-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), together with associated genes (cas), form the CRISPR–cas adaptive immune system, which can provide resistance to viruses and plasmids in bacteria and archaea. Here, we use mathematical models, population dynamic experiments, and DNA sequence analyses to investigate the host–phage interactions in a model CRISPR–cas system, Streptococcus thermophilus DGCC7710 and its virulent phage 2972. At the molecular level, the bacteriophage-immune mutant bacteria (BIMs) and CRISPR–escape mutant phage (CEMs) obtained in this study are consistent with those anticipated from an iterative model of this adaptive immune system: resistance by the addition of novel spacers and phage evasion of resistance by mutation in matching sequences or flanking motifs. While CRISPR BIMs were readily isolated and CEMs generated at high rates (frequencies in excess of 10−6), our population studies indicate that there is more to the dynamics of phage–host interactions and the establishment of a BIM–CEM arms race than predicted from existing assumptions about phage infection and CRISPR–cas immunity. Among the unanticipated observations are: (i) the invasion of phage into populations of BIMs resistant by the acquisition of one (but not two) spacers, (ii) the survival of sensitive bacteria despite the presence of high densities of phage, and (iii) the maintenance of phage-limited communities due to the failure of even two-spacer BIMs to become established in populations with wild-type bacteria and phage. We attribute (i) to incomplete resistance of single-spacer BIMs. Based on the results of additional modeling and experiments, we postulate that (ii) and (iii) can be attributed to the phage infection-associated production of enzymes or other compounds that induce phenotypic phage resistance in sensitive bacteria and kill resistant BIMs. We present evidence in support of these hypotheses and discuss the implications of these results for the ecology and (co)evolution of bacteria and phage.
Published in the journal: The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity. PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003312
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003312Summary
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), together with associated genes (cas), form the CRISPR–cas adaptive immune system, which can provide resistance to viruses and plasmids in bacteria and archaea. Here, we use mathematical models, population dynamic experiments, and DNA sequence analyses to investigate the host–phage interactions in a model CRISPR–cas system, Streptococcus thermophilus DGCC7710 and its virulent phage 2972. At the molecular level, the bacteriophage-immune mutant bacteria (BIMs) and CRISPR–escape mutant phage (CEMs) obtained in this study are consistent with those anticipated from an iterative model of this adaptive immune system: resistance by the addition of novel spacers and phage evasion of resistance by mutation in matching sequences or flanking motifs. While CRISPR BIMs were readily isolated and CEMs generated at high rates (frequencies in excess of 10−6), our population studies indicate that there is more to the dynamics of phage–host interactions and the establishment of a BIM–CEM arms race than predicted from existing assumptions about phage infection and CRISPR–cas immunity. Among the unanticipated observations are: (i) the invasion of phage into populations of BIMs resistant by the acquisition of one (but not two) spacers, (ii) the survival of sensitive bacteria despite the presence of high densities of phage, and (iii) the maintenance of phage-limited communities due to the failure of even two-spacer BIMs to become established in populations with wild-type bacteria and phage. We attribute (i) to incomplete resistance of single-spacer BIMs. Based on the results of additional modeling and experiments, we postulate that (ii) and (iii) can be attributed to the phage infection-associated production of enzymes or other compounds that induce phenotypic phage resistance in sensitive bacteria and kill resistant BIMs. We present evidence in support of these hypotheses and discuss the implications of these results for the ecology and (co)evolution of bacteria and phage.
Introduction
The experimental demonstrations, in 2007 and 2008, that the Clustered Regularly Interspaced Short Palindromic Repeats that abound in the genomes of the vast majority of archaea and nearly half of bacteria can serve as part of an adaptive immune system (CRISPR–cas) that protects these prokaryotes against lytic phage [1] and conjugative plasmids [2] raised many intriguing questions and stimulated a great deal of research. Between 2007 and 2012, according to PubMed, the number of articles with the acronym CRISPR in their title and/or body has been doubling every 20 months. Understandably, the majority of these articles are reports or reviews of the genetic, biochemical, and molecular mechanisms by which (i) the CRISPR–cas system acquires the 26–72 base pair sequences (spacers) of DNA from infecting phage, generating Bacteriophage Insensitive Mutants (BIMs), or from invading plasmids, forming Plasmid Interfering Mutants (PIMs); (ii) the small RNAs coded for by these spacers abort subsequent infections by phage and plasmids with DNA sequences corresponding to those in these spacers (protospacers) [3], [4], [5], [6]; (iii) the phage can evade this resistance and generate CRISPR Escape Mutants (CEMs) by modifying the DNA sequences of protospacers or protospacer adjacent motifs (PAM) [3], [7], [8], [9]; (iv) the Cas proteins participate in these processes [1], [8], [9]; and (v) the existence, distribution and genetic diversity of CRISPR–cas systems in the archaea and bacteria came about [10], [11], [12]
Some of this research has been devoted to the practical applications of CRISPR and the CRISPR–cas system. Long before the immunological role of CRISPR in these microorganisms was recognized, differences in the number of these palindromic repeats had been used as markers for studies of the genetic (molecular) epidemiology and forensics of pathogenic bacteria [13], [14], [15], [16]. Isolates that would be classified as identical by standard typing procedures, like Serotyping, Multi-Locus Sequence Typing, and Pulse Field Gel Electrophoresis, can differ in their CRISPR regions, particularly in their spacer sequences [17], [18]. Genes, and clusters of genes, borne on plasmids, phage, or chromosomal genes, and acquired by horizontal transfer, code for the virulence and antibiotic resistance of many pathogenic bacteria. And, as one might anticipate, strains of at least some species of pathogenic bacteria that lack functional CRISPR–cas systems are more likely to be virulent and resistant to antibiotics than those that bear these systems [19]. A number of biotechnological and industrial processes rely on the use of bacteria that are subject to infection by phage, and therefore risk disruption of production, low-quality products and economic loss. Can CRISPR–cas systems be harnessed to counter antibiotic resistance and virulence of pathogenic bacteria and/or prevent phage contamination of industrially important bacteria (8)?
Central to understanding these practical implications and potential applications of CRISPR–cas systems are the population and evolutionary dynamics of bacteria bearing these adaptive immune systems and those of the phage and plasmids that infect them. Prior to the experimental demonstration that CRISPR is part of an adaptive immune system, and motivating those experiments, were observations that the spacer sequences in the CRISPR regions of bacteria and archaea from natural communities were identical to sequences in the genomes of viruses in their communities [15], [20], [21]. These and more recent studies of bacteria and archaea and their viruses [10], [11], [12], [22], and metagenomic studies of microbiomes [23], provided compelling support for the hypothesis that CRISPR–cas systems play an active role in the population dynamics and co-evolution of these organisms in natural communities. However, the inferences one can draw from these investigations are limited. They provide little of the necessary quantitative information about the nature and contribution of CRISPR–cas to these dynamics and co-evolution and its consequential role in determining the structure of their communities.
Studies using mathematical models and computer simulations suggest that bacteria with CRISPR–cas systems can invade communities of phage and bacteria without this immune system and can also prevent phage from invading their communities [24]. Modeling studies also suggest that, in theory, a CRISPR–cas-mediated BIM-CEM co-evolutionary arms race can maintain considerable diversity in the interacting populations of bacteria and phage [25] and account for spacer changes in the CRISPR elements in the metagenomic structure of natural communities of archaea and bacteria and their phage [26]. But, as compelling as these mathematical models and computer simulation studies may be, their limitations are also apparent. The models used are based on simplifying assumptions about the phage infection process and CRISPR–cas adaptive immunity that have not been formally tested in the systems under study. The computer simulations and other numerical analyses of the properties of these models use parameter values for which there are few independent estimates.
In this study we use mathematical models, computer simulations, parameter estimation, and population dynamic and other microbiological experiments with the Gram-positive bacteria Streptococcus thermophilus and its virulent phage 2972 (Siphoviridae family) to explore the nature and contribution of CRISPR–cas adaptive immunity to the population and evolutionary dynamics of bacteria and phage. The results of these experiments and the molecular characterization of the bacteria and phage employed demonstrate that the qualitative Lamarckian (iterative acquisition of spacers) [27] and Darwinian (mutation in protospacers) genetic conditions for a CRISPR–mediated arms race are met with this phage-host system. BIMs are readily formed and CEMs capable of growing on these BIMs are generated at high rates. On the other hand, our experimental results and theoretical analyses indicate that even at a qualitative level, models based on existing assumptions about phage infection and CRISPR–mediated resistance cannot account for the population and evolutionary dynamics of the interactions between S. thermophilus and phage 2972. Findings contrary to what is anticipated from the analysis of the properties of these models include: (i) the invasion of phage into populations of BIMs resistant due to the acquisition of one (but not two) spacers; (ii) the survival of substantial numbers of sensitive bacteria despite the presence of high densities of phage; and (iii) the maintenance of phage-limited communities due to the failure of even two-spacer BIMs to become established in populations with wild type bacteria and phage. We postulate that the latter two results can be attributed to the phage infection-associated production of enzymes or other compounds that kill resistant BIMs and generate phenotypically phage-resistant “persister” populations of sensitive bacteria. By modifying the model to account for these chemical processes we can explain the observed dynamics. The predictions of this extended model are supported by longer-term population dynamic experiments.
Results
Theoretical framework
Overview
We open this report with a mathematical model that is based on the classical view of the process of infection of bacteria with lytic phage [28] and the current perspective on CRISPR–cas-mediated immunity to phage infection [3]. The roles of this model and the computer simulations used to analyze its properties are to (i) evaluate the contributions of the major parameters governing the dynamics of phage infection of bacteria with CRISPR–cas immune systems; (ii) generate testable hypotheses about these dynamics; (iii) guide the design of the experiments testing these hypotheses; and (iv) interpret the results of these experiments. This opening model is in essence a quantitative straw person. It is not intended to provide quantitatively precise descriptions of these dynamics, but rather to identify major qualitative deviations between the results of our experiments and a theory based on the canonical view of phage infection and CRISPR–cas immunity. For our analysis of the properties of this model, we use semi-stochastic simulations employing parameter values estimated experimentally in cultures of S. thermophilus strain DGCC7710 and phage 2972.
The model
The bacteria and phage are of three possible orders, with densities and designations, respectively, B0, B1, and B2, as well as P0, P1, and P2 (Figure 1). Phage are able to adsorb to bacteria of all orders, but are only able to replicate on bacteria of states with the same or lower index numbers (earlier states). For example, phage P1 can adsorb to and replicate on B0 and B1 and adsorb to but not replicate on B2. Phage that adsorb to bacteria on which they cannot replicate are lost from their population. Phage resistance (the generation of BIMs) is acquired by the addition of DNA sequences (spacers) into the CRISPR locus that correspond to sequences borne by the infecting phage (protospacers). Although we realize that there are at least two active CRISPR–cas systems in this S. thermophilus strain, for simplicity, we assume there is a unique functional CRISPR–cas system. Spacers accumulate without loss of earlier spacers. By mutation in a phage protospacer corresponding to a host spacer, phage are able to replicate on bacteria with that spacer. These CEMs accumulate protospacer (or PAM) mutations without loss of those acquired earlier and thereby can replicate on bacteria with spacers providing resistance to all earlier incarnations of that phage. For example a phage P1 with protospacer mutation that enables it to replicate on bacteria B1 can also replicate on B0. For simplicity and tractability, we assume that there is a single bacterial and phage genotype within each order as defined by the number of spacers. In reality BIMs may be generated by the acquisition of many different spacers. We consider the implications of this simplifying assumption in the Discussion.
