeres in Drosophila germline stem cells, and become paired during the following four mitosis of the differentiating daughter cell. Surprisingly, we further demonstrate that components of the central region of the synaptonemal complex are already expressed in the mitotic region of the ovaries, localize close to centromeres, and promote de novo association of centromeres. Our results thus show that meiotic proteins and meiotic organization of centromeres, which are key features to ensure reductional segregation, are laid out in amplifying germ cells, before meiosis has started.
Introduction
In Drosophila females, mitosis and meiosis occur sequentially throughout adult life in two distinct regions of the germarium at the tip of each ovary [1]. In the mitotic zone (called region 1), germline stem cells (GSCs) generate a precursor cell called a cystoblast (CB), which undergoes exactly four mitoses to produce a germline cyst made of 16 cells (Figure 1A). These mitoses are not complete and all 16 sister cells remain connected through ring canals and by an organelle called the fusome, which links all cells. The branched shape of the fusome is a useful marker to distinguish each stage, GSC, CB, 2-, 4-, 8 - and 16-cell cyst (cc) in the mitotic zone [2]. After the last mitosis, cysts enter region 2 and all 16 cells start meiosis [3]. During differentiation in region 2, only one cell per cyst, however, remains in meiosis, while the 15 others exit meiosis and endoreplicate their DNA [4].
Fig. 1. Dispersed centromeres in germline stem cells become paired during cyst divisions. 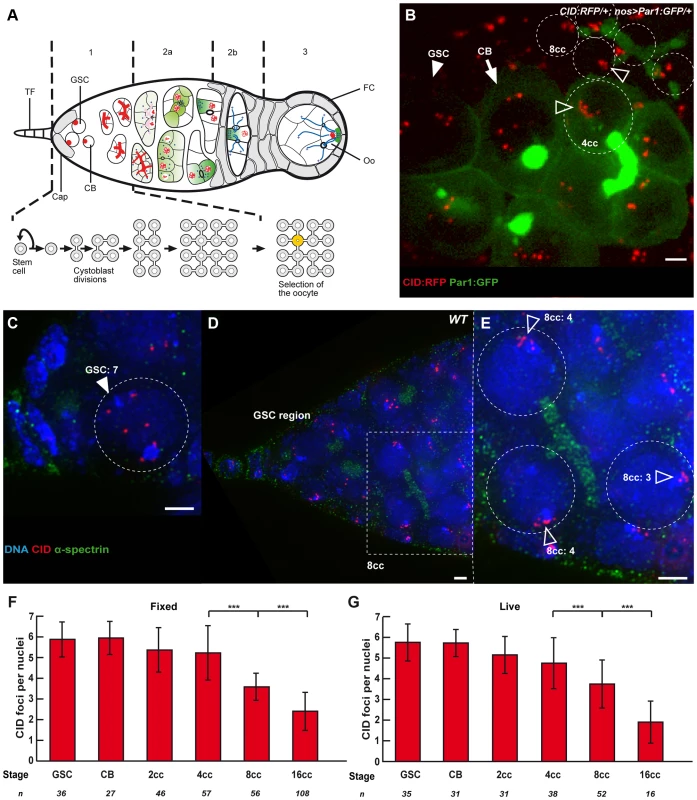
(A) A schematic for developmental stages during Drosophila female meiosis. The egg chambers are produced in a specialized structure, called the germarium, at the anterior of the ovariole. The germarium is divided into four morphological regions along the anterior–posterior axis. The germline stem cells (GSCs) reside at the anterior tip of the germarium (left) and divide to produce cystoblasts, which divide four more times in region 1 to produce 16 cell germline cysts that are connected by ring canals. The stem cells and cystoblasts (CB) contain a spectrosome (red circles), which develops into a branched structure called the fusome, which orients each division of the cyst. In early region 2a, the synaptonemal complex (red lines) forms along the chromosomes of the two cells with four ring canals (pro-oocytes, yellow) as they enter meiosis. The synaptonemal complex then appears transiently in the two cells with three ring canals, before becoming restricted to the pro-oocytes in late region 2a. By region 2b, the oocyte (Oo) has been selected, and is the only cell to remain in meiosis. Terminal Filament (TF), Cap cells (Cap), Follicle Cells (FC). Blue dots are centrosomes migrating into the oocyte. Green shade shows the progressive restriction of some proteins or mRNAs into the oocyte. (B) Projection of Z-sections obtained by time lapse microscopy (spinning disc) of a living CID::RFP/+; nos>Par1::GFP/+ germarium expressing the centromere marker CID::RFP (red) and the fusome marker Par1::GFP (green). One Germline Stem Cell (GSC, arrowhead) is identified by its position close to the cap cells (not shown), one cystoblast (CB, arrow) is identified by a round bright fusome, and a 4-cell cyst (4cc), whose cells are linked by a typical U-shaped fusome, demonstrating that they are from the same cyst. One nucleus of a 4-cell cyst (4cc) and 5 nuclei of an 8-cell cyst (8cc), with fusome GFP, are surrounded by dotted lines. Open arrows indicate centromeres. Scale bar represents 2 µm. (C–E) C is a projection of z-sections, D and E are single sections, all obtained by DV microscopy of a wild-type fixed germarium stained for centromere (CID, red), fusome (α-spectrin, green), and DNA (DAPI, blue). Close up on a GSC (C, GSC) and on 3 nuclei of an 8-cell cyst (E, 8cc) from the corresponding boxed region in D are shown. 7 centromeres can be distinguished in the GSC, while centromeres are clustered (open arrows) in 8-cell cyst nuclei. The number of distinguishable centromeres is indicated for each 8cc nucleus. Note that the GSC close up in C includes more projected images than the corresponding area in D to show the exact number of centromeres. Scale bars represent 2 µm. (F,G) Developmental changes in the number of CID foci for each cell stage in region 1 in fixed wild-type germaria (F) or in living germaria (G). The number of analyzed cells is indicated under each stage. *** p≤0.0005 (two-tailed Student's t-test comparing 4cc with 8cc and 8cc with 16cc). How homologous chromosomes find each other is a central question in reproductive biology. In most organisms, this process involves pairing of homologous chromosomes, formation of the synaptonemal complex (SC), genetic recombination and formation of crossing-over [5]–[6]. In flies, homologous chromosomes are always paired in somatic cells, a phenomenon called “somatic pairing” [7]. Meiotic pairing is thus viewed as an extension of a pre-existing somatic pairing. Live-imaging provided strong support for this hypothesis in males, and