-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
Aberrant protein modifications play an important role in the pathophysiology of many human diseases, in terms of both dysfunction of physiological modifications and the formation of pathological modifications by reaction of proteins with endogenous electrophiles. Recent studies have identified a chemical homolog of lysine acetylation, N6-formyllysine, as an abundant modification of histone and chromatin proteins, one possible source of which is the reaction of lysine with 3′-formylphosphate residues from DNA oxidation. Using a new liquid chromatography-coupled to tandem mass spectrometry method to quantify all N6-methyl-, -acetyl - and -formyl-lysine modifications, we now report that endogenous formaldehyde is a major source of N6-formyllysine and that this adduct is widespread among cellular proteins in all compartments. N6-formyllysine was evenly distributed among different classes of histone proteins from human TK6 cells at 1–4 modifications per 104 lysines, which contrasted strongly with lysine acetylation and mono-, di-, and tri-methylation levels of 1.5-380, 5-870, 0-1400, and 0-390 per 104 lysines, respectively. While isotope labeling studies revealed that lysine demethylation is not a source of N6-formyllysine in histones, formaldehyde exposure was observed to cause a dose-dependent increase in N6-formyllysine, with use of [13C,2H2]-formaldehyde revealing unchanged levels of adducts derived from endogenous sources. Inhibitors of class I and class II histone deacetylases did not affect the levels of N6-formyllysine in TK6 cells, and the class III histone deacetylase, SIRT1, had minimal activity (<10%) with a peptide substrate containing the formyl adduct. These data suggest that N6-formyllysine is refractory to removal by histone deacetylases, which supports the idea that this abundant protein modification could interfere with normal regulation of gene expression if it arises at conserved sites of physiological protein secondary modification.
Published in the journal: Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases. PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003328
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003328Summary
Aberrant protein modifications play an important role in the pathophysiology of many human diseases, in terms of both dysfunction of physiological modifications and the formation of pathological modifications by reaction of proteins with endogenous electrophiles. Recent studies have identified a chemical homolog of lysine acetylation, N6-formyllysine, as an abundant modification of histone and chromatin proteins, one possible source of which is the reaction of lysine with 3′-formylphosphate residues from DNA oxidation. Using a new liquid chromatography-coupled to tandem mass spectrometry method to quantify all N6-methyl-, -acetyl - and -formyl-lysine modifications, we now report that endogenous formaldehyde is a major source of N6-formyllysine and that this adduct is widespread among cellular proteins in all compartments. N6-formyllysine was evenly distributed among different classes of histone proteins from human TK6 cells at 1–4 modifications per 104 lysines, which contrasted strongly with lysine acetylation and mono-, di-, and tri-methylation levels of 1.5-380, 5-870, 0-1400, and 0-390 per 104 lysines, respectively. While isotope labeling studies revealed that lysine demethylation is not a source of N6-formyllysine in histones, formaldehyde exposure was observed to cause a dose-dependent increase in N6-formyllysine, with use of [13C,2H2]-formaldehyde revealing unchanged levels of adducts derived from endogenous sources. Inhibitors of class I and class II histone deacetylases did not affect the levels of N6-formyllysine in TK6 cells, and the class III histone deacetylase, SIRT1, had minimal activity (<10%) with a peptide substrate containing the formyl adduct. These data suggest that N6-formyllysine is refractory to removal by histone deacetylases, which supports the idea that this abundant protein modification could interfere with normal regulation of gene expression if it arises at conserved sites of physiological protein secondary modification.
Introduction
In addition to physiological secondary modifications, proteins are subjected to reactions with endogenous electrophiles generated by oxidative stress, inflammation, and normal cell metabolic processes [1]–[5]. These adventitious or pathological modifications typically arise by reaction of the nucleophilic side chains of lysine, histidine, and cysteine with reactive electrophiles such as malondialdehyde, 4-hydroxynonenal (HNE), and glyoxal generated by oxidation of polyunsaturated fatty acids and carbohydrates, among other biomolecules [2]–[4], [6], [7]. The resulting adducts, which can alter protein function and lead to protein degradation, are associated with a variety of pathological processes and human diseases [1]–[5], [8]. Among these pathological adducts, N6-formylation of lysine has recently emerged as an abundant protein modification [5], [9]–[11]. While originally described in chromatin proteins [9]–[11], it has since been identified as an adduct arising in proteins subjected to nitrosative and oxidative stresses [5]. In chromatin proteins, N6-formyllysine has the potential to interfere with the functions of other post-translational modifications that perform signaling functions [12]–[15], such as acetylation, methylation, phosphorylation, ubiquitylation, and ADP ribosylation, with some locations modified in more than one way (e.g., refs. [16]–[18]). The chemical similarities of N6-formyllysine and N6-acetyllysine suggest a disruptive role for the former in signaling by histone acetylation. Indeed, N6-formyllysine has been detected at conserved sites of lysine acetylation and methylation in histones [10], [11].
While N6-formyllysine adducts are now well recognized as abundant protein modifications in cells, the source of these pathological adducts remains unclear. We recently showed that some portion of N6-formyllysine arises in chromatin proteins by reaction of lysine side chains with the 3′-formylphosphate residue derived from 5′-oxidation of 2-deoxyribose in DNA in cells (Figure 1) [9]. However, the observation of this adduct in proteins treated with the biological oxidant, peroxynitrite, suggests other sources for the formyl species [5]. Considering that formaldehyde reacts with amines to give a carbinolamine intermediate that is only one oxidation state away from a formamide functional group (Figure 1), we hypothesized that endogenous formaldehyde could serve as a source of N6-formyllysine residues in histone and other proteins. In addition to environmental and occupational sources [19]–[21], formaldehyde arises from cellular processes such as oxidative demethylation of nucleic acid and histone proteins, as well as biosynthesis of purines, thymidine, and some amino acids [20], [22], [23], making it a relatively abundant metabolite at concentrations ranging from 13 to 97 µM in human plasma [20]. To test this hypothesis and to clarify the cellular locations and quantities of N6-formyllysine relative to other major histone modifications, we developed a novel liquid chromatography-coupled electrospray tandem mass spectrometry (LC-MS/MS) method to quantify all N6-methyl-, -acetyl-, and -formyl-lysine modifications. Application of this method reveals that endogenous formaldehyde is a major source of N6-formyllysine, that this adduct is widespread among proteins in all cellular compartments, and that, in chromatin proteins, it is refractory to removal by histone deacetylases.
