-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
Heterotrimeric G proteins are an important group of signaling molecules found in eukaryotes. They function with G-protein-coupled-receptors (GPCRs) to transduce various signals such as steroid hormones in animals. Nevertheless, their functions in plants are not well-defined. Previous studies suggested that the heterotrimeric G protein α subunit known as D1/RGA1 in rice is involved in a phytohormone gibberellin-mediated signaling pathway. Evidence also implicates D1 in the action of a second phytohormone Brassinosteroid (BR) and its pathway. However, it is unclear how D1 functions in this pathway, because so far no partner has been identified to act with D1. In this study, we report a D1 genetic interactor Taihu Dwarf1 (TUD1) that encodes a functional U-box E3 ubiquitin ligase. Genetic, phenotypic, and physiological analyses have shown that tud1 is epistatic to d1 and is less sensitive to BR treatment. Histological observations showed that the dwarf phenotype of tud1 is mainly due to decreased cell proliferation and disorganized cell files in aerial organs. Furthermore, we found that D1 directly interacts with TUD1. Taken together, these results demonstrate that D1 and TUD1 act together to mediate a BR-signaling pathway. This supports the idea that a D1-mediated BR signaling pathway occurs in rice to affect plant growth and development.
Published in the journal: The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice. PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003391
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003391Summary
Heterotrimeric G proteins are an important group of signaling molecules found in eukaryotes. They function with G-protein-coupled-receptors (GPCRs) to transduce various signals such as steroid hormones in animals. Nevertheless, their functions in plants are not well-defined. Previous studies suggested that the heterotrimeric G protein α subunit known as D1/RGA1 in rice is involved in a phytohormone gibberellin-mediated signaling pathway. Evidence also implicates D1 in the action of a second phytohormone Brassinosteroid (BR) and its pathway. However, it is unclear how D1 functions in this pathway, because so far no partner has been identified to act with D1. In this study, we report a D1 genetic interactor Taihu Dwarf1 (TUD1) that encodes a functional U-box E3 ubiquitin ligase. Genetic, phenotypic, and physiological analyses have shown that tud1 is epistatic to d1 and is less sensitive to BR treatment. Histological observations showed that the dwarf phenotype of tud1 is mainly due to decreased cell proliferation and disorganized cell files in aerial organs. Furthermore, we found that D1 directly interacts with TUD1. Taken together, these results demonstrate that D1 and TUD1 act together to mediate a BR-signaling pathway. This supports the idea that a D1-mediated BR signaling pathway occurs in rice to affect plant growth and development.
Introduction
Brassinosteroids (BRs) are a class of polyhydroxylated sterol derivatives, structurally similar to animal steroids, which appear to be ubiquitously distributed throughout the plant kingdom [1]. As a group of growth-promoting steroid hormones, they play pivotal roles in promoting cell expansion and division, regulating senescence, male fertility, fruit ripening and modulating plant responses to various environmental signals [1], [2]. Extensive studies in Arabidopsis have identified a nearly complete BR signaling pathway starting with BRI1 (BRASSINOSTEROID INSENSITIVE 1) as the cell membrane receptor which perceives and binds to BRs [3], initiates a phosphorylation-mediated cascade involving BSK1 (BR SIGNALING KINASE 1), BSU1 (BRI1-SUPPRESSOR1), BIN2 (BRASSINOSTEROID INSENSITIVE2), and PP2A (Protein phosphates 2A) and which subsequently transduces the extracellular steroid signal to the transcription factor BZR1 (BRASSINAZOLE RESISTANT 1) [1], [2], [4], [5]. In rice (O.sativa), the BRI1-mediated BR pathway appears to be conserved with Arabidopsis as several important components of this signaling pathway such as OsBRI1, OsBZR1 and 14-3-3 have the same function as their Arabidopsis orthologs [6], [7], [8]. Further, numerous BR-insensitive mutants in tomato, barley and pea have been identified as mutations in BRI1 orthologs [9], [10], [11], [12], indicating that the BRI1 pathway is conserved in flowering plants [7], [8].However, a major question for both Arabidopsis and rice remains; how do G proteins fit into this cascade? Several previous reports indicate that the canonical heterotrimeric Gα of rice and Arabidopsis are involved in a BR response but are apparently not related to the BRI pathway [13], [14], [15].
Heterotrimeric G proteins consist of three subunits, Gα, Gβ and Gγ, which play essential roles in various biological processes in many eukaryotes [16]. Once a ligand binds to a GPCR, the GPCR undergoes a conformational change which activates the G proteins by promoting the exchange of GDP/GTP associated with the Gα subunit, leading to its dissociation from Gβ/Gγ. Subsequently, Gα acts in its cascade while Gβ/Gγ regulate their own downstream effectors [17], [18]. In contrast to mammals that possess 16 Gα, 5 Gβ and 14 Gγ genes [19], Arabidopsis has 1 Gα, 1 Gβ and 3 Gγ (1 atypical) genes, while rice has 1 Gα, 1 Gβ and 5 Gγ (3 atypical) genes [18], [19], [20]. Despite this limited number of plant G protein members, their functions are diverse and have multiple roles in various hormone responses [16], [21]. In particular, a loss-of-function mutant in the rice Gα gene D1/RGA1, displays dwarfism, erect leaves, compact panicle and small round seeds. The d1 mutant was originally identified as a gibberellic acid signaling mutant and exhibited reduced growth and a highly hypersensitive response to infection by a fungus [22], [23], suggesting that D1 is involved in both GA signaling pathway and disease resistance. However, several recent studies have shown that the phenotypic characteristic of the d1 mutants are more similar to that of BR-deficient mutants, displaying shortened second internodes, erect leaves, constitutive photomorphogenic growth in darkness and decreased sensitivity to the brassinosteroid 24-epibrassinolide (24-eBL) [13]. Importantly, double mutants obtained from crossing d1-7 and d61-1 (an OsBRI1 allelic mutant) showed no epistasis in many organs except in seed length and seed weight [14], [24]. In addition, the expression patterns of several BR biosynthetic genes were not altered by brassinosteroid in d1 mutants. These results indicated that there may exist a BR signaling pathway in rice which involves Gα, but which is different from the canonical BRI1 pathway [25]. This idea is in agreement with the results for the Arabidopsis Gα gene (GPA1). The mutant gpa1 shows less sensitivity to 24-eBL and double mutants between Gα-deficient mutants and BR-deficient mutants had additive effects in many organs and tissues [15]. Thus, it is important to understand this potentially novel Gα-mediated BR pathway and to show how it controls BR-mediated growth responses.
Recent studies have shown that the ubiquitin-proteasome system (UPS) is an integral part of auxin, GA, jasmonic acid (JA), ethylene and abscisic acid signaling or biosynthetic pathways [26]. UPS is regarded as one of the most prominent mechanisms which regulates protein degradation to modulate protein levels in plants to efficiently alter their proteomes and so ensure proper developmental responses and environmental adaptations [27]. Ubiquitin is a 76 amino acids polypeptide that is covalently attached to a protein target through an enzymatic cascade comprising a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). The E3s are key factors that define substrate specificity. In plants four main types of E3s have been classified according to their mechanisms of action and subunit composition: HECT, RING, U-box and Cullin-RING ligases (CRLs) [26]. U-box E3 ligases are grouped based on a conserved 70 amino acid motif, that lacks characteristic zinc-chelating cysteine and histidine residues, and so uses intramolecular interactions to maintain the U-box scaffold [28], [29]. Yeast and humans have 2 and 21 U-box genes, respectively. In contrast, Arabidopsis and rice have 64 and 77 U-box genes, respectively [30], [31]. The expansion of the plant U-box gene family suggests that they are key in controlling diverse cellular processes, with possibly many being specific to plants. The biological functions of over 25 U-box E3s have been reported, involving hormone responses, biotic stress, abiotic stress and self-incompatibility [32], [33], [34], [35]. Here we characterize a new U-box E3 function. We have identified the U-box E3 ligase gene, TAIHU DWARF1 (TUD1) and shown its genetic interactions with the rice Gα subunit D1. TUD1 is a functional E3 ligase and acts as a BR signaling activator. Furthermore, TUD1 and D1 physically interact and together define a D1-mediated BR signaling pathway which may parallel or partly overlap with the canonical BRI1 pathway.