in females, it is known that homologous chromosomes are already paired when cysts enter region 2 [8]–[12]. Recently, an additional organization of meiotic chromosomes was described in Drosophila females, whereby centromeres of all chromosomes aggregate into one or two clusters [13]–[14]. This organization is reminiscent of the clustering of telomeres during the bouquet stage in other species. It was further shown that formation of the synaptonemal complex (synapsis) initiates at centromere clusters, and that SC components, such as C(3)G and Corona, were required for centromere clustering in addition to synapsis [13]–[14]. In this study, we set out to explore the organization of chromosomes in the earliest stages of adult germ cell development before the onset of meiosis.
Results
Unpaired centromeres in germline stem cells
To distinguish the different stages of region 1, we marked the fusome with an antibody against α-spectrin, a membrane skeletal protein, and to visualize centromeres, we used an anti-CID antibody marking the Drosophila homologue of Cenp-A, a histone H3 variant present only in centromeric regions [15] (Figure 1C–E). Drosophila diploid cells have eight chromosomes representing 4 pairs of homologues, therefore when homologues are all paired, one should distinguish 4 dots of CID. When centromeres are not all paired, one should see more than 4 dots, and if centromeres are clustered, one should see 1 or 2 dots [13]–[14]. We used the same methodology as published in (Takeo et al., 2011). CID foci were scored as unpaired as soon as a single “black” pixel could be distinguished between the two foci. Overlapping foci were all considered paired. We found in GSCs and CBs an average of 6 dots of CID (GSCs: 5.9±0.8, n = 36; CBs: 5.9±0.8, n = 27), which indicates that centromeres were not all paired (Figure 1F). This result is in contrast with most cell types examined so far by us and others in Drosophila males or females. We also found that centromeres became more associated, as cysts underwent more divisions. Indeed, 2-cell cysts showed an average of 5.3±1.1 CID foci, 4-cell cysts 5.2±1.3 CID foci and it dropped to 3.6±1.3 dots of CID in 8-cell cysts and 2.4±0.9 in 16-cell cysts (Figure 1F). Thus, there was a decrease of almost 50% in the number of CID dots from GSCs to 8-cell cysts, indicating that most 8-cell cysts had already paired their centromeres in the mitotic region. We also observed the appearance of classes of nuclei with only 1 or 2 dots of CID in 8-cell cysts (19,6%, n = 56 ; Figure S1A) indicating a tendency for centromeres to cluster already at this stage. To confirm that our fixation procedure preserved the 3D organization of nuclei in the germarium, we repeated our experiments using live-imaging on intact tissue with CID-RFP and Par-1-GFP to label the fusome (Figure 1B). We found the same numbers of CID dots at every stage confirming our previous results with fixed tissue (Figure 1G). We further found that centromere movements were highly dynamic whether as single, paired or clustered centromeres (Movie S1). Even when centromeres were clustered, it was not a static nor passive maintenance of a Rabl organization [16]. We concluded that several centromeres were unpaired in GSCs, but that most centromeres became associated before the onset of meiosis.
Centromeres of chromosome II and III become associated in the mitotic region, whereas X-chromosome centromeres are always paired
We next investigated the pairing behaviour of individual chromosomes to test if pre-meiotic centromere pairing occurred between homologous chromosomes. We used the dodeca, the AACAC and the 359 probes to label the pericentromeric regions of chromosome III, II and X, respectively [17]. We combined fluorescence in situ hybridisation (FISH) with immunostaining against the fusome marker α-spectrin (Figure 2). We considered that chromosomes were paired when only one focus was visible or when two foci were visible separated by a distance ≤0.70 µm [9]–[10]. We found that the peri-centromeric region of chromosome III and II became paired individually in region 1, with the same timing as observed with the centromere marker CID. In GSCs and CBs only 20.0% (n = 40) and 24.1% (n = 29) of nuclei displayed pairing of the peri-centromeric region of chromosome III (Figure 2A, C). The peri-centromeric region of chromosome II was paired in only 2.9% (n = 34) of GSCs and 10.0% (n = 30) of CBs (Figure 2D, F). In 8-cell cysts 66.2% (n = 71) of nuclei displayed pairing with the dodeca probe, and 71% (n = 51) in 16-cell cysts (Figure 2B, C). Similarly, with the AACAC probe, 64.4% (n = 45) of 8-cell cysts were paired, and 82.9% (n = 41) of 16-cell cysts (Figure 2E, F). In contrast, the peri-centromeric region of the X chromosome appeared mostly paired already in GSCs (82%, n = 33), and only slightly increased to 86% (n = 43) of pairing in 8-cell cysts and 100% (n = 42) of pairing in 16-cell cysts (Figure 2G–I). We thus concluded that while the X-chromosome peri-centromeric region is always paired, there is de novo pairing for centromeres of chromosome II and III in the mitotic region.
