-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
NTRK3 is a member of the neurotrophin receptor family and regulates cell survival. It appears to be a dependence receptor, and thus has the potential to act as an oncogene or as a tumor suppressor gene. NTRK3 is a receptor for NT-3 and when bound to NT-3 it induces cell survival, but when NT-3 free, it induces apoptosis. We identified aberrantly methylated NTRK3 in colorectal cancers through a genome-wide screen for hypermethylated genes. This discovery led us to assess whether NTRK3 could be a tumor suppressor gene in the colon. NTRK3 is methylated in 60% of colon adenomas and 67% of colon adenocarcinomas. NTRK3 methylation suppresses NTRK3 expression. Reconstitution of NTRK3 induces apoptosis in colorectal cancers, if NT-3 is absent. Furthermore, the loss of NTRK3 expression associates with neoplastic transformation in vitro and in vivo. We also found that a naturally occurring mutant NTRK3 found in human colorectal cancer inhibits the tumor suppressor activity of NTRK3. In summary, our findings suggest NTRK3 is a conditional tumor suppressor gene that is commonly inactivated in colorectal cancer by both epigenetic and genetic mechanisms whose function in the pathogenesis of colorectal cancer depends on the expression status of its ligand, NT-3.
Published in the journal: Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer. PLoS Genet 9(7): e32767. doi:10.1371/journal.pgen.1003552
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003552Summary
NTRK3 is a member of the neurotrophin receptor family and regulates cell survival. It appears to be a dependence receptor, and thus has the potential to act as an oncogene or as a tumor suppressor gene. NTRK3 is a receptor for NT-3 and when bound to NT-3 it induces cell survival, but when NT-3 free, it induces apoptosis. We identified aberrantly methylated NTRK3 in colorectal cancers through a genome-wide screen for hypermethylated genes. This discovery led us to assess whether NTRK3 could be a tumor suppressor gene in the colon. NTRK3 is methylated in 60% of colon adenomas and 67% of colon adenocarcinomas. NTRK3 methylation suppresses NTRK3 expression. Reconstitution of NTRK3 induces apoptosis in colorectal cancers, if NT-3 is absent. Furthermore, the loss of NTRK3 expression associates with neoplastic transformation in vitro and in vivo. We also found that a naturally occurring mutant NTRK3 found in human colorectal cancer inhibits the tumor suppressor activity of NTRK3. In summary, our findings suggest NTRK3 is a conditional tumor suppressor gene that is commonly inactivated in colorectal cancer by both epigenetic and genetic mechanisms whose function in the pathogenesis of colorectal cancer depends on the expression status of its ligand, NT-3.
Introduction
Colorectal cancer (CRC) arises through the accumulation of gene mutations and epigenetic alterations that result in the transformation of normal colon epithelial cells into adenocarcinomas [1]. One of the most common epigenetic changes observed in CRC is the aberrant methylation of CpG islands in the promoter region of genes. Aberrant CpG island methylation is associated with gene silencing and can inactivate tumor suppressor genes in the colon, which promotes tumor formation through the deregulation of various cellular processes including proliferation and apoptosis, among others [1], [2].
In order to identify methylated tumor suppressor genes that play a role in the formation of CRC, we conducted a genome-wide screen for methylated genes in colorectal cancers and matched normal colon epithelium tissue samples. Through this screen, we found a set of novel methylated genes, which included neurotrophin tyrosine kinase receptor 3 (NTRK3), a gene that has been found to be hypermethylated in esophageal adenocarcinoma [3]. The identification of methylated NTRK3 in CRC was unexpected given that NTRK3 has been shown to be an oncogene in breast cancer and possibly hepatocellular carcinoma [4], [5]. However, NTRK3 has also been shown to be a tumor suppressor gene in neuroblastomas [6]. Thus, our findings raised the question of whether NTRK3 acts as an oncogene or tumor suppressor gene in the pathogenesis of CRC.
NTRK3 is a member of the NTRK neurotrophin receptor family, which includes NTRK1 (TRKA), NTRK2 (TRKB) and NTRK3 (TRKC). NTRK family members and their ligands, nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3) and NT4/5, are crucial to the development of the nervous system and have poorly defined roles in other tissues [7]. NTRK1 is the receptor for NGF, and NTRK2 preferentially binds BDNF and NT4/5. NT-3 is the only known physiologically relevant ligand for NTRK3. The NTRKs have been shown to play oncogenic roles in certain cancers, such as breast cancer and liver cancer [8]. For example, a fusion of ETV6 (ETS translocation variant 6) to TRKC leads to the constitutive activation of TRKC tyrosine kinase, which promotes tumor formation and progression in human breast carcinoma [9].
However, rather than being classic tyrosine kinase receptors, recent data suggests that NTRK1 and NTRK3 may be dependence receptors [6], [10], [11]. Dependence receptors are characterized by their ability to induce opposing biological effects depending on the availability of their ligands. In the presence of the receptor's ligand, a positive cellular differentiation or survival signal is transduced, whereas lack of the ligand results in cleavage of a death-domain peptide and induction of apoptosis [12]. Indirect support for the role of NTRK3 as a dependence receptor and conditional tumor suppressor gene is provided by the observation that NTRK3 is a favorable-prognostic factor in a variety of cancers, such as melanoma [13] and medulloblastomas [14]. These findings suggested that NTRK3 might likewise serve as a conditional tumor suppressor gene in colorectal cancer.
With regards to the possibility that NTRK3 could act as a tumor suppressor gene in the colon rather than as an oncogene, somatic inactivating mutations of NTRK3 have been identified in CRC [15], as well as in other cancers including breast, lung, and pancreatic [16]. These mutations are missense mutations that are predicted to inhibit the function of NTRK3 [16] (See Table S1). Thus, the discovery of mutant, as well as methylated, NTRK3 in CRC suggested the possibility of NTRK3 being a CRC tumor suppressor gene. Consequently, we carried out a series of studies to determine the effect of aberrant DNA methylation on the expression of NTRK3 and to determine if NTRK3 had oncogene or tumor suppressor activities in colorectal cancer cell lines.
Results
Aberrant methylation of NTRK3 is common in colorectal adenomas and cancers
DNA from CRCs and normal colon mucosa samples was subjected to analysis using Infinium HumanMethylation450 BeadChip arrays (Illumina). After filtering the data as described previously [3], we identified a number of genes that were aberrantly methylated in the CRCs. One of these methylated genes was NTRK3, which was methylated in all the CRCs and in none of the normal colon epithelium samples. In light of the preferential methylation of NTRK3 in CRCs and because of its role as a neurotrophin receptor, which suggested it could have a functional role in the formation of colorectal cancer, we carried out a series of studies to further assess the effect of NTRK3 methylation on CRCs.
The promoter region of NTRK3 (NM_002530) contains a dense CpG island located from nucleotides −96 to +179 relative to the transcription start site (TSS; Figure 1A). After observing methylated NTRK3 in the colorectal cancers run on the HumanMethylation450 arrays, we assessed the methylation status of NTRK3 in a second independent set of normal colon mucosa, colon adenomas, and CRCs using a quantitative methylation-specific PCR assay (qMSP; MethyLight) designed to assess the promoter region of NTRK3. We first established that a Percent of Methylated Reference (PMR) threshold of 13.7% had a specificity of ∼90% for cancer vs. normal tissue. Using this PMR threshold, we detected NTRK3 promoter methylation in 67% of colorectal cancers (N = 76) (See Text S1 for methods used to determine optimal PMR. Figure S11 and Table S4). Using this same PMR threshold, NTRK3 promoter methylation was found in 60% of adenomas (N = 55) and 10% of the normal colon samples (N = 98; normal versus cancer: p<0.0001; normal versus adenoma: p<0.0001). The frequency of normal colon mucosa cases adjacent to CRC with NTRK3 promoter methylation did not differ significantly from that observed in the normal mucosa of cancer-free individuals (Table 1). We also assessed the status of NTRK3 in a panel of colon cancer cell lines (N = 9) and found that all the cell lines had methylated NTRK3 using the 13.7% PMR threshold.
