-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
Common genetic variants contribute to the observed variation in breast cancer risk for BRCA2 mutation carriers; those known to date have all been found through population-based genome-wide association studies (GWAS). To comprehensively identify breast cancer risk modifying loci for BRCA2 mutation carriers, we conducted a deep replication of an ongoing GWAS discovery study. Using the ranked P-values of the breast cancer associations with the imputed genotype of 1.4 M SNPs, 19,029 SNPs were selected and designed for inclusion on a custom Illumina array that included a total of 211,155 SNPs as part of a multi-consortial project. DNA samples from 3,881 breast cancer affected and 4,330 unaffected BRCA2 mutation carriers from 47 studies belonging to the Consortium of Investigators of Modifiers of BRCA1/2 were genotyped and available for analysis. We replicated previously reported breast cancer susceptibility alleles in these BRCA2 mutation carriers and for several regions (including FGFR2, MAP3K1, CDKN2A/B, and PTHLH) identified SNPs that have stronger evidence of association than those previously published. We also identified a novel susceptibility allele at 6p24 that was inversely associated with risk in BRCA2 mutation carriers (rs9348512; per allele HR = 0.85, 95% CI 0.80–0.90, P = 3.9×10−8). This SNP was not associated with breast cancer risk either in the general population or in BRCA1 mutation carriers. The locus lies within a region containing TFAP2A, which encodes a transcriptional activation protein that interacts with several tumor suppressor genes. This report identifies the first breast cancer risk locus specific to a BRCA2 mutation background. This comprehensive update of novel and previously reported breast cancer susceptibility loci contributes to the establishment of a panel of SNPs that modify breast cancer risk in BRCA2 mutation carriers. This panel may have clinical utility for women with BRCA2 mutations weighing options for medical prevention of breast cancer.
Published in the journal: Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk. PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003173
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003173Summary
Common genetic variants contribute to the observed variation in breast cancer risk for BRCA2 mutation carriers; those known to date have all been found through population-based genome-wide association studies (GWAS). To comprehensively identify breast cancer risk modifying loci for BRCA2 mutation carriers, we conducted a deep replication of an ongoing GWAS discovery study. Using the ranked P-values of the breast cancer associations with the imputed genotype of 1.4 M SNPs, 19,029 SNPs were selected and designed for inclusion on a custom Illumina array that included a total of 211,155 SNPs as part of a multi-consortial project. DNA samples from 3,881 breast cancer affected and 4,330 unaffected BRCA2 mutation carriers from 47 studies belonging to the Consortium of Investigators of Modifiers of BRCA1/2 were genotyped and available for analysis. We replicated previously reported breast cancer susceptibility alleles in these BRCA2 mutation carriers and for several regions (including FGFR2, MAP3K1, CDKN2A/B, and PTHLH) identified SNPs that have stronger evidence of association than those previously published. We also identified a novel susceptibility allele at 6p24 that was inversely associated with risk in BRCA2 mutation carriers (rs9348512; per allele HR = 0.85, 95% CI 0.80–0.90, P = 3.9×10−8). This SNP was not associated with breast cancer risk either in the general population or in BRCA1 mutation carriers. The locus lies within a region containing TFAP2A, which encodes a transcriptional activation protein that interacts with several tumor suppressor genes. This report identifies the first breast cancer risk locus specific to a BRCA2 mutation background. This comprehensive update of novel and previously reported breast cancer susceptibility loci contributes to the establishment of a panel of SNPs that modify breast cancer risk in BRCA2 mutation carriers. This panel may have clinical utility for women with BRCA2 mutations weighing options for medical prevention of breast cancer.
Introduction
The lifetime risk of breast cancer associated with carrying a BRCA2 mutation varies from 40 to 84% [1]. To determine whether common genetic variants modify breast cancer risk for BRCA2 mutation carriers, we previously conducted a GWAS of BRCA2 mutation carriers from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) [2]. Using the Affymetrix 6.0 platform, the discovery stage results were based on 899 young (<40 years) affected and 804 unaffected carriers of European ancestry. In a rapid replication stage wherein 85 discovery stage SNPs with the smallest P-values were genotyped in 2,486 additional BRCA2 mutation carriers, only published loci associated with breast cancer risk in the general population, including FGFR2 (10q26; rs2981575; P = 1.2×10−8), were associated with breast cancer risk at the genome-wide significance level among BRCA2 mutation carriers. Two other loci, in ZNF365 (rs16917302) on 10q21 and a locus on 20q13 (rs311499), were also associated with breast cancer risk in BRCA2 mutation carriers with P-values<10−4 (P = 3.8×10−5 and 6.6×10−5, respectively). A nearby SNP in ZNF365 was also associated with breast cancer risk in a study of unselected cases [3] and in a study of mammographic density [4]. Additional follow-up replicated the findings for rs16917302, but not rs311499 [5] in a larger set of BRCA2 mutation carriers. To seek additional breast cancer risk modifying loci for BRCA2 mutation carriers, we conducted an extended replication of the GWAS discovery results in a larger set of BRCA2 mutation carriers in CIMBA, which represents the largest, international collection of BRCA2 mutation carriers.
Materials and Methods
Ethics statement
Each of the host institutions (Table S1) recruited under ethically-approved protocols. Written informed consent was obtained from all subjects.
Study subjects
The majority of BRCA2 mutation carriers were recruited through cancer genetics clinics and some came from population or community-based studies. Studies contributing DNA samples to these research efforts were members of the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) with the exception of one study (NICCC). Eligible subjects were women of European descent who carried a pathogenic BRCA2 mutation, had complete phenotype information, and were at least 18 years of age. Harmonized phenotypic data included year of birth, age at cancer diagnosis, age at bilateral prophylactic mastectomy and oophorectomy, age at interview or last follow-up, BRCA2 mutation description, self-reported ethnicity, and breast cancer estrogen receptor status.
