-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Two Plant Viral Suppressors of Silencing Require the Ethylene-Inducible Host Transcription Factor RAV2 to Block RNA Silencing
RNA silencing is a highly conserved pathway in the network of interconnected defense responses that are activated during viral infection. As a counter-defense, many plant viruses encode proteins that block silencing, often also interfering with endogenous small RNA pathways. However, the mechanism of action of viral suppressors is not well understood and the role of host factors in the process is just beginning to emerge. Here we report that the ethylene-inducible transcription factor RAV2 is required for suppression of RNA silencing by two unrelated plant viral proteins, potyvirus HC-Pro and carmovirus P38. Using a hairpin transgene silencing system, we find that both viral suppressors require RAV2 to block the activity of primary siRNAs, whereas suppression of transitive silencing is RAV2-independent. RAV2 is also required for many HC-Pro-mediated morphological anomalies in transgenic plants, but not for the associated defects in the microRNA pathway. Whole genome tiling microarray experiments demonstrate that expression of genes known to be required for silencing is unchanged in HC-Pro plants, whereas a striking number of genes involved in other biotic and abiotic stress responses are induced, many in a RAV2-dependent manner. Among the genes that require RAV2 for induction by HC-Pro are FRY1 and CML38, genes implicated as endogenous suppressors of silencing. These findings raise the intriguing possibility that HC-Pro-suppression of silencing is not caused by decreased expression of genes that are required for silencing, but instead, by induction of stress and defense responses, some components of which interfere with antiviral silencing. Furthermore, the observation that two unrelated viral suppressors require the activity of the same factor to block silencing suggests that RAV2 represents a control point that can be readily subverted by viruses to block antiviral silencing.
Published in the journal: Two Plant Viral Suppressors of Silencing Require the Ethylene-Inducible Host Transcription Factor RAV2 to Block RNA Silencing. PLoS Pathog 6(1): e32767. doi:10.1371/journal.ppat.1000729
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000729Summary
RNA silencing is a highly conserved pathway in the network of interconnected defense responses that are activated during viral infection. As a counter-defense, many plant viruses encode proteins that block silencing, often also interfering with endogenous small RNA pathways. However, the mechanism of action of viral suppressors is not well understood and the role of host factors in the process is just beginning to emerge. Here we report that the ethylene-inducible transcription factor RAV2 is required for suppression of RNA silencing by two unrelated plant viral proteins, potyvirus HC-Pro and carmovirus P38. Using a hairpin transgene silencing system, we find that both viral suppressors require RAV2 to block the activity of primary siRNAs, whereas suppression of transitive silencing is RAV2-independent. RAV2 is also required for many HC-Pro-mediated morphological anomalies in transgenic plants, but not for the associated defects in the microRNA pathway. Whole genome tiling microarray experiments demonstrate that expression of genes known to be required for silencing is unchanged in HC-Pro plants, whereas a striking number of genes involved in other biotic and abiotic stress responses are induced, many in a RAV2-dependent manner. Among the genes that require RAV2 for induction by HC-Pro are FRY1 and CML38, genes implicated as endogenous suppressors of silencing. These findings raise the intriguing possibility that HC-Pro-suppression of silencing is not caused by decreased expression of genes that are required for silencing, but instead, by induction of stress and defense responses, some components of which interfere with antiviral silencing. Furthermore, the observation that two unrelated viral suppressors require the activity of the same factor to block silencing suggests that RAV2 represents a control point that can be readily subverted by viruses to block antiviral silencing.
Introduction
Plants have a complex interconnected system of defense and stress pathways [1],[2] that receives incoming stimuli, transduces the signal and initiates the appropriate response. The process is orchestrated by a variety of plant hormones and small signaling molecules, and the final shape of the response is refined by crosstalk among different pathways in the network. Evidence emerging over the last decade has made it clear that RNA silencing and endogenous small RNA pathways constitute a major response to a variety of biotic and abiotic stresses [3],[4],[5]. Surprisingly, however, although many of the components of the silencing machinery are known, little is yet known about how silencing is regulated or how it is integrated into the network of other defense and stress pathways.