Fig. 2. Centromeres of chromosome II and III become associated in the mitotic region, whereas X-chromosome centromeres are always paired. 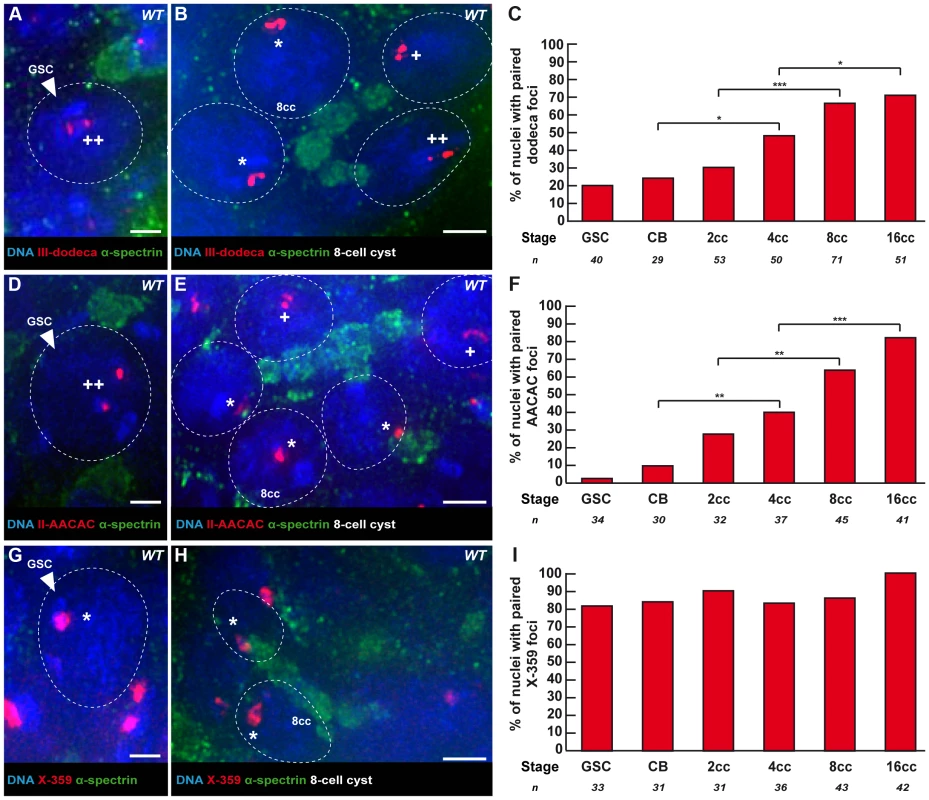
Fusome (α-spectrin, green) and DNA (DAPI, blue). (A, B) Projection of Z-sections obtained by DV microscopy of a wild-type germarium stained for the chromosome III centromere (dodeca probe, red), fusome, and DNA. (A) GSC showing 2 dodeca foci separated by a distance >0,7 µm. (B) 4 nuclei of a 8-cell cyst showing foci separated by a distance <0,7 µm. Nuclei with * show one focus, thus centromeres are paired, nuclei with one + display two foci separated by a distance ≤0,7 µm thus they are considered paired and nuclei with ++ display two foci separated by a distance >0,7 µm and are thus considered unpaired. Scale bars represent 2 µm. (C) Developmental changes in the percentage of paired chromosome III centromeres for each cell stage in region 1 using the dodeca probe. The number of cells analyzed is indicated under each stage. * p≤0.05, ** p≤0.005, *** p≤0.0005 (khi2 test comparing CB with 4cc, 2cc with 8cc and 4cc with 16cc). (D, E) Projection of Z-sections obtained by DV microscopy of a wild-type germarium stained for the chromosome II centromere (AACAC probe, red), fusome, and DNA. (D) GSC showing 2 AACAC foci separated by a distance >0,7 µm. (E) 5 nuclei of a 8-cell cyst showing foci separated by a distance <0,7 µm. Nuclei with * show one foci, thus centromeres are paired, nuclei with one + display two foci separated by a distance ≤0,7 µm thus they are considered paired and nuclei with ++ display two foci separated by a distance >0,7 µm and are thus considered unpaired. Scale bars represent 2 µm. (F) Developmental changes in the percentage of paired chromosome II centromeres for each cell stage in region 1 using the AACAC probe. The number of cells analyzed is indicated under each stage. * p≤0.05, ** p≤0.005, *** p≤0.0005 (khi2 test comparing CB with 4cc, 2cc with 8cc and 4cc with 16cc). (G, H) Projection of Z-sections obtained by DV microscopy of a wild-type germarium stained for the chromosome X centromere (359 probe, red), fusome, and DNA. (G) GSC showing one 359 focus. (H) 2 nuclei of an 8-cell cyst showing paired foci. Nuclei with * show one focus, thus centromeres are paired. Scale bars represent 2 µm. (I) Developmental changes in the percentage of paired X-chromosome centromeres for each cell stage in region 1 using the 359 probe. The number of cells analyzed is indicated under each stage. As indicated previously, we found that all centromeres showed a tendency to cluster already in 8-cell cysts (Figure S1A), raising the possibility of interactions between centromeres of non-homologous chromosomes. We thus measured for each centromere the percentage of interaction with a non-homologous centromere at each stage. At the 8-cell stage, for example, we found significant interactions between centromeres of chromosome III and X (28.7%, n = 108 and 38%, n = 108; Figure S2A and S2C). These percentages were, however, well below homologous interactions at the same stage (66.2% for the chromosome III and 86% for the chromosome X). In contrast, centromeres of chromosome II did not associate strongly with any of the other centromeres (highest interaction at any stage was 20% with centromere III, Figure S2B). We thus concluded that the decrease in the number of CID foci (Figure 1) mainly resulted from homologous interactions, but that non-homologous interactions also contributed to this reduction.
