-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
The kinetochore is the macromolecular complex that assembles onto centromeric DNA and orchestrates the segregation of duplicated chromosomes. More than 60 components make up the budding yeast kinetochore, including inner kinetochore proteins that bind to centromeric chromatin and outer proteins that directly interact with microtubules. However, little is known about how these components assemble into a functional kinetochore and whether there are quality control mechanisms that monitor kinetochore integrity. We previously developed a method to isolate kinetochore particles via purification of the conserved Dsn1 kinetochore protein. We find that the Mub1/Ubr2 ubiquitin ligase complex associates with kinetochore particles through the CENP-CMif2 protein. Although Mub1/Ubr2 are not stable kinetochore components in vivo, they regulate the levels of the conserved outer kinetochore protein Dsn1 via ubiquitylation. Strikingly, a deletion of Mub1/Ubr2 restores the levels and viability of a mutant Dsn1 protein, reminiscent of quality control systems that target aberrant proteins for degradation. Consistent with this, Mub1/Ubr2 help to maintain viability when kinetochores are defective. Together, our data identify a previously unknown regulatory mechanism for the conserved Dsn1 kinetochore protein. We propose that Mub1/Ubr2 are part of a quality control system that monitors kinetochore integrity, thus ensuring genomic stability.
Published in the journal: The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein. PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003216
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003216Summary
The kinetochore is the macromolecular complex that assembles onto centromeric DNA and orchestrates the segregation of duplicated chromosomes. More than 60 components make up the budding yeast kinetochore, including inner kinetochore proteins that bind to centromeric chromatin and outer proteins that directly interact with microtubules. However, little is known about how these components assemble into a functional kinetochore and whether there are quality control mechanisms that monitor kinetochore integrity. We previously developed a method to isolate kinetochore particles via purification of the conserved Dsn1 kinetochore protein. We find that the Mub1/Ubr2 ubiquitin ligase complex associates with kinetochore particles through the CENP-CMif2 protein. Although Mub1/Ubr2 are not stable kinetochore components in vivo, they regulate the levels of the conserved outer kinetochore protein Dsn1 via ubiquitylation. Strikingly, a deletion of Mub1/Ubr2 restores the levels and viability of a mutant Dsn1 protein, reminiscent of quality control systems that target aberrant proteins for degradation. Consistent with this, Mub1/Ubr2 help to maintain viability when kinetochores are defective. Together, our data identify a previously unknown regulatory mechanism for the conserved Dsn1 kinetochore protein. We propose that Mub1/Ubr2 are part of a quality control system that monitors kinetochore integrity, thus ensuring genomic stability.
Introduction
Accurate chromosome segregation is essential to avoid the aneuploidy that is associated with cancer and birth defects [1]. Segregation is directed by the kinetochore, the macromolecular protein complex that assembles onto the centromeric region of each chromosome and that interacts with spindle microtubules during mitosis and meiosis [2]–[5]. The inner part of the kinetochore binds to centromeric DNA, whereas the outer portion interacts with microtubules. Although greater than 60 kinetochore components have been identified in the budding yeast kinetochore, it remains unclear how the individual proteins assemble onto the centromere to form the macromolecular kinetochore structure [6].
At the base of the kinetochore, the CENP-A centromeric histone H3 variant forms a specific chromatin environment essential for recruiting other kinetochore proteins [7], [8]. Components of the CCAN (constitutive centromere-associated network, e.g. CENP-C) closely associate with CENP-A [9], [10]. These inner kinetochore components are essential for the assembly of the outer kinetochore [11]–[13]. The outer kinetochore possesses microtubule-binding activity mediated through the KNL1 and Ndc80 complexes in the KMN (KNL1, Mis12, Ndc80 complexes) network [14]. Although the Mis12 complex does not directly bind to microtubules, it is important for the assembly of the KMN and may be a keystone to promote outer kinetochore assembly [14]–[20]. Recently, the conserved centromere-binding protein CENP-CMif2 has been shown to link the inner and outer portions of the kinetochore by binding directly to the Mis12 complex [21], [22].
Ubiquitin (Ub)-mediated proteolysis is a widely used cellular system to monitor the quality and quantity of numerous proteins [23], [24]. Ubiquitylation of a substrate requires multiple enzymes. Ubiquitin is first activated by an Ub-activating enzyme (E1), transferred to an Ub-conjugating enzyme (E2), and then conjugated to a substrate via an E3 ligase. Although the E3 enzymes largely dictate substrate specificity, efficient ubiquitylation of target proteins requires additional adaptor or cofactor proteins in some cases (e.g. [25], [26]). Proteolysis of kinetochore proteins appears to be important to ensure genomic stability. In budding yeast and flies, the CENP-A protein that is the foundation of centromeric chromatin and directs kinetochore formation is degraded to ensure its exclusive localization at the centromere [27], [28]. In addition, ubiquitin-mediated degradation of components of the CBF3 and Mis12 complexes regulates kinetochore assembly and function [18], [29], [30].
Ubr2 was identified as an E3 ligase that regulates the level of the transcription factor Rpn4 through ubiquitylation [31]. Ubiquitylation of Rpn4 by Ubr2 requires an additional factor Mub1 [32], so we will refer to the complex as Mub1/Ubr2 throughout this paper. A recent study showed that a ribonucleotide reductase inhibitor, Sml1, is also targeted by Mub1/Ubr2 [33]. Although mub1Δ and ubr2Δ mutants are viable, ubr2Δ mutant cells are sensitive to increased proteasome activity [31] and defective in the degradation of unfolded cytoplasmic protein [34]. It is not clear whether the Mub1/Ubr2 ligase complex targets additional proteins or cellular processes.
Here, we used a recently developed kinetochore purification method to show that the Mub1/Ubr2 E3 ligase complex interacts with kinetochore particles through the conserved CENP-CMif2 protein that links inner and outer kinetochore proteins [15]. We found that Mub1/Ubr2 mediate Dsn1 ubiquitylation and regulate its protein levels, especially when the Dsn1 protein is mutated. In addition, Mub1/Ubr2 become important for viability when kinetochore function is compromised. Taken together, these data suggest that Mub1/Ubr2 regulate Dsn1 levels to ensure kinetochore integrity.
Results
Kinetochore particles associate with the E3 ubiquitin ligase Ubr2 through Mub1
We previously developed a technique to isolate functional kinetochore particles via the purification of the Dsn1 kinetochore protein [15]. Although the particles contain components spanning the inner to outer kinetochore, they lack the centromere-binding complex CBF3 and some other inner kinetochore proteins. These data suggest that the particles are not bound to centromeric DNA after purification. Most of the proteins in the preparations that are visible by silver-stained SDS-PAGE are kinetochore components. However, a ∼190 kDa band that did not correspond to any candidate kinetochore proteins consistently co-purified with the particles (Figure 1A). Notably, mass spectrometry (MS) analysis of purified particles from asynchronously growing cells identified the E3 ubiquitin ligase Ubr2 and its cofactor protein Mub1 that had not previously been implicated in kinetochore function [15]. Because the predicted molecular weight of Ubr2 is 216 kDa, we tested whether the unknown candidate is Ubr2 by purifying kinetochore particles from cells expressing GFP-Ubr2. The band shifted upward when kinetochores were analyzed by silver-stained SDS-PAGE, confirming its identity as Ubr2 (Figure 1A). The Mub1 protein overlaps with non-specifically co-purifying proteins that migrate around 70 kDa (indicated by * in Figure 1A), so we performed an immunoblot to confirm that Mub1 also specifically binds to Dsn1-derived kinetochore particles (Figure 1B). The association between Ubr2 and Dsn1 was abolished in the absence of Mub1 (Figure 1C), suggesting that Mub1 acts as an adaptor protein that recruits Ubr2 onto kinetochore particles. Although this result appears to contrast previous studies that showed that Mub1 is not required for Ubr2 to interact with the Rpn4 protein [26], the interaction with Rpn4 was only tested in the presence of high Ubr2 levels.
