-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
Alternative splicing is commonly used by the Metazoa to generate more than one protein from a gene. However, such diversification of the proteome by alternative splicing is much rarer in fungi. We describe here an ancient fungal alternative splicing event in which these two proteins are generated from a single alternatively spliced ancestral SKI7/HBS1 gene retained in many species in both the Ascomycota and Basidiomycota. While the ability to express two proteins from a single SKI7/HBS1 gene is conserved in many fungi, the exact mechanism by which they achieve this varies. The alternative splicing was lost in Saccharomyces cerevisiae following the whole-genome duplication event as these two genes subfunctionalized into the present functionally distinct HBS1 and SKI7 genes. When expressed in yeast, the single gene from Lachancea kluyveri generates two functionally distinct proteins. Expression of one of these proteins complements hbs1, but not ski7 mutations, while the other protein complements ski7, but not hbs1. This is the first known case of subfunctionalization by loss of alternative splicing in yeast. By coincidence, the ancestral alternatively spliced gene was also duplicated in Schizosaccharomyces pombe with subsequent subfunctionalization and loss of splicing. Similar subfunctionalization by loss of alternative splicing in fungi also explains the presence of two PTC7 genes in the budding yeast Tetrapisispora blattae, suggesting that this is a common mechanism to preserve duplicate alternatively spliced genes.
Published in the journal: Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes. PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003376
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003376Summary
Alternative splicing is commonly used by the Metazoa to generate more than one protein from a gene. However, such diversification of the proteome by alternative splicing is much rarer in fungi. We describe here an ancient fungal alternative splicing event in which these two proteins are generated from a single alternatively spliced ancestral SKI7/HBS1 gene retained in many species in both the Ascomycota and Basidiomycota. While the ability to express two proteins from a single SKI7/HBS1 gene is conserved in many fungi, the exact mechanism by which they achieve this varies. The alternative splicing was lost in Saccharomyces cerevisiae following the whole-genome duplication event as these two genes subfunctionalized into the present functionally distinct HBS1 and SKI7 genes. When expressed in yeast, the single gene from Lachancea kluyveri generates two functionally distinct proteins. Expression of one of these proteins complements hbs1, but not ski7 mutations, while the other protein complements ski7, but not hbs1. This is the first known case of subfunctionalization by loss of alternative splicing in yeast. By coincidence, the ancestral alternatively spliced gene was also duplicated in Schizosaccharomyces pombe with subsequent subfunctionalization and loss of splicing. Similar subfunctionalization by loss of alternative splicing in fungi also explains the presence of two PTC7 genes in the budding yeast Tetrapisispora blattae, suggesting that this is a common mechanism to preserve duplicate alternatively spliced genes.
Introduction
Gene duplication is thought to be a major source of evolutionary innovation. Although the fate of duplicated genes is incompletely understood, it is thought to fit one of three patterns: nonfunctionalization, neofunctionalization or subfunctionalization. Of these nonfunctionalization is thought to be the most common. Immediately after duplication, duplicated genes are typically redundant. Thus, purifying selection cannot provide selective pressure to maintain both. The absence of selective pressure generally leads to loss of function mutations (nonfunctionalization) in one of the copies, followed by loss of that copy of the gene. In neofunctionalization, one of the duplicated copies acquires a new advantageous function that is different from the ancestral function maintained by the other copy [1]. Subfunctionalization can occur when an ancestral gene carries out more than one function. If one duplicated copy mutates so that it loses one of the functions, and the other copy mutates so that it loses a separate function, selective pressure can subsequently maintain both copies by selecting for both functions [2], [3]. Multiple functions in this context can mean being expressed in multiple cell types, encoding proteins localized to different compartments, encoding proteins with distinct biochemical activities, etc.
Saccharomyces cerevisiae is an excellent model organism to study the fate of duplicated gene pairs because an ancestor underwent a whole genome duplication (WGD) approximately 100 million years ago resulting in a transient increase in genome size from around 5000 protein coding genes to 10,000 [4], [5], [6]. Following genome duplication most duplicated genes were lost (nonfunctionalized), but 544 duplicated gene pairs that arose from WGD remain [4]. The genomes of many related species have been sequenced, which revealed through synteny which genes were duplicated as part of the WGD [6], [7], [8]. The related genomes also provide a large amount of sequence information on the duplicated genes and their non-duplicated homologs. The pattern of gene retention in these genomes revealed that nonfunctionalization after WGD is random such that different post-WGD species retained different subsets of duplicated genes [9]. In addition, gene function can be easily assayed in S. cerevisiae. Using these advantages, we have previously shown that subfunctionalization is a major mechanism by which duplicated S. cerevisiae genes were retained, which was confirmed by others [10], [11], .
One example of subfunctionalized genes resulting from the WGD event is provided by SKI7 and HBS1 [11]. The S. cerevisiae Ski7 and Hbs1 proteins both recognize ribosomes stalled during translation and initiate degradation of the mRNA. However, they recognize different stalled ribosomes and initiate mRNA degradation differently. When an mRNA lacks an in frame stop codon, the ribosome is thought to translate until it reaches the 3′ end of that mRNA [13]. The stalled ribosome is then recognized by Ski7, which recruits the RNA exosome to degrade that mRNA. In contrast, Hbs1 recognizes ribosomes stalled within the coding region, for example due to a structure or damage in the mRNA [14]. Recognition by Hbs1 causes cleavage of the mRNA in an RNA exosome-independent manner [14], [15]. Although the SKI7 and HBS1 genes of S. cerevisiae perform distinct functions, we have previously shown that the single ancestral gene performed both functions [11]. The related budding yeast Lachancea kluyveri diverged from S. cerevisiae before WGD and thus contains a single ortholog to SKI7 and HBS1, which we will call SKI7/HBS1. Our key finding was that when the L. kluyveri SKI7/HBS1 gene was introduced into S. cerevisiae it could complement the defects caused by both ski7Δ and hbs1Δ, thus indicating that this single gene carried out both functions [11].
Since the function of duplicated genes can diverge from each other through neo - or subfunctionalization, gene duplication may be one way to generate a more diverse proteome. The proteome can also be diversified through alternative splicing, where one gene generates multiple distinct mRNAs that each encode a distinct protein. Although alternative splicing is important to diversify the proteome in metazoans, it is much rarer in the fungal kingdom. Most fungal alternative splicing events that have been described are of the intron retention type, where the spliced mRNA encodes a functional protein, and the unspliced mRNA is nonfunctional. For example, transcriptome sequencing of Aspergillus oryzae identified only 8.6% of the genes as alternatively spliced, which is 10-fold lower than in humans and 92% of the Aspergillus alternative splicing was intron retention [16]. A well-studied and typical example of fungal intron retention is the S. cerevisiae CYH2 gene. The CYH2 mRNA encodes a 17 KDa ribosomal protein. The intron in the CYH2 pre-mRNA is retained approximately 50% of the time, which results in an mRNA that codes for a 2 KDa peptide with no known function. Furthermore, intron-retained mRNAs are typically very rapid degraded by the nonsense-mediated mRNA decay pathways [17]. In these cases instead of diversifying the proteome, intron retention may function to regulate gene expression. Similarly, the S. cerevisiae SRC1 gene is alternatively spliced using alternative 5′ splice sites, but only the longer splice isoform has been shown to be functional [18], [19]. To the best of our knowledge, the only case in which intron retention or alternative splicing leads to two functional mRNAs in S. cerevisiae is in PTC7, which contains one intron and encodes a protein phosphatase subunit. If this intron is spliced out, the mRNA is translated into a protein that is imported into the mitochondria, while after intron retention the mRNA is translated into a protein that is inserted into the nuclear envelope [20]. A few other cases have been described were fungi use alternative splicing to target a protein to multiple locations [21], [22], [23].