GWAS discovery stage samples
Details of these samples have been described previously [2]. Data from 899 young (<40 years) affected and 804 older (>40 years) unaffected carriers of European ancestry from 14 countries were used to select SNPs for inclusion on the iCOGS array.
Samples genotyped in the extended replication set
Forty-seven studies from 24 different countries (including two East-Asian countries) provided DNA from a total of 10,048 BRCA2 mutations carriers. All eligible samples were genotyped using COGs, including those from the discovery stage.
Genotyping and quality control
BRCA2 SNP selection for inclusion on iCOGS
The Collaborative Oncological Gene-Environment Study (COGS) consortium developed a custom genotyping array (referred to as the iCOGS array) to provide efficient genotyping of common and rare genetic variants to identify novel loci that are associated with risk of breast, ovarian, and prostate cancers as well as to fine-map known cancer susceptibility loci. SNPs were selected for inclusion on iCOGS separately by each participating consortium: Breast Cancer Association Consortium (BCAC) [6], Ovarian Cancer Association Consortium (OCAC) [7], Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) [8], and CIMBA. SNP lists from a BRCA1 GWAS and SNPs in candidate regions were used together with the BRCA2 GWAS lists to generate a ranked CIMBA SNP list that included SNPs with the following nominal proportions: 55.5% from the BRCA1 GWAS, 41.6% from the BRCA2 GWAS and fine mapping, 2.9% for CIMBA candidate SNPs. Each consortium was given a share of the array: nominally 25% of the SNPs each for BCAC, PRACTICAL and OCAC; 17.5% for CIMBA; and 7.5% for SNPs from commonly researched pathways (e.g., inflammation). For the CIMBA BRCA2 GWAS, we used the iCOGS array as the platform to genotype the extended replication set of the discovery GWAS stage [2]. SNPs were selected on the basis of the strength of their associations with breast cancer risk in the discovery stage [2], using imputed genotype data for 1.4 M SNPs identified through CEU+TSI samples on HapMap3, release 2. A ranked list of SNPs was based on the 1-df trend test statistic, after excluding highly correlated SNPs (r2>0.4). The final list included the 39,015 SNPs with the smallest p-values. An additional set of SNPs were selected for fine mapping of the regions surrounding the SNPs found to be associated with breast cancer in the discovery GWAS stage: rs16917302 on 10q21 and rs311499 on 20q13, including SNPs with a MAF >0.05 located 500 kb in both directions of the SNP, based on HapMap 2 data. The final combined list of SNPs for the iCOGS array comprised 220,123 SNPs. Of these, 211,155 were successfully manufactured onto the array. The present analyses are based on the 19,029 SNPs selected on the basis of BRCA2 GWAS and fine mapping that were included on the iCOGS array.
Genotyping
The genotyping was performed on DNA samples from 10,048 BRCA2 mutation carriers at the McGill University and Génome Québec Innovation Centre (Montreal, Canada). As a quality control measure, each plate included DNA samples from six individuals who were members of two CEPH trios. Some plates also contained three duplicate pairs of quality control samples. Genotypes were called using GenCall [9]. Initial calling was based on a cluster file generated using 270 samples from Hapmap2. To generate the final calls, we first selected a subset of 3,018 individuals, including samples from each of the genotyping centers in the iCOGS project, each of the participating consortia, and each major ethnicity. Only plates with a consistent high call rate in the initial calling were used. We also included 380 samples of European, African, and Asian ethnicity genotyped as part of the Hapmap and 1000 Genomes project, and 160 samples that were known positive controls for rare variants on the array. This subset was used to generate a cluster file that was then applied to call the genotypes for the remaining samples.
Quality control of SNPs
Of the 211,155 SNPs on the iCOGS array, we excluded SNPs for the following reasons (Table S2): on the Y-chromosome, call rate <95%, deviations from Hardy-Weinberg equilibrium (P<10−7) using a stratified 1-d.f. test [10], and monomorphic. SNPs that gave discrepant genotypes among known duplicates were also excluded. After quality control filtering, 200,908 SNPs were available for analysis (Table S2); 18,086 of which were selected on the basis of the discovery BRCA2 GWAS [2]. Cluster plots of all reported SNPs were inspected manually for quality (Figure S1).
Description of imputation
Genotypes for SNPs identified through the 1000 Genomes Phase I data (released Jan 2012) [11] were imputed using all SNPs on the iCOGS chip in a region of 500 kb around the novel modifier locus at 6p24. The boundaries were determined according to the linkage disequilibrium (LD) structure in the region based on HapMap data. The imputation was carried out using IMPUTE 2.2 [12]. SNPs with imputation information/accuracy r2<0.30 were excluded in the analyses.
Quality control of DNA samples
Of 10,048 genotyped samples (Table S2), 742 were excluded because they did not meet the phenotypic eligibility criteria or had self-reported non-CEU ethnicity. Samples were then excluded for the following reasons: not female (XXY, XY), call rate <95%, low or high heterozygosity (P<10−6), discordant genotypes from previous CIMBA genotyping efforts, or discordant duplicate samples. For duplicates with concordant phenotypic data, or in cases of cryptic monozygotic twins, only one of the samples was included. Cryptic duplicates for which phenotypic data indicated different individuals were all excluded. Samples of non-European ancestry were identified using multi-dimensional scaling, after combining the BRCA2 mutation carrier samples with the HapMap2 CEU, CHB, JPT and YRI samples using a set of 37,120 uncorrelated SNPs from the iCOGS array. Samples with >19% non-European ancestry were excluded (Figure S2). A total of 4,330 affected and 3,881 unaffected BRCA2 mutation carrier women of European ancestry from 42 studies remained in the analysis (Table S1), including 3,234 breast cancer cases and 3,490 unaffected carriers that were not in the discovery set.
BRCA1 and BCAC samples
Details of the sample collection, genotyping and quality control process for the BRCA1 and BCAC samples, are reported elsewhere [13], [14].