Fig. 1. CRISPR–mediated arms race. 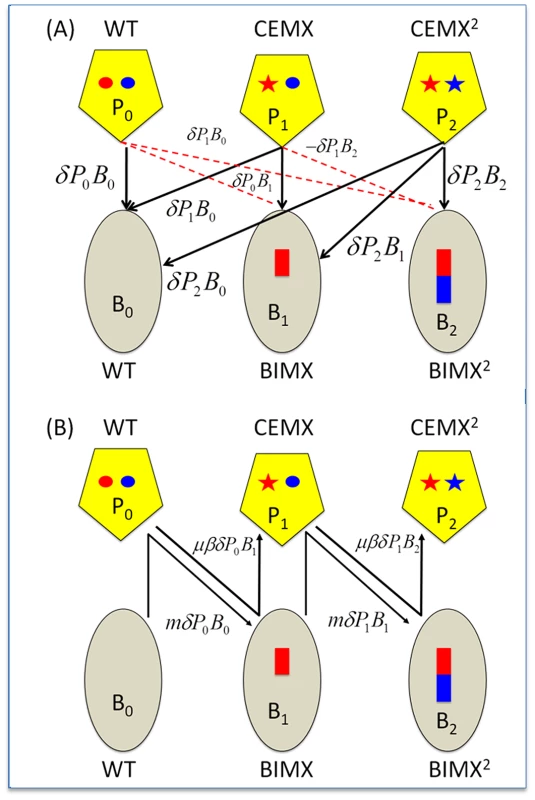
Ellipses - bacteria; Pentagons - phage; red and blue rectangles - acquired spacers; red and blue circles, regions of the phage genome corresponding to the acquired spacers (protospacers); stars, mutated protospacers generating CEMs. WT, Wild type bacteria and phage; BIMX and CEMX, bacteria with spacers for acquired resistance to WT phage and the first-order CRISPR Escape mutants (CEMX), respectively; BIMX2 and CEMX2, bacteria with spacers for acquired resistance to CEMX and the second-order CRISPR–escape mutants, respectively. Panel A: Infection and phage replication relationships. Solid black lines, phage adsorption and replication; broken red lines - phage adsorption and loss. Panel B: Changes in state. BIMX are produced by WT phage infecting WT cells and BIMX2 are produced by CEMX infecting BIMX. CEMX are produced by P0 infecting and replicating on B0 and CEMX2 are produced by CEMX infecting and replicating on BIMX. The rate of growth of the bacteria is a monotonic increasing function of the concentration of a limiting resource r (µg/ml), , where v (per hour) is the maximum growth rate and k (µg) is the concentration of the resource where the growth rate is half its maximum value (v/2) [29]. Resources are taken up at a rate proportional to the densities of the bacteria, their growth rates, and a conversion efficiency parameter e (µg) which is the amount of resource needed to produce a new cell [30].
The phage adsorb to bacteria of all states with a rate parameter δ (ml per cell per phage per hour) and produce β phage per infection (burst size). As in [24], [25], [31], we assume phage infection is a mass action process that occurs at a rate proportional to the product of the densities of the populations of bacteria and phage and the rate constant, δ. Spacers are incorporated only from phage capable of replicating on bacteria of a given state. The parameter m is the probability that a bacterium infected by a phage that can replicate on it will add a spacer for immunity to that phage. CEM phage of state P1 are produced by Po during the course of replication on B0 with a probability μ per phage per infection. To account for the latent period of length λ hours, we assume that infected bacteria enter a fated state, Mij, for bacteria of state i infected with phage of state j. At time t, a fraction (1-m) of the cells infected λ hours earlier (at time t-λ) burst and produce β-1 particles. The (1-m) is to account for the fraction of infected B0 and B1 cells forming BIMs (B1 and B2, respectively); the β-1 new phage particles accounts for the loss of the infecting phage. CEMs of a given order are formed by mutation occurring during infections of sensitive bacteria by phage of the previous order. For example, phage of state P1 are produced as a fraction μ of the phage particles generated upon burst of B0 cells infected with P0. With these definitions and assumptions, the rates of change in the concentration of the resource and densities of phage are given by:
Computer simulations
In our numerical solutions to these equations - computer simulations - the acquisition of spacers and CEM mutations are random processes. We use the Euler method for solving the differential equations and a Monte Carlo protocol for the generation of BIMs and CEMs. In any time interval Δt, the probability that a BIM is formed is proportional to the number of sensitive cells infected with phage during that time interval. For example, in a given interval Δt, the probability that a P1 infecting a B1 will produce a single B2 is approximately mM1(t-λ)Δt where t is time in hours. If a random number x (0<x<1) from a uniform distribution is less than this probability, a single B2 will enter the B2 population. In any time interval, the probability that a sensitive cell infected with phage will produce a CEM is proportional to the number of phage. For example, the probability that a P1 CEM will be produced by a P0 infecting a B0 in an interval Δt is approximately μM0(t-λ)(β-1)Δt. If a random number x is less than this product, a single phage particle will enter the P1 population and be removed from the P0 population. The interval size Δt is chosen so that at any time these probabilities are less than 1.0, as they should be. These simulations were programmed in Berkeley Madonna. The program used for the analysis of the properties of this model can be obtained at www.eclf.net/programs.
Parameter values and short-term population dynamics
Bacterial growth
The maximum growth rate of the wild type S. thermophilus strain DGCC7710 in LM17 medium (the parameter v) was estimated as the regression coefficient of the changes in the natural log of the optical density (OD 600 nm) of growing culture: 1.37±0.27 (95% confidence interval). Since the bacteria were grown in broth rather than a medium with a defined limiting resource, we cannot directly estimate the Monod constant (k) or the conversion efficiency (e). For the former, we arbitrarily assume a value of 1 µg/ml. Although we cannot directly estimate the parameter e, for our simulations we use values of e and a maximum resource concentration R that, along with the estimated v and assumed k, provide a reasonable fit to the population growth and saturation (stationary phase density) of wild type S. thermophilus.
Phage infection
The latent period λ and burst size β were estimated from one-step growth experiments [28]. A late-log culture of wild-type S. thermophilus was mixed with an equal volume of LM17Ca for a total volume of 0.9 ml and incubated at 42°C for 10 minutes, at which time 0.1 ml of a LM17Ca-diluted wild-type lysate of 2972 was added. The cell concentration was approximately 2×108 per ml while the phage titer was 106 phage per ml. The culture was incubated for 15 minutes at 42°C and then serially diluted in LM17Ca broth for cell densities and phage titers of approximately (a) 2×105 and 103, (b) 2×104 and 102, and (c) 2×103 and 101. At periodic intervals, 100 µl samples from (a), (b), and (c) were each incubated with 1 ml of a late-log culture of wild type S. thermophilus and the number of phage particles was estimated from soft LM17Ca agar lawns. Based on this protocol, the latent period ended between 25 and 30 minutes, thus λ is approximately 0.4 hour. Previous studies [7], [32] estimated the latent period of phage 2972 between 34 to 40 minutes. For the burst size, we use the difference between the mean estimated phage densities in the time intervals 15–25 minutes (before the burst) and 40–60 minutes (after the burst). β was estimated to be approximately 80 particles per infected cell. A previous study estimated the burst size of phage 2972 at 190±33 new virions per infected cell [7]. Presumably these discrepancies can be attributed to differences in the growth conditions, including media.
There are a variety of ways to obtain independent estimates of the adsorption rate constant δ, but it is usually through the decline in the titers of free phages in bacterial cultures [28]. These methods require separating plaque-forming units (which include infected cells) from free phages. While we tried various protocols, the results were variable, presumably due to the production of exopolysaccharides by the strain DGCC7710 [33], [34]. Moreover, since relatively high densities of cells were required for these estimates (which in theory could be made with BIMs as well as sensitive cells), the physiological state of these bacteria may be different from that of the rapidly growing cultures. Thus, instead of independently estimating this parameter, we used the value of δ that visually provides a good fit for this model for short-term bacterial growth and phage replication experiments with wild-type phage and bacteria. For this fit, we use the above estimates of maximum growth rate (v), latent period, and burst size. In this way, δ is the only fitted phage infection parameter (Figure 2).
Fig. 2. Short-term bacterial and phage growth dynamics in the absence of immunity: change in the density of WT bacteria and WT phage. 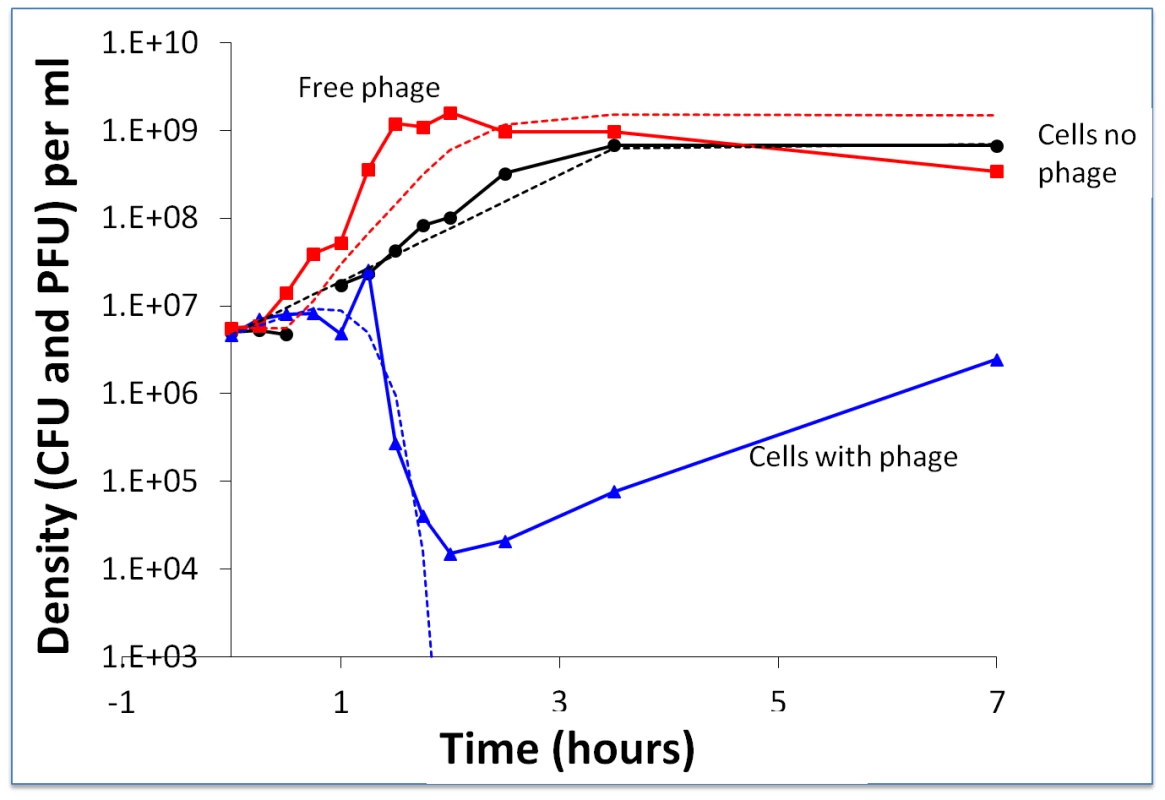
The broken lines are the densities predicted by the above model with the following parameters: v = 1.4 hr−1 per hour, e = 5×10−7 µg per cell, R (initial resource concentration) = 350 µg/ml, δ = 8×10−8 ml per cell per phage per hour, β = 80 phage particles per infected cell and, λ = 0.4 hours. At 24 hours, the estimated densities of bacteria and phage in these cultures were, respectively, 5×108 cells per ml for the control and 3.2×105 CFU and 6.2×107 PFU for the bacteria and phage in the mixed culture. While the observed and predicted dynamics of the decline in the density of bacteria and the increase in phage titer during the first two hours were similar, from the perspective of the longer-term population dynamics, the most significant difference between the observed and predicted dynamics was the re-ascent and persistence of the bacterial population. In accordance with the model, all of the bacteria should have been killed after two hours of exposure to the phage. The results of the spot tests, made with WT phage and cultures derived from single bacterial colonies recovered from the above at 7–9 and 24–26 hours, showed that these bacteria were still sensitive to the WT phage (more than 20 independent colonies from separate experiments). Stated another way, there was no evidence for BIMs evolving and ascending to dominance in these cultures. This qualitative deviation from what was anticipated under the canonical perspective of the dynamics of lytic phage infection and bacteria with CRISPR–cas immunity set the stage for experiments described in the following.