Fig. 1. Bisulfite sequencing results of representative methylated and unmethylated samples as determined by the NTRK3 MethyLight (qMSP) assay. 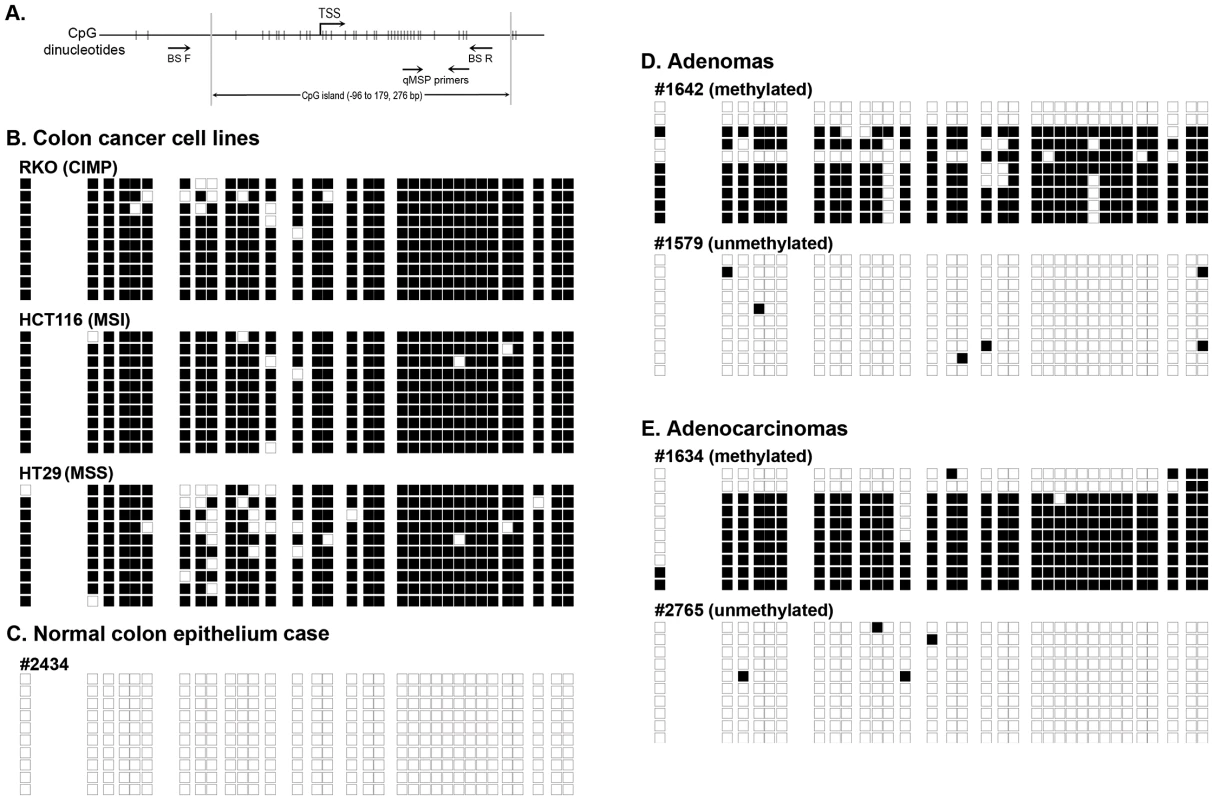
The black boxes indicate methylated CpG dinucleotides, and the white boxes represent unmethylated CpG dinucleotides. Each row depicts the sequencing results for a single clone. A. Schematic diagram of the 5′ end of the NTRK3 gene showing the location of the qMSP and bisulfite sequencing primers (TSS: transcriptional start site). B. Results of bisulfite sequencing of DNA from colon cancer cell lines representative of different molecular subtypes: RKO is a CpG island methylator phenotype (CIMP) cell line, HCT116 is microsatellite unstable (MSI) line, and HT29 is a microsatellite stable (MSS) line. These cell lines have dense methylation of NTRK3. C. Representative case of normal colon, which carries unmethylated NTRK3. D. and E. Representative cases of adenomas and adenocarcinomas that carry either methylated or unmethylated NTRK3 are shown. Tab. 1. NTRK3 promoter methylation in colorectal adenocarcinoma, adenoma and normal colon epithelium. 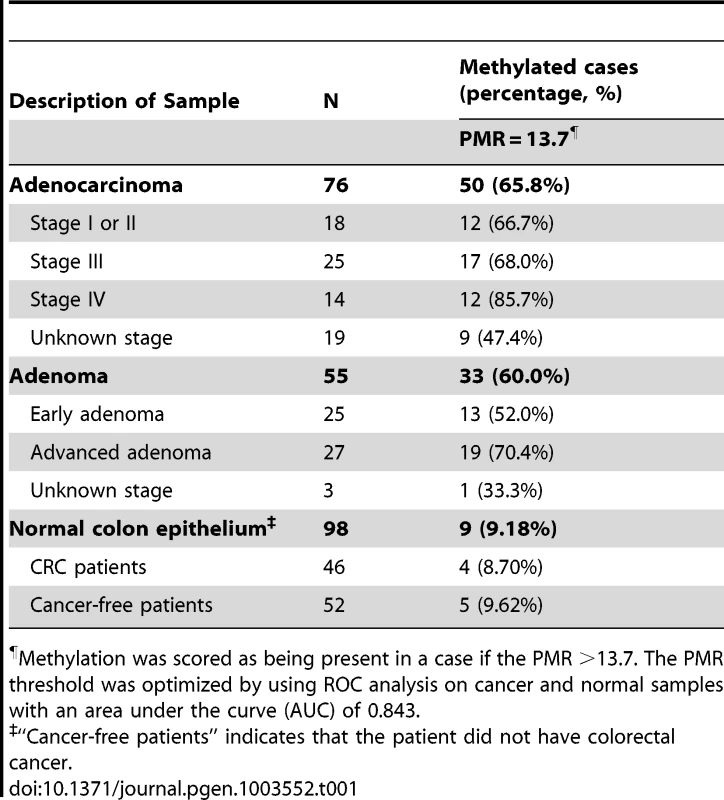
Methylation was scored as being present in a case if the PMR >13.7. The PMR threshold was optimized by using ROC analysis on cancer and normal samples with an area under the curve (AUC) of 0.843. In addition, we performed bisulfite sequencing of the promoter region of NTRK3 in representative cases of normal colon epithelium, adenomas, and adenocarcinomas (5 samples/group) and correlated these results with those of the NTRK3 qMSP assay. The bisulfite sequencing results correlated well with the qMSP results (Figure 1B–E).
Methylation of NTRK3 is independent of CIMP status, MSI status, KRAS mutations and BRAFV600E mutations in colorectal cancer
Colorectal cancer can be classified into molecular classes, which include the Microsatellite Unstable (MSI), Chromosome Unstable (CIN, also known as Microsatellite Stable, MSS), and CpG Island Methylator Phenotype (CIMP) [1]. These classes of CRC appear to have unique pathogenic mechanisms that give rise to the CRCs. We assessed the association of methylated NTRK3 with these classes of CRC and with mutations that are commonly found in CRC. As shown in Table 2, methylated NTRK3 is more frequent in tumors in women, and is independent of CIMP and MSI status, as well as KRAS, BRAFV600E, TP53, PIK3CA or APC mutations. In addition, as shown in Figure S1, methylated NTRK3 appears to be independent of other genes that are frequently methylated in CRC, such as MLH1, CDKN2A/p16 or RASSF1A.
Tab. 2. Clinical, pathological, and molecular characteristics of CRCs that carry methylated <i>NTRK3</i> and unmethylated <i>NTRK3</i>. 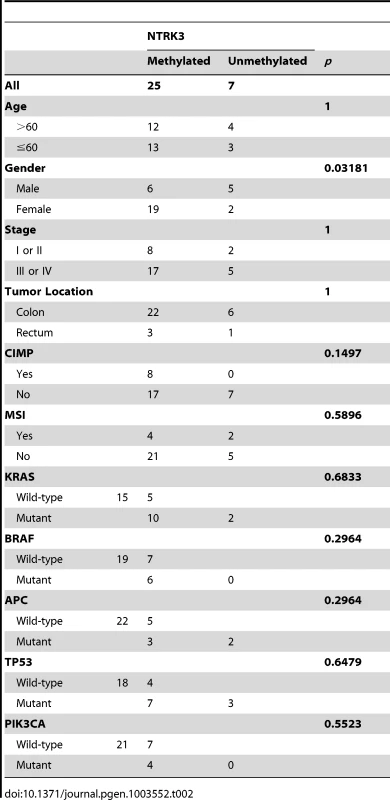
Methylation of NTRK3 silences NTRK3 expression
As mentioned above, we noticed that all nine of the colon cancer cell lines analyzed carried methylated NTRK3. Consistent with methylation silencing NTRK3 expression, we did not detect NTRK3 mRNA expression in any of these cell lines. Next, the CRC cell lines RKO and HCT116, which carry methylated NTRK3, were treated with 5-aza-2′-deoxycytidine (5-AZA), which inhibits DNA methyltransferase1 (DNMT1), to determine if demethylation of the NTRK3 promoter would induce NTRK3 expression. Following 5-AZA treatment, NTRK3 mRNA expression was induced in both HCT116 and RKO cells (Figure 2A). We next assessed NTRK3 mRNA expression in normal colon mucosa samples, colorectal adenomas, and primary colon adenocarcinomas. NTRK3 mRNA expression was significantly lower in colorectal adenocarcinomas and adenomas as compared to the matched normal colon mucosa, which carried unmethylated NTRK3 (Figure 2B). Moreover, the expression of NTRK3 was significantly higher in the primary colon tumors that carry unmethylated NTRK3 compared to the tumors that carry methylated NTRK3 (Figure 2C).