Statistical methods
The associations between genotype and breast cancer risk were analyzed within a retrospective cohort framework with time to breast cancer diagnosis as the outcome [15]. Each BRCA2 carrier was followed until the first event: breast or ovarian cancer diagnosis, bilateral prophylactic mastectomy, or age at last observation. Only those with a breast cancer diagnosis were considered as cases in the analysis. The majority of mutation carriers were recruited through genetic counseling centers where genetic testing is targeted at women diagnosed with breast or ovarian cancer and in particular to those diagnosed with breast cancer at a young age. Therefore, these women are more likely to be sampled compared to unaffected mutation carriers or carriers diagnosed with the disease at older ages. As a consequence, sampling was not random with respect to disease phenotype and standard methods of survival analysis (such as Cox regression) may lead to biased estimates of the associations [16]. We therefore conducted the analysis by modelling the retrospective likelihood of the observed genotypes conditional on the disease phenotypes. This has been shown to provide unbiased estimates of the associations [15]. The implementation of the retrospective likelihoods has been described in detail elsewhere [15], [17]. The associations between genotype and breast cancer risk were assessed using the 1degree of freedom score test statistic based on the retrospective likelihood [15]. In order to account for non-independence between relatives, an adjusted version of the score test was used in which the variance of the score was derived taking into account the correlation between the genotypes [18]. P-values were not adjusted using genomic control because there was little evidence of inflation. Inflation was assessed using the genomic inflation factor, λ. Since this estimate is dependent on sample size, we also calculated λ adjusted to 1000 affected and 1000 unaffected samples. Per-allele and genotype-specific hazard-ratios (HR) and 95% confidence intervals (CI) were estimated by maximizing the retrospective likelihood. Calendar-year and cohort-specific breast cancer incidences for BRCA2 were used [1]. All analyses were stratified by country of residence. The USA and Canada strata were further subdivided by self-reported Ashkenazi Jewish ancestry. The assumption of proportional hazards was assessed by fitting a model that included a genotype-by-age interaction term. Between-country heterogeneity was assessed by comparing the results of the main analysis to a model with country-specific log-HRs. A possible survival bias due to inclusion of prevalent cases was evaluated by re-fitting the model after excluding affected carriers that were diagnosed ≥5 years prior to study recruitment. The associations between genotypes and tumor subtypes were evaluated using an extension of the retrospective likelihood approach that models the association with two or more subtypes simultaneously [19]. To investigate whether any of the significant SNPs were associated with ovarian cancer risk for BRCA2 mutation carriers and whether the inclusion of ovarian cancer patients as unaffected subjects biased our results, we also analyzed the data within a competing risks framework and estimated HR simultaneously for breast and ovarian cancer using the methods described elsewhere [15]. Analyses were carried out in R using the GenABEL libraries [20] and custom-written software. The retrospective likelihood was modeled in the pedigree-analysis software MENDEL [21], as described in detail elsewhere [15].
TCGA analysis
Affymetrix SNP 6.0 genotype calls for normal (non-tumor) breast DNA were downloaded for all available individuals from The Cancer Genome Atlas in September 2011. Analyses were limited to the 401 individuals of European ancestry based on principal component analysis. Expression levels in breast tumor tissue were adjusted for the top two principal components, age, gender (there are some male breast cancer cases in TCGA), and average copy number across the gene in the tumor. Linear regression was then used to test for association between the SNP and the adjusted gene expression level for all genes within one megabase.
Gene set enrichment analysis
To investigate enrichment of genes associated with breast cancer risk, the gene-set enrichment approach was implemented using Versatile Gene-based Association Study [22] based on the ranked P-values from retrospective likelihood analysis. Association List Go Annotator was also used to prioritize gene pathways using functional annotation from gene ontology (GO) [23] to increase the power to detect association to a pathway, as opposed to individual genes in the pathway. Both analyses were corrected for LD between SNPs, variable gene size, and interdependence of GO categories, where applicable, based on imputation. 100,000 Monte Carlo simulations were performed in VEGAS and 5000 replicate gene lists using random sampling of SNPs and 5000 replicate studies (sampling with replacement) were performed to estimate P-values.
Predicted absolute breast cancer risks by combined SNP profile
We estimated the absolute risks of developing breast cancer based on the joint distribution of SNPs associated with breast cancer for BRCA2 mutation carriers. The methods have been described elsewhere [24]. To construct the SNP profiles, we considered the single SNP from each region with the strongest evidence of association in the present dataset. We included all loci that had previously been found to be associated with breast cancer risk through GWAS in the general population and demonstrated associations with breast cancer risk for BRCA2 mutation carriers, and loci that had GWAS level of significance in the current study. We assumed that all loci in the profile were independent (i.e. they interact multiplicatively on BRCA2 breast cancer risk). Genotype frequencies were obtained under the assumption of Hardy-Weinberg Equilibrium. For each SNP, the effect of each allele was assumed to be consistent with a multiplicative model (log-additive). We assumed that the average, age-specific breast cancer incidences, over all associated loci, agreed with published breast cancer risk estimates for BRCA2 mutation carriers [1].
Results
The genomic inflation factor (λ) based on the 18,086 BRCA2 GWAS SNPs in the 6,724 BRCA2 mutation carriers who were not used in the SNP discovery set was 1.034 (λ adjusted to 1000 affected and 1000 unaffected: 1.010, Figure S3). Multiple variants were associated with breast cancer risk in the combined discovery and replication datasets (Figure S4). SNPs in three independent regions had P-values<5×10−8; one was a region not previously associated with breast cancer.