Rate of BIM formation (spacer acquisition)
The failure of BIMs to ascend in the experiment depicted in Figure 2 could be due to the rate of BIM formation being too low for these resistant cells to be present in the WT population. Our results suggest this is not the case. BIMs can be readily isolated from lawns initiated with mixtures of 108 phage and 107 cells, and as we demonstrate below. Even when BIMs resistant to the phage are present, they do not ascend in populations dominated by bacteria sensitive to the phage. The exact rate at which BIMs are generated is not clear from our results.
On first consideration, it would seem straightforward to estimate the probability that an infection of a sensitive bacterium with a virulent phage would result in immunity (the acquisition of a spacer) rather than a lytic infection (this probability is the parameter m in the above model). Thus, by incubating a mixture of WT cells and WT phage for a defined period, plating the mixture and counting the number of colonies, from the estimated P0, B0, and δ, it should be possible to estimate m. When we did this experiment, the number of surviving colonies varied with the numbers of cells and phage plated, but not in a way anticipated from this model if all of the BIMs were formed in liquid. For example, there were more surviving (resistant) colonies when lower densities of the WT phage and bacteria were plated. We interpret the results of these experiments and some of those that follow as support for the hypothesis that processes other than those considered in the above model affect the rate of BIM formation. In particular, the generation and ascent of BIMs depends on the experimental conditions in ways that we consider below.
Frequency of CEM production
In our model, we assume that CEM phage capable of growing on BIMs are produced upon burst of an infected bacterium. While, in our experiments, we did not estimate the rate at which these CEMs are generated, we estimated their frequencies in lysates. Estimating mutation rates, rather than frequencies, in phage requires more information about the way these viruses replicate in infected cells than we currently have for phage 2972 and its S. thermophilus host. To estimate the frequency of CEMs capable of replicating on the first-order BIMs, we prepared independent single-plaque lysates of WT phage and estimated the number of particles per ml that formed plaques on 10 different BIMs (BIM1 through BIM10). These 10 BIMs are different from each other based on their newly acquired spacers (see below). With the exception of one WT lysate and the lawns of BIM7 and BIM8, the relative frequency of first-order CEM phage in WT phage lysates was between 5×10−7 and 5×10−5 (Figure S1). We also obtained single estimates of the frequencies of second-order CEMs from the ratio of the numbers of plaques formed by first-order CEMs on lawns of second-order BIMs and WT lawns, respectively. In a similar experiment we did with the five second-order BIMs and their corresponding first-order CEM phage, the frequency of second-order CEMs also ranged from 5×10−7 to 4×10−6 (Figure S1).
Molecular characterization of the BIMs and CEMs
Genetic analysis of the two active CRISPR loci of ten first-order BIMs revealed that they each had acquired a novel spacer in either the CRISPR1 or CRISPR3 locus. Five BIMs acquired one unique novel spacer in the CRISPR1 locus and 5 BIMs acquired one unique novel spacer in the CRISPR3 locus (Figure 3). Similar analysis of the five second-order BIMs revealed that they each had also acquired an additional novel spacer in the CRISPR1 or CRISPR3 locus. Two of these second-order BIMs acquired one additional unique novel spacer in the CRISPR1 locus, indicating iterative addition into the CRISPR1 locus (BIM32 and BIM52 in Figure 3). Likewise, three second-order BIMs acquired novel spacers in the CRISPR3 locus, including two variants with iterative build-up of the CRISPR3 locus (BIM22 and BIM82 in Figure 3). Interestingly, BIM72 acquired a CRISPR1 spacer to become a first-order CEM and a CRISPR3 spacer to become a second-order CEM, illustrating the duality of active CRISPR loci in Streptococcus thermophilus.
Fig. 3. Graphical representation of spacers across the two CRISPR loci for S. thermophilus BIMs. 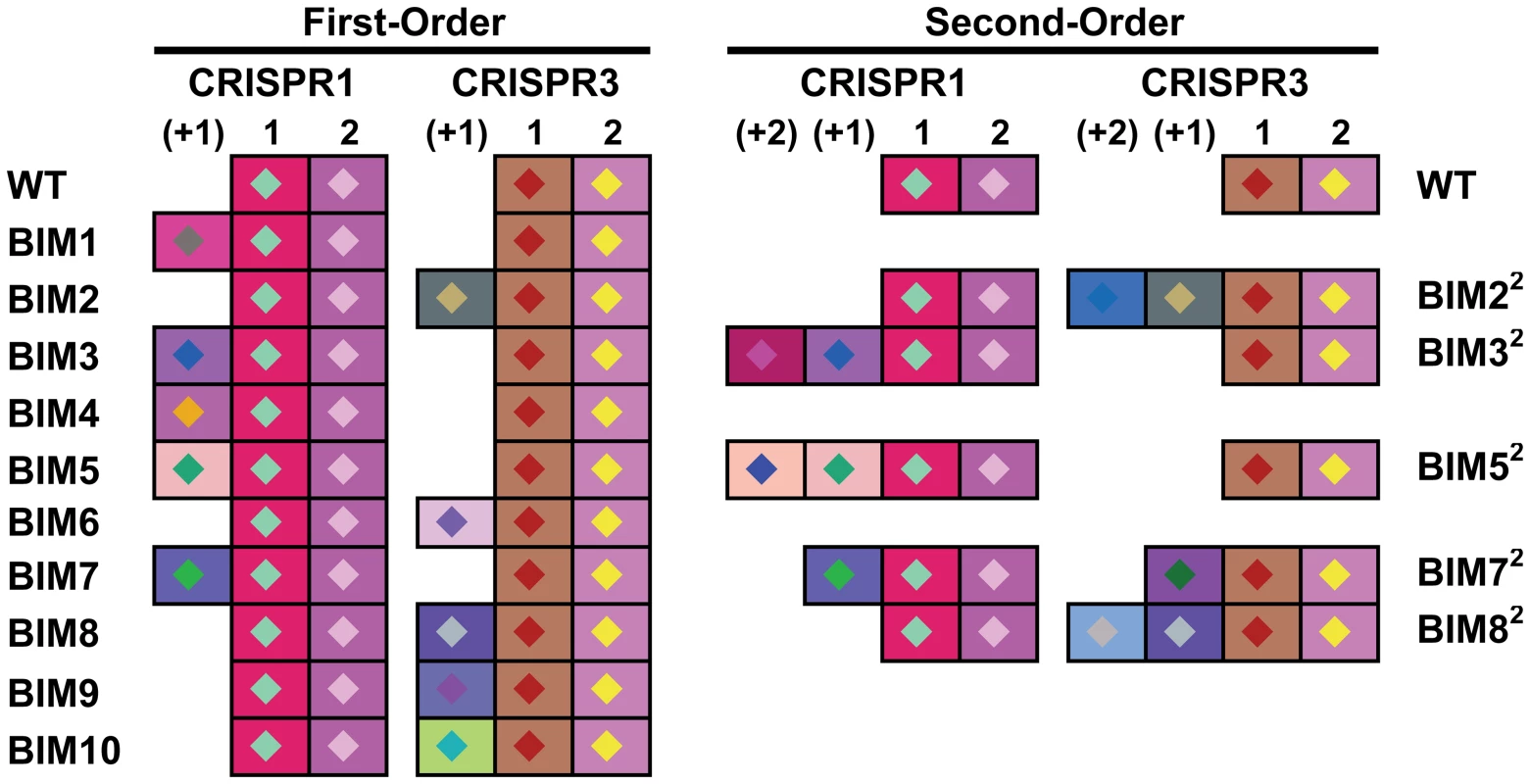
Repeats are not included; only spacers are represented. Each spacer is represented by a combination of one select character in a particular color, on a particular background color, as previously described [36]. The color combination allows unique representation of a particular spacer. Similar color schemes (combination of character color and background color) represent identical spacers, whereas different color combinations represent distinguishable spacers. Sequence analysis of all the CRISPR1 and CRISPR3 spacers acquired in the first and second rounds of phage challenges indicated perfect matches to the WT genome of phage 2972 used in the challenge (Table S1). Further, all new CRISPR1 and CRISPR3 spacers were associated with protospacer-adjacent motifs (PAMs): NNAGAAW and NGGNG, respectively (Table S1). These findings are consistent with previous results indicating that spacer sequences are directly derived from phage DNA and are associated with PAMs [7], [8], [35], [36].
The results of our sequence analysis of the CEM protospacer and PAM regions are fully consistent with previous findings that single mutations in the matching protospacer or PAM are sufficient to circumvent CRISPR–mediated immunity (Figure 4). Of the 10 CEMs that evaded the immunity of the first-order BIMs, one had a mutation in the protospacer region and the remaining nine CEMs had mutations in PAM sequences (four in the CRISPR1 PAM and five in the CRISPR3 PAM). Each of the five second-order CEMs that evaded the immunity of the second-order BIMs had acquired a second mutation, as expected. One had acquired a second mutation in a protospacer, two had mutations in the PAM, one contained one mutation in the protospacer and another in the PAM, and the last one had an insertion in the PAM. Taken together, these sequencing data are consistent with previous results indicating that CEMs are the product of mutations in the protospacers and/or PAMs which enable these viruses to circumvent CRISPR–cas acquired immunity [7], [8].
Fig. 4. Nucleotide sequences in wild-type and mutant phages that correspond to the newly acquired spacers by the S. thermophilus BIM strains. 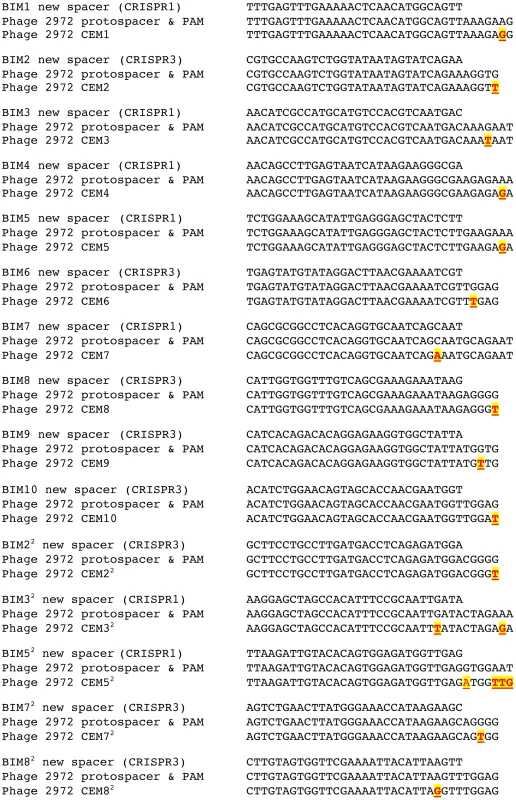
Each mutant is highlighted in red and yellow, bold and underlined. Population dynamics
Optical density data and the criteria for resource - and phage-limited cultures
Although we estimated cell and phage densities for specific samples by serial dilution and plating, much of our inference about the population dynamics of these bacteria and phage comes from optical density (OD 600 nm) data. In these experiments, we follow the changes in OD of cultures of bacteria and phage with frequent sampling for 8 or so hours. To illustrate how the changes in optical density are related to changes in cell density and what we would anticipate for these changes in resource - and phage-limited cultures, we inoculated 4 ml broth with 40 µl of an overnight culture of WT cells (for a final concentration of approximately 2×106 cells per ml), with different initial concentrations of phage, and without phage for the control.
As anticipated (intuitively as well as from the model), the time before the bacterial density declines due to the phage is inversely proportional to the initial density of these viruses (Figure S2). As predicted by the model, the optical densities of the cultures with phage converge to a level well below that of the phage-free control. However, the times required for these convergences to occur in our experiments are greater than predicted.
Phage and sensitive bacteria
To determine whether the failure of BIMs to emerge when sensitive bacteria are confronted with phage is a general result, we mixed 2×106 WT cells, first-order BIMs, and second-order BIMs with approximately 2×106 WT phage, first-order CEMs, and second-order CEMs, respectively, and followed the changes in OD for two transfers. The results of these experiments are presented in Figure 5.