Fig. 1. Sources of N6-formyllysine. 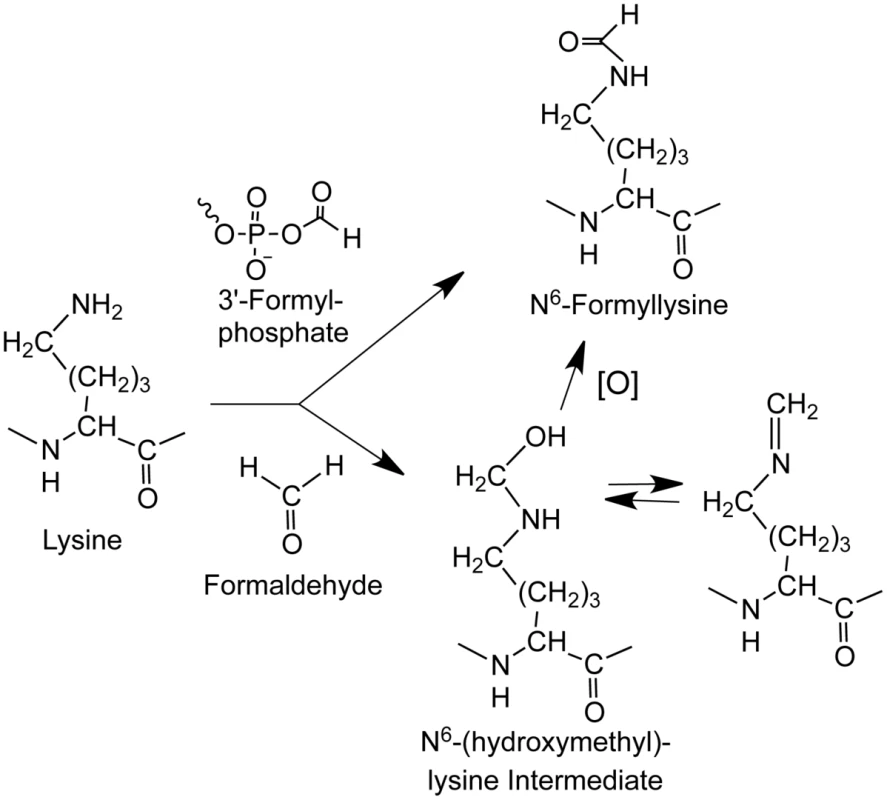
The adduct can be generated in chromatin proteins from reaction of lysine with 3′-formylphosphate residue derived from 5′-oxidation of 2-deoxyribose in DNA or from reaction of lysine with endogenous or exogenous formaldehyde. Formaldehyde reacts with amines to give a carbinolamine intermediate (N6-(hydroxymethyl)-lysine) that is in equilibrium with a Schiff base and that is one oxidation state away from the formamide functional group of N6-formyllysine. Results
Quantification of N6-lysine modifications in proteins
Our previous method for quantifying N6-formyllysine in proteins involved proteinase K-mediated hydrolysis of proteins, derivatization of the resulting amino acids with phenylisothiocyanate (PITC), HPLC pre-purification of amino acid derivatives, and final LC-MS/MS analysis of the derivatized amino acids [9]. This method proved to be relatively insensitive and biased as a result of using proteinase K, which produced only partial hydrolysis of some proteins when used in small quantities to minimize background autolysis. To resolve these problems, we used Streptomyces griseus protease at ratio of 1 µg enzyme per 10 µg proteins, which resulted in efficient and complete digestion of all proteins as judged by comparing the measured amount of lysine released per µg of purified histone proteins to the theoretical lysine content of the proteins. In addition, the method was optimized to eliminate HPLC pre-purification step, and the need for PITC derivatization to achieve chromatographic resolution of amino acids was obviated by use of aqueous normal phase HPLC with a diamond-hydride column. This chromatographic system resolved N6-acetyllysine, mono-, di-, and tri-N6-methyllysines, as well as N6-formyllysine and lysine, as shown in Figure 2. With isotopically labeled internal standards added prior to protease digestion, identification and quantification of amino acids were accomplished by HPLC-coupled to tandem quadrupole mass spectrometry in positive ion mode, using multiple reaction monitoring (MRM) transitions. With a 2% precision for technical replicates, the limits of detection were found to be 1 fmol for N6-formyl - and N6-acetyllysine, 10 fmol for lysine, and 50 fmol for each of N6-mono-, di-, and tri-methyl lysine. Data for the various lysine modifications are expressed here as proportions of the total number of lysines in the sample.
Fig. 2. Different lysine species detected in purified histone H4 from TK6 cells. 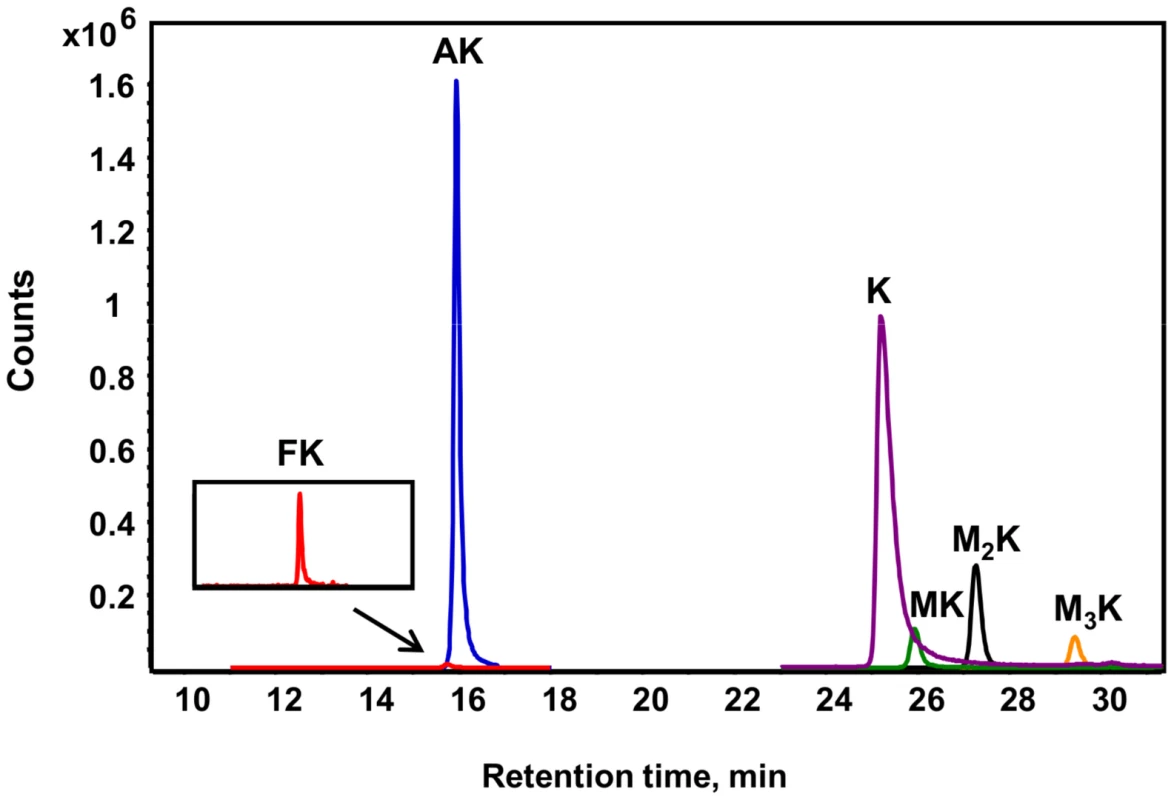
Lysine adducts were monitored by tandem mass spectrometry, as described in Materials and Methods. Abbreviations: FK, N6-formyllysine; AK, N6-acetyllysine; K, lysine; MK, N6-mono-methyllysine; M2K, N6-di-methyllysine; M3K, N6-tri-methyllysine. To validate the new analytical method for lysine modifications, we compared the frequency of N6-formyllysine among different classes of histone proteins extracted from TK6 cells and resolved by reversed-phase HPLC. As shown in Figure S1A, all of the major histone classes were separated, with further resolution of variant forms of histones H1 and H3 (SDS-PAGE verification in Figure S1B), which is consistent with previous observations in cultured human cells [10]. N6-Formyllysine was detected in all histone classes at a frequency of 1–4 modifications per 104 lysines. This 3 - to 4-fold variation among histone classes stands in contrast to the 10 - to 100-fold variation in the frequency of other modifications (Table 1). The data in Table 1 represent the first absolute quantification of the various lysine acetyl and methyl modifications in histone proteins, and are consistent with published studies of relative quantities of histone modifications using immunologic and radiolabeling techniques [10], [24]–[26]. Histone modification-based signaling involves the location and number of specific modification targets within a histone protein, as well as the frequency of modification of a target among all copies of a particular histone protein. Our data provide some insight into this issue. For example, we observed low-level acetylation and methylation in histone H1, which is consistent with studies using radiolabeled acetate [24], while this low level of modification maps to specific sites in the globular domain and N-terminal tail of histone H1 [10]. This low-level of acetylation and methylation in histone H1 stands in contrast to the high level of acetylation of H2, H3 and H4 (Table 1), which is again supported by studies using radiolabeled acetate [24].