The most significant associations were observed for known breast cancer susceptibility regions, rs2420946 (per allele P = 2×10−14) in FGFR2 and rs3803662 (P = 5.4×10−11) near TOX3 (Table 1). Breast cancer risk associations with other SNPs reported previously for BRCA2 mutation carriers are summarized in Table 1. In this larger set of BRCA2 mutation carriers, we also identified novel SNPs in the 12p11 (PTHLH), 5q11 (MAP3K1), and 9p21 (CDKN2A/B) regions with smaller P-values for association than those of previously reported SNPs. These novel SNPs were not correlated with the previously reported SNPs (r2<0.14). For one of the novel SNPs identified in the discovery GWAS [2], ZNF365 rs16917302, there was weak evidence of association with breast cancer risk (P = 0.01); however, an uncorrelated SNP, rs17221319 (r2<0.01), 54 kb upstream of rs16917302 had stronger evidence of association (P = 6×10−3).
Tab. 1. Per allele hazard ratios (HR) and 95% confidence intervals (CI) of previously published breast cancer loci among BRCA2 mutation carriers from previous reports and from the iCOGS array, ordered by statistical significance of the region. 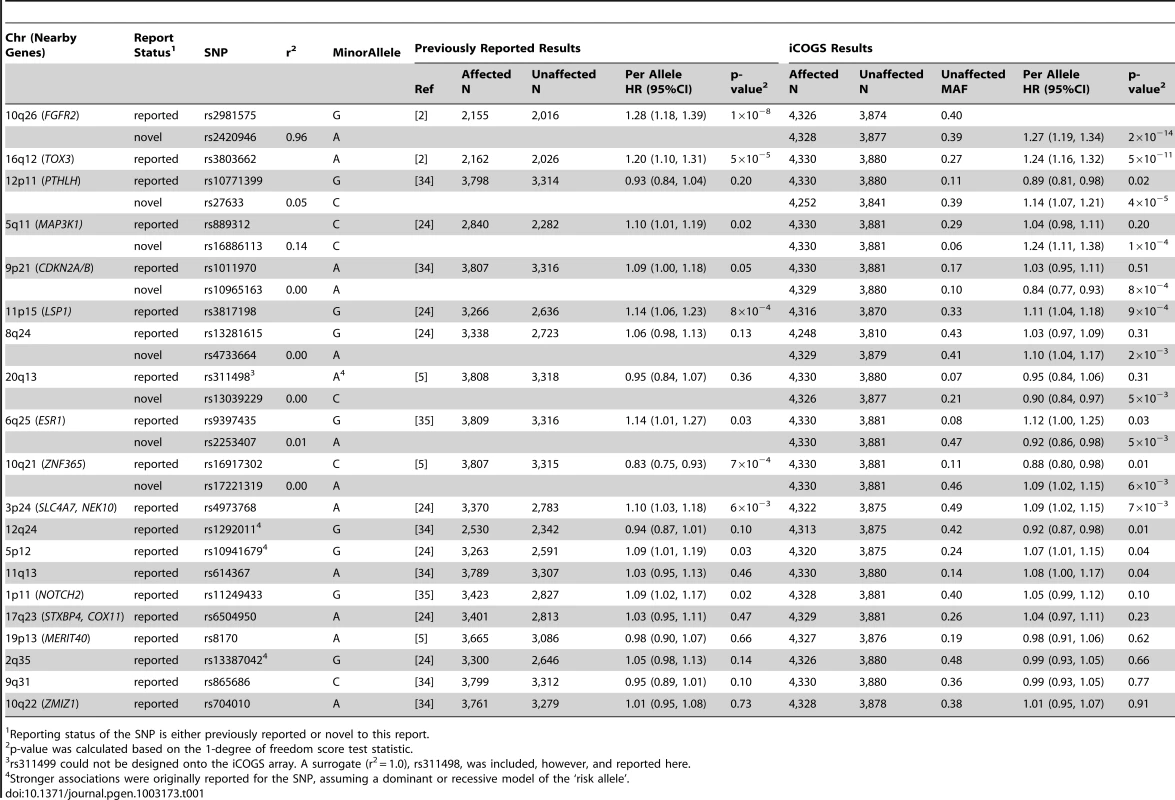
Reporting status of the SNP is either previously reported or novel to this report. One SNP, rs9348512 at 6p24 not known to be associated with breast cancer, had a combined P-value of association of 3.9×10−8 amongst all BRCA2 samples (Table 2), with strong evidence of replication in the set of BRCA2 samples that were not used in the discovery stage (P = 5.2×10−5). The minor allele of rs9348512 (MAF = 0.35) was associated with a 15% decreased risk of breast cancer among BRCA2 mutation carriers (per allele HR = 0.85, 95% CI 0.80–0.90) with no evidence of between-country heterogeneity (P = 0.78, Figure S5). None of the genotyped (n = 68) or imputed (n = 3,507) SNPs in that region showed a stronger association with risk (Figure 1; Table S3), but there were 40 SNPs with P<10−4 (pairwise r2>0.38 with rs9348512, with the exception of rs11526201 for which r2 = 0.01, Table S3). The association with rs9348512 did not differ by 6174delT mutation status (P for difference = 0.33), age (P = 0.39), or estrogen receptor (ER) status of the breast tumor (P = 0.41). Exclusion of prevalent breast cancer cases (n = 1,752) produced results (HR = 0.83, 95% CI 0.77–0.89, P = 3.40×10−7) consistent with those for all cases.