Fig. 5. Changes in optical density of cultures with S. thermophilus and the corresponding phages to which these WT cells or BIMs are sensitive. 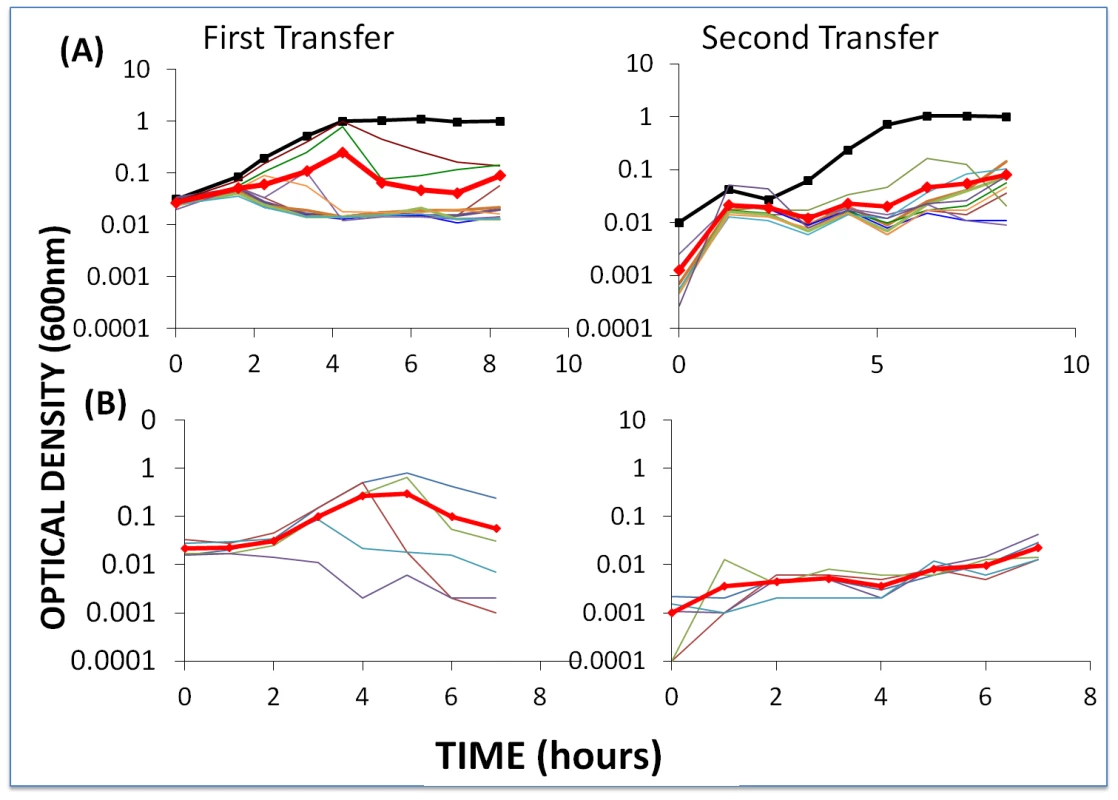
The first transfer was initiated with 40 µl of an overnight culture (for approx. 2×106 cells per ml) and phage (approx. 2×106 per ml) in 4 ml LM17Ca medium. The second transfer was initiated by adding 40 µl of the 24-hour first-pass cultures to 4 ml fresh LM17Ca. The heavy black lines with square tick marks indicate phage-free WT controls. The heavy red lines with diamond tick marks are the arithmetic mean ODs for the cultures with bacteria and phage. Panel A: WT S. thermophilus and WT phage (light blue) and nine first-order BIMs and their corresponding CEMs. Panel B: five second-order BIMs and their corresponding CEMs. During the first transfers, the OD of the cultures with phage initially rose and then declined. In the second transfer, these ODs remained at least an order of magnitude less than those of the phage-free controls. The initial densities of phage in the first transfer cultures ranged from 1.2×106 pfu/ml (BIM4) to 3.4×107 pfu/ml (BIM10). With the exception of BIM1, the estimated cell densities in these cultures with phage were between 6.0×104 cfu/ml (BIM7) and 1×107 cfu/ml (BIM6). In the absence of phage, the average 24-hour density of wild type cells and BIMs exceeded 2×108 cfu/ml. This experiment was repeated at least three times and, qualitatively, the same results were obtained; the cultures with phage remained at optical densities in the range we consider to be phage - rather than resource - limited.
To determine whether BIMs resistant to the phage in these cultures emerged, a similar experiment was performed without frequent sampling. Single colonies were taken from each of the first - and second-order BIM-CEM cultures at end of the first transfer. These colonies were re-streaked and grown up in liquid culture and, as with the wild type bacteria, were spot tested for resistance to the corresponding CEMs. While we can't rule out minority populations of BIMs resistant to these phages, all of the colonies tested were sensitive to the phages.
Resource - or phage-limited densities
In the preceding, we use the phrase “phage-limited” to describe situations where the ODs of bacterial cultures with phage remain substantially less than those they would achieve were the phage not there and the bacteria limited by the availability of resources. Implicit in the phrase “phage-limited” is the assumption that there is an abundance of resources that could be used by the bacteria for growth were their densities not limited by the phage or products of phage replication. To test this phage-limited hypothesis, we inoculated batch cultures with wild type, first-order, or second-order BIMs and phage capable of growing on these bacteria. After 24 hours, we transferred the cultures to fresh medium (1/100 dilutions) and maintained the original and diluted cultures, estimating the optical density each day. All were in the “phage-limited” range (approximately 0.2 or lower). At 72 hours, we divided these 4 ml cultures in half. To one half we added 40 µl of an overnight culture of another industrial S. thermophilus strain (SMQ-301), which is unrelated to DGCC7710 and is insensitive to phage 2972. After 24 hours of incubation, we estimated the OD of the cultures with and without SMQ-301. At that time, the original culture was 72 hours old and the diluted culture 48 hours old.
The ODs of the cultures with SMQ-301 are substantially greater than those without these distinct cells. The extent of this residual growth is inversely proportional to the density of the culture to which these SMQ-301 cells are added. The linear regression coefficient for 31 pairs of cultures with and without SMQ-301 is −2.53, (F(1,29) = 24.6, p<2.7×10−5). We interpret these results as direct evidence that the phage, rather than resources limit the density of bacteria in cultures maintained at lower optical densities (see Table S2).
Population dynamics of phage and BIMs with single-spacer resistance
As suggested earlier, one reason BIMs do not ascend and take over cultures with sensitive bacteria and phage (Figure 2 and Figure 5) might be that they are not generated in the sensitive population (i.e. the rate of BIM formation is too low). If BIMs are originally present in cultures of bacteria sensitive to the dominant population of phage, it would seem that unless they are countered by phage mutants (CEMs) capable of replicating on them, these resistant BIMs would ascend to dominate the cultures. Moreover, because the phage would adsorb to these resistant BIMs, phage densities would be expected to wane and eventually the culture should be cleared of phage. If, however, CEM phages are initially present or are generated during the course of replication, these phages would be expected to ascend and the BIM population would be reduced and potentially eliminated. To illustrate these predicted results, we use our model and consider two situations: one in which a BIM is the only bacterial population and is resistant to all but a single phage particle, and one where there are no CEM phage and initially equal densities of the BIM and WT cells upon which the phage can grow and produce CEMs (Figure S3).
First scenario: in the absence of bacteria upon which it can replicate, the density of P0 phage continues to decline because of adsorption to the immune B1. The rare P1 population, which can grow on B0 and B1, ascends and kills off the B1 population (Figure S3A). Second scenario: although they are not initially present, during the course of replication on B0, the CEM phage P1 arise and ascend. Both populations of bacteria are eliminated and both wild type and CEM phage persist (Figure S3B).
To test these hypotheses for situations where the bacteria are resistant to the dominant population of phage, we inoculated cultures with first-order BIMs and WT phage and second-order BIMs with their corresponding first-order CEMs, e.g. CEM2 with BIM22 (approx. 2×106 phage and cells per ml). As in the experiment depicted in Figure 5, we followed the changes in optical density and at 24 hours diluted the cultures 100 fold in LM17Ca and followed the changes in OD for this second transfer. The results of this experiment are presented in Figure 6.
Fig. 6. Changes in optical density of cultures with S. thermophilus and high initial densities of phage (approx. 5×106/ml) to which the BIMs were resistant. 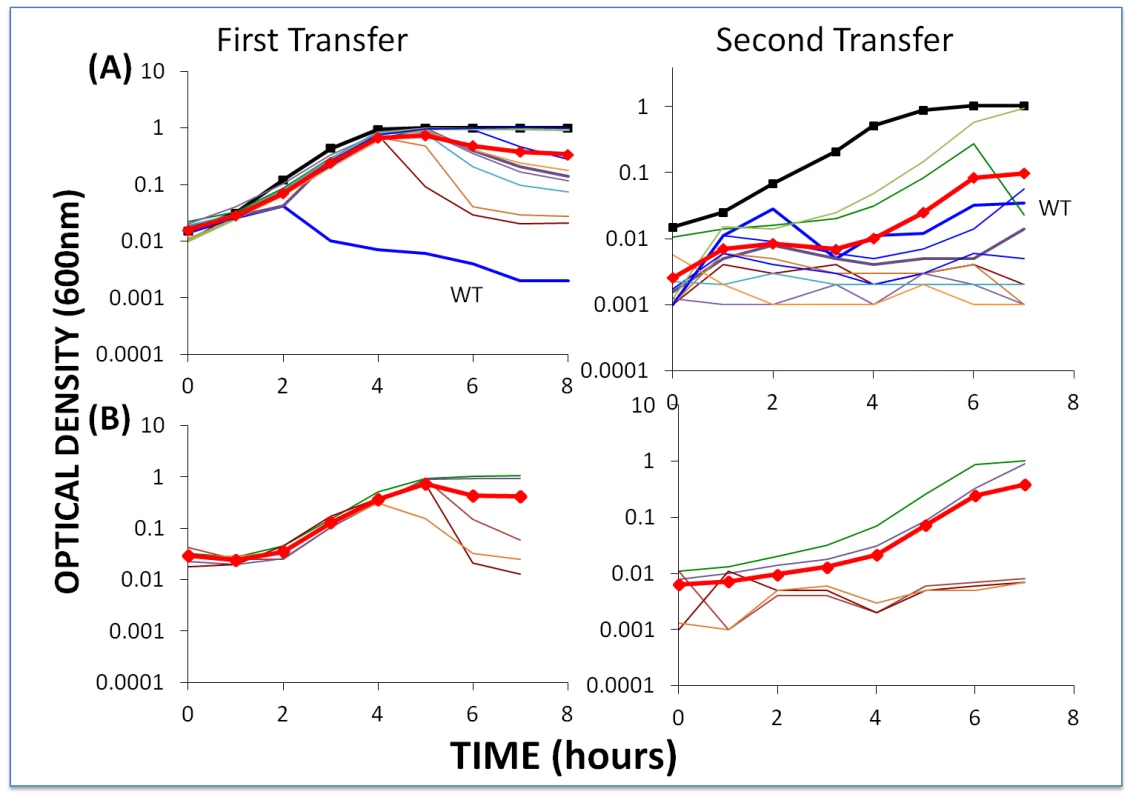
The first transfers (left) were initiated with 40 µl of overnight bacteria and phage in 4 ml LM17Ca medium. The second transfers (right) were initiated by adding 40 µl of the 24-hour first-pass cultures to 4 ml fresh LM17Ca. Panel A: WT phage with WT cells (medium-weight blue line) and 10 first-order BIMs. Panel B: five second-order BIMs and their corresponding first-order CEM phages. The heavy black lines with square tick marks are phage-free WT controls and the heavy red lines with diamond tick marks are the arithmetic mean densities of all the cultures with phage. Two lines of evidence support the hypothesis that the failure of these BIMs to ascend to the OD level of phage-free cultures and the decline in their densities can, at least in part, be attributed to the emergence of CEM phage capable of replicating on these resistant cells. First, based on our estimates of the frequency of CEMs in WT lysates (Figure S1), we would anticipate one or more CEMs in the original inoculum. Second, and more directly, the phage recovered at 48 hours generated clear zones on lawns of WT cells and lawns of their respective BIMs. For example, clear zones were noted when lysates from the culture initiated with WT phage and BIM1 were spotted on BIM1 lawns and when cultures initiated with CEM2 and BIM22 were spotted on BIM22 lawns.