As mentioned above, there are multiple ways a gene can be multifunctional in the context of subfunctionalization. A corollary of that is that subfunctionalization can occur through distinct molecular changes. In yeast, subfunctionalization through changes in the coding region seem to be common [10], [11], [12]. In these cases a single amino acid change can be responsible for subfunctionalization [10], [12]. In multicellular organisms, genes that are expressed in multiple cell types, in response to multiple stimuli, or by multiple transcription factors can be subfunctionalized through changes in expression pattern [e.g. ref 3]. Subfunctionalization through changes in splicing patterns have been described in a few cases [e.g. refs 24], [25]. In these cases, an alternatively spliced gene upon duplication results in two genes where one gene follows one ancestral splicing pattern and the other follows another ancestral splicing pattern. However, the function of these alternative splicing isoforms is often not clear. Thus while loss of alternative splicing happens at the same time as some gene duplications, whether they cause subfunctionalization has not been experimentally demonstrated.
Here we show that the pre-WGD ancestor of SKI7 and HBS1 was alternatively spliced. We also show that in most extant fungi, including ascomycetes and basidiomycetes, the SKI7/HBS1 gene is still alternatively spliced, thereby describing the by far most conserved fungal alternative splicing event. The L. kluyveri alternative splicing isoforms are functionally distinct, such that one spliced mRNA encodes a functional Hbs1, while an alternatively spliced mRNA encodes a functional Ski7. Sequence analysis indicates that a very similar subfunctionalization occurred in an ancestor of the Schizosaccharomyces genus. Finally, while the S. cerevisiae PTC7 gene encodes two differently localized proteins through intron retention, in a related species this gene is replaced by a pair of duplicated genes that arose form WGD. Thus, evolution of a fungal ancestral alternatively spliced gene into two subfunctionalized genes occurred at least three times: twice for the SKI7/HBS1 gene, and once for PTC7. This further suggests that alternative splicing and gene duplication are not independent mechanisms to diversify the proteome, but instead are interrelated.
Results
The Lachancea kluyveri SKI7/HBS1 gene provides a rare example of producing two fungal proteins through alternative splicing
Several genomes of Saccharomycetaceae have been sequenced, but incompletely annotated. Upon careful analysis of these sequences we noticed that the SKI7/HBS1 genes in five pre-WGD Saccharomycetaceae each have a potential intron (Figure 1A). In contrast, S. cerevisiae and five other post-WGD species lack introns in both SKI7 and HBS1. Furthermore, each pre-WGD gene has two potential 3′ splice sites, resulting in the potential to encode two different conserved proteins. In the case of L. kluyveri these proteins are predicted to be 70 and 96 KDa (Figure 1A). Although alternative splicing is rare in fungi, we speculated that the SKI7/HBS1 gene may be alternatively spliced. We used rt-PCR to show that both predicted splice sites are indeed used. Use of the proximal 3′ splice site was confirmed using rt-PCR with a primer upstream of the 5′ splice site and a primer downstream of the proximal 3′ splice site and sequencing the resulting PCR product (Figure 1B left panel). Use of the distal 3′ splice site was similarly confirmed using a primer downstream of the distal 3′ splice site (Figure 1B right panel). Thus, the L. kluyveri gene is indeed alternatively spliced through the use of alternative 3′ splice sites.
Fig. 1. The SKI7/HBS1 gene in pre-whole-genome duplication Saccharomycetaceae encodes two different proteins. 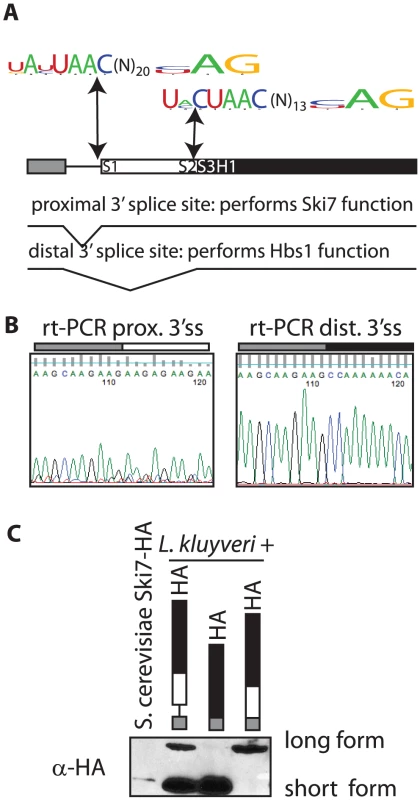
A: Shown is the structure of the L. kluyveri gene drawn to scale. Exon 1 is indicated as a grey box, the exon that is only included in the long splice isoform is indicated as a white box, and the sequence downstream of the distal 3′ splice site is indicated as a black box. The sequence logos depict alternative 3′ splice sites shared with five pre-WGD Saccharomycetaceae. B: Sequencing rtPCR products confirmed that both L. kluyveri SKI7/HBS1 3′ splice sites are used. C: Western blotting shows that translation of the two splice isoforms of L. kluyveri SKI7/HBS1 results in two distinct proteins. The first lane is a control S. cerevisiae strain with its SKI7 locus HA-tagged. The second, third and fourth lane contain plasmids with either an alternatively spliced L. kluyveri SKI7/HBS1 gene, or a modified gene with the intron removed as indicated. To determine whether both spliced mRNAs were used to generate a protein, we generated a plasmid that introduced the HA epitope at C-terminus of the L. kluyveri ORF. A western blot shows that two proteins of the expected size are indeed made in L. kluyveri (Figure 1C; second lane). We further modified the HA-tagged plasmid by deleting the intron. In one construct we deleted sequences between the 5′ and distal 3′ splice sites, such that only the short 70 KDa splice isoform could be expressed. Figure 1C (third lane) shows that the encoded protein comigrates precisely with the smaller of the two species seen when the intron is included. Conversely, in another plasmid (fourth lane) we deleted sequences between the 5′ and proximal 3′ splice sites and as expected only the large 96 KDa isoform was made. Therefore, the rt-PCR and Western blot data show that the single SKI7/HBS1 gene of L. kluyveri is used to generate two distinct proteins through use of alternative 3′ splice sites.
The two proteins encoded by L. kluyveri SKI7/HBS1 are functionally distinct
We have previously shown that the L. kluyveri SKI7/HBS1 can carry out both the Ski7 and Hbs1 functions by showing that the L. kluyveri gene can complement both a ski7Δ and an hbs1Δ in S. cerevisiae [11]. To test whether both L. kluyveri proteins were generated in this context, we introduced the same HA-tagged constructs describe above into a wild-type S. cerevisiae strain. Western blot analysis indicates that when the L. kluyveri SKI7/HBS1 gene is introduced into S. cerevisiae, both L. kluyveri proteins are made (data not shown). To determine whether one splice isoform carries out the Ski7 function and the other splice isoform carries out the Hbs1 function, plasmids expressing one or both proteins were introduced into both a dcp1-2 ski7Δ strain and an rps30aΔ hbs1Δ strain. A ski7Δ by itself does not result in a growth phenotype, but in combination with dcp1-2, results in a failure to grow at 30°C [26]. This ski7Δ phenotype can be complemented by the unmodified L. kluyveri gene (Figure 2A third row top panel) and by the long splice isoform (fourth row) but not by the short splice isoform (fifth row). Thus, only the long splice isoform can perform the Ski7 function. hbs1Δ by itself does not result in a growth phenotype but in combination with rps30aΔ results in slow growth at room temperature [27]. This hbs1Δ phenotype can be complemented by the unmodified L. kluyveri gene (Figure 2B third row bottom panel) and by the short splice isoform (5th row), but not by the long splice isoform (4th row). We conclude that alternative splicing generates two functionally distinct polypeptides, with the long splice isoform functioning similar to Ski7 and the short splice isoform similar to Hbs1.