RNA silencing is a sequence specific RNA degradation mechanism that serves an important antiviral role in plants [6]. Antiviral silencing is triggered by double stranded RNA (dsRNA) that arises during virus infection. The dsRNA trigger is processed by DICER-LIKE (DCL) ribonucleases into primary short interfering RNAs (siRNAs), which incorporate into an ARGONAUTE (AGO) protein-containing effector complex and guide it to complementary target RNAs. The destruction of target RNAs can be amplified via a process called transitive silencing, in which the target RNA serves as template for host RNA-dependent RNA polymerases (RDRs) to produce additional dsRNA that is subsequently processed into secondary siRNAs. In addition to these RDRs, a number of other genes, including DCL2, AGO1 and SUPPRESSOR OF GENE SILENCING 3 (SGS3), are required for transitive silencing, but not for primary silencing [7],[8],[9]. The primary and transitive silencing pathways work together to limit the accumulation of viral RNAs during both the initial and systemic phases of infection.
In addition to antiviral silencing and related pathways that target invading nucleic acids, there are endogenous small RNA pathways that regulate gene expression by directing cleavage of target RNA, inhibition of mRNA translation, or modification of chromatin structure. The best studied of the endogenous small RNAs are the microRNAs (miRNAs), which play major roles in development and in response to a variety of stresses [10],[11],[12]. Although different small RNA mediated pathways have unique genetic requirements, all make use of an overlapping set of genes for their biogenesis (four DCL genes) and function (ten AGO genes), and there is growing evidence that these pathways are interconnected and compete with one another. For example, DCL1, the Dicer that produces most miRNAs, represses antiviral silencing by down-regulation of DCL3 and DCL4 [13] and, when over-expressed, blocks silencing induced by a sense transgene [14]. In addition, many viral suppressors of RNA silencing also interfere with the biogenesis and/or function of endogenous small RNAs such as miRNAs and trans-acting small interfering RNAs (tasiRNAs) [15],[16]. However, the mechanisms that regulate and integrate the various small RNA pathways are just beginning to be elucidated.
Plant viruses have evolved a variety of effective counter-defensive strategies to suppress silencing. Numerous plant viruses encode proteins that block some aspect of RNA silencing [15],[16]. These viral proteins are highly diverse in primary sequence and protein structure, though they may share certain mechanistic features. For example, the ability to bind small RNAs is a feature of many viral suppressors of silencing, including the two used in the present work. Indeed, it has been proposed that most viral suppressors of silencing work by binding and sequestering small RNAs, thereby blocking their activity [17],[18]. However, the physiological significance of small RNA binding is not yet clear in many cases [6], and some suppressors manipulate silencing via interaction with host proteins that are either components of the silencing machinery [19],[20],[21],[22] or proposed regulators of the pathway [23]. Thus, the mechanism of action of viral suppressors is likely both diverse and complex and is not yet fully understood.
Our studies have focused on understanding the mechanism of action of HC-Pro, a potent viral suppressor of silencing that blocks both primary and transitive silencing. Our approach has been to identify host proteins that physically interact with HC-Pro and examine the effect of altering the levels of these proteins on both RNA silencing and the ability of HC-Pro to block silencing [23]. Using this approach, we find that RAV2/EDF2 (hereafter referred to as RAV2), an HC-Pro-interacting protein that is a member of the RAV/EDF family of transcription factors, is required for suppression of silencing not only by potyvirus HC-Pro, but also by carmovirus P38, the silencing suppressor from a virus family unrelated to potyviruses. Interestingly, RAV2 is required exclusively for blocking the activity of primary siRNAs, whereas suppression of transitive silencing and effects on the endogenous microRNA pathway are RAV2-independent. Whole genome tiling microarray experiments were used to characterize HC-Pro-mediated changes in host expression and identify which, if any, were RAV2-dependent. The results raise the interesting possibility that HC-Pro-suppression of silencing is not caused by decreased expression of genes that are required for silencing, but instead, by induction of stress and defense pathways that interfere with antiviral silencing.
Results
Ectopic expression of a RAV/EDF transcription factor delays the onset of transgene-induced RNA silencing in tobacco
In previous work we used a yeast two-hybrid screen to identify Nicotiana tabacum proteins that interact with Tobacco Etch Potyvirus (TEV) HC-Pro [23]. One of the proteins identified in this way was named ntRAV because of its relatedness to the Arabidopsis thaliana RAV/EDF family of transcription factors. The RAV/EDF protein family has six members, and these are unique among transcription factors in having two unrelated DNA binding domains (AP2 and B3) [24]. Members of this family are responsive to numerous biotic and abiotic stresses [25],[26],[27],[28] and are inducible by the plant hormone ethylene [29], which controls many aspects of plant physiology, including defense against pathogens [30],[31].