Fig. 2. NTRK3 methylation correlates with reduced NTRK3 mRNA expression. 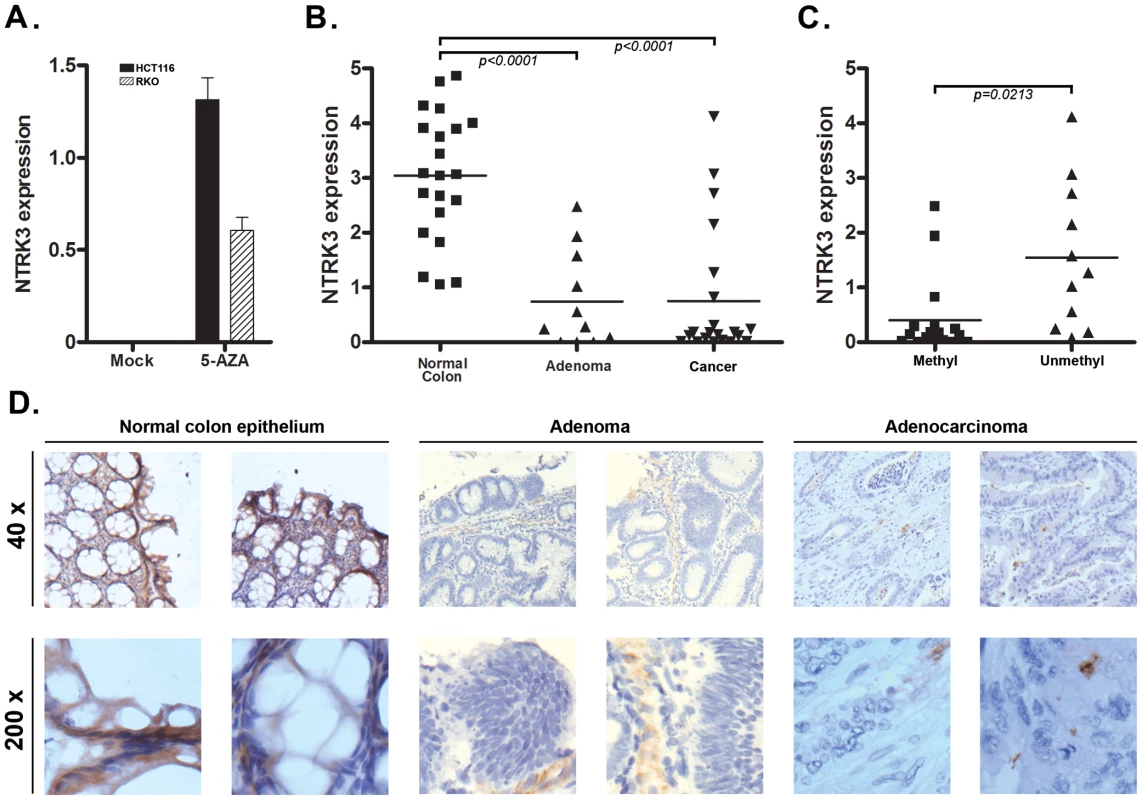
A. NTRK3 mRNA expression after treatment with 5-aza-2′-deoxycytidine (5-AZA). Colon cancer cell lines HCT116 and RKO, which carry methylated NTRK3, have no detectable NTRK3 mRNA expression when treated with the vehicle only (Mock). The expression of NTRK3 is increased significantly after 5-AZA treatment in both lines. B. Expression of NTRK3 mRNA in primary normal colon mucosa, colon adenomas, and colon adenocarcinomas (cancer). When compared to normal colon mucosa (average expression = 3.04±0.25, N = 21), NTRK3 expression is significantly lower in the colon adenomas (average expression = 0.75±0.27, N = 11) and cancers (average expression = 0.75±0.27, N = 21), presumably related to the high prevalence of colorectal adenomas and adenocarcinomas that carry methylated NTRK3 (P<0.0001, one way ANOVA). C. NTRK3 mRNA expression is higher in the adenomas and cancers that carry unmethylated NTRK3 (0.40±0.17, N = 17) compared to tumors that carry methylated NTRK3 (1.55±0.40, N = 11; P = 0.0213, 2-sided student t test). (The units on the Y-axis in A–C are relative expression units.) D. NTRK3 immunostaining of representative cases of normal colon mucosa, adenomas and adenocarcinomas (40×, upper panels; 200×, lower panels). NTRK3 is expressed in the normal colon epithelium cells, however, NTRK3 is reduced or absent in the adenomas or adenocarcinomas. The adenocarcinomas and adenomas shown all carry methylated NTRK3. We also assessed NTRK3 protein expression in normal colon mucosa and in adenomas and colorectal cancer by immunostaining. The normal colon mucosa showed heterogeneous membrane and cytoplasmic staining using an anti-NTRK3 monoclonal antibody, whereas almost no expression was detected in most adenoma and cancer cases (N = 30). Among the 20 adenoma and cancer samples, only 6 samples showed weak or moderate NTRK3 expression, while 9 out of 10 normal samples showed strong or moderate NTRK3 expression (Figure 2D). Taken together, these data provide support for the aberrant methylation of the NTRK3 promoter silencing NTRK3 expression in colon neoplasms.
Expression of NT-3 in colon cancer cell lines and primary tissues
Because NTRK3 has been shown to function as a dependence receptor in certain tissues, we also assessed the expression of NTRK3's ligand, NT-3, in CRC cell lines and primary CRCs. NTRK3's preferred ligand is NT-3, and NT-3 has been shown to inhibit NTRK3 mediated apoptosis and to induce NTRK3-mediated activation of signaling pathways involved in cell proliferation, apoptosis and motility [6], [17]. Therefore, we assessed the expression levels of NT-3 in the panel of colon cancer cell lines previously assessed for methylated NTRK3. No NT-3 expression was detected in RKO, HCT116, FET, Vaco400 and HT-29, whereas NT-3 was expressed at a low (although relatively high level in relation to the other CRC cell lines) in SW480. NT-3 expression was present at low levels in Lovo, LS174T, and AAC1/SB10 (Figure S2A). We next assessed the expression of NT-3 in primary CRC tissues and in matched normal colon mucosa specimens. NT-3 expression was significantly lower in the CRC's when compared to the normal colon (Figure 3A). Interestingly, we found a direct correlation between NT-3 expression and NTRK3 expression in the normal colon and in the CRC's (r2 = 0.81, Pearson's correlation P<0.0001), suggesting that the presence of NT-3 relieved the selective pressure to silence NTRK3 (Figure 3B).
Fig. 3. NT3 expression and the relationship of NTRK3 and NT3 mRNA expression in primary colon tissues. 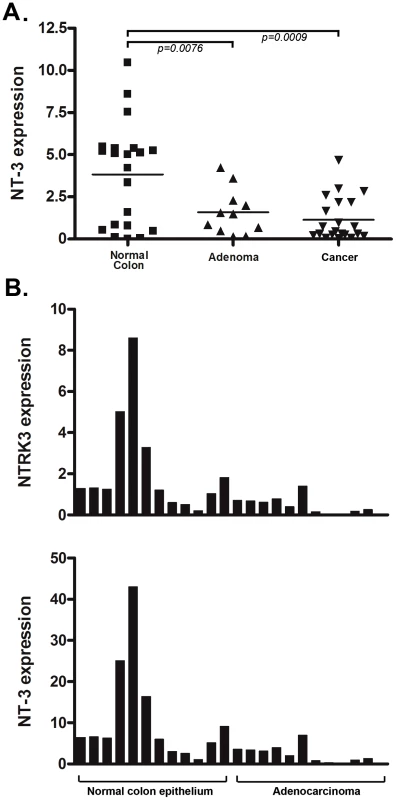
A. NT3 mRNA expression is significantly lower in both the adenoma and adenocarcinoma samples compared to the matched normal colon epithelium (both p<0.01) (Please see methods for units on Y axis). B. There is a direct correlation between NT3 and NTRK3 expression in normal colon and adenocarcinomas (r2 = 0.81, Pearson's correlation P<0.0001). The units on the Y-axis are relative expression units and each bar on the X-axis represents one sample. The bar graphs are aligned so that the samples correspond in the upper and lower graphs. In order to determine the mechanism responsible for loss of NT-3 expression, we assessed the methylation status of the promoter region of NT3 using an NT3 MSP assay and correlated these results with the NT3 mRNA expression levels. We found that the colon cancer cell lines lacking NT3 expression have aberrantly methylated NT3, whereas those that express NT3 mRNA carry unmethylated NT3 (Figure S2C). We also found that 5-AZA treatment of two cell lines that carry methylated NT3, HCT116 and RKO, induces the expression of NT3 (Figure S2B). Therefore, we conclude that the methylation of NT3 can repress the expression of NT3.
NTRK3 is a dependence receptor in CRC
NTRK3 has been shown to be a dependence receptor in certain tumors and can trigger caspase-based apoptosis when not bound by NT-3 [6], [10]. In the presence of NT-3, NTRK3 induces differentiation, guidance or survival in neurons; however, NTRK3 can alternatively induce apoptotic cell death in the absence of NT-3 in neuroblastoma cells and presumably other cell types [6]. The dependence receptor aspect of the biological effects of NTRK3 suggests it has the potential to be either an oncogene or a tumor suppressor gene, depending on the presence of NT-3. In order to assess the effect of NTRK3 on colorectal cancer, NTRK3 was transfected into the HCT116 (MSI), RKO (CIMP) and HT29 (CIMP/MSS) cell lines, which lack NTRK3 and NT3 mRNA expression (Figure S3A and B). In these cell lines, NTRK3 reconstitution increased caspase activity by 2–3 fold compared to the control vector transfected cells. Furthermore, the addition of NT-3 (100 ng/mL) suppressed apoptosis induced by NTRK3 reconstitution (Figure 4A, B and C). These results were confirmed using an independent assay that assesses apoptosis by detecting apoptosis specific DNA:histone complexes (Cell Death Detection Assay (Roche); Figure S4 A, B and C).
Fig. 4. Assessment of normalized caspase 3 and 7 activity after reconstitution of NTRK3 in HCT116 (A), RKO (B) and HT29 (C) cells. 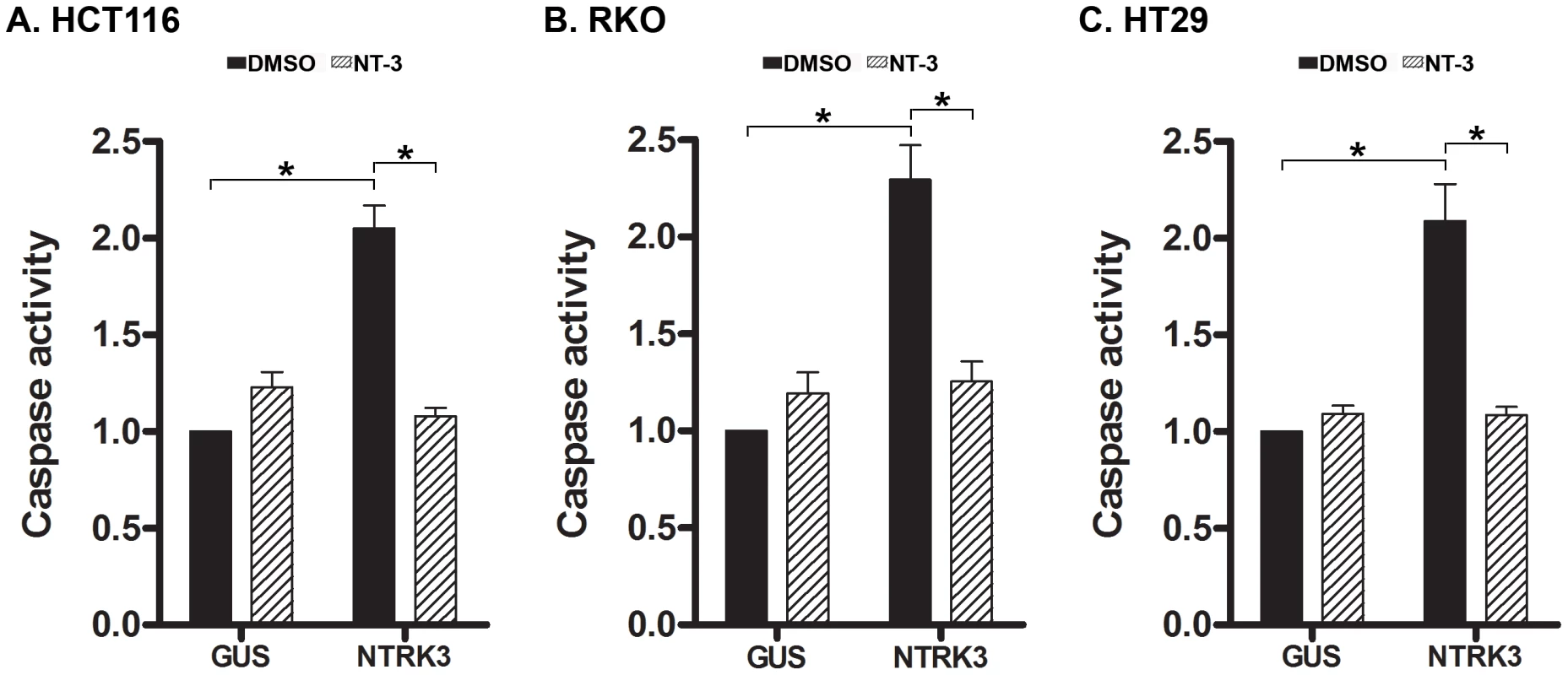
NTRK3 induces caspase activity in HCT116 (MSI), RKO (CIMP) and HT29 (MSS) cells, and NT-3 (100 ng/mL) inhibits this effect in all three cell lines. DMSO is the vehicle control for NT-3, and GUS is the control vector (pDEST27-GUS) used to normalize for nonspecific effects of the transfection on caspase activity. The caspase activity in the GUS transfected cells treated with DMSO was used to normalize the results from the other experimental groups. The asterisks indicate statistically significant differences, p<0.05 as determined by a 2-sided Mann-Whitney rank sum test. Somatic NTRK3 mutations occur in primary colorectal cancer and can inactivate NTRK3
Somatic mutations of NTRK3 have been identified in primary colorectal cancers [15]. In order to determine the effect of the mutant NTRK3 genes on the behavior of colorectal cancers, we constructed plasmids that express the following NTRK3 mutants: NTRK3-G608S, NTRK3-I695V and NTRK3-L760I [15]. The mutant NTRK3 constructs were then transfected into the CRC cell line RKO. Transfection of NTRK3-L760I into the RKO cells did not induce apoptosis (Figure 5B), but the wild-type NTRK3, NTRK3-G608S or NTRK3-I695V alleles did induce apoptosis. Moreover, inhibition of colony formation by NTRK3 was not induced by NTRK3-L760I, but was induced by NTRK3-G608S and NTRK3-I695V (Figure 5C). These findings demonstrate that NTRK3-L760I inactivates NTRK3 with regards to its apoptosis and colony formation ability. However, the other mutant alleles do not affect the function of NTRK3 and are presumably passenger mutations. (Of note, the mutation status of all the constructs was confirmed by direct sequencing, see Figure S5). Also, we did not observe any change in NT-3 expression after transfection with the wild-type or mutant NTRK3 constructs. These findings suggest NTRK3 is a tumor suppressor gene in the colon that can be inactivated by both epigenetic and genetic mechanisms. The identification of both methylated NTRK3 and inactivating NTRK3 mutations in colorectal cancers provides evidence that inactivation of NTRK3 promotes tumor formation in the colon.
Fig. 5. Assessment of caspase activity and colony formation after reconstitution with mutant NTRK3 in RKO. 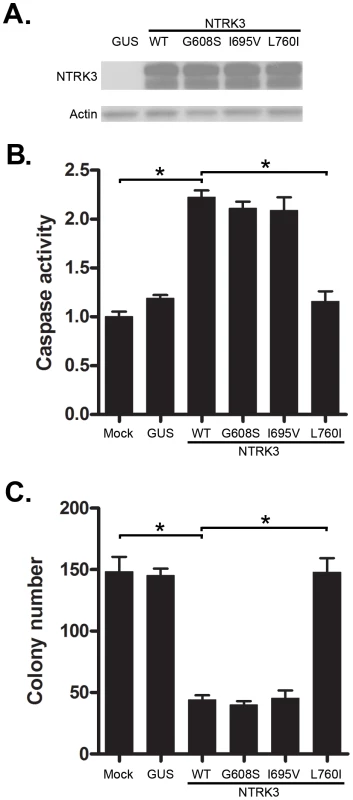
The three mutant NTRK3 constructs contain NTRK3 mutants found in primary human CRC. A. The expression of reconstituted wild-type NTRK3 (WT), NTRK3-G608S (G608S), NTRK3-I695V (I695V) and NTRK3-L760I (L760I) was confirmed by western blotting. B. Apoptosis was assessed by normalized caspase 3 and 7 activity in the RKO cell line 48 hours after transfection with WT, G608S, I695V and L760I NTRK3 constructs. C. Soft agar colony formation was assessed in stably-transfected RKO cells after 2 weeks. Results are plotted as the mean colony numbers from three independent experiments. L760I did not induce apoptosis (B) or suppress colony formation (C), whereas WT, G608S and I695V did. Thus, the G608S and I695V mutations appear to be passenger mutations. GUS was used as the control vector to normalize for nonspecific effects of the transfection on apoptosis. The asterisks indicate statistically significant differences, p<0.05 as determined by a 2-sided Mann-Whitney rank sum test. Loss of NTRK3 leads to perturbed MAPK signaling pathway
As shown in the experiments above, NTRK3 can act as a tumor suppressor gene in colon cancer cell lines and can induce apoptosis in CRC cell lines through the activation of caspase 3 or caspase 7. We next assessed the signaling pathways that are affected by NTRK3, which have been shown to include the MAPK/Erk, NF-κB and PI3K/Akt pathways, to determine if they may be mediating NTRK3 induced apoptosis in the colon cancer cell lines [18]. We initially assessed the activation status of the MAPK/Erk pathway in the HCT116 and RKO cell lines. HCT116 and RKO cells reconstituted with NTRK3 show increased activation of the MAPK/Erk pathway as determined by increased phospho-Erk1/2 (p-Erk1/2) expression. This increase in p-Erk1/2 was accompanied by increased caspase3/7 activity. The NTRK3-induced apoptosis was inhibited by the MAPK inhibitor U0126 (Figure 6A and B). In order to confirm that increased p-Erk1/2-induced apoptosis was specific to NTRK3, we used 16% FBS as an extracellular stimulus to induce increased p-Erk1/2. Not surprisingly, cells treated with 16% FBS showed significant increased p-Erk1/2, which was not accompanied by increased apoptosis (Figure S6). Since the addition of NT-3 inhibited apoptosis induced by NTRK3 expression, we assessed whether the introduction of NT-3 affected the activation status of the MAPK/Erk pathway. Interestingly, the addition of NT-3 decreased NTRK3 protein expression and decreased p-Erk1/2 levels (Figure S7). These findings suggest that at least part of NTRK3's pro-apoptotic effects occur through the MAPK signaling pathway in colon cancer cell lines.
Fig. 6. NTRK3 expression induces phosphorylation of Erk1/2 (Thr202/Tyr204), and the inhibition of phosphorylation of Erk1/2 correlates with decreased apoptosis induced by NTRK3. 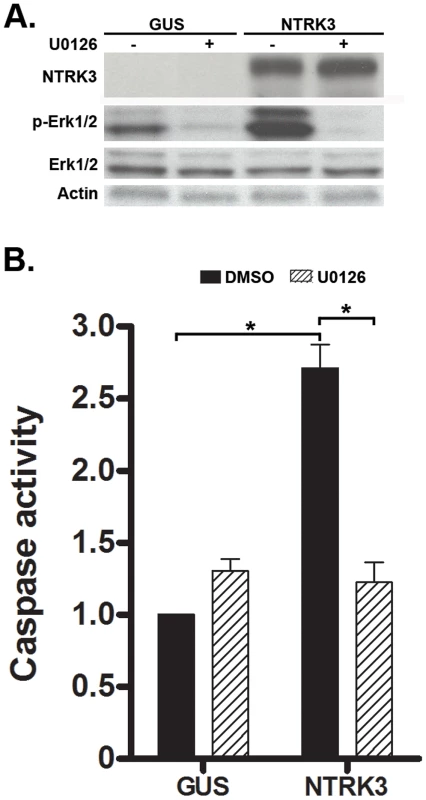
HCT116 cells were transfected with either GUS or NTRK3, and then 24 hours after the transfection, were treated with the MAPK inhibitor U0126 (10 µM) for an additional 24 hours. A. NTRK3 mediated phosphorylation of Erk1/2 is inhibited by U0126. B. U0126 reverses the caspase activation induced by NTRK3 expression in HCT116 cells. DMSO was used as a vehicle control. The asterisks indicate statistically significant differences (p<0.05; 2-sided Mann-Whitney rank sum test). Caspase activity was normalized to the GUS vector transfected HCT116 cells treated with DMSO. In light of the prior reports implicating a fusion gene involving NTRK3 affecting TGF-β signaling and TGF-β mediated cell behavior, we also assessed the TGF-β, BMP signaling and EMT markers in colon cancer cells after transfection with NTRK3 [4], [5]. However, we did not observe a significant change in the expression of any of these proteins (Figure S8).
Suppression of NTRK3 induces transformed behavior in colon epithelial cells
Since NTRK3 is frequently methylated in colorectal adenomas, we carried out a series of studies to determine if loss of NTRK3 could induce transformed behavior in normal colon epithelial cells. We knocked down the expression of Ntrk3 in an immortalized murine colon epithelial cell line (YAMC) and then assessed the cells for transformed behavior using a soft agar colony formation assay. As shown in Figure 7, the knockdown of Ntrk3 was ∼80% as measured by RT-PCR, and this level of knockdown promoted anchorage independent growth in the YAMC cells. Of note, the parental YAMC cells grow slowly in soft agar. These findings suggest that loss of NTRK3 could be an early event in CRC formation.
Fig. 7. Knock-down of Ntrk3 expression contributes to neoplastic transformation in normal colon epithelial cells. 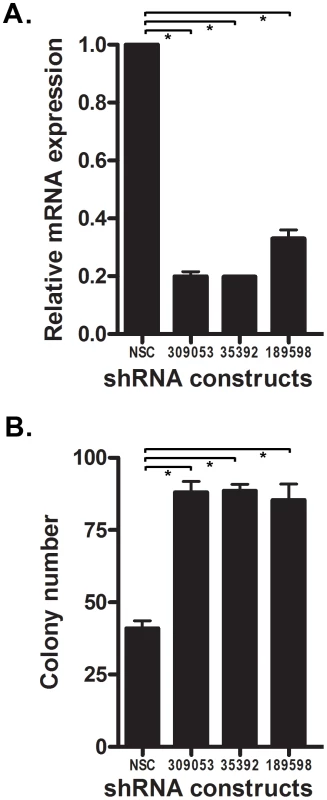
A. Ntrk3 mRNA expression was efficiently knocked down by 3 different shRNA constructs (309053, 35392 and 189598) in the immortalized mouse colon epithelial cell line, YAMC. B. Suppression of Ntrk3 promotes soft agar colony formation in the YAMC cells. The non-silencing control (NSC) is a control short-hairpin construct for the knockdown experiments. The asterisks indicate statistically significant differences. (p<0.05; two-sided student t test). NTRK3 suppresses the tumorigenic behavior of colon cancer cell lines both in vitro and in vivo
We also performed studies on anchorage independent growth and tumor xenograft formation in established CRC cell lines to assess the putative tumor suppressor role of NTRK3 in CRC. First, we assessed the effect of NTRK3 expression on soft agar colony formation in HCT116 (MSI), RKO (CIMP) and HT29 (CIMP/MSS) cells. Transfection of full-length NTRK3 induced a nearly 5 - to 10-fold reduction in colony number of HCT116 (Figure 8A), RKO and HT29 cells (Figure S9A and B). We also assessed the tumor-suppressor activity of NTRK3 in xenografts in immunodeficient nu/nu nude mice. NTRK3 reconstitution significantly suppressed tumor growth compared to xenografts containing a control vector (Figure 8B). Twenty-one days after subcutaneous injection of the cells, the mice were sacrificed and the tumors were excised and measured. We found that both the size and weight of the NTRK3-expressing tumors were significantly reduced compared to the control xenografts (Figure 8C and Figure S10). NTRK3 expression in the xenografts from the cells transfected with NTRK3 was confirmed by IHC (Figure 8D). Taken together, these results provide support for a tumor suppressor role for NTRK3 in CRC.
Fig. 8. NTRK3 suppresses in vitro soft agar colony formation (A) and tumor xenograft growth (B–D) of colon cancer cell lines. 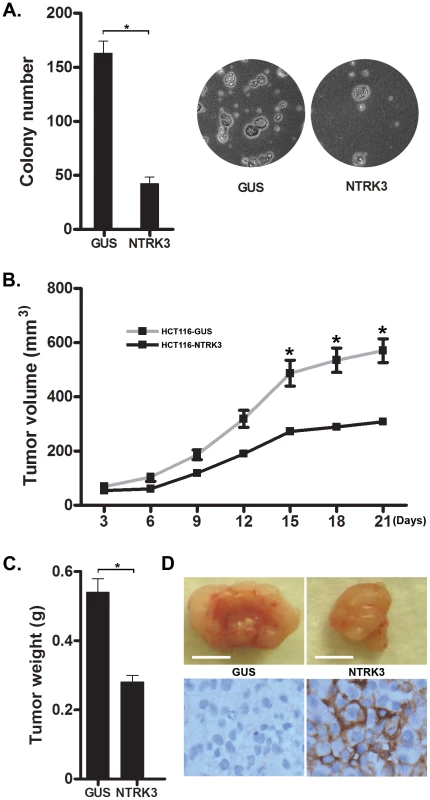
A. The GUS and NTRK3 stably transfected HCT116 cells were grown in soft agar for 2 weeks. Results are plotted as the mean colony numbers from three independent experiments. The asterisks indicate statistically significant differences (p<0.05; two-sided student t test). Representative fields depicting colonies of HCT116 cells grown in soft agar are shown. B. The growth of the HCT116-NTRK3 and HCT116-GUS xenograft tumors in the nu/nu mice was measured over 3 weeks. The mean tumor volume is indicated, and the asterisks indicate statistically significant differences between the mean tumor volume of the NTRK3-reconstituted and control tumors (two-sided student t test). C. Tumors were removed 21 days after subcutaneous injection. The final tumor weight in NTRK3-expressing tumors was significantly less than the weight of the control tumors (p = 0.0021, n = 10). D. Representative tumors are shown (scale bar: 5 mm) along with confirmatory NTRK3 immunostaining. Discussion
The aberrant methylation of CpG islands in the promoter regions of genes is a common event in many cancers [19]. The average colon cancer genome contains 1,000–3,000 abnormally methylated genes [20]. In many cases, the aberrant methylation of these genes can silence the expression of tumor suppressor genes and consequently promote tumor formation. However, it is also apparent that the hypermethylation of many genes in cancer has no effect on the expression of the methylated gene and does not influence tumor formation. This later class of methylated genes is felt to represent passenger events in tumorigenesis [20]. Through a genome-wide screen for methylated genes in colon cancers, we identified methylated NTRK3 in colon adenomas and adenocarcinomas. Methylated NTRK3 was found in 67% of colorectal adenocarcinomas and 60% of adenomas. With regards to the functional significance of this epigenetic alteration, we found that the aberrant methylation of NTRK3 suppressed NTRK3 expression, which suggested NTRK3 might act as a tumor suppressor gene in colon cancer. Our findings are in contrast to other studies in breast cancer that have demonstrated that NTRK3 is oncogenic [4]. These opposing results appear to be a consequence of NTRK3 being a dependence receptor, which means that it can induce proliferation when it binds its ligand, NT-3, but induces apoptosis when NT-3 is not available [6]. Because NT-3 is expressed in the colon epithelium but not in colon neoplasms, our findings suggest that silencing of NTRK3 releases colon cancer cells from NTRK3-mediated apoptosis. These findings suggest that NTRK3 might function as a novel conditional tumor suppressor gene in CRC.
Although somatic mutations of NTRK3 that are predicted to inactivate function have been observed in CRC, NTRK3's role as a tumor suppressor gene in CRC has not been clearly demonstrated to date [15]. In the present study, we have provided evidence that NTRK3 can have conditional tumor suppressor activities in CRC. A similar role for NTRK3 in neuroblastomas has recently been shown [6]. Reconstitution of NTRK3 in the absence of NT-3, the ligand for NTRK3, induced caspase-related apoptosis and cell death in the colon cancer cell lines RKO, HT29 and HCT116. We found that the effects on apoptosis could be suppressed by the treatment of the NTRK3 expressing cell lines with NT-3. Perhaps most importantly, NTRK3 inhibited colony formation in soft agar colony formation assays and suppressed the growth of tumor xenografts, which are hallmark in vitro effects of tumor suppressor genes. In addition, we have shown that the naturally occurring NTRK3-L760I mutation impairs NTRK3's ability to induce apoptosis and suppress anchorage independent growth. These findings suggest that NTRK3 is a CRC tumor suppressor gene that is inactivated by both genetic and epigenetic mechanisms.
The demonstration of NTRK3 as a potential conditional tumor suppressor gene in the colon suggests NTRK3 may be the latest member of a class of dependence receptors that suppress colon cancer formation. Other conditional tumor suppressor genes identified in CRC and other cancers, include DCC, UNC5C, p75NTR and MET [12], [21]. The dependence receptor model purports that some receptors induce different biological effects on cells depending on whether they are in a ligand-bound or ligand-free state. These receptors can induce caspase-mediated apoptosis in the absence of ligand, but induce proliferation when bound by their ligands. Therefore, one of the critical aspects of this study is the assessment of the expression of the NTRK3 ligand NT-3 in the colon. NTRK3's preferred ligand, NT-3, was found to be substantially suppressed in both colorectal adenomas and adenocarcinomas, presumably secondary to hypermethylation of the NT3 promoter region. It is plausible that the loss of NT-3 expression precedes the loss of NTRK3, which would create a clonal survival advantage for those CRC cells that silence NTRK3. Our studies suggest that inactivation of NTRK3 occurs early in the polyp→cancer sequence and that it contributes to the transformation of colon epithelial cells.
With regards to the results of our studies, it is also important to consider the effects of loss of NT-3 and NTRK3 in the context of the entire neurotrophin receptor and ligand families because cross-talk between the ligand and receptor family members can occur. It has been shown that a precursor of NT-3, proNT-3, can activate p75NTR and that NT-3 can activate NTRK1 or NTRK2, although this happens with low efficiency [18]. However, despite the potential for cross-talk, we did not observe any effects on colon cancer cells that lacked NTRK3 after being treated with NT-3. Therefore, our findings suggest that NTRK3 is the primary and perhaps only receptor for NT-3 in the colon and in colon neoplasms.
When bound to NT-3, NTRK3 functions as a typical receptor tyrosine kinase. Its activation is stimulated by neurotrophin-mediated dimerization and transphosphorylation of an activation loop tyrosine [22]. The major pathways activated by the NTRKs are MAPK, PI3K and PLC-γ1, among others [18], [22], [23]. Activation of the NTRKs and p75NTR promote activation of NF-κB, and p75NTR can activate the JNK pathway [18], [24], [25]. Previous studies have demonstrated that the activation of the MAPK and PI3K pathways by NTRK3 promotes cell differentiation, which in turn affects tumor progression [4], [23]. In this study, we also found that NTRK3 expression can activate the MAPK pathway. However, in this context the activation of ERK1/2 appears to be involved in the apoptotic response in colon cancer cells. There is a possibility that the MAPK activation we observed in this setting is an indirect effect of NTRK3 and a consequence of unopposed activation of p75NTR [18]. Our studies do not allow us to exclude this possibility, although even if such a mechanism was present, it would not change the interpretation of NTRK3 as being a colorectal cancer tumor suppressor gene.
In summary, we have identified NTRK3 as a novel conditional tumor suppressor gene in the colon that is inactivated by epigenetic and genetic mechanisms. We have provided evidence that NTRK3 can trigger apoptosis and inhibit tumor growth in the absence of its ligand NT-3 and that these effects are reversed by the addition of NT-3. We also showed that suppression of NTRK3 can induce transformed behavior in immortalized colon epithelial cells. Our studies provide further insight into the complex relationship between NTRK3 and NT-3 in cancers as well as into dependence receptor biology in the colon. This class of tumor suppressor genes may offer new therapeutic strategies in the colon.
Materials and Methods
Cell lines, tissues and nucleic acid extraction
All studies in this manuscript have been approved by the FHCRC IRB committee and the IACUC committee. The studies of human tissues were all done on anonymous samples. The IRB protocol covering this study is IRB 1989 and is available upon request.
Nine human colorectal cancer cell lines (SW480, Vaco400, LS174T, HT29, Vaco576, RKO, Vaco503, HCT116 and Lovo) representing the spectrum of CRC molecular subtypes [MSI, CIN (aka MSS) and CIMP] were used. The cell lines were either purchased from ATCC or were kindly provided by Sanford Markowitz (Case Western Reserve University School of Medicine and Case Medical Center, Cleveland, OH). All cell lines had their identity confirmed by DNA genotyping. Some of the cell lines were treated with the DNMT1 inhibitor (5 µM) 5-aza-2′-deoxycytidine (5-AZA; Sigma) in the experiments in this study.
Primary tissue samples used in the methylation array studies (N = 8 CRC's) were obtained from the ColoCare CRC cohort study (Fred Hutchinson Cancer Research Center, Seattle, WA) and from healthy individuals undergoing screening colonoscopy at the University of Washington Medical Center (Seattle, WA) (N = 6). Detailed information on these samples is shown in Table S2.
Formalin-fixed, paraffin-embedded (FFPE) and fresh-frozen colon neoplasms and normal colon tissue samples were obtained from the pathology archives at Vanderbilt University Medical Center (Nashville, TN), the Department of Veterans Affairs Tennessee Valley Health Care System, Meharry Medical Center (Nashville, TN), and the University Hospital of Cleveland (Cleveland, OH) following IRB approved protocols at each institution. Colorectal cancers, colon adenomas, and adjacent normal tissue samples were also provided by the Cooperative Human Tissue Network. In total, these samples included 52 cases of histologically normal colonic mucosa from individuals without cancer or inflammatory bowel disease (IBD) and 25 samples of histological normal colonic mucosa from individuals who had undergone colon resection for CRC or colon adenomas.
DNA and RNA were extracted from these samples as previously described [26].
Methylation array studies
These studies were conducted using Infinium HumanMethylation450 BeadChip arrays (Illumina) with DNA from CRCs (N = 8) and normal colon epithelium samples from cancer-free individuals (N = 6). Specific details regarding the platform, sample preparation and data filtering strategies have been described in our previous studies [3].
Sodium bisulfite conversion of genomic DNA
Bisulfite conversion of DNA was performed as described previously [27], [28].
Bisulfite sequencing
For sequencing, bisulfite-converted DNA was PCR amplified, and the amplicons were then subjected to direct sequencing. The primers are described in detail in Table S3. The sequencing was conducted as described previously [27].
Quantitative methylation-specific PCR
Quantitative methylation-specific PCR (qMSP; MethyLight) was performed using an ABI Prism 7700 detection system (Applied Biosystems). Detailed methods are provided in the Text S1 as well as in previous publications [27]. The primers and probes targeting NTRK3 are described in detail in Table S3.
Molecular characterization of colorectal neoplasms
The CpG Island Methylator Phenotype (CIMP) status and Microsatellite instability (MSI) status of a subset of the colorectal neoplasms were determined as described previously [27]. The gene mutation status of KRAS, BRAF, APC, TP53 and PIK3CA was assessed by using the qBiomarker Somatic Mutation PCR System Arrays/Human Colon Cancer (Qiagen) following the manufacturer's protocol.
Cell culture, plasmid constructs and transfection
Human CRC cell lines (SW480, Vaco400, LS174T, HT29, Vaco576, RKO, Vaco503, HCT116, and Lovo) were grown in Dulbecco's Modified Eagle media (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen). The YAMC (Young Adult Mouse Colon) cell line was a kind gift from Dr. Robert H. Whitehead (Vanderbilt University School of Medicine, Nashville, TN) and was cultured as described previously [29]. To investigate the effects of re-expression of NTRK3, HCT116 and RKO cells were incubated for 72 hours with 5 µM 5′-AZA (Sigma). The media was replaced every 24 hours with fresh 5′-AZA. After 72 hours of treatment, the cells were washed twice with PBS and then grown in drug-free media for another 72 hours before harvesting.
The full-length NTRK3 cDNA (IOH54159, Invitrogen) was subcloned into the pDEST27 Vector (Invitrogen) to create pDEST27-NTRK3. The correct orientation of the insert was confirmed by restriction digest.
The pDEST27-based plasmids containing NTRK3-G608S, NTRK3-I695V and NTRK3-L760I were constructed based on the wild-type NTRK3 expression vector by using the GENEART site-direct mutagenesis system (Invitrogen) following the manufacturer's protocol. Successful mutagenesis was confirmed by direct sequencing (Figure S5). The sequencing primers are described in Table S3.
pGIPz-based short hairpin RNA (shRNA) constructs specifically targeting mouse Ntrk3 mRNA were purchased from Thermo Scientific/Open Biosystems Mouse shRNAmir Libraries maintained by the Genomics Shared Resource Core at the Fred Hutchinson Research Center (FHCRC, Seattle, WA). The details regarding the target regions of each shRNA are shown in Table S3. All 3 shRNA constructs were confirmed by direct sequencing (Table S3). Lentivirus containing the shRNA constructs were generated by co-transfecting pGIPz-shRNA with the packaging plasmid psPAX2 and the envelope plasmid pMD2.G (kindly provided by Michael Davis, FHCRC, Seattle, WA) into 293T packaging cells. Virus supernatant was filtered through a 0.2-µm filter and stored at −80°C until use. shRNA-mediated knock-down studies were performed by infecting YAMC cells with shRNA lentivirus for 48 hours, followed by an additional 72 hours of puromycin selection (4 µg/mL; Invitrogen). The stable YAMC cells were maintained in media containing 2 µg/mL puromycin. pGIPz-shNSC (Thermo Scientific/Open Biosystems), which expresses a scrambled shRNA with no known target sequence in the mouse genome, was used as a control for the transduction procedure.
HCT116 (MSI), RKO (CIMP) and HT29 (CIMP/MSS) cells were transiently transfected with pDEST27-based constructs (Invitrogen) using the XtremeGENE 9 DNA transfection reagent (Roche) following the manufacturer's protocol. Transfected cells were grown for 10–14 days in media containing 2400 µg/mL G418 (RKO cells) (Invitrogen) or 1200 µg/mL G418 (HCT116 and HT29 cells). For the experiments using NT-3, recombinant human neurotrophin-3 (NT-3) (R&D System) was added into the culture media 24 hours after transfection and incubated for another 24 hours.
Quantitative RT-PCR
Total RNA was isolated from CRC cell lines and primary tissues using TRIzol (Ambion) and was purified with the RNeasy Mini kit (Qiagen) according to the manufacturers' protocols. For human samples, TaqMan On-Demand primers and probes (Applied Biosystems) were used to determine the relative expression levels of NTRK3 (Hs00176797_m1) and NT-3 (Hs01548350_m1). GUSB (Hs99999908_m1) was used as a RNA input loading control. For YAMC cells (mouse) related RT-PCR, Ntrk3 (Mm00456222_m1) and Gusb (Mm01197698_m1) were used. All reactions were run in triplicate on an ABI Prism 7700 detection system.
Immunohistochemistry
FFPE sections of CRCs, adenomas and matched normal colonic mucosa were subjected to immunostaining with a rabbit anti-human TrkC monoclonal antibody (C44h5, Cell Signaling Technology). Briefly, 4 µm tissue sections were deparaffinized, rehydrated, and subjected to antigen retrieval by boiling in sodium citrate buffer (10 mmol/L, pH 6.0). The sections were incubated for 60 minutes with TrkC primary antibody (1∶50), stained with 3,3-diaminobenzidine, counterstained with hematoxylin, and mounted as described previously [30].
In vitro apoptosis assays
Programmed cell death was analyzed using the Cell Death Detection ELISA (Roche Molecular Biochemicals) following the manufacturer's instructions. Briefly, 48 hours after transfection or 24 hours after treatment with NT-3 or a selective ERK inhibitor, the cells were lysed, and the lysates analyzed using the ELISA kit.
Caspase-3 and caspase-7 activity was measured using the Caspase-Glo3/7 Assay (Promega) following the manufacturer's protocol. Relative caspase activation was calculated as the ratio of the caspase activity of the NTRK3- transfected cells and the negative control GUS-transfected cells.
Soft agar colony formation assay
The soft agar colony formation assays were performed as previously described [26], [27]. Briefly, 6,000 stably transfected HCT116, RKO or YAMC cells were mixed with 0.4% Sea Plaque agarose containing tissue culture media (top layer) and pipetted onto a solidified layer of 0.8% Sea Plaque agarose containing tissue culture media (bottom layer) in 35 mm petri dishes in triplicate. The tissue culture media was exchanged every 2∼3 days. After 14 days, colonies larger than 50 µm in diameter were counted using a phase-contrast microscope equipped with a reticule in 4 randomly selected fields in the three replicate dishes. The experiment was performed in triplicate.
Western immunoblotting
Western blotting experiments were conducted as described previously [30]. Briefly, cells were lysed using RIPA buffer containing Phosphatase Inhibitor Cocktail (P5726, Sigma) at 0°C. The protein extracts (50 µg/sample) were subjected to electrophoresis through 12% Bis-Trispolyacrylamide gels (BioRad) and then transferred to PVDF membranes. Antibodies used were purchased from Cell Signaling Technology: anti-phospho-Akt (Ser473, #9271), anti-phospho-NF-κB (Ser536, #3033), anti-phospho-Erk1/2 (Thr202/Tyr204, #4370), anti-phospho-Smad1 (Ser463/465)/Smad5 (Ser463/465)/Smad8 (Ser426/428) (#9511), anti-phospho-Smad2 (Ser465/467, #3101), anti-phospho-Smad3 (C25A9, Ser423/425, #9520), anti-Smad2/3 (#3102), anti-TrkC (C44h5, #3376), anti-BMPR2 (#6979), and Epithelial-Mesenchymal Transition (EMT) antibodies, anti-N-Cadherin (#4061), anti-Vimentin (D21H3, #5741), anti-E-Cadherin (24E10, #3195), anti-ZO-1 (D7D12, #8193), anti-Snail (C15D3, #3879). Immunoreactive proteins were then visualized by incubating the PVDF membranes with ECL plus detection reagents, followed by imaging of chemiluminescence on an imager (X-ray film-based).
Tumor xenograft studies
The care and use of the mice was conducted following protocols approved by the Fred Hutchinson Cancer Research Center IACUC. The IACUC protocol covering this work is IACUC 1624 and is available upon request. All IACUC protocols at the FHCRC require that animal suffering is eliminated unless required for the studies, in which case extensive justification is required.
Three to four week-old female athymic nu/nu mice were obtained from Harlan Laboratories. The mice were housed for one week in a pathogen-free animal facility prior to tumor cell injection. 1×107 GUS or NTRK3 stably transfected HCT116 cells in 200 µL DMEM and Matrigel (1∶1 mix) (BD Biosciences) were injected subcutaneously into the right flank of each mouse. The tumor sizes were measured using a caliper, and the tumor volume was calculated as follows: 0.5(length×width2) [6]. The mice were assessed every three days and sacrificed at three weeks after injection.
Statistical analyses
Receiver operating characteristic (ROC) curves and area under the curve (AUC) for NTRK3 methylation frequency of primary tissues were constructed on the basis of methylation levels. The Chi-squared test was used to compare the frequency of methylated NTRK3 between cancer and normal samples. The Fisher's exact test was used to test the association between the NTRK3 methylation status and clinical/molecular characteristics of CRC patients. Student t test or analysis of variance (ANOVA) was used to analyze the RT-PCR data. The Mann-Whitney rank sum test was used to analyze the data obtained from the cell death and apoptosis assays. Statistical analysis was performed using SPSS 13.0 software. All p values are two-sided, and a p value<0.05 was considered statistically significant.
Supporting Information
Zdroje
1. GradyWM, CarethersJM (2008) Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135 : 1079–1099.
2. JonesPA, BaylinSB (2007) The epigenomics of cancer. Cell 128 : 683–692.
3. KazAM, WongCJ, LuoY, VirginJB, WashingtonMK, et al. (2011) DNA methylation profiling in Barrett's esophagus and esophageal adenocarcinoma reveals unique methylation signatures and molecular subclasses. Epigenetics 6 : 1403–1412.
4. JinW, KimGM, KimMS, LimMH, YunC, et al. (2010) TrkC plays an essential role in breast tumor growth and metastasis. Carcinogenesis 31 : 1939–1947.
5. JinW, LeeJJ, KimMS, SonBH, ChoYK, et al. (2011) DNA methylation-dependent regulation of TrkA, TrkB, and TrkC genes in human hepatocellular carcinoma. Biochem Biophys Res Commun 406 : 89–95.
6. Bouzas-RodriguezJ, CabreraJR, Delloye-BourgeoisC, IchimG, DelcrosJG, et al. (2010) Neurotrophin-3 production promotes human neuroblastoma cell survival by inhibiting TrkC-induced apoptosis. J Clin Invest 120 : 850–858.
7. LutherJA, BirrenSJ (2009) Neurotrophins and target interactions in the development and regulation of sympathetic neuron electrical and synaptic properties. Auton Neurosci-Basic 151 : 46–60.
8. NakagawaraA (2001) Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett 169 : 107–114.
9. TognonC, KnezevichSR, HuntsmanD, RoskelleyCD, MelnykN, et al. (2002) Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2 : 367–376.
10. Tauszig-DelamasureS, YuLY, CabreraJR, Bouzas-RodriguezJ, Mermet-BouvierC, et al. (2007) The TrkC receptor induces apoptosis when the dependence receptor notion meets the neurotrophin paradigm. Proc Natl Acad Sci U S A 104 : 13361–13366.
11. NikoletopoulouV, LickertH, FradeJM, RencurelC, GiallonardoP, et al. (2010) Neurotrophin receptors TrkA and TrkC cause neuronal death whereas TrkB does not. Nature 467 : 59–63.
12. GoldschneiderD, MehlenP (2010) Dependence receptors: a new paradigm in cell signaling and cancer therapy. Oncogene 29 : 1865–1882.
13. XuX, TahanSR, PashaTL, ZhangPJ (2003) Expression of neurotrophin receptor Trk-C in nevi and melanomas. J Cutan Pathol 30 : 318–322.
14. SegalRA, GoumnerovaLC, KwonYK, StilesCD, PomeroySL (1994) Expression of the neurotrophin receptor TrkC is linked to a favorable outcome in medulloblastoma. Proc Natl Acad Sci U S A 91 : 12867–12871.
15. BardelliA, ParsonsDW, SillimanN, PtakJ, SzaboS, et al. (2003) Mutational analysis of the tyrosine kinome in colorectal cancers. Science 300 : 949.
16. WoodLD, CalhounES, SillimanN, PtakJ, SzaboS, et al. (2006) Somatic mutations of GUCY2F, EPHA3, and NTRK3 in human cancers. Hum Mutat 27 : 1060–1061.
17. KaplanDR, MillerFD (2000) Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol 10 : 381–391.
18. ReichardtLF (2006) Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci 361 : 1545–1564.
19. KimMS, LeeJ, SidranskyD (2010) DNA methylation markers in colorectal cancer. Cancer Metastasis Rev 29 : 181–206.
20. IssaJP (2004) CpG island methylator phenotype in cancer. Nat Rev Cancer 4 : 988–993.
21. ThibertC, TeilletMA, LapointeF, MazelinL, Le DouarinNM, et al. (2003) Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science 301 : 843–846.
22. HuangEJ, ReichardtLF (2003) Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem 72 : 609–642.
23. PorterAC, VaillancourtRR (1998) Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene 17 : 1343–1352.
24. FoehrED, LinX, O'MahonyA, GeleziunasR, BradshawRA, et al. (2000) NF-kappa B signaling promotes both cell survival and neurite process formation in nerve growth factor-stimulated PC12 cells. J Neurosci 20 : 7556–7563.
25. WootenMW, VandenplasML, SeibenhenerML, GeethaT, Diaz-MecoMT (2001) Nerve growth factor stimulates multisite tyrosine phosphorylation and activation of the atypical protein kinase C's via a src kinase pathway. Mol Cell Biol 21 : 8414–8427.
26. KazAM, LuoY, DzieciatkowskiS, ChakA, WillisJE, et al. (2012) Aberrantly methylated PKP1 in the progression of Barrett's esophagus to esophageal adenocarcinoma. Genes Chromosomes Cancer 51 : 384–393.
27. LuoY, TsuchiyaKD, Il ParkD, FauselR, KanngurnS, et al. (2013) RET is a potential tumor suppressor gene in colorectal cancer. Oncogene 32 : 2037–2047.
28. GradyWM, RajputA, LutterbaughJD, MarkowitzSD (2001) Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res 61 : 900–902.
29. WhiteheadRH, VaneedenPE, NobleMD, AtaliotisP, JatPS (1993) Establishment of Conditionally Immortalized Epithelial-Cell Lines from Both Colon and Small-Intestine of Adult H-2kb-Tsa58 Transgenic Mice (Vol 90, Pg 587, 1993). P Natl Acad Sci USA 90 : 6894–6894.
30. LuoYX, CuiJ, WangL, ChenDK, PengJS, et al. (2009) Identification of cancer-associated proteins by proteomics and downregulation of beta-tropomyosin expression in colorectal adenoma and cancer. Proteomics Clin Appl 3 : 1397–1406.
Štítky
Genetika Reprodukčná medicína
Článek Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and MammalsČlánek The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and DevelopmentČlánek Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative ElementsČlánek Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine ExposureČlánek Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 7- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Solution for Crossover Formation
- Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, , as a Genetic Component of Neural Tube Defects in Humans
- Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
- Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
- Genomic Analysis of Natural Selection and Phenotypic Variation in High-Altitude Mongolians
- Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
- Role of CTCF Protein in Regulating Locus Transcription
- Gene Set Signature of Reversal Reaction Type I in Leprosy Patients
- Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups
- Is Required for Formation of the Genital Ridge in Mice
- Monopolin Subunit Csm1 Associates with MIND Complex to Establish Monopolar Attachment of Sister Kinetochores at Meiosis I
- Recombination Dynamics of a Human Y-Chromosomal Palindrome: Rapid GC-Biased Gene Conversion, Multi-kilobase Conversion Tracts, and Rare Inversions
- Mechanisms of Protein Sequence Divergence and Incompatibility
- Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack
- Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies
- Combinatorial Regulation of Meiotic Holliday Junction Resolution in by HIM-6 (BLM) Helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 Nucleases
- The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
- The Role of Interruptions in polyQ in the Pathology of SCA1
- Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in
- Fine Time Course Expression Analysis Identifies Cascades of Activation and Repression and Maps a Putative Regulator of Mammalian Sex Determination
- Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90
- Oxidative Stress and Replication-Independent DNA Breakage Induced by Arsenic in
- A Moonlighting Enzyme Links Cell Size with Central Metabolism
- Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
- The Conserved Intronic Cleavage and Polyadenylation Site of CstF-77 Gene Imparts Control of 3′ End Processing Activity through Feedback Autoregulation and by U1 snRNP
- The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in through Maintaining a Progenitor-like Cell State
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- The RNA-binding Proteins FMR1, Rasputin and Caprin Act Together with the UBA Protein Lingerer to Restrict Tissue Growth in
- Pattern Dynamics in Adaxial-Abaxial Specific Gene Expression Are Modulated by a Plastid Retrograde Signal during Leaf Development
- A Network of HMG-box Transcription Factors Regulates Sexual Cycle in the Fungus
- Bacterial Adaptation through Loss of Function
- ENU-induced Mutation in the DNA-binding Domain of KLF3 Reveals Important Roles for KLF3 in Cardiovascular Development and Function in Mice
- Interplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
- FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription
- The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression
- Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative Elements
- The Conserved ADAMTS-like Protein Lonely heart Mediates Matrix Formation and Cardiac Tissue Integrity
- The cGMP-Dependent Protein Kinase EGL-4 Regulates Nociceptive Behavioral Sensitivity
- RBM5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility
- Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of Expression at the 6q23.2 Coronary Heart Disease Locus
- Yeast Pol4 Promotes Tel1-Regulated Chromosomal Translocations
- A Dual Role for SOX10 in the Maintenance of the Postnatal Melanocyte Lineage and the Differentiation of Melanocyte Stem Cell Progenitors
- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle,
- Prediction of Complex Human Traits Using the Genomic Best Linear Unbiased Predictor
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
- Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine Exposure
- Exquisite Light Sensitivity of Cryptochrome
- miR-133a Regulates Adipocyte Browning In Vivo
- Strabismus Promotes Recruitment and Degradation of Farnesylated Prickle in Planar Polarity Specification
- Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
- Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
- Joint Molecule Resolution Requires the Redundant Activities of MUS-81 and XPF-1 during Meiosis
- The Mating Competence of Geographically Diverse Strains in Their Natural and Unnatural Sand Fly Vectors
- Defective Repair of Oxidative Base Lesions by the DNA Glycosylase Nth1 Associates with Multiple Telomere Defects
- Effective Blocking of the Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function
- Trans-Ancestral Studies Fine Map the SLE-Susceptibility Locus
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Bacterial Adaptation through Loss of Function
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