Fig. 2. The two proteins encoded by L. kluyveri SKI7/HBS1 are functional. 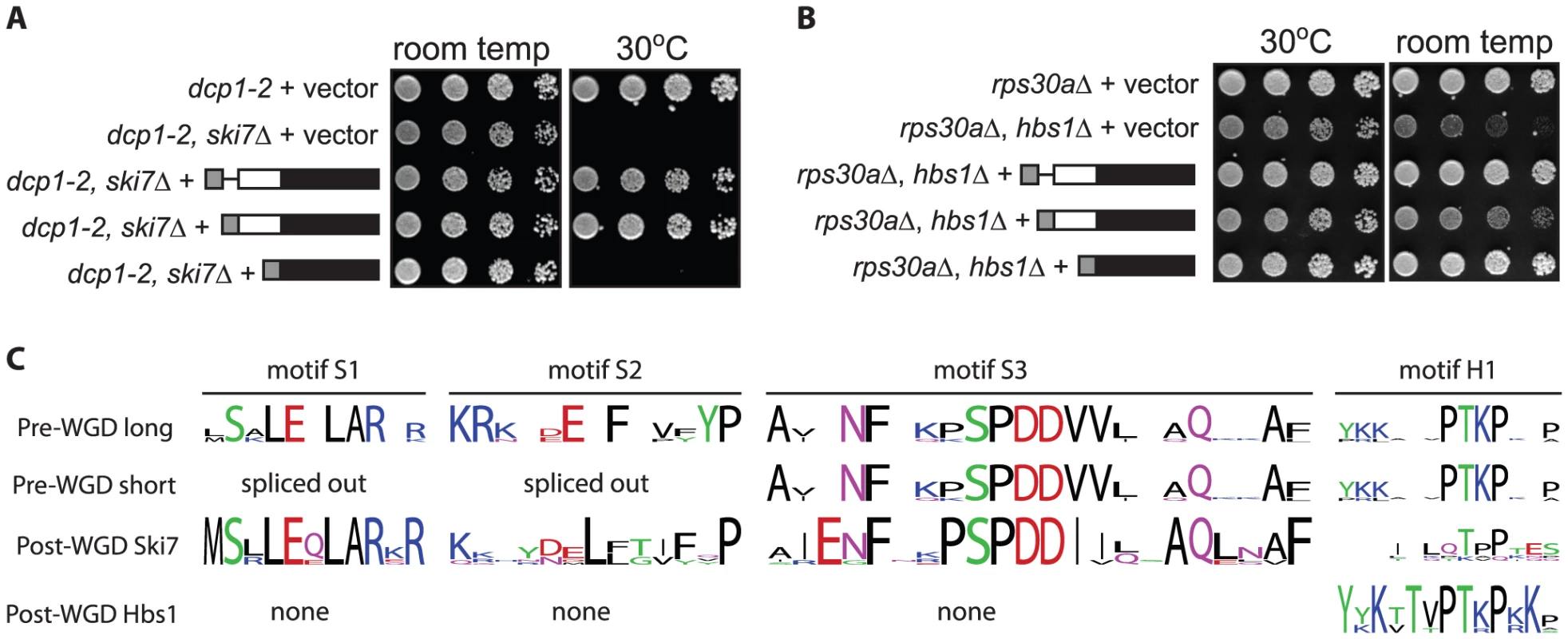
A: As shown before, L. kluyveri SKI7/HBS1 can complement both the temperature sensitive phenotype of dcp1-2 ski7Δ (compare rows 2 and 3). In contrast, a plasmid encoding only the long splice isoform complements ski7Δ (Row 4), but the short isoform does not. B: Conversely, the cold sensitive phenotype of rps30AΔ hbs1Δ is complemented by the short isoform, but not the long isoform C: The pre-WGD Saccharomycetaceae SKI7/HBS1 genes encode both sequences conserved in post-WGD SKI7 but not in post-WGD HBS1 (indicated as motifs S1, S2, and S3) and sequences conserved in post-WGD HBS1 (N-terminal domain encoded by exon 1 and a short sequence motif indicated as motif H1) but not in post-WGD SKI7 genes. Multiple sequence alignment (Figure S1) identified several sequence elements that correlated with Hbs1 function in short splice isoforms of pre-WGD Saccharomycetaceae SKI7/HBS1 genes and post-WGD HBS1 genes. The structure of S. cerevisiae Hbs1 has been solved, which shows a structured N-terminal domain and a C-terminal GTPase domain connected by a flexible linker [28], [29], [30]. The structured N-terminal domain is conserved in other post-WGD Hbs1 proteins but not in post-WGD Ski7. In the alternatively spliced pre-WGD homologs, this domain is encoded by exon 1, and thus present in both splice isoforms. The unstructured linker of Hbs1 is poorly conserved, with the exception of one sequence motif (Motif H1 in Figure 2C). A similar sequence motif is encoded in pre-WGD SKI7/HBS1 genes, but has diverged very much in post-WGD SKI7 genes. The GTPase domain is also highly conserved in post-WGD Hbs1 proteins and pre-WGD Ski7/Hbs1 proteins (Motifs G1 to G5 in Figure S1), consistent with previous findings that Hbs1 GTPase activity is important for its function [31], [32], [33]. In contrast, Ski7 has not been shown to be an active GTPase and the domain has diverged rapidly post-WGD. Specifically, a catalytically important His residue in motif G3 is changed to Ser, Asn or Asp in post-WGD Ski7s.
The structure of Ski7 has not been experimentally determined, but the N-terminus is known to be important for interaction with the RNA exosome and three other Ski proteins [34]. Multiple sequence alignment indicated that although the Ski7 N-terminus is generally poorly conserved, it contains three conserved sequence motifs (Figure 2C and Figure S1; motif S1, S2, and S3). Alignment of pre-WGD Ski7/Hbs1 sequences shows that these motifs are also conserved in the pre-WGD species. Motif S1 and S2 are encoded between the two alternative 3′ splice sites and thus are only present in the longer splice form. These observations suggest that the short isoform of pre-WGD SKI7/HBS1 genes may fail to carry out Ski7 function because they lack motifs S1 and S2.
Usage of alternative 3′ splice sites in SKI7/HBS1 is conserved in diverse ascomycetes
Most ascomycetes contain a single SKI7/HBS1 gene. To determine whether alternative splicing of SKI7/HBS1 was restricted to pre-WGD Saccharomycetaceae such as L. kluyveri or is a more ancient feature, we next looked at the more distantly related ascomycetes. The phylum Ascomycota can be divided in three subphyla, the Saccharomycotina (which includes Saccharomyces and Lachancea), the Pezizomycotina and the Taphrinomycotina. We therefore used the same rt-PCR and sequencing approach described above to analyze SKI7/HBS1 splicing in Aspergillus nidulans and Saitoella complicata, which are members of the Pezizomycotina and Taphrinomycotina, respectively. Figure 3A shows that Aspergillus nidulans also uses alternative 3′ splice sites in SKI7/HBS1 to generate two distinct mRNAs. The single A. nidulans SKI7/HBS1 gene contains four introns. The second intron contains two predicted alternative 3′ splice sites, and rt-PCR and sequencing indicated that both are used. Similarly, the single SKI7/HBS1 gene in Saitoella complicata contains seven introns, and the second intron contains two predicted 3′ splice sites. Figure 3B shows that both 3′ splice sites are used. Notably, the alternative spliced introns in Lachancea, Aspergillus and Saitoella are in the same position, just upstream of motif S3 characteristic of Ski7. Therefore, the capacity to use alternative 3′ splice sites in SKI7/HBS1 is conserved throughout the phylum Ascomycota.
