-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
The superiority of hybrids has long been exploited in agriculture, and although many models explaining “heterosis” have been put forth, direct empirical support is limited. Particularly elusive have been cases of heterozygosity for single gene mutations causing heterosis under a genetic model known as overdominance. In tomato (Solanum lycopersicum), plants carrying mutations in SINGLE FLOWER TRUSS (SFT) encoding the flowering hormone florigen are severely delayed in flowering, become extremely large, and produce few flowers and fruits, but when heterozygous, yields are dramatically increased. Curiously, this overdominance is evident only in the background of “determinate” plants, in which the continuous production of side shoots and inflorescences gradually halts due to a defect in the flowering repressor SELF PRUNING (SP). How sp facilitates sft overdominance is unclear, but is thought to relate to the opposing functions these genes have on flowering time and shoot architecture. We show that sft mutant heterozygosity (sft/+) causes weak semi-dominant delays in flowering of both primary and side shoots. Using transcriptome sequencing of shoot meristems, we demonstrate that this delay begins before seedling meristems become reproductive, followed by delays in subsequent side shoot meristems that, in turn, postpone the arrest of shoot and inflorescence production. Reducing SFT levels in sp plants by artificial microRNAs recapitulates the dose-dependent modification of shoot and inflorescence production of sft/+ heterozygotes, confirming that fine-tuning levels of functional SFT transcripts provides a foundation for higher yields. Finally, we show that although flowering delays by florigen mutant heterozygosity are conserved in Arabidopsis, increased yield is not, likely because cyclical flowering is absent. We suggest sft heterozygosity triggers a yield improvement by optimizing plant architecture via its dosage response in the florigen pathway. Exploiting dosage sensitivity of florigen and its family members therefore provides a path to enhance productivity in other crops, but species-specific tuning will be required.
Published in the journal: Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture. PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004043
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004043Summary
The superiority of hybrids has long been exploited in agriculture, and although many models explaining “heterosis” have been put forth, direct empirical support is limited. Particularly elusive have been cases of heterozygosity for single gene mutations causing heterosis under a genetic model known as overdominance. In tomato (Solanum lycopersicum), plants carrying mutations in SINGLE FLOWER TRUSS (SFT) encoding the flowering hormone florigen are severely delayed in flowering, become extremely large, and produce few flowers and fruits, but when heterozygous, yields are dramatically increased. Curiously, this overdominance is evident only in the background of “determinate” plants, in which the continuous production of side shoots and inflorescences gradually halts due to a defect in the flowering repressor SELF PRUNING (SP). How sp facilitates sft overdominance is unclear, but is thought to relate to the opposing functions these genes have on flowering time and shoot architecture. We show that sft mutant heterozygosity (sft/+) causes weak semi-dominant delays in flowering of both primary and side shoots. Using transcriptome sequencing of shoot meristems, we demonstrate that this delay begins before seedling meristems become reproductive, followed by delays in subsequent side shoot meristems that, in turn, postpone the arrest of shoot and inflorescence production. Reducing SFT levels in sp plants by artificial microRNAs recapitulates the dose-dependent modification of shoot and inflorescence production of sft/+ heterozygotes, confirming that fine-tuning levels of functional SFT transcripts provides a foundation for higher yields. Finally, we show that although flowering delays by florigen mutant heterozygosity are conserved in Arabidopsis, increased yield is not, likely because cyclical flowering is absent. We suggest sft heterozygosity triggers a yield improvement by optimizing plant architecture via its dosage response in the florigen pathway. Exploiting dosage sensitivity of florigen and its family members therefore provides a path to enhance productivity in other crops, but species-specific tuning will be required.
Introduction
More than a century ago, simple garden studies by Darwin revealed a remarkable phenomenon in which crossing related varieties of plants produced hybrid progeny with superior growth and fecundity compared to their parents [1]. Understanding this hybrid vigor began with population genetics theories postulating that outcrossing facilitates adaptation and improves fitness by shuffling allelic diversity to thwart inbreeding depression [2]. However, it was the agricultural exploitation of hybrid vigor, or “heterosis,” in both crop and animal breeding that propelled efforts to dissect its genetic and molecular bases [3]–[10]. Maize geneticists noted early on that inbreeding prior to hybridization drives yield heterosis, and heterotic effects generally improve with greater genetic distance between parental lines [3]. These observations led to the notion that heterosis derives from genome-wide masking of independently accrued deleterious recessive mutations. Extensive quantitative genetic, transcriptomic, and genomic sequencing studies in crop and model plants have provided widespread indirect support for a “dominance complementation” model [2], [6], [11]; however, there is lingering evidence that a model known as overdominance might also contribute to heterosis [5]–[8]. Overdominance has long been an appealing explanation, because theoretically heterozygosity at only a single gene is needed to cause heterotic effects, presumably from intra-locus allelic interactions functionally superseding any one allelic form. However, the relevance of overdominance for yield and whether allelic interactions are the underlying cause remains controversial, primarily because quantitative trait locus (QTL) mapping studies reporting overdominant QTL have failed to pinpoint responsible genes [12]–[16]. Importantly, though, there have been scattered reports of single gene overdominance over the years, and among these have been several unexplained examples from yeast, plants, and animals involving heterozygosity for single gene loss-of-function mutations [17]–[24].
We previously reported a dramatic case of overdominance for tomato yield in multiple environments and planting densities resulting from loss-of-function mutations in the gene SINGLE FLOWER TRUSS (SFT) encoding the generic flowering hormone florigen [25]. Tomato yield, on both a per plant basis and in the context of tons per acre, depends partly on fruit size, but is mainly driven by the production of dozens of multi-flowered inflorescences and resulting fruit clusters that develop according to the “sympodial” growth habit [26]. The defining feature of sympodial plants is the shoot apical meristem (SAM) ends growth by differentiating into a terminal flower after producing a set number of leaves, and growth then renews from a specialized axillary (i.e. sympodial) meristem (SYM) that, in tomato, produces just three leaves before undergoing its own flowering transition and termination. Indefinite reiteration of three-leaf sympodial flowering events results in an “indeterminate” plant that continuously produces equally spaced inflorescences (Figure 1A). In homozygous sft mutants, reduced florigen signals delay the transition to reproductive growth and cause a substantial loss of flower production and yield due to loss of sympodial growth and conversion of inflorescences into leafy vegetative shoots producing scattered flowers [27]. Counter-intuitively, sft/+ heterozygotes generate more inflorescences, flowers, and harvestable ripe fruits compared to parental controls in the same growing period, but these effects are limited to “determinate” tomato types in which sympodial shoot and inflorescence production ends prematurely due to a classical mutation in the gene SELF PRUNING (SP) (Figure 1A) [25], [26]. Notably, SP is a flowering repressor and a known florigen antagonist in the SFT gene family, implying that SFT-dependent yield heterosis is likely directly linked to the flowering transition, and specifically to the opposing functional relationship of SP to SFT.
Fig. 1. Precocious shoot termination in determinate tomatoes is partially suppressed by sft/ 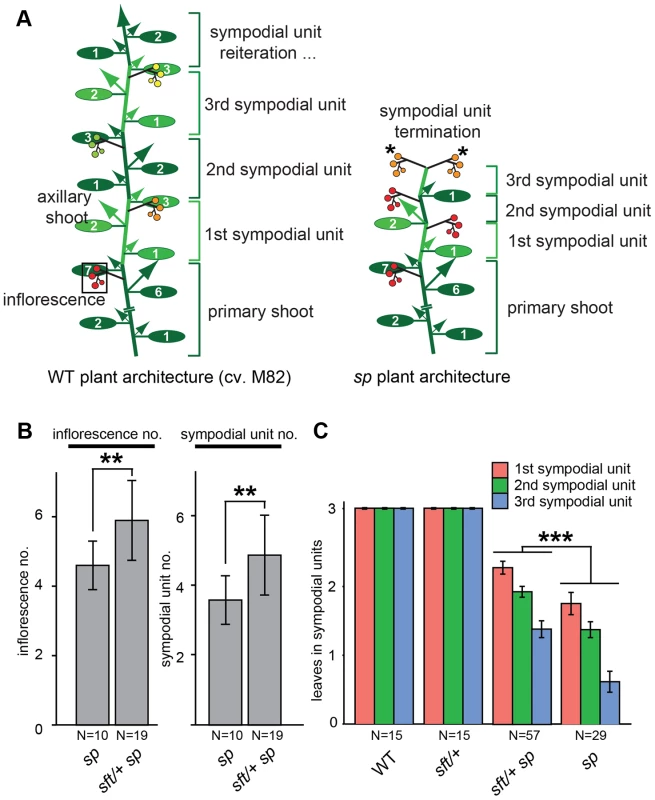
+ mutant heterozygosity. (A) Schematic diagrams showing shoot architecture of a wild type (WT) indeterminate tomato plant (left) and an sp determinate mutant (right). In WT M82 plants the primary shoot meristem (PSM) from the embryo gives rise to 7–9 leaves before terminating in the first flower of the first multi-flowered inflorescence (boxed). A specialized axillary meristem called a sympodial meristem (SYM) in the axil of the last leaf on primary shoot then generates three leaves before terminating in the first flower of the next inflorescence. In indeterminate tomatoes, this process continues indefinitely (left). In sp mutants (right), sympodial cycling accelerates progressively on all shoots causing leaf production to decrease in successive units until growth ends in two juxtaposed inflorescences (asterisks). Alternating colored groups of three ovals represent leaves within successive sympodial units numbered at right. Colored circles represent fruits and flowers within each inflorescence (red: fully ripe fruit; orange: ripening fruit; green: unripe fruit; yellow: flowers) and arrows represent canonical axillary shoots. (B) Compared to sp mutants alone, sft/+ sp plants produce more inflorescences (left) and sympodial units (right) before sympodial cycling terminates on the main shoot. Genotypes and sample sizes are shown below, and standard deviations of averages are presented. (C) Compared to sp alone, sft/+ sp plants produce more leaves in the first three sympodial units, indicating a delay in precocious termination. Colored bars indicate average leaf numbers within sympodial units with standard deviations. Statistical significance in B and C was tested by Wilcoxon rank sum test, and significance levels are indicated by asterisks (*P<0.05, **P<0.01, ***P<0.001). Tomato breeding goals are multifaceted and shift according to the needs and desires of growers (e.g. improved pest resistances) and consumers (e.g. better quality), but one unwavering aim is to improve yield. Indeterminate cultivars are grown commercially to enable continuous market delivery of “round,” “roma,” “cocktail,” “grape,” and “cherry” tomato types that are eaten fresh and command a premium price. Indeterminate tomatoes are primarily grown in greenhouses where successively ripening clusters are harvested by hand multiple times over an extended period, in some cases up to a year, to maximize yield on plants that must be pruned to one or two main shoots to enable efficient greenhouse growth and maintain fresh market quality [28]. While the necessary pruning of indeterminate tomatoes facilitates agronomic practices that maximize quality, such as size, shape, and flavor, it also limits yield [29]. In contrast, tomatoes grown for sauces, pastes, juices, or other processed can or jar products where fruit quality is less relevant, must be managed agronomically to produce maximum yields (per acre) through once-over mechanical harvests to be economically justified [28]. Maximal yields for processing tomatoes are achieved by growing determinate sp mutants in the open field to their full potential, because sequential sympodial shoots transition to flowering progressively faster in sp plants, which results in a compact bush-like form where fruits ripen uniformly (Figure 1A) [26]. Thus, sp varieties lend themselves to once-over mechanical harvesting and have therefore come to dominate the processing tomato industry, although determinate varieties have also been bred for fresh market production [28]. In a parallel to the physical pruning of indeterminate tomatoes, one drawback of sp-imposed determinate growth is that inflorescence and fruit production is restricted, because of a genetic pruning that causes sympodial cycling to stop. Thus, strategies to improve processing tomato yield are limited, primarily because the most logical approach of simply increasing sympodial flowering events would lead back to indeterminate growth and large plants that perform poorly in the field from competition and a loss of uniform ripening. Thus, maximizing inflorescence and fruit production while simultaneously minimizing shoot production for the processing tomato industry has remained a challenging goal. To explore how interactions between mutations in SP and SFT affect tomato flowering to create a new optimum for fruit yield, we explored tomato sft heterosis from a developmental and molecular context of the reproductive transition and its impact on plant architecture and inflorescence production.