Synaptonemal components are expressed in the mitotic region and localize close to centromeres
It is known that the formation of the SC initiates at the clustered centromeres as soon as cysts enter region 2, and that SC proteins are required for the clustering of centromeres in the same region [13]–[14]. Could SC components, such as C(3)G and Corona, also be required to initiate the pairing and clustering of centromeres in region1? C(3)G and Corona are transverse filaments and central element components of the SC, respectively, and are required for SC formation [18]–[19]. Surprisingly for meiosis-specific proteins, we found that C(3)G and Corona were expressed in germ cells of the mitotic zone and associated with centromeres [18]–[19] (Figure 3A, B and data not shown). No staining was detected in nuclei of either wild-type follicle cells or c(3)G mutant germ cells attesting to the specificity of the signal (data not shown). Using wide-field deconvolution microscopy and confocal microscopy, we found that C(3)G signals were partially overlapping with CID at centromeres (Figure 3A). However, using super-resolution microscopy, we could separate CID foci from C(3)G dots (Figure 3B). It showed that C(3)G localized very close to the centromeres, but not at centromeres, at least marked by CID. We further found an increase in the association of C(3)G with centromeres from GSCs to 8-cell cysts. In GSCs, 25.7% (n = 152) of centromeres overlapped or touched a C(3)G staining, while there was a dramatic increase in 8-cell cysts, where 72.0% of centromeres (n = 164) overlapped with dots of C(3)G (Figure 3C). Furthermore, we found that most dots of C(3)G in each nucleus were associated with centromeres at every stage (Figure 3D). The accumulation of C(3)G at centromeres correlates with an increase in centromere interactions, suggesting that SC components may play an active role in this process.
Fig. 3. The Synaptonemal Complex component C(3)G is present near some centromeres in the mitotic region. 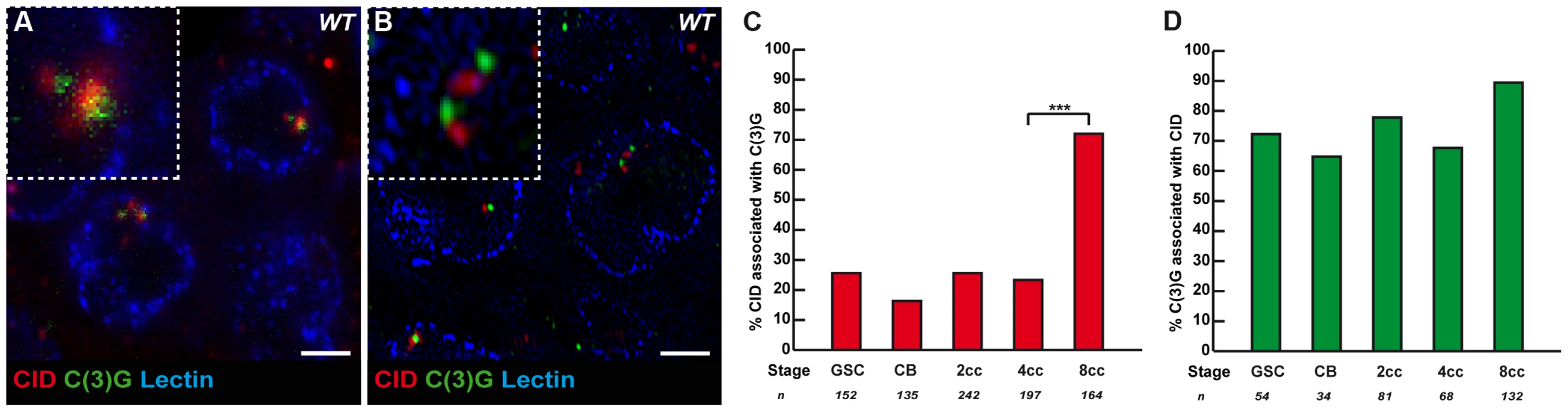
(A) Projection of Z-sections obtained by confocal microscopy of a wild-type 2cc stained for centromere (CID, red), C(3)G (green) and nuclear membrane (Lectin, blue). Scale bar represents 2 µm. (B) Projection of Z-sections obtained by OMX of a wild-type 2cc stained for centromere (CID, red), C(3)G (green) and nuclear membrane (Lectin, blue). Scale bar represents 2 µm. (C) Developmental changes in the percentage of centromeres associated with C(3)G for each cell stage in region 1. CID and C(3)G are associated when the 2 foci touch or overlap in a nucleus. Cysts were staged according to the shape of the fusome marked by α-spectrin (not shown). (D) Developmental changes in the percentage of C(3)G associated with centromeres for each cell stage in region 1. CID and C(3)G are associated when the 2 foci touch or overlap in a nucleus. Cysts were staged according to the shape of the fusome marked by α-spectrin (not shown). Synaptonemal components promote centromere clustering in the mitotic zone
To test whether the presence of SC components at centromeres in region 1 had a function in centromere association, we counted the number of CID dots in c(3)G68 and conaf04903/conaA12 mutants in both live and fixed tissue. We found that the association of centromeres was dramatically affected in both mutant backgrounds at the 8-cell and 16-cell stages (Figure 4). c(3)G mutant 8-cell cysts show an average of 5.2±1.4 CID foci as compared to 3.6±1.3 in the wild-type situation (two-tailed Student's t-test, p<0,0005). Instead the number of CID dots in mutant 8-cell cysts is similar to wild-type GSCs (5.9±0.9) (Figure 4B, C). Furthermore, we never observed nuclei with only 1 or 2 dots of CID in mutant 8-cell cysts (0%, n = 49; Figure S1), indicating that C(3)G and Corona were also required for centromere clustering in region 1. We have analysed centromere pairing and clustering in ord5/ord10 mutant females both on fixed and live germarium (Figure 4B and 4C). Ord is a cohesion protein, which localizes to the Lateral Elements (LEs) of the SC. We found that the number of CID foci was significantly higher in ord mutant females from GSC