Fig. 3. The use of alternative 3′ splice sites in SKI7/HBS1 is conserved in the two other subphyla of Ascomycota. 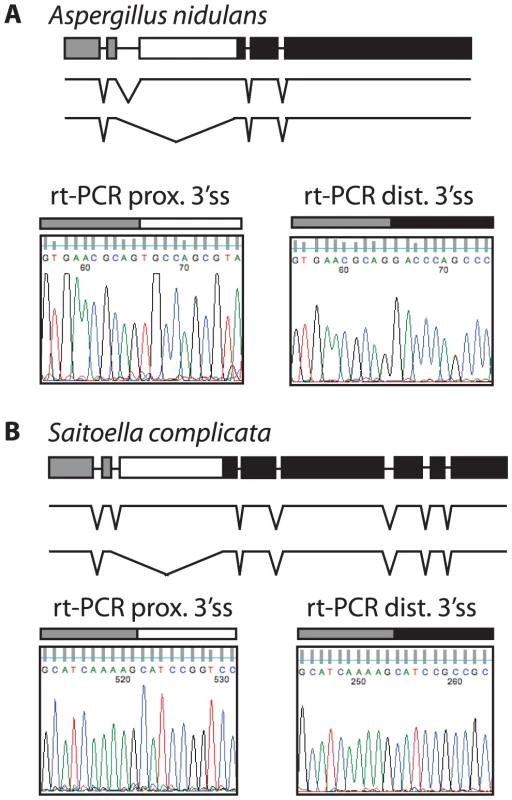
Shown are schematics of the SKI7/HBS1 gene of Aspergillus nidulans (A; a representative of the Pezizomycotina) and of Saitoella complicata (B; a representative of the Taphrinomycotina), along with parts of the sequencing results of rt-PCR products confirming use of alternative 3′ splice sites. As in Figure 1, the alternative exon is indicated as a white box, while the upstream and downstream exons are indicated as grey and black boxes, respectively. Some fungi use different mechanisms to express distinct mRNAs from a single SKI7/HBS1 gene
Although we were able to predict alternative 3′ splice sites in the single SKI7/HBS1 gene of many other fungi, we noted four notable differences in the Schizosaccharomyces genus, a subset of the CTG clade (Saccharomycetales that use CUG as a serine codon instead of the canonical leucine), and the basidiomycetes Cryptococcus neoformans and Ustilago maydis. Species within the Schizosaccharomyces genus have duplicated SKI7 and HBS1 genes (see next section), while species in the CTG clade and the two basidiomycetes each contain a single SKI7/HBS1 gene, but these genes lack obvious alternative 3′ splice sites.
The CTG clade can be divided into two smaller clades. One clade contains Candida guilliermondii, Debaryomyces hansenii, and Candida lusitaniae. In all three species there are two potential 3′ splice sites in locations similar to L. kluyveri (Figure S2). Thus, these three species appear to use the same mechanism as other ascomycetes to express two splice isoforms from a single SKI7/HBS1 gene. The other clade includes C. albicans, C. dubliniensis, C. tropicalis, and C. parapsilosis. These four Candida species also contain a predicted intron within their single HBS1/SKI7 gene, however we only detected one potential 3′ splice site, which corresponds to the distal 3′ splice site of other ascomycetes. rt-PCR and sequencing confirmed that this 3′ splice site is used to generate an mRNA that is equivalent to the short splice isoform of L. kluyveri (Figure 4A). Although a proximal 3′ splice site is absent in these species, the capacity to encode Ski7-like sequence upstream of the distal 3′ splice site is conserved in these four species. In all four species motif S1 starts with a methionine. Since motif S1 is at the extreme N-terminus of the protein in post-WGD SKI7 genes, we tested the hypothesis that the four Candida species generated a distinct mRNA that uses the AUG codon at the beginning of motif S1 as start codon. Figure 4A shows that 5′RACE indeed identified an mRNA with a 5′ end five nucleotides upstream of the conserved Ski7 motif S1. Thus, C. albicans uses alternative transcription start sites/first exons instead of alternative 3′ splice sites to generate two distinct mRNAs from the single SKI7/HBS1 gene.
Fig. 4. Some fungi use alternate methods to generate distinct SKI7 and HBS1 mRNAs from a single gene. 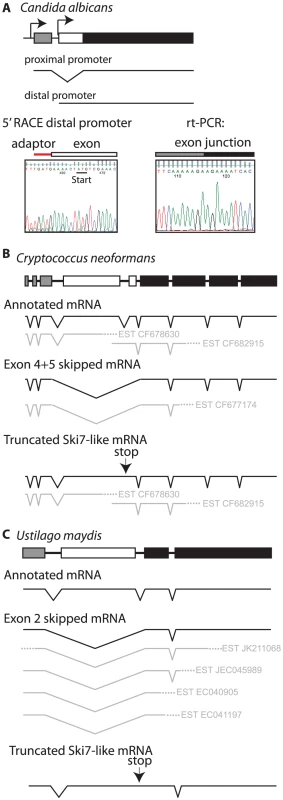
Diagrammed are the gene structure of Candida albicans (A), Cryptococcus neoformans (B), and Ustilago maydis (C). Sequencing of rtPCR and 5′ RACE products confirms that Candida albicans generates distinct mRNAs through the use of alternative promoters, while EST sequences below the gene diagram show that basidiomycetes generates distinct mRNAs through exon skipping. As in Figure 1, the alternative exon is indicated as a white box, while the upstream and downstream exons are indicated as grey and black boxes, respectively. The single SKI7/HBS1 gene in basidiomycetes also appears to be alternatively spliced, although the details differ from the ascomycete situation. The Cryptococcus neoformans and Ustilago maydis genes have 9 and 4 annotated exons, respectively. Several EST sequences indicate that exons 4 and 5 in C. neoformans and exon 2 in U. maydis are skipped to generate an Hbs1-like protein (Figure 4B and 4C). Existing annotations suggest that a Ski7-like protein can be encoded by inclusion of these exons. However, EST, RNA sequencing and protein sequence similarity suggests an alternative where the annotated intron 4 of C. neoformans (and intron 2 of U. maydis) is not a true intron (See Text S1). This alternative mechanism encodes a truncated protein that resembles the N-terminus of Ski7, but is missing the GTPase domain (Figure 4B and 4C). This mechanism is strikingly similar to potential alternative splicing of the metazoan homolog (See Text S1 and Figure S5). Overall, while it is clear that the basidiomycetes use alternative splicing of their single SKI7/HBS1 gene, it is not entirely clear which mechanism they use to generate a Ski7-like protein.
Independent SKI7/HBS1 duplication and loss of alternative splicing in the Schizosaccharomyces lineage
The fourth exception to conserved alternative 3′ splice sites in SKI7/HBS1 occurs in the Schizosaccharomyces genus. The Hbs1 protein of Schizosaccharomyces pombe has been previously studied and appears to function similarly to S. cerevisiae Hbs1 [29]. In addition we found an uncharacterized paralog in S. pombe (Systematic name SPAP8A3.05) that encodes amino acid sequence motifs characteristic of Ski7p (labeled S1, S1′ and S3 in Figure S3). We therefore refer to this S. pombe gene as SKI7. Thus, an ancestor to S. pombe must have independently duplicated its SKI7/HBS1 gene. The other three Schizosaccharomyces species with sequenced genomes each contain one clear ortholog of HBS1 and one clear ortholog of SKI7 (Figure S3), which suggests this duplication occurred before the Schizosaccharomyces species diverged from each other. Of the sequenced fungal genomes, the most closely related species with a single SKI7/HBS1 gene is S. complicata. As discussed above, S. complicata has an alternatively spliced SKI7/HBS1 gene, and thus the duplication in the Schizosaccharomyces genus appears to have occurred after it diverged from S. complicata, but before the Schizosaccharomyces species diverged from each other.