Tab. 1. Quantification of lysine modifications in HPLC-purified histone proteins.1 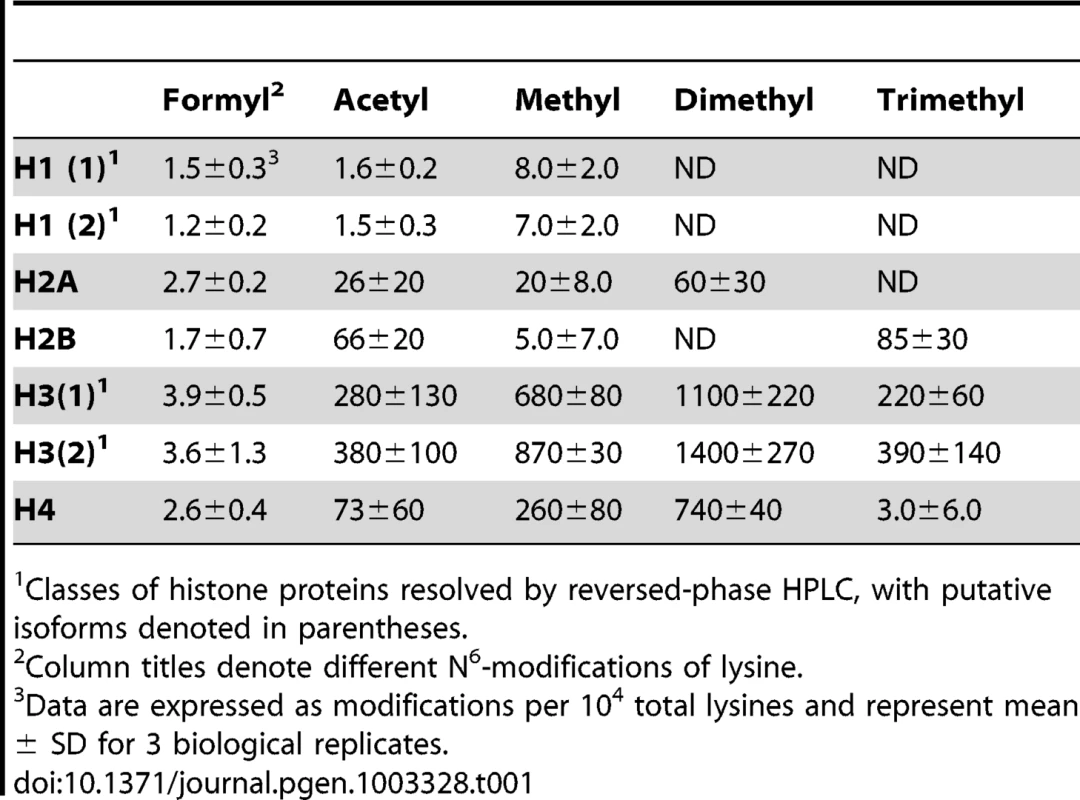
Classes of histone proteins resolved by reversed-phase HPLC, with putative isoforms denoted in parentheses. The new analytical method was next applied to quantify N6-formyllysine in non-histone proteins. We had previously observed N6-formyllysine mainly in histone proteins [9], perhaps as a result of biased proteolysis or subsequent steps in the technique. However, using the new method, we are now able to detect N6-formyllysine modifications in a variety of different proteins, as shown in Table 2. Further, an analysis of proteins in nuclear, cytosolic, and membrane compartments in bovine liver revealed the presence of N6-formyllysine in all three locations (Table 2). These observations are consistent with a source for N6-formyllysine other than the 3′-formylphosphate residues of DNA oxidation previously identified for histone proteins [9].
Tab. 2. Quantification of N<sup>6</sup>-formyllysine in different proteins. 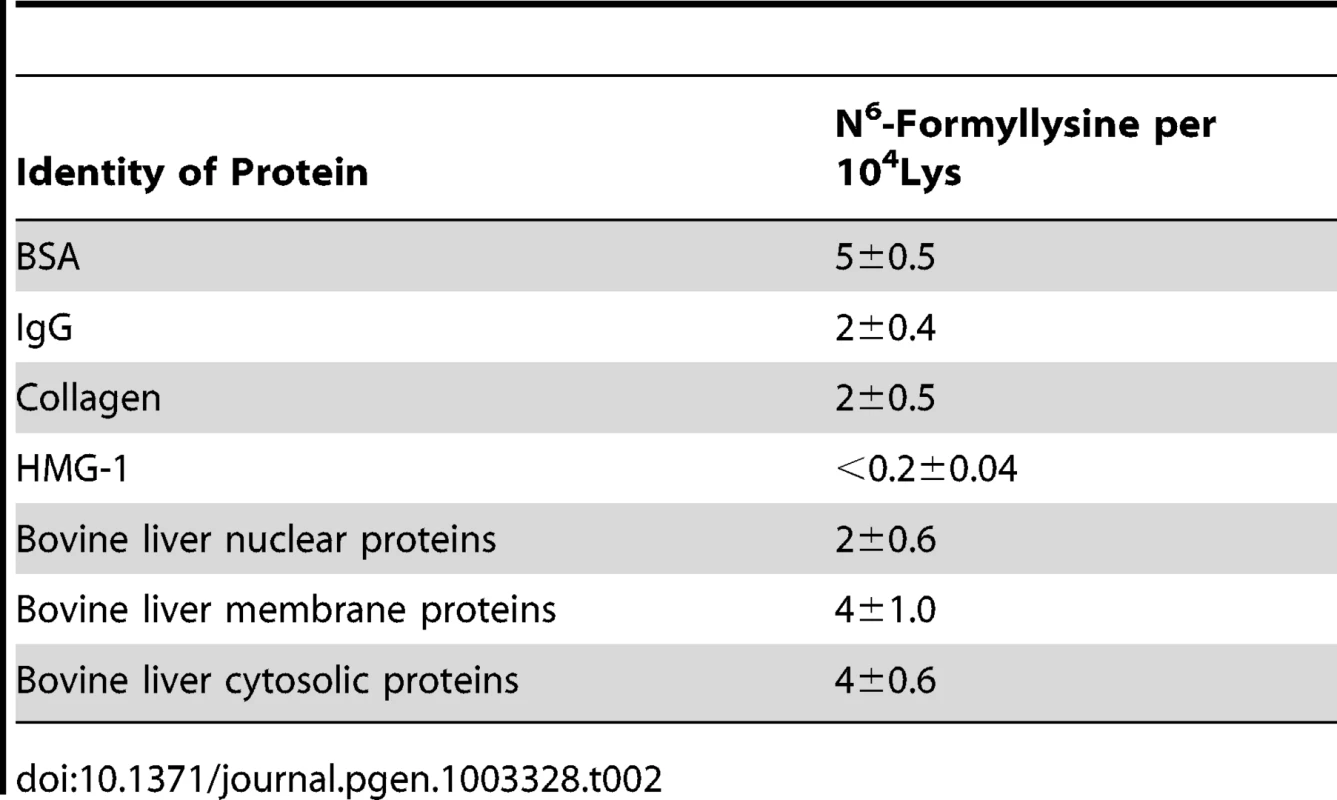
Formaldehyde as a source of N6-formyllysine
One alternative to 3′-formylphosphate residues as a source of N6-formyllysine is oxidation of the carbinolamine intermediate in the reaction of formaldehyde with side chain amine of lysine (Figure 1; N6-(hydroxymethyl)-lysine). To test this hypothesis, we performed a series of experiments, starting with an in vitro reaction of L-lysine with different concentrations of formaldehyde and quantification of N6-formyllysine. As shown in Figure 3A, there was a concentration-dependent formation of N6-formyllysine in reactions with formaldehyde, presumably as a result of oxidation of the carbinolamine adduct by the background of reactive oxygen species generated by trace metals and dissolved oxygen in the solution. The oxygen dependence of formaldehyde-induced N6-formyllysine was verified by bubbling 100% oxygen (4 h) into the solution of 1 mM lysine and 10 mM formaldehyde, which caused a 2.2 (±0.4)-fold increase in the level of N6-formyllysine after 12 h of incubation at 37°C. The dose-response relationship for formaldehyde-induced N6-formyllysine formation was also observed in histone proteins extracted from TK6 cells exposed to formaldehyde for 2 h at 37°C (Figure 3B), with 10 mM formaldehyde producing roughly the same fold-change of N6-formyllysine in both in vitro and cellular studies.