to 8-cell cysts, similar to c(3)G mutant (Figure 4B and 4C). However, the increase in CID foci at the 8-cell stage became weaker in ord mutant females than in c(3)G mutants (4.3±1.6 CID foci in ord5/ord10 compared to 5.2±1.4 in c(3)G68 and 3.6±1.3 in WT). By the 16-cell stage, centromeres were well clustered in ord mutant germaria (2.2±0.9 CID foci in ord5/ord10 compared 2.4±0.9 in WT). Surprisingly, later on in region 2a, the number of CID foci rose dramatically again (data not shown), as published in (Takeo et al, 2011). In conclusion, ord mutant germ cells initially behaved like c(3)G and corona mutants, then were “rescued” in young 16-cell cysts, and finally centromeres separated again in region 2a.
Fig. 4. Synaptonemal Complex components promote centromere association in the mitotic zone. 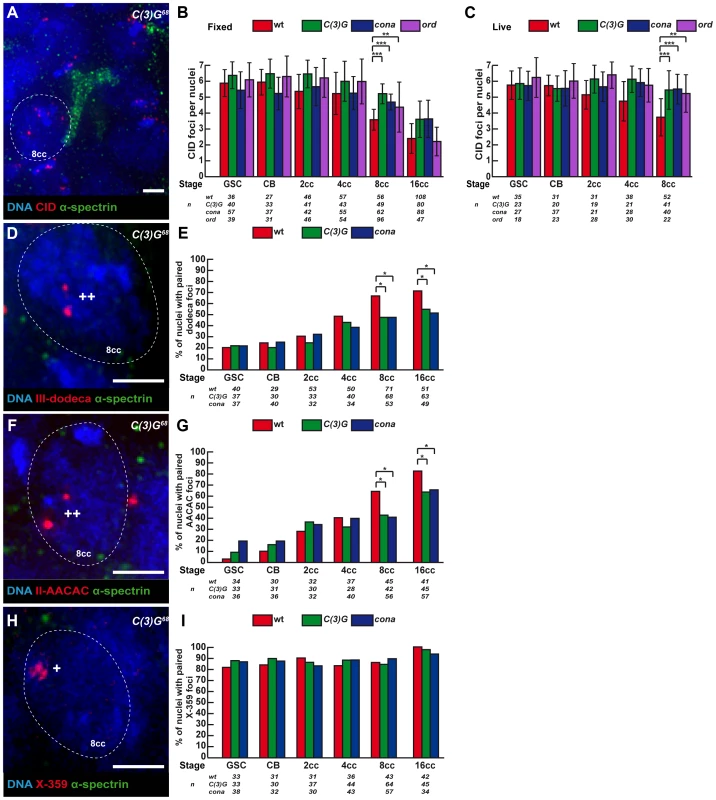
Fusome (α-spectrin, green) and DNA (DAPI, blue). (A) Projection of Z-sections obtained by DV microscopy of a c(3)G68 germarium stained for centromere (CID, red), fusome, and DNA. One nucleus of an 8-cell cyst with a branched fusome is shown. Scale bar represent 2 um. (B,C) Developmental changes in the number of CID foci for each cell stage in region 1 in wild-type, c(3)G68, conaf04903/conaA12and ord5/ord10 fixed germaria (B) or in living germaria (C). The number of analyzed cells is indicated under each stage. * p≤0.05, ** p≤0.005, *** p≤0.0005, (two-tailed Student's t-test comparing wild-type with c(3)G68 and conaf04903/conaA12). (D) Projection of Z-sections obtained by DV microscopy of a c(3)G68 germarium stained for dodeca (red), fusome , and DNA. One nucleus of an 8-cell cyst (8cc), surrounded by dotted lines, displays two foci separated by a distance ≥0,7 µm (++) indicating unpaired centromeres. Scale bar represents 2 µm. (E) Developmental changes in the percentage of paired chromosomes III for each cell stage in region 1 using the dodeca probe in wild-type, c(3)G68 and conaf04903/conaA12 fixed germaria. Chromosome III centromeres are considered paired when the distance between the 2 foci ≤0,7 µm in a nucleus. The number of analysed cells is indicated under each stage. * p≤0.05 (khi2 test comparing wild-type with c(3)G68 and conaf04903/conaA12). (F) Projection of Z-sections obtained by DV microscopy of a c(3)G68 germarium stained for AACAC (red), fusome, and DNA. One nucleus of an 8-cell cyst (8cc), surrounded by dotted lines, displays two foci separated by a distance ≥0,7 µm (++) indicating unpaired centromeres. Scale bar represents 2 µm. (G) Developmental changes in the percentage of paired chromosomes II for each cell stage in region 1 using the AACAC probe in wild-type, c(3)G68 and conaf04903/conaA12 fixed germaria. Chromosome II centromeres are considered paired when the distance between the 2 foci ≤0,7 µm in a nucleus. The number of analysed cells is indicated under each stage. * p≤0.05 (khi2 test comparing wild-type with c(3)G68 and conaf04903/conaA12). (H) Projection of Z-sections obtained by DV microscopy of a c(3)G68 germarium stained for X-359 (red), fusome, and DNA. One nucleus of an 8-cell cyst (8cc), surrounded by dotted lines, displays one focus (*) indicating pairing. Scale bar represents 2 µm. (I) Developmental changes in the percentage of paired X-chromosomes for each cell stage in region 1 using the 359 probe in fixed wild-type, c(3)G68 and conaf04903/conaA12fixed germaria. Chromosome X centromeres are considered paired when the distance between the 2 foci ≤0,7 µm in a nucleus. The number of analyzed cells is indicated under each stage. We then investigated how the pairing of individual centromeres is affected in our mutants in different cysts by combining FISH with immunostaining against the fusome (Figure 4D, F, H). We found that in c(3)G68 and conaf04903/conaA12 mutant 8-cell cysts, the number of paired chromosomes III at the level of the centromeric regions dropped from 66% (wt, n = 71) to 47% (c(3)G68: n = 68, conaf04903/conaA12: n = 53; khi2, p<0,02) (Figure 4E); and for chromosome II, pairing went from 64% (wt, n = 45) to 42.9% (c(3)G68: n = 42; khi2, p<0,05) and to 41.1% (conaf04903/conaA12: n = 56; khi2, p<0,02) (Figure 4G). In contrast, X-chromosome centromere pairing is unaffected in both c(3)G68 and conaf04903/conaA12 mutant germaria (Figure 4H, I). Thus, the SC components C(3)G and Cona are important for pairing of chromosome II and III, but not for chromosome X pairing, at the level of the centromeric regions in the mitotic zone.