PTC7 provides a second example of post-WGD duplicate retention and loss of splice-isoforms
Our above observations indicate that S. cerevisiae SKI7 and HBS1 evolved from a single alternatively spliced ancestral gene and that they correspond to the different splice isoforms of the pre-WGD ancestor. This conclusion suggests that a similar mechanism may apply to other alternatively spliced genes. The only S. cerevisiae gene known to use splicing to generate two different functional proteins is PTC7 [20]. The PTC7 gene contains an intron that can either be spliced out or retained. Both the spliced and unspliced mRNAs encode type 2C protein phosphatases (PP2C) [20]. It has previously been noted that an intron of 3n nucleotides without any in frame stop codons is conserved in the PTC7 gene of twelve species within the Saccharomycetaceae, both pre - and post-WGD ([20]; Figure 5 and Figure S4). We searched for PTC7 genes in additional yeast genomes and noticed that the only species within the Saccharomycetaceae that did not follow this pattern is Tetrapisispora blattae. The genome of this species contains two PTC7 genes (which we will call PTC7a and PTC7b). The synteny pattern (http://wolfe.gen.tcd.ie/ygob/) indicates that after WGD the T. blattae lineage maintained both copies of PTC7, while one copy was lost in the S. cerevisiae lineage. We searched for potential introns in PTC7a and PTC7b, but failed to find one in the PTC7a gene, while PTC7b contains a 103 nucleotide intron. The spliced PTC7b mRNA is predicted to encode a functional protein. In contrast to other post-WGD species, translation of the PTC7b unspliced mRNA does not encode a functional PP2C: translation starting from the normal start codon would end after 20 amino acids at a stop codon within the intron, while the only other in frame AUG codon is only 8 amino acid upstream of the normal stop codon. Thus, unlike other Saccharomycetaceae that encode two proteins from one alternatively spliced PTC7 gene, the T. blattae PTC7a and PTC7b genes each can only encode a single protein.
Fig. 5. The PTC7 gene is subfunctionalized through loss of alternative splicing in Tetrapisispora blattae. 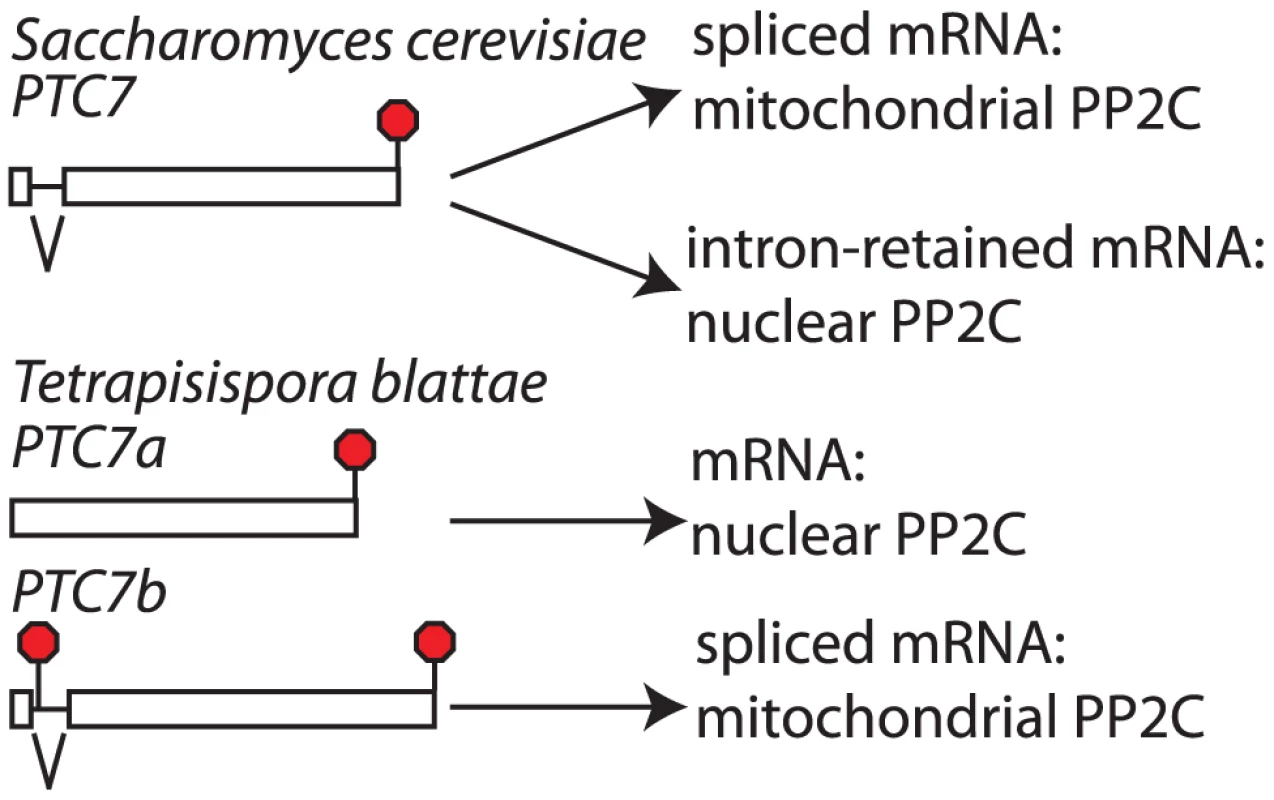
In Saccharomyces cerevisiae and most other Saccharomycetaceae the PTC7 gene produces a nuclear envelope associated protein phosphatase through translation of the pre-mRNA and a mitochondrial protein phosphatase through translation of the spliced mRNA. In the post-WGD species T. blattae, both duplicated copies were maintained and one encodes a predicted nuclear envelope associated protein phosphatase, while the other encodes a predicted mitochondrial protein phosphatase. The two splice isoforms of S. cerevisiae PTC7 are targeted to different compartments. The spliced S. cerevisiae PTC7 mRNA encodes a protein that is localized to the mitochondria, while the intron-retained mRNA encodes a protein localized to the nuclear envelope. Targeting to the nuclear envelope has been attributed to a predicted trans-membrane helix (TM) that is encoded by the retained intron [20]. We used the TMHMM 2.0 server (http://www.cbs.dtu.dk/services/TMHMM/) to predict TMs in Ptc7 proteins of various Saccharomycetaceae. Each of the intron-retained mRNAs from post-WGD species and the T. blattae PTC7a gene encodes a single predicted TM near the N-terminus, suggesting that all of these proteins are targeted to the nuclear envelope. In contrast, none of the spliced mRNAs or PTC7b encode a predicted TM. We also used the PSORT II server (http://psort.hgc.jp/form2.html) to predict TMs and protein localization. The TM results agreed with the TMHMM server. In addition, all of the spliced isoforms and PTC7b were predicted to contain a mitochondrial targeting sequence that was absent from the unspliced isoforms. Thus, T. blattae PTC7a encodes a single PP2C that is predicted to be targeted to the nuclear envelope, like the protein encoded by intron-retained PTC7 mRNA in S. cerevisiae, while T. blattae PTC7b gene appears to encode a single PP2C that is predicted to be targeted to the mitochondria, like the protein encoded by spliced PTC7 mRNA in S. cerevisiae. While separate functions of the S. cerevisiae PTC7 splice isoforms have not been defined, these results strongly suggests that the PTC7a and PTC7b genes are subfunctionalized, and thus that subfunctionalization by loss of splicing isoforms is not restricted to SKI7/HBS1.