In vitro pull-down experiments were used to confirm a physical interaction between TEV HC-Pro and ntRAV. 35S-methionine-labeled ntRAV produced in a coupled in vitro transcription/translation system co-purified with an HC-Pro-GST fusion protein isolated from recombinant bacteria, but not with GST alone (Fig. 1A and see also Fig. S1 and Text S1). This result validates the HC-Pro-ntRAV interaction initially identified in the yeast two-hybrid system.
Fig. 1. ntRAV Interacts with HC-Pro and Delays the Onset of Sense Transgene Silencing when Over-expressed in Tobacco. 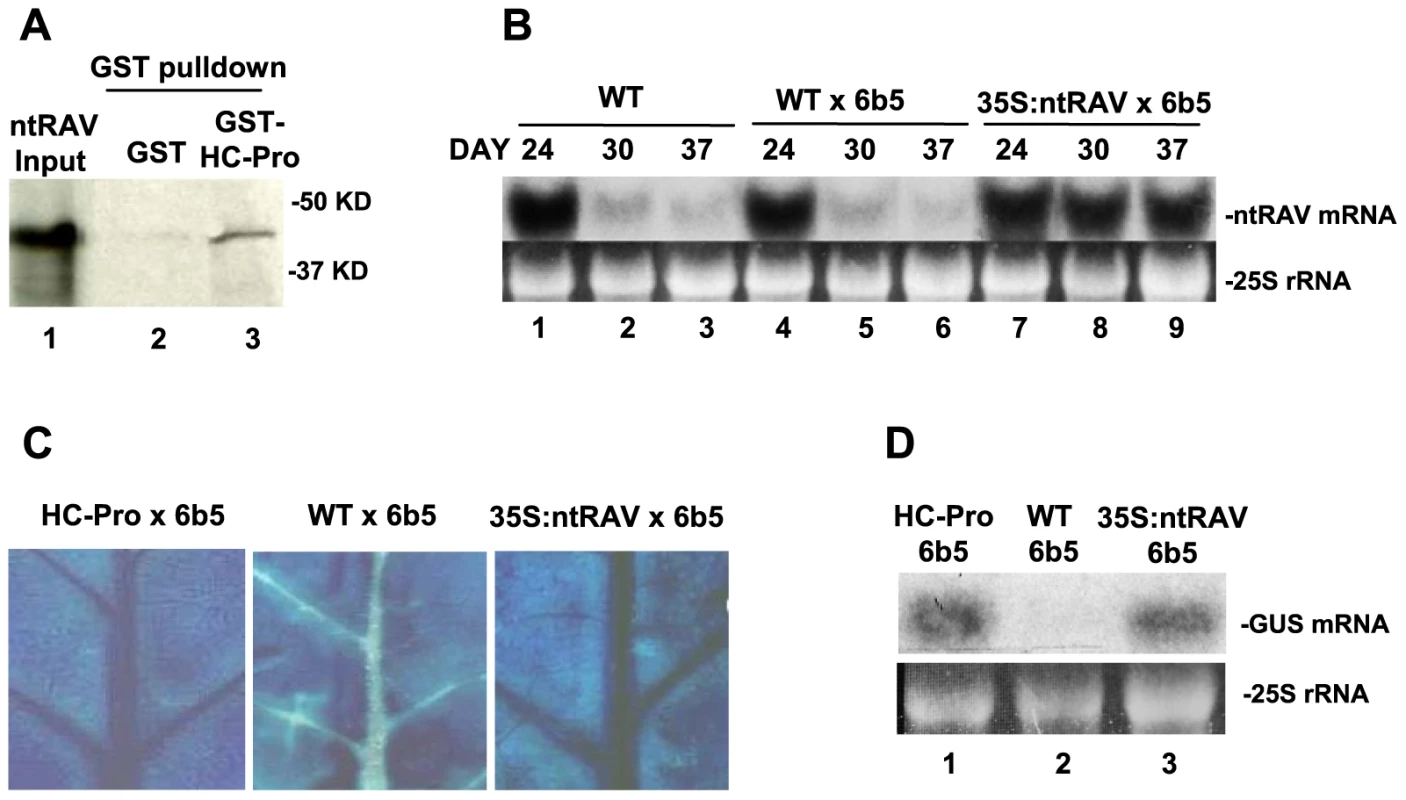
(A) Tobacco ntRAV interacts with TEV HC-Pro in in vitro pulldown experiments. 35S-labelled ntRAV co-purifies with HC-Pro-GST (lane 3), but not with GST (lane 2). Lane 1 shows the amount of input 35S-labelled ntRAV protein used in the pulldown experiments. (B) The accumulation of ntRAV mRNA at 24, 30 and 37 days after germination in whole leaves of wild type (WT) tobacco plants (lanes 1–3), plants heterozygous for the silenced 6b5 GUS transgene (WT X 6b5) (lanes 4–6), and plants heterozygous for the silenced 6b5 GUS transgene and expressing the 35S:ntRAV transgene (lanes 7–9). (C) Histochemical staining of leaves from HC-Pro X 6b5 (left panel), WT X 6b5 (center panel) and 35S:ntRAV X 6b5 leaves (right panel) at 26 days after germination. (D) GUS mRNA levels in the veins of leaves of HC-Pro X 6b5 (lane 1), WT X 6b5 (lane 2) and 35SntRAV X 6b5 plants (lane 3) at 26 days after germination. To determine if ntRAV plays a role in RNA silencing, we evaluated the effect of ntRAV over-expression on transgene-induced silencing. In tobacco, ntRAV is normally expressed at high levels throughout fully expanded healthy leaves of young plants, but expression decreases greatly starting at about 24 days after germination (Fig. 1B, lanes 1–6). In contrast, a tobacco line that ectopically expresses ntRAV from the constitutive Cauliflower mosaic virus (CaMV) 35S promoter maintains high level expression of ntRAV (Fig. 1B, lanes 7–9). We crossed the 35S:ntRAV transgenic line, as well as wild type and HC-Pro-expressing