Fig. 1. Dsn1 associates with an E3 ubiquitin ligase, Ubr2, via its adaptor, Mub1. 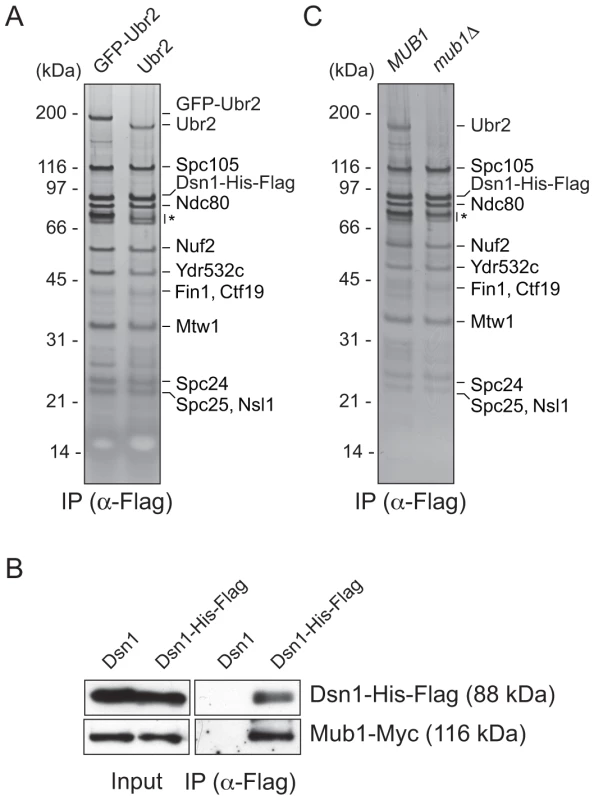
(A) Dsn1 associates with Ubr2. Dsn1-His-Flag was purified from cells containing either pGAL-GFP-UBR2 (SBY8605) or UBR2 (SBY8253) after 2 hours of growth in galactose and analyzed via silver staining. The prominent band migrating ∼190 kDa is shifted up when GFP-Ubr2 is expressed, confirming that it is Ubr2 (Ubr2: 217 kDa, GFP: 27 kDa). Note that Mub1 runs at the same position as background bands (indicated by asterisk). (B) Co-immunoprecipitation confirms that Dsn1 associates with Mub1. Proteins were purified with anti-Flag antibodies from cells containing Mub1-Myc that express either Dsn1-His-Flag (SBY8550) or untagged Dsn1 (SBY8590) and analyzed by immunoblot. Note that the Dsn1-His-Flag band overlaps with a background signal in the input. (C) The association between Dsn1 and Ubr2 requires Mub1. Dsn1-His-Flag was purified from cells in the presence (SBY8253) or absence (SBY8480) of MUB1. The band corresponding to Ubr2 is absent in mub1Δ cells. Background bands are indicated by asterisk. CENP-C recruits Mub1/Ubr2 onto kinetochore particles
To identify the kinetochore protein that recruits Mub1/Ubr2, we purified kinetochore particles from various kinetochore mutants and assayed for the presence of Ubr2 on a silver-stained gel. Ubr2 associated normally with kinetochore particles purified from outer kinetochore mutants ndc80-1 (Ndc80 complex), spc105-15 (KNL1 complex), and dad1-1, ask1-3 (Dam1 complex), as well as inner kinetochore mutants mcm21Δ, okp1-5 (CCAN/COMA complex), and cse4-323 (CENP-A) (Figure S1 and [15]). In contrast, the association of Mub1/Ubr2 with kinetochore particles was almost completely abolished in the mif2-3 (CENP-C) mutant (Figure 2A and 2B). To test whether CENP-CMif2 and Mub1/Ubr2 closely associate, we purified the CENP-CMif2-Flag protein. A silver-stained SDS-PAGE gel of purified CENP-CMif2 revealed a band that migrates around 190 kDa in addition to the Mif2-Flag band (Figure 2C). MS analysis of the purified sample showed that Mub1 and Ubr2 are among the most abundant proteins in the sample, suggesting that the 190 kDa band is most likely Ubr2 (Figure 2D and Table S1). Conversely, when Mub1-Flag was purified, several kinetochore proteins including CENP-CMif2 were detected (Figure S2 and Table S2). Our purifications of other kinetochore proteins (e.g. Nuf2, KNL-1Spc105, CENP-ACse4, Fin1) did not result in significant enrichment of the Mub1/Ubr2 proteins [15], [35], suggesting that the association of Mub1/Ubr2 depends on a unique feature of CENP-CMif2. The interaction between Mif2 and Ubr2 requires Mub1 (Figure 2E), whereas the interaction between CENP-CMif2 and Mub1 is not dependent on Ubr2 (Figure 2F). These data show that Mub1 requires CENP-CMif2 to recruit the Ubr2 ligase onto kinetochore particles.