To further test the hypothesis that CEMs will emerge and ascend only when single-spacer resistant BIMs are exposed to phage populations that are large enough to have CEMs bearing mutations in the protospacer (or PAM) targeted by the BIMs, we performed the above experiment with low initial densities of phage. Based on the estimated frequencies of CEMs, with the low initial numbers of phage in these cultures (approximately 2×10−4) we would not anticipate preexisting CEMs with the needed mutations. As a positive control, we performed these experiments with second-order BIMs and low densities of WT phage (Figure 7). Since second-order BIMs are protected by two spacers, the generation of second order CEMs – which would require mutations in two different protospacers in order to permit infection of second-order BIMs – is not anticipated. The results of this assay are presented in Figure 7B.
Fig. 7. Changes in optical density of cultures with S. thermophilus and low initial densities of WT phage. 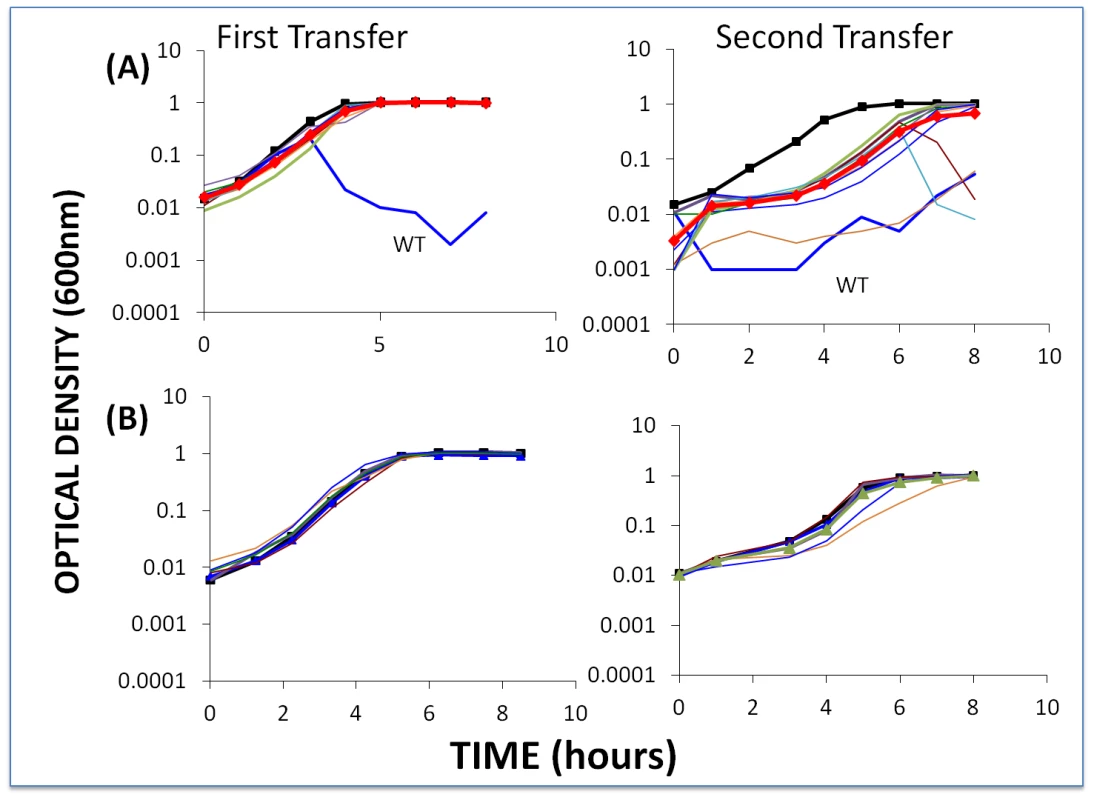
The first transfer was initiated with 40 µl of an overnight culture of bacteria (approx. 2×106 cells per ml) and WT phage (approx. 2×104 ml) in 4 ml LM17Ca medium. The second transfer was initiated by adding 40 µl of the 24-hour first-pass cultures to 4 ml fresh LM17Ca. Panel A: WT phage with WT cells (medium-weight blue line) and 10 first-order BIMs. Panel B: WT phage with five second-order BIMs. The heavy black lines with square tick marks are phage-free WT controls. The heavy red lines with diamond tick marks are the arithmetic mean densities of all BIM cultures with phage. The second-order BIMs with low densities of WT phage (Figure 7A) behaved as anticipated. Moreover, in our enrichment experiments with 100 µl of overnight cultures of sensitive bacteria added to growing cultures of second-order BIMs, we were unable to find CEMs capable of growing on the second-order BIMs. And due to adsorption to the second-order BIMs (and intracellular cleavage of their genome by the CRISPR–Cas system), by the end of the second transfer there were few (less than 102) WT phage in these cultures. Indeed, for three of the five second-order BIMs, we were unable to recover any phage at the end of the second transfer in enrichment experiments when 100 µl filtrates added to cultures with WT cells. In essence, phage cannot initiate an arms race when confronting BIMs that have acquired two or more spacers matching parts of the phage genome, despite the high rate of CEM generation.
The results obtained with low densities of WT phage and first-order BIMs are inconsistent with what our model predicted. Although there was no evidence for CEMs evolving in the majority of cultures with first-order BIMs and low densities of WT phage, they did emerge in two of the 10 first-order BIM cultures (BIM2 and BIM8). Furthermore, in a replicate of this experiment, CEMs emerged in 3 out of 10 cultures with first-order BIMs (BIM2, BIM3 and BIM7). In all cases where these CEMs emerged, the cultures became phage - rather than resource-limited. At this juncture we do not know why, in the second transfer, the growth of the bacteria in cultures for which we did not isolate CEM mutants (Figure 7A) was delayed relative to the phage-free controls. One possibility is that CEM mutants in these cultures delayed the replication of the bacteria, but were undetectable at 8 hours.
Population dynamics with first - and second-order BIMs with low densities of WT phage and WT cells
It seems reasonable to assume that if bacteria with CRISPR–cas immunity were invading a habitat with phage present, there would also be sensitive bacteria upon which these phage would be replicating. To mimic this situation, we performed experiments with low initial densities of phage but also with WT cells at densities similar to those of the BIMs. The results of one of these experiments are presented in Figure 8.
Fig. 8. Changes in optical density of cultures with S. thermophilus first- and second-order BIMs and WT cells with low initial densities of WT phage. 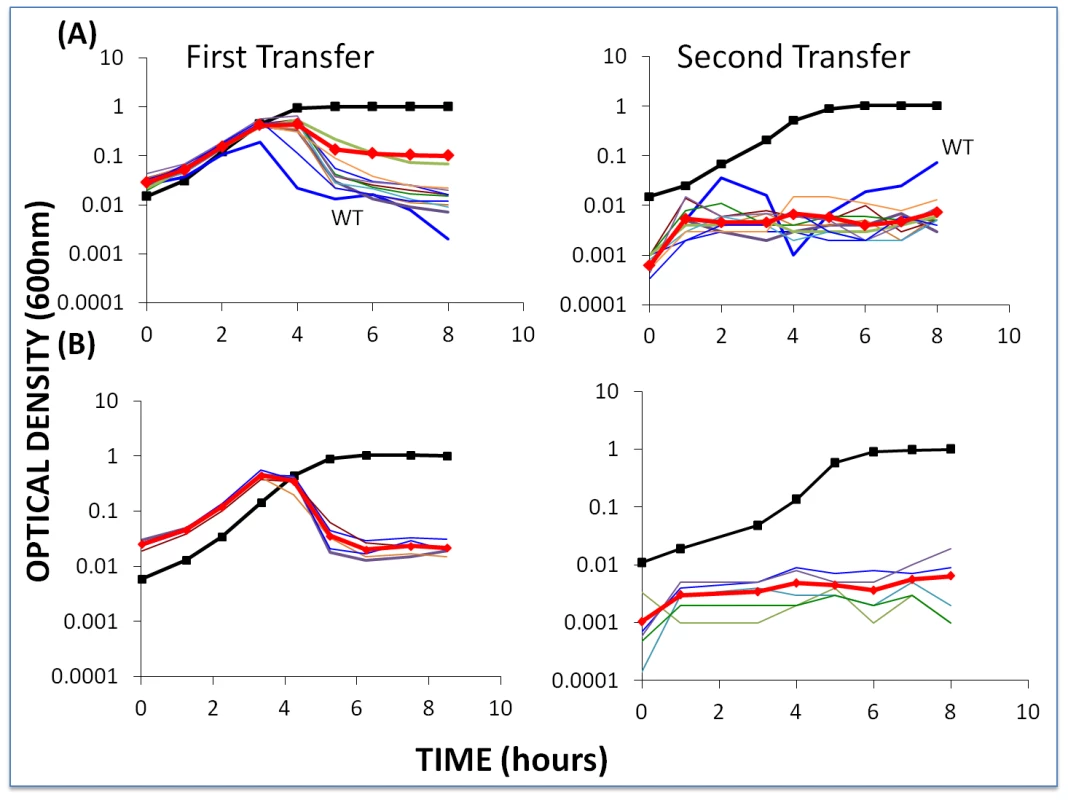
The first transfer was initiated with 40 µl of an overnight culture of each BIM (approx. 2×106 per ml) and WT cells (approx. 2×106 per ml) and low densities (approx. 2×104 per ml) of WT phage in 4 ml LM17Ca medium. The second transfer was initiated by adding 40 µl of the 24-hour first-pass cultures to 4 ml fresh LM17Ca. Panel A: WT phage with WT cells and 10 first-order BIMs. Panel B: low densities of WT phage (approx. 2×104 per ml) with WT cells and five second-order BIMs. The heavy black lines with square tick marks are the phage-free controls (WT cells in (A) and BIM22 in (B)). The heavy red lines with diamond tick marks are the arithmetic mean densities of all the BIM cultures with phage. Within the first transfer for both first - and second-order BIMs, the density of the bacteria declined to phage-limited levels. In accordance with our model, we would anticipate this result for the first-order BIMs due to the generation of CEMs in the course of replication of the phage on the wild-type host. Even if they were not present at the start of the experiment, first-order CEMs are likely to emerge whilst the phage are replicating on the WT cells. On the other hand, they would not be expected to emerge in the cultures with second-order BIMs. And, indeed, we have been unable to detect the presence of second-order CEMs at the end of the 2nd transfer of the above experiment. As we discuss below, processes other than the emergence of second-order CEMs are responsible for the failure of second-order BIMs to ascend in these cultures with WT phage and WT cells. It should, however, be noted that, at least for the second-order BIMs, the phage-limited state observed for the first 8 hours of the second transfer was not maintained. At 26 hours, two of the five cultures in this second transfer were already as turbid as the phage-free controls and the remaining three were on their way to being so.
Accounting for the deviations from the predictions of the model
At best, the mathematical model and computer simulations employed here are simplistic caricatures of the population dynamic and other processes occurring in these experiments. They are not intended or anticipated to provide quantitatively precise predictions of the changes in density of bacteria and phage observed in our experiments. Some quantitative deviation between experiments and theory is expected. Not expected, however, are major qualitative deviations. There were two of these. One is the failure of the phage to eliminate the sensitive bacteria. The other is the failure of BIMs to ascend and dominate cultures with an abundance of resources. We postulate these failures can be attributed to either the phage or the products of phage replication generating phenotypic immunity among sensitive cells (persistence) [37] and killing emerging BIMs.
One line of evidence in support of the hypothesis that something(s) in the phage lysate is preventing BIMs from ascending and also generating persisters comes from results obtained with 24-hour cell-free (filtered, rather than chloroformed) lysates of WT phage. S. thermophilus cells resistant to the phage (BIM22 and SMQ-301) were added to these lysates and the optical and viable cell densities were followed with frequent sampling. In this experiment, 40 µl of BIM22 or SMQ-301 from overnight cultures were added to 4 ml of the phage lysate. As phage-free controls, these bacteria were added to 4 ml LM17Ca (Figure 9). Cells of SMQ-301 were able to grow in these WT phage lysates, thereby indicating that there were plenty of resources. However, the viable cell density of the BIM22 cells declined. At 24 and 31 hours, the optical density of the BIM22 culture in the phage lysate was less than 0.02 and the viable cell densities were, respectively, 4.7×105 cfu/ml and 1.2×106 cfu/ml. At 48 and 54 hours, these viable cell densities were, respectively, 5.6×105 cfu/ml and 2.1×105 cfu/ml. The sensitivity of BIM22 to the phage and/or molecules in the WT lysates was not shown by SMQ-301.