Fig. 1. Associations between SNPs in the region surrounding rs9348512 on chromosome 6 and breast cancer risk. 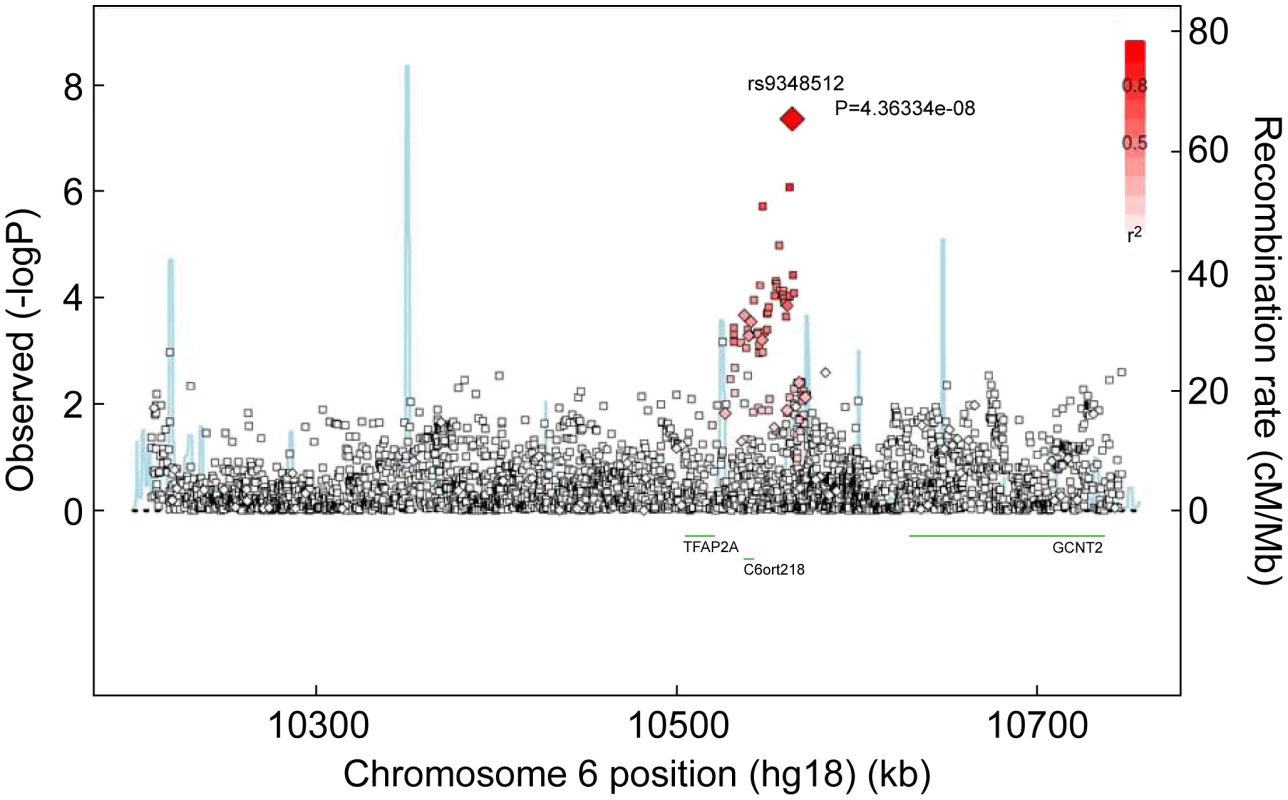
Results based on imputed and observed genotypes. The blue spikes indicate the recombination rate at each position. Genotyped SNPs are represented by diamonds and imputed SNPs are represented by squares. Color saturation indicates the degree of correlation with the SNP rs9348512. Tab. 2. Breast cancer hazard ratios (HR) and 95% confidence intervals (CI) of novel breast cancer loci with P-values of association <10−5 among BRCA2 mutation carriers. 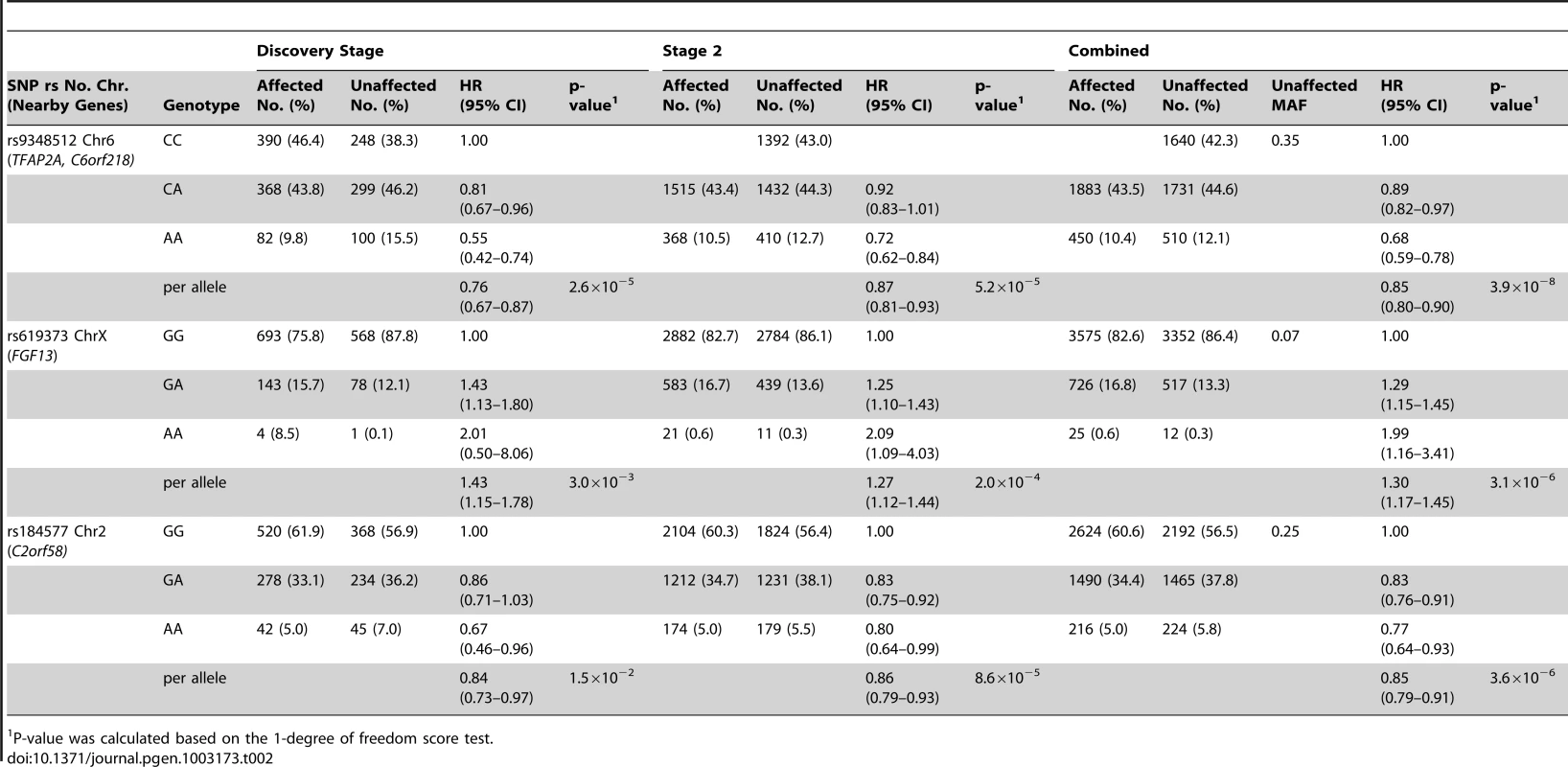
P-value was calculated based on the 1-degree of freedom score test. SNPs in two additional regions had P-values<10−5 for breast cancer risk associations for BRCA2 mutation carriers (Table 2). The magnitude of associations for both SNPs was similar in the discovery and second stage samples. In the combined analysis of all samples, the minor allele of rs619373, located in FGF13 (Xq26.3), was associated with higher breast cancer risk (HR = 1.30, 95% CI 1.17–1.45, P = 3.1×10−6). The minor allele of rs184577, located in CYP1B1-AS1 (2p22–p21), was associated with lower breast cancer risk (HR = 0.85, 95% CI 0.79–0.91, P = 3.6×10−6). These findings were consistent across countries (P for heterogeneity between country strata = 0.39 and P = 0.30, respectively; Figure S6). There was no evidence that the HR estimates for rs619373 and rs184577 change with age of the BRCA2 mutation carriers (P for the genotype-age interaction = 0.80 and P = 0.40, respectively) and no evidence of survival bias for either SNP (rs619373: HR = 1.35, 95% CI 1.20–1.53, P = 1.5×10−6 and rs184577: HR = 0.86, 95% CI 0.79–0.93, P = 2.0×10−4, after excluding prevalent cases). The estimates for risk of ER-negative and ER-positive breast cancer were not significantly different (P for heterogeneity between tumor subtypes = 0.79 and 0.67, respectively). When associations were evaluated under a competing risks model, there was no evidence of association with ovarian cancer risk for SNPs rs9348512 at 6p24, rs619373 in FGF13 or rs184577 at 2p22 and the breast cancer associations were virtually unchanged (Table S4).
Gene set enrichment analysis confirmed that strong associations exist for known breast cancer susceptibility loci and the novel loci identified here (gene-based P<1×10−5). The pathways most strongly associated with breast cancer risk that contained statistically significant SNPs included those related to ATP binding, organ morphogenesis, and several nucleotide bindings (pathway-based P<0.05).