Fig. 3. Formaldehyde is a source of N6-formyllysine. 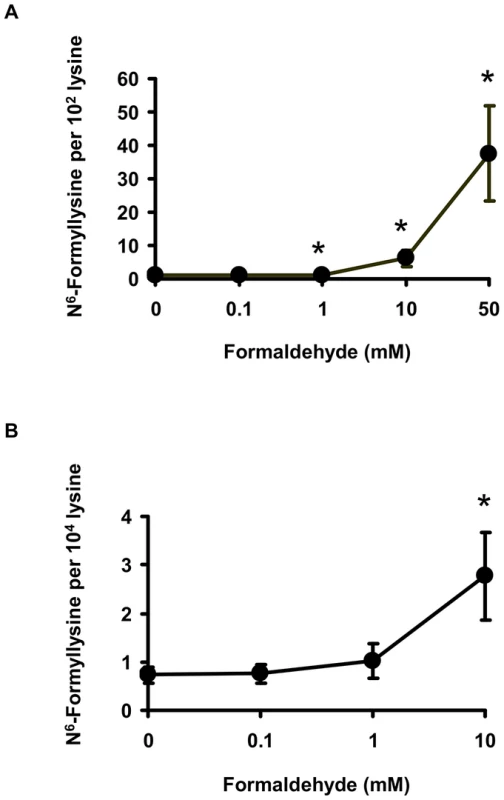
Formation of N6-formyllysine in (A) in vitro reactions of 1 mM L-lysine with formaldehyde for 2 h at 37°C, and in (B) TK6 cells exposed to formaldehyde, as described in Materials and Methods. Data represent mean ± SD for N = 3, with asterisks denoting statistically significant differences by Student's t-test (p<0.05). The relatively high endogenous levels of N6-formyllysine in histone and other proteins (Table 1 and Table 2) raised the question of the contribution of exogenous formaldehyde exposures to the total load of N6-formyllysine in the cells. To address this issue, we exposed TK6 cells to [13C,2H2]-labeled formaldehyde, which led to the formation of N6-[13C,2H]-formyllysine that is 2 mass units heavier than the endogenous adducts (Figure 4A). Following extraction of the histone proteins from formaldehyde-treated TK6 cells (2 h, 37°C), both endogenous and exogenous N6-formyllysine were quantified by monitoring the transitions m/z 175→112 and m/z 177→114, respectively (Figure 4A), with a third transition (m/z 179→116) for the 4,4,5,5-[2H]-N6-formyllysine internal standard. As shown in Figure 4B, levels of endogenous (unlabeled) N6-formyllysine remained constant at all concentrations of [13C,2H2]-formaldehyde, while N6-[13C,2H]-formyllysine increased as a function of the concentration of labeled formaldehyde.
Fig. 4. Addition of [13C,2H2]-formaldehyde to TK6 cells distinguishes exogenous from endogenous sources of N6-formyllysine. ![Addition of [<sup>13</sup>C,<sup>2</sup>H<sub>2</sub>]-formaldehyde to TK6 cells distinguishes exogenous from endogenous sources of N<sup>6</sup>-formyllysine.](https://www.prelekara.sk/media/cache/resolve/media_object_image_small/media/image/7ee658d11d6003ed9982c6012f79a481.png)
(A) LC-MS/MS analysis showing signals for the three isotopomeric N6-formyllysine species, as described in Materials and Methods. (B) Plot of N6-formyllysine levels as a function of exposure to [13C,2H2]-formaldehyde. Data represent mean ± SD for N = 3. Lysine demethylation as a source of N6-formyllysine
The enzymatic demethylation of N6-methyllysine modifications represents another possible source of N6-formyllysine in histone proteins, given both the carbinolamine intermediate known to form during the process of lysine demethylation and the ultimate release of the methyl group as formaldehyde [23]. Adventitious oxidation of the carbinolamine intermediate or secondary reaction of the released formaldehyde could result in the formation of N6-formyllysine locally. To test these hypotheses, TK6 cells were grown in customized RPMI medium containing L-methionine with a [13C,2H3]-methyl group for 20 days to label all methyl groups in N6-methyllysine species, and histone proteins were extracted for analysis every 2 days. If N6-formyllysine is a product of disrupted lysine demethylation in histones and is formed via oxidation of the carbinolamine intermediate known to form during the process of lysine demethylation [23], or by reaction of lysine with the formaldehyde released at the last step of successful lysine demethylation [23], then one would expect to see an increase of 2 mass units corresponding to formation of N6-[13C, 2H]-formyllysine (m/z 177→114 transition). In order to increase the signal-to-noise ratio for N6-[13C,2H]-formyllysine, N6-formyllysine was HPLC-pre-purified in all samples before LC-MS/MS analysis. Figure 5 depicts an example of the analysis using the day 6 sample. As shown in Figure 5, N6-mono-methyllysine and N6-di-methyllysine are predominately labeled (>90%) with heavy isotope methyl groups (i.e.,[13C,2H3]-methyl). In contrast to methyllysines, the level of N6-[13C,2H]-formyllysine did not increase beyond the natural isotope abundance level of ∼0.7% for the [M+2] ion of N6-formyllysine (Figure 5C and Figure S2). Note that the HPLC gradient was changed here to fully resolve a contaminant signal from the TK6 cells (identified as the [M+1] ion of citrulline) that otherwise co-eluted with N6-formyllysine and produced an m/z value similar to the [M+2] isotopomer of N6-formyllysine.
Fig. 5. Analysis of lysine demethylation as a source of N6-formyllysine. 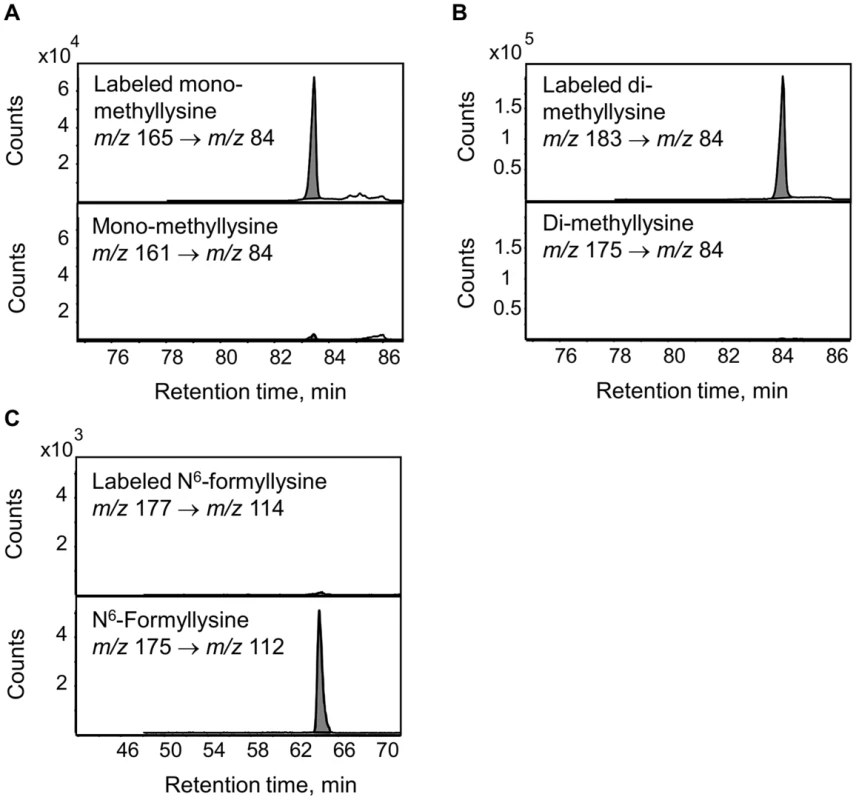
Methyl groups in N6-methyllysine species in TK6 cells were labeled using L-methionine-([13C,2H3]-methyl) and N6-formyllysine and N6-methyllysine species were quantified by LC-MS/MS as described in Materials and Methods. Panels A and B: N6-mono-methyllysine and N6-di-methyllysine are predominately labeled (>90%) with heavy isotope methyl groups (mass increase of 4 m/z and 8 m/z, respectively), with <10% of the modifications containing unlabeled methyl groups. Panel C: the level of N6-[13C, 2H]-formyllysine (177 m/z→114 m/z transition) in histones did not show an increase beyond the natural isotope abundance level of ∼0.7% for [M+2] ion of N6-formyllysine. Persistence of N6-formyllysine in cells
The chemical similarity of N6-formyllysine and N6-acetyllysine suggested that the former might be subject to removal by lysine deacetylases that recognize and remove N6-acetyllysine from histone and other proteins [16], [27]–[30]. Lysine deacetylases fall into several classes, including classes I and II that share a common hydrolytic mechanism and are all inhibited by suberoylanilidehydroxamic acid (SAHA), and the class III enzymes (i.e., sirtuins) that are NAD+-dependent deacetylases [31], [32]. In order to assess the activity of lysine deacetylases with N6-formyllysine substrates, TK6 cells were treated with SAHA and the levels of N6-acetyllysine and N6-formyllysine were quantified. The results shown in Figure 6A reveal that, while SAHA caused a 3-fold increase in the level of N6-acetyllysine (4 to 14 per 103 lysines), lysine formylation was not affected. To assess sirtuin activity against N6-formyllysine, we performed in vitro reactions of SIRT1 with a consensus peptide (GGAKRHR) containing N6-formyllysine or N6-acetyllysine, and the quantities of the modified and unmodified peptides were analyzed by LC-MS/MS. As shown in Figure 6B, SIRT1 removed the acetyl modification completely to generate the unmodified peptide, while only ∼10% (±4%) of N6-formyllysine was removed.
