-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Retrotransposon Activates Ectopic Expression: A Short Tail
article has not abstract
Published in the journal: Retrotransposon Activates Ectopic Expression: A Short Tail. PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003331
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003331Summary
article has not abstract
In 1925, Charles Danforth wrote about “mice with six legs [that] appeared about two years ago in a stock which had descended from five individuals and had been inbred for several generations” [1]. Danforth studied this “duplicitas posterior” (including duplication of internal and external urogenital organs, with quadrilateral symmetry) and its genetic transmission for several years [2], but over time he derived a line in which the principal characteristic was dominant transmission of a short, kinky, or absent tail, eponymously described as the Danforth's short tail (Sd) mutation [3] (Figure 1A and 1B). Internal caudal regression phenotypes are more serious and include defects in the axial skeleton caused by early degeneration of the notochord; small, malformed or absent kidneys; and hindgut abnormalities. Homozygotes die shortly after birth with more severe phenotypes, including complete loss of the tail, loss of both kidneys, lack of innervation along sections of colon, imperforate anus, and persistence of an abnormally small cloaca—a developmentally transient structure in mammals important for urogenital development. Interestingly, analysis of chimeras indicated cell autonomy for defects in the spine and hindgut, but not in kidney [4].
Fig. 1. The tale of Danforth's short tail. 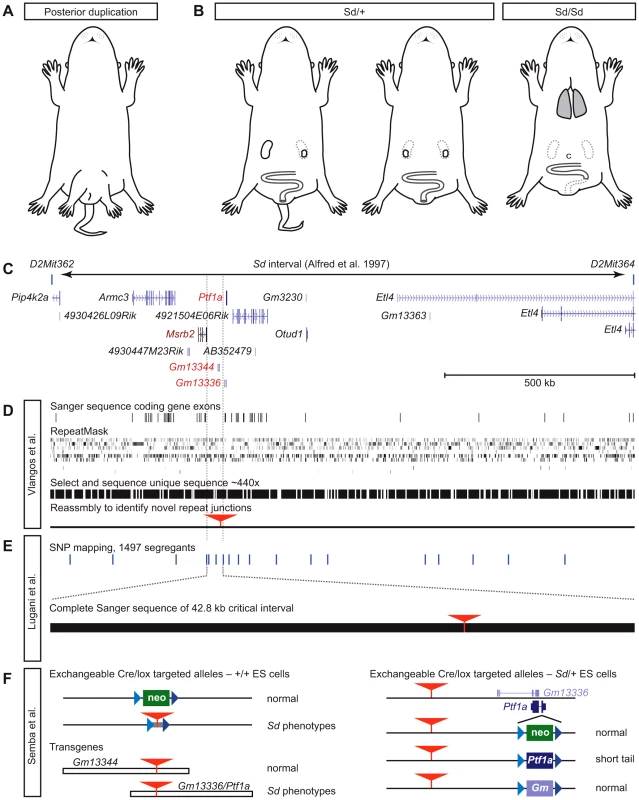
(A) Mice from Danforth's original stock showed posterior duplications, including duplicated hind limbs and a pelvic bulge, in addition to kinked and sometimes shortened tails. Drawing idealized from photographs in [2]. (B) Sd/+ mice show a strain-dependent range of caudal phenotypes, including kinked, shortened, or absent tail and reduction or loss of one kidney, but without pattern duplications. Sd/Sd animals die at birth with caudal regression, including malformation of vertebrae, absent tail, loss of both kidneys, persistent cloaca (c), aganglionosis of the hindgut, and absence of an anal opening. Semba et al. [7] also show that lungs have not inflated. (C) The Sd interval of Alfred et al. [11], re-interpreted with gene annotation from the UCSC Genome Browser, shows parts of seven protein-coding genes and seven non-coding transcripts as positional candidate genes. (D) Vlangos et al. [5] report no mutations in exons (horizontal lines), nor in unique sequences obtained by oligonucleotide selection of RepeatMasked genomic sequence (black bars). Reassembly to identify unique sequences whose paired-end reads might identify repetitive sequences not present in the reference genome identifies a novel ETn insertion (red triangle) in an intergenic region 5′ to Ptf1a. (E) Lugani et al. [6] report high-resolution recombination mapping with known SNPs (horizontal lines) to limit the Sd critical region to 42.8 kb. Exploded view shows the completely sequenced region, the ETn (red triangle) as the only variant in Sd. (F) Semba et al. [7] demonstrate both the functional significance of the ETn and the requirement for a Ptf1a open reading frame using serial gene targeting of wild type–and Sd/+-derived ES cells and conventional transgenic mice. Blue triangles, variant loxP sites; red triangles, ETn insertion; colored boxes, neo (green), Ptf1a (deep purple), and Gm13336 (light purple) replacement cassettes. Despite continued interest over several decades, Sd resisted efforts to identify the causal gene—until now. In this issue of PLOS Genetics, three laboratories independently identify the Sd mutation as an 8.5 kb ETn retrotransposon insertion 12.3 kb upstream from the Ptf1a gene [5]–[7]. Ptf1a encodes a cell type–restricted basic helix-loop-helix transcription factor required for development of the pancreas and cerebellum [8]–[10]. In the new work, all three groups conclude that ectopic expression of Ptf1 is the causal event in Sd mice.