Results
Identification of a genetic interactor with D1
To dissect additional components involved in D1-dependent BR responses [13], [14], we took a genetic approach to identify mutants similar to d1. Among 250 reduced plant height mutants of rice, a dwarf mutant similar to d1 was identified and subsequently shown to be non-allelic to d1. We named this dwarf mutant taihu dwarf1 (tud1). Five allelic mutants (tud1-1 to -5) were further identified from our dwarf mutant collection. To examine the genetic relationship of d1 and tud1, we crossed a weak d1 allele with a strong tud1 allele (tud1-5) with dm-type dwarfism (see below) in the same background of Nipponbare [36]. The phenotype of the F1 was normal, suggesting that they were non-allelic. The phenotype of the double mutant in the F2 was similar to tud1-5 with a specific reduction of the second internode length, erect leaves and shortened grain lengths (Figure 1A–1C), indicating that tud1-5 was epistatic to d1-c (Table S1). In addition, tud1-4 and d61-2 (an OsBRI1 allelic mutant) were crossed and the double mutant showed that tud1 and d61 had an additive effect on rice growth and development (9∶3∶3∶1,χ2:0.677, p>0.05 Figure 1D). These results showed that TUD1 acts in the same genetic pathway as D1 but different from that involving the rice BRI1 ortholog D61.
Fig. 1. tud1 is epistatic to d1, but additive to d61. 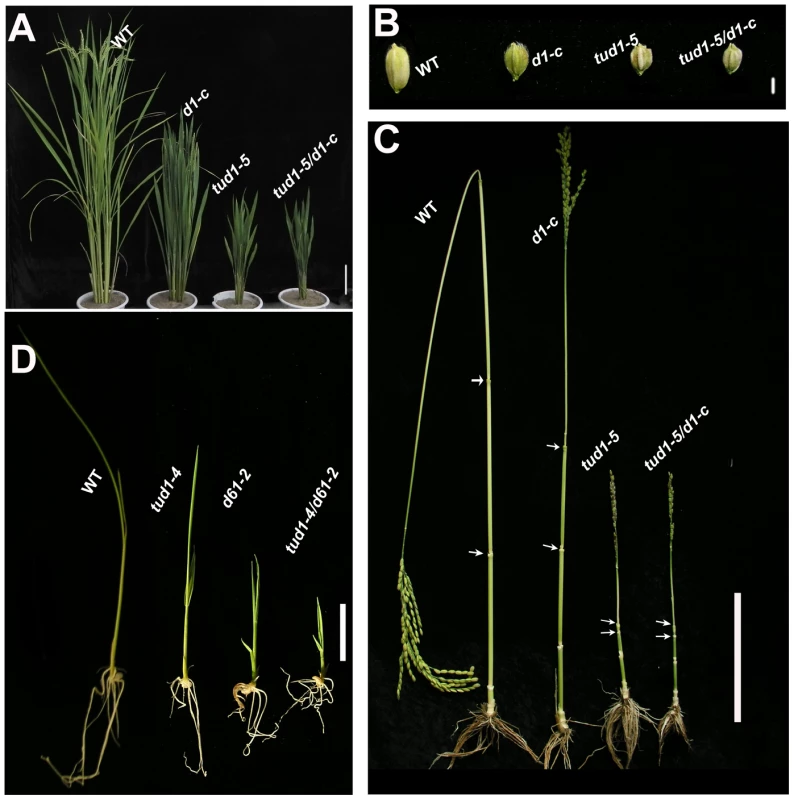
(A) Adult plant morphology of wild type (WT) (Nipponbare), d1-c, tud1-5 and d1-c/tud1. Both tud1-5 and d1-c mutants were derived from Nipponbare. The plant height of tud1 is similar to d1-c/tud1, showing that tud1 is epistatic to d1 in plant height. Bar: 10 cm. (B) The grain morphology of wild type (WT), d1-c, tud1-5 and d1-c/tud1 mutants. The vertical grain lengths of tud1-5, d1-c and d1-c/tud1 were shortened compared to wild type. The vertical grain length of tud1-5 is similar to d1-c/tud1, showing that tud1 is also epistatic to d1 in grain length. (C) The patterns of internode elongation in wild type (WT), d1-c, tud1-5 and d1-c/tud1. d1-c is characterized by a dn-type dwarfism, but tud1 and d1-c/tud1 are of dm-type dwarfism and their second internode elongations are specifically inhibited. Arrows indicate start and end of the second internode. Bar: 10 cm. (D) Plant heights for wild type (WT), tud1-4, d61-2 and tud1/d61, showing that these mutants are additive in the reduction of plant height. Bar: 2 cm. tud1 is a pleiotropic dwarf mutant
To examine the type of dwarfism of tud1, we compared the gross morphology of 9-week-old wild-type and tud1 plants (Figure 2A). The plant heights of tud1 mutants were significantly shorter than their corresponding wild type, and tud1-5 showed a severe dwarf phenotype. The internode elongation was inhibited in all of the mutants. Lengths of the individual internodes of plants were measured and expressed as a relative value (Figure 2B). Among them, tud1-1, tud1-2, and tud1-5 showed a specific internodal inhibition; the second internode was severely shortened relative to other internodes. This pattern was typically classified as a dm-type elongation pattern [36]. In tud1-3 and tud1-4, the lengths of each internode were almost uniformly shortened, resulting in an elongation pattern similar to that of wild type. According to Takeda [36], tud1-3 and tud1-4 showed dn-type dwarfism (Figure S1). To our knowledge, rice mutants showing different patterns of inhibition of internode elongation were only reported in BR-insensitive mutants of d61 and d1 [6], [14], suggesting that tud1 is also involved in a BR pathway. In addition, either unhulled or hulled seeds were specifically shortened in the vertical direction in tud1 mutants (Figure 2C). Compared with their corresponding wild type, the grain lengths of tud1 mutants were reduced by 30 to 44%. Their leaves were also shortened, erect and dark-green, similar to d1 (24), but their severe rugose (curled) nature appeared to be different from d1 (Figure 2D).
Fig. 2. tud1 is a pleiotropic dwarf mutant. 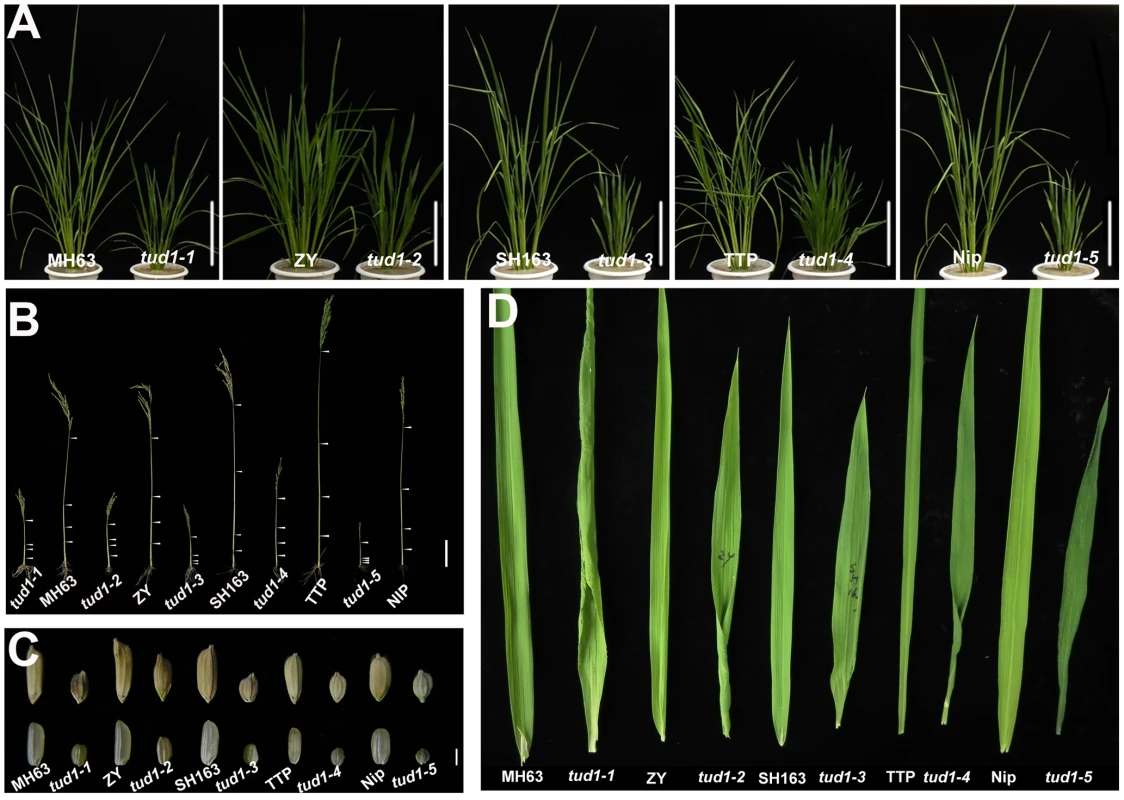
(A) Adult plant morphology. Comparison of five allelic mutants of tud1 (tud1-1 to -5) with their corresponding wild rice varieties MH63, ZY, SH163, TTP and Nip, respectively. Bar: 10 cm. (B) Comparison of culm (stem) elongation of the tud1 mutant alleles with their corresponding wild types. According to Takeda (1977), tud1-1, tud1-2 and tud1-5 showed dm-type dwarfism, and tud1-3 and tud1-4 showed dn type. Arrowheads indicate the positions of nodes. Bar: 10 cm. (C) Grain morphology. All of the mutants have shortened grains. The upper panel and the lower panel represent the unhulled and hulled seeds, respectively. Bar: 2 mm. (D) Leaf morphology. The leaf blades of tud1 mutants all are dark-green and rugose. To determine whether dwarfism in tud1 was due to cell division, cell elongation or both, we measured the cell length and number in the third leaf sheath, the third internode and the lemma of tud1-2 and its wild type (Figure 3A–3C). Overall we found that the total number of cells, in all given organs, was reduced in the mutant compared to wild type (Table S2). The length of the third leaf sheath in tud1-2 was decreased by 57% compared to wild type, while the average cell length was similar. Thus, the estimated cell number in the third leaf sheath was reduced by about 43% compared with the wild type, indicating that the shortened leaf sheath in tud1-2 was due to a reduction in cell number rather than cell length (Figure 3D, 3E). Similarly, the calculated cell number of the third internode and the lemma in tud1-2 was reduced by about 67% and 36% respectively (Figure 3F–3H). These results indicated that tud1 has a significantly reduced cell number in these aerial plant organs.