Discussion
Conserved alternative splicing of fungal SKI7/HBS1 genes
We describe alternative splicing in fungal SKI7/HBS1 genes that is unusual in two respects. First, unlike in Metazoa, most fungal alternative splicing events do not produce two different proteins, but instead either have no known function or function to quantitatively regulate gene expression. Both of the mRNAs that are produced through SKI7/HBS1 alternative splicing are predicted to encode functional proteins, western blot analysis indicates that both predicted proteins are produced, and complementation of S. cerevisiae mutants shows that the two proteins are functionally distinct. Second, most alternative splicing events that have been described in fungi are not widely conserved but instead have only been described in one species [MDH1, Ref. 22], genus [GND1, Ref. 21] or family [PTC7 Ref. 20]. Besides SKI7/HBS1, the most conserved fungal alternative splicing events were recently reported for PGK1 in the Ascomycota and GAPDH in the Basidiomycota [23]. In contrast, alternative splicing of SKI7/HBS1 most likely arose before the divergence of the Ascomycota from the Basidiomycota, and it might even have arisen before fungi and animals diverged (Text S1 and Figure S5). Thus, this alternative splicing event has been maintained for at least 500 million years. It has been suggested that the Ski7 function is peculiar to S. cerevisiae and close relatives [6], [35]. The finding of ancient alternative splicing indicates that Ski7 function is much older than appreciated and suggests that the ability to produce both Hbs1 and Ski7 is very important to fungi. Interestingly, one of the reasons why conserved alternative splicing in fungi has not been previously reported is that S. cerevisiae and S. pombe have been chosen somewhat arbitrarily as model fungi, and in both of these species the alternatively spliced SKI7/HBS1 gene has been replaced with duplicate genes.
Although alternative splicing of SKI7/HBS1 is conserved in diverse fungi, we have characterized changes in expression strategies for Ski7 and Hbs1, which are summarized in Figure 6A. The common ancestor of the ascomycetes and basidiomycetes appears to have had an alternatively spliced SKI7/HBS1 gene. Although the exact nature of SKI7/HBS1 alternative splicing event in basidiomycetes remains to be determined, it is clear that the mechanism by which a Ski7-like protein is expressed is different. Fully characterizing this event will require additional data from the basidiomycetes and/or additional early branching fungi. Independent duplications in the Saccharomyces and Schizosaccharomyces lineages allowed loss of alternative splicing (Figure 6 events 2). In the Saccharomyces lineage this duplication was part of a WGD, but in the Schizosaccharomyces lineage this duplication appears to be restricted to a single gene. The fourth evolutionary change occurred in the Candida clade in which a single SKI7/HBS1 gene gained an alternative initiation codon for Ski7 (Figure 6 event 3). Interestingly, after duplication, the S. cerevisiae, and S. pombe SKI7 genes also appear to have gained a new initiation codon (see below).
Fig. 6. Changes in SKI7, HBS1, and PTC7 gene structure during fungal diversification. 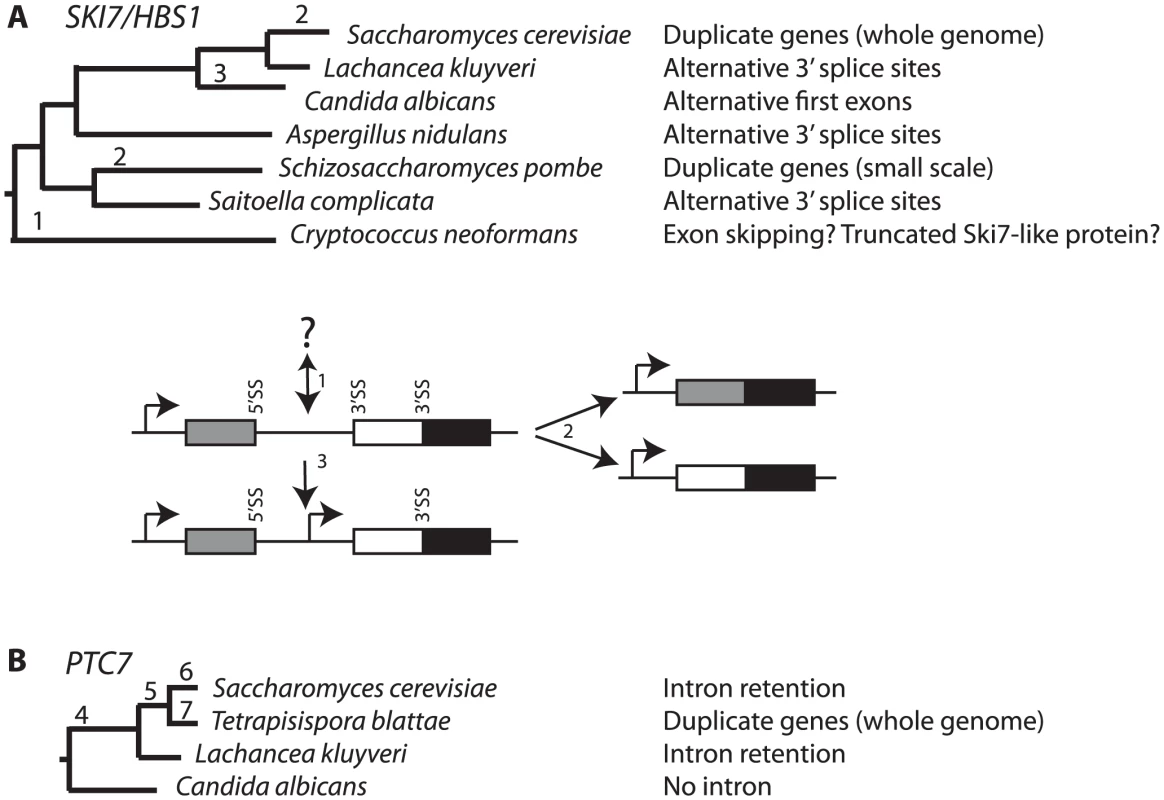
SKI7, HBS1 (A) and PTC7 (B). See discussion for details. As in Figure 1, the alternative exon is indicated as a white box, while the upstream and downstream exons are indicated as grey and black boxes, respectively. Parallel precise post-duplication loss of the HBS1 intron in budding and fission yeasts
A major mechanism for intron loss in S. cerevisiae involves a transposon-encoded reverse transcriptase that converts spliced mRNA into cDNA. This cDNA then recombines with the gene, resulting in precise deletion of the intron [36]. Multiple sequence alignment shows that the intron in post-WGD Saccharomycetaceae is precisely deleted (Figure S1), consistent with it being deleted by this mechanism. Similarly, the short isoform from Saitoella complicata aligns very well with Hbs1 sequences from four Schizosaccharomyces species, indicating that the alternatively spliced intron was precisely deleted (Figure S3). The Saitoella SKI7/HBS1 gene contains seven introns. Recombination with a cDNA preferentially deletes introns near the 3′ end of the gene while introns near the 5′ end are more likely to be retained [36]. Consistent with intron loss by recombination with cDNA, HBS1 genes from all four sequenced Schizosaccharomyces species contain two introns that correspond to the first two S. complicata introns. Thus, the alternatively spliced intron was precisely deleted in both the Saccharomyces and Schizosaccharomyces lineage, possibly by reverse transcription of the mRNA into cDNA and recombination.