Catherine Keegan and colleagues (Vlangos et al.) took advantage of the published Sd map location [11] (Figure 1C) and powerful genomic tools to isolate the mutation [5]. Finding no mutations after Sanger sequencing all the exons of annotated coding genes in the interval, they developed a custom oligonucleotide array to capture all non-repetitive sequences from the interval for massively parallel sequencing, using paired-end reads and requiring both ends to map correctly in the interval to ensure high-quality assembly (Figure 1D). This allowed the group to analyze more than half of the nucleotides in the critical region at a read depth of 440×, but still provided no plausible candidate mutations for Sd. Because de novo insertions of repetitive sequence are a frequent source of mutations in mice [12], the group reassembled paired-end samples that had been discarded in the first assembly pipeline because one end landed in a repeat. Using this new algorithm, they identified exactly one novel insertion, not present in a reference genome that shares extensive haplotype with the Sd chromosome, 12.3 kb proximal to Ptf1a. This candidate insertion was not found in any modern strains tested. RT-qPCR experiments showed a striking, dose-dependent increase in Ptf1a expression in Sd/+ and Sd/Sd embryos. Genomic and cDNA-based transgenes provide preliminary evidence that broad overexpression of Ptf1a causes embryonic lethality, possibly as an extreme example of the severe Sd/Sd phenotype.
Ali Gharavi and co-workers (Lugani et al.) took a complementary approach, performing SNP-based linkage mapping in 1,497 segregants to refine the Sd interval to a remarkably tidy 42.8 kb intergenic region [6] (Figure 1E). Complete Sanger sequencing of this interval by the group yielded a single DNA change relative to reference sequences: the 8.5 kb ETn element. The complete, high-confidence sequencing of the critical region provides a rigorous demonstration that the ETn insertion must be the Sd mutation. Both qPCR and in situ hybridization assays again confirmed strong ectopic expression of Ptf1a, encompassing all Sd-affected tissues. No other protein-coding genes in the broader region near the insertion site showed any similar change. A limited analysis of known Ptf1a transcriptional target genes failed to identify upregulation induced by the spread of Ptf1a in Sd embryos, but key Ptf1a targets in ectopic tissues need not be the same as those in its normal sites of expression.
Ken-ichi Yamamura's group (Semba et al.) also began with a conventional positional cloning approach. By physical mapping with an Sd/Sd cosmid library constructed for that purpose, they found an unexpectedly large fragment containing the 8.5 kb ETn element as a candidate mutation [7]. To test its functional relevance to Sd, the group performed a true tour-de-force of mouse genetics, producing a series of targeted and transgenic alleles to determine which gene products were functionally important to the Sd phenotype in the context of the ETn (Figure 1F). As the first step of a serial targeting strategy [13], a neomycin resistance cassette flanked by mutant loxP sites was introduced at the same position as the ETn insertion. This did not induce an Sd-related phenotype, suggesting that simple disruption of a cis-acting sequence is unlikely to explain the defect. However, replacing the neo cassette with a fragment containing the Sd ETn produced an allele with dosage-sensitive short tail phenotypes, indicating that the insertion of the ETn at this location is sufficient to create an Sd-like mouse. However, in addition to Ptf1a, this group found overexpression in Sd embryos of two adjacent non-coding RNAs (ncRNAs), Gm13344 and Gm13336. A transgene including the ETn, Ptf1a, and Gm13336 (which overlaps Ptf1a on the opposite strand) was sufficient to induce caudal phenotypes, but a similar construct containing the ETn and the other ncRNA was not, narrowing the list of functional candidate genes to two. The team then created germline-competent ES cells from Sd/+ embryos and serially targeted the Ptf1a/Gm13336 overlap. Integration of a floxed neo cassette on the ETn haplotype creates a Ptf1a null with the expected pancreatic agenesis, but no tail or other Sd-like effects. However, replacing the neo cassette with Ptf1a, but not Gm13336, does phenocopy Sd, demonstrating that it is specifically the ETn-dependent expression of Ptf1a that triggers the developmental abnormalities that have been studied in Sd mice for more than 70 years.
Through what effectors does Ptf1a ectopic expression act? Semba et al. provide an initial answer by profiling RNA in both Sd and ETn-Ptf1a transgenic mice relative to controls. They find down-regulation of Cdx2, another key transcription factor, along with three of its known activation targets, Cyp26a1, T, and Wnt3a. While it is not yet clear whether ectopic Ptf1a acts physically at Cdx2 in ectopic tissues, rather than indirectly through intervening factors, these profiling results provide clues to important pathways whose expression is disrupted as a consequence of Ptf1a-Sd. The results from all three groups, along with analyses of target pathways activated or repressed by Ptf1a in target tissues, will now allow us to ask how well, or in which aspects, Sd accurately models human caudal malformation and regression syndromes.
Whether the ETn acts on Ptf1a by creating a broad enhancer or by blocking an endogenous silencing element remains to be determined. Either answer might provide insight into other ETn-induced regulatory mutations [14], [15] or more broadly for integration of multiple cis-regulatory sites in the presence of retroelements. Additional serial targeting constructs that test activity of specific sequences in the ETn or perhaps chromatin conformation capture methods (3C or its more sophisticated derivatives) might help to resolve the details here. In addition, most mutagenic ETn elements are much smaller [16], [17] than that reported here, and the serial targeting strategy could be used to test whether more typical ETn elements confer a similar property to this locus. The discovery of the Sd mutation after so many decades might also prompt us to ask how often regulatory mutations might account for the remaining classical alleles that have been refractory to intragenic-centered analysis and exome sequencing.
The unusual nature of the Sd mutation also raises a final question: What relationship—if any—does Sd have to the original posterior duplication reported by Danforth, for which several specimens included completely duplicated hindlimbs, kidneys, gonads, phalli, and external urogenital openings? Did this stock contain multiple mutations, a more complex retrotransposon-mediated event that resolved into Sd, or other mutations unrelated to Sd? Perhaps that is another tail.
Zdroje
1. DanforthCH (1925) Hereditary doubling suggesting anomalous chromatin distribution in the mouse. Proc Soc Exp Biol Med 23 : 145–147.
2. DanforthCH (1930) Developmental anomalies in a special strain of mice. Am J Anat 45 : 275–287.
3. DunnLC, Gluecksohn-SchoenheimerS, BrysonV (1940) A new mutation in the mouse affecting spinal column and urogenital system. J Hered 31 : 343–348.
4. MattmanR, ZachgoJ, GosslerA (1997) The Danforth's short tail mutation acts cell autonomously in notochord cells and ventral hindgut endoderm. Development 124 : 4019–4028.
5. VlangosCN, SiuniakAN, RobinsonD, ChinnaiyanAM, LyonsRH, et al. (2013) Next generation sequencing identifies the Danforth's short tail mouse mutation as a retrotransposon insertion affecting Ptf1a expression. PLoS Genet 9: e1003205 doi:10.1371/journal.pgen.1003205
6. LuganiF, AroraR, PapetaN, PatelA, ZhengZ, et al. (2013) A retrotransposon insertion in the 5′ regulatory domain of Ptf1a results in ectopic gene expression and multiple congenital defects in Danforth's short tail mouse. PLoS Genet 9: e1003206 doi:10.1371/journal.pgen.1003206
7. SembaK, ArakiK, MatsumotoK, SudaH, AndoT, et al. (2013) Ectopic expression of Ptf1a induces spinal defects, urogenital defects, and anorectal malformations in Danforth's short tail mice. PLoS Genet 9: e1003204 doi:10.1371/journal.pgen.1003204
8. SellickGS, BarkerKT, Stolte-DijkstraI, FleischmannC, ColemanRJ, et al. (2004) Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet 36 : 1301–1305.
9. HoshinoM, NakamuraS, MoriK, KawauchiT, TeraoM, et al. (2005) Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47 : 201–213.
10. GlasgowSM, HenkeRM, MacdonaldRJ, WrightCV, JohnsonJE (2005) Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development 132 : 5461–5469.
11. AlfredJB, RanceK, TaylorBA, PhillipsSJ, AbbottCM, et al. (1997) Mapping in the region of Danforth's short tail and the localization of tail length modifiers. Genome Res 7 : 108–117.
12. HamiltonBA, FrankelWN (2001) Of mice and genome sequence. Cell 107 : 13–16.
13. ArakiK, ArakiM, YamamuraK (2002) Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res 30: e103.
14. SidowA, BulotskyMS, KerrebrockAW, BirrenBW, AltshulerD, et al. (1999) A novel member of the F-box/WD40 gene family, encoding dactylin, is disrupted in the mouse dactylaplasia mutant. Nat Genet 23 : 104–107.
15. KanoH, KurahashiH, TodaT (2007) Genetically regulated epigenetic transcriptional activation of retrotransposon insertion confers mouse dactylaplasia phenotype. Proc Natl Acad Sci U S A 104 : 19034–19039.
16. RibetD, DewannieuxM, HeidmannT (2004) An active murine transposon family pair: retrotransposition of “master” MusD copies and ETn trans-mobilization. Genome Res 14 : 2261–2267.
17. ZhangY, MaksakovaIA, GagnierL, van de LagemaatLN, MagerDL (2008) Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet 4: e1000007 doi:10.1371/journal.pgen.1000007
Štítky
Genetika Reprodukčná medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