Fig. 6. Effect of lysine deacetylases on N6-formyllysine. 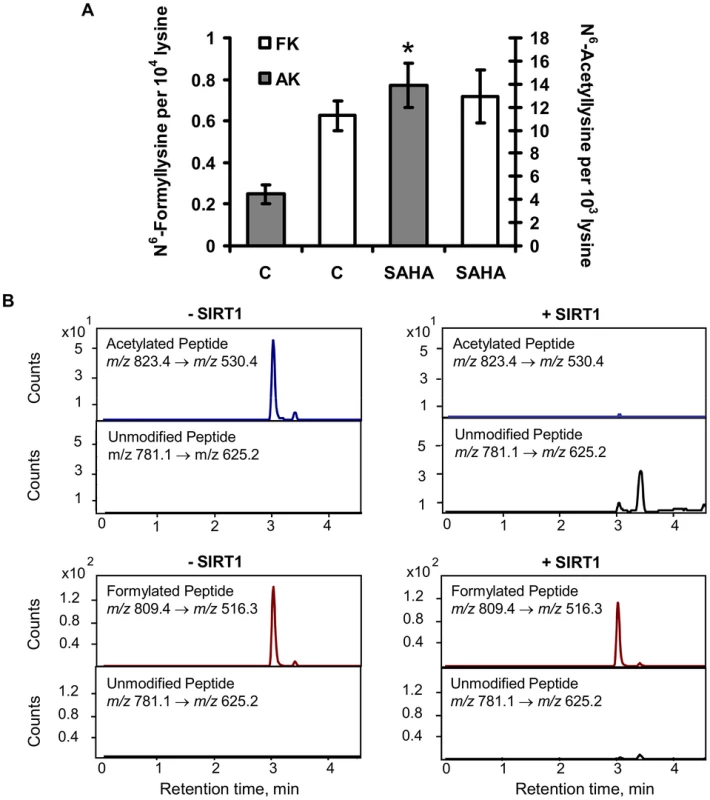
(A) TK6 cells were treated with the class I and class II histone deacetylase inhibitor, SAHA, as described in Materials and Methods. Data represent mean ± SD for N = 3, with asterisks denoting statistically significant differences by Student's t-test (p<0.05). (B) Treatment of peptide substrates containing N6-acetyllysine or N6-formyllysine with the class III histone deacetylase, SIRT1. Discussion
N6-Formyllysine was first reported in 1985 in reactions of lysine with formaldehyde in vitro [33] and, more recently, it was shown to form during in vitro silver-staining procedures that involve the use of formaldehyde [34]. The recent discovery of N6-formyllysine as a relatively abundant endogenous posttranslational modification of histones and other nuclear proteins in cells [9]–[11] has raised questions about its mechanism of formation and its potential for interfering with the regulatory function of lysine N6-acetylation. With respect to formation, we previously presented evidence that N6-formyllysine in histones could arise from reactions with 3′-formylphosphate residues derived from DNA oxidation [9]. However, formaldehyde emerged as an alternative source given the high reactivity of formaldehyde toward primary amines, such as the side chain of lysine, and the potential for endogenous oxidation to convert a formaldehyde-derived carbinolamine to a stable formamide (Figure 1). We have now demonstrated in vitro and in cells that formaldehyde exposure leads to formation of N6-formyllysine residues in proteins. The fact that this modification arises in proteins other than chromatin proteins and in cellular compartments other than the nucleus (Table 2) suggests that 3′-formylphosphate residues in oxidized DNA do not account for all N6-formyllysine adducts. This is consistent with recent studies in which N6-formyllysine was detected in a protein oxidized with peroxynitrite in vitro [5]. The absence of detectable N6-formyllysine arising from demethylation of N6-methyllysine species (Figure 5C and Figure S2) suggests that interruption of histone demethylation reactions to form the carbinolamine precursor of N6-formyllysine occurs at low frequency, or that the formaldehyde produced by complete lysine demethylation [23] does not occur at concentrations high enough to drive formylation of lysine or cause substantial changes in N6-formyllysine levels detected by our current analytical method. Furthermore, there is the possibility that the formaldehyde released during lysine demethylation may be scavenged before it could react with lysines in histones. A recent study reports that lysine-specific demethylase 1 (LSD1) is a folate binding protein [35], which led the authors to hypothesize that the biological function of folate is to trap the formaldehyde that is generated during lysine demethylation [35]. In addition, the observation that the formaldehyde equivalent derived from histone demethylation might not account for a significant portion of formyllysine residues is not surprising in light of the abundance of formaldehyde from other cellular processes, such as nucleic acid demethylation and biosynthesis of purines, thymidine and some amino acids [20], [22], [23]. This is clear from the high steady-state concentrations of formaldehyde in human plasma (13–97 µM) [20], though the contribution of long-term, low-level exposure from environmental sources of formaldehyde cannot be ruled out.
The relative abundance of N6-formyllysine in histone and other proteins (Table 1 and Table 2) [9]–[11] and the persistence of these adducts in histone proteins provides insights into both their source and their potential effects on cell function. The N6-formyllysine residues are relatively evenly distributed among different classes of histone proteins (Table 1), while the other functional modifications show very biased distributions over a large frequency range, which is consistent with the known function and conserved locations for lysine methylation and acetylation [16]–[18]. This random distribution of formyllysine adducts in histone proteins suggests that they are adventitious and not physiological. The fact that N6-formyllysine levels are similar in histone and non-nuclear proteins and in all cell compartments also suggests that the sources of this protein modification are equally balanced in the various compartments and proteins, or that there is a single dominant source that distributes uniformly throughout the cell. With regard to its persistence in cells, there is still no evidence supporting the enzymatic removal of N6-formyllysine. Our investigation reveals that N6-formyllysine adducts appear to be refractory to removal by histone deacetylase enzymes, which suggests that they will persist throughout the lifetime of individual histone proteins. Although our results are consistent with lysine N6-formylation as a stable protein modification, we cannot rule out the possibility of an enzyme that removes this modification from selected conserved lysine sites in histone proteins, resulting in small changes in the population level of formyllysine that are not detectable by our current analytical method. Figure 7 summarizes our findings presented here on N6-formyllysine adducts.