Fig. 2. CENP-C recruits Mub1/Ubr2 onto Dsn1-derived kinetochore particles. 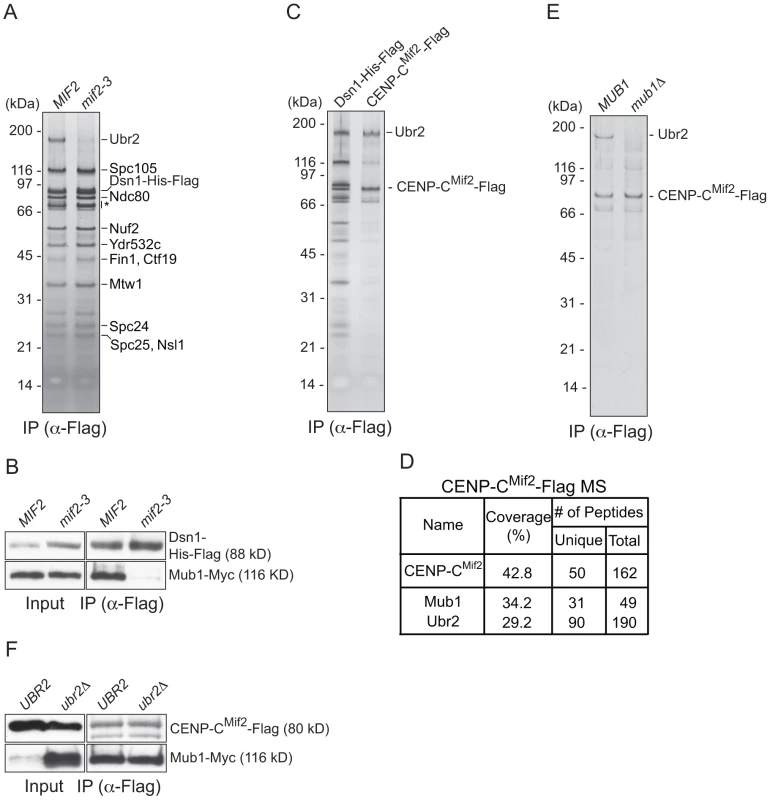
(A) Dsn1 association with Ubr2 requires CENP-CMif2. Dsn1-His-Flag was immunoprecipitated from cells with either MIF2 (SBY8253) or mif2-3 (SBY8405) and analyzed via SDS-PAGE and silver staining. Note that CENP-CMif2 runs at the same position as background bands (indicated by asterisk). (B) Dsn1 association with Mub1 requires CENP-CMif2. Dsn1-His-Flag was immunoprecipitated from cells containing Mub1-Myc and either MIF2 (SBY8550) or mif2-3 (SBY8551). (C) CENP-CMif2 associates with Ubr2. Dsn1-His-Flag (SBY8253) and CENP-CMif2-Flag (SBY8519) were immunoprecipitated and analyzed via SDS-PAGE and silver staining. (D) CENP-CMif2-Flag MS summary table. See Table S1 for all proteins identified by MS. (E) CENP-CMif2 association with Ubr2 requires Mub1. CENP-CMif2-Flag was immunoprecipitated from cells in the presence (SBY8519) or absence (SBY8911) of MUB1 and analyzed via SDS-PAGE and silver staining. (F) The association between CENP-CMif2 and Mub1 does not require Ubr2. CENP-CMif2-Flag was immunoprecipitated from cells containing Mub1-Myc in the presence (SBY8546) or absence (SBY8572) of UBR2 and analyzed via immunoblot. The Mub1 protein level in the input is higher in ubr2Δ due to the lack of Ubr2-dependent proteolysis [26]. Note that the CENP-CMif2-Flag band overlaps with a background signal in the input. Mub1/Ubr2 mediate Dsn1 ubiquitylation and degradation
Our identification of a ubiquitin ligase that associates with kinetochore particles suggests there might be a relevant kinetochore target. Although CENP-C protein is ubiquitylated by the viral immediate-early protein ICP0 upon Herpes Simplex Virus infection [36], we found no evidence that the wild-type budding yeast CENP-CMif2 protein is ubiquitylated in vivo despite its close association with the Mub1/Ubr2 ligase complex (data not shown). We therefore considered Dsn1 as a potential target because we isolated an unstable mutant while studying its phosphoregulation (manuscript in preparation). Dsn1 contains two conserved Aurora B kinase consensus sites (S240 and S250) [19], [37], [38] that we mutated to analyze the corresponding phenotypes. When both sites were mutated to alanine to block phosphorylation (dsn1-S240A,S250A), the cells were inviable (Figure 3A). In contrast, the phospho-mimic mutant (dsn1-S240D,S250D) is viable. To analyze the corresponding protein levels, myc-tagged Dsn1 phospho-mutants were expressed from the endogenous DSN1 promoter in the presence of wild-type DSN1 to keep the cells alive. Although the protein levels of wild-type Dsn1 and the phospho-mimic mutant were similar, there were lower levels of the Dsn1-S240A,S250A mutant (Figure 3B). To test whether the inviability of dsn1-S240A,S250A is a result of low protein levels, we expressed dsn1-S240A,S250A from a high copy plasmid, which led to much higher protein levels (Figure 3C). The overexpressed dsn1-S240A,S250A mutant complements dsn1Δ (Figure 3D), supporting the idea that the inviability is at least partially due to reduced Dsn1 protein levels.
Fig. 3. Dsn1-S240A,S250A levels correlate with viability. 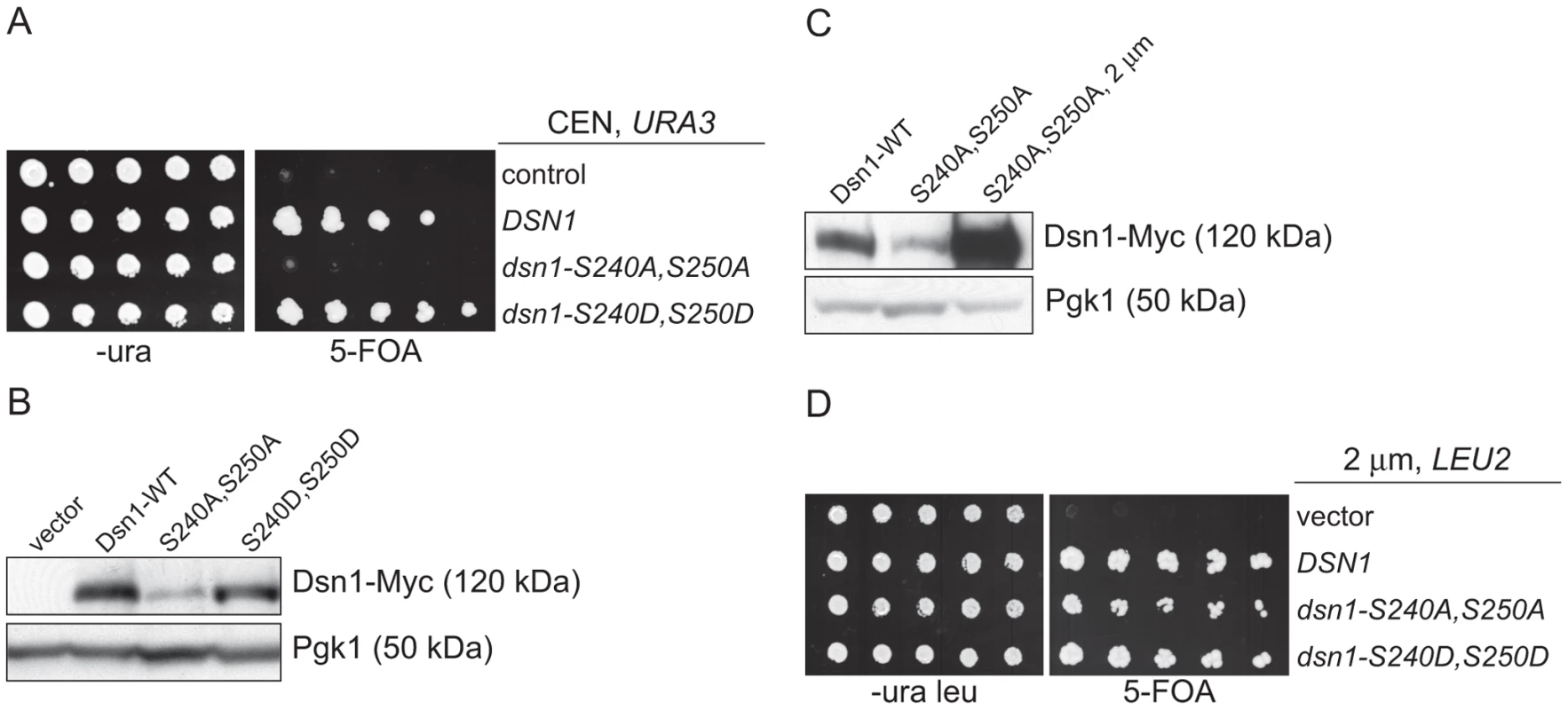
(A) Dsn1-S240A,S250A cells are inviable. Serial dilutions (3-fold) of dsn1Δ cells containing DSN1 on a URA3, CEN vector and the indicated integrated point mutants (SBY2318, SBY5948, SBY5949, SBY5950) were plated. Cells that need to maintain the URA3, CEN vector for viability are sensitive to 5-FOA. (B) Dsn1-S240A,S250A protein levels are reduced. Whole cell extracts of Dsn1 and the indicated point mutants (SBY2153, SBY7864, SBY7865, SBY7867) were prepared and analyzed via immunoblot. (C) Overexpression of Dsn1-S240A,S250A restores protein levels. Whole cell extracts of the indicated Dsn1 mutants (SBY8766, SBY8521, SBY7373) were prepared and analyzed via immunoblot. (D) Overexpression of dsn1-S240A,S250A restores