Results
sft/+ heterozygosity suppresses sympodial shoot termination in determinate tomatoes
The discovery that sft/+ heterozygosity in an sp background (sft/+ sp) dramatically increases fruit production while only modestly increasing plant size was remarkable, but explaining this single gene overdominant effect was limited to showing that the yield boost mostly came from sft/+ sp plants having altered sympodial architectures that lead to more inflorescences [25]. sft mutant phenotypes are epistatic over sp [27], leading us to speculate that having only one functional allele of SFT might result in a dose-dependent partial suppression of sp determinacy. Indeed, heterosis disappears in a functional SP background [25]; yet, how the sft/+ sp genetic constitution affects the flowering process to create a new optimum for yield has not been resolved. To address this, we grew sp and sft/+ sp plants in controlled greenhouse conditions to precisely compare inflorescence production and flowering times of recurring sympodial shoots on the main axis (i.e. derived from the primary shoot; Figure 1A). We found an average of 1.5 more inflorescences and sympodial units on sft/+ sp plants, confirming a delay in sympodial termination (Figure 1B). To determine whether this was based on a delay in the flowering transition of each sympodial shoot, we measured leaf number in the first three units and observed a modest, but significant, increase in leaf production (Figure 1C). Importantly, and as expected [25], these delays required the sp background, as sft/+ heterozygosity alone produced three-leaf sympodial units like WT (Figure 1C). Importantly, delays in flowering time and sympodial termination were also observed on side shoots (Figure S1A–C), indicating a whole plant effect from sft/+ heterozygosity that explains the increase in total inflorescence number (Figure S1D) [25]. Thus, postponement of sympodial termination in sp mutants from sft/+ heterozygosity is based on recurring weak delays of all main and side shoot sympodial flowering transitions.
sft/+ heterozygosity weakly delays the primary flowering transition
Initiation and perpetuation of tomato sympodial growth depends on a gradual flowering transition culminating in PSM termination in a process mediated in part by accumulating florigen product from SFT counterbalancing repressive signals from SP. Regardless of whether SP is mutated, mutations in SFT cause late flowering and produce vegetative inflorescences, and strong alleles fail to initiate sympodial growth (Figure 2A) [27]. Our observation that precocious sympodial termination was delayed in sft/+ sp plants beginning with the first sympodial shoot (Figure 1C) led us to ask whether the flowering delay might commence in the PSM where sft homozygous mutant phenotypes first manifest. Surprisingly, whereas flowering time of sft/+ heterozygotes alone was not significantly different from sp mutants and WT, sft/+ sp plants were slightly later flowering (Figure 2A). We pinpointed this weak semi-dominant effect more precisely by evaluating developmental progression (ontogeny) of meristems. Like vegetative shoots, multi-flowered inflorescences of tomato are based on sympodial growth [26]. Just before the PSM transitions to a terminal floral meristem (FM), a sympodial inflorescence meristem (SIM) initiates perpendicularly, and this process reiterates several times to produce the characteristic zigzag inflorescence [30]. At 20 days after germination (DAG), we quantified SIM production in the primary inflorescence and found that sft/+ sp plants were on average one SIM behind sp mutants (Figure 2B–D). At this same point, while the first SYM of sp plants had already given rise to the first or second FM-SIM pair of the second inflorescence, most sft/+ sp SYMs were still in the reproductive transition (no FM evident morphologically) or starting the development of the first SIM-FM pair (Figure 2E–G). Thus, having only one fully functional allele of SFT delays the flowering transitions of both primary and sympodial shoots in sp mutants.
Fig. 2. sft/+ heterozygosity induces weak semi-dominant delays in both primary and sympodial flowering transitions. 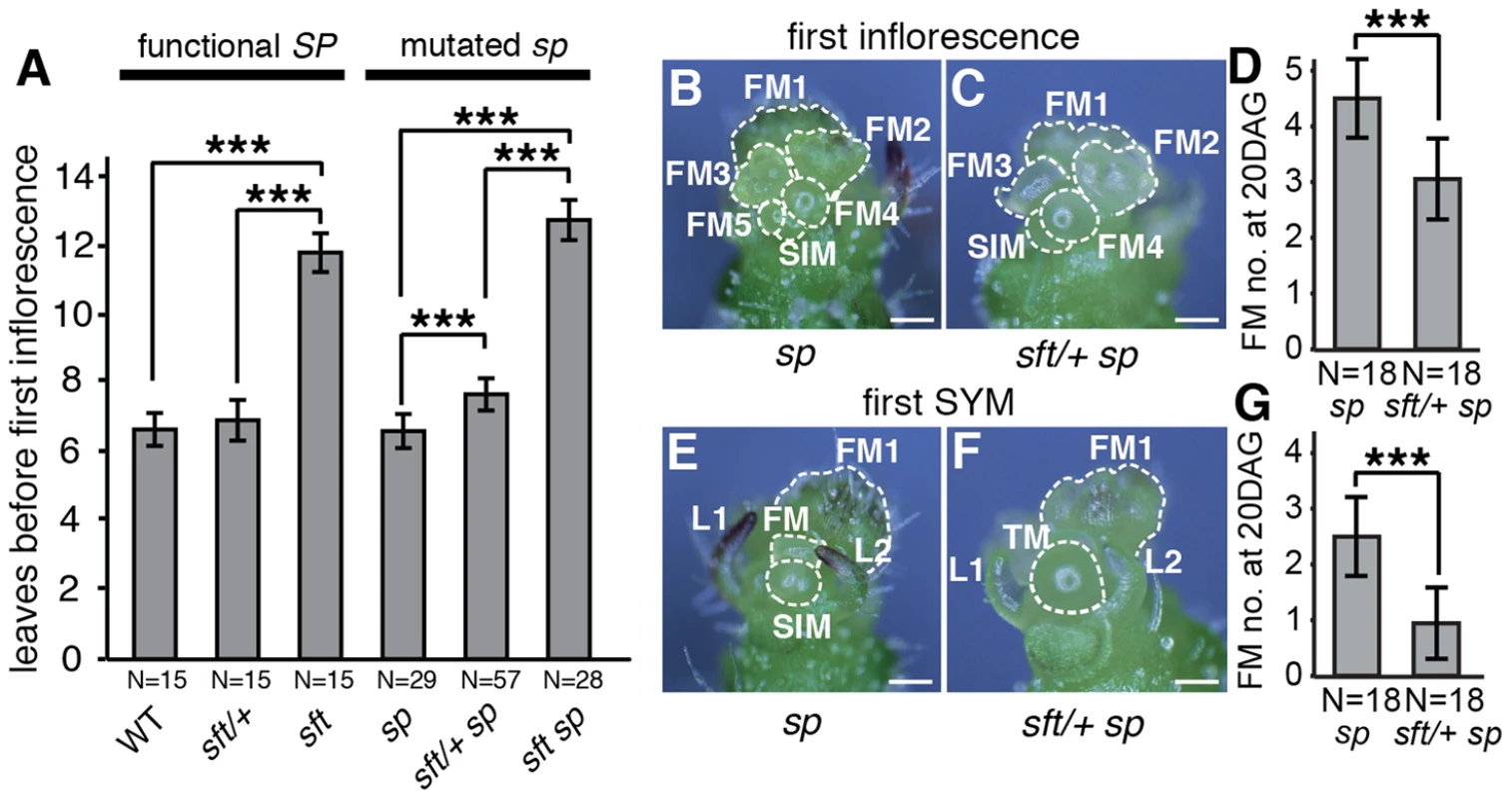
(A) sft/+ sp plants show slightly delayed primary shoot flowering time compared to sp as measured by leaf production before formation of the first inflorescence. Note the extremely delayed flowering of sft sp double mutants, indicating a weak semi-dominant effect for sft/+ heterozygosity. Bars indicate average leaf numbers with standard deviations. Genotypes and sample sizes are shown below. Statistical differences were tested by Wilcoxon rank sum tests and significance levels are marked by asterisks (***P<0.001). (B–G) Representative images and quantification of developmental progression (ontogeny) of meristems in the first inflorescence and sympodial shoot meristems (SYM) of sp (left images) and sft/+ sp plants (right images) at 20th DAG. Both sp (B) and sft/+ sp (C) PSMs have completed the primary flowering transition and generated a series of floral meristems (FM) and sympodial inflorescence meristems (SIM) [26], [30]. sft/+ sp plants are consistently one SIM behind ontogenically, consistent with a weak delay in flowering from sft/+ heterozygosity (D). Developmental progression of the first SYM in sp (E) and sft/ + sp (F) plants at the same time point as in B–C. While the SYM of sp mutants has already completed the flowering transition and differentiated into the first or second FM and initiated the next SIM, the SYM of sft/+ sp plants is still transitioning or initiating the first SIM, indicating a developmental delay parallel to the PSM of sft/+ sp plants (G). In D and G, bars indicate average numbers of initiated FMs with standard deviations. Genotypes and sample sizes are shown below. Statistical differences were tested by Wilcoxon rank sum tests and significance levels are marked by asterisks (***P<0.001). Scale bar: 100 um. sft/+ heterozygosity delays seedling development and primary shoot meristem maturation
Our developmental findings suggested that sft/+ overdominance and yield increases might commence with a semi-dominant delay of the primary flowering event. The flowering transition is paralleled by a maturation of seedlings marked by changes in morphological complexity and molecular states (e.g. transcriptomes) of leaves [27], [31]. As leaves of sft/+ sp plants are indistinguishable from those of WT and sp, we captured global gene expression patterns of the 6th expanding (3 cm) leaf, which is when differences in meristem ontogeny first appear (Figure 2B–G, Figure 3A and Dataset S1). In comparing sp single and sft sp double mutant leaf mRNA-Seq generated transcriptomes with those of sft/+ sp plants, we found 838 differentially expressed genes among all genotypes. Previous studies comparing gene expression between hybrids and parents involved whole genome heterozygosity and reported thousands of differentially expressed genes representing all modes of gene action (e.g. dominant, recessive, additive, overdominant, etc.) [6], [8], [32]. Surprisingly, despite having heterozygosity at only a single gene in an otherwise homozygous background, we observed expression changes in all directions (Dataset S2). One possible explanation among many for this complexity is that SFT is involved in multiple feedback loops and regulates major signaling cascades [33]. However, our primary interest was not to classify and compare these expression differences to whole genome heterozygotes or to dissect transcriptional regulatory networks controlled by SP or SFT, but rather to use the RNA-Seq data as a quantitative molecular phenotyping tool to determine if there are changes in seedling maturation caused by sft/+ heterozygosity before gross morphological differences in shoot architecture become apparent.