Fig. 7. Summary of findings on N6-formyllysine in histones. 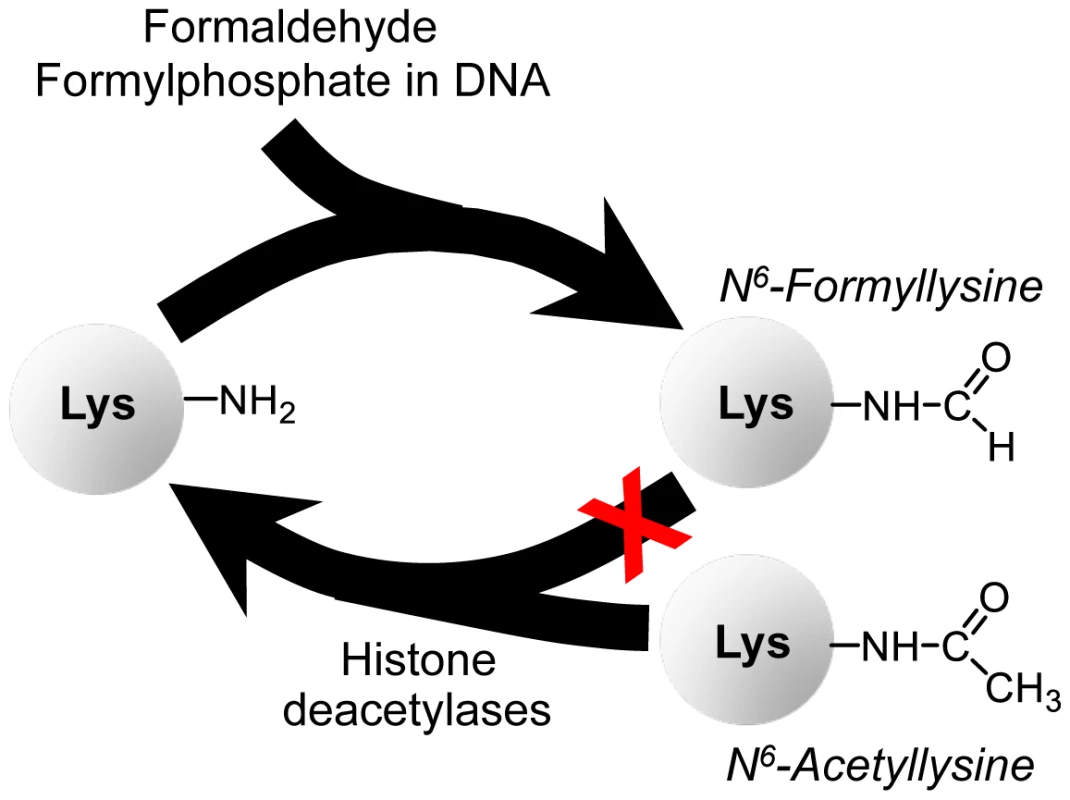
N6-formyllysine can arise from reaction of lysine with the 3′-formyl phosphate residue derived from 5′-oxidation of 2-deoxyribose in DNA or from reaction of lysine with formaldehyde. Furthermore, our data suggest that N6-formyllysine is refractory to removal by histone deacetylases, which is consistent with the persistence of this pathological adduct throughout the life of individual histone proteins. The high abundance of lysine N6-formylation in histone proteins (Table 1) as well as its occurrence at many of the known conserved functional locations for lysine acetylation and methylation in histones [10], [11] suggests that N6-formyllysine could interfere with signaling processes associated with physiological histone modifications [16], [27]. Mann and co-workers found formylation of core and linker histones at multiple lysines in both the N - and C-terminal tails and in the globular domains [10], which is consistent with our observation of a random distribution of N6-formyllysines among the different histone classes. Several of the formylated lysines occurred at functionally important sites. For example, the observed N6-formylation of lysine 12 in histone H4 could interfere with BRD2 bromodomain-dependent transcriptional activation that occurs when the lysine is acetylated [10]. Furthermore, formylation was observed at several lysines known to make contact with the DNA backbone in nucleosomes, which could interfere with nucleosome organization given the conserved acetylation and methylation at these sites [10]. Our observation that N6-formyllysine is refractory to removal by several histone deacetylases suggests that the adducts could interfere with the epigenetic regulatory processes mediated by acetylation and methylation. While this potential epigenetic mechanism of disruption of cell function may contribute to the toxicity and carcinogenicity associated with formaldehyde [20], [36]–[40], the association of N6-formyllysine with a variety of different cell and organismal processes, including metabolism and the oxidative and nitrosative stresses of inflammation [5], [9], suggests that this adduct may play a role in many pathophysiological processes in humans.
Materials and Methods
Materials
Unlabeled and [13C,2H2]-labeled formaldehyde were purchased as 37% and 20% aqueous solutions from Amresco (Solon, OH) and Isotec (Miamisburg, OH), respectively. 4,4,5,5-[2H]-Lysine was purchased from Cambridge Isotope Laboratories (Andover, MA). 4,4,5,5-[2H]-N6-Formyllysine was synthesized from 4,4,5,5-[2H]-lysine according to Jiang et al. [9]. 3,3,4,4,5,5,6,6-[2H]-N6-Acetyllysine were obtained from CDN Isotopes (Pointe-Claire, Quebec, Canada). L-Methionine-([13C,2H3]-methyl) was obtained from Isotec (Miamisburg, OH). L-Lysine, N6-formyllysine, N6-acetyllysine, bovine serum albumin, human recombinant HMG-1, human IgG, Streptomyces griseus protease, and protease inhibitor cocktail (for use with mammalian cell and tissue extracts) were obtained from Sigma-Aldrich (St. Louis, MO). N6-Mono-methyllysine, N6-di-methyllysine, and N6-tri-methyllysine were purchased from Bachem Bioscience Inc. (King of Prussia, PA). Nonidet P-40 was from Roche Diagnostic Corporation (Indianapolis, IN). Suberoylanilidehydroxamic acid (SAHA) and SIRT1 (human recombinant) enzyme were purchased from Cayman chemical (Ann Arbor, MI). Peptide substrates for SIRT1 (GGAKRHR and its lysine-acetylated and -formylated forms) were synthesized at Massachusetts Institute of Technology Biopolymers Laboratory. The human lymphoblastoid TK6 cell line was a generous gift of Prof. Gerald Wogan (Massachusetts Institute of Technology).
TK6 cell culture, exposure, and labeling
TK6 cells were cultured in RPMI 1640 medium (Cellgro, Manassas, VA) supplemented with 10% heat-inactivated horse serum (Atlanta Biologicals, Lawrenceville, GA), 10,000 U penicillin/ml and 10,000 µg streptomycin/ml (Lonza, Walkersville, MD), and 2 mM L-glutamine (Lonza, Walkersville, MD) at 37°C in a 5% CO2 atmosphere. For formaldehyde exposure studies, TK6 cells were pelleted, washed, and resuspended in RPMI 1640 medium without any supplements, prior to addition of formaldehyde to the medium. Following addition of formaldehyde, cells were incubated at 37°C for 2 h with occasional mixing prior to extracting chromatin proteins. Histones were extracted from ∼107 cells after exposure and the quantity of formyllysine, as a percentage of total lysine, was measured as described below. For lysine demethylation studies, TK6 cells were grown in a customized RPMI-1640 medium identical to the traditional medium (e.g., supplemented with horse serum, antibiotics, and L-glutamine), except for the presence of labeled methionine (L-methionine-([13C,2H3]-methyl)) instead of non-labeled methionine. Histones (from ∼107 cells) were extracted every 2 d for 20 d in order to investigate the formation of N6-[13C,2H]-formyllysine. For histone deacetylase studies, TK6 cells were incubated with the histone deacetylase inhibitor, suberoylanilidehydroxamic acid (SAHA), for 20 h at 37°C in a 5% CO2 atmosphere prior to histone extraction. SAHA was dissolved in a 50∶50 solution of DMSO∶PBS prior to addition to cell media. Control cells (∼107) were treated with the DMSO∶PBS vehicle.
Histone extraction from TK6 cells and subcellular protein fractionation from tissue
Extraction of histones was performed according to Boyne et al. [41], with modifications. Cells (∼107 per sample) were pelleted by centrifugation at 1000× g for 5 min at 4°C and the pellet was washed once with PBS. Cell pellets were then lysed by resuspension in ice-cold lysis buffer (15 mM Tris-HCl, pH 7.5, 15 mM NaCl, 60 mM KCl, 1 mM CaCl2, 5 mM MgCl2, 250 mM sucrose, 1 mM dithiothreitol, 10 mM sodium butyrate) containing a 100∶1 v∶v dilution of protease inhibitor cocktail in the presence of 0.03% Nonidet P-40 and incubation on ice for 5 min with occasional gentle mixing. Nuclei were pelleted by centrifugation at 600× g for 5 min at 4°C, and the pellet was washed twice with ice-cold lysis buffer without Nonidet P-40. Histones were extracted by addition of ice-cold 0.4 N H2SO4 and incubation overnight on ice. The solution was centrifuged at 3000× g for 5 min and proteins in supernatant were precipitated by addition of 20% v/v trichloroacetic acid and overnight incubation at 4°C. Samples were then centrifuged at 14000× g for 10 min at 4°C, washed once with ice-cold acetone containing 0.1% HCl, and once with ice-cold acetone. The extracts were air-dried and stored at −20°C until use. For collecting membrane, cytosolic, and nuclear fractions, 20 mg of bovine liver tissue was cut into small pieces and washed with PBS, and proteins were fractionated using the Subcellular Protein Fractionation Kit from Thermo Scientific (Waltham, MA) and a Kontes all-glass Dounce homogenizer (10 strokes with a type B pestle). Proteins in subcellular extracts were precipitated by addition of 20% v/v trichloroacetic acid and overnight incubation at 4°C. Samples were then centrifuged at 14000× g for 10 min at 4°C, washed once with ice-cold acetone containing 0.1% HCl, and once with ice-cold acetone. The extracts were air-dried and stored at −20°C until use.