control lines, to the well-characterized tobacco transgenic line 6b5 [32], which is post-transcriptionally silenced for a transgene encoding β-glucuronidase (GUS). Silencing of the GUS locus in line 6b5 reinitiates every generation, starting in the vascular tissue of the oldest leaves and then spreading throughout the leaf. The expression of GUS in F1 progeny of these crosses was assayed histochemically in leaves (Fig. 1C) and by northern blots of RNA from the vascular tissue (Fig. 1D) at 26 days after germination. In these young plants, ectopic expression of ntRAV blocked silencing of GUS in vascular tissue of fully expanded, healthy leaves about as well as HC-Pro (Fig. 1C and D). However, unlike HC-Pro, which completely blocks silencing over the lifetime of the plant, ectopic expression of ntRAV only delayed the onset of silencing, and GUS was eventually silenced throughout the leaf (data not shown). These results, together with those showing a physical interaction between ntRAV and TEV HC-Pro proteins, raised the possibility that ntRAV plays a role in HC-Pro-mediated suppression of silencing.
Experiments in the model plant, Arabidopsis thaliana
To further investigate the role of ntRAV in HC-Pro suppression of silencing, we switched from tobacco to Arabidopsis thaliana, in order to take advantage of the numerous genetic tools available in that model system. Our experiments focused on a RAV gene family member closely related to the tobacco ntRAV, Arabidopsis RAV2 (At1g68840), which had already been cloned and characterized, and for which a validated T-DNA insertional knockout line was available [29]. The change in experimental system also necessitated a change from the HC-Pro encoded by TEV to that encoded by turnip mosaic virus (TuMV), a related potyvirus that infects Arabidopsis. Like the TEV HC-Pro transgene in tobacco, expression of the TuMV HC-Pro in transgenic Arabidopsis plants has been shown to suppress both virus - and transgene-induced RNA silencing [14],[33]. The TuMV HC-Pro transgenic line used in our experiments expresses HC-Pro at a high level and is highly phenotypic [14].