Fig. 3. Transcriptome profiling reveals an early semi-dominant delay on seedling development from sft/+ heterozygosity. 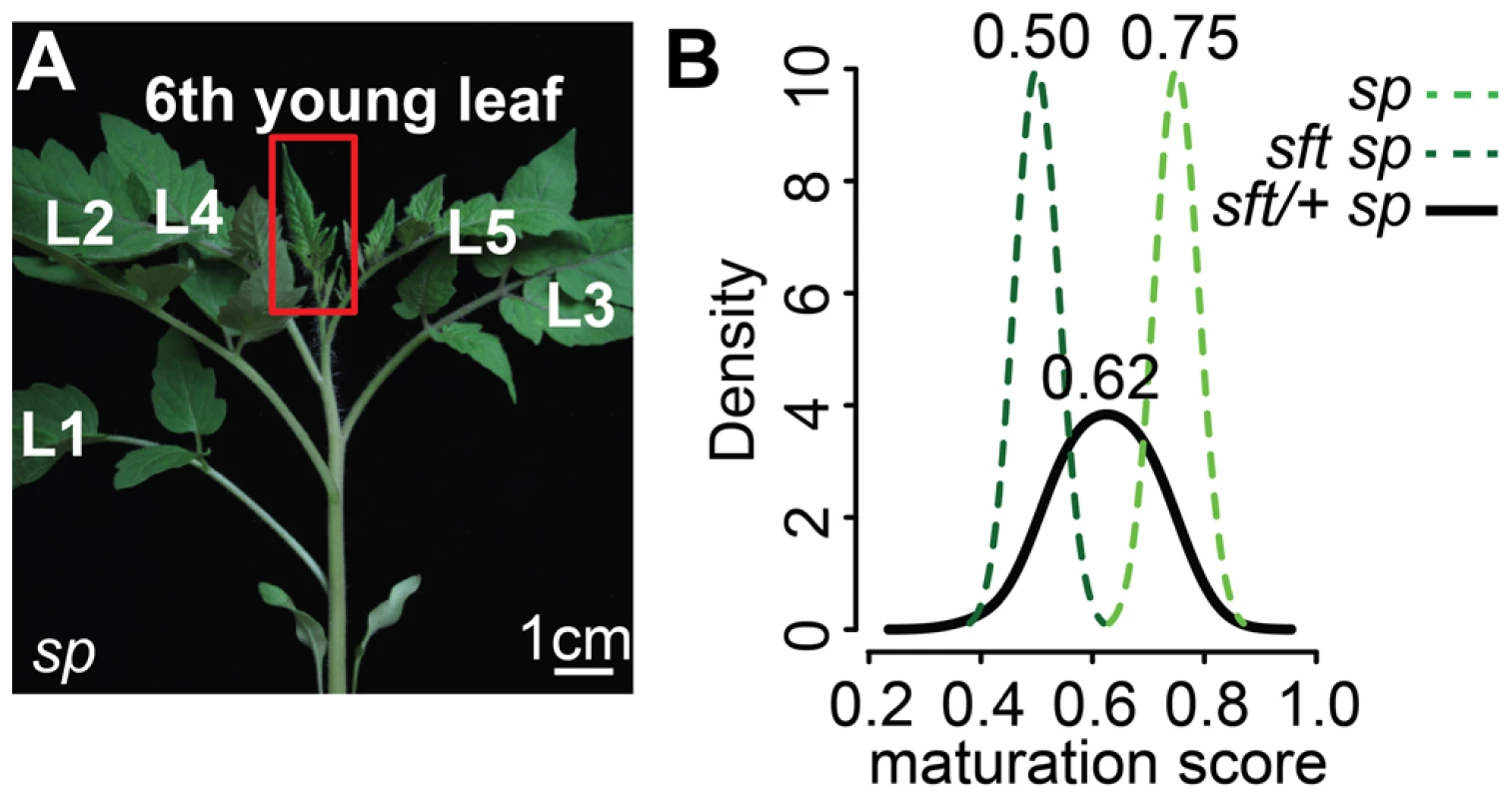
(A) Representative 6th expanding leaf from sp mutants. The same leaf and stage (3 cm long) was profiled by RNA-Seq for sft/+ sp and sft sp genotypes. (B) Molecular quantification of leaf maturation using the DDI algorithm [31]. Given that seedling development of sft sp is delayed compared to sp based on extreme late flowering, the sft sp 6th expanding leaf was designated an early leaf calibration point. Dark and light green curves indicate sft sp and sp maturation score distributions based on 124 DDI-defined marker genes. The black curve for the sft/+ sp 6th leaf indicates an intermediate maturation state. Numbers above indicate average maturation scores. The Digital Differentiation Index (DDI) algorithm identifies transcriptional marker genes whose expressions peak at chosen reference stages to identify stage-enriched marker genes and then queries these marker genes from transcriptomes of “unknown” tissues to predict their maturation states relative to the references [31]. DDI revealed that sft/+ sp 6th leaf maturity was in between sft sp and sp, indicating that sft/+ heterozygosity delays maturation of sp plants already as young seedlings (Figure 3B, Dataset S3). We next asked whether the change in SFT dosage might be sensed in the PSM before it transitioned to flowering. We previously captured and quantified transcriptomes of five developmental stages of PSM maturation, which revealed a meristem maturation clock underlies a gradual transition of the PSM to a reproductive state [34]. The transition meristem (TM) stage of this clock is marked by increasing expression of flowering transition genes [34], and we therefore chose this stage for molecular phenotyping and comparison (Dataset S2 and S3). Importantly, TMs can be collected at precisely matched ontogenetic points, defined by initiation of the last leaf and indistinguishable meristem morphologies of tall round domes (Figure 4A–C) [34]. As expected based on the primary inflorescence of sft mutants reverting into a vegetative shoot, and consistent with sft epistatic over sp, DDI revealed that the TM of sft sp double mutants exhibited a severely delayed maturation, most closely matching a vegetative meristem state (Figure 4D). In contrast, whereas sp TM maturity was indistinguishable from WT, the sft/+ sp TM was delayed relative to sp and therefore intermediate between sp single and sft sp double mutants (Figure 4D). Importantly, we also profiled the first SYM from sp and sft/+ sp plants (sft sp plants fail to form a SYM) (Figure 4E and F), and found that, like in the PSM, the sft/+ sp SYM was also delayed relative to sp (Figure 4G). Altogether, these expression data suggest an early semi-dominant effect on the PSM flowering transition is the triggering event for sft/+ yield increases, and that all subsequently formed vegetative meristems in sp plants become equally sensitive to reduced dosage of SFT as they transition to a reproductive state.