Discussion
In most species, early meiotic prophase can be characterized by the pairing of homologous chromosomes, the appearance of synaptonemal complex components or the re-organisation of chromosomes in bouquet or similar conformations [5]–[6]. We found that all three processes initiate in the mitotic region of the Drosophila germarium and predominantly in 8-cell cysts, i.e. well before the onset of meiotic prophase. Our finding that centromeres of chromosome II and III are not paired in GSCs further indicates that “somatic” pairing stricto sensu does not occur in these cells. Thus, one can conclude that meiotic pairing is not an extension of somatic pairing present in GSCs. However, we note that most centromeres are paired and clustered in 8-cell cysts, indicating that the meiotic nuclear organization in region 2a, i.e. when centromeres are fully paired and clustered, can still be viewed as an extension of a pre-existing state. In an interesting twist, we show that pairing and clustering of centromeres in the mitotic region depend partially on meiotic proteins such as C(3)G and Corona. Therefore, although meiotic pairing is the continuation of mitotic pairing, we show that mitotic pairing in germ cells depends on several meiotic proteins.
We found an average of 6 foci of CID in GSCs, which can be explained by half of the centromeres being paired and the other half being unpaired. Centromeres of the X chromosome appear always paired, while centromeres of the chromosome II and III are mostly unpaired. It suggests that the remaining chromosome 4 is mostly paired. Further experiments are however required to demonstrate this hypothesis. In addition, we found some non-homologous interactions between centromeres of chromosome X and III. It is also known that chromosomes X and 4 tend to be associated in different contexts. We can thus speculate that centromeres of chromosome X, III and 4 are preferentially associated together when centromere clustering becomes complete in region 2a, while centromeres of chromosome II remain distinct. This model would be compatible with the average of 2 dots of CID observed in region 2a [13]–[14]. One large dot would correspond to centromeres of chromosome X, III and 4, while the smaller dot would often correspond to chromosome II centromere.
Our data show that chromosome X (and probably chromosome 4) behaves differently and appears always paired throughout oogenesis. This pairing is independent of C(3)G and Corona, and the underlying mechanisms remain to be identified. Since both X-chromosomes have centromeric clusters of rDNA repeats that localize to the same nucleolus, one could speculate that mechanisms similar to Drosophila males X-Y pairing may help pairing in this centromeric region [20]. It is likely that additional mechanisms also facilitate pairing of autosomal homologues in germline cysts. Indeed, although we found that centromeric pairing is dramatically affected in 4 - and 8-cell cysts mutant for c(3)G, there remains a tendency to pair in region 1 and also later in region 2 (Figure S3 and [13]). In addition, the lab of Ting Wu (Harvard Medical School) found the same pairing behavior for the chromosome arms, as we did for centromeric regions (Eric Joyce, Nicholas Apostolopoulos, Brian J. Beliveau, and C.-ting (Ting) Wu, personal communication). Using oligopaints probes targeting euchromatic and heterochromatic sequences on each chromosome arm, they found that the chromosome X was always paired, and that chromosomes II and III only became paired later on during cyst divisions. We found that C(3)G and Corona localize only close to the centromeres and peri-centromeric regions. It remains unknown whether pairing along chromosome arms also depends on C(3)G and Corona. We can thus hypothesize that other mechanisms independent of C(3)G and Corona are also at play to promote pairing even for chromosomes II and III.