To begin to determine the functional effect of rs9348512, we examined associations of expression levels of any nearby gene in breast tumors with the minor A allele. Using data from The Cancer Genome Atlas, we found that the A allele of rs9348512 was strongly associated with mRNA levels of GCNT2 in breast tumors (p = 7.3×10−5).
The hazard ratios for the percentiles of the combined genotype distribution of loci associated with breast cancer risk in BRCA2 mutation carriers were translated into absolute breast cancer risks under the assumption that SNPs interact multiplicatively. Based on our results for SNPs in FGFR2, TOX3, 12p11, 5q11, CDKN2A/B, LSP1, 8q24, ESR1, ZNF365, 3p24, 12q24, 5p12, 11q13 and also the 6p24 locus, the 5% of the BRCA2 mutation carriers at lowest risk were predicted to have breast cancer risks by age 80 in the range of 21–47% compared to 83–100% for the 5% of mutation carriers at highest risk on the basis of the combined SNP profile distribution (Figure 2). The breast cancer risk by age 50 was predicted to be 4–11% for the 5% of the carriers at lowest risk compared to 29–81% for the 5% at highest risk.
Fig. 2. Predicted breast cancer risks for BRCA2 mutation carriers by the combined SNP profile distributions. 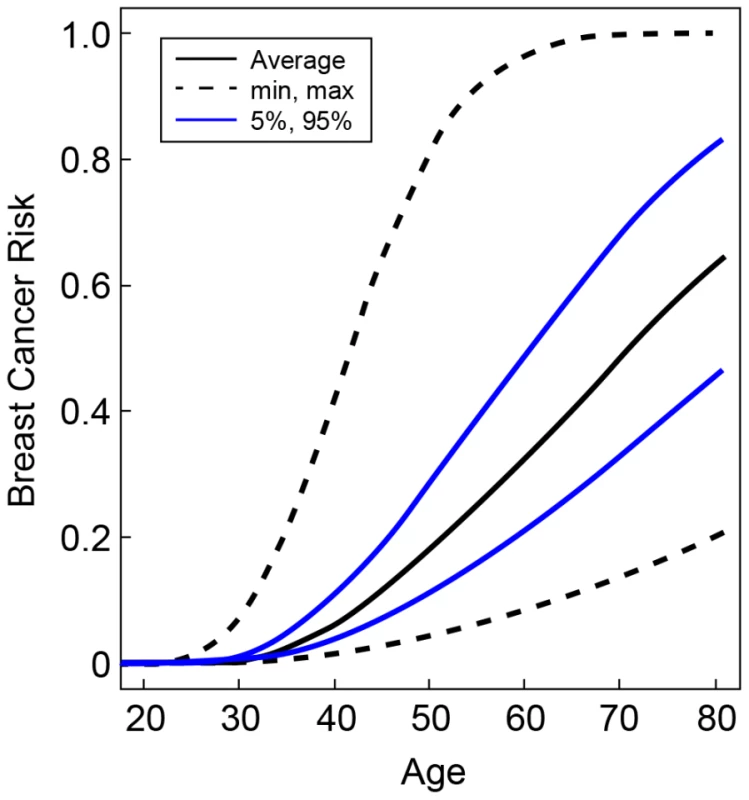
Based on the known breast cancer susceptibility loci at FGFR2, TOX3, 12p11, 5q11, CDKN2A/B, LSP1, 8q24, ESR1, ZNF365, 3p24, 12q24, 5p12, 11q13 and the newly identified BRCA2 modifier locus at 6p24. The figure shows the risks at the 5th and 95th percentiles of the combined genotyped distribution as well as minimum, maximum and average risks. Discussion
In the largest assemblage of BRCA2 mutation carriers, we identified a novel locus at 6q24 that is associated with breast cancer risk, and noted two potential SNPs of interest at Xq26 and 2p22. We also replicated associations with known breast cancer susceptibility SNPs previously reported in the general population and in BRCA2 mutation carriers. For the 12p11 (PTHLH), 5q11 (MAP3K1), and 9p21 (CDKN2A/B), we found uncorrelated SNPs that had stronger associations than the originally identified SNP in the breast cancer susceptibility region that should be replicated in the general population. In BRCA2 mutation carriers, evidence for a breast cancer association with genetic variants in PTHLH has been restricted previously to ER-negative tumors [25]; however, the novel susceptibility variant we reported here was associated with risk of ER+ and ER - breast cancer.