Parallel post-duplication loss of SKI7 N-terminal exons and gain of a novel initiation codon in budding and fission yeasts
In contrast to HBS1, the intron in post-duplication SKI7 genes was not precisely deleted. Multiple sequence alignment (Figure S1) of post-duplication Saccharomycetaceae showed that although the Ski7 N-terminus is generally poorly conserved, it contains three conserved sequence motifs (motif S1, S2, and S3). In post-WGD SKI7 genes in both the Saccharomycetaceae and in Schizosaccharomyces, motif S1 is located at the extreme N-terminus of Ski7, starting with the Met translated from the start codon. This is most consistent with the model that after duplication S. cerevisiae SKI7 lost exon 1 and gained a new initiation codon. Similarly, Schizosaccharomyces Ski7 appears to have lost exons 1 to 4, and gained a new initiation codon. This deletion of both the SKI7 intron and the first exon(s) in Saccharomyces and Schizosaccharomyces is inconsistent with simple recombination with a cDNA.
It has previously been noted that a significant number of the genes that were duplicated and retained in S. cerevisiae after WGD are also duplicated in S. pombe [37], suggesting parallel subfunctionalization events in the two species. Our observations provide striking similarities of the evolution of the Hbs1 protein in Saccharomycetaceae and Schizosaccharomyces and of the Ski7 protein in these same clades and in Candida albicans. Thus, after independent duplication in these lineages, a similar sequence of changes occurred, intron deletion through recombination (HBS1) or generation of an alternative start site (SKI7).
Generation of two Ptc7 proteins through splicing or gene duplication in Saccharomycetaceae
The only known example of a S. cerevisiae gene encoding functional proteins from both unspliced and spliced mRNA is PTC7. This capacity to encode two proteins is conserved in most Saccharomycetaceae, but not in other Saccharomycetales (including Candida, Pichia and Yarrowia species) [20]. The time of divergence of the Saccharomycetaceae has not been carefully defined, but estimates indicate that it preceded divergence of mice from humans 75 million year ago. Strikingly, only about 30% of alternative splicing events are conserved from mice to human. Therefore, although alternative splicing of PTC7 is not nearly as well conserved as that of SKI7/HBS1, it still has been conserved for a longer period than many human alternative splicing events.
Since PTC7 homologs outside the Saccharomycetaceae lack an intron in the same position, the alternatively spliced PTC7 intron appears to have been gained by an ancestor of the Saccharomycetaceae (Figure 6B event 4). The PSORT server (http://psort.hgc.jp/form2.html) predicts that Ptc7 proteins outside the Saccharomycetaceae (i.e. from Candida, Pichia, and Yarrowia species) localize to the mitochondria. Thus, the gain of an intron and the alternative splicing of this intron provides the Saccharomycetaceae with a PP2C localized to the nuclear envelope. After WGD (Figure 6 event 5), one copy of PTC7 was lost in the Saccharomyces lineage and the remaining copy maintained the capacity to encode two proteins (Figure 6 event 6). In contrast, in T. blattae both duplicated PTC7 genes were maintained, but each lost the ability to encode two PP2C splice isoforms (Figure 6 event 7) and subfunctionalized into one gene for a mitochondrial PP2C and one gene for a PP2C in the nuclear envelope.
Subfunctionalization of duplicated genes by loss of alternative splicing
Our combined bioinformatic and experimental analysis shows that alternative splicing and gene duplication may be interrelated events in a cycle that diversifies the proteome. In this cycle, gain of alternative splicing, duplication, and loss of alternative splicing and subfunctionalization result in functionally distinct paralogs. Although there have been some previous descriptions of subfunctionalization by loss of alternative splicing [e.g. ref 24], our findings extend these descriptions in three important ways. First, previous descriptions are generally limited to two closely related species and thus cover only part of the evolutionary history of the gene. The ever-increasing number of sequenced fungal genomes allowed us to analyze SKI7/HBS1 and PTC7 gene structure and expression in diverse fungi thereby identifying when alternative splicing arose and was lost. The whole cycle of gain of an alternative splicing event, duplication, and loss of alternative splicing can be observed in the PTC7 gene of Saccharomycetaceae. In contrast, although we have not been able to identify when alternative splicing of SKI7/HBS1 arose, we have described independent subfunctionalization events by loss of alternative splicing in the Schizosaccharomyces and Saccharomyces lineages.
Second, our observations suggest that subfunctionalization by loss of alternative splicing occurred very similarly in the Saccharomyces and Schizosaccharomyces lineages. Thus, unlike previously described isolated examples this phenomenon appears to have occurred multiple times.
Third, in most previously described cases of loss of alternative splicing in duplicated genes it was not clear whether the splicing isoforms have distinct functions, and thus it is not clear in those cases that subfunctionalization and loss of alternative splicing are causally linked. Similarly, lack of one PTC7 splice isoform does not cause an easily identifiable phenotype under lab conditions, making it impossible to test whether the duplicate T. blattae genes can substitute for one but not the other splice isoform. In contrast, SKI7 and HBS1 have well-described functions, allowing us to demonstrate that the splice isoforms of L. kluyveri SKI7/HBS1 are functionally distinct.
Materials and Methods
Strains
The S. cerevisiae, C. albicans, and wild-type L. kluyveri strains have been described [11], [38]. The L. kluyveri ura3 mutant strain FM628 was a kind gift of Mark Johnston. The S. complicata type strain Y-17804 was obtained from the USDA ARS culture collection.
Identification of a single SKI7/HBS1 gene of S. complicata and closing a gap in the draft genome sequence
A draft sequence of the S. complicata genome is available [39]. BLAST analysis using the S. pombe Hbs1 identified two non-overlapping contigs that encoded N - and C-terminal parts of a S. complicata homolog. Extensive BLAST analysis with other queries did not reveal additional homologs. We hypothesized that these contigs represented different parts of the same gene. We used PCR to close the gap between the contigs and sequenced the PCR product directly. The assembled sequence of the S. complicata SKI7/HBS1 gene has been submitted to Genbank (Accession number JQ928880). Since exons proved difficult to predict due to their small size, we sequenced rt-PCR products to determine the gene structure depicted in Figure 3B, which was then used to align the encoded protein with Schizosaccharomyces homologs.
RNA isolation
L. kluyveri and S. complicata were grown in YPD and RNA was extracted using our standard method for S. cerevisiae. C. albicans growth and RNA extraction was performed as described [40]. Aspergillus RNA isolated from strain R21 was a kind gift from Taylor Schoberle and Greg May (UT MD Anderson Cancer Center). When contamination with genomic DNA proved to be a problem, we treated the RNA with DNase (Promega).
PCR, rt-PCR, and 5′RACE
rt-PCR was done using a commercial kit per manufacturer's instructions (Sigma-Aldrich). To close the gap between the two S. complicata contigs, we used the same kit but omitted the reverse transcriptase to amplify genomic DNA. 5′ RLM-RACE was done using a commercial kit per manufacturer's instructions (Invitrogen). All PCR, rt-PCR and RACE products were sequenced directly (Genewiz) and exactly confirmed the predicted splice sites.
Plasmids
pAv231 has been described [11]. It contains the L. kluyveri SKI7/HBS1 gene, including the promoter, intron and 3′UTR sequences. pAv844 and pAv847 have the long form and short form of the intron removed, respectively. They were generated by overlap PCR using the forward overlap oligonucleotides oAv963 (tgctcaaccaaagcaagaag aagagaagaaattatctaaactgg) for pAv844 and oAv965 (tgctcaaccaaagcaagaag ccaaaaaacaagctatctctaatttc) for pAv847 and their reverse complements oAv964 and oAv966. To generate the HA-tagged SKI7/HBS1 gene, a plasmid encoding the C-terminus and triple HA tag was chemically synthesized (by Genewiz). This fragment was used to replace the Bcl I to Bgl II restriction enzyme fragment of pAv231, generating pAv888, which contains the entire L. kluyveri SKI7/HBS1 gene with a C-terminal triple HA tag. A Bam HI Xba I fragment of pAv888 was then used to replace the Bam HI Xba I fragment of pAv844 and pAv846 to generate pAv903 and pAv905. Thus, pAv903 encodes a C-terminally HA-tagged version of the long Ski7-like isoform of L. kluyveri SKI7/HBS1, while pAv905 encodes the tagged short Hbs1-like isoform.