Fig. 4. Transcriptome profiling reveals a semi-dominant delay in meristem maturation from sft/+ heterozygosity. 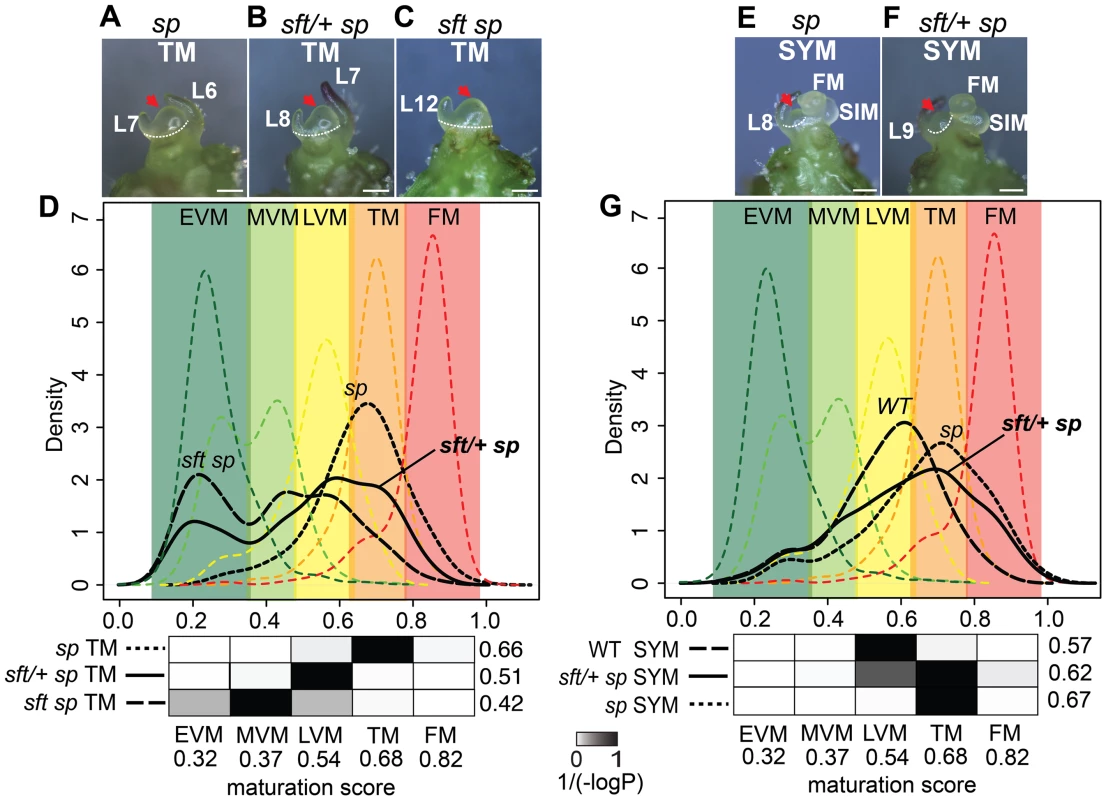
(A–C) Stereoscope images showing morphology and dissection (white dashed line) of the TM stage used for mRNA-Seq from sp (A), sft/+ sp (B) and sft sp (C) genotypes. Scale bar: 100 um. Red arrows highlight identical TM morphologies. L: leaf primordium number. The additional leaf primordium at the sft/+ sp TM is consistent with the one leaf delay in primary shoot flowering time (Figure 2). (D) DDI quantification of maturation scores for sp, sft sp, and sft/+ sp predicted from the WT PSM meristem maturation atlas [34]. Colored dashed curves indicate maturation stages for the 5 PSM stages used for calibration EVM, MVM, LVM, TM and FM [the Early, Middle, and Late Vegetative Meristems, Transition Meristem and the Flower Meristem]. Colored areas define boundaries of these stages estimated from the curves. Maturation scores are derived from 637 DDI-selected marker genes (Dataset S3). Student's t-tests are presented as heat-maps of scaled 1/(−log10P) values below each graph, and associated numbers to the right indicate average maturation scores for the predicted meristems. Darker color indicates greater similarity in maturation state. Note the statistically intermediate TM maturation state of sft/+ sp relative to sft sp and sp, indicating sft/+ heterozygosity causes a semi-dominant delay in the primary flowering transition. The presence of more than one peak along the curves of the sft sp and sft/+ sp genotypes reflect mixed maturation states for these TMs, as different subsets of marker genes are driving different maturation stage estimates that translate to less uniform maturation patterns. (E–F) Stereoscope images showing morphology and dissection of the first sympodial shoot meristem (SYM) used for mRNA-Seq profiling in sp (E) and sft/+ sp genotypes (F). Meristems and leaf primordia are marked as in Figure 2. (G) DDI quantification of SYM maturation scores from sp, sft/+ sp, and WT using the PSM stages as calibrations. Maturation scores for sft/+ sp, sp and WT indicate an intermediate maturation state for the SYM of sft/+ sp plants, mirroring the delay in the PSM. P-value heat maps are shown below along with average maturation scores to the right. Suppression of SFT by artificial microRNA phenocopies the dosage effects of sft/+ heterozygosity
Our findings that sft single gene overdominance traced back to cumulative delays on recurring flowering transitions led us to reason that the dosage effects of sft/+ heterozygosity might be recapitulated by simply partially reducing levels of functional SFT transcripts. We tested this by over-expressing artificial microRNAs against SFT (35S::amirSFT) in the sp background [35], [36]. In addition to SFT, the artificial microRNAs were designed to target the Arabidopsis thaliana SFT ortholog, FLOWERING LOCUS T (FT), to assess their broad efficacy, and were incorporated into two different Arabidopsis pre-microRNA templates, At pre-mir164b and At pre-mir319a, to guard against differential amir backbone efficiencies (Figure 5A). In Arabidopsis, 35S:amiR-SFT/FTAt164b and 35S:amiR-SFT/FTAt319a transformants exhibited late flowering phenotypes equivalent to ft mutants (Figure S2B–C). In tomato, six of eight first generation (T1) transformants showed sp suppression phenotypes, and we selected three lines representing the range of observed suppression for further analysis. SFT transcript abundance was evaluated in these lines by quantitative RT-PCR, revealing a range of knockdown levels by the artificial microRNAs (Figure 5B). We evaluated progenies from two 35S:amiR-SFT/FTAt164b (referred to as amirSFTa and amirSFTb) and one 35S:amiR-SFT/FT At319a (referred to as amirSFTc) transformants, and found that the amirSFTa produced an average of one additional sympodial unit and inflorescence compared to non-transformed sp mutants, closely resembling the dosage effects of sft/+ heterozygosity (Figure 5C). amirSFTc showed greater suppression, terminating sympodial growth after producing often more than two additional units, while amirSFTb fully suppressed sp to indeterminacy like WT plants (Figure 5C). Notably, the level of suppression of sp determinacy corresponded with the level of knockdown of SFT; e.g. the indeterminate line, amirSFTb, showed the greatest reduction of SFT transcripts (Figure 5B–C). In all six lines, we failed to find strong sft sp double mutant phenotypes of reverted inflorescences or loss of sympodial growth, suggesting only weak alleles of SFT were created with the 35S::amirSFT transgene – an effect that is also consistent with often observed weak target knockdown by artificial microRNAs [35], [36]. Importantly, we found delayed flowering time in successive sympodial units like in sft/+ sp heterozygotes, and all three amirSFT progeny populations exhibited delayed primary shoot flowering time (Figure 5D). Thus, tuning SFT dosage transgenically mimics the effects of sft/+ heterozygosity, further illustrating that a classical epistasis relationship between the sft and sp mutants is ultimately responsible for the overdominant effect on yield.
Fig. 5. Reducing SFT transcripts with artificial microRNAs mimics the dosage effects of sft/+ heterozygosity. 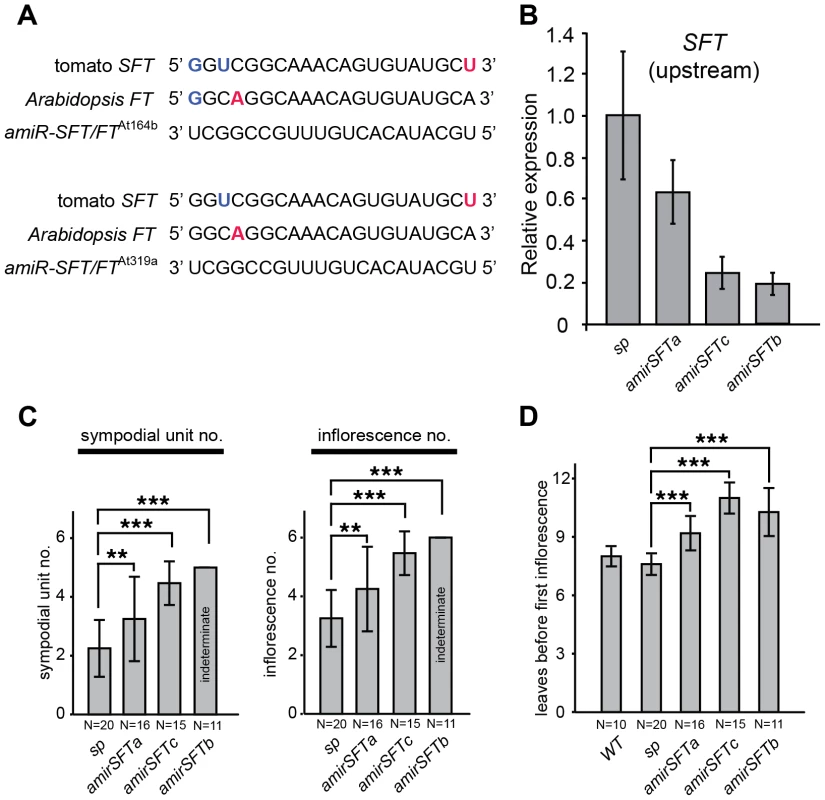
(A) Artificial microRNAs targeting tomato SFT and Arabidopsis FT. Shown are alignments of amiR-SFT/FTAt164b and amiR-SFT/FTAt319a with the complementary region of SFT and FT. G–U wobbles and mismatches between the two amiR-SFT/FTs and the target are highlighted in the target sequence with bold blue and red, respectively. (B) Quantitative RT-PCR measurements of tomato SFT transcript levels in amirSFT plants showing knock down. Results shown are from using primers targeting SFT transcripts 5′ to the amiRNA binding site, consistent with reports of primer-dependent transitivity occurring at the 3′ to 5′ direction upon the initial target cleavage, resulting in degradation of the 5′ cleaved product of the target but not the 3′ product [80], [81] (Figure S2). Bars indicate relative expression level and error bars indicate standard deviation among replicates. (C) Depending on the strength of suppression, amirSFT plants produce at least one additional sympodial unit and inflorescence compared to sp alone, indicating that reducing SFT transcript levels by artificial microRNA partially suppresses sp sympodial termination, mimicking the dosage effect of sft/+ heterozygosity. Note that some amirSFTc progeny plants showed indeterminacy, whereas amirSFTb progeny plants were always indeterminate, indicating that a stronger suppression of SFT completely suppresses the sp phenotype and reverts the plants to normal sympodial cycling. Differences in sympodial unit and inflorescence numbers between amirSFT and sp plants were tested by Wilcoxon rank sum test and significance levels are marked by asterisks (* P<0.05, ** P<0.01, *** P<0.001). (D) amirSFT plants have delayed primary shoot flowering time compared to sp and WT controls, similar to sft/+ heterozygosity. Bars indicate average leaf numbers with standard deviations. Genotypes and sample sizes are shown below. Differences in leaf numbers between amirSFT and sp plants were tested by Wilcoxon rank sum test and significance levels are marked by asterisks (* P<0.05, ** P<0.01, *** P<0.001). A dosage effect from florigen mutant heterozygosity is conserved in Arabidopsis, but does not cause heterosis
As florigen is a universal inductive signal for flowering that several flowering pathways converge upon [37], [38], we wondered if and how florigen mutant heterozygosity in a different system might affect growth, and specifically whether heterosis would result. We tested this by creating orthologous mutant combinations in Arabidopsis thaliana, which is a monopodial plant in which a single flowering event converts the SAM into a continuously growing inflorescence meristem (IM) that produces flowers laterally, in contrast to the tomato sympodial growth habit in which multiple flowering transitions occur. Despite this difference, Arabidopsis ft (sft) mutants are likewise late flowering [39] and completely epistatic over the early flowering and precocious termination of inflorescence meristems of tfl (sp) mutants [40]. To evaluate potential dosage effects of ft/+ heterozygosity, we phenotyped progeny from ft-2/+ tfl1-2 plants, in which the ft-2 mutation, a strong allele, segregates in the tfl1 background (Figure 6A). We measured flowering time by counting rosette leaves and found a clear dosage effect in ft-2/+ tfl1 plants compared to tfl1 single and ft-2 tfl1 double mutants (Figure S3A). We next tested for heterosis by quantifying yield related traits, including plant height, number of axillary shoots, and, as a parallel to tomato yield, the number of siliques, flowers, and flower buds (Figure 6B–C and Figure S3B–D). Surprisingly, ft/+ tfl plants showed semi-dominance for plant height and total yield (Figure 6B and C), and similar effects were observed for a moderate second allele of ft (Figure S3E). Thus, whereas the dosage effect on flowering time from florigen mutant heterozygosity is conserved in the monopodial growth habit of Arabidopsis, it does not translate to heterosis.