The novel SNP rs9348512 (6p24) is located in a region with no known genes (Figure 1). C6orf218, a gene encoding a hypothetical protein LOC221718, and a possible tumor suppressor gene, TFAP2A, are within 100 kb of rs9348512. TFAP2A encodes the AP-2α transcription factor that is normally expressed in breast ductal epithelium nuclei, with progressive expression loss from normal, to ductal carcinoma in situ, to invasive cancer [26], [27]. AP-2α also acts as a tumor suppressor via negative regulation of MYC [28] and augmented p53-dependent transcription [29]. However, the minor allele of rs9348512 was not associated with gene expression changes of TFAP2A in breast cancer tissues in The Cancer Genome Atlas (TCGA) data; this analysis might not be informative since expression of TFAP2A in invasive breast tissue is low [26], [27]. Using the TCGA data and a 1 Mb window, expression changes with genotypes of rs9348512 were observed for GCNT2, the gene encoding the enzyme for the blood group I antigen glucosaminyl (N-acetyl) transferase 2. GCNT2, recently found to be overexpressed in highly metastatic breast cancer cell lines [30] and basal-like breast cancer [31], interacts with TGF-β to promote epithelial-to-mesenchymal transition, enhancing the metastatic potential of breast cancer [31]. An assessment of alterations in expression patterns in normal breast tissue from BRCA2 mutation carriers by genotype are needed to further evaluate the functional implications of rs9348512 in the breast tumorigenesis of BRCA2 mutation carriers.
To determine whether the breast cancer association with rs9348512 was limited to BRCA2 mutation carriers, we compared results to those in the general population genotyped by BCAC and to BRCA1 mutation carriers in CIMBA. No evidence of an associations between rs9348512 and breast cancer risk was observed in the general population (OR = 1.00, 95% CI 0.98–1.02, P = 0.74) [14], nor in BRCA1 mutation carriers (HR = 0.99, 95% CI 0.94–1.04, P = 0.75) [13]. Stratifying cases by ER status, there was no association observed with ER-subtypes in either the general population or among BRCA1 mutation carriers (BCAC: ER positive P = 0.89 and ER negative P = 0.60; CIMBA BRCA1: P = 0.49 and P = 0.99, respectively). For the two SNPs associated with breast cancer with P<10−5, neither rs619373, located in FGF13 (Xq26.3), nor rs184577, located in CYP1B1-AS1 (2p22-p21), was associated with breast cancer risk in the general population [14] or among BRCA1 mutation carriers [13]. The narrow CIs for the overall associations in the general population and in BRCA1 mutation carriers rule out associations of magnitude similar to those observed for BRCA2 mutation carriers. The consistency of the association in the discovery and replication stages and by country, the strong quality control measures and filters, and the clear cluster plot for rs9348512 suggest that our results constitute the discovery of a novel breast cancer susceptibility locus specific to BRCA2 mutation carriers rather than a false positive finding. Replicating this SNP in an even larger population of BRCA2 mutation carriers would be ideal, but not currently possible because we know of no investigators with appropriate data and germline DNA from BRCA2 mutation carriers who did not contribute their mutation carriers to iCOGS. However, CIMBA studies continue to recruit individuals into the consortium.
rs9348512 (6p24) is the first example of a common susceptibility variant identified through GWAS that modifies breast cancer risk specifically in BRCA2 mutation carriers. Previously reported BRCA2-modifying alleles for breast cancer, including those in FGFR2, TOX3, MAP3K1, LSP1, 2q35, SLC4A7, 5p12, 1p11.2, ZNF365, and 19p13.1 (ER-negative only) [18], [32], [33], are also associated with breast cancer risk in the general population and/or BRCA1 mutation carriers. Knowledge of the 6p24 locus might provide further insights into the biology of breast cancer development in BRCA2 mutation carriers. Additional variants that are specific modifiers of breast cancer risk in BRCA2 carriers may yet be discovered; their detection would require assembling larger samples of BRCA2 mutation carriers in the future.
While individually each of the SNPs associated with breast cancer in BRCA2 mutation carriers are unlikely to be used to guide breast cancer screening and risk-reducing management strategies, the combined effect of the general and BRCA2-specific breast cancer susceptibility SNPs might be used to tailor manage subsets of BRCA2 mutation carriers. Taking into account all loci associated with breast cancer risk in BRCA2 mutation carriers from the current analysis, including the 6p24 locus, the 5% of the BRCA2 mutation carriers at lowest risk were predicted to have breast cancer risks by age 80 in the range of 21–47% compared to 83–100% for the 5% of mutation carriers at highest risk on the basis of the combined SNP profile distribution. These results might serve as a stimulus for prospective trials of the clinical utility of such modifier panels.