Fig. 3. Cell numbers are reduced in aerial organs of tud1-2. 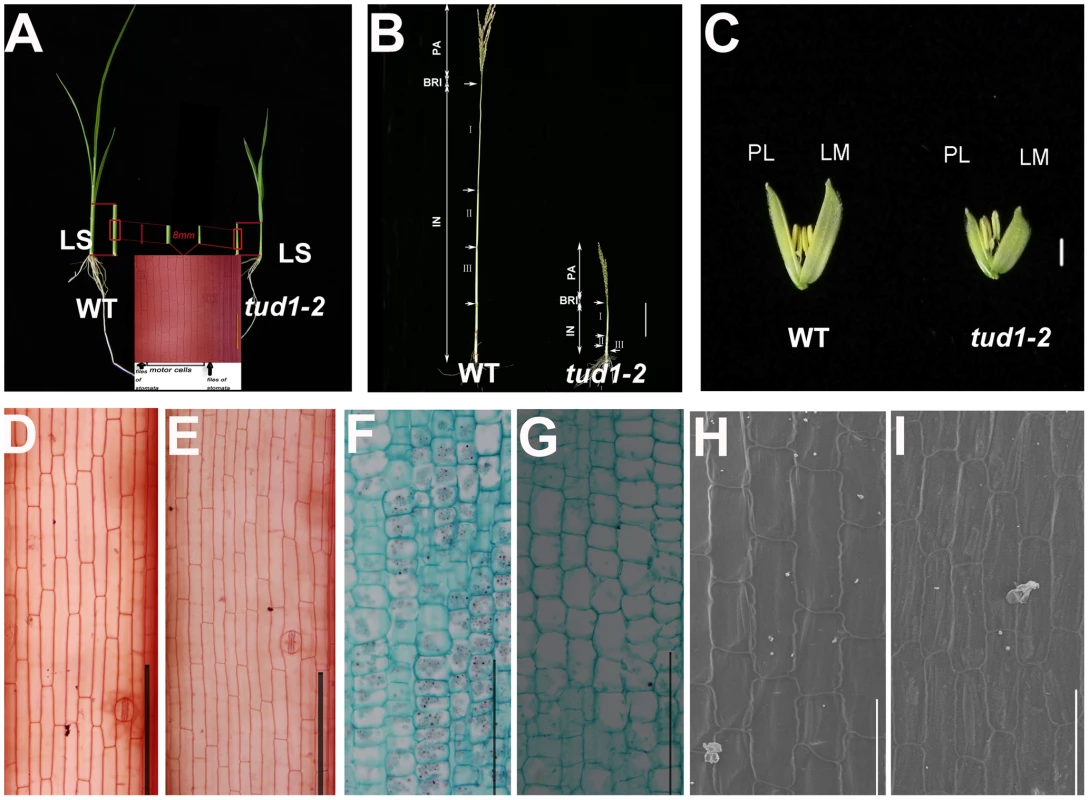
(A) Aerial parts of three-week old wild type (WT) (upper-left) and tud1-2 (upper-right). Schematic illustration of the third leaf sheaths is shown in middle bottom. Central parts of the leaf sheaths were cut off and the lengths of motor cells between files of stomata were measured. (B) Panicles and internodes of wild type (WT) and tud1-2. PA, panicle; BRI, basic rachis internode; IN, internode. Internode I, II and III are the first, second and third internodes, respectively. Bar: 10 cm. (C) Lemma and palea of WT and tud1-2. LM: Lemma; PL: Palea. Bar: 2 mm. (D,E) Longitudinal sections of the third leaf sheath from one-month old seedlings of wild type (WT) (D) and tud1-2 (E). Bar: 200 µm. (F,G) Longitudinal sections of the third internode from one-month old seedlings of wild type (WT) (F) and tud1-2 (G). Bar: 200 µm. (H,I) Inner epidermal cells of lemma from one-month old seedlings of wild type (WT) (H) and tud1-2(I) observed by SEM. Bar: 100 µm. We analyzed more closely the changes in the second internode and adaxial leaf surfaces. In the wild type, cells in the second internode were elongated and well-organized into longitudinal files, but in tud1-2 and tud1-1, cell files were disorganized and not elongated (Figures S2, S3). In the wild type, the epidermal cells of leaf blade almost run parallel to the vertical vascular tissues, but in tud1-2, all of the vertical tissues were waved and the leaf epidermal cells had a disorganized arrangement (Figure S4). However, it was clear that the extent of the overall disorganization of leaf epidermal cells was less severely affected in tud1-2 than in the second internodal cells. These results showed that the deficiency of cell division and the arrangement of poorly organized cell files leads to dwarfism in tud1 plants.
TUD1 is involved in BR responses but not in gibberelin or cytokinin responses
Because of its dwarf phenotype, d1 was initially classified as a gibberellin (GA)-insensitive mutant [22]. To examine whether tud1 was a GA-deficient mutant, we performed GA and paclabutrazol (PAC, an inhibitor GA biosynthesis) application assays for tud1-2 and wild type. Treatment of tud1-2 plants with 1 µM GA3 or 30 µM PAC had effects similar to those seen in wild type (Figure 4A). Similarly, the effect of different concentration of GA3 on increase of seedling plant height was also similar between the wild type and tud1-2 (Figure 4B), indicating that tud1-2 plants have a normal sensitivity to GA. To further confirm this, we performed an immunoblot analysis of SLENDER1 (SLR1) protein, which is a repressor in the GA signaling pathway [37], [38], and found that SLR1 protein levels remained similar in tud1-2 and wild type plants treated by either GA3 or PAC (Figure 4C). Meanwhile, an α-amylase activity assay (diagnostic for GA responses) also showed that wild type and tud1-2 responded similarly to GA (Figure 4D). Additionally, a new eui1-d (elongation uppermost internode1-d) mutant, which accumulated exceptionally large amount of biologically active GAs in the uppermost internode [39] in tud1-2 background, was recovered and showed additive phenotypes (Figure 4E), indicating that tud1 did not influence the GA biosynthesis. Furthermore, a tud1-2/slr1-l double mutant exhibited additive plant height phenotypes (Figure 4F). Taken together, these results showed that tud1-2 is not a GA-deficient mutant.