Purification of individual histones
HPLC purification of total histones was performed according to Boyne et al. [41] with modifications. Total histones (≤50 µg) were dissolved in 0.1% trifluoroacetic acid (TFA) in water and fractionated by HPLC on an Agilent 1100 series instrument (Agilent Technologies, Santa Clara, CA), using a Source 5RPC C18 reversed-phase column (4.6×150 mm, 5 µm particles; GE Healthcare Life Sciences). The mobile phase flow rate was 1 mL/min and the solvent system was 0.1% TFA in water (A) and 0.094% TFA in acetonitrile (B) with the elution starting at 0% B, linearly increasing to 28% B over 28 min, reaching 37% B at 70 min, 38% B at 100 min, 60% B at 150 min, and finally 100% B at 151 min, before the column was re-equilibrated to 0% B for 10 min. Protein elution was monitored by UV absorbance at 214 nm and histones in each fraction were tentatively identified by resolution on a 13% SDS-PAGE gel with Coomassie Blue staining (see Figure S1).
Enzymatic hydrolysis of proteins
Histones extracted from TK6 cells and other protein samples were dissolved in 50 µL of 100 mM ammonium bicarbonate buffer (pH 8.5), 4,4,5,5-[2H]-lysine (2 nmol),4,4,5,5-[2H]-N6-formyllysine (1 pmol), and 3,3,4,4,5,5,6,6-[2H]-N6-acetyl lysine (10 pmol) were added as internal standards, and the proteins hydrolyzed by addition of S. griseus protease (freshly prepared solution each time) with incubation at 37°C for ≥16 h. Streptomyces griseus was used at a ratio of 1 µg of enzyme per 10 µg of protein. Samples were then dried under vacuum and resuspended in 50 µL of water before mass spectrometry analysis.
Quantification of amino acids
N6-Formyllysine and other amino acids were quantified as a percentage of the total quantity of lysine, by liquid chromatography-coupled mass spectrometry (LC-MS/MS). HPLC was performed with an Agilent 1100 series instrument. Adducts of interest in the resuspended protein hydrolysates were separated using an aqueous normal phase Cogent diamond hydride column (2.1×150 mm, 4 µm) from MicroSolv Technology Corporation (Eatontown, NJ). The mobile phase flow rate was 400 µL/min, and the column temperature was maintained at 20°C. The solvent system was 0.1% acetic acid in water (A) and 0.1% acetic acid in acetonitrile (B), with the elution starting at 100% B, the gradient linearly decreased to 25% B over 30 min, stayed at 25% B for 3 additional min before the column was re-equilibrated at 100% B for 7 min. In order to separate a co-eluting contaminant from pre-purified lysine demethylation study samples, an extended chromatography run time was used, with the elution starting at 100% B, the gradient linearly decreased to 75% B over 75 min, further decreased to 25% B over the next 3 min, reached 15% by 83 min before the column was re-equilibrated at 100% B for 7 min. The species of interest were then analyzed using Agilent 6410 triple quadrupole mass spectrometer (MS/MS) equipped with an electrospray ionization (ESI) source operated in positive ion mode. The operating parameters were as follows: ESI capillary voltage, 4000 V; gas temperature, 350°C; drying gas flow, 12 L/min; and nebulizer pressure, 30 psi. Selected reaction monitoring (SRM) transitions are summarized in Table 3. Note that in addition to chromatographic separation (Figure 2) and presence of internal standards, the unique product ions of 112 m/z and 126 m/z for formyl and acetyl lysines, respectively, distinguish them from their isobaric compounds di - and tri-methyl lysines. The fragmentor voltage and collision energy were optimized in order to maximize the signal of each product ion monitored (see Figure S3) and are summarized in Table 3. 4,4,5,5-[2H]-Lysine, 4,4,5,5-[2H]-N6-formyllysine, and 3,3,4,4,5,5,6,6-[2H]-N6-acetyl lysine were used as internal standards. Calibration curves for the labeled and unlabeled forms of these analytes were constructed by plotting the ratios of the areas of the MS signals for the labeled and unlabeled forms against their corresponding concentration ratios (Figure S4). N6-Methylated lysine species were quantified using the deuterated acetyl lysine internal standard signal (Figure S4).
Tab. 3. Summary of mass spectrometry parameters for each species. 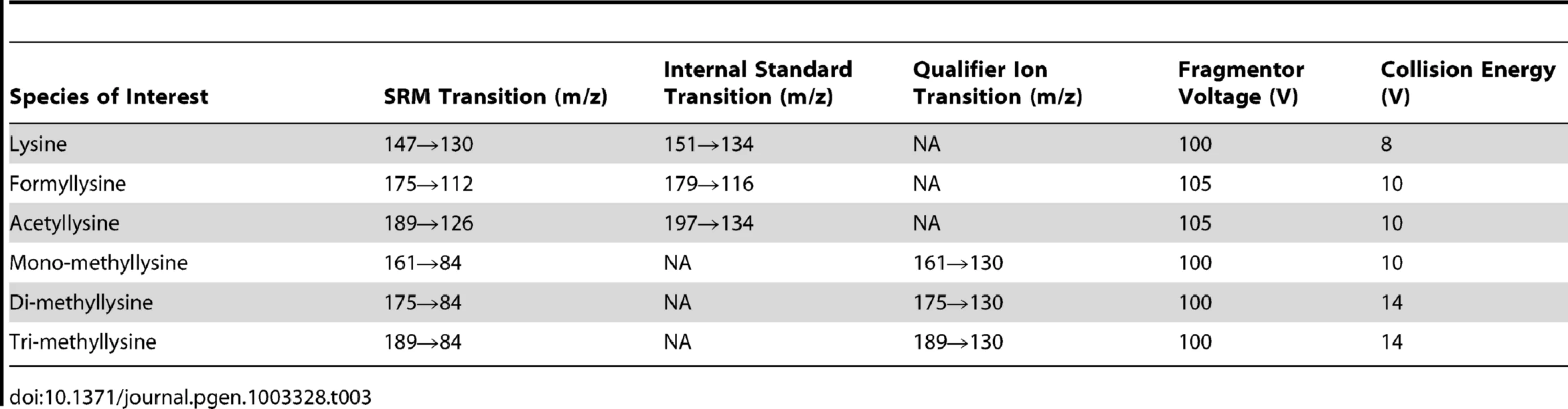
SIRT1 peptide experiment
SIRT1 peptide substrates (GGAKRHR, GGAKacetylRHR, and GGAKformylRHR) were HPLC purified on an Agilent 1100 series instrument using Vydac 218TP52 C18 reverse-phase column (2.1×250 mm, 5 µm) from Grace Vydac (Hesperia, CA). The mobile phase flow rate was 200 µL/min, and the column temperature was maintained at 30°C. The solvent system was 0.05% trifluoroacetic acid in water (A) and 0.05% trifluoroacetic acid in 80% acetonitrile (B), with the isocratic elution of 5% B for the first 5 min, then a linear increase to 42% B over 25 min, reaching 100% B at 31 min followed by column re-equilibration at 5% B for 10 min. Each purified SIRT1 peptide substrate (100 pmol) were incubated overnight (12 h) at 25°C with 1 µg SIRT1, in 50 mM Tris-HCl (pH 8) buffer containing 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, and 6 mM NAD+. The removal of acetyl and formyl groups from SIRT1 peptide substrates was determined using liquid chromatography-coupled mass spectrometry. HPLC was performed on an Agilent 1100 series instrument using Agilent Exclipse XDB C18 reverse-phase column (4.6×150 mm, 5 µm). The mobile phase flow rate was 200 µL/min, and the column temperature was maintained at 40°C. The solvent system was 0.1% acetic acid in water (A) and 0.1% acetic acid in acetonitrile (B), with the elution starting at 20% B, the gradient linearly increased to 50% B over 5 min, reached 100% B at 6 min, and kept at 100% B for 9 minutes before the column was re-equilibrated at 20% B for 10 min. The species of interest were then analyzed using the Agilent 6410 MS/MS system equipped with an electrospray ionization (ESI) source operated in positive ion mode. The operating parameters were as follows: ESI capillary voltage, 3500 V; gas temperature, 345°C; drying gas flow, 8 L/min; and nebulizer pressure, 30 psi. Multiple reaction monitoring (MRM) transitions were as follow: GGAKRHR peptide, m/z 781.1→625.2; GGAKformylRHR peptide, m/z 809.4→516.3; and GGAKacetylRHR peptide, m/z 823.4→530.4. The fragmentor voltage and collision energy were 200 V and 40 V for GGAKRHR peptide, respectively; 100 V and 46 V for GGAKformylRHR peptide; and 100 V and 52 V for GGAKacetylRHR peptide.