Fig. 9. Changes in optical and viable cell density of S. thermophilus in cell-free lysates of wild-type phage. 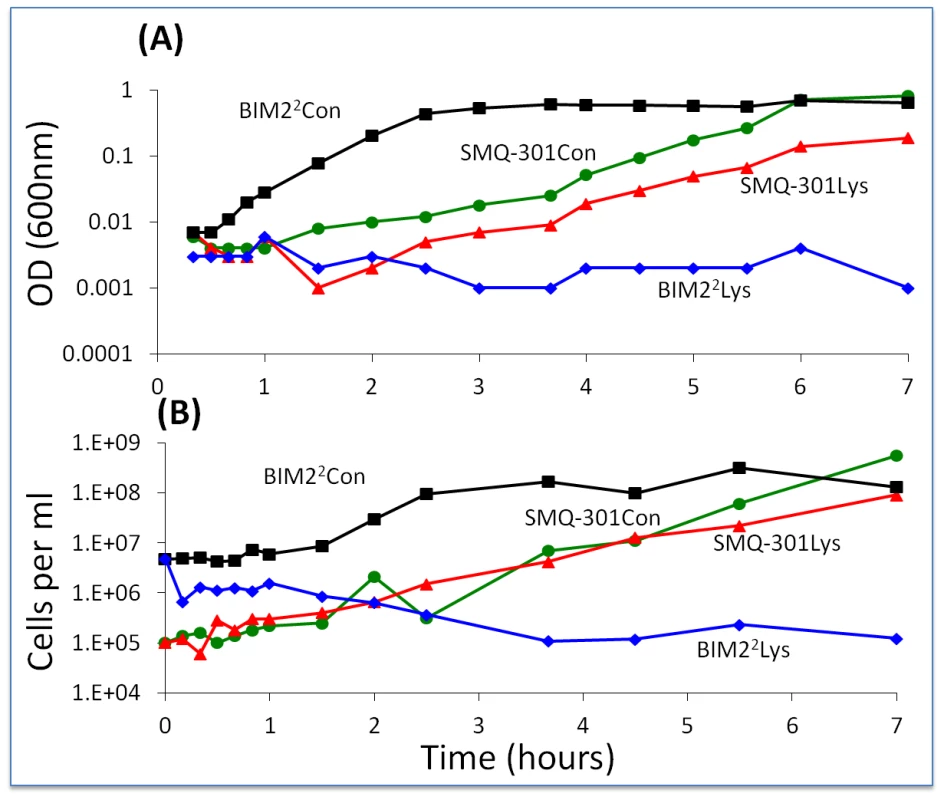
BIM22 and SMQ-301 in a full concentration phage lysate (1.5×108 pfu/ml) and an LM17Ca control (1/100 dilution). Con – LM17Ca control; Lys – cell-free phage lysate. Panel A: optical density. Panel B: estimated cell density. A second line of evidence in support of the hypothesis that phage or the products of their replication or lysis of infected cells prevent the ascent of BIMs is the results of our experiments with WT phage in cultures with first and second-order BIMs. Batch cultures inoculated with low densities of WT phage and bacteria that are resistant to those phages by one or two spacers become resource-limited. However, if we allow for considerable phage replication by adding WT cells, cultures with first - and second-order BIMs become phage-limited (Figure S4).
We have yet to characterize the agent(s) responsible for killing and/or inhibiting the growth of infected BIMs. However, preliminary results from experiments with lysates prepared from French Press extracts suggest that this agent is likely to be present within the cells and need not be a product of phage infection. The growth rates of resistant BIMs are markedly reduced when cultured in mixtures of medium and French Press extracts of phage-free cultures of wild type cells (Text S1). In this interpretation, this agent is released as a product of lysis of the cells by the phage.
An extended model
Never believe an experiment until it has been confirmed by a theory. (attributed to Sir Arthur Eddington)
To explore, in a more quantitative way, whether the mechanisms postulated above, where products of phage replication generate phenotypically resistant persisters and kill BIMs, can account for the observed dynamics, we use an extension of the basic model described in the Theoretical Framework. In this extension, we consider three bacterial populations with densities B0, B1, and B2, and two populations of phage, P0 and P1, respectively corresponding to WT, first - and second - order BIM bacteria, and WT and first-order CEM phage. For convenience, we assume there are agents of collective concentration LY µg/ml that kill resistant cells and provide transient immunity to sensitive bacteria (persistence effect [38], [39]). Since the effects of LY in killing resistant cells and generating persisters are governed by separate parameters, they can be at least two different compounds. These agents are produced at a rate proportional to the product of the densities of replicating phage and the corresponding sensitive bacteria:
where vL is a rate parameter for the production of LY. We assume that the rates of growth of the bacteria now depend on the concentration of LY, as well as the resources, R. For example, the per capita growth of B0 is now where ν is the maximum growth rate of this strain, k the Monod constant, η the maximum rate of killing by LY, and KL the concentration of LY where killing is at its maximum rate. In the simulation LY/KL is set equal to 1 when LY exceeds KL. To account for phenotypically resistant B0 and B1 bacteria, we assume there are populations of non-dividing, persistent cells of densities BP0 and BP1 bacteria per ml that are produced from their respective dividing populations at a rate proportional to LY/KL and a constant (g) and return to the dividing state at a constant rate (h). For example, the rate of change in the density of the wild type (B0) population is now The Berkeley Madonna simulation used for this extended model can be obtained from www.eclf.net/programs.In Figure 10, we present the results of simulation experiments initiated with wild type phage (P0), wild type bacteria (B0), and a second-order BIM (B2). The population growth, initial resource concentration and phage infection parameters used in this simulation are same as those used in earlier simulations and thus in the range estimated in these experiments. The parameters for the production of LY and its action are arbitrary and chosen to illustrate the principle rather than fit specific data in any but a qualitative way.
Fig. 10. Simulation results of the extended model. 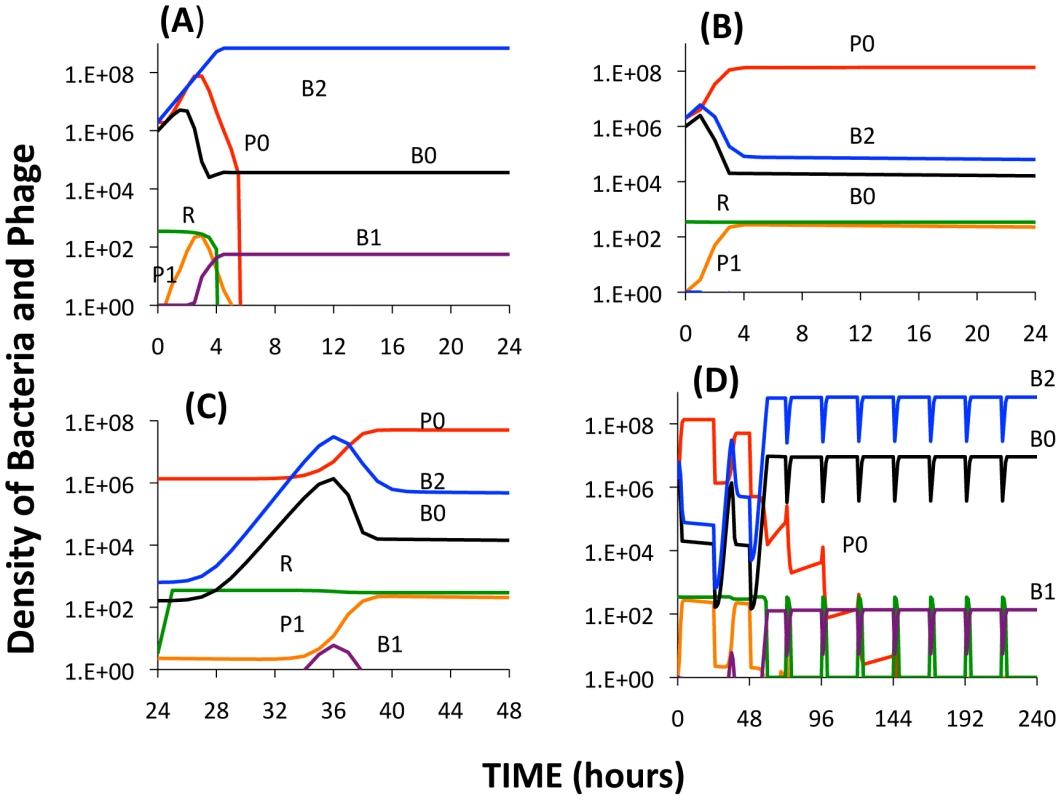
Changes in the densities of bacteria and phage in cultures inoculated with WT phage (P0), second-order BIMs (B2), and WT cells upon which the phage can replicate (B0). The first-order BIMs (B1) and first-order phage (P1) are respectively produced by spacers added to the CRISPR region and generated by mutation, with probabilities of 10−6 per cell or phage per hour. The variable R is the concentration of the limiting resource. The bacterial growth and phage infection parameters in these simulations are the same as those in Figure 2. Save for (A) the parameters for the production and action of LY and persistence are as follows: vL = 10−6, KL = 106, η = 1.4, g = h = 0.01. The initial concentration of the resource at the start of a transfer is 350 µg/ml. The cultures were initiated with 2×106 B0 and B2 cells and 2×106 P0 phage. Panel A: No LY produced (vL = 0). Panel B: The first transfer of the culture. Panel C: The second transfer. Panel D: sequential transfers (approx. 2×104). In the absence of the killing effect, Figure 10A, the B2 population ascends and the phage are eliminated. The B0 population continues to be maintained, B1 bacteria and P1 phage are produced, and within short order the population becomes resource-limited.
During the first transfer of the simulations with the killing effect (Figure 10B), the P0 phage ascend and the densities of the B0 and B2 populations decline and the community becomes phage limited. While the second transfer (Figure 10C) also ends in a phage - limited state, the relative density of B2 is now greater. In subsequent transfers (Figure 10D), B2 ascends to dominance, phage are eliminated, and the community becomes resource-limited. As a consequence of the serial passages, the molecule killing BIMs is diluted out and the BIMs ascend. For the arms race to continue, the phage would have to generate CEMs capable of replicating on the dominant population of BIMs.
Preliminary supporting experimental results
While data may be a crutch to the insecure, really self-confident scientists subject their hypotheses to tests that will reject them. (www.eclf.net)
If the above postulated effect of dilution in serial passage is valid, in successive transfers of experiments initiated with WT cells and WT phage, BIMs should emerge. And, unless they are countered by CEM phage, they will ascend to dominance and the phage will be lost. To test this hypothesis, we mixed WT cells with phage and each day transferred the culture to fresh medium (40 µl into 4 ml). We initiated six separate cultures from the same mixture of approximately 2×106 cells and phage. As a control we serially transferred a phage-free WT culture. At the end of each transfer, we estimated optical densities of these cultures and, by serial dilution and plating, the viable densities of bacteria and free phage. The results of this experiment are presented in Figure 11.