viability. Serial dilutions (3-fold) of dsn1Δ strains containing DSN1 on a URA3, CEN vector and a 2 µm LEU2 plasmid with the indicated point mutants (SBY7368, SBY7362, SBY7363, SBY7364 were plated on –ura leu and 5-FOA plates. We did not detect any obvious defect in cells overexpressing wild-type or mutant Dsn1 proteins. We asked whether Dsn1 is a target of Mub1/Ubr2 by testing whether a deletion of MUB1 or UBR2 could stabilize the Dsn1-S240A,S250A mutant protein. Strikingly, Dsn1-S240A,S250A protein levels were restored to near wild-type in both mub1Δ and ubr2Δ strains (Figure 4A). The levels of WT Dsn1 were also increased in the mub1Δ and ubr2Δ strains, suggesting that Mub1/Ubr2 mediate the degradation of WT Dsn1 protein. We therefore analyzed Dsn1 stability by adding cycloheximide to repress translation. The Dsn1 protein levels were higher in the mub1Δ strain at the start of the experiment and did not decrease as much as in WT cells (Figure 4B), although there is still some degradation in the absence of Mub1/Ubr2. Because Mub1/Ubr2 target the Dsn1-S240A,S250A protein that lacks Aurora B phosphorylation sites, we also analyzed Dsn1 stability in a budding yeast Aurora B mutant, ipl1-321. We found that Dsn1 is somewhat less stable in an ipl1-321 mutant relative to WT cells and that the degradation is at least partially dependent on Mub1/Ubr2 (Figure 4C and 4D). These data are consistent with the lack of phosphorylation on residues 240 and 250 leading to the degradation of Dsn1.
Fig. 4. Mub1/Ubr2 mediate Dsn1 ubiquitylation and regulate protein levels. 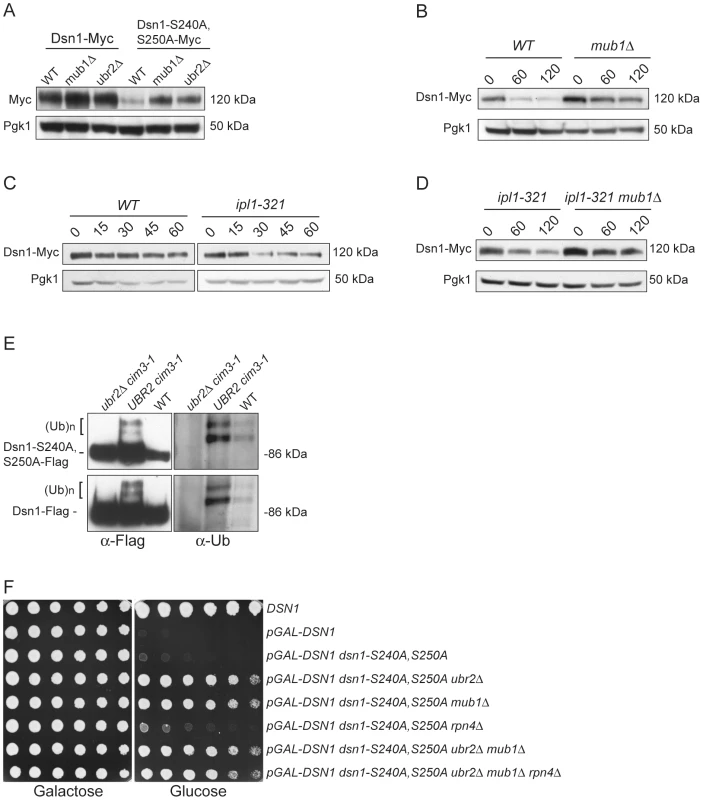
(A) Deleting MUB1 and UBR2 restores Dsn1-S240A,S250A protein levels. Whole cell extracts were prepared from WT (SBY8766, SBY8521), mub1Δ (SBY10959, SBY8164), and ubr2Δ cells (SBY10960, SBY8265). Dsn1-Myc and Dsn1-S240,S250A-Myc levels were monitored by immunoblot. (B) Mub1 regulates Dsn1 stability. WT (SBY8766) and mub1Δ (SBY10959) cells containing Dsn1-myc were treated with cycloheximide and analyzed for Dsn1 protein levels at the indicated time points (min). (C) Aurora B regulates Dsn1 stability. WT (SBY8766) and ipl1-321 (SBY8150) cells containing Dsn1-myc were shifted to 37°C and treated with cycloheximide. Cells were analyzed for Dsn1 protein levels at the indicated time points (min). (D) Mub1/Ubr2 regulate Dsn1 stability in an Aurora B mutant. Ipl1-321 (SBY8150) and ipl1-321 mub1Δ (SBY9428) cells containing Dsn1-myc were shifted to 37°C and treated with cycloheximide. Cells were analyzed for Dsn1 protein levels at the indicated time points (min). (E) Ubr2 ubiquitylates Dsn1-S240A,S250A and wild-type Dsn1. Flag epitope-tagged Dsn1-S240A,S250A or Dsn1-WT was immunoprecipitated from cim3-1 ubr2Δ cells (SBY8703, SBY8705), cim3-1 UBR2 cells (SBY8704, SBY8706) and WT cells (SBY8615, SBY7441). Samples were analyzed via immunoblot with anti-Flag and anti-Ub antibodies. (F) Deleting MUB1 and UBR2 restores viability to Dsn1-S240A,S250A cells. Serial dilutions (3-fold) of pGAL-DSN1 cells containing integrated dsn1-S240A,S250A with the indicated deletions (SBY8264, SBY8262, SBY8844, SBY8469, SBY8842, SBY8901) were plated on either glucose or galactose media. A WT strain (SBY3) and a pGAL-DSN1 strain lacking the integrated point-mutant (SBY7948) were included as controls. We next tested whether Mub1/Ubr2 directly mediate the ubiquitylation of Dsn1. When purified from a cim3-1 proteasome mutant, upper forms of WT Dsn1 as well as the Dsn1-S240A,S250A mutant protein that are characteristic of ubiquitylation were apparent on an immunoblot (Figure 4E). Indeed, upper bands specific to a Dsn1 immunoprecipitation were recognized by anti-ubiquitin antibodies and were more abundant in cim3-1 mutant cells. Importantly, the ubiquitylation of Dsn1 was dependent on the Ubr2 E3 ligase (Figure 4E). Taken together, these data strongly suggest that the Mub1/Ubr2 E3 ligase targets the Dsn1 kinetochore protein for degradation.