Supporting Information
Zdroje
1. AntoniouAC, CunninghamAP, PetoJ, EvansDG, LallooF, et al. (2008) The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer 98 : 1457–1466.
2. GaudetMM, KirchhoffT, GreenT, VijaiJ, KornJM, et al. (2010) Common genetic variants and modification of penetrance of BRCA2-associated breast cancer. PLoS Genet 6: e1001183 doi:10.1371/journal.pgen.1001183.
3. TurnbullC, AhmedS, MorrisonJ, PernetD, RenwickA, et al. (2010) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 42 : 504–507.
4. LindstromS, VachonCM, LiJ, VargheseJ, ThompsonD, et al. (2011) Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet 43 : 185–187.
5. CouchFJ, GaudetMM, AntoniouAC, RamusSJ, KuchenbaeckerKB, et al. (2012) Common variants at the 19p13.1 and ZNF365 loci are associated with ER subtypes of breast cancer and ovarian cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol Biomarkers Prev 21 : 645–657.
6. Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. J Natl Cancer Inst 98 : 1382–1396.
7. GaytherSA, SongH, RamusSJ, KjaerSK, WhittemoreAS, et al. (2007) Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res 67 : 3027–3035.
8. Kote-JaraiZ, EastonDF, StanfordJL, OstranderEA, SchleutkerJ, et al. (2008) Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev 17 : 2052–2061.
9. Kermani BG (2008) Artificial intelligence and global normalization methods for genotype.
10. RobertsonA, HillWG (1984) Deviations from Hardy-Weinberg proportions: sampling variances and use in estimation of inbreeding coefficients. Genetics 107 : 703–718.
11. A map of human genome variation from population-scale sequencing. Nature 467 : 1061–1073.
12. HowieBN, DonnellyP, MarchiniJ (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529 doi:10.1371/journal.pgen.1000529.
13. CouchFJ, WangX, McGuffogL, LeeA, OlswoldC, et al. (2012) Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. Nat Genet under review
14. MichailidouK, HallP, Gonzalez-NeiraA, GhoussainiM, DennisJ, et al. (2012) Large-scale genotyping identifies 38 new breast cancer susceptibility loci. Nat Genet under review
15. Barnes D, Lee A, Embrace, Easton D, Antoniou AC (2012) Evaluation of association methods for analyzing modifiers of disease risk in carriers of high-risk mutations. Genet Epidemiol in press.
16. AntoniouAC, GoldgarDE, AndrieuN, Chang-ClaudeJ, BrohetR, et al. (2005) A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet Epidemiol 29 : 1–11.
17. AntoniouAC, SinilnikovaOM, SimardJ, LeoneM, DumontM, et al. (2007) RAD51 135G–>C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet 81 : 1186–1200.
18. AntoniouAC, WangX, FredericksenZS, McGuffogL, TarrellR, et al. (2010) A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet 42 : 885–892.
19. MulliganAC, CouchFJ, BarrowdaleD, DomchekSM, EcclesD, et al. (2011) Common breast cancer susceptibility alleles are associated with tumour subtypes in BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Res 13: R110.
20. AulchenkoYS, RipkeS, IsaacsA, van DuijnCM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23 : 1294–1296.
21. LangeK, WeeksD, BoehnkeM (1988) Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 5 : 471–472.
22. LiuJZ, McRaeAF, NyholtDR, MedlandSE, WrayNR, et al. (2010) A versatile gene-based test for genome-wide association studies. Am J Hum Genet 87 : 139–145.
23. AshburnerM, BallCA, BlakeJA, BotsteinD, ButlerH, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25 : 25–29.
24. AntoniouAC, BeesleyJ, McGuffogL, SinilnikovaOM, HealeyS, et al. (2010) Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res 70 : 9742–9754.
25. AntoniouAC, KuchenbaeckerKB, SoucyP, BeesleyJ, ChenX, et al. (2012) Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast Cancer Res 14: R33.
26. FriedrichsN, JagerR, PaggenE, RudlowskiC, Merkelbach-BruseS, et al. (2005) Distinct spatial expression patterns of AP-2alpha and AP-2gamma in non-neoplastic human breast and breast cancer. Mod Pathol 18 : 431–438.
27. GeeJM, RobertsonJF, EllisIO, NicholsonRI, HurstHC (1999) Immunohistochemical analysis reveals a tumour suppressor-like role for the transcription factor AP-2 in invasive breast cancer. J Pathol 189 : 514–520.
28. GaubatzS, ImhofA, DoschR, WernerO, MitchellP, et al. (1995) Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J 14 : 1508–1519.
29. McPhersonLA, LoktevAV, WeigelRJ (2002) Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53. J Biol Chem 277 : 45028–45033.
30. ZhangH, MengF, LiuG, ZhangB, ZhuJ, et al. (2011) Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res 71 : 1292–1301.
31. ZhangH, MengF, WuS, KreikeB, SethiS, et al. (2011) Engagement of I-branching {beta}-1, 6-N-acetylglucosaminyltransferase 2 in breast cancer metastasis and TGF-{beta} signaling. Cancer Res 71 : 4846–4856.
32. AntoniouAC, SpurdleAB, SinilnikovaOM, HealeyS, PooleyKA, et al. (2008) Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet 82 : 937–948.
33. AntoniouAC, SinilnikovaOM, McGuffogL, HealeyS, NevanlinnaH, et al. (2009) Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet 18 : 4442–4456.
34. AntoniouAC, KuchenbaeckerKB, SoucyP, BeesleyJ, ChenX, et al. (2012) Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast Cancer Res 14: R33.
35. AntoniouAC, KartsonakiC, SinilnikovaOM, SoucyP, McGuffogL, et al. (2011) Common alleles at 6q25.1 and 1p11.2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet 20 : 3304–3321.
Štítky
Genetika Reprodukčná medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