Supporting Information
Zdroje
1. LevineRL (2002) Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic Biol Med 32 : 790–796.
2. JacobsAT, MarnettLJ (2010) Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res 43 : 673–683.
3. ThornalleyPJ (2008) Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems–role in ageing and disease. Drug Metabol Drug Interact 23 : 125–150.
4. DedonPC (2008) The chemical toxicology of 2-deoxyribose oxidation in DNA. Chem Res Toxicol 21 : 206–219.
5. VanaL, KanaanNM, HakalaK, WeintraubST, BinderLI (2011) Peroxynitrite-induced nitrative and oxidative modifications alter tau filament formation. Biochemistry 50 : 1203–1212.
6. CodreanuSG, ZhangB, SobeckiSM, BillheimerDD, LieblerDC (2009) Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics 8 : 670–680.
7. TallmanKA, KimHY, JiJX, SzapacsME, YinH, et al. (2007) Phospholipid-protein adducts of lipid peroxidation: synthesis and study of new biotinylated phosphatidylcholines. Chem Res Toxicol 20 : 227–234.
8. PrasadA, BekkerP, TsimikasS (2012) Advanced Glycation Endproducts and Diabetic Cardiovascular Disease. Cardiol Rev 20 : 177–183.
9. JiangT, ZhouX, TaghizadehK, DongM, DedonPC (2007) N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci U SA 104 : 60–65.
10. WisniewskiJR, ZougmanA, MannM (2008) Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res 36 : 570–577.
11. LeRoyG, WestonJT, ZeeBM, YoungNL, Plazas-MayorcaMD, et al. (2009) Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics 8 : 2432–2442.
12. PesaventoJJ, KimYB, TaylorGK, KelleherNL (2004) Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J Am Chem Soc 126 : 3386–3387.
13. FelsenfeldG, GroudineM (2003) Controlling the double helix. Nature 421 : 448–453.
14. ChiP, AllisCD, WangGG (2010) Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10 : 457–469.
15. RandoOJ (2012) Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev 22 : 148–55.
16. KouzaridesT (2007) Chromatin modifications and their function. Cell 128 : 693–705.
17. SiutiN, RothMJ, MizzenCA, KelleherNL, PesaventoJJ (2006) Gene-specific characterization of human histone H2B by electron capture dissociation. J Proteome Res 5 : 233–239.
18. GarciaBA, BarberCM, HakeSB, PtakC, TurnerFB, et al. (2005) Modifications of human histone H3 variants during mitosis. Biochemistry 44 : 13202–13213.
19. LuK, MoellerB, Doyle-EiseleM, McDonaldJ, SwenbergJA (2011) Molecular dosimetry of N2-hydroxymethyl-dG DNA adducts in rats exposed to formaldehyde. Chem Res Toxicol 24 : 159–161.
20. ZhangL, FreemanLE, NakamuraJ, HechtSS, VandenbergJJ, et al. (2010) Formaldehyde and leukemia: epidemiology, potential mechanisms, and implications for risk assessment. Environ Mol Mutagen 51 : 181–191.
21. Le CurieuxF, PluskotaD, MunterT, SjoholmR, KronbergL (2000) Identification of fluorescent 2′-deoxyadenosine adducts formed in reactions of conjugates of malonaldehyde and acetaldehyde, and of malonaldehyde and formaldehyde. Chem Res Toxicol 13 : 1228–1234.
22. BegleyTJ, SamsonLD (2003) AlkB mystery solved: oxidative demethylation of N1-methyladenine and N3-methylcytosine adducts by a direct reversal mechanism. Trends Biochem Sci 28 : 2–5.
23. ShiY, WhetstineJR (2007) Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 25 : 1–14.
24. PasqualiniJR, MercatP, GiambiagiN (1989) Histone acetylation decreased by estradiol in the MCF-7 human mammary cancer cell line. Breast Cancer Res Treat 14 : 101–105.
25. ByvoetP, BarberM, AmideiK, LowellN, TrudeauW (1986) Effect of exogenous histone H5 on integration of histone H1 in rat liver chromatin. Correlations with aberrant epsilon-N-methylation of histone H1. Biochim Biophys Acta 867 : 163–175.
26. WuM, AllisCD, RichmanR, CookRG, GorovskyMA (1986) An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc Natl Acad Sci USA 83 : 8674–8678.
27. StrahlBD, AllisCD (2000) The language of covalent histone modifications. Nature 403 : 41–45.
28. de RuijterAJ, van GennipAH, CaronHN, KempS, van KuilenburgAB (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370 : 737–749.
29. HildmannC, RiesterD, SchwienhorstA (2007) Histone deacetylases–an important class of cellular regulators with a variety of functions. Appl Microbiol Biotechnol 75 : 487–497.
30. HakeSB, XiaoA, AllisCD (2004) Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br J Cancer 90 : 761–769.
31. ChenL (2011) Medicinal chemistry of sirtuin inhibitors. Curr Med Chem 18 : 1936–1946.
32. DokmanovicM, ClarkeC, MarksPA (2007) Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res 5 : 981–989.
33. TyihakE, TrezlL, KolonitsP (1985) The isolation of Nepsilon-formyl-L-lysine from the reaction between formaldehyde and L-lysine and its identification by OPLC and NMR spectroscopy. J Pharm Biomed Anal 3 : 343–349.
34. Oses-PrietoJA, ZhangX, BurlingameAL (2007) Formation of epsilon-formyllysine on silver-stained proteins: implications for assignment of isobaric dimethylation sites by tandem mass spectrometry. Mol Cell Proteomics 6 : 181–192.
35. LukaZ, MossF, LoukachevitchLV, BornhopDJ, WagnerC (2011) Histone demethylase LSD1 is a folate-binding protein. Biochemistry 50 : 4750–4756.
36. Humans IWGotEoCRt (2006) Formaldehyde, 2-butoxyethanol and 1-tert-butoxypropan-2-ol. IARC Monogr Eval Carcinog Risks Hum 88 : 1–478.
37. ZhangL, TangX, RothmanN, VermeulenR, JiZ, et al. (2010) Occupational exposure to formaldehyde, hematotoxicity, and leukemia-specific chromosome changes in cultured myeloid progenitor cells. Cancer Epidemiol Biomarkers Prev 19 : 80–88.
38. MonticelloTM, SwenbergJA, GrossEA, LeiningerJR, KimbellJS, et al. (1996) Correlation of regional and nonlinear formaldehyde-induced nasal cancer with proliferating populations of cells. Cancer Res 56 : 1012–1022.
39. SwenbergJA, KernsWD, MitchellRI, GrallaEJ, PavkovKL (1980) Induction of squamous cell carcinomas of the rat nasal cavity by inhalation exposure to formaldehyde vapor. Cancer Res 40 : 3398–3402.
40. KernsWD, PavkovKL, DonofrioDJ, GrallaEJ, SwenbergJA (1983) Carcinogenicity of formaldehyde in rats and mice after long-term inhalation exposure. Cancer Res 43 : 4382–4392.
41. BoyneMT2nd, PesaventoJJ, MizzenCA, KelleherNL (2006) Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J Proteome Res 5 : 248–253.
Štítky
Genetika Reprodukčná medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