Fig. 4. tud1-2 is unlikely a GA-related mutant. 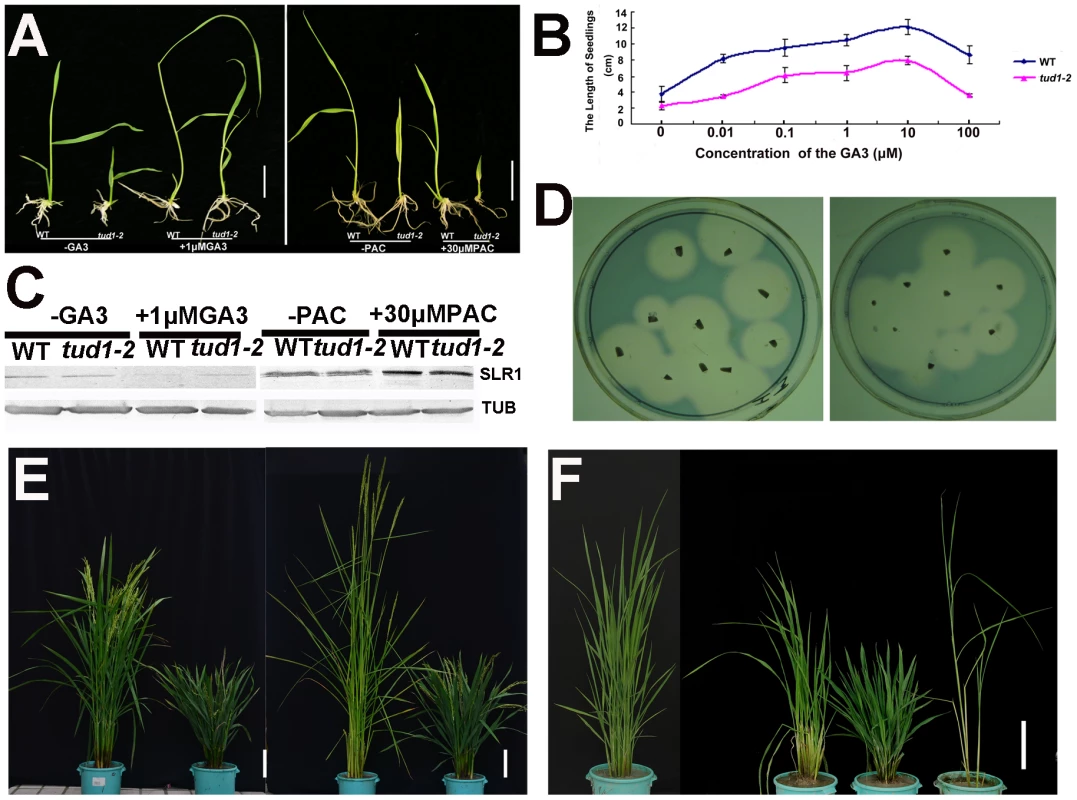
(A) Seeds of wild type (TUD1) and the dwarf mutant (tud1-2) were germinated on agar plates in the presence (+) or absence (−) of 1 µM GA3 (Left), or 30 µM of PAC (Right), and seedlings were examined 10 days after germination. (B) Seedling lengths (heights) in response to GA3 treatment in the wild type (blue) and tud1-2 (pink). (C) Changes in SLR1 protein levels triggered by GA3 or PAC treatment. Young seedlings of the wild type (WT) and tud1-2 were treated with (+) 1 µM GA3, 30 µM PAC or control solution (−), as described in Figure 4A and 4B, and their extracts subjected to western blot analysis by anti-Tubulin and anti-SLR1 antibodies, respectively. (D) A plate assay for α-amylase induction. Left: wild type (WT); Right: tud1-2. (E) Gross morphology of tud1-2 and a new eui1 mutant in the tud1-2 background. From left to right; wild type (WT), tud1-2, eui1-h,tud1-2/eui1 (right). Bar: 10 cm. (F) Gross morphology of tud1-2, slr1 and tud1-2/slr1 mutants. From left to right; wild type (WT), tud1-2/slr1, tud1-2 and slr1 (right). Bar: 15 cm. To ascertain whether or not the tud1 is a cytokinin (CK)-related mutant, the seeds of tud1-2 and its wild type were germinated on agar plates with or without the cytokinin 6-benzyladenine (6-BA). The growth of the seminal (main) root of both wild type and tud1-2 were inhibited and the degree of reduction in root length was similar in both. Also, the relative expression of three CK-related genes OsIP4, OsRR1, and OsHP2 (Table S4) [40] were not significantly altered between wild type and tud1-2 (Figure S5). Additionally, the degree of the alteration in the relative expression level of OsIP4, OsRR1, OsHP2 were also found to be similar in wild type and tud1-2 with treatment of 6-BA when compared to no treatment (Figure S6A, S6B, S6C). These results showed that tud1-2 is not a CK-deficient mutant and its phenotype is not directly associated with CK.
To examine whether tud1 was involved in BR responses, we first performed a mesocotyl elongation experiment in the dark. The tud1-2 mutant showed a typical deetiolated phenotype, similar to the BR-insensitive mutant d61-2 [6] (Figure 5A and 5B). Next, seeds of wild type and tud1-2 mutant plants were germinated on agar plates containing different concentrates of 24-eBL and examined for the length of the seminal root after one week. The growth of the seminal root of wild type was inhibited by 24-eBL in a dose-dependent manner, but the tud1-2 mutant showed a significantly reduced response to 24-eBL (Figure S7A and S7B). The growth of the seminal root was inhibited 43% in the wild type, but only 27% in the tud1-2 mutant in the presence of 10−6 M 24-eBL (Figure S7B), suggesting that tud1 is much less sensitive to exogenous BL than the wild type. Furthermore, a lamina joint test was performed using tud1-2 and wild type [41]. In wild type, the lamina joint bending was increased in a dose-dependent manner, from 14.2° to 178.5° with 0 to 1000 ng 24-eBL (Figure 5C and 5D). In tud1-2 plants, the degree of bending of the leaf lamina also increased with increased concentrations of 24-eBL, but the effect of the bent angles was much smaller than that of wild type under the same condition (Figure 5C and 5D). We compared the effects of 24-eBL on the size of cells in the lamina regions of the wild type and tud1-2. Under no treatment of 24-eBL, the adaxial cell number in the lamina regions of the wild type was similar to that in the tud1-2. However, in the presence of 24-eBL, the adaxial cells in this region of wild type were greatly expanded while the cells in tud1-2 were only expanded slightly (Figure S8). These results indicated that the cells of lamina region in tud1-2 were less insensitive to 24-eBL than that in the wild type. Therefore, these results show that tud1 is a BR insensitive mutant.