We used in vivo pull-down experiments to determine whether the TuMV HC-Pro and RAV2 proteins interact, as would be expected if RAV2 were a functional homolog of ntRAV. In these experiments, the homozygous rav2 knockout line [29] was transformed with a construct designed to express a transgene encoding FLAG-tagged RAV2. A transformant that expressed the FLAG-RAV2 transgene was crossed to our TuMV HC-Pro transgenic line [14], and expression of both transgenes in the F1 offspring was confirmed by RNA gel blot analysis (data not shown). Pull-down experiments using antiserum specific to the FLAG tag, followed by western blot analysis, showed that TuMV HC-Pro co-immunoprecipitates with the Flag-tagged RAV2 (Fig. 2), indicating that RAV2 and TuMV HC-Pro interact in planta in Arabidopsis. This result confirms that RAV2 is a functional homolog of ntRAV and also provides evidence that the interaction between potyviral HC-Pro and host RAV-like transcription factors is a conserved feature of these proteins.
Fig. 2. In vivo Interaction of RAV2 and TuMV HC-Pro in Arabidopsis. 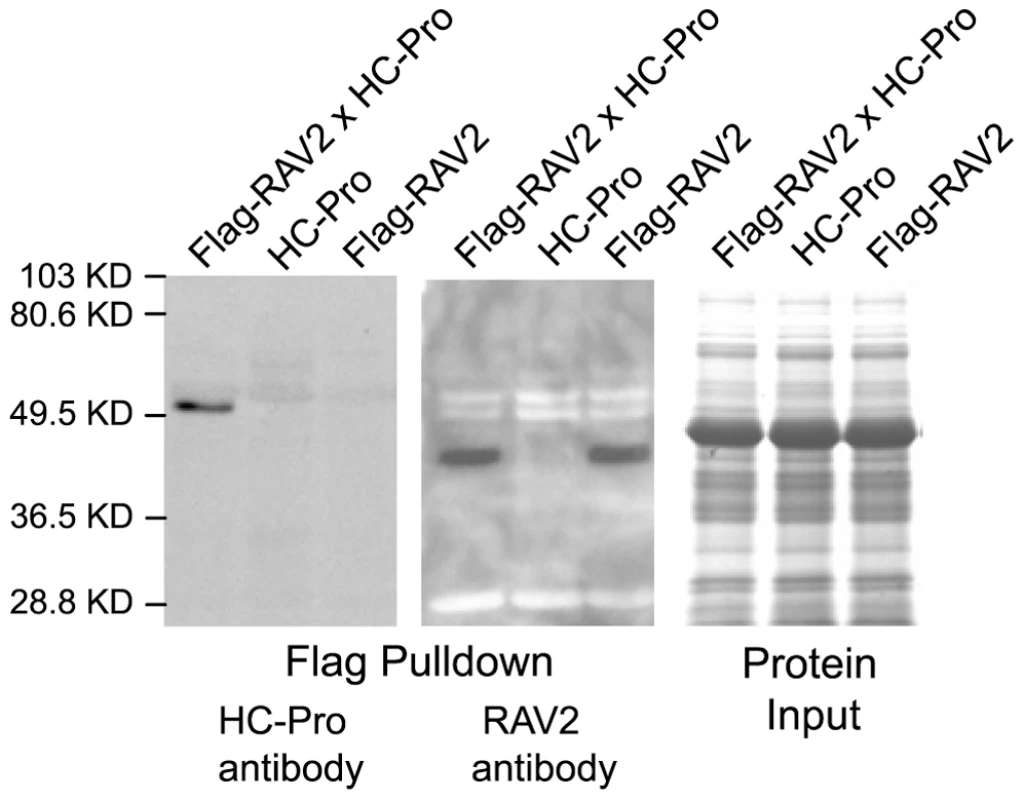
Proteins isolated from plants expressing either FLAG-tagged RAV2 (Flag-RAV2) alone, TuMV HC-Pro alone or both Flag-RAV2 and TuMV HC-Pro were incubated with anti-FLAG agarose beads. The bound protein was fractionated on acrylamide gels and subjected to western blot analysis using either HC-Pro antiserum (left panel) or RAV2 antiserum (center panel). The far right panel shows the relative input amounts of protein used in the pulldown experiments as determined by Coomassie blue staining. RAV2 is required for HC-Pro suppression of virus induced gene silencing (VIGS)
Our initial experiments to examine the role of RAV2 in HC-Pro suppression of silencing focused on VIGS. These experiments used the well characterized geminivirus silencing vector, cabbage leaf curl virus (CaLCV), which carried a portion of the endogenous CHLORATA42 (CH42) gene [34]. CH42 is required for chlorophyll accumulation, and VIGS of CH42 in wild type plants results in extensive chlorosis and marked reduction in the level of CH42 mRNA. These changes are accompanied by a pronounced accumulation of 24-nt siRNAs that derive from the CH42 sequences