Fig. 11. Serial transfer experiment initiated with WT phage and WT cells. 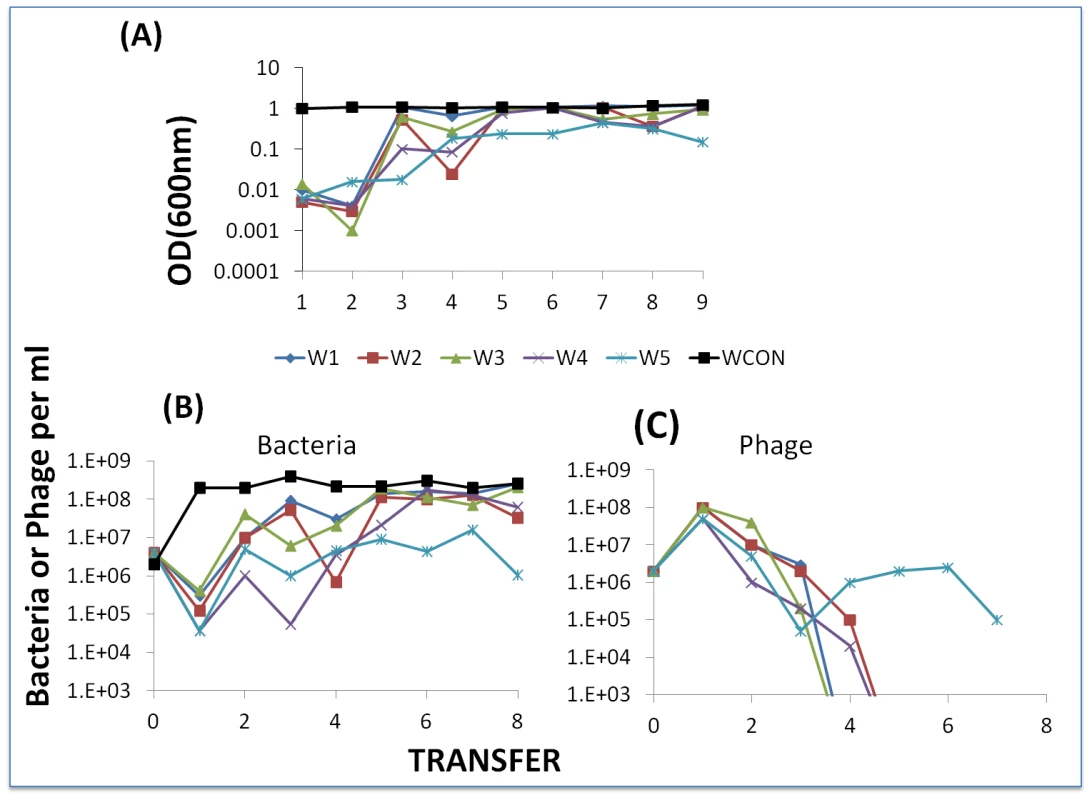
Panel A: Optical densities at the end of successive passages. Panel B: estimated viable cell densities at the end of successive transfer. Panel C: estimated free phage densities at the end of successive transfers. We interpret the results of this experiment to be consistent with the hypothesis that as a consequence of successive dilutions of the compound killing BIMs, the resistant mutants ascend. Moreover, in 5 of the 6 cultures in which BIMs emerged, phage were lost and could not be recovered even with enrichment with WT cells (i.e. the arms race was terminated). Additional serial transfer experiments should be performed with different starting conditions and possible different media (perhaps milk) to explore the generality of this observation about the limit to a BIM-CEM arms race.
Discussion
We interpret the results of these experiments with S. thermophilus DGCC7710 and its virulent phage 2972, and the associated computer simulations, as support for the hypotheses that the CRISPR–cas systems can (i) prevent the invasion of phage into established communities of bacteria; (ii) enable a population of bacteria to become established and be maintained in communities with phage; and (iii) promote an arms race between phage-mediated selection for CRISPR–cas acquired immunity (BIMs) and immunity-mediated selection for CEMs capable of replicating on these BIMs. However, these experimental and computer simulation results indicate that the conditions under which these ecological and (co)evolutionary outcomes occur are restrictive in ways that are not predicted by the canonical view of phage infection and current understanding of CRISPR–cas immunity.
Although not surprising to the molecular biologist (RB) and microbiologist (SM) among us, it was particularly gratifying for the population-evolutionary biologists on this team (BRL and MB) to realize that the perspective of CRISPR–cas adaptive immunity one gleans from the simplified diagrams of reviews is a reality for the S. thermophilus DGCC7710 – phage 2972 system. Bacterial mutants resistant to WT phage (BIMs) were readily isolated. The resistance of all 10 BIMs examined can be accounted for by the acquisition of unique 30 base pair sequences (spacers) that correspond to regions of the phage genome. Phage capable of replicating on these first-order BIMs (CRISPR Escape Mutants, or CEMs) were also readily isolated. As anticipated from the canonical view of CRISPR–mediated immunity, the host range of these resistant viruses was restricted to the BIMs on which they evolved and their wild type ancestors, and can be attributed to mutations in protospacers (or PAMs) corresponding to the DNA sequences in the spacers acquired by their respective first-order BIMs. This iterative process of CRISPR–cas acquired immunity in the bacteria and CRISPR–escape mutation in the phage was also observed for the five second-order BIMs isolated from these first-order BIMs and their respective second-order CEM phage. How many orders of BIMs and CEMs can be generated by this iterative process of spacer acquisition by the bacteria and protospacer mutation in the phage remains unclear at this time and is a subject for future study.
Other than to say that first and second-order BIMs “are readily isolated,” for the reasons discussed below, we cannot provide estimates of the actual rate at which this phage resistance is acquired. What is clear is that CEMs are found at frequencies of 10−6 or greater in lysates. This seemingly high rate of CRISPR–escape mutation is particularly surprising because only a small fraction of the total sequence space in the protospacers-PAM region is responsible for generating these CEMs. It is tempting to speculate that as part of their adaptation to CRISPR–cas these phage have evolved a mechanism to elevate mutation rates.
On the other side, the results of our phage-host interaction experiments as well as our computer simulations indicate that there is more to the population and (co)evolutionary dynamics of phage and bacteria with CRISPR–mediated acquired immunity than can be inferred from the current perspective on this adaptive immune system. Moreover, qualitatively as well as quantitatively, there is more to these dynamics than can be inferred from models of the population dynamics of the interactions of bacteria and lytic phage based on the traditional view of phage infection [24], [25], [40].
First, although S. thermophilus BIMs can readily be picked up experimentally, even when present, these resistant bacteria do not reach measurable densities in liquid cultures with phage. For BIMs resistant through single spacers, one reason for this failure to ascend is the high rate of mutation to CEMs capable of replicating on and killing them. Most interestingly, CEMs emerge when the number of phage is too low to anticipate their presence in the original lysate or their generation during replication on sensitive cells. We postulate that the reason for this is that resistance due to a single spacer is not absolute and a fraction of the phage adsorbing to resistant cells replicate and produce CEMs. This interpretation is consistent with the previous observation that not all phage DNA molecules are cleaved by CRISPR–cas when infecting BIMs [8].
Another result not predicted by our simple model is that substantial densities of sensitive bacteria (in excess of 104 cells per ml) survive their encounter with phage. We postulate that the survival of these genetically phage-sensitive cells can be attributed to phenotypic resistance [37] induced by a product of phage infection. A phenomenon of this type has been demonstrated in Shigella dysentariae infected with the phage T7 [38], [39]. In the interpretation of this process presented in the review by Barksdale [39], a phage enzyme is released as a product of phage infection and strips the phage receptors off of the surviving bacteria, thereby making them phenotypically immune to phage infection. Whether a similar mechanism is responsible for the phenotypic immunity to phage 2972 of S. thermophilus remains to be seen, and will constitute a worthy study in its own right.
The results of our experiments indicate that BIMs resistant to phage due to the acquisition of two spacers can establish communities where phage are present and prevent the invasion of phage mutants into established populations of bacteria. The conditions for these outcomes are, however, restrictive. If the community includes sensitive bacteria upon which the phage are replicating, neither first - nor second-order BIMs will become established. We postulate that this can be attributed to a product of phage infection that kills and/or inhibits the replication of BIMs. As a consequence of phage replicating on WT bacteria, there would be high densities of phage and correspondingly high concentrations of this product. While we have yet to characterize this product, there is evidence for phage 2972 bearing genes that code for an endolysin [34]. On the other hand, the results of additional experiment we performed indicate that the agent(s) responsible for inhibiting the growth of BIMs are not phage-encoded products, but rather released by the lysed bacteria (see the Text S1).
Models of the population dynamics of bacteria and phage that incorporate the persistence of sensitive cells and the killing of BIMs due to chemicals generated during the course of phage infection can account for the qualitative inconsistencies between the results predicted by the simple model and those observed experimentally. This extended model predicts that in the course of successive transfers, the concentration of this chemical would be reduced and BIMs resistant to the phage would ascend to dominate the culture. This prediction is supported by the results of our longer-term serial transfer experiments initiated with WT bacteria. Also consistent with this interpretation are the results of a recent metagenomic analysis of experimental populations of S. thermophilus and the phage 2972 [41].
Hypotheses, caveats, and limitations
The results of this jointly theoretical and experimental study make at least five predictions about the contribution of CRISPR–cas immunity to the population and community ecology of bacteria and phage and the (co)evolution of these organisms.
-
A CRISPR–cas immune system can protect established populations of bacteria from invasion by phage, but only if all of the bacteria carry at least two spacers for resistance to the invading viruses.
-
Although CRISPR–mediated immunity can enable bacteria to invade established communities of bacteria and phage, the conditions for this outcome are restrictive: (i) the invading population of bacteria has to be resistant to the established population of phage by at least two spacers and (ii) the concentrations of inhibitory compound in the established community have to be low enough for the invading population of bacteria to replicate.
-
As a consequence of phenotypic resistance (persistence), communities of bacteria and phage will contain populations of bacteria that are sensitive to the phage.
-
Although BIMs and CEMs are readily generated at relatively high rates, a CRISPR–mediated BIM-CEM (co)evolutionary arms race may not ensue; BIMs to which CEMs are not or cannot be generated may ascend and dominate the bacterial population.
-
The acquisition of multiple spacers could be leveraged to effectively develop phage-resistant strains for industrial, biotechnological or food processes to protect against specific phage contamination.
Taken at large, this study predicts that the role of CRISPR–cas systems in the day-to-day ecology and evolution of natural communities of prokaryotes and their viruses is at best modest and more restrictive than anticipated from existing theory.
As compelling as we may consider these predictions to be, at this juncture we only see them as hypotheses that have not been fully tested. We have restricted this study to the dynamics of the early phase of the interactions between single populations of phage and bacteria with very active CRISPR–cas immune systems. While our models and experiments are appropriate for this phase of the association between these organisms, they are not sufficient for making predictions about the longer-term population and (co)evolutionary dynamics of these interactions. Most importantly, our models and experiments do not formally consider the potential diversity of BIMs and CEMs that can be generated at each level and the interactions between them (see, for example, [25]). How this diversity will play into the longer-term population and (co)evolutionary dynamics of these interactions and whether the bacterial populations will be limited by phage rather than resources remain to be seen. Particularly important in this regard is whether bacteria with other mechanisms of resistance to the phage will emerge and whether the phage will be able to generate mutants capable of replicating on these resistant cells (see [42] but also [43]). It may well be that the failure to observe bacteria with other resistance mechanisms in these experiments is due to the relatively high rate at which CRISPR–mediated immunity is generated in S. thermophilus.
Finally, in support of the prediction that CRISPR–mediated immunity plays a limited role in the day-to-day (rather than historical) ecology and evolution of prokaryotes and their viruses, is our choice of experimental system. S. thermophilus and the phage 2972 may well be an exceptional system for in vitro study of the population dynamics and evolution of phage and bacteria with CRISPR–mediated immunity. Not only is it arguably (surely, to those who work with it) the best-characterized naturally occurring CRISPR–phage system; with respect to the rate of acquisition of spacers and possibly the rate of generation of CEMs, it is the most active. It is an ideal system in which to observe CRISPR playing a prominent role in the ecology and evolution of prokaryotes and their viruses. To our knowledge, there is no evidence for naturally occurring CRISPR–cas systems of other organisms being as active in generating BIMs and CEMs as those of S. thermophilus and its phages. For these reasons, we conjecture that the CRISPR–cas systems of other bacteria, archaea and their phages would play an even less active role in the day-to-day ecology and evolution than anticipated from this study.
Materials and Methods
Bacteria and phage
For our experimental model, we selected the strain S. thermophilus DGCC7710 because it contains arguably the best-characterized CRISPR–cas functional system. S. thermophilus DGCC7710 [1], which bears four different CRISPR–cas systems, two of which are active (CRISPR1 and CRISPR3), was used as a host for the virulent phage 2972 [34]. We also used S. thermophilus SMQ-301 [44], which is resistant to 2972 as a non-host strain.