Our finding that Mub1/Ubr2 regulate Dsn1 protein levels led us to test whether mub1Δ and ubr2Δ deletion mutants suppress the lethality of dsn1-S240A,S250A. As predicted, there was significant suppression by deletion of either Mub1 or Ubr2 (Figure 4F). The mub1Δ ubr2Δ double mutant suppressed to a similar extent as the individual mutants, consistent with Mub1 and Ubr2 functioning in a complex to control Dsn1 protein levels. It was previously found that mub1Δ and ubr2Δ mutants exhibit increased proteasome activity because the Rpn4 transcription factor is not degraded [26]. As expected, suppression of the dsn1-S240A,S250A lethality is not due to increased proteasome activity, because suppression occurred even in the mub1Δ ubr2Δ rpn4Δ triple mutant. Taken together, these results show that the dsn1-S240A,S250A mutant is lethal because it has reduced protein levels due to Mub1/Ubr2-dependent proteolysis. Our kinetochore particles also interact with another E3 ligase, Psh1 [15], but its deletion does not suppress dsn1-S240A,S250A lethality (data not shown), consistent with its specific regulation of CENP-A [39].
Mub1/Ubr2 are important when kinetochore function is compromised
The regulation of Dsn1 by Mub1/Ubr2 prompted us to test whether they have a role in chromosome segregation. Because Mub1 mediates the Mub1/Ubr2 interaction with kinetochores, we analyzed a mub1Δ strain. However, chromosome biorientation in metaphase cells (as judged by the characteristic bi-lobed formation of the Mtw1 kinetochore protein [40], Figure 5A) and chromosome segregation (monitored by segregation of a pair of marked sister chromatids [41]) (data not shown) appeared normal in mub1Δ mutant cells. Interestingly, we failed to identify Mub1/Ubr2 by MS or immunoblot when we purified centromeric minichromosomes [35], suggesting they may not be core kinetochore components. To determine whether Mub1/Ubr2 localize to endogenous kinetochores, we performed chromatin immunoprecipitation (Figure 5B) and chromosome spread experiments (data not shown). We did not detect an association of Mub1/Ubr2 with endogenous kinetochores, indicating that Mub1/Ubr2 are not structural components of kinetochores. However, they may transiently associate with kinetochores.
Fig. 5. Mub1/Ubr2 cannot be detected at endogenous centromeres but are important for kinetochore function. 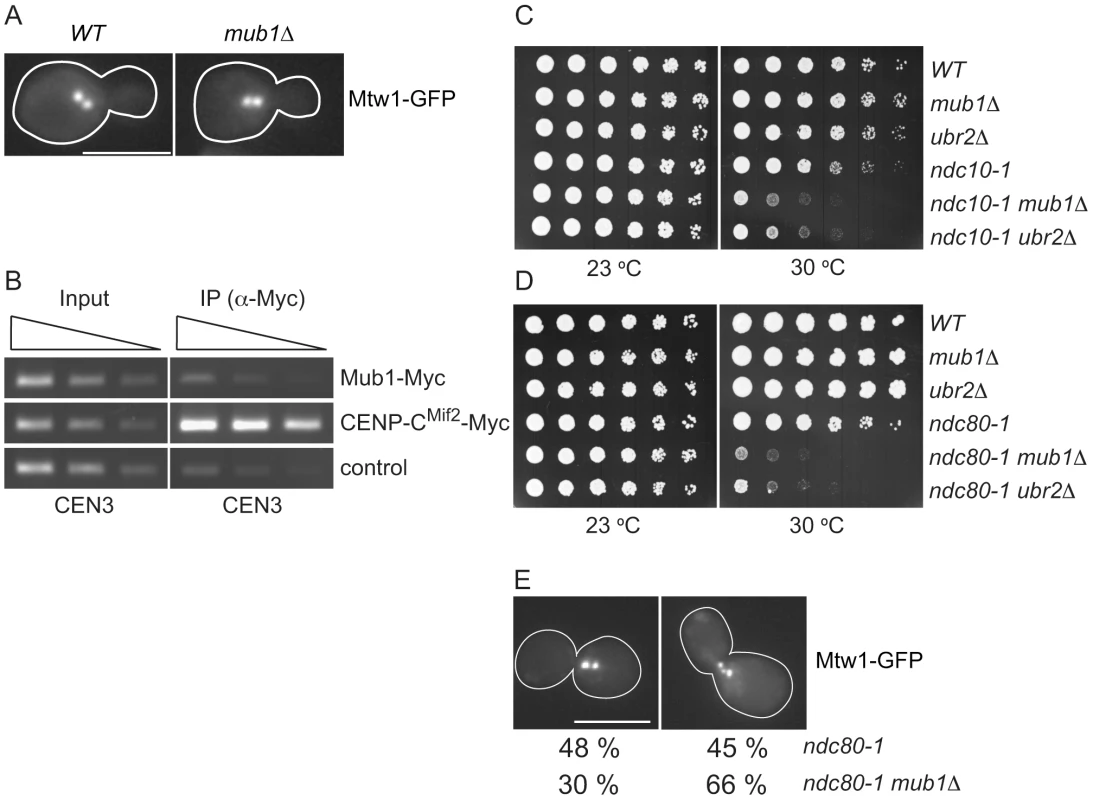
(A) Mub1 is dispensable for kinetochore biorientation. Mtw1-3GFP was monitored in either MUB1 (SBY3798) or mub1Δ (SBY8026) cells for biorientation defects. A representative cell from each strain is shown. Bar, 5 µm. (B) Mub1 cannot be detected at centromeres. Chromatin immunoprecipitation was carried-out on Mub1-Myc ubr2Δ (SBY8572), Mif2-Myc (SBY1566), and untagged control (SBY3) cells using a probe for CEN3. We obtained similar results using Mub1-Myc UBR2 cells (data not shown). (C, D) Mub1Δ and ubr2Δ exhibit negative genetic interactions with kinetochore mutants. Serial dilutions (5-fold) of ndc10-1 (SBY3, SBY7793, SBY7851, SBY164, SBY8613, SBY8773) and ndc80-1 (SBY3, SBY7793, SBY7851, SBY1117, SBY8436, SBY8432) kinetochore mutants with mub1Δ and ubr2Δ were plated on YPD and incubated at the indicated temperatures to examine genetic interactions. (E) Ndc80-1 mub1Δ double mutants exhibit an increase in declustered kinetochores. Ndc80-1 (SBY3934) and ndc80-1 mub1Δ (SBY8670) cells containing Mtw1-3GFP were released from G1 to 30 degrees and kinetochores were visualized at 180′. The percent of clustered (left panel) vs. unclustered (right panel) was quantified. Bar, 5 µm. Because we did not detect a major defect in segregation in the absence of Mub1/Ubr2, we considered the possibility that Mub1/Ubr2 becomes important when kinetochore function is compromised. We analyzed the ndc10-1 temperature sensitive mutant that is completely defective in kinetochore assembly [42] as well as the ndc80-1 mutant that assembles a kinetochore with defects specifically outer kinetochore function [43]. Strikingly, deletion of Mub1/Ubr2 decreased the semi-permissive temperature of these mutants, indicating that their function is important for viability when kinetochores are defective (Figure 5C and 5D). To determine whether the growth defect in ndc80-1 mub1Δ strains is due to defective kinetochore function, we monitored kinetochore clustering in the double mutant. Although kinetochores in wild-type yeast cells cluster into two distinct foci at metaphase [40], [44], [45], cells with defects in microtubule-kinetochore attachments exhibit three or more declustered foci [46]. We visualized kinetochores using GFP epitope-tagged Mtw1 kinetochore protein in ndc80-1 and mub1Δ ndc80-1 mutant strains. Cells were released from G1 into the cell cycle at 30 degrees and monitored for kinetochore foci after 140 min (Figure 5E). When the number of kinetochore foci were quantified, 45% of the ndc80 mutant cells exhibited declustered kinetochores (greater than 3 foci) compared to 66% in the ndc80 mub1 cells. These data are consistent with greater than 75% of the cells being arrested in metaphase with high Pds1 levels due to spindle checkpoint activation. Taken together, these data suggest that kinetochore integrity is further compromised by the lack of Mub1 in the ndc80-1 mutant cells.