Fig. 6. Dose-dependent suppression of tfl1 (sp) by ft/+ (sft/+) heterozygosity is conserved in Arabidopsis thaliana. 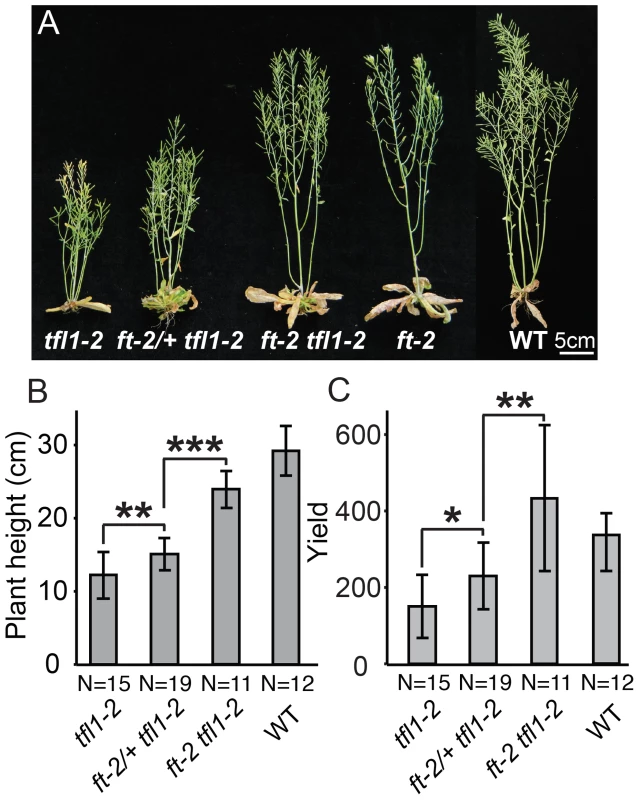
(A) Representative plants from left to right of: tfl1-2 single mutants, ft-2/+ tfl1-2, ft-2 tfl1-2 double mutants, ft-2 single mutants and wild type Ler-0 (WT) showing the intermediate height of ft-2/+ tfl1-2 plants compared to tfl1-2 and ft-2 tfl1-2 genotypes. (B–C) Statistical comparisons among all genotypes for plant height and flower/fruit yield showing semi-dominant effects from ft-2/+heterozygosity in the tfl1-2 background. Bars indicate average values with standard deviation. Genotypes and sample size are shown below. Differences between genotypes were tested by a Wilcoxon rank sum test and significance levels are marked by asterisks (*P<0.05, **P<0.01, ***P<0.001). (B) ft-2 heterozygosity in a tfl1-2 mutant background partially suppresses the early flowering and early termination phenotype of the tfl1-2 mutation in a semi-dominant manner, resulting in plant height in between tfl1-2 and ft-2 tfl1-2 mutant parental lines. (C) Unlike tomato, ft/+ heterozygosity in a tfl1-2 mutant background does not drive heterosis for yield (number of total siliques and floral buds) in Arabidopsis. Rather, yield in the ft-2/+ tfl1-2 plants is intermediate to tfl1-2 and ft-2 tfl1-2 double mutants. Discussion
Crop yields derive from a complex integration of fitness-related traits founded on developmental and physiological mechanisms for organ production and biomass accumulation. Thus, studying heterosis inevitably involves a broad analysis of the myriad mechanisms controlling plant growth. It is therefore perhaps not surprising that recent gathering of vast genetic, phenotypic, and molecular data on cases of heterosis from diverse systems has suggested that multiple non-mutually exclusive system-specific mechanisms are likely at work [8]–[10], [41]. Looking at heterosis from the developmental perspective, it would be reasonable to assume a priori that flowering would have a major role given that selection of allelic variation for flowering time regulators has been a major contributor to adaptation, domestication, and maximizing crop yields through classical and modern breeding [42]. In rice, for example, alleles of strong effect from various flowering regulators, many showing epistatic interactions, were selected to enable growth at different climates and day lengths [43]–[45]. The same was achieved in maize, but, instead, dozens of loci of small additive effect were found to be involved [46]. In both rice and maize, and as occurred during the domestication and breeding of many crops, this selection enabled a shift from an extended period of flowering in wild populations to uniform flowering, which provided sudden bursts of yield that facilitated agronomic practices, particularly harvesting [42]. Interestingly, the genetic path leading to high yielding tomatoes has differed from other major crops in that domestication has mostly acted on fruit size to increase yield with little evidence for selection on flowering [47]–[50]. Indeed, while there is certainly flowering time and architectural variation among distantly related wild tomato species [51], cultivated tomatoes and their wild progenitor, S. pimpinellifolium, share nearly identical flowering times and indeterminate growth habits, suggesting there was little or no standing genetic variation for artificial selection to act upon [52]. Only with the relatively recent discovery of sp did a change in flowering provide a major agronomic shift in how tomato was grown in the field, enabling a burst of flower production and yield on compact plants grown at high density, which gave rise to the processing tomato industry [26]. In this regard, in contrast to maize where altered flowering times are frequently observed in hybrids [10], [53], cultivated tomato hybrids do not differ substantially from their parental inbreds for flowering time, inflorescence production, or overall plant architectures. Only upon introgressing quantitative trait loci (QTL) from distantly related wild species are heterotic effects on yield observed, a subset of which have been tied to changes in flowering and plant architecture, but the causative genes have not been identified [54]. Thus, our dissection of sft heterosis is the first to expose a direct link to flowering and resolve the underlying mechanism.
Our combined developmental and molecular phenotyping of sft/+ overdominance has exposed a novel principle for how tomato plant architecture and yields might be further optimized by taking advantage of the surprising and remarkable level of dosage sensitivity within florigen and the florigen pathway. The genetically induced reduction in dosage of florigen from sft/+ heterozygosity causes a slight delay in the transition to reproductive growth that, in the context of recurring flowering events of the sympodial habit and the sp mutant background, translates to cumulative overdominance. Indeed, this heterosis example, like many others [20]–[24], [55], is conditional. Yet, it is this genetic and developmental conditionality that suggests sft/+ heterosis could be considered less about heterozygosity and heterosis per se and more about the potential to genetically fine-tune SFT expression levels to manipulate yield in a way that domestication and breeding efforts have not yet capitalized on, perhaps because standing allelic diversity for florigen and members of its pathway is limited. In this respect, we propose that additional directed quantitative manipulation of the relative doses of SFT to SP might enable further fine-tuning of flowering, sympodial cycling, and inflorescence production. For example, as yet undiscovered, or artificially created [56], [57], transcriptional or loss-of-function alleles of SFT and SP of various strengths could be combined in different genetic constitutions to pinpoint an even higher optimum of plant architecture to maximize yield. In an even simpler scenario, homozygosity for very weak mutant alleles of SFT in a strong sp background, or homozygosity for weaker mutant alleles of SP alone, could potentially match or exceed fruit production of the sft/+ sp genotype. Finally, beyond tweaking SP and SFT, partial suppression of sp determinacy by generating mutations in other pathway genes, especially those encoding components of the florigen activating complex [58], could provide novel alleles and breeding germplasm that natural variation might not be able to provide.