Fig. 5. tud1-2 is a BR insensitive mutant. 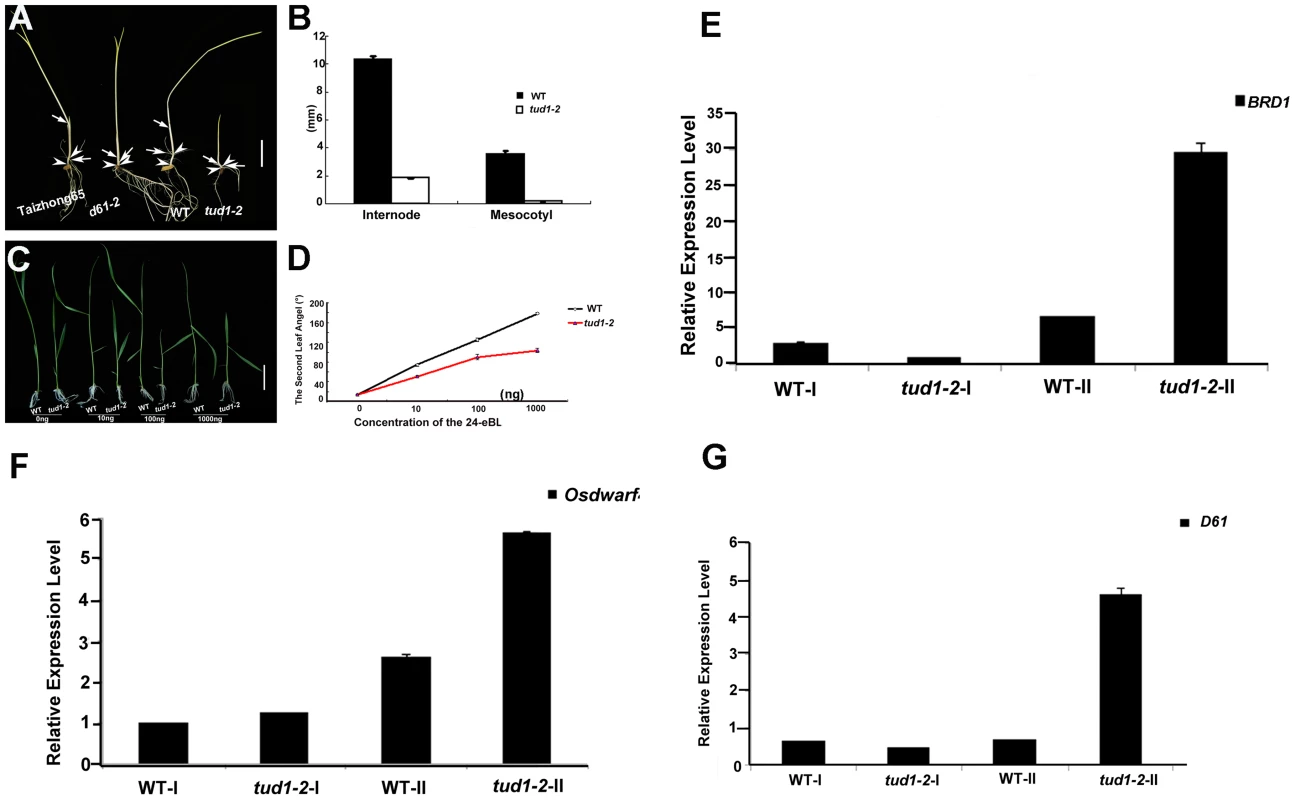
(A,B) Photomorphogenic phenotypes of tud1 grown in the dark. Plants of Taizhong 65 (WT), d61-2 (BR-insensitive mutant), wild type (WT) and tud1-2 were grown in complete darkness (A). Arrows indicate nodes and arrowheads. Bar: 2 cm. The lengths of internodes and mesocotyls were measured after ten days growth. Data presented are the means of results from five plants (B). Error Bars = SD. (C,D) Effects of 24-eBL on the degree of inclination of the leaf lamina in wild type (WT) and tud1 plants. Typical responses of the second leaf lamina joint from wild type and tud1-2 plants to 24-eBL at 0 ng, 10 ng, 100 ng or 1000 ng. Bar: 2 cm (E). The dose responses to 24-eBL, for the bending angle, for WT and tud1-2 (F). Data presented are the means of results from a total of five plants. Error Bars = SD. (E,F,G) Relative expression levels of BRD1 (E), OsDWARF4 (F) and D61 (G) in different internodes. WT-I and tud1-2-I represent the uppermost internode, WT-II and tud1-2-II represent the second elongated internode, respectively (from top to bottom). Negative feedback regulation of BR-related genes in the second internode and seedling leaf blade in the tud1-2 mutant
The results of histological analysis above showed that the main cause of dwarfism in the second internode and leaf blade appeared to be different from that in most of the other aerial organs of tud1-2. To understand what is the molecular mechanism of retardation in these two type organs, we checked the relative expression levels of several BR-related genes as tud1-2 is affected in BR responses. We analyzed the uppermost (first) and second internode of tud1-2 and its wild type, and the seedling leaf blade of wild type, d1-c, tud1-5 and d1-c/tud1-5 with and without treatment of 24-eBL. The q-RT-PCR results showed that the amount of BRD1, OsDWARF4 and D61 [6], [42], [43], [44] expression differed significantly between the uppermost internode and the second internode; expression of all BR-related genes was stronger in the second internode of tud1-2 than that in the uppermost internode (Figure 5E, 5F, 5G). Similarly, the q-RT-PCR results also indicated that relative expression level of D61 was higher in the seedling leaf blade of the d1-c/tud1-5 double mutant compared with its parents (Figure S9). The expression level of D61 was also found to be higher in the second internode and seedling leaf blade in tud1-2. These results suggested that mutation in TUD1 impairs the OsBRI1-mediated BR pathway signal transduction. This affect has consequences with altered feedback regulation of the expression of BRD1 and OsDWARF4 occurring in the second internode of tud1-2 mutant.
TUD1 encodes a functional U-box E3 ligase
To examine its molecular function, we isolated TUD1 by map-based cloning. The TUD1 locus was first mapped to the short arm of chromosome 3 between markers s1193411 and s32681 (Figure 6A, Table S5). TUD1 was further localized to an 18.457 kb region containing three open reading frames (Figure 6A, Table S5). These reading frames were sequenced and the second one, annotated as a U-box protein (LOC_03g13010, Os03g0232600), was found to be mutated in all the five allelic mutants (Figure 6A). To further confirm the identity of TUD1, a DNA fragment of ∼6.4 kb in size including the entire sequence of the putative gene was introduced into tud1-2 by Agrobacterium-mediated transformation [45]. Transformants with the TUD1 gene-containing vector showed phenotypes similar to wild-type plants, while transformants with the control vector containing no target gene did not (Figure 6B). Thus, tud1 was caused by a loss-of-function mutation in a U-box gene.
Fig. 6. TUD1 is a functional E3 ligase. 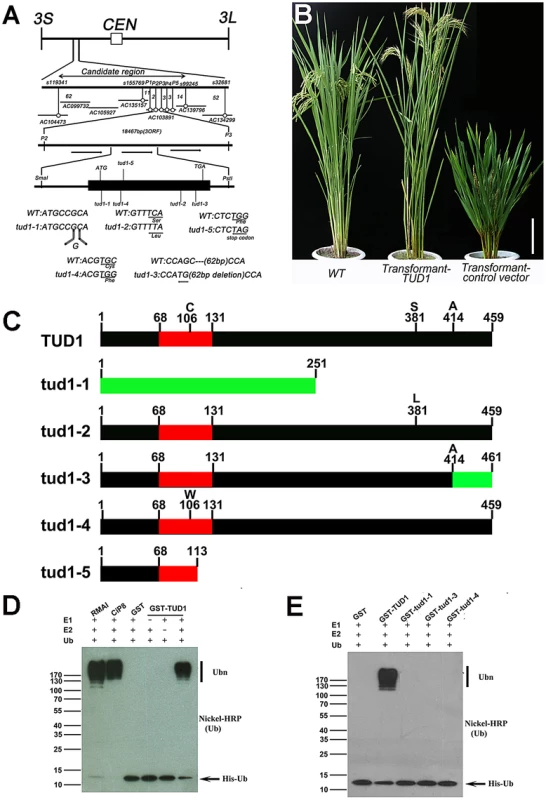
(A) High resolution linkage and physical map of tud1 locus. Horizontal lines represent chromosome 3 and vertical bars the molecular markers. The numbers of recombinant plants are indicated between the markers. The physical map of the tud1 locus was constructed using 7 BAC clones and the candidate region of the tud1 mutation was found between markers P2 and P3. The genomic structure of the TUD1 gene and the positions of mutations in tud1 alleles are also shown. The black box indicates the single exon. The mutated DNA sequences of tud1 alleles are shown in the bottom. The tud1-1 has one-base insertion near the initiation codon. The tud1-2, tud1-4 and tud1-5 have different one-base substitutions in the coding sequence. The tud1-3 has a one-base substitution and 62 bp base deletion in the exon. CEN:centromere, 3S: the short arm of chromosome 3, 3L: the long arm of chromosome 3. (B) Phenotypic complementation by introduction of the TUD1 gene. Nipponbare was the parent for tud1-2 and used as the wild-type plant. Left, the wild type (Nipponbare); center, transformant-TUD1; right, transformant-control vector. Bar: 10 cm. (C) Schematic structures of TUD1 protein and its mutant alleles. Black bars indicate TUD1 coding regions. Red bars depict U-box motifs. Green bars represent the aberrant truncated protein of tud1-1 and partial amino acids residues of tud1-3 due to a frameshift mutation. Numbers represent the amino acids of protein sequences. (D) Ubiquitination assays with GST-TUD1. GST-TUD1, GST, and positive controls of RMAI and CiP8 were assayed for E3 activity in the presence of E1 (from wheat), E2 (UBCh5b), and 6×His tag ubiquitin (Ub). The numbers at left denote the molecular mass in kilodaltons. Samples were resolved by 8% SDS-PAGE. The nickel-horseradish peroxidase was used to detect His tag ubiquitin. (E) Ubiquitination assays with GST-TUD1 and its mutant variants. GST-TUD1 and its mutant forms (GST-tud1-1, GST-tud1-3 and GST-tud1-4 fusion proteins) were assayed for E3 activity. The reaction conditions were the same as in (D). On the basis of public data (www.tigr.org and http://cdna01.dna.affrc.go.jp/cDNA/) and our results of 3′ RACE (3′ end rapid amplification cDNA ends), we found that TUD1 is an intronless gene corresponding to an ORF(open reading frame) of 1380 bp, which is predicted to encode a protein containing 459 amino acids residues with a U-box motif near its N-terminus (Figure 6C). All five allelic mutants were shown to be mutated in the coding sequence (CDS), but at different locations. Despite their differences in genetic background, the tud1-5, tud1-1 and tud1-2 were easily grouped as strong alleles by their mutant phenotypes, showing the smallest leaf angles and most dwarfism in elongation of internodes when compared to wild type. In contrast, tud1-3 and tud1-4 exhibited mild mutant phenotypes, with respect to the elongation pattern of internodes. The degree of mutant phenotype strength appeared to be correlated to the severity of mutation in TUD1. tud1-5 was identified as a null allele due to the change of 341G into 341A leading to a premature stop codon. tud1-1 has a “G” inserted into the CDS of TUD1, near the initiator codon (ATG), and generates an aberrant truncated protein compared to TUD1 (Figure 6A, 6C). Although there is a single base substitution mutation both in tud1-2 and tud1-4, the mutant phenotype of tud1-2 was more severe than that of tud1-4, suggesting that the 381S amino acid in TUD1 is very important (Figure 6A, 6C). tud1-3 has a mild phenotype, likely due to a mild defective function in TUD1 with a 62 bp deletion in the CDS of TUD1, not entirely abolishing its function in vivo, but showing no in vitro ubiquitination activity (see below).
Blast searches revealed that the deduced TUD1 protein sequence has high similarity to several sequences in other plant species: 90% identity to 01g042180 protein from Sorghum bicolor, 90% to LOC100281502 and 88% to LOC100383857 proteins from Zea mays, 60% to AT3G49810 and 56% to AT5G65920 proteins from Arabidopsis (Figure S10). Therefore, the function of TUD1 may be conserved in higher plants.
We examined whether TUD1 possesses a ubiquitination E3 ligase activity as a predicted U-box protein. Ubiquitination activity was observed for the purified GST-TUD1, compared to 2 well-characterised E3 ligases RMA1 and CIP8 proteins [46] (Figure 6D). In addition, tud1-1, tud1-3 and tud1-4 proteins did not possess any apparent E3 ligase activity (Figure 6E), showing that the ubiquination activity of TUD1 is essential for its function.
TUD1 predominantly localizes to the plasma membrane and physically interacts with D1
To investigate the subcellular localization of TUD1, we conducted an in-vivo targeting experiment using fusions of TUD1 with synthetic green fluorescent protein (sGFP) as a fluorescent marker in a transient transfection assay. The TUD1::sGFP fusion protein in rice protoplasts was mainly associated with the plasma membrane (Figure 7A), similar to that of D1 [20].
Fig. 7. TUD1 is predominantly associated with the plasma membrane and physically interacts with D1. 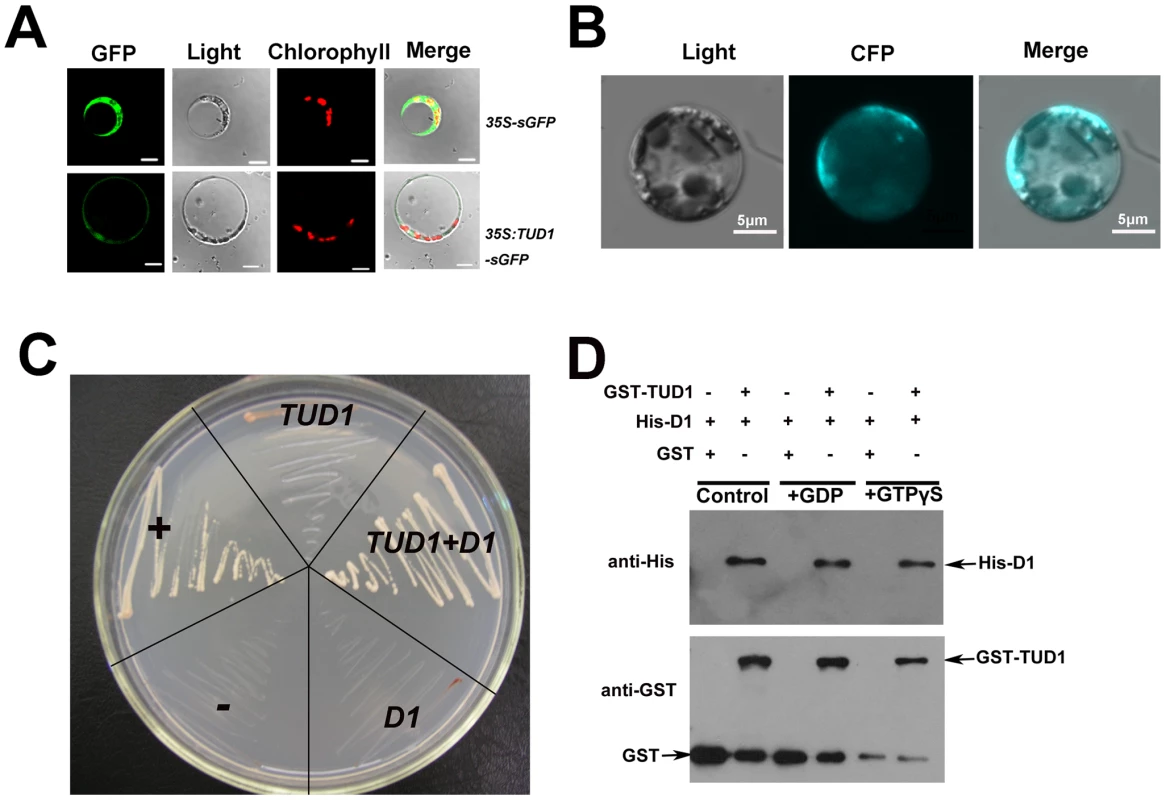
(A) Expression of the TUD1-GFP fusion protein in rice protoplasts. The 35S:sGFP and 35S:TUD1-sGFP constructs were transformed into mesophyll protoplasts prepared from rice seedlings and the expression of the introduced genes was viewed after 16 h by confocal microscopy under dark-field or light-field conditions. Bar: 10 µm. (B) BiFC detection of the TUD1-D1 interaction in rice protoplasts. CFP: cyan fluorescence protein. Bar: 5 µm. (C) Interaction between TUD1 and D1 detected by yeast two-hybrid assays. The full-length D1 and TUD1 cDNAs were cloned into pGADT7 and pGBKT7, respectively. Yeast AH109 cells were transformed with the vectors indicated and are shown after three days on selective medium. “+” and “−” represent positive and negative control, respectively. D1 or TUD1 alone shows weak self-activation. TUD1+D1 show yeast cells transformed with pGBKT-TUD1 and pGADT7-D1 together. (D) Pull-down assay for TUD1−D1 interaction. Purified D1 fusion protein was incubated in buffer with either GDP or GTPγS or blank (control) for 2 hours before adding the GST-TUD1 to the binding assay buffer, then precipitated with glutathione-agarose beads. The precipitates were separated by gel electrophoresis and probed with anti-His or anti-GST, respectively. Together with the result that tud1 is epistatic to d1, we wondered whether TUD1 would physically interact with D1. We examined this possibility in three ways. First, we used a biomolecular fluorescence complementation (BiFC) assay [47] to test the interaction between the TUD1 and D1 in rice protoplasts. Cyan fluorescence protein (CFP) fluorescence was reconstituted when the full-length TUD1 and D1 proteins were co-expressed in rice protoplasts (Figure 7B), showing that they physically interact with each other in vivo. Second, yeast two-hybrid assays [48] were performed using the D1 or TUD1 and the interaction between TUD1 and D1 was subsequently detected (Figure 7C). Third, we performed a glutathione S-transferase pull-down assay [48] for which we also determined whether TUD1 preferentially interacts with the GTP-bound form of D1. Thus, D1-tagged His and TUD1-tagged GST were expressed and purified from E. coli by nickel and glutathione S-transferase columns, respectively. Purified D1 fusion protein was incubated in buffer with either GDP or GTPγS or blank for 2 hours before adding the GST-TUD1 to the binding assay buffer. We subsequently detected that both the GDP - and GTPγS-bound forms of D1 have similar binding ability to GST-TUD1, whereas no binding occurred to GST alone (Figure 7). Based on these results, we concluded that TUD1 physically interacts with D1.
Discussion
TUD1 functions together with D1 in the plasma membrane
Heterotrimeric G proteins, consisting of α, β and γ subunits (Gα, Gβ and Gγ), function as signal mediators at the cell plasma membrane in mammals and higher plants [16], [17]. These G protein complexes dissociate into an α subunit and the βγ dimers upon activation of the complex by signal perception in mammals, yeast and higher plants [49].
In rice, previous studies demonstrated that Gα(D1),Gβ, Gγ1, and Gγ2 are not only localized in the plasma membrane, but are also present in a large protein complex (ca. 400 kDa) [19], [20]. As the molecular mass of the αβγ trimer is 100 kDa, the rice 400 kDa G protein complex should contain additional proteins [20]. TUD1 is also localized in the plasma membrane and directly functions with D1 in the plasma membrane. TUD1, therefore, could be associated with the large G protein complex in rice. It is well known that the active Gα forms are free from βγ dimmers, and Gα-GTP monomer and a Gβγ dimmer regulate their own downstream effectors, respectively. However, in our study, pull-down assays showed that TUD1 physically interacts with both the GDP - and GTPγS-bound form of D1, indicating that TUD1 may interact with either inactive or active form of D1. To better understand how D1 and TUD1 interact with each other in vivo, further studies including Co-IP (co-immuoprecipitation) and expressing a constitutively active form of D1 (Q233L) in the tud1 mutant would be interesting.
Gα subunits and E3 ligases act as integral components of signaling pathways; Gα in the G protein complex and E£ ligase in the ubiquitin-proteasome system (UPS) [17], [23], [27], [28], [32], [33], [34], [35]. However, there was no previous evidence to link Gα with an E3 ligase in any signaling pathway in plants. Now, in our study, we have demonstrated that first link; TUD1 is a functional U-box E3 ligase and directly acts downstream of the Gα subunit D1. Further, TUD1 and D1 mutants show impairment in BR responses. Together these results suggest we have uncovered a novel signaling pathway controlling rice growth, whereby a BR signal, mediated by heterotrimeric G protein, is then potentiated into the UPS. It is well-known that a signal perceived by G proteins is generally primary, and so we suggest that the D1-TUD1 interaction could mediate a very early BR response. This proposal is also consistent with the observation of the defective phenotypes in d1 and tud1 being at early developmental stages. As yet, we cannot discern whether the BR signal mediated by D1-TUD1 originates from the OsBRI1 - mediated BR signaling pathway or other unknown BR receptor(s). It will be interesting to isolate the target(s) of TUD1 to further define this pathway.
Possible overlaps of the D1-TUD1 - and OsBRI1-mediated BR signaling pathways
The well-characterized BRI-mediated BR signaling pathway is conserved among several plant species [1], [2]. Gα also appears to be involved in a BR signaling pathway, but as yet has not been linked to BRI1 [13], [14], [15]. In rice, it is presently regarded as the D1-mediated BR pathway. One possibility is that the D1-mediated BR pathway is parallel to the BRI-mediated BR signaling pathway. The second possibility is that D1 may amplify some BR responses that are initiated by OsBRI1 [14].
In our study, tud1 mutant was classified as a BR-deficient mutant in the broad sense, with its phenotype more close to d1 than other reported BR-deficient mutants, such as d61, brd1, and d2 [6], [42], [44]. The tud1 and d1 mutants have common, characteristic phenotypes of BR-related mutants, but also show short, compact panicles and more specifically decreased length in the vertical direction of grain shape. Histological observation showed that the dwarfism in most of aerial organs of tud1 was mainly due to a decreased cell proliferation, which is similar to the cause of dwarfism in d1. Furthermore, tud1 is completely epistatic to d1 and TUD1 functions together with D1 in vivo. Based on these results, we conclude that TUD1 is a direct downstream factor of D1 signaling and mediates a G-protein signaling pathway for BR.
Despite TUD1 and D1 showing differences to BRI1, we found that the dwarfism in the second internode and leaf blade of tud1-2 (due to disorganized cell files and failure of normal cell elongation) is similar to the phenotypes in BR-deficient mutants, such as d61 and brd1. In addition, q-RT-PCR results showed that mutations in TUD1 may lead to an impairment of the OsBRI1-mediated BR pathway transduction. Thus, the D1-TUD1-mediated BR signaling pathway might have an overlapping function with the OsBRI1-mediated BR pathway in the second internode and leaf blade.
In rice, dm-type mutants are commonly identified as BR-deficient mutants [6], [14], [42], [43], [44], but why the second internode is specifically inhibited remains unknown. It is difficult to explain if it is simply due to differential OsBRI1 expression in different internodes, leading to differences in BR signal strength, through only one OsBRI1-mediated BR pathway. Rather, it suggests there should be additional signal pathway(s) involved in the second internode elongation [6]. Our results support this idea, as we show that the OsBRI1 - and D1-TUD1-mediated BR signaling pathways may both have important roles in regulating the second internode elongation in rice. However, there are many questions remaining. For example, how do these two BR signaling pathways function together in the second internode? How are the BR signals perceived and transduced by the D1-TUD1-mediated pathway if not via BRI1? Is the D1-TUD1-mediated BR pathway involved in the signal amplification of some responses that originate from OsBRI1?
Potential application of the D1-TUD1-mediated signaling pathway in enhancing rice yield
Despite the simple repertoire of G protein signaling elements in plants, multiple signals can be propagated through the G-proteins, to mediate diverse physiological responses [19], [20]. It is well-known that some physiological responses are mainly accounted for by Gα, whereas others are predominantly mediated by Gβγ. In particular, any given G protein component may have multiple differential roles during plant growth and may have different responses to biotic and abiotic stress in a cell-type - or developmental-stage-specific manner [50], [51]. In this context, two important rice yield QTLs, GS3 and DEP1, were recently identified as atypical G protein Gγsubunits [52], [53], [54]. This could serve as a good example for selection and their uses in practical cereal breeding.
In our study, D1 and TUD1 function together not only to promote plant height, panicle development and seed length increase, but also to control a hypersensitive response to infection by avirulent races of a rice blast fungus (our unpublished data). Based on the D1-TUD1-mediated pathway having dual roles in promoting plant growth and resistance to rice blast, it may be feasible to manipulate the components of this pathway to enhance rice yield. Furthermore, similar genes to TUD1 are found in both Arabidopsis and several cereal species, such as Sorghum bicolor and Zea mays (Figure S10). As the D1-TUD1-mediated BR signaling pathway is likely conserved across flowering plants, it may provide a way to manipulate the G protein pathway that leads to increased plant productivity.
Materials and Methods
Plant materials and growth conditions
The dwarf1 (d1-c), slender1 (slr1-l), d61-2 mutants were kindly provided by Dr. Chengcai Chu, Da Luo and Makoto Mastsuoka, respectively. Among them, d1-c and slr1-l were new allelic mutants. The tud1-1 was derived from tissue culture of an indica cultivar-MH63 (Oryza sativa cv. MingHui63), and tud1-2, tud1-3, and tud1-4 were isolated from spontaneous mutations of indica cultivars, ZhangYe (ZY), SHuHui163 (SH163), and TeTePu (TTP), respectively. The tud1-5 was derived from chemical mutagenesis with ethyl methylsulfonate of japonica cultivar Nipponbare.
Double mutants were isolated by phenotype observation and verified by genotyping (the primers used for genotyping are listed in Table S3). For ascertaining the genetic relationship between tud1-5 and d1-c, we successively investigated the segregation ratio of mutations in F2 and F3 populations of tud1-5/d1-c. The phenotypic segregation ratio of F2 fitted to 9 (normal)∶3 (d1-c)∶4 (tud1-5). This result was further confirmed by analyzing the F3 seeds from homozygous d1-c and tud1-5 plants. F3 progeny of d1-c plants occasionally segregated the tud1-5 phenotype in 1∶3 ratio, but F3 progeny of tud1-5 never showed the d1-c phenotype (Table S1). Mutants and wild-type rice plants were planted in paddy fields under natural conditions or greenhouse at 30°C (day) and 24°C (night).
Histological analysis and microscopy observation
Longitudinal sections of the third leaf sheaths and the third internodes were analyzed by light microscopy. Cells of the inner epidermal tissues of lemma were analyzed by scanning electron microscopy. Cell lengths were measured in each organ, both in WT and in tud1-2 and the total cell number in each organ was estimated. For light microscopy, leaf sheaths and internodes were fixed with formalin: glacial acetic acid: 70% ethanol (1 ∶ 1 ∶ 18) and then dehydrated in a graded ethanol series. Fixed tissues were embedded in paraplast (Sigma), and cut using a microtome into 12 µm thick sections and then applied to glass slides. The sections were counter-stained by 0.005% (w/v) safranin. Tissues were observed under a light microscope (BX51; Olympus, Tokyo, Japan). The inner epidermal cells of lemma were observed by scanning electron microscopy (SEM) (S-3000N; Hitachi, Tokyo, Japan). The samples were photographed under the microscopes and the size of >50 cells in each tissue were measured.
Real-time RT–PCR
Total RNA was extracted from the third leaf, the uppermost and the second internode using a Invtrogen Extraction kit. Total RNA was treated with RNase-free DNase (Promega; http://www.promega.com) and first strand cDNA was synthesized using SuperScriptII reverse transcriptase (Invtrogene). Real-time PCR was performed using 2×SYBR Green PCR Master Mix (Applied Biosystems) on an Applied Biosystems 7900HT Real-Time PCR System with at least three PCR replicates for each sample. The PCR conditions were 2 min at 50°C, then 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, and 1 min at 60°C.
In addition, methods for GA and BR sensitivity test, map-based cloning, and assays for E3 ubiquitin ligase, subcellular localization, yeast two-hybrid, BiFC and pull-down are provided in Text S1.
Supporting Information
Zdroje
1. ClouseSD (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23 : 1219–1230.
2. YeH, LiL, YinY (2011) Recent Advances in the Regulation of Brassinosteroid Signaling and Biosynthesis Pathways. Journal of Integrative Plant Biology 53 : 455–468.
3. SheJ, HanZ, KimTW, WangJ, ChengW, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474 : 472–476.
4. KimTW, WangZY (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61 : 681–704.
5. YuX, LiL, ZolaJ, AluruM, YeH, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65 : 634–646.
6. YamamuroC, IharaY, WuX, NoguchiT, FujiokaS, et al. (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12 : 1591–1606.
7. BaiMY, ZhangLY, GampalaSS, ZhuSW, SongWY, et al. (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci U S A 104 : 13839.
8. TongH, ChuC (2011) Brassinosteroid Signaling and Application in Rice. J Genet Genomics 39 (1) 3–9 doi:10.1016/j.jgg.2011.12.001
9. MontoyaT, NomuraT, FarrarK, KanetaT, YokotaT, et al. (2002) Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14 : 3163–3176.
10. ChonoM, HondaI, ZeniyaH, YoneyamaK, SaishoD, et al. (2003) A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant physiol 133 : 1209–1219.
11. NomuraT, BishopGJ, KanetaT, ReidJB, ChoryJ, et al. (2003) The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J 36 : 291–300.
12. BishopGJ (2003) Brassinosteroid Mutants of Crops. J Plant Growth Regul 22 : 325–335.
13. WangL, XuYY, MaQB, LiD, XuZH, et al. (2006) Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res 16 : 916–922.
14. OkiK, InabaN, KitagawaK, FujiokaS, KitanoH, et al. (2009) Function of the alpha subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol 50 : 161–172.
15. GaoY, WangS, AsamiT, ChenJG (2008) Loss-of-function mutations in the Arabidopsis heterotrimeric G-protein α subunit enhance the developmental defects of brassinosteroid signaling and biosynthesis mutants. Plant and Cell Physiol 49 : 1013.
16. AssmannSM (2002) Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 14: S355–S373.
17. Van EpsN, PreiningerAM, AlexanderN, KayaAI, MeierS, et al. (2011) Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci U S A 108 : 9420.
18. UtsunomiyaY, SamejimaC, TakayanagiY, IzawaY, YoshidaT, et al. (2011) Suppression of the rice heterotrimeric G protein beta-subunit gene, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J 67 : 907–916.
19. TempleBRS, JonesAM (2007) The plant heterotrimeric G-protein complex. Annu Rev Plant Biol 58 : 249–266.
20. KatoC, MizutaniT, TamakiH, KumagaiH, KamiyaT, et al. (2004) Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J 38 : 320–331.
21. Perfus-BarbeochL, JonesAM, AssmannSM (2004) Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol 7 : 719–731.
22. Ueguchi-TanakaM, FujisawaY, KobayashiM, AshikariM, IwasakiY, et al. (2000) Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci U S A 97 : 11638–11643.
23. SuharsonoU, FujisawaY, KawasakiT, IwasakiY, SatohH, et al. (2002) The heterotrimeric G protein α subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci U S A 99 : 13307.
24. OkiK, InabaN, KitanoH, TakahashiS, FujisawaY, et al. (2009) Study of novel d1 alleles, defective mutants of the alpha subunit of heterotrimeric G-protein in rice. Genes Genet Syst 84 : 35–42.
25. Nakagawa H, Tanaka A, Mori M (2011) Brassinosteroid signaling in rice. In: Hayat S, Ahmad A, editors. Brassinosteroids: A Class of Plant Hormone: Springer Science+Business Media B.V. pp. 83–117.
26. VierstraRD (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nature Reviews Molecular Cell Biol 10 : 385–397.
27. MoonJ, ParryG, EstelleM (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16 : 3181–3195.
28. AndersenP, KragelundBB, OlsenAN, LarsenFH, ChuaNH, et al. (2004) Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem 279 : 40053–40061.
29. AzevedoC, Santos-RosaMJ, ShirasuK (2001) The U-box protein family in plants. Trends Plant Sci 6 : 354–358.
30. YeeD, GoringDR (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60 : 1109–1121.
31. ZengLR, ParkCH, VenuRC, GoughJ, WangGL (2008) Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant 1 : 800–815.
32. AmadorV, MonteE, PratS (2001) Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106 : 343–354.
33. ZengLR, QuS, BordeosA, YangC, BaraoidanM, et al. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16 : 2795–2808.
34. ChoSK, RyuMY, SongC, KwakJM, KimWT (2008) Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20 : 1899–1914.
35. StoneSL, AndersonEM, MullenRT, GoringDR (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15 : 885–898.
36. TakedaK (1977) Internode elongation and dwarfism in some gramineous plants. Gamma Field Sym 16 : 1–18.
37. IkedaA, Ueguchi-TanakaM, SonodaY, KitanoH, KoshiokaM, et al. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 : 999–1010.
38. ItohH, SasakiA, Ueguchi-TanakaM, IshiyamaK, KobayashiM, et al. (2005) Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant and Cell Physiol 46 : 1392.
39. ZhuY, NomuraT, XuY, ZhangY, PengY, et al. (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18 : 442–456.
40. TsaiYC, WeirNR, HillK, ZhangW, KimHJ, et al. (2012) Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol 158 : 1666–1684.
41. WadaK, MarumoS, IkekawaN, MorisakiM, MoriK (1981) Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant and Cell Physiol 22 : 323–325.
42. HongZ, Ueguchi-TanakaM, Shimizu-SatoS, et al. (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, c-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32 : 495–508.
43. SakamotoT, MorinakaY, OhnishiT, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nature Biotech 24 : 105–109.
44. HongZ, Ueguchi-TanakaM, UmemuraK, et al. (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome p450. Plant Cell 15 : 2900–2910.
45. HieiY, OhtaS, KomariT, KumashiroT (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 : 271–282.
46. XieQ, GuoHS, DallmanG, FangS, WeissmanAM, et al. (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 : 167–170.
47. WaadtR, SchmidtLK, LohseM, HashimotoK, BockR, et al. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56 : 505–516.
48. QiaoH, WangF, ZhaoL, ZhouJ, LaiZ, et al. (2004) The F-box protein AhSLF-S2 controls the pollen function of S-RNase–based self-incompatibility. Plant Cell 16 : 2307–2322.
49. ChenJG, GaoY, JonesAM (2006) Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol 141 : 887–897.
50. Perfus-BarbeochL, JonesAM, AssmannSM (2004) Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol 7 : 719–731.
51. ChenJG (2008) Heterotrimeric G-proteins in plant development. Front Biosci 13 : 3321–3333.
52. FanC, XingY, MaoH, LuT, HanB, et al. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112 : 1164–1171.
53. HuangX, QianQ, LiuZ, SunH, HeS, et al. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41 : 494–497.
54. BotellaJR (2012) Can heterotrimeric G proteins help to feed the world? Trends Plant Sci 17 : 563–568.
Štítky
Genetika Reprodukčná medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