within the viral vector [14],[34]. HC-Pro transgenic plants become infected when bombarded with the CH42 VIGS vector and, although high levels of siRNAs accumulate in the plants, the CH42 gene is not silenced as evidenced by accumulation of CH42 mRNA and the absence of chlorosis [14]. To determine if RAV2 is required for HC-Pro suppression of VIGS, plants expressing HC-Pro in either the wild type or the rav2 knockout background, along with control plants, were bombarded with the CH42 VIGS vector. Wild type control plants as well as rav2 knockout plants exhibited chlorosis of infected tissues (Fig. 3A, top two panels) accompanied by reduction in CH42 mRNA levels and the concomitant accumulation of siRNAs, as expected for VIGS (Fig. 3B, lanes 1–4). HC-Pro transgenic plants were suppressed for VIGS of CH42, remaining green (Fig. 3A, bottom left panel) and accumulating wild type levels of CH42 mRNA as previously reported (Fig. 3B, lanes 5 and 6). In contrast, HC-Pro transgenic plants in the rav2 knockout background were competent for VIGS of CH42 as evidenced by systemic chlorosis (Fig. 3A, bottom right panel) accompanied by reduction in CH42 mRNA levels (Fig. 3B, lanes 7–9). This result indicates that RAV2 is required for HC-Pro suppression of VIGS.
Fig. 3. RAV2 is Required for HC-Pro Suppression of Virus Induced Gene Silencing (VIGS). 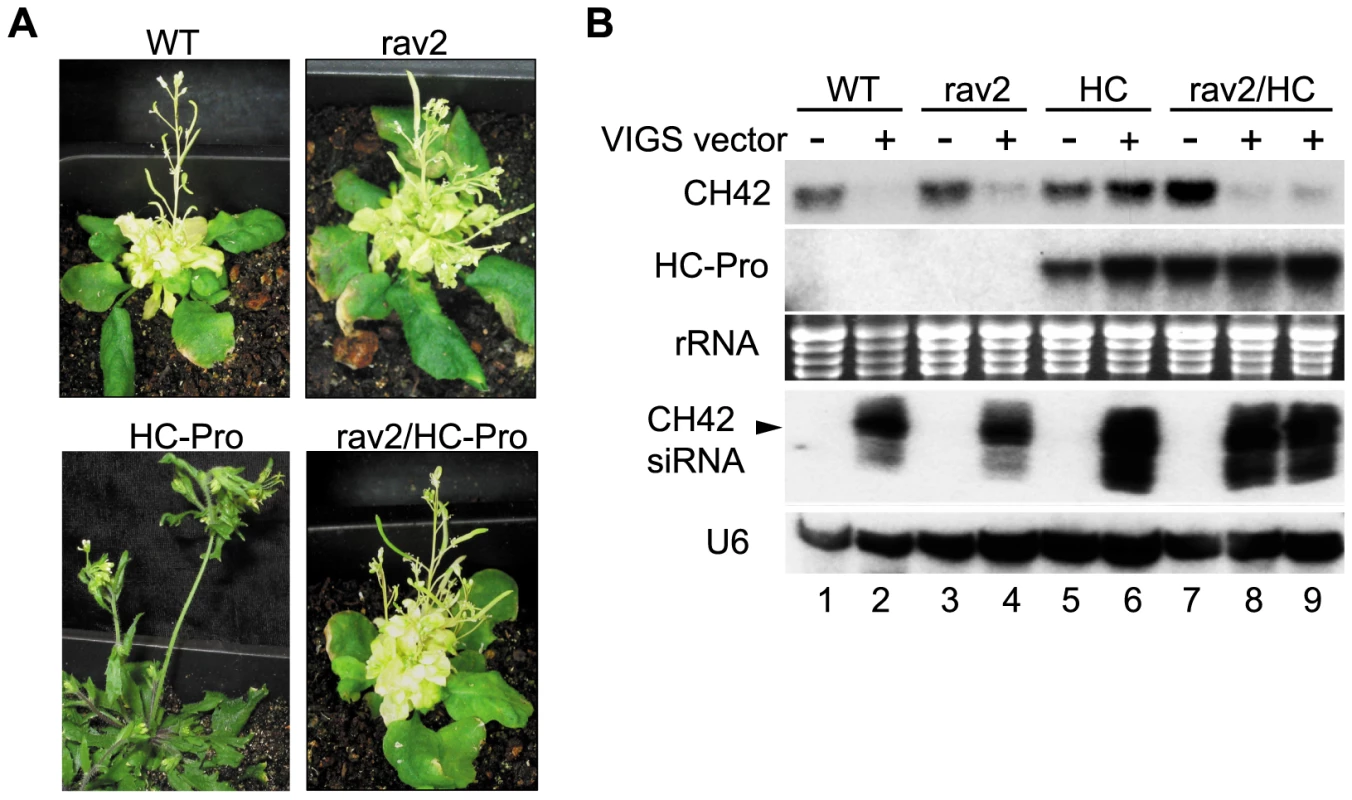
(A) Phenotype of plants bombarded with CaLCV vector carrying a portion of the endogenous CH42 gene. VIGS of CH42 results in pronounced yellowing in wild type or rav2 knockout plants (upper left and right panels, respectively). Plants expressing HC-Pro are suppressed for VIGS and therefore remain green (lower left panel); whereas HC-Pro plants in the rav2 knockout background fail to block silencing and display yellowing typical of wild type plants (lower right panel). (B) RNA gel blot analysis of CH42 mRNA, HC-Pro and CH42 siRNA levels in wild type (lanes 1 and 2), rav2 knockout (lanes 3 and 4), HC-Pro plants (lanes 5 and 6) and HC-Pro plants in the rav2 background (lanes 7–9) either uninfected (lanes 1, 3, 5 and 7) or after bombardment with the CH42 VIGS vector (lanes 2, 4, 6, 8 and 9). Ethidium staining of rRNA is shown as the loading control for the high molecular weight blots and the hybridization signal for U6 is shown as the loading control for the small RNA blot. The migration of 24 nt siRNAs is marked by an arrow. RAV2 is required for HC-Pro-suppression of the primary, but not the transitive, branch of hairpin transgene-induced RNA silencing