Importantly, although there is tremendous diversity among angiosperms in when and where inflorescences and flowers form, the SFT/SP system is highly conserved [38], [59], [60], suggesting the aforementioned concepts could be applicable to other plants. Yet, our findings in Arabidopsis imply that while dosage effects on flowering time from florigen mutant heterozygosity will be broadly conserved, yield benefits might not be, and species-specific outcomes will likely trace back to differences in growth habits. The lack of meristem termination and recurring flowering events in the monopodial growth habit of Arabidopsis means that florigen mutant heterozygosity is sensed only once during development, and that no compounding of the semi-dominant dosage effect is possible. Indeed, increasing yield in Arabidopsis simply requires a larger plant, which can be achieved by delaying and prolonging flowering either environmentally through short day growth conditions or genetically through mutations in flowering regulators like FT. Consistent with this, we found that homozygous ft mutants were the highest yielding of all genotypes (Figure 6). At first glance, this would suggest limited possibilities for exploiting our findings beyond tomato; however, for some breeding goals, such as improving biomass, delaying flowering quantitatively and predictably through an allelic series of florigen mutants in either the homozygous or heterozygous condition could prove valuable to customize plant architecture and size for particular agronomic needs. Remarkably, yield benefits from heterozygous mutations in florigen orthologs have been found in at least one plant that lacks sympodial growth. In a strikingly similar example to tomato, a major domestication QTL for flowering time in sunflower traces back to a deletion in a duplicated paralogous FT gene that causes heterosis for both seed size and weight when heterozygous under short day conditions [61]. In another example, a classical report of overdominance for sorghum yield involves heterozygosity for an as yet uncharacterized late flowering mutant that has all the hallmarks of being defective in florigen or a florigen pathway component [62]. Thus, heterozygosity for florigen mutants holds potential for broadly improving crop yields, which, in hindsight, is perhaps not surprising given that selection for beneficial alleles of various strengths in florigen family genes, especially orthologs of SFT and SP, was key for the domestication of barley [63], beets [64], beans [65], [66], grape [67], potatoes [68], roses [69], soybeans [70], [71], sunflower [61], tobacco [72], and likely many other plants. With these examples in mind, and considering our findings in Arabidopsis, we suggest that sft/+ heterozygosity in a dose-dependent epistatic relationship with sp may represent only one of several ways to genetically tailor florigen levels, and that hunting for new alleles in existing germplasm or engineering custom alleles could allow an optimal fine-tuning of florigen and its pathway to maximize flowering, inflorescence production, and other yield components in these and other crops. The potential to broadly manipulate agronomic traits by florigen and its family members in diverse plant species stems not only from roles in flowering time, but also as general coordinators of diverse physiological processes affecting multiple aspects of plant growth and fertility [38]. Thus, parallel to how mutations in biosynthesis genes for the hormone gibberellin created the dwarf mutants that propelled the Green Revolution [73], our findings provide compelling evidence that manipulating florigen family genes can provide a new path to meet current breeding challenges associated with a rapidly changing climate.
Materials and Methods
Tomato plant growth conditions, genotyping, and phenotyping
The sp mutant was first reported more than 80 years ago and arose spontaneously, and the strong sft mutant allele used in this study, sft-7187, was isolated from a fast neutron mutagenesis screen performed in tomato cultivar M82, and has a two nucleotide deletion that truncates the C-terminal portion of the protein [26], [27], [74]. All mutants were backcrossed to M82 at least four times to eliminate background mutations prior to the original yield trials [25]. For all experiments in this study, plants were grown in controlled greenhouse conditions at Cold Spring Harbor Laboratory. Greenhouses were supplemented with artificial light from high-pressure sodium bulbs (50 µmol/m2/sec; 16 h/8 h) and daytime temperature was 78°F and nighttime temperature was 65°F, with a relative humidity of 40–60%. Tomato F2 generation seeds derived from self fertilization of an sft/+ sp F1 plant were grown in 72-cell insert flats and transplanted after four weeks into 2 gallon pots (three plants per pot) for quantitative phenotyping. Young leaf tissue was collected from each F2 individual at the time of transplanting for DNA extraction and genotyping. Total genomic DNA was extracted using a standard cetrimonium bromide (CTAB) DNA extraction protocol. Genomic fragments of the SFT locus were amplified using the PCR primers: “sft-7187 full exon F2” 5′-GGGCAAGAAATAGTGAGCTAT-3′ and “sft-7187 full exon R2” 5′-TTCAAATAAATTGAGAGGAAGA-3′ and the following PCR program: initial denaturation at 94°C for 3 minutes, then 35 cycles at 94°C for 30 seconds, annealing at 52°C for 30 seconds, extension at 72°C for 1 minute, and a final extension at 72°C for 10 minutes. The PCR products were subjected to enzyme digestion with TseI at 60°C for 6 hours, resulting in two bands for wild type, one band for sft mutant and three bands for sft/+ after running on a 3% agarose gel at 150 V for 40 minutes. The number of leaves in the primary shoot prior to the first inflorescence and leaves within three successive sympodial units were counted for each individual at 8–12 weeks after germination. This same phenotyping scheme was applied to two axillary shoots: the lower (basal) axillary shoot originating from the axil of the first leaf on the primary shoot and the uppermost (proximal) axillary shoot originating from the axil of the last leaf formed before the first inflorescence. Quantitative measurements for inflorescence number, sympodial unit number, primary and lateral shoot flowering time, and leaf number in three sympodial units were evaluated for the shape of each phenotype's distribution and subjected to two-tailed Wilcoxon rank sum tests between genotypes and Kruskal–Wallis one-way analysis of variance across all genotypes. To quantitatively compare the progression of sympodial inflorescence meristem (SIM) and floral meristem (FM) initiation on the first developing inflorescence of sp and sft/+ sp plants, we germinated 18 plants for both genotypes at the same time and counted the number of differentiated FMs on both primary and sympodial shoots at 20th days after germination (DAG). The FM numbers were subjected to two-tailed Wilcoxon rank sum tests between genotypes. To image live meristems, shoot apices were dissected from seedlings, and older leaf primordia (>150 µm) were removed under a Nikon SMZ1500 stereomicroscope. The meristem images were taken immediately after dissection with an integrated Nikon digital camera, recaptured by Z-series manually, and merged to create focused images.
Arabidopsis plant growth conditions, genotyping, and phenotyping
Arabidopsis thaliana plants were grown in the greenhouse under long day (16 h light, 8 hr dark) conditions in 32-cell flats with two plants per cell. Individual seeds were delivered to the corner of each cell to avoid growth competition during germination. The seeds were stratified at 4°C for 4 days before transferring to a long day greenhouse maintained at 21°C. All mutant lines were acquired from the Arabidopsis Biological Resource Center (ABRC) and originated from EMS mutagenesis in the Landsberg erecta (Ler) background. Homozygous tfl1-2 mutant plants were crossed to a moderate (ft-1) and strong (ft-2) allele of ft. Individual F1 plants from each cross were self-fertilized to generate F2 populations segregating for both tfl1-2 and ft mutants. Plants homozygous for the tfl1-2 mutation and heterozygous for the ft-2 mutation were self-fertilized to generate F3 populations fixed for the tfl1-2 mutation and segregating for the ft-2 mutation. Tissue was harvested from young rosette leaves and DNA was extracted using a standard CTAB DNA extraction protocol. The tfl1-2 and ft-2 mutations were detected using derivative CAPS (dCAPS) assays. A fragment of TFL1 was amplified by PCR using the primers “tfl1-2 dCAPS-F” 5′ - AAACGTCTCACTTCCTTTTCCTC-3′ and “tfl1-2 dCAPs-R2” 5′ - AAATGAAAAGAAAGAATAAATAAATTAAAGGTAC-3′ and a fragment of FT was amplified using “ft-2 dCAPS-F2” 5′ - CCCTGCTACAACTGGAACAACCTTTGGTG-3′ and “ft-2 dCAPS-R2” 5′ - AAACTCGCGAGTGTTGAAGTTCTGGGGC-3′. Both TFL1 and FT fragments were amplified using a touchdown PCR program: initial denaturation at 95°C for 3 minutes, then 10 cycles at 95°C for 20 seconds, 65°C for 30 seconds (decreased by −0.5°C/cycle), 72°C for 30 seconds followed by an additional 30 cycles at 95°C for 20 seconds, 52°C for 30 seconds, 72°C for 30 seconds and ending with a final extension at 72°C for 10 minutes. Underlined nucleotides in the aforementioned sequences introduce a new restriction site in the wild type PCR amplicons. TFL1 PCR amplicons were digested using KpnI for 3 hours at 37°C, which cuts wild type but not mutant sequences. FT PCR amplicons were digested using HaeIII for 3 hours at 37°C, which cuts wild type but not the ft-2 mutant sequences. Wild type versus mutant banding patterns was resolved on a 3% half MetaPhor agarose-half regular agarose gel. Phenotyping was completed in the F3 generation, and we compared tfl1-2 ft-2 double, tfl1-2 ft-2/+ and tfl1-2 single mutants. Homozygous single mutants and wild type Ler-0 were grown at the same time for comparison. Phenotyping and imaging was performed when the plants completed flowering and inflorescence meristems stopped growing (6–8 weeks after germination). The height of each plant was measured along the main shoot of the plant from where the base emerged from the rosette to top of the shoot. The number of rosette leaves, axillary shoots, siliques, open flowers, and floral buds were also recorded as measures of flowering time and yield. For each measured trait, the mean and standard deviation was calculated for each genotype. The means were compared using a Student's t-test (Wilcoxon rank sum test when the phenotypic distribution was not normal).
Global gene expression profiling (mRNA-Seq) of tomato leaves and meristems
Tomato homozygous sp mutants, sft sp double mutants and F1 single gene heterozygotes of sft/+ sp plants were used for leaf and meristem expression profiling experiments. All sft/+ sp plants originated from F1 seeds of direct crosses between the sp and sft sp parents, and a subset of F1 plants were confirmed by PCR genotyping to ensure 100% sft/+ heterozygosity. Seeds were germinated in petri plates on water-soaked Whatman paper at 28°C for 72 hours until the root radicles emerged. The germinated seeds were then transplanted to 72-cell insert flats with pre-wet soil and placed in the greenhouse. The plants used for leaf expression profiling were transplanted to two-gallon pots (three plants per pot), and tissue from the 6th young expanding leaf from each plant was collected and immediately frozen in liquid Nitrogen when the leaves reached 3 cm in length. Total RNA was extracted using a Qiagen RNeasy mini total RNA extraction kit according to the manufacturer's protocol. Growth of seedlings for meristem expression profiling was monitored daily under a dissecting microscope using the meristem morphological cues marking previously defined maturation stages [34]. At the transition maturation (TM) stage, the cotyledons and leaves were removed from seedlings and the shoot apices with 3 cm hypocotyl attached were collected and stored in 100% acetone followed by vacuum infiltration for 30 minutes. Meristem tissue was dissected from the fixed stems using a surgical blade following the lines shown in Figure 4A–C and E–F under a dissecting microscope after confirming the morphology that marks the TM stage. Total RNA was extracted from the dissected meristem tissues with an Arcturus PicoPure total RNA extraction kit (Life Technologies). Except for the sp SYM, which is difficult to capture in high numbers because of a rapid termination, for all genotypes, tissue was harvested and prepared for mRNA-Seq construction for two biological replicates, and sp SYM was subjected to two technical replicates. As reported previously [34], two replicates were sufficient to quantify meristem maturation states using the DDI algorithm, which was our primary goal in the expression analysis.