Western blot analysis
Plasmids carrying the HA-tagged L. kluyveri SKI7/HBS1 gene, or empty vector controls, were transformed into S. cerevisiae or L. kluyveri strains using a standard method [41]. Transformants were selected on SC-URA, and then grown overnight in SC-URA. Total protein was isolated using the glass bead method and analyzed by western blotting using anti-HA antibodies (Roche). As a control for the western blotting we used a S. cerevisiae strain with the HA epitope integrated at the C-terminus of the endogenous SKI7 locus.
Sequence analysis
Fungal SKI7 and HBS1 homologs were initially identified by BLAST in the sequenced genomes and the predicted proteomes from 14 Saccharomycetaceae, 10 species from the CTG clade, 4 Pezizomycotina, 5 Taphrinomycotina and 2 basidiomycetes. In none of the cases was the gene annotated as alternatively spliced, and in a number of cases introns were not annotated or incorrectly annotated and were corrected based on our rt-PCR analysis. Multiple sequence alignments of various subsets of protein sequences were generated with the help of the ClustalW (http://www.ch.embnet.org/software/ClustalW.html) BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html), and WebLogo (weblogo.berkeley.edu/) servers. The species trees in Figure 6 and Figures S2 and S3 are adapted from [42], [43], and [44].
Supporting Information
Zdroje
1. Ohno S (1970) Evolution by gene duplication: Springer-Verlag.
2. HughesAL (1994) The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond B Biol Sci 256 : 119–124.
3. ForceA, LynchM, PickettFB, AmoresA, YanYL, et al. (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 : 1531–1545.
4. GordonJL, ByrneKP, WolfeKH (2009) Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet 5: e1000485 doi:10.1371/journal.pgen.1000485
5. OhEigeartaighSS, ArmisenD, ByrneKP, WolfeKH (2011) Systematic discovery of unannotated genes in 11 yeast species using a database of orthologous genomic segments. BMC genomics 12 : 377.
6. KellisM, BirrenBW, LanderES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428 : 617–624.
7. ByrneKP, WolfeKH (2005) The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome research 15 : 1456–1461.
8. Proux-WeraE, ArmisenD, ByrneKP, WolfeKH (2012) A pipeline for automated annotation of yeast genome sequences by a conserved-synteny approach. BMC bioinformatics 13 : 237.
9. ScannellDR, FrankAC, ConantGC, ByrneKP, WoolfitM, et al. (2007) Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proceedings of the National Academy of Sciences of the United States of America 104 : 8397–8402.
10. FroydCA, RuscheLN (2011) The duplicated deacetylases Sir2 and Hst1 subfunctionalized by acquiring complementary inactivating mutations. Molecular and cellular biology 31 : 3351–3365.
11. van HoofA (2005) Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics 171 : 1455–1461.
12. FinniganGC, Hanson-SmithV, StevensTH, ThorntonJW (2012) Evolution of increased complexity in a molecular machine. Nature 481 : 360–364.
13. van HoofA, FrischmeyerPA, DietzHC, ParkerR (2002) Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295 : 2262–2264.
14. DomaMK, ParkerR (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440 : 561–564.
15. SchaefferD, van HoofA (2011) Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc Natl Acad Sci U S A 108 : 2366–2371.
16. WangB, GuoG, WangC, LinY, WangX, et al. (2010) Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic acids research 38 : 5075–5087.
17. HeF, PeltzSW, DonahueJL, RosbashM, JacobsonA (1993) Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1 - mutant. Proc Natl Acad Sci U S A 90 : 7034–7038.
18. GrundSE, FischerT, CabalGG, AntunezO, Perez-OrtinJE, et al. (2008) The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. The Journal of cell biology 182 : 897–910.
19. Rodriguez-NavarroS, IgualJC, Perez-OrtinJE (2002) SRC1: an intron-containing yeast gene involved in sister chromatid segregation. Yeast 19 : 43–54.
20. JuneauK, NislowC, DavisRW (2009) Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics 183 : 185–194.
21. StrijbisK, van den BurgJ, VisserWF, van den BergM, DistelB (2012) Alternative splicing directs dual localization of Candida albicans 6-phosphogluconate dehydrogenase to cytosol and peroxisomes. FEMS yeast research 12 : 61–68.
22. KabranP, RossignolT, GaillardinC, NicaudJM, NeuvegliseC (2012) Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA research : an international journal for rapid publication of reports on genes and genomes 19 : 231–244.
23. FreitagJ, AstJ, BolkerM (2012) Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485 : 522–525.
24. AltschmiedJ, DelfgaauwJ, WildeB, DuschlJ, BouneauL, et al. (2002) Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics 161 : 259–267.
25. CusackBP, WolfeKH (2007) When gene marriages don't work out: divorce by subfunctionalization. Trends in genetics : TIG 23 : 270–272.
26. van HoofA, StaplesRR, BakerRE, ParkerR (2000) Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol Cell Biol 20 : 8230–8243.
27. Carr-SchmidA, PfundC, CraigEA, KinzyTG (2002) Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol Cell Biol 22 : 2564–2574.
28. BeckerT, ArmacheJP, JaraschA, AngerAM, VillaE, et al. (2011) Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nature structural & molecular biology 18 : 715–720.
29. ChenL, MuhlradD, HauryliukV, ChengZ, LimMK, et al. (2010) Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nature structural & molecular biology 17 : 1233–1240.
30. van den ElzenAM, HenriJ, LazarN, GasME, DurandD, et al. (2010) Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nature structural & molecular biology 17 : 1446–1452.
31. ShoemakerCJ, EylerDE, GreenR (2010) Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330 : 369–372.
32. PisarevaVP, SkabkinMA, HellenCU, PestovaTV, PisarevAV (2011) Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. The EMBO journal 30 : 1804–1817.
33. ShoemakerCJ, GreenR (2011) Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proceedings of the National Academy of Sciences of the United States of America 108: E1392–1398.
34. ArakiY, TakahashiS, KobayashiT, KajihoH, HoshinoS, et al. (2001) Ski7p G protein interacts with the exosome and the Ski complex for 3′ - to-5′ mRNA decay in yeast. EMBO J 20 : 4684–4693.
35. AtkinsonGC, BaldaufSL, HauryliukV (2008) Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC evolutionary biology 8 : 290.
36. FinkGR (1987) Pseudogenes in yeast? Cell 49 : 5–6.
37. HughesAL, FriedmanR (2003) Parallel evolution by gene duplication in the genomes of two unicellular fungi. Genome research 13 : 794–799.
38. FonziWA, IrwinMY (1993) Isogenic strain construction and gene mapping in Candida albicans. Genetics 134 : 717–728.
39. NishidaH, HamamotoM, SugiyamaJ (2011) Draft genome sequencing of the enigmatic yeast Saitoella complicata. The Journal of general and applied microbiology 57 : 243–246.
40. RamirezMA, LorenzMC (2009) The transcription factor homolog CTF1 regulates {beta}-oxidation in Candida albicans. Eukaryotic cell 8 : 1604–1614.
41. GietzRD, WoodsRA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350 : 87–96.
42. LiuY, LeighJW, BrinkmannH, CushionMT, Rodriguez-EzpeletaN, et al. (2009) Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Molecular biology and evolution 26 : 27–34.
43. ButlerG, RasmussenMD, LinMF, SantosMA, SakthikumarS, et al. (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459 : 657–662.
44. RhindN, ChenZ, YassourM, ThompsonDA, HaasBJ, et al. (2011) Comparative functional genomics of the fission yeasts. Science 332 : 930–936.
Štítky
Genetika Reprodukčná medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