It also remains possible that C(3)G and Corona do not play a direct role in promoting homologous pairing for chromosome II and III. These SC components could be mainly required to facilitate centromere association both homologous and non-homologous, which would indirectly increase the efficiency of homologous pairing. In support of this view, it is interesting to note that the 20% of 8-cell cysts with fully clustered centromeres (1 or 2 dots of CID, Figure S1A, S1E) disappeared in the absence of C(3)G (Figure S1B, S1F), indicating that C(3)G is absolutely required for clustering of centromeres. This absence of clustering could account for the 30% decrease in homologous pairing in C(3)G mutant (Figure 4). In this model C(3)G and Corona would not have a role in centromere homology assessment, and an independent homolog pairing process would act in parallel.
Our results in Drosophila reveal surprising similarities with the initiation of meiosis in budding yeast. Indeed, centromeres in S. cerevisiae also become associated or “coupled” before meiotic prophase [21]. This early association depends on Zip1, a central component of the SC functionally similar to C(3)G and SYCP1 in mouse. This similarity with our results further extend to the localization of Zip1, which partially overlaps with yeast centromeres at this early stage, like C(3)G and CID in flies [21]. The resemblance between C(3)G and Zip1 localization and function is surprising, as it is well known that SC formation requires double strand breaks (DSBs) in yeast but not in Drosophila [22]–[24]. Pre-meiotic pairing of homologues is thus present in a wide range of organisms, although the underlying mechanisms remain unknown in most cases [25]. Our results open the possibility to test whether the molecular mechanisms identified here in flies facilitate pre-meiotic pairing in other organisms.
Materials and Methods
Fly stocks and genetics
For all experiments on fixed germaria the following strains were used: w1118 was used as the wild-type strain. For testing meiotic mutants, the following null alleles were used: c(3)G68 e ca/c(3)G68 e [18], [26], conaA12/conaf04903 [19], ord5/ord10 [27]. For experiments on live germaria CID::RFP [28], nos-Gal4, UASp::Par1:GFP/+ (gift from Daniel St Johnston) was used as a wild-type strain. Similarly the following mutant strains were used: CID::RFP, c(3)G68 e/Par1::GFP, c(3)G68 e ca and CID::RFP, conaA12/Par1-GFP, conaf04903.
Immunohistochemistry and FISH
For immunostaining, ovaries were dissected in PBS, fixed in 4%PFA-PEPS, permeabilized in PBT (0,2%Triton) for 30 min, left overnight with primary antibodies in PBS at 4°C , washed 4×30 min in PBS, left with secondary antibody for 2 hr at room temperature, washed 4×30 min in PBS where DAPI (1∶500) was added during the last wash and mounted in Cityfluor. For FISH experiments followed by immunostaining, ovaries were dissected in PBS fixed in 4% PFA in 1× fix buffer (100 mm potassium cacodylate, 100 mm sucrose, 40 mm sodium acetate, and 10 mm EGTA). Ovaries were then rinsed three times in 2× SSCT and hybridization with the dodeca and 359 probes targeting the pericentromeric regions of the 3rd and X chromosome respectively was performed as previously described [11]. Following hybridization ovaries were rinsed in 2× SSCT, rinsed two times in PBST and the immunostaining protocol was resumed as above. We used the following primary antibodies: mouse anti-c(3)G (1/500) (gift from Scott Hawley, Stowers Institute, USA), rat anti-cid (1∶1000) (gift from Claudio E. Sunkel, Universidade do Porto, Portugal). We used rabbit anti-spectrin (1∶1000 for regular immunostaining, 1∶500 following FISH) (gift from R. Dubreuil) to label the fusome and identify the cyst stages. Secondary antibodies conjugated with Cy3, Cy5 and FITC (Jackson laboratories) were used at 1/200.
Image acquisition and data analysis
DV Images of fixed germaria were collected under a DeltaVision deconvolution microscope system (Applied Precision) equipped with an Olympus 1670 inverted microscope and high-resolution CCD camera. All images were acquired with the PlanApo 60×/1,42 oil objective lens with 1.5× auxiliary magnification at 0.2 µm intervals along z-axis and deconvolved using the softWoRx v.3.5.1 software (Applied Precision).
Confocal Images of fixed germaria were collected under a Zeiss LSM 780 NLO confocal. All images were acquired with a PlanApo 63×/1,46 oil objective at 0.6 um along z-axis and operated by ZEN 2012 software.
When structured illumination was performed we used an OMX v3 system (Applied Precision - gehealthcare), equipped with 3 EMCCD, evolve cameras (photometrics). Signal from all channels were realigned using fluorescent beads prior to each session of image acquisition. Registration was done using UnwarpJ in ImageJ. All images were acquired with a PlanApo 100×/1,4 oil objective at 125 nm along z-axis. Pixel size is 40 nm along xy-axis after reconstruction.