RNA-Seq library preparation
For all tissues, poly-A containing mRNA was purified from total RNA using Invitrogen oligo-dT DynaBeads for mRNA-Seq library construction using the ScriptSeq v2 RNA library preparation kit (Epicentre). The maximum amount of mRNA input (50 ng) was used when possible to maximize the library output. The final PCR enrichment step was carried out following the standard protocol with 15 cycles and primers with barcode indices supplied by Epicentre to create barcoded mRNA-Seq libraries. The quantity and size distribution of each individual barcoded mRNA-Seq library was detected with a High Sensitivity DNA Chip on a Bioanalyzer 2100 machine (Agilent). The final concentration of each library was verified by qPCR using a KAPA library quantification kit and based on these results, four to six barcoded libraries were pooled together with equal concentration for one lane of Illumina paired-end (PE) 100 bp sequencing on an Illumina HiSeq sequencing machine (Dataset S1). All reads files were deposited to SGN (ftp://ftp.solgenomics.net/transcript_sequences/by_species/Solanum_lycopersicum/libraries/illumina/LippmanZ/) and the mean RPKM values of meristems are visualized on an eFP browser (http://tomatolab.cshl.edu/efp/cgi-bin/efpWeb.cgi, SFT heterosis panel).
Read mapping and analysis
All mRNA-Seq reads were trimmed to 50 bp to remove the bases with low qualities and mapped using Bowtie [75] to the tomato reference CDS [76] with paired-end relationships maintained. Trimming the reads to 50 bp also made the libraries comparable to our previous mRNA-Seq libraries [34] for combined DDI analyses. The lack of size selection step in the Epicentre ScriptSeq v2 mRNA-Seq library preparation protocol allowed lower initial mRNA input but produced a larger insert size range (150 bp∼1000 bp), which lowered the successful mapping with proper distance between paired-end reads. Mapping to predicted CDS also reduced the mapping rate due to failed mapping of reads coming from 5′ and 3′ UTR regions. However, the higher total read number from Illumina HiSeq compensated for the relatively lower mapping rates, yielding comparable mapped read numbers and sequencing depth to previous mRNA-Seq libraries that allowed for differential expression analysis and molecular phenotyping by DDI [34]. The resulting bam alignments were sorted and indexed by SAMtools [77], and the number of reads mapped to each CDS was counted to calculate the raw counts for all libraries. The raw counts from leaf and TM tissues across three genotypes were normalized using the TMM method. The distribution of gene expression levels were modeled following a negative-binomial distribution and tag-wise dispersion were estimated based on two replicates. Finally, exact tests for differential expression were conducted based on the replicates in pairwise comparisons. All normalization and differential expression tests were conducted using the edgeR package [78], [79]. Although only two replicates were performed, we classified gene expression patterns from comparing sft/+ sp heterozygotes and homozygous parents into 12 categories belonging to five major classes: additive, recessive, dominant, overdominant and underdominant (Dataset S2) using a threshold of two-fold change and P-value< = 0.01. Numbers of genes in each category were counted and their proportions in each category relative to all differential expressed genes were calculated for the 6th young leaf and TM, respectively, revealing all categories of gene expression changes were detected (Dataset S2).
Digital Differentiation Index analyses
Raw counts for the leaf expression profiles (including sp, sft/+sp and sft sp 6th young leaves) were incorporated into a master leaf data set. Raw counts for the meristem expression profiles (including sp and sft/+sp TM and SYM) were incorporated into a master meristem data set that includes all raw counts from our previous meristems profiling experiments [34]. For both master data sets, all raw counts were then summarized over replicates and normalized against number of mapped reads and CDS lengths to calculate RPKM values for DDI analyses [31]. DDI selects samples with known or pre-determined maturation states in the whole data set as calibration points, and then identifies marker genes that show maximum expression at each calibration point. These genes characterize the calibration points molecularly. DDI checks the marker gene expressions in the samples that are submitted to query (the ‘unknown’ samples) and quantifies the ‘unknown’ samples' maturation states relative to the calibration points. For each marker gene, DDI compares expression levels between ‘unknown’ samples and each calibration point and calculates a ‘maturation score’. Collectively, all marker genes generate a distribution of maturation scores for the ‘unknown’ sample [31]. Importantly, curves showing multiple ‘peaks’ reflect a mixed molecular maturation state for the queried tissue, as different marker genes give different maturation estimates. This is most evident in sft sp double mutants that still transition to flowering, but at a much slower rate compared to wild type and with vegetative reversion of the inflorescence, indicative of a mixed vegetative-reproductive state. At the same time, a Student's t-test of average maturation score difference between calibration and unknown samples was conducted for each unknown meristem sample, yielding a P-value for the significance of the maturation state difference. For each prediction, this P-value was obtained for comparisons between the unknown sample and temporarily successive calibration points, in order to generate a ‘gradient’ of meristem similarity (plotted in heat-maps in the form of scaled 1/(−log10P)). For example, to predict the maturation state of sft/+ sp SYM using the first replicate of WT EVM, MVM, LVM, TM and FM [the Early, Middle, and Late Vegetative Meristems (EVM: 5th leaf initiated; MVM: 6th leaf initiated; LVM: 7th leaf initiated), the Transition Meristem (TM: 8th leaf initiated), and the Flower Meristem (FM)] as calibration points, P-values were calculated for maturation state comparisons SYM vs. EVM, SYM vs. MVM, SYM vs. LVM, SYM vs. TM and SYM vs. FM, respectively. The P-values were then transformed into 1/(−log10P) and scaled across five values into a zero to one range (scaling was done for each prediction independently). Because smaller P-values indicate larger differences in maturation scores, the scaled 1/(−log10P) values quantify the relative similarity of the sft/+ sp SYM to each of the five calibration points. With the master leaf data set, DDI analyses were conducted using sft sp and sp 6th young leaves as two calibration points to predict maturation stages of sft/+sp leaf maturation. With the master meristem data set, DDI analyses were conducted using five WT primary shoot meristem (PSM) stages as calibration points to predict maturation stages of sp, sft/+sp and sft sp meristems. As in [34], one replicate of calibration samples was used for marker gene identification (Dataset S3), a second replicate of calibration samples treated as unknowns was predicted and plotted to set the boundaries of maturation stages (colored curves and boxes in Figure 4D and Figure 4G), and averaged RPKM values of predicting leaves and meristems were used to generate and plot the predicted distribution of maturation scores. All parameters for DDI analyses were as previously described [34]. All DDI analyses were carried out using modified R scripts as described previously [34].
Artificial microRNA construction and transformation
Artificial microRNAs were designed to repress both tomato SFT and Arabidopsis FT with two different backbones (Figure S1) [35]. The artificial microRNA amiR-SFT/FTAt164b and amiR-SFT/FT At319a were synthesized by DNA2.0 and Bio S&T, respectively, and transformed into both tomato and Arabidopsis plants and phenotyped for repression of SFT and FT, respectively (Figure 5B, Figure S2). Tomato plants carrying mirSFT transgenes were measured for sympodial unit and inflorescence number, and phenotyping stopped after counting five or more sympodial units with two or more leaves in each unit and classified as indeterminate. The means of phenotypes were compared using a Student's t-test (Wilcoxon rank sum test when phenotype distribution is not a normal distribution).
For quantitative RT-PCR of SFT transcript abundance in the amirRNA lines, cotyledon tissue was collected from two-week old seedlings for total RNA extraction with Qiagen RNeasy mini total RNA extraction kit including DNase treatment with RNase-free DNase (Qiagen) according to the manufacturer's instructions. First-strand cDNA was then synthesized using the SuperScript III First-Strand Synthesis System with oligo dT (Invitrogen). Ubiquitin mRNA (Solyc01g056940) was used as the reference for normalization in quantifying cDNA. 5′ mRNA (upstream, Figure 5B) and 3′ mRNA (downstream, Figure S2A) of SFT transcript (Solyc03g063100) from the amirSFT binding site were quantified with 1 ul of cDNA using Phusion High-fidelity DNA polymerase (NEB), iQTM SYBR Green Supermix (Bio-Rad). A loss of transcripts was detected 5′ to the amiRNA binding site, consistent with reports of primer-dependent transitivity occurring at the 3′ to 5′ direction upon the initial target cleavage, resulting in degradation of the 5′ cleaved product of the target but not the 3′ product [80], [81]. Primers pairs used were: 5′-CGTGGTGGTGCTAAGAAGAG-3′ and 5′ - ACGAAGCCTCTGAACCTTTC-3′ for Ubiquitin (UBI); 5′-GCTTAGGCCTTCCCAAGTTA-3′ and 5′-GGGTCCACCATAACCAAAGT-3′ for 5′ mSFT (upstream); 5′-GACAATTAGGTCGGCAAACA-3′ and 5′-AGCAGCAACAGGTAAACCAA - 3′ for 3′mSFT (downstream). Two biological replicates of qRT-PCR were performed on the CFX96TM Real-time PCR System (Bio-Rad). qRT-PCR data were calculated from the number of PCR cycles needed to reach the linear phase for each SFT transcript from amirSFT lines and normalized against Ubiquitin using the qbase PLUS Data-Analysis Software.
Supporting Information
Zdroje
1. Darwin C (1868) The Variation of Animals and Plants under Domestication. 2.
2. CharlesworthD, WillisJH (2009) The genetics of inbreeding depression. Nat Rev Genet 10 : 783–796.
3. ShullGH (1908) The composition of a field of maize. Am Breed Assn Rep 4 : 269–301.
4. CrowJF (1948) Alternative Hypotheses of Hybrid Vigor. Genetics 33 : 477–487.
5. HochholdingerF, HoeckerN (2007) Towards the molecular basis of heterosis. Trends Plant Sci 12 : 427–432.
6. LippmanZB, ZamirD (2007) Heterosis: revisiting the magic. Trends Genet 23 : 60–66.
7. SpringerNM, StuparRM (2007) Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res 17 : 264–275.
8. ChenZJ (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet 14 : 471–482.
9. SchnablePS, SpringerNM (2013) Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol 64 : 71–88.
10. BirchlerJA, YaoH, ChudalayandiS, VaimanD, VeitiaRA (2010) Heterosis. The Plant cell 22 : 2105–2112.
11. McMullenMD, KresovichS, VilledaHS, BradburyP, LiH, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325 : 737–740.