To examine the role of RAV2 in HC-Pro-suppression of transgene silencing, we used a well-characterized system in which silencing occurs through both the primary and transitive branches of the silencing pathway [7],[35]. This system is composed of two transgenes, the 306 and 6b4 loci (Fig. 4A). The 6b4 locus encodes an expressing GUS transgene that includes the entire GUS coding sequence, while the 306 locus encodes a hairpin construct designed to silence GUS expression. The GUS sequence in the 306 locus has a 231 nucleotide deletion in the coding region (Fig. 4A, shown in green) so that RNAs originating from the 6b4 transcript can be unambiguously distinguished. The primary and transitive branches of silencing can be easily differentiated in this system. Basically, primary siRNAs derive only from the stem of the 306 hairpin transcript (Fig. 4A, shown in red, probe 1), whereas secondary siRNAs arise from either locus during an RDR6-dependent process called transitive silencing. In the case of the 306 transgene, siRNAs that arise from the loop of hairpin transcript are secondary siRNAs (Fig. 4A, shown in blue, probe 3). In contrast to the 306 hairpin transcript, the 6b4 mRNA produces only RDR6-dependent secondary siRNAs (Fig. 4A, shown in red, green and blue; [7]. Thus, in the 306/6b4 system, 6b4 mRNA can be degraded by two mechanisms. It can be targeted by a RISC complex directed by siRNAs, or it can be a substrate for RDR6, producing dsRNA that is subsequently processed by DCL to produce secondary siRNAs via transitive silencing. HC-Pro suppresses silencing in the 306/6b4 system, but has different effects on primary and secondary siRNAs: accumulation of secondary siRNAs is eliminated, as shown by the failure to detect any siRNAs when using either probe 2 or probe 3 [7]. In contrast, high levels of primary siRNAs accumulate, but are unable to mediate degradation of the 6b4 target RNA [7].