For live imaging ovaries were dissected in oil (10S, Halocarbon, Sigma). The muscular sheath around each ovariole was removed and germaria were made to stick to coverslips in oil.
Movies were collected with an Inverted Spinning disk Confocal Microscope (Roper/Nikon) operated by Metamorph on an inverted Nikon Eclipse Ti microscope coupled to a CoolSNAP HQ2 camera (Photometrics) and temperature control chamber. All images were acquired with the Planapo 60×/1,4 oil objective lens with 1.5× auxiliary magnification. Single position movies in the germarium were acquired for 30–40 min at 25±1°C, with a 30 sec temporal resolution (7 slices Z-stack, 0.7 µm/slice).
The use of α-spectrin and Par1:GFP on fixed and live germaria respectively allowed to precisely identify the different cyst stages. For quantification of CID foci on fixed germaria, we counted the number of distinguishable CID foci within each single nucleus, and for live germaria we took into account the number of distinguishable CID foci at a given (t) projection where the number of CID foci was maximal. In all figures the images of fixed germaria shown are the projection of all z-series which cover a region ranging from the first CID foci until the last CID foci seen. For live germaria images shown are the projection of all z-series of a single (t) projection.
To measure the distances between foci of the dodeca and 359 probes a macro designed by Olivier Leroy from the unit imaging facility was used that computes 3D distances between selected foci. Peri-centromeric regions of chromosomes were considered as paired when only one foci was visible or when two foci were visible but separated by a distance ≤0,70 µm [10].
Supporting Information
Zdroje
1. Spradling A (1993) Developmental genetics of oogenesis. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. New-York: Cold Spring Harbor Laboratory Press. pp. 1–70.
2. de CuevasM, SpradlingAC (1998) Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development 125 : 2781–2789.
3. CarpenterA (1975) Electron microscopy of meiosis in Drosophila melanogaster females. I Structure, arrengement, and temporal change of the synaptonemal complex in wild-type. Chromosoma 51 : 157–182.
4. HuynhJR, St JohnstonD (2004) The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr Biol 14: R438–449.
5. LakeCM, HawleyRS (2012) The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes. Annu Rev Physiol 74 : 425–451.
6. ZicklerD (2006) From early homologue recognition to synaptonemal complex formation. Chromosoma 115 : 158–174.
7. McKeeBD (2004) Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta 1677 : 165–180.
8. GrellRF, DayJW (1970) Chromosome pairing in the oogonial cells of Drosophila melanogaster. Chromosoma 31 : 434–445.
9. BlumenstielJP, FuR, TheurkaufWE, HawleyRS (2008) Components of the RNAi machinery that mediate long-distance chromosomal associations are dispensable for meiotic and early somatic homolog pairing in Drosophila melanogaster. Genetics 180 : 1355–1365.
10. GongWJ, McKimKS, HawleyRS (2005) All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet 1: e67.
11. SherizenD, JangJK, BhagatR, KatoN, McKimKS (2005) Meiotic recombination in Drosophila females depends on chromosome continuity between genetically defined boundaries. Genetics 169 : 767–781.
12. VazquezJ, BelmontAS, SedatJW (2002) The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr Biol 12 : 1473–1483.
13. TakeoS, LakeCM, Morais-de-SaE, SunkelCE, HawleyRS (2011) Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr Biol 21 : 1845–1851.
14. TannetiNS, LandyK, JoyceEF, McKimKS (2011) A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr Biol 21 : 1852–1857.
15. BlowerMD, KarpenGH (2001) The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol 3 : 730–739.
16. RablC (1865) Uber Zelltheilung. Morphol Jahrb 10 : 214–330.
17. JoyceEF, WilliamsBR, XieT, WuCT (2012) Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet 8: e1002667.
18. PageSL, HawleyRS (2001) c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev 15 : 3130–3143.
19. PageSL, KhetaniRS, LakeCM, NielsenRJ, JeffressJK, et al. (2008) Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLoS Genet 4: e1000194.
20. McKeeBD, KarpenGH (1990) Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell 61 : 61–72.
21. TsubouchiT, RoederGS (2005) A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308 : 870–873.
22. KlecknerN (1996) Meiosis: how could it work? Proc Natl Acad Sci U S A 93 : 8167–8174.
23. McKimKS, Green-MarroquinBL, SekelskyJJ, ChinG, SteinbergC, et al. (1998) Meiotic synapsis in the absence of recombination. Science 279 : 876–878.
24. RoederGS (1995) Sex and the single cell: meiosis in yeast. Proc Natl Acad Sci U S A 92 : 10450–10456.
25. StewartMN, DawsonDS (2008) Changing partners: moving from non-homologous to homologous centromere pairing in meiosis. Trends Genet 24 : 564–573.
26. JeffressJK, PageSL, RoyerSK, BeldenED, BlumenstielJP, et al. (2007) The formation of the central element of the synaptonemal complex may occur by multiple mechanisms: the roles of the N - and C-terminal domains of the Drosophila C(3)G protein in mediating synapsis and recombination. Genetics 177 : 2445–2456.
27. WebberHA, HowardL, BickelSE (2004) The cohesion protein ORD is required for homologue bias during meiotic recombination. J Cell Biol 164 : 819–829.
28. SchuhM, LehnerCF, HeidmannS (2007) Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17 : 237–243.
Štítky
Genetika Reprodukčná medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