Media and sampling methods
A single medium designated LM17Ca was used for all of the experiments. M17 (Oxoid) was supplemented with 0.5% lactose (L) and 0.05 M Calcium Borogluconate (Ca). For the soft/top agar and hard/bottom agar, LM17Ca was supplemented with 0.6% and 1.5% agar, respectively. Cell densities were estimated by dilution in 0.85% saline and plating on LM17Ca. To estimate phage titers, serially diluted samples were added to 1 ml of a late-log culture of S. thermophilus, put into 3 ml soft agar and poured onto LM17Ca plates.
Isolation of BIMs and CEM phages
About 5×108 S. thermophilus DGCC7710 cells were mixed with about 2.5×107 of wild type (WT) phage 2972 and held for about 10 minutes. These mixtures were then added to 3 ml of LM17Ca soft agar, which was then poured over LM17Ca plates. Ten experiments identical to the above were run in parallel. All the plates were incubated at 42°C for 30 hours. Single colonies were removed from each plate and streaked twice to purify the BIM clones and remove contaminating phages. Resistance to WT phage 2972 was confirmed by spotting 10 µl (at least 5×105 pfu) of the diluted lysate on soft agar lawns of the BIMs. Clear zones were scored as sensitive while turbid zones or few separate plaques were scored as resistant. From these experiments, we selected 10 BIMs (one per experiment), designated BIM1, BIM2, … BIM9, and BIM10. A similar procedure was used to randomly isolate five second-order BIMs from five of these first-order BIMs by spotting CEMs capable of replicating on these first-order bacteria. These were designated BIM22, BIM32, BIM52, BIM72, and BIM82. CEM phages were isolated by mixing 100 µl of phage 2972 lysate with 1 ml of overnight cultures of first - or second-order BIMs. After 10 minutes these mixtures were added into 3 ml soft agar, poured onto LM17Ca plates, and incubated overnight at 42°C. Single phage plaques were picked from the plates the following day and added to 3 or 10 ml of LM17Ca. A few drops of chloroform were added to each tube to kill the remaining bacteria. These lysates were then vortexed and centrifuged. These single plaque lysates were then filtered (0.45 µm) and stored at 4°C. This single plaque isolation process was repeated two times to obtained purified phages. The CEM phages for the first-order BIMs (BIM1 through BIM10) were designated CEM1 through CEM10. The CEM phages for the second-order BIMs were designated CEM22, CEM32, CEM52, CEM72, and CEM82.
Assays for the presence of WT phage and CEMs
Two procedures were used to assay liquid cultures for the presence of WT phage and specific CEMs. One was a spot test: 10 µl of filtered or chloroform-treated cultures were either spotted directly or after serial dilution onto soft agar lawns of WT cells or first - or second-order BIMs. Either clear zones or turbid zones with specific plaques were considered to indicate the presence of phage capable of replicating on that bacterial cell line. In cases where negative results were obtained with this spot test, we used an enrichment procedure where 100 µl of the lysate was added to 4 ml of medium with 100 µl of overnight culture bacteria. The optical densities of these enrichment cultures were followed at 42°C and those that cleared were considered positive for phage capable of replicating on those host bacteria.
Assays for the presence of BIMs
Two procedures were also used to test for the presence of bacteria resistant to wild type or CEM phages. One was a spot test: single colonies were streaked onto fresh medium to purify them from the phage in the cultures from whence they came. Single colonies were grown overnight at 42°C in liquid medium (1 ml) and then put into soft agar to be used as a lawn for spot testing with phage. For this we used the procedure and criteria for sensitivity/resistance described in the preceding paragraph. In some cases, the presence of resistant cells in samples was tested directly by adding 100 µl of phage and 100 µl of the cell culture to soft agar and looking for the presence of colonies.
Molecular characterization of the BIMs and CEMs
The active S. thermophilus CRISPR loci, namely CRISPR1 and CRISPR3, were subjected to PCR, in order to test whether novel spacers had been acquired in BIMs following exposure to phage. Primers targeting the leader sequence and a unique WT spacer at the trailer end of CRISPR1, as well as a primer pair targeting the leader and the trailer end of CRISPR3, were used for PCR amplification. The names and sequences of these primers are, respectively: yc70 5′-TGCTGAGACAACCTAGTCTCTC-3′ and 89R6 5′-TCAGCAGATTGTCAAATCGG-3′ for CRISPR1 and F1 5′CTGAGATTAATAGTGCGATTACG-3′ and R2 5′-GCTGGATATTCGTATAACATGTC-3′ for CRISPR3. PCR amplifications were carried out as previously described [36], with Ambion Mastermix and the following PCR cycling: denaturation for 7 minutes at 95 C; followed by 35 cycles of 30 sec denaturation at 95C ; 30 sec hybridization at 53 C and 1 min extension at 72 C; followed by a final round of 5 min extension at 72 C. PCR amplicons were subjected to agarose gel electrophoresis for size analysis, purified using a Qiagen Kit, and subsequently subjected to Sanger sequencing using the same primer sets used for amplification. The sequencing results were visualized in EXCEL as previously described [36] in order to assess CRISPR repeat occurrence and visualize spacer content. In order to assess the protospacer sequences in the CEMs, targeted PCR amplifications followed by Sanger sequencing of the PCR amplicons were carried out for first and second generation CEMs as described previously [7], [8].
Supporting Information
Zdroje
1. BarrangouR, FremauxC, DeveauH, RichardsM, BoyavalP, et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 : 1709–1712.
2. MarraffiniLA, SontheimerEJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322 : 1843–1845.
3. DeveauH, GarneauJE, MoineauS (2010) CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64 : 475–493.
4. HaleCR, ZhaoP, OlsonS, DuffMO, GraveleyBR, et al. (2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139 : 945–956.
5. BrounsSJ, JoreMM, LundgrenM, WestraER, SlijkhuisRJ, et al. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321 : 960–964.
6. DeltchevaE, ChylinskiK, SharmaCM, GonzalesK, ChaoY, et al. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 : 602–607.
7. DeveauH, BarrangouR, GarneauJE, LabonteJ, FremauxC, et al. (2008) Phage response to CRISPR–encoded resistance in Streptococcus thermophilus. J Bacteriol 190 : 1390–1400.
8. GarneauJE, DupuisME, VillionM, RomeroDA, BarrangouR, et al. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468 : 67–71.
9. SapranauskasR, GasiunasG, FremauxC, BarrangouR, HorvathP, et al. (2011) The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39 : 9275–9282.
10. AnderssonAF, BanfieldJF (2008) Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320 : 1047–1050.
11. TysonGW, BanfieldJF (2008) Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol 10 : 200–207.
12. SnyderJC, BatesonMM, LavinM, YoungMJ (2009) Use of cellular CRISPR (clusters of regularly interspaced short palindromic repeats) spacer-based microarrays for detection of viruses in environmental samples. Appl Environ Microbiol 76 : 7251–7258.
13. BrudeyK, DriscollJR, RigoutsL, ProdingerWM, GoriA, et al. (2006) Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 6 : 23.
14. CuiY, LiY, GorgeO, PlatonovME, YanY, et al. (2008) Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS ONE 3: e2652 doi:10.1371/journal.pone.0002652.
15. PourcelC, SalvignolG, VergnaudG (2005) CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151 : 653–663.
16. VergnaudG, LiY, GorgeO, CuiY, SongY, et al. (2007) Analysis of the three Yersinia pestis CRISPR loci provides new tools for phylogenetic studies and possibly for the investigation of ancient DNA. Adv Exp Med Biol 603 : 327–338.
17. LiuF, BarrangouR, Gerner-SmidtP, RibotEM, KnabelSJ, et al. (2011) Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl Environ Microbiol 77 : 1946–1956.
18. BarrangouR, HorvathP (2012) CRISPR: New Horizons in Phage Resistance and Strain Identification. Annu Rev Food Sci Technol 3 : 143–162.
19. PalmerKL, GilmoreMS (2010) Multidrug-resistant enterococci lack CRISPR–cas. MBio 1: e00227–10 doi:10.1128/mBio.00227-10.
20. BolotinA, QuinquisB, SorokinA, EhrlichSD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151 : 2551–2561.
21. MojicaFJ, Diez-VillasenorC, Garcia-MartinezJ, SoriaE (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60 : 174–182.
22. LintnerNG, KerouM, BrumfieldSK, GrahamS, LiuH, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J Biol Chem 286 : 21643–21656.
23. RhoM, WuYW, TangH, DoakTG, YeY (2012) Diverse CRISPRs evolving in human microbiomes. PLoS Genet 8: e1002441 doi:10.1371/journal.pgen.1002441.
24. LevinBR (2010) Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining CRISPR–mediated adaptive immunity in bacteria. PLoS Genet 6: e1001171 doi:10.1371/journal.pgen.1001171.
25. ChildsL, HeildN, YoungMJ, WhitakerRJ, WeitzJS (2012) Mult-scale model of CRISRP-induced coevolutionary dynamics: Diversification at the inteface of Lamarck and Darwin. Evolution doi:10.1111/j.1558-5646.2012.01595.x.
26. WeinbergerA, SunC, PlucińskiM, DeneV, ThomasB, et al. (2012) Persisting Viral Sequences Shape Microbial CRISPR–based Immunity. PLoS Comput Biol 8: e1002475 doi:10.1371/journal.pcbi.1002475.
27. ValePF, LittleTJ (2010) CRISPR–mediated phage resistance and the ghost of coevolution past. Proc Biol Sci doi:10.1098/rspb.2010.0055.
28. Adams MH (1959) Bacteriophages. New York: Wiley-Interscience.
29. MonodJ (1949) The growth of bacterial cultures. Annual Review of Microbiology 3 : 371–394.
30. StewartFM, LevinBR (1973) Resource partitioning and the outcome of interspecific competition: a model and some general considerations. American Naturalist 107 : 171–198.
31. LevinBR, StewartFM, ChaoL (1977) Resource-Limited Growth, Competition, and Predation - a Model and Experimental Studies with Bacteria and Bacteriophage. American Naturalist 111 : 3–24.
32. DuplessisM, RussellWM, RomeroDA, MoineauS (2005) Global gene expression analysis of two Streptococcus thermophilus bacteriophages using DNA microarray. Virology 340 : 192–208.
33. DeveauH, Van CalsterenMR, MoineauS (2002) Effect of exopolysaccharides on phage-host interactions in Lactococcus lactis. Appl Environ Microbiol 68 : 4364–4369.
34. LevesqueC, DuplessisM, LabonteJ, LabrieS, FremauxC, et al. (2005) Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl Environ Microbiol 71 : 4057–4068.
35. BarrangouR, FremauxC, DeveauH, RichardsM, BoyavalP, et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 : 1709–1712.
36. HorvathP, RomeroDA, Coute-MonvoisinAC, RichardsM, DeveauH, et al. (2008) Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus.. J Bacteriol 190 : 1401–1412.
37. BiggerJW (1944) Treatment of staphylococcal infections with penicillin - By intermittent sterilisation. Lancet 2 : 497–500.
38. LiK, BarksdaleL, GarmiseL (1961) Phenotypic alterations associated with the bacteriophage carrier state of Shigella dysenteriae. J Gen Microbiol 24 : 355–367.
39. BarksdaleL, ArdenSB (1974) Persisting bacteriophage infections, lysogeny, and phage conversions. Annu Rev Microbiol 28 : 265–299.
40. StewartFM, LevinBR (1977) The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics 87 : 209–228.
41. SunCL, BarrangouR, ThomasBC, HorvathP, FremauxC, et al. (2012) Phage mutations in response to CRISPR diversification in a bacterial population. Environ Microbiol 15 (2)
463–70 doi:10.1111/j.1462-2920.2012.02879.x.
42. LenskiRE, LevinBR (1985) Constraints on the Coevolution of Bacteria and Virulent Phage - a Model, Some Experiments, and Predictions for Natural Communities. American Naturalist 125 : 585–602.
43. MeyerJR, DobiasDT, WeitzJS, BarrickJE, QuickRT, et al. (2012) Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 335 : 428–432.
44. TremblayDM, MoineauS (1999) Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology 255 : 63–76.
Štítky
Genetika Reprodukčná medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy






