-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
Breaks at common fragile sites (CFS) are a recognized source of genome instability in pre-neoplastic lesions, but how such checkpoint-proficient cells escape surveillance and continue cycling is unknown. Here we show, in lymphocytes and fibroblasts, that moderate replication stresses like those inducing breaks at CFSs trigger chromatin loading of sensors and mediators of the ATR pathway but fail to activate Chk1 or p53. Consistently, we found that cells depleted of ATR, but not of Chk1, accumulate single-stranded DNA upon Mre11-dependent resection of collapsed forks. Partial activation of the pathway under moderate stress thus takes steps against fork disassembly but tolerates S-phase progression and mitotic onset. We show that fork protection by ATR is crucial to CFS integrity, specifically in the cell type where a given site displays paucity in backup replication origins. Tolerance to mitotic entry with under-replicated CFSs therefore results in chromosome breaks, providing a pool of cells committed to further instability.
Published in the journal: Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity. PLoS Genet 9(7): e32767. doi:10.1371/journal.pgen.1003643
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003643Summary
Breaks at common fragile sites (CFS) are a recognized source of genome instability in pre-neoplastic lesions, but how such checkpoint-proficient cells escape surveillance and continue cycling is unknown. Here we show, in lymphocytes and fibroblasts, that moderate replication stresses like those inducing breaks at CFSs trigger chromatin loading of sensors and mediators of the ATR pathway but fail to activate Chk1 or p53. Consistently, we found that cells depleted of ATR, but not of Chk1, accumulate single-stranded DNA upon Mre11-dependent resection of collapsed forks. Partial activation of the pathway under moderate stress thus takes steps against fork disassembly but tolerates S-phase progression and mitotic onset. We show that fork protection by ATR is crucial to CFS integrity, specifically in the cell type where a given site displays paucity in backup replication origins. Tolerance to mitotic entry with under-replicated CFSs therefore results in chromosome breaks, providing a pool of cells committed to further instability.
Introduction
Accurate genome duplication is required at each cell generation to maintain genetic information. However, mammalian genomes contain regions that challenge the replication process, such as common fragile sites (CFS). CFSs are loci that recurrently exhibit breaks on mitotic chromosomes following moderate slowing of replication fork movement [1]. To date, there is a consensus considering that such stresses delay completion of CFS replication more than the rest of the genome, and that breaks occur at under-replicated sequences upon chromosome condensation at mitotic onset. This delay was believed to result from replication fork blockage arising when forks encounter secondary structures formed at particular nucleotide sequences, notably AT-rich repeats [1]. However, the instability of FRA3B, the most active CFS in human lymphocytes, was recently shown to result from paucity in initiation events along a large region overlapping the most instable part of the site. This paucity forces replication forks emanating from flanking origins to cover long distances before merging in late S or G2 phases, leaving the sites incompletely replicated upon fork slowing [2]. Strikingly, FRA3B is weakly fragile in fibroblasts, in which initiation events are evenly distributed all along the locus [2]. Conversely, the two major CFSs in fibroblasts, that are not fragile in lymphocytes, display origin paucity in fibroblasts and a normal distribution of initiation events in lymphocytes [3]. Thus, the tissue-dependent organization of replication initiation controls the epigenetic setting of CFSs [4].
CFSs are a recognized source of the genomic instability driving oncogenesis from early steps of the process [5]. Indeed, CFS instability was repeatedly observed in pre-neoplasic lesions [5], [6], [7]. How pre-neoplasic cells, that generally retain wild-type checkpoints, escape surveillance by the DNA damage response (DDR) remains unclear. Central to DDR are two related protein kinases, ATM and ATR, that respectively sense double strand breaks (DSB) and RPA-coated single stranded DNA (ssDNA) accumulated upon fork slowing [8]. ATR and ATM activation then leads to phosphorylation of a large panel of substrates, including Chk1 and Chk2, which triggers a second wave of phosphorylations that amplifies and spreads the signal [9]. Among these downstream targets is the major tumour suppressor p53, a transcription factor that integrates signals from many different pathways [10]. Not surprisingly, inactivation of key DDR components leads to various diseases, including cancer [11].
In vertebrate cells, like in yeasts, the ATR/Mec1 pathway was mostly studied under conditions imposing a complete block to fork progression. Among other effects, such stresses lead, in cis, to stabilization of damaged replication forks and, in trans, to delayed mitotic onset [12]. In contrast, little is known about the cell response to moderate stresses such as treatments with low concentrations of aphidicolin, a well-known inhibitor of DNA polymerases, commonly used to induce breaks at CFSs. Several reports suggested that the frequency of breaks at CFSs increases in cells deprived of ATR, TopBP1, Hus1 or Chk1 [4]. However, while the role of ATR has been largely confirmed, notably in vivo in human patients and in mutant mice [13], [14], the impact of other proteins, including Chk1, in the maintenance of CFS integrity remains more controversial.
Here we compared the response of human lymphoblastoid cells and normal fibroblasts to various levels of fork slowing. We showed that a two - to ten-fold reduction of fork speed (called below moderate stress conditions) leads to global chromatin recruitment of sensors and mediators of the ATR pathway without substantial activation of Chk1, ATM or p53. Analysis of the phenotype of cells depleted of ATR or Chk1 and submitted to moderate levels of stress shows that ATR, but not Chk1, is crucial to fork protection and CFS integrity specifically in cell types where the site is fragile. These observations shed light on how pre-neoplastic cells continue cycling under inappropriate conditions.
Results
Chromatin loading of sensors and mediators of the ATR pathway upon fork slowing
We used DNA combing to determine how increasing concentrations of aphidicolin impact fork movement in JEFF cells (B lymphocytes immortalized by Eptsein-Barr virus) and MRC-5 cells (normal embryonic human fibroblasts) (Figure 1A). In untreated JEFF cells, forks progress at approximately 1.85 kb/min. In cells grown with aphidicolin 1.2 or 2.4 µM or with HU 1 mM, fork movement is too slow to be accurately measured with the labelling conditions we used. These treatments are considered below to block fork progression. For aphidicolin concentrations between 0.038 and 0.6 µM, the medians of fork speeds range between 1 and 0.2 kb/min (Figures 1B upper panel and S1), which is considered as moderate speed reduction. Similar results were obtained upon treatment of MRC-5 cells (Figure S2A).