Discussion
Here, we show that the Mub1/Ubr2 E3 ligase complex regulates the levels of the conserved Dsn1 kinetochore protein via ubiquitin-mediated proteolysis. Mub1/Ubr2 have previously been implicated in proteasome function through the regulation of Rpn4 [26], [31], as well as regulation of dNTP levels through Sml1 degradation [33]. Our data suggest that Mub1/Ubr2 also regulate kinetochore function through proteolysis of the Dsn1 protein. Although Mub1/Ubr2 requires the Rad6 E2 protein to target Rpn4 and Sml1 [26], [33], a rad6Δ does not suppress the lethality of dsn1-S240A,S250A (data not shown), suggesting that Mub1/Ubr2 utilizes a different or additional E2 to target the Dsn1 kinetochore protein. Identification of the E2 that facilitates the ubiquitylation of Dsn1 should shed additional light on the mechanism by which Mub1/Ubr2 target Dsn1 for degradation.
Our data suggest that CENP-CMif2 is the receptor for Mub1/Ubr2 on kinetochore particles. Kinetochores purified from mif2 mutants lacked Mub1/Ubr2, and the purification of CENP-CMif2, but not other kinetochore proteins (e.g. Nuf2, KNL-1Spc105, CENP-ACse4, Fin1), resulted in a signification enrichment of the Mub1/Ubr2 proteins [15], [35]. Therefore, a unique feature of CENP-CMif2 or a CENP-CMif2-binding protein is important for the interaction of Mub1/Ubr2 with kinetochore particles. However, despite the close association of Mub1/Ubr2 with CENP-CMif2, we did not detect ubiquitylation of CENP-CMif2 or a CENP-CMif2 mutant allele ([47] and data not shown). Because we could not detect a stable association of Mub1/Ubr2 with endogenous kinetochores, Mub1/Ubr2 likely bind to kinetochore particles during the purification process. This raises the possibility that Mub1/Ubr2 only transiently associate with kinetochores to regulate Dsn1, and/or that they regulate a pool of Dsn1 that is not associated with kinetochores.
Although Mub1/Ubr2 are not essential during mitotic growth, a deletion of mub1 or ubr2 can restore both the protein levels and inviability of the dsn1-S240A,S250A mutant. These data indicate that Mub1/Ubr2 have a physiological role in regulating kinetochore function in vivo despite their lack of enrichment at endogenous kinetochores. In addition, deletions in Mub1/Ubr2 exhibited negative genetic interactions with the ndc80-1 and ndc10-1 mutants. In the ndc80 mutant cells, kinetochore-microtubule attachments and biorientation defects were exacerbated by the lack of Mub1. This is reminiscent of the spindle checkpoint proteins that are also not essential in budding yeast but become critical to ensure genomic stability when kinetochore-microtubule interactions are compromised [48], [49]. Mub1 and Ubr2 mutants display stronger meiotic phenotypes [50], and we note that the monopolin complex that joins sister kinetochores during meiosis I to ensure that sister chromatids segregate to the same pole associates with kinetochores via its interaction with CENP-C and Dsn1 [51]. It will be interesting to determine if the Mub1/Ubr2 complex regulates monopolin binding to kinetochores in meiosis. In addition, although monopolin components are sequestered in the nucleolus during mitosis, Mub1/Ubr2 may provide a back up system to prevent them from linking sister kinetochores during mitosis. An important future goal will be to understand the cellular location of the Mub1/Ubr2 ubiquitylation of Dsn1.
We speculate that Mub1/Ubr2 monitor kinetochore integrity and only become important for mitosis when kinetochores are defective. We were not able to detect a significant Mub1/Ubr2-dependent change in Dsn1 stability in ndc10-1 and ndc80-1 kinetochore mutants (data not shown), possibly because there is only a small pool of Dsn1 that is regulated by Mub1/Ubr2. In mammalian cells, Dsn1 is regulated by the SCF-Skp1 ubiquitin ligase [18], so it will be important to test whether Skp1-mediated degradation also regulates Dsn1 in budding yeast. We also detected ubiquitylation and regulation of WT Dsn1 protein levels by Mub1/Ubr2, although the effect was mild. We presume that WT cells would not have a need for robust regulation of Dsn1 by Mub1/Ubr2. A deletion of Mub1/Ubr2 led to a strong increase in the levels of the Dsn1-S240A,S250A mutant lacking Aurora B phosphorylation sites. Degradation of mutant alleles is a hallmark of quality control systems. Some temperature sensitive mutants, although possessing normal or near normal activity, are recognized as aberrant and destroyed by quality control mechanisms [52]–[56]. Inhibition of the degradation suppresses the temperature sensitivity, similar to the suppression of dsn1-S240A,S250A inviability by deletion of Mub1/Ubr2. One possibility is that kinetochore quality control may be needed to ensure that defective kinetochores do not assemble, or that they are turned over in an attempt to assemble functional kinetochores. For example, ndc80-1 and ndc10-1 mutant cells could accumulate aberrant kinetochores in mub1Δ and ubr2Δ mutants, enhancing their temperature sensitivity. It was recently shown that CENP-CMif2 mediates the interaction between the centromere and outer kinetochore via its interaction with the Mis12 complex that contains Dsn1 [21], [22]. One possibility is that Mub1/Ubr2 binding to CENP-CMif2 regulates kinetochore assembly by preventing stable association of the outer kinetochore via Dsn1-mediated degradation. Consistent with this model, Dsn1 mutants lacking Aurora B phosphorylation sites in other organisms exhibit defects in kinetochore assembly [57], [58]. We speculate that this mechanism would help to avoid the formation of ectopic kinetochores outside of the centromere, as well as play a key role in preventing aberrant kinetochores from stably assembling at centromeres.
There are numerous examples in mammalian studies where mutant kinetochore proteins show decreased protein levels, so it will be important to determine whether a similar quality control system operates in other eukaryotes [16], [59]. Mub1 has a conserved MYND domain that is implicated in mediating protein-protein interactions [60], and MYND-containing proteins are implicated in protein ubiquitylation in human cells [61], [62]. Importantly, many of the MYND-containing proteins are associated with cancer and other diseases (e.g. ZMYND10, ZMYND11/BS69, ETO (eight-twenty-one)/MTG (myeloid translocation gene) family members, DEAF1, and Suppressin (reviewed in [63]). In addition, the stability of Dsn1 orthologs appears to be carefully regulated by chaperones and proteolysis [16], [18], so it will be critical to understand how these MYND proteins function and whether they also regulate kinetochores in mammalian cells. Further studies of potential kinetochore quality control mechanisms could shed light on the process of kinetochore assembly as well as the maintenance of genomic stability in all organisms.
Materials and Methods
Yeast strains, plasmids, and microbial techniques
Media and genetic and microbial techniques were essentially as described [64]. Yeast strains and plasmids used in this study are listed in Tables S3 and S4. The mif2-3 [65], cim3-1 [66], ndc10-1 [42], ndc80-1 [43], , okp1-5 [46], [68], dad1-1 [69], ask1-3 [70], and cse4-323 [71] alleles were crossed to make strains for this study. Deletions, as well as 3Flag, 13Myc, GFP epitope tags were made using a PCR-based integration system and confirmed by PCR [72]–[74]. Specific primer sequences are listed in Table S5.
Plasmid construction
pSB1097 (DSN1, HIS3, integrating vector) was constructed by subcloning DSN1 with its endogenous promoter from pSB624 [75] into pRS303 (HIS3, integrating vector) [76] using EcoRI and XhoI. pSB1322 (DSN1-12myc, LEU2, 2 micron plasmid) was constructed by subcloning DSN1-12myc with the endogenous DSN1 promoter of pSB1110 (DSN1-12myc, URA3, integrating vector) [15], [77] into pRS425 (LEU2, 2 micron plasmid) using XhoI and SacII. Phospho-mutants were made by Quickchange site-directed mutagenesis (Stratagene).
Protein and immunological techniques
Immunoprecipitation was performed using BH/0.15 (25 mM HEPES pH 8.0, 2 mM MgCl2, 0.1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.1% NP-40, 150 mM KCl, 15% glycerol) containing protease inhibitors, phosphatase inhibitors as described [35]. Immunoblotting was performed as described [78]. Anti-Flag antibodies (M2, Sigma-Aldrich) were used at 1∶3,000, anti-Myc antibodies at 1∶10,000 (9E10, Covance), and anti-Pgk1 (Invitrogen) at 1∶10,000. Anti-Mif2 (OD2, 1∶6,000) antibodies were kind gifts from Arshad Desai [35]. To detect ubiquitylation of Dsn1, 10 mM N-ethyl maleimide was added to the buffer throughout purifications. In addition, the nitrocellulose membrane was autoclaved after the transfer to increase sensitivity [79], and anti-Ubiquitin antibodies (Zymed) were used at 1∶500. Silver-staining was performed on 4–12% NuPAGE Novex Bis-Tris gels (Invitrogen) using a SilverQuest silver-staining kit according to instructions (Invitrogen). Chromatin immunoprecipitation was performed using anti-c-Myc antibodies (A14, Santa Cruz Biotechnology) as described [75]. To identify co-purifying proteins, associated proteins were eluted with detergent and analyzed by MS with LTQ-Orbitrap as described [35]. Stability experiments were performed by adding 50 microgram/ml cycloheximide.
Microscopy
Analysis of Mtw1-3GFP and fluorescently marked sister chromatids were performed as described using a Nikon microscope [46]. Data was collected with two second exposures using Metamorph software. At least 300 cells were analyzed for all reported experiments.
Supporting Information
Zdroje
1. HollandAJ, ClevelandDW (2012) Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep 13 : 501–514.
2. WestermannS, DrubinDG, BarnesG (2007) Structures and functions of yeast kinetochore complexes. Annu Rev Biochem 76 : 563–591.
3. CheesemanIM, DesaiA (2008) Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9 : 33–46.
4. SantaguidaS, MusacchioA (2009) The life and miracles of kinetochores. EMBO J 28 : 2511–2531.
5. BloomK, JoglekarA (2010) Towards building a chromosome segregation machine. Nature 463 : 446–456.
6. LampertF, WestermannS (2011) A blueprint for kinetochores - new insights into the molecular mechanics of cell division. Nat Rev Mol Cell Biol 12 : 407–412.
7. AllshireRC, KarpenGH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9 : 923–937.
8. BlackBE, ClevelandDW (2011) Epigenetic Centromere Propagation and the Nature of CENP-A Nucleosomes. Cell 144 : 471–479.
9. McAinshAD, TytellJD, SorgerPK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19 : 519–539.
10. TakeuchiK, FukagawaT (2012) Molecular architecture of vertebrate kinetochores. Exp Cell Res 318 : 1367–1374.
11. TanakaK, ChangHL, KagamiA, WatanabeY (2009) CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell 17 : 334–343.
12. OkadaM, CheesemanIM, HoriT, OkawaK, McLeodIX, et al. (2006) The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8 : 446–457.
13. CarrollCW, StraightAF (2006) Centromere formation: from epigenetics to self-assembly. Trends Cell Biol 16 : 70–78.
14. CheesemanIM, ChappieJS, Wilson-KubalekEM, DesaiA (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127 : 983–997.
15. AkiyoshiB, SarangapaniKK, PowersAF, NelsonCR, ReichowSL, et al. (2010) Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468 : 576–579.
16. KlineSL, CheesemanIM, HoriT, FukagawaT, DesaiA (2006) The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 173 : 9–17.
17. CheesemanIM, HoriT, FukagawaT, DesaiA (2008) KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell 19 : 587–594.
18. DaviesAE, KaplanKB (2010) Hsp90-Sgt1 and Skp1 target human Mis12 complexes to ensure efficient formation of kinetochore-microtubule binding sites. J Cell Biol 189 : 261–274.
19. MaskellDP, HuXW, SingletonMR (2010) Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol 190 : 823–834.
20. PetrovicA, PasqualatoS, DubeP, KrennV, SantaguidaS, et al. (2010) The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol 190 : 835–852.
21. PrzewlokaMR, VenkeiZ, Bolanos-GarciaVM, DebskiJ, DadlezM, et al. (2011) CENP-C is a structural platform for kinetochore assembly. Curr Biol 21 : 399–405.
22. ScrepantiE, De AntoniA, AlushinGM, PetrovicA, MelisT, et al. (2011) Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol 21 : 391–398.
23. DeshaiesRJ, JoazeiroCA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78 : 399–434.
24. HershkoA (2005) The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ 12 : 1191–1197.
25. KavsakP, RasmussenRK, CausingCG, BonniS, ZhuH, et al. (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell 6 : 1365–1375.
26. JuD, WangX, XuH, XieY (2008) Genome-wide analysis identifies MYND-domain protein Mub1 as an essential factor for Rpn4 ubiquitylation. Mol Cell Biol 28 : 1404–1412.
27. Moreno-MorenoO, Torras-LlortM, AzorinF (2006) Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res 34 : 6247–6255.
28. CollinsKA, FuruyamaS, BigginsS (2004) Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol 14 : 1968–1972.
29. KaplanKB, HymanAA, SorgerPK (1997) Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91 : 491–500.
30. KopskiKM, HuffakerTC (1997) Suppressors of the ndc10-2 mutation: a role for the ubiquitin system in Saccharomyces cerevisiae kinetochore function. Genetics 147 : 409–420.
31. WangL, MaoX, JuD, XieY (2004) Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J Biol Chem 279 : 55218–55223.
32. JuD, WangL, MaoX, XieY (2004) Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem Biophys Res Commun 321 : 51–57.
33. AndresonBL, GuptaA, GeorgievaBP, RothsteinR (2010) The ribonucleotide reductase inhibitor, Sml1, is sequentially phosphorylated, ubiquitylated and degraded in response to DNA damage. Nucleic Acids Res 38 : 6490–6501.
34. NillegodaNB, TheodorakiMA, MandalAK, MayoKJ, RenHY, et al. (2010) Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell 21 : 2102–2116.
35. AkiyoshiB, NelsonCR, RanishJA, BigginsS (2009) Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev 23 : 2887–2899.
36. EverettRD, EarnshawWC, FindlayJ, LomonteP (1999) Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J 18 : 1526–1538.
37. WestermannS, CheesemanIM, AndersonS, YatesJR3rd, DrubinDG, et al. (2003) Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol 163 : 215–222.
38. WelburnJP, VleugelM, LiuD, YatesJR3rd, LampsonMA, et al. (2010) Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell 38 : 383–392.
39. RanjitkarP, PressMO, YiX, BakerR, MacCossMJ, BigginsS (2010) An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40 : 455–464.
40. GoshimaG, YanagidaM (2000) Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100 : 619–633.
41. StraightAF, BelmontAS, RobinettCC, MurrayAW (1996) GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol 6 : 1599–1608.
42. GohPY, KilmartinJV (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol 121 : 503–512.
43. WiggePA, JensenON, HolmesS, SouesS, MannM, et al. (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol 141 : 967–977.
44. HeX, AsthanaS, SorgerPK (2000) Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101 : 763–775.
45. PearsonCG, YehE, GardnerM, OddeD, SalmonED, et al. (2004) Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol 14 : 1962–1967.
46. PinskyBA, KungC, ShokatKM, BigginsS (2006) The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol 8 : 78–83.
47. PengY, WongCC, NakajimaY, TyersRG, SarkeshikAS, et al. (2011) Overlapping kinetochore targets of CK2 and Aurora B kinases in mitotic regulation. Mol Biol Cell 22 : 2680–2689.
48. LiR, MurrayAW (1991) Feedback control of mitosis in budding yeast. Cell 66 : 519–531.
49. HoytMA, TotisL, RobertsBT (1991) S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66 : 507–517.
50. MarstonAL, ThamWH, ShahH, AmonA (2004) A genome-wide screen identifies genes required for centromeric cohesion. Science 303 : 1367–1370.
51. CorbettKD, YipCK, EeLS, WalzT, AmonA, et al. (2010) The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell 142 : 556–567.
52. BettingJ, SeufertW (1996) A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin - and proteasome-dependent pathway. J Biol Chem 271 : 25790–25796.
53. BordalloJ, PlemperRK, FingerA, WolfDH (1998) Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 9 : 209–222.
54. BaysNW, GardnerRG, SeeligLP, JoazeiroCA, HamptonRY (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol 3 : 24–29.
55. BrewCT, HuffakerTC (2002) The yeast ubiquitin protease, Ubp3p, promotes protein stability. Genetics 162 : 1079–1089.
56. GardnerRG, NelsonZW, GottschlingDE (2005) Degradation-mediated protein quality control in the nucleus. Cell 120 : 803–815.
57. EmanueleMJ, LanW, JwaM, MillerSA, ChanCS, et al. (2008) Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol 181 : 241–254.
58. YangY, WuF, WardT, YanF, WuQ, et al. (2008) Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J Biol Chem 283 : 26726–26736.
59. KiyomitsuT, MurakamiH, YanagidaM (2011) Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol Cell Biol 31 : 998–1011.
60. AmannJM, NipJ, StromDK, LutterbachB, HaradaH, et al. (2001) ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol 21 : 6470–6483.
61. IsobeT, UchidaC, HattoriT, KitagawaK, OdaT, et al. (2006) Ubiquitin-dependent degradation of adenovirus E1A protein is inhibited by BS69. Biochem Biophys Res Commun 339 : 367–374.
62. ChoiKO, LeeT, LeeN, KimJH, YangEG, et al. (2005) Inhibition of the catalytic activity of hypoxia-inducible factor-1alpha-prolyl-hydroxylase 2 by a MYND-type zinc finger. Mol Pharmacol 68 : 1803–1809.
63. HessonLB, CooperWN, LatifF (2007) Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene 26 : 7283–7301.
64. Rose MD, Winston F, Heiter P (1990) Methods in yeast genetics. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press. p. 198 p.
65. BrownMT, GoetschL, HartwellLH (1993) MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J Cell Biol 123 : 387–403.
66. GhislainM, UdvardyA, MannC (1993) S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366 : 358–362.
67. NekrasovVS, SmithMA, Peak-ChewS, KilmartinJV (2003) Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell 14 : 4931–4946.
68. OrtizJ, StemmannO, RankS, LechnerJ (1999) A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev 13 : 1140–1155.
69. Enquist-NewmanM, CheesemanIM, Van GoorD, DrubinDG, MeluhPB, et al. (2001) Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol Biol Cell 12 : 2601–2613.
70. LiY, BachantJ, AlcasabasAA, WangY, QinJ, et al. (2002) The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev 16 : 183–197.
71. BigginsS, BhallaN, ChangA, SmithDL, MurrayAW (2001) Genes Involved in Sister Chromatid Separation and Segregation in the Budding Yeast Saccharomyces cerevisiae. Genetics 159 : 453–470.
72. LongtineMS, McKenzieA3rd, DemariniDJ, ShahNG, WachA, et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 : 953–961.
73. GelbartME, RechsteinerT, RichmondTJ, TsukiyamaT (2001) Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol 21 : 2098–2106.
74. GoldsteinAL, McCuskerJH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553.
75. PinskyBA, TatsutaniSY, CollinsKA, BigginsS (2003) An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell 5 : 735–745.
76. SikorskiRS, HieterP (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 : 19–27.
77. AkiyoshiB, BigginsS (2010) Cdc14-dependent dephosphorylation of a kinetochore protein prior to anaphase in Saccharomyces cerevisiae. Genetics 186 : 1487–1491.
78. BigginsS, SeverinFF, BhallaN, SassoonI, HymanAA, et al. (1999) The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev 13 : 532–544.
79. SwerdlowPS, FinleyD, VarshavskyA (1986) Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal Biochem 156 : 147–153.
Štítky
Genetika Reprodukčná medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