12. StuberCW, LincolnSE, WolffDW, HelentjarisT, LanderES (1992) Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 132 : 823–839.
13. LiZK, LuoLJ, MeiHW, WangDL, ShuQY, et al. (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158 : 1737–1753.
14. LuoLJ, LiZK, MeiHW, ShuQY, TabienR, et al. (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158 : 1755–1771.
15. StuparRM, SpringerNM (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173 : 2199–2210.
16. IshikawaA (2009) Mapping an overdominant quantitative trait locus for heterosis of body weight in mice. J Hered 100 : 501–504.
17. SchulerJF (1954) Natural Mutations in Inbred Lines of Maize and Their Heterotic Effect. I. Comparison of Parent, Mutant and Their F(1) Hybrid in a Highly Inbred Background. Genetics 39 : 908–922.
18. MukaiT, BurdickAB (1959) Single Gene Heterosis Associated with a Second Chromosome Recessive Lethal in Drosophila Melanogaster. Genetics 44 : 211–232.
19. RedeiGP (1962) Single Locus Heterosis. Zeitschrift Fur Vererbungslehre 93 : 164–&.
20. EfronY (1973) Specific differences in maize alcohol dehydrogenase: possible explanation of heterosis at the molecular level. Nat New Biol 241 : 41–42.
21. HallJG, WillsC (1987) Conditional overdominance at an alcohol dehydrogenase locus in yeast. Genetics 117 : 421–427.
22. GrobetL, MartinLJ, PonceletD, PirottinD, BrouwersB, et al. (1997) A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 17 : 71–74.
23. MosherDS, QuignonP, BustamanteCD, SutterNB, MellershCS, et al. (2007) A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet 3: e79.
24. DelneriD, HoyleDC, GkargkasK, CrossEJ, RashB, et al. (2008) Identification and characterization of high-flux-control genes of yeast through competition analyses in continuous cultures. Nat Genet 40 : 113–117.
25. KriegerU, LippmanZB, ZamirD (2010) The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet 42 : 459–463.
26. PnueliL, Carmel-GorenL, HarevenD, GutfingerT, AlvarezJ, et al. (1998) The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125 : 1979–1989.
27. LifschitzE, EviatarT, RozmanA, ShalitA, GoldshmidtA, et al. (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci U S A 103 : 6398–6403.
28. Saltveit ME (2005) Post harvest biology and handling. In: Heuvelink E, editor. Tomatoes. Wallingford, U.K.: CABI Publishing. pp. 305–325.
29. Peet MM, Welles G (2005) Greenhouse tomato production. In: Heuvelink E, editor. Tomatoes. Wallingford, U.K.: CABI Publishing. pp. 257–304.
30. LippmanZB, CohenO, AlvarezJP, Abu-AbiedM, PekkerI, et al. (2008) The making of a compound inflorescence in tomato and related nightshades. PLoS Biol 6: e288.
31. EfroniI, BlumE, GoldshmidtA, EshedY (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20 : 2293–2306.
32. Swanson-WagnerRA, JiaY, DeCookR, BorsukLA, NettletonD, et al. (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci U S A 103 : 6805–6810.
33. JaegerKE, PullenN, LamzinS, MorrisRJ, WiggePA (2013) Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25 : 820–833.
34. ParkSJ, JiangK, SchatzMC, LippmanZB (2012) Rate of meristem maturation determines inflorescence architecture in tomato. Proc Natl Acad Sci U S A 109 : 639–644.
35. AlvarezJP, PekkerI, GoldshmidtA, BlumE, AmsellemZ, et al. (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18 : 1134–1151.
36. SchwabR, OssowskiS, RiesterM, WarthmannN, WeigelD (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 : 1121–1133.
37. WiggePA (2011) FT, a mobile developmental signal in plants. Curr Biol 21: R374–378.
38. ShalitA, RozmanA, GoldshmidtA, AlvarezJP, BowmanJL, et al. (2009) The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc Natl Acad Sci U S A 106 : 8392–8397.
39. KoornneefM, HanhartCJ, van der VeenJH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular & general genetics : MGG 229 : 57–66.
40. ShannonS, Meeks-WagnerDR (1991) A Mutation in the Arabidopsis TFL1 Gene Affects Inflorescence Meristem Development. The Plant cell 3 : 877–892.
41. GoffSA (2011) A unifying theory for general multigenic heterosis: energy efficiency, protein metabolism, and implications for molecular breeding. New Phytol 189 : 923–937.
42. DoebleyJF, GautBS, SmithBD (2006) The molecular genetics of crop domestication. Cell 127 : 1309–1321.
43. IzawaT, TakahashiY, YanoM (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Current opinion in plant biology 6 : 113–120.
44. ZhaoK, TungCW, EizengaGC, WrightMH, AliML, et al. (2011) Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nature communications 2 : 467.
45. TsujiH, TaokaK, ShimamotoK (2011) Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Current opinion in plant biology 14 : 45–52.
46. BucklerES, HollandJB, BradburyPJ, AcharyaCB, BrownPJ, et al. (2009) The genetic architecture of maize flowering time. Science 325 : 714–718.
47. TanksleySD (2004) The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. The Plant cell 16 Suppl S181–189.
48. CongB, BarreroLS, TanksleySD (2008) Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nature genetics 40 : 800–804.
49. DoganlarS, FraryA, KuHM, TanksleySD (2002) Mapping quantitative trait loci in inbred backcross lines of Lycopersicon pimpinellifolium (LA1589). Genome/National Research Council Canada = Genome/Conseil national de recherches Canada 45 : 1189–1202.
50. GrandilloS, TanksleySD (1996) QTL analysis of horticultural traits differentiating the cultivated tomato from the closely related species Lycopersicon pimpinellifolium. Theoretical and Applied Genetics 92 : 935–951.
51. Jimenez-GomezJM, Alonso-BlancoC, BorjaA, AnastasioG, AngostoT, et al. (2007) Quantitative genetic analysis of flowering time in tomato. Genome/National Research Council Canada = Genome/Conseil national de recherches Canada 50 : 303–315.
52. PeraltaIE, SpoonerDM (2005) Morphological characterization and relationships of wild tomatoes (Solanum L. Section Lycopersicon). Monogr Syst Bot Missouri Bot Gard 227–257.
53. Flint-GarciaSA, BucklerES, TiffinP, ErsozE, SpringerNM (2009) Heterosis is prevalent for multiple traits in diverse maize germplasm. PLoS One 4: e7433.
54. SemelY, NissenbaumJ, MendaN, ZinderM, KriegerU, et al. (2006) Overdominant quantitative trait loci for yield and fitness in tomato. Proceedings of the National Academy of Sciences of the United States of America 103 : 12981–12986.
55. SchuelkeM, WagnerKR, StolzLE, HubnerC, RiebelT, et al. (2004) Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350 : 2682–2688.
56. JiangW, ZhouH, BiH, FrommM, YangB, et al. (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic acids research 41 (20) e188 doi: 10.1093/nar/gkt780
57. ShanQ, WangY, LiJ, ZhangY, ChenK, et al. (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nature biotechnology 31 : 686–688.
58. TaokaK, OhkiI, TsujiH, KojimaC, ShimamotoK (2013) Structure and function of florigen and the receptor complex. Trends in plant science 18 : 287–294.
59. LifschitzE, EshedY (2006) Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. Journal of experimental botany 57 : 3405–3414.
60. MengX, MuszynskiMG, DanilevskayaON (2011) The FT-like ZCN8 Gene Functions as a Floral Activator and Is Involved in Photoperiod Sensitivity in Maize. Plant Cell 23 : 942–960.
61. BlackmanBK, StrasburgJL, RaduskiAR, MichaelsSD, RiesebergLH (2010) The role of recently derived FT paralogs in sunflower domestication. Curr Biol 20 : 629–635.
62. QuinbyJR, KarperRE (1946) Heterosis in sorghum resulting from the heterozygous condition of a single gene that affects duration of growth. Am J Bot 33 : 716–721.
63. ComadranJ, KilianB, RussellJ, RamsayL, SteinN, et al. (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44 : 1388–1392.
64. PinPA, BenllochR, BonnetD, Wremerth-WeichE, KraftT, et al. (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330 : 1397–1400.
65. KwakM, VelascoD, GeptsP (2008) Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J Hered 99 : 283–291.
66. RepinskiSL, KwakM, GeptsP (2012) The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor Appl Genet 124 : 1539–1547.
67. FernandezL, TorregrosaL, SeguraV, BouquetA, Martinez-ZapaterJM (2010) Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. Plant J 61 : 545–557.
68. NavarroC, AbelendaJA, Cruz-OroE, CuellarCA, TamakiS, et al. (2011) Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478 : 119–122.
69. IwataH, GastonA, RemayA, ThouroudeT, JeauffreJ, et al. (2012) The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J 69 : 116–125.
70. TianZ, WangX, LeeR, LiY, SpechtJE, et al. (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci U S A 107 : 8563–8568.
71. LiuB, WatanabeS, UchiyamaT, KongF, KanazawaA, et al. (2010) The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol 153 : 198–210.
72. HarigL, BeineckeFA, OltmannsJ, MuthJ, MullerO, et al. (2012) Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J doi: []10.1111/j.1365-313X.2012.05125.x [Epub ahead of print].
73. HeddenP (2003) The genes of the Green Revolution. Trends in genetics : TIG 19 : 5–9.
74. MendaN, SemelY, PeledD, EshedY, ZamirD (2004) In silico screening of a saturated mutation library of tomato. The Plant journal : for cell and molecular biology 38 : 861–872.
75. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
76. Tomato GenomeC (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485 : 635–641.
77. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
78. RDC T (2011) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
79. RobinsonMD, McCarthyDJ, SmythGK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140.
80. AllenE, HowellMD (2010) miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Seminars in cell & developmental biology 21 : 798–804.
81. MoissiardG, ParizottoEA, HimberC, VoinnetO (2007) Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 13 : 1268–1278.
Štítky
Genetika Reprodukčná medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