Fig. 1. Moderate fork slowing triggers chromatin loading of sensors and mediators of the ATR pathway. 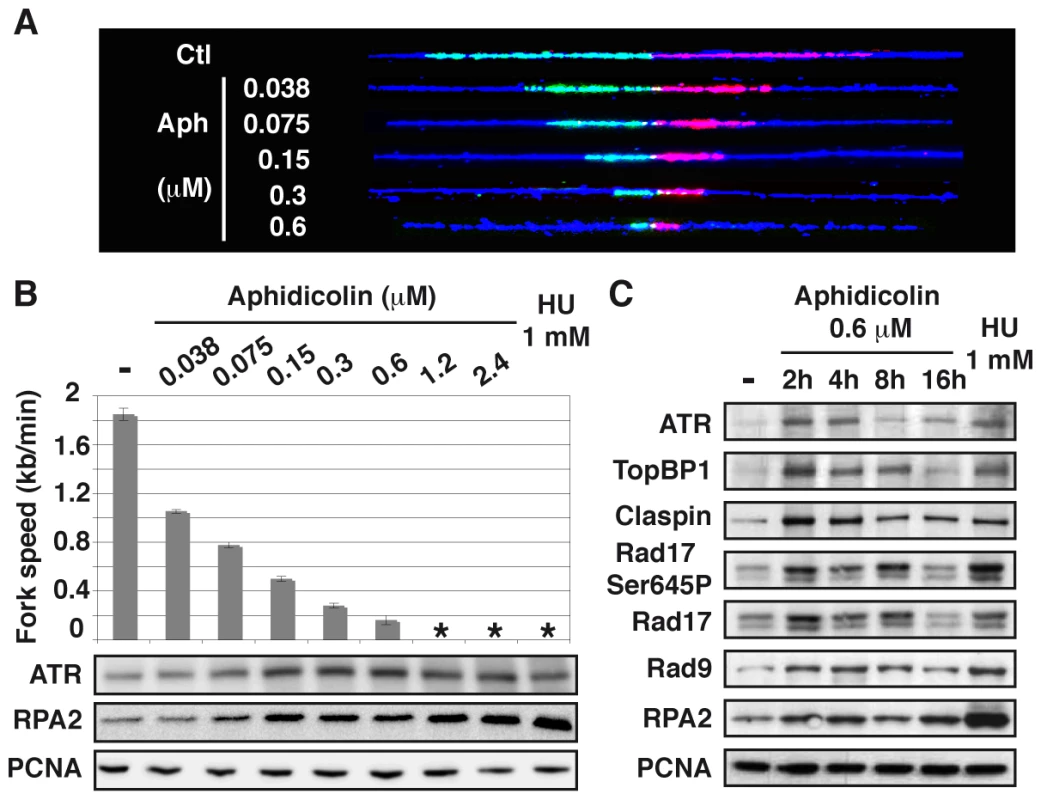
(A) Measurement of replication fork speed. Typical examples of combed DNA molecules displaying replications forks in control cells (Ctl) and cells treated with aphidicolin. Cells were pulse-labelled with IdU then CldU before DNA was purified and combed (see Methods). The analogues were revealed on stretched molecules as previously described [44] and fork speed was determined by measuring the length of IdU (red) and CldU (green) tracks. Blue: counterstaining of the DNA used to select unbroken IdU and CldU tracks (B) Mean fork speed (upper panel), kb per min ± standard error of the mean (s.e.m.), and chromatin recruitment of checkpoint proteins (lower panel). Mean fork speed and chromatin recruitment of ATR and RPA2 were studied after 4 h of treatment with the indicated aphidicolin concentrations or 1 h of treatment with HU 1 mM. Asterisks indicate that fork speed cannot be measured. (C) Western blot analysis of chromatin extracts from exponentially growing cells, untreated (-) or treated as indicated. The status of the ATR pathway was determined by western blot analysis of chromatin-bound ATR and RPA2 in cells treated for 4 h with different concentrations of aphidicolin or HU 1 mM (Figure 1B lower panels). We found that chromatin loading of ATR starts to increase in cells treated with aphidicolin 0.075 µM and reaches a maximum at 0.15 µM, namely when fork speed is reduced approximately by a factor of two. Noticeably, the amount of chromatin-bound RPA2 also starts to increase upon treatment with aphidicolin 0.075 µM, but remains lower in cells treated with up to 2.4 µM of the drug than in cells grown with HU 1 mM. The study of MRC-5 cells also shows that a two-fold decrease in fork speed triggers recruitment of ATR to the chromatin and that, like in JEFF cells, ATR binding behaves essentially as an all-or-nothing phenomenon (Figure S2B).
A time course analysis of the status of the ATR pathway in JEFF cells treated with aphidicolin 0.6 µM shows that ATR, TopBP1, Claspin, RPA2 and Rad9, a subunit of the 9-1-1 complex, are rapidly loaded on the chromatin (Figure 1C). Noticeably, Rad17 is loaded and phosphorylated on Ser645, a recognized ATR phosphorylation site [15]. We observed that the amount of chromatin-bound proteins decreases upon prolonged treatment, a phenomenon previously observed by others [16], [17]. Short treatments with aphidicolin 0.6 µM or HU 1 mM lead to comparable levels of recruitment for all proteins but RPA2, which chromatin amount remains stable and lower in the presence of aphidicolin than in the presence of HU. Thus, chromatin loading of checkpoint sensors and mediators of the ATR pathway surprisingly appears poorly correlated to the amount of chromatin-bound RPA.
To reinforce this conclusion, we studied ssDNA accumulation in JEFF cells treated as above using the procedure schematized in Figure 2A. As expected, no CldU labelling was observed in untreated JEFF cells (Figure 2B). Following 1 h of treatment with HU 1 mM, 104 out of 110 (95%) cells in S-phase, identified by the presence of PCNA foci, display CldU foci while cells not in S-phase remain unlabelled. In addition, these CldU foci generally co-localize with PCNA foci (Figure 2C), showing that ssDNA forms at blocked forks. Strikingly, CldU foci were absent from the vast majority of S-phase cells following up to 16 h of treatment with aphidicolin 0.6 µM. Thus, under moderate speed reduction, the amount of sensors and mediators of the ATR pathway loaded on the chromatin is not proportional to the amount of RPA-coated ssDNA exhibited at the forks.
Fig. 2. Moderate fork slowing is not associated with formation of ssDNA foci. 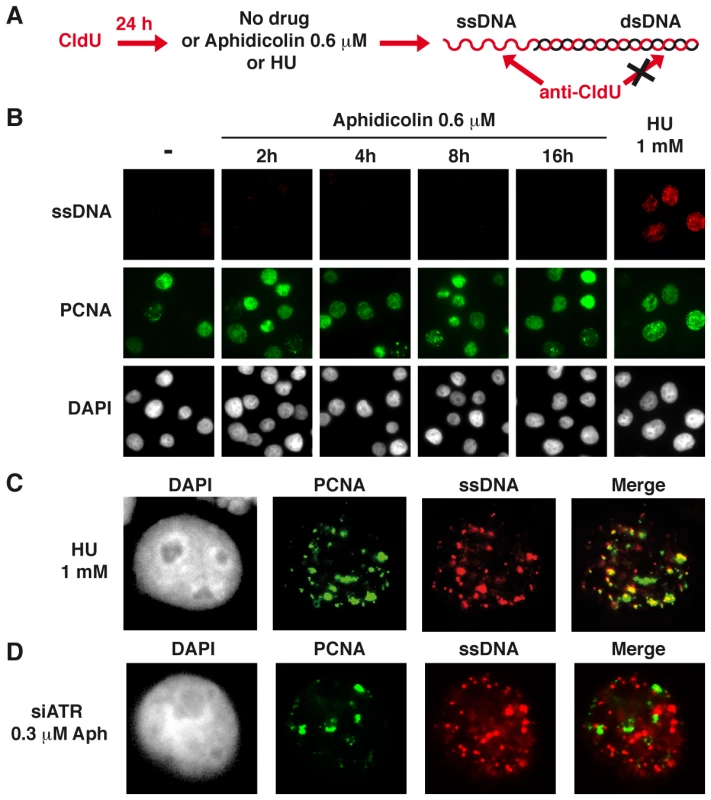
(A) ssDNA detection scheme. Cells hemi-substituted throughout their genome after growth in the presence of CldU for about 1.5 cell generations were treated or not with aphidicolin or HU for different periods of time prior to fixation. CldU was immuno-detected without DNA denaturation, which only permits visualization of substituted and single-stranded regions. (B) Co-detection of ssDNA (red) and chromatin bound PCNA (green) by immunostaining of untreated cells (-) and cells treated as indicated. Nuclei were counterstained with DAPI. (C) Typical pattern of PCNA and CldU foci in cells treated with HU 1 mM during 1 h. (D) Typical pattern of PCNA and CldU foci in cells depleted of ATR and treated with 0.3 µM of aphidicolin for 4 h. Several ATR targets, including Chk1, are not activated upon moderate fork slowing
Chk1 phosphorylation on Ser317 and Ser345 was analyzed (Figure 3A). Both residues appear phosphorylated in JEFF cells treated with aphidicolin 2.4 µM or HU 1 mM but not, or weakly, in cells treated with up to 0.6 µM of aphidicolin. Chk1 status was confirmed by western blot analysis of extracts from S-phase cells (Figure S3A) and by immunofluorescence (Figure S3B). Similar observations were made with MRC-5 cells (Figure S2C). These results agree with some reports [17], [18] but others found Chk1 phosphorylated in some cancer cell lines upon treatment with aphidicolin 0.6 µM [19]. These discrepancies most probably reflect differences in genetic backgrounds leading to cell-type variations in aphidicolin sensitivity. Unfortunately, the absence of fork speed measurement in previous works prevents further comparison of the results.
Fig. 3. Moderate fork slowing does not trigger phosphorylation of ATR targets. 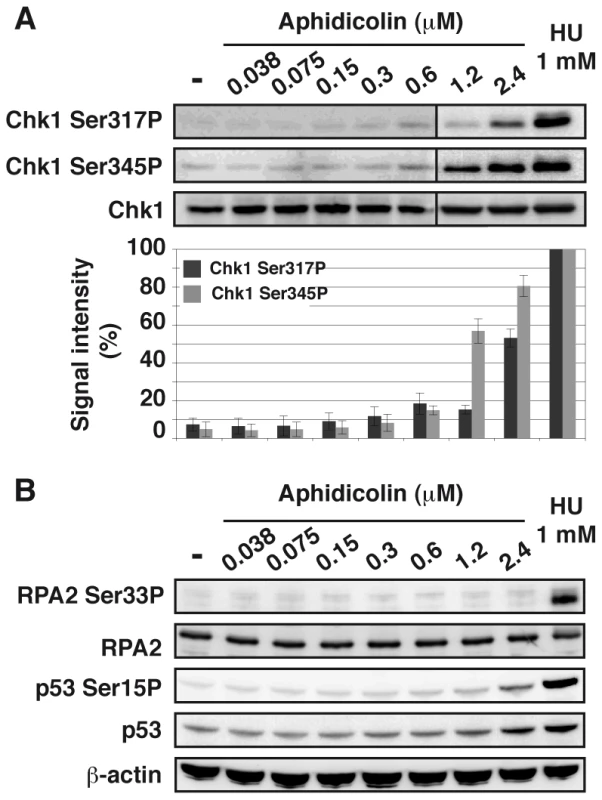
(A) Western blot detection of Chk1-Ser317 and Ser345 phosphorylations in total extracts of cells treated 4 h with aphidicolin or 1 h with HU 1 mM (upper panel). Quantification of Chk1-Ser317P and -Ser345P from 3 independent experiments (lower panel). Results (mean ± s.e.m) are expressed as percentage of the level of phosphorylation found in cells treated with 1 mM HU. Lanes 1–6 and lanes 7–9 correspond to different blots. (B) Western blot detection of RPA2-Ser33 phosphorylation, p53 and p53-Ser15 phosphorylation in total extracts of cells treated for 4 h with aphidicolin or 1 h with HU 1 mM. Loading control: β-actin. In addition, we observed that phosphorylation of p53 on Ser15 and of RPA2 on Ser33 remains undetectable under moderate replication stress (Figure 3B). Therefore, chromatin-bound ATR fails to trigger activation of several DDR effectors under these conditions.
Moderate fork speed reduction does not activate the ATM pathway
To further analyze the cellular response to replication stress, we studied the status of the ATM pathway (Figure 4A). We found that a slight phosphorylation of ATM on Ser1981 starts to appear upon treatment with aphidicolin 2.4 µM, but remains much lower than in cells treated with HU 1 mM. Chromatin accumulation of ATM and phosphorylation of histone variant H2AX (γH2AX) occur only in cells treated with HU 1 mM. Time course analyses of ATM (Figure S3C) and H2AX (Figures 4B and S3D) in cells treated with aphidicolin 0.6 µM confirmed these observations. Thus, the ATM pathway is not triggered in cells experiencing moderate stress for up to 16 h.
Fig. 4. Moderate fork slowing triggers neither γ-H2AX foci nor ATM activation. 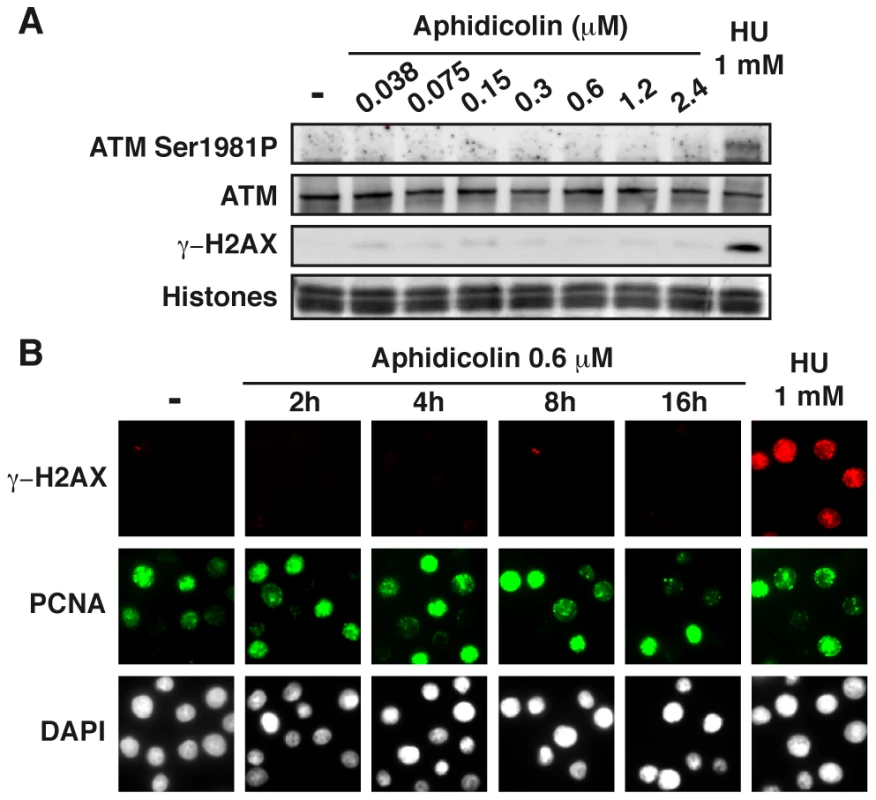
(A) Western blot analysis of ATM-Ser1981 phosphorylation and γ-H2AX in exponentially growing cells, untreated (-) or treated as indicated. (B) Immunostaining of γ-H2AX together with chromatin-bound PCNA in untreated cells (-) and in cells treated with aphidicolin 0.6 µM for the indicated periods of time, or for 1 h with HU 1 mM. Moderate fork speed reduction results in Chk1-independent decrease in mitotic flow
The fact that moderate stresses fail to activate the whole DDR cascade raises the question as whether mitotic onset is restrained under these conditions. The mitotic flow was determined following treatment with various aphidicolin concentrations and nocodazole to block mitotic exit, in DDR-proficient cell populations and in populations of cells depleted of either ATR or Chk1 by RNA silencing, (Figure 5A). We found that the percentages of JEFF cells entering mitosis decrease upon treatment with increasing aphidicolin concentrations in the three genetic backgrounds and that only ATR depletion modestly impacts these percentages (Figure 5B). Similar results were obtained in the presence of z-VAD fmk, a pan-caspase inhibitor that blocks apoptosis (Figure S3E). Thus, the deficit in mitotic cells we observed upon aphidicolin treatment does not result from mitotic death but rather from delayed mitotic entry.
Fig. 5. Moderate fork slowing decreases mitotic flow independently of ATR and Chk1. 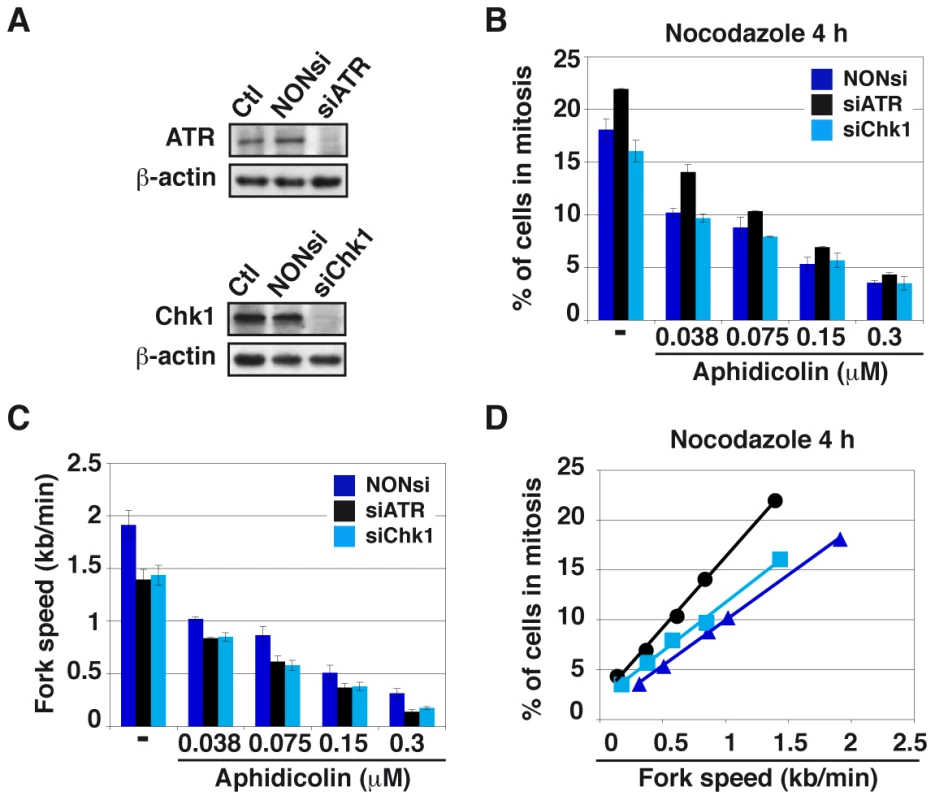
(A) Western blot analyses of total cell extracts 48 h post-transfection with a NONsi RNA or siRNAs specific to ATR (left panel) or Chk1 (right panel). Loading control: β-actin. (B) The percentages of cells in mitosis were measured by flow cytometry with MPM2 immuno-detection 48 h post-transfection with the indicated RNAs. Cells were treated for 4 hr with aphidicolin, then for 4 hr with both aphidicolin and nocodazole to block mitotic exit. Aphidicolin concentrations are indicated. (C) Measurement of replication speed in each condition of transfection and treatment. (D) Correlations between fork speed and the percentage of cells in mitosis. Data presented in B and C were used to plot the percentage of cells in mitosis against fork speed. We then determined how mitotic flow correlates with the degree of fork slowing. In good agreement with previous works [20], [21], we observed that ATR or Chk1 depletion “per se” reduces fork speed by a mechanism not yet elucidated. Not surprisingly, aphidicolin treatment further reduces fork movement (Figure 5C). Plotting the percentage of mitotic cells against fork speed in the different conditions reveals a linear relationship between the two parameters, regardless of the transfection conditions (Figures 5D and S3F). In addition, the curve obtained for Chk1-depleted cells aligns with that of control cells, indicating that the decrease in mitotic flow resulting from Chk1 depletion is completely accounted for by fork slowing. Therefore, Chk1 plays no direct role in the control of mitotic onset under these conditions, which agrees with the absence of Chk1 phosphorylation upon moderate fork speed reduction. The curve corresponding to ATR-depleted cells does not strictly align with the other two, which suggests that ATR plays a role, though modest, in the control of mitotic entry. The prominent mechanism that correlates mitotic flow to fork speed in these conditions is presently unknown.
ATR, but not Chk1, is crucial for chromosome stability upon moderate fork slowing
To evaluate the contribution of the ATR pathway to chromosome maintenance under moderate stress, we determined the percentage of metaphase plates displaying chromosome breaks in DDR-proficient cells and in cells depleted of either ATR or Chk1 following 16 h of treatment with various aphidicolin concentrations (Figures 6A and S4A). We found that this percentage is the highest in cells depleted of ATR, although Chk1 depletion also alters chromosome stability. The percentage of metaphases displaying chromosome breaks was then plotted against fork speed for the different conditions of cell depletion and treatment (Figure 6B). The curve obtained for Chk1-depleted cells is identical to that of control cells, indicating that the increase in chromosome breakage resulting from Chk1 depletion is completely accounted for by fork slowing. Strikingly, the curve for ATR-depleted cells coincides with the other two only when fork speed stands higher than 0.85 kb/min. Further slowing leads to percentages of cells with broken chromosomes greatly exceeding those observed in control or Chk1-depleted cells.
Fig. 6. ATR depletion compromises genome stability. 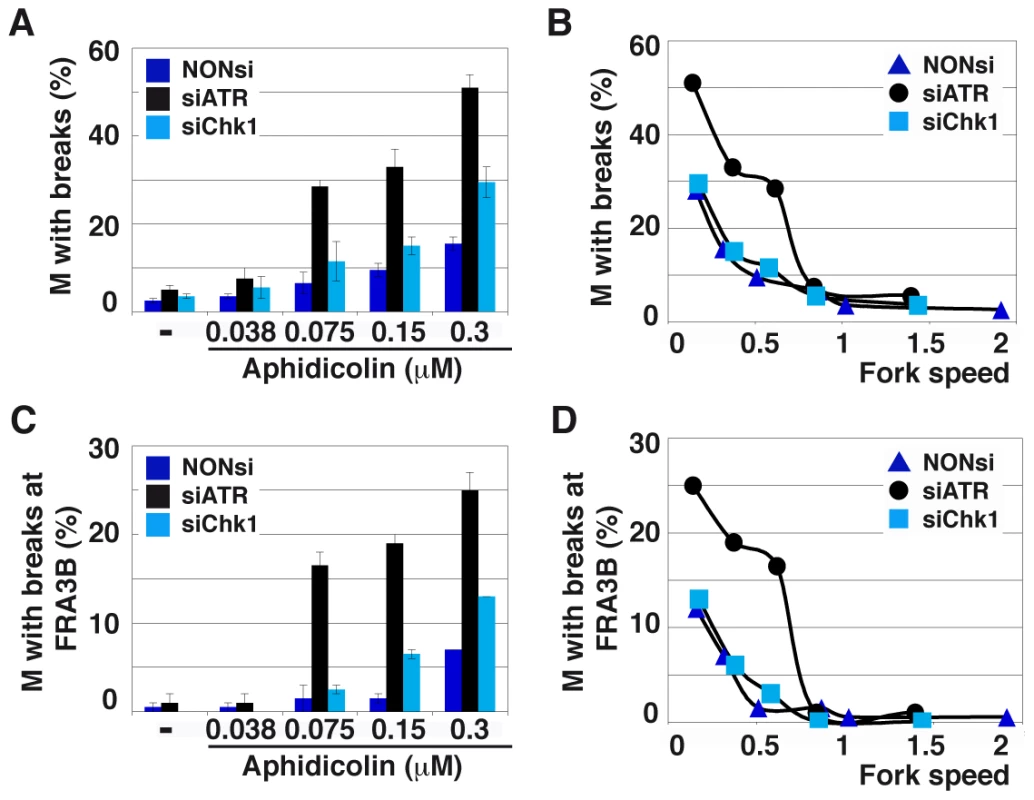
(A) Quantification of metaphases (M) with breaks in cells transfected as above and treated for 16 h with the indicated aphidicolin concentrations. Mean ± s.e.m are presented. (B) Correlations between replication fork speed and chromosome instability. The data presented in A and Figure 5C were used to plot the percentage of metaphase plates with chromosome breaks against fork speed in each condition of transfection and treatment. (C) Breaks occurring at FRA3B quantified as in C. (D) Correlations between replication speed and FRA3B instability evaluated as in B. Breaks at CFSs usually account for most of the breaks induced by low aphidicolin concentrations. We thus used fluorescent in situ hybridization (FISH) (Figure S4B) to determine the percentage of metaphase plates displaying chromosome breaks at FRA3B (Figures 6C and 6D), the most active CFS in JEFF cells [2]. In either transfection conditions, breaks at FRA3B represent approximately 50% of total breaks and evolve like total breaks. Thus, ATR plays a major role in the maintenance of CFS stability when fork speed falls below a threshold of 0.85 kb/min. This value is consistent with the threshold for chromatin recruitment of ATR (Figure 1B). Noticeably, this high level of CFS instability is not explained by the weak effect of ATR depletion on mitotic entry (Figure 5D).
CFSs co-localize with ssDNA foci in ATR-depleted and moderately stressed cells
To determine whether break frequencies in the different genetic contexts correlate with the presence of ssDNA at the fork, we studied cells displaying foci of ssDNA and/or PCNA in the three conditions of transfection (Figure 7A). In the absence of aphidicolin treatment, we found that the percentage of cells displaying foci is low (approximately 2%) in either condition. Upon treatment with aphidicolin 0.3 µM, some 2% of DDR-proficient and 5% of Chk1-depleted cells exhibit CldU foci. This percentage reaches 15% in ATR-depleted cells and those foci strikingly do not co-localize with PCNA foci (Figure 2D), suggesting that ssDNA takes place at collapsed forks. To determine whether these foci result from resection of collapsed forks by the Mre11-Rad50-Nbs1 (MRN) complex, the cells were depleted of Mre11 by RNA silencing or treated with mirin, an inhibitor of Mre11 nuclease activity [22]. Strikingly, compared to ATR depleted cells treated with mirin 100 µM or depleted of Mre11, ATR-depleted cells treated with both aphidicolin 0.3 µM and mirin or co-depleted of ATR and Mre11 show similar percentages of cells with ssDNA foci (Figure 7B). These results suggest that ssDNA foci we observed result from Mre11-mediated resection of collapsed forks. This conclusion agrees with the results of a recent work showing that MRN activity leads to the formation of RPA foci in checkpoint deficient U2OS cells [23].
Fig. 7. ATR-depletion enhances formation of ssDNA foci and fork asymmetry. 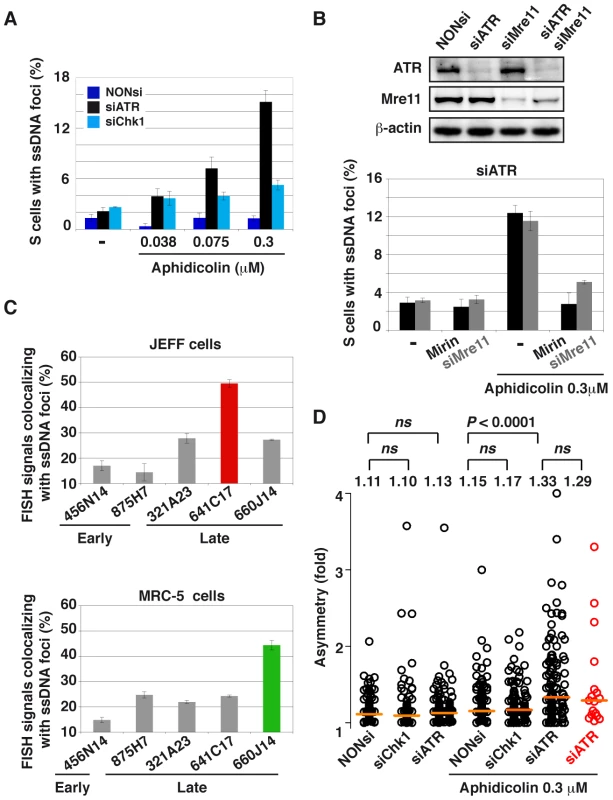
(A) Percentage of S-phase cells with ssDNA foci in the indicated conditions of transfection and aphidicolin treatment. Mean ± s.e.m are presented. (B) Upper panel: Western blot analyses of total cell extracts 48 h post-transfection with a NONsi RNA or siRNAs specific to ATR or/and Mre11. Loading control: β-actin. Lower panel: percentage S-phase cells (mean ± s.e.m) displaying ssDNA foci in populations of cells depleted of ATR and treated or not with aphidicolin 0.3 µM. Mre11 status is indicated (siMre11 or treatment with Mirin: 100 µM). (C) Percentage of FISH signal co-localizing with ssDNA foci in cells depleted of ATR and treated with 0.3 µM aphidicolin. The immunostaining of ssDNA and PCNA was combined with FISH with BAC probes. Upper panel: JEFF lymphoblastoid cells, the BACs probe early replicating regions (875H7 and 456N14) and late replicating regions (321A23, 660J14, and 641C17 that corresponds to FRA3B, the major CFS in lymphocytes). Lower panel: MRC-5 fibroblasts, BAC 456N14 probe for an early replicating region, other BACs correspond to late replicating regions (875H7, 321A23, 641C17, and 660J14 which corresponds to the major CFS in fibroblast mapping at 3q13.3). (D) Distributions of fork asymmetry in cells transfected and treated as indicated. Forks travelling in the FRA3B locus are represented by red circles (n = 17) and in the bulk genome by black circles (untreated: NONsi, n = 128; siCHk1, n = 103; siATR, n = 147; with aphidicolin: NONsi, n = 121; siCHk1, n = 132; siATR, n = 123). Horizontal orange lines represent the medians of fork distributions. Medians and P values are indicated above the distributions (not significant, ns). We then asked whether ssDNA foci formed in ATR-depleted cells treated with aphidicolin 0.3 µM co-localize with CFSs (Figure S4C). We found that FRA3B co-localizes more often with ssDNA foci (50%) than non-fragile regions replicating late (25%) or replicating early (15%) (Figure 7C). The relative co-localization of CFSs and ssDNA foci was also studied in MRC-5 fibroblasts where FRA3B is not fragile. In these cells the major CFS lies at 3q13.3 [3]. We found that this CFS also associates at higher frequency with ssDNA foci (45%) than other tested sequences, including FRA3B that behaves like non-fragile and late replicating sequences (Figure 7C). Noticeably, the non-fragile region identified by BACs 875H7, that replicates early in JEFF cells and late in MRC-5 cells [24], displays a percentage of co-localization with ssDNA foci that reflects its replication timing in each cell type, 12% and 24% respectively. The frequency of co-localization of a sequence with ssDNA foci is thus timing-dependent and, in the case of CFSs, correlates with their level of fragility in the tissue under study.
Forks collapse and/or stall similarly along FRA3B and in the bulk genome
Finally, we asked whether the preferential association of FRA3B with ssDNA foci in JEFF cells reflects the fact that forks collapse or stall more often along the site than in the bulk genome. Collapse and/or stalling should lead to asymmetrical forks, namely to individual forks presenting unequal IdU and CldU tracks [2]. Therefore, we calculated asymmetry as the ratio of the longest to the shortest track in cells treated or not with aphidicolin 0.3 µM and depleted or not of ATR or Chk1 (Figures 7D and S5). At the genome-wide level, fork asymmetry increases significantly only in cells depleted of ATR and treated with aphidicolin. We also studied fork asymmetry along FRA3B in the latter conditions. Strikingly, the distributions and the medians of fork asymmetries along FRA3B and in the bulk genome appear remarkably similar.
Discussion
We determine here how various levels of fork speed reduction impact DDR activation and genome integrity in human lymphoblastoid and fibroblastic cells. Surprisingly, ATR is loaded on the chromatin at roughly similar levels whether forks are completely blocked or only slowed by a factor of 2 to 3. This loading thus behaves as an all-or-nothing phenomenon. Other sensors and mediators of the ATR pathway accumulate on the chromatin with similar kinetics while a modest increase in RPA-coated ssDNA is seen in cells experiencing moderate stress. These results suggest a model in which speed thresholds dictate the DDR status. Upon moderate fork impediment, stretches of RPA-coated ssDNA may be generated at a small fraction of the forks, which would escape detection by the techniques we used. Fork impediment may occur at random in the genome, or preferentially along regions difficult to replicate such as micro - and mini-satellites [8], [25], [26], telomeric repeats [27], [28], [29], [30], [31] or highly transcribed genes [32], [33], [34], [35]. We postulate that blocked forks elicit an alarm signal that commits cells to take steps against replisome disassembly, notably by recruiting sensors and mediators of the ATR pathway at all on-going forks.
ATR contributes importantly to the stabilization of blocked forks in the yeast S. cerevisiae [36], [37], [38] and in higher eukaryotes [36]. We show here that ATR is also crucial to stabilize forks experiencing moderate slowing. Indeed, ssDNA foci form at high frequency in S-phase cells depleted of ATR and treated with low doses of aphidicolin. Those ssDNA foci do not co-localize with PCNA foci, which strikingly contrasts with the co-localization of ssDNA and PCNA foci we observed in DDR-proficient cells upon fork blockage. In addition, we observed that Mre11 depletion or Mre11 inhibition by mirin suppresses ssDNA foci formation in cells depleted of ATR and treated with aphidicolin. Together, these observations show that moderate stresses strongly impact fork stability in ATR-deficient cells, then resection of collapsed forks by the Mre11 nuclease activity gives rise to large amounts of ssDNA that form the foci we observe.
Although phosphorylation of Chk1, p53 or RPA2 does not occur in DDR-proficient cells submitted to moderate stress, the observation that Rad17 is phosphorylated indicates that chromatin-bound ATR is active. In addition, neither formation of γ-H2AX foci nor activation of ATM and/or Chk2 takes place under these conditions. How DDR is committed to complete activation remains unknown. Considering the many physiological situations that may lead to global or local replication fork slowing, such adaptation of the checkpoint response to the degree of stress might be crucial to cell proliferation, but this tolerance is detrimental to CFS integrity because mitotic entry with under-replicated sites triggers DNA breaks [39].
In agreement with the fact that Chk1 is not activated under moderate replication stress, the increase in chromosome instability observed in Chk1-depleted cells does not rely on its classical checkpoint function. Indeed, the frequencies of cells displaying ssDNA foci and breaks at CFSs are similar in checkpoint-proficient and in Chk1-depleted cells for similar fork speed reduction. In contrast, the frequencies of cells displaying ssDNA foci and chromosome breaks, notably at CFSs, increase considerably in cells depleted of ATR and submitted to moderate stress. This increase is accounted for neither by fork slowing nor by unscheduled mitotic entry. Noticeably, the specific increase in CFS breakage starts at the exact aphidicolin concentration that triggers chromatin recruitment of ATR in checkpoint-proficient cells. Together, these results confirm that ATR, and possibly other sensors and mediators of the pathway, plays a major role to prevent CFS instability under moderate fork slowing.
These results do not fit with a previous model postulating that, upon replication stress, helicases tend to travel uncoupled from polymerases along CFS, giving rise to long stretches of ssDNA. In sub-regions able to adopt secondary structures, ssDNA would evolve into fork barriers that cause DNA breaks. In this model, checkpoint-proficient cells are supposed to be protected against these deleterious events because local accumulation of ssDNA triggers the ATR signalling pathway, resulting in delayed mitotic onset and activation of the repair machinery [1]. We show here that under moderate stress conditions, similar to those used to induce breaks at CFSs, forks stall and/or collapse in ATR-deficient cells at the same frequency along FRA3B as in the bulk genome. These results support data previously obtained in checkpoint-proficient cells showing that forks do not encounter sequence-specific obstacles along the sites [2]. In addition, we show here that, in ATR-deficient cells, ssDNA foci result from Mre11-dependent resection of collapsed forks, suggesting that long stretches of ssDNA are a consequence rather than a cause of CFS instability. Together, these results strongly argue against the model above. In contrast, the remarkable sensitivity of CFSs to moderate replication stress in ATR-deficient cells is well explained by paucity in initiation events along large regions nested in the sites [2], [3]. Indeed, the lack of backup initiation events might prevent fast rescue of forks collapsing at random and normal frequency along CFSs, favouring extensive resection. This conclusion is supported by the fact that co-localization of ssDNA and CFSs increases specifically in the cell type in which a given site displays paucity in initiation events, namely FRA3B in lymphocytes and 3q13.3 in fibroblasts [2], [3].
Tumor suppressor genes are rarely mutated in sporadic cancers, notably in preneoplasic lesions [5]. Surprisingly, chromosome instability has been repeatedly observed in cells of these lesions, while DDR activation should block their proliferation. We show here in two cellular models that a moderate reduction of fork speed does not fully activate the DDR, fails to block mitotic entry and elicits breaks at under-replicated CFSs. Strikingly, some of the genes hosted by CFSs behave as tumor suppressor genes in human and mouse models [40]. FRA3B, for example, hosts the FHIT gene that has been involved in the cellular response to DNA damage. Thus, FRA3B expression not only triggers local instability but also favors genome-wide DNA damages through FHIT inactivation [41], [42]. The origin of the replication stress leading to CFS expression in pre-neoplastic lesions is still debated. It has been proposed to result from oncogene activation [7], from alteration of metabolic process or from exposure to environmental stress [42], [43]. Whatever the cause of CFS instability, we propose that imperfect repair of these damages in cells of pre-neoplastic lesions creates a pool of cells committed to further instability, from which the selection of cells with mutations affecting genes causally implicated in cancer development is facilitated.
Materials and Methods
Cell culture and transfection
Lymphoblastoid cells were grown in RPMI 1640+GlutaMAX-I medium (GIBCO) and MRC-5 cells in MEM plus Earle's salts without L-glutamine medium (GIBCO), 1% MEM nonessential amino acids (GIBCO), 1 mM sodium pyruvate (GIBCO) and 2 mM L-glutamine (GIBCO). In addition, all cells were grown with 10% foetal calf serum (PAN-Biotech GmbH) and 100 µg/mL of penicillin and streptomycin (GIBCO). For transfections, 2×106 lymphoblastoid cells were resuspended in 100 µL of Nucleofector C solution (Lonza Cologne AG) with 0.6 µM RNAi and transfected with the Z-001 program according to manufacturer's instructions. MRC-5 cells were resuspended in Nucleofector R solution (Lonza Cologne AG) and transfected with the V-020 program. A mixture of 3 RNAi directed against ATR (HSS100876, HSS100877, HSS100878, Invitrogen) or Chk1 (HSS101854, HSS101855, HSS101856, Invitrogen) was used for transfection. The AllStars Negative Control siRNA (1027281, QIAGEN) was used for NONsi.
DNA combing and image acquisition
Combing was performed as described [2], [44], [45]. An epifluorescence microscope (Axio Imager.Z2; Carl Zeiss) equipped with a 63× objective lens (PL APO, NA 1.4 Oil DIC M27) connected to a charge-coupled device camera (Cool-SNAP HQ2; Roper Scientific), and MetaMorph software (Roper Scientific) was used for image acquisition.
Immunochemistry and metaphase spreading
Metaphase spreading and DNA fluorescent in situ hybridization for FRA3B detection were performed as described [39]. BACs were selected from the human genome project RP11 library. FRA3B has been assigned to band 3p14.2 and was probed with BAC 641C17. Cells were spread on slide with a cytospin (Shandon) and processed for immunostaining as previously described [46]. Combined immunofluorescence and DNA fluorescent in situ hybridization was performed according to Chaumeil et al. [47]. Microscopic images were acquired using an upright motorized microscope (Axio Imager.Z2; Carl Zeiss). Acquisitions were performed using an oil immersion objective 100× (PL APO, NA 1.46 Oil DIC M27) and a high-sensitive cooled interlined CCD camera (Cool-SNAP HQ2; Roper Scientific). For colocalization studies, rapid and precise Z-positioning was accomplished by a piezoelectric motor (P-725.1CD ; Physik Instrumente) mounted underneath the objective lens. Image stacks were acquired without camera binning, with a plane spacing of 0.2 µm, and MetaMorph software (Roper Scientific) was used for image acquisition.
Cell extracts
Chromatin fractionation was performed as described [48]. For total extract of cells in S-phase, DNA was stained with 5 µg/ml Hoechst 33342 (Molecular Probes). Cells were sorted with a standard FACSVantage DiVa (Becton Dickinson Immunocytometry Systems, San Jose, CA) equipped with a 488 nm laser used at 250 mW and a multiline UV (351–363 nm) laser used at 200 mW. Linear Hoechst fluorescence was acquired using a 424/44 filter. Doublets were excluded using pulse area vs width. 105 sorted cells were washed in cold PBS, re-suspended in 50 µL 1X SDS sample buffer (New England BioLabs Inc.), sonicated 5 min (Bioruptor - Diagenode) and boiled before loading for western blotting.
Mitotic flow
Cells were treated with aphidicolin and nocodazole as indicated and washed with buffer GM (1.1 g/L Glucose, 8 g/L NaCl, 0.4 g/L KCl, 0.37 g/L Na2HPO4, 0.15 g/L KHPO4, 0.5 mM EDTA). The cells were re-suspended in 1 mL of buffer GM and fixed by addition of 3 mL of ethanol 100%. The cell pellet was incubated with a mouse anti-MPM2 (Mitotic Protein Monoclonal 2) antibody (05-368, Upstate) in buffer PBT (PBS 1X, 1% BSA and 0.05% Tween20), washed with PBS and incubated with a goat Alexa488-conjugated anti-mouse antibody (Molecular Probes) in PBT. Cells were washed with PBS and finally re-suspended in PBS containing 50 ng/µL propidium iodide and 40 µg/mL RNase A (USB). The quality of the mitotic cell preparations was verified by FACScan analysis of MPM2-positive cells with CellQuest software (Beckton Dickinson).
Antibodies
The following primary antibodies were used in this study: mouse anti-PCNA (PC10) antibody (MAB424, Chemicon International), goat anti-ATR (N-19) antibody (sc-1887, Santa Cruz Biotechnology, Inc.), rabbit anti-Claspin antibody (A300-266A, Bethyl Laboratories, Inc.), mouse anti-Rad17 (sc-17761, Santa Cruz Biotechnology, Inc.), rabbit anti-phospho-Rad17 (Ser645) antibody (#3421, Cell Signaling Technology), rabbit anti-Rad9 (sc-8324, Santa Cruz Biotechnology, Inc.), goat anti-Hus1 antibody (sc-30543, Santa Cruz Biotechnology, Inc.), rabbit anti-RPA32 antibody (GTX70258, GeneTex, Inc.), mouse anti-Chk1 antibody (sc-8408, Santa Cruz Biotechnology, Inc.), rabbit anti-phospho-Chk1 (Ser317) antibody (#2344, Cell Signaling Technology), rabbit anti-phospho-Chk1 (Ser345) antibody (#2348, Cell Signaling Technology), rabbit γ-H2AX (phospho S139) antibody (ab2893, Abcam). In immunochemistry experiments, primary antibodies were visualized with Alexa594-conjugated goat anti-rabbit and Alexa488-conjugated goat anti-mouse antibodies (Molecular Probes). Detection of ssDNA was performed as previously described [49], with a rat anti-BrdU (OBT0030, AbD Serotec) and Alexa594-conjugated goat anti-rat antibodies (Molecular Probes).
Supporting Information
Zdroje
1. DurkinSG, GloverTW (2007) Chromosome fragile sites. Annu Rev Genet 41 : 169–192.
2. LetessierA, MillotGA, KoundrioukoffS, LachagesAM, VogtN, et al. (2011) Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470 : 120–123.
3. Le TallecB, DutrillauxB, LachagesAM, MillotGA, BrisonO, et al. (2011) Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol 18 : 1421–1423.
4. DebatisseM, Le TallecB, LetessierA, DutrillauxB, BrisonO (2012) Common fragile sites: mechanisms of instability revisited. Trends Genet 28 : 22–32.
5. NegriniS, GorgoulisVG, HalazonetisTD (2010) Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11 : 220–228.
6. JacomeA, Fernandez-CapetilloO (2011) Lac operator repeats generate a traceable fragile site in mammalian cells. EMBO Rep 12 : 1032–1038.
7. BesterAC, RonigerM, OrenYS, ImMM, SarniD, et al. (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145 : 435–446.
8. BranzeiD, FoianiM (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11 : 208–219.
9. MatsuokaS, BallifBA, SmogorzewskaA, McDonaldER3rd, HurovKE, et al. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316 : 1160–1166.
10. MenendezD, IngaA, ResnickMA (2009) The expanding universe of p53 targets. Nat Rev Cancer 9 : 724–737.
11. JacksonSP, BartekJ (2009) The DNA-damage response in human biology and disease. Nature 461 : 1071–1078.
12. CicciaA, ElledgeSJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40 : 179–204.
13. CasperAM, DurkinSG, ArltMF, GloverTW (2004) Chromosomal instability at common fragile sites in Seckel syndrome. Am J Hum Genet 75 : 654–660.
14. MurgaM, BuntingS, MontanaMF, SoriaR, MuleroF, et al. (2009) A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet 41 : 891–898.
15. MedhurstAL, WarmerdamDO, AkermanI, VerwayenEH, KanaarR, et al. (2008) ATR and Rad17 collaborate in modulating Rad9 localisation at sites of DNA damage. J Cell Sci 121 : 3933–3940.
16. FreireR, van VugtMA, MamelyI, MedemaRH (2006) Claspin: timing the cell cycle arrest when the genome is damaged. Cell Cycle 5 : 2831–2834.
17. ShimuraT, MartinMM, TorresMJ, GuC, PluthJM, et al. (2007) DNA-PK is involved in repairing a transient surge of DNA breaks induced by deceleration of DNA replication. J Mol Biol 367 : 665–680.
18. SchwartzM, OrenYS, BesterAC, RahatA, SfezR, et al. (2009) Impaired replication stress response in cells from immunodeficiency patients carrying Cernunnos/XLF mutations. PLoS ONE 4: e4516.
19. DurkinSG, ArltMF, HowlettNG, GloverTW (2006) Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene 25 : 4381–4388.
20. PetermannE, Maya-MendozaA, ZachosG, GillespieDA, JacksonDA, et al. (2006) Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol 26 : 3319–3326.
21. Maya-MendozaA, PetermannE, GillespieDA, CaldecottKW, JacksonDA (2007) Chk1 regulates the density of active replication origins during the vertebrate S phase. Embo J 26 : 2719–2731.
22. DupreA, Boyer-ChatenetL, SattlerRM, ModiAP, LeeJH, et al. (2008) A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol 4 : 119–125.
23. ThompsonR, MontanoR, EastmanA (2012) The mre11 nuclease is critical for the sensitivity of cells to chk1 inhibition. PLoS One 7: e44021.
24. HansenRS, ThomasS, SandstromR, CanfieldTK, ThurmanRE, et al. (2010) Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A 107 : 139–144.
25. McMurrayCT (2010) Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet 11 : 786–799.
26. LopesJ, PiazzaA, BermejoR, KriegsmanB, ColosioA, et al. (2011) G-quadruplex-induced instability during leading-strand replication. EMBO J 30 : 4033–4046.
27. GilsonE, GeliV (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8 : 825–838.
28. RizzoA, SalvatiE, PorruM, D'AngeloC, StevensMF, et al. (2009) Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res 37 : 5353–5364.
29. SfeirA, KosiyatrakulST, HockemeyerD, MacRaeSL, KarlsederJ, et al. (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138 : 90–103.
30. YeJ, LenainC, BauwensS, RizzoA, Saint-LegerA, et al. (2010) TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142 : 230–242.
31. BoscoN, de LangeT (2012) A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma 121 : 465–474.
32. AzvolinskyA, GiresiPG, LiebJD, ZakianVA (2009) Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34 : 722–734.
33. TuduriS, CrabbeL, ContiC, TourriereH, Holtgreve-GrezH, et al. (2009) Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11 : 1315–1324.
34. BermejoR, LaiMS, FoianiM (2012) Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell 45 : 710–718.
35. SabouriN, McDonaldKR, WebbCJ, CristeaIM, ZakianVA (2012) DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev 26 : 581–593.
36. ErricoA, CostanzoV (2012) Mechanisms of replication fork protection: a safeguard for genome stability. Crit Rev Biochem Mol Biol 47 : 222–235.
37. BermejoR, CapraT, JossenR, ColosioA, FrattiniC, et al. (2011) The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146 : 233–246.
38. LabibK, De PiccoliG (2011) Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philos Trans R Soc Lond B Biol Sci 366 : 3554–3561.
39. El AchkarE, Gerbault-SeureauM, MulerisM, DutrillauxB, DebatisseM (2005) Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc Natl Acad Sci U S A 102 : 18069–18074.
40. DruscoA, PekarskyY, CostineanS, AntenucciA, ContiL, et al. (2011) Common fragile site tumor suppressor genes and corresponding mouse models of cancer. J Biomed Biotechnol 2011 : 984505.
41. OkumuraH, IshiiH, PichiorriF, CroceCM, MoriM, et al. (2009) Fragile gene product, Fhit, in oxidative and replicative stress responses. Cancer Sci 100 : 1145–1150.
42. SaldivariJC, MiumaS, BeneJ, HosseiniSA, ShibataH, et al. (2012) Initiation of genome Instability and preneoplastic processes through loss of Fhit expression. PLoS Genetics 8(11): e1003077.
43. CoquelleA, ToledoF, SternS, BiethA, DebatisseM (1998) A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol Cell 2 : 259–265.
44. AnglanaM, ApiouF, BensimonA, DebatisseM (2003) Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114 : 385–394.
45. TecherH, KoundrioukoffS, AzarD, WilhelmT, CarignonS, et al. (2013) Replication Dynamics: Biases and Robustness of DNA Fiber Analysis. J Mol Biol [epub ahead of print]. doi:10.1016/j.jmb.2013.03.040
46. GreenCM, AlmouzniG (2003) Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. Embo J 22 : 5163–5174.
47. ChaumeilJ, AuguiS, ChowJC, HeardE (2008) Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol Biol 463 : 297–308.
48. MendezJ, StillmanB (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20 : 8602–8612.
49. RaderschallE, GolubEI, HaafT (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci U S A 96 : 1921–1926.
Štítky
Genetika Reprodukčná medicína
Článek Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and MammalsČlánek The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and DevelopmentČlánek Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative ElementsČlánek Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine ExposureČlánek Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2013 Číslo 7- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Solution for Crossover Formation
- Genome-Wide Association Mapping in Dogs Enables Identification of the Homeobox Gene, , as a Genetic Component of Neural Tube Defects in Humans
- Independent Evolution of Transcriptional Inactivation on Sex Chromosomes in Birds and Mammals
- Stepwise Activation of the ATR Signaling Pathway upon Increasing Replication Stress Impacts Fragile Site Integrity
- Genomic Analysis of Natural Selection and Phenotypic Variation in High-Altitude Mongolians
- Modification of tRNA by Elongator Is Essential for Efficient Translation of Stress mRNAs
- Role of CTCF Protein in Regulating Locus Transcription
- Gene Set Signature of Reversal Reaction Type I in Leprosy Patients
- Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups
- Is Required for Formation of the Genital Ridge in Mice
- Monopolin Subunit Csm1 Associates with MIND Complex to Establish Monopolar Attachment of Sister Kinetochores at Meiosis I
- Recombination Dynamics of a Human Y-Chromosomal Palindrome: Rapid GC-Biased Gene Conversion, Multi-kilobase Conversion Tracts, and Rare Inversions
- Mechanisms of Protein Sequence Divergence and Incompatibility
- Histone Methyltransferase DOT1L Drives Recovery of Gene Expression after a Genotoxic Attack
- Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies
- Combinatorial Regulation of Meiotic Holliday Junction Resolution in by HIM-6 (BLM) Helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 Nucleases
- The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development
- The Role of Interruptions in polyQ in the Pathology of SCA1
- Dietary Restriction Induced Longevity Is Mediated by Nuclear Receptor NHR-62 in
- Fine Time Course Expression Analysis Identifies Cascades of Activation and Repression and Maps a Putative Regulator of Mammalian Sex Determination
- Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90
- Oxidative Stress and Replication-Independent DNA Breakage Induced by Arsenic in
- A Moonlighting Enzyme Links Cell Size with Central Metabolism
- Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A for Timely Mitotic Progression
- The Conserved Intronic Cleavage and Polyadenylation Site of CstF-77 Gene Imparts Control of 3′ End Processing Activity through Feedback Autoregulation and by U1 snRNP
- The BTB-zinc Finger Transcription Factor Abrupt Acts as an Epithelial Oncogene in through Maintaining a Progenitor-like Cell State
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- The RNA-binding Proteins FMR1, Rasputin and Caprin Act Together with the UBA Protein Lingerer to Restrict Tissue Growth in
- Pattern Dynamics in Adaxial-Abaxial Specific Gene Expression Are Modulated by a Plastid Retrograde Signal during Leaf Development
- A Network of HMG-box Transcription Factors Regulates Sexual Cycle in the Fungus
- Bacterial Adaptation through Loss of Function
- ENU-induced Mutation in the DNA-binding Domain of KLF3 Reveals Important Roles for KLF3 in Cardiovascular Development and Function in Mice
- Interplay between Structure-Specific Endonucleases for Crossover Control during Meiosis
- FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription
- The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression
- Selective Pressures to Maintain Attachment Site Specificity of Integrative and Conjugative Elements
- The Conserved ADAMTS-like Protein Lonely heart Mediates Matrix Formation and Cardiac Tissue Integrity
- The cGMP-Dependent Protein Kinase EGL-4 Regulates Nociceptive Behavioral Sensitivity
- RBM5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility
- Disease-Related Growth Factor and Embryonic Signaling Pathways Modulate an Enhancer of Expression at the 6q23.2 Coronary Heart Disease Locus
- Yeast Pol4 Promotes Tel1-Regulated Chromosomal Translocations
- A Dual Role for SOX10 in the Maintenance of the Postnatal Melanocyte Lineage and the Differentiation of Melanocyte Stem Cell Progenitors
- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle,
- Prediction of Complex Human Traits Using the Genomic Best Linear Unbiased Predictor
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
- Reassembly of Nucleosomes at the Promoter Initiates Resilencing Following Decitabine Exposure
- Exquisite Light Sensitivity of Cryptochrome
- miR-133a Regulates Adipocyte Browning In Vivo
- Strabismus Promotes Recruitment and Degradation of Farnesylated Prickle in Planar Polarity Specification
- Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis
- Is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer
- Joint Molecule Resolution Requires the Redundant Activities of MUS-81 and XPF-1 during Meiosis
- The Mating Competence of Geographically Diverse Strains in Their Natural and Unnatural Sand Fly Vectors
- Defective Repair of Oxidative Base Lesions by the DNA Glycosylase Nth1 Associates with Multiple Telomere Defects
- Effective Blocking of the Enhancer Requires Cooperation between Two Main Mechanisms Suggested for the Insulator Function
- Trans-Ancestral Studies Fine Map the SLE-Susceptibility Locus
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- SLC26A4 Targeted to the Endolymphatic Sac Rescues Hearing and Balance in Mutant Mice
- Bacterial Adaptation through Loss of Function
- The Cohesion Protein SOLO Associates with SMC1 and Is Required for Synapsis, Recombination, Homolog Bias and Cohesion and Pairing of Centromeres in Drosophila Meiosis
- Gene × Physical Activity Interactions in Obesity: Combined Analysis of 111,421 Individuals of European Ancestry
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