Fig. 4. RAV2 is Required for Suppression of Hairpin Transgene Silencing by Two Unrelated Viral Suppressors. 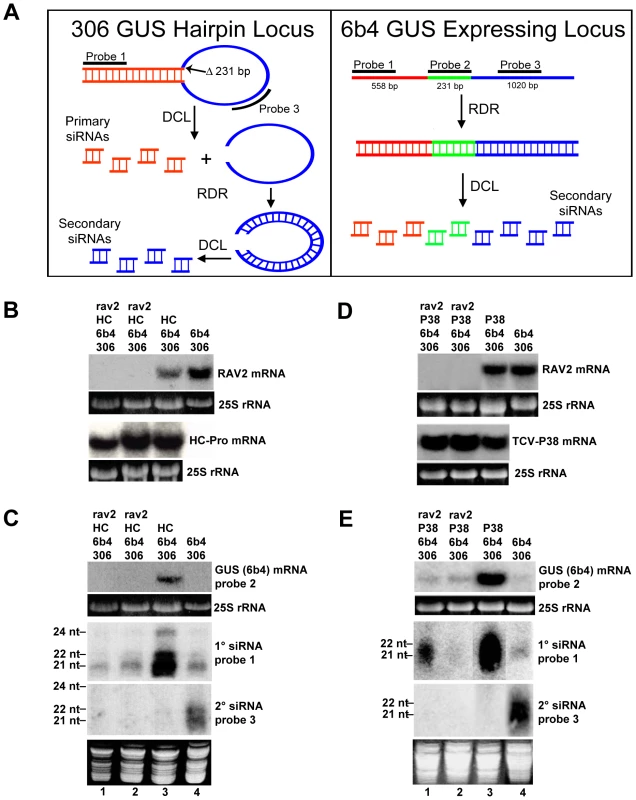
(A) Diagrams showing the structures of the 6b4 and 306 transgene loci. The 6b4 locus is an expressing locus which encodes a functional GUS protein. The 306 locus produces a GUS hairpin RNA that acts in trans to silence the 6b4 locus. The locations of the hybridization probes used in parts B, C and D are indicated. (B and D) The accumulation of RAV2, TCV-P38 and/or TuMV HC-Pro mRNA in plants of the genotypes indicated at the top of the lanes. (C and E) The top panel of each shows the accumulation of 6b4 GUS mRNA in plants of the genotypes indicated at the top of the lanes, and the bottom two panels show the accumulation of primary and secondary siRNAs in the same samples. The size of 21-, 22- and 24-nt marker RNAs are indicated to the left of the small RNA panels and the probes used are indicated to the right of each panel. To determine if RAV2 is required for HC-Pro suppression of hairpin transgene silencing, we crossed the homozygous rav2 knockout line to a transgenic line homozygous for the 306 and 6b4 loci and hemizygous for the TuMV HC-Pro locus. F1 offspring of this cross were allowed to self-fertilize, producing an F2 population that was segregating for all four loci. F2 plants were genotyped, and individuals containing the 306/6b4/HC-Pro loci in the homozygous rav2 mutant background were identified, along with control plants containing all three loci in the wild type RAV2 background. The absence of RAV2 mRNA in rav2 knockout plants was verified by RNA gel blot analysis (Fig. 4B). Initial analysis of the 306/6b4/HC-Pro plant lines addressed the possibility of transcriptional gene silencing (TGS) of the three transgenes involved, all of which are under the control of the CaMV 35S promoter. This was especially important because it has been shown that T-DNA insertion mutants that carry 35S promoter sequences, such as the rav2 knockout line used in this work, can induce TGS of other 35S promoters in the genome [36] and because HC-Pro cannot suppress silencing at the transcriptional level [37],[38]. RNA gel blot analysis showed that the level of HC-Pro mRNA was similar in all plants carrying the HC-Pro transgene (Fig. 4B), arguing against transcriptional silencing of 35S promoter sequences in the plants. In addition, the presence of siRNAs that derive from the GUS transcripts (Fig. 4C) indicates that the observed silencing of the GUS transgenes is at the post-transcriptional rather than the transcriptional level.
The role of RAV2 in HC-Pro suppression of hairpin transgene silencing was assayed using northern blot analysis to measure the accumulation of 6b4 GUS target mRNA as well as that of GUS primary and secondary siRNAs (Fig. 4C). As previously reported [7], HC-Pro blocked target RNA degradation when 306/6b4/HC-Pro transgenic plants were wild type for RAV2, showing the characteristic absence of secondary siRNAs accompanied by high levels of nonfunctional primary siRNAs (Fig. 4C, compare lanes 3 and 4). In contrast, HC-Pro failed to prevent degradation of the 6b4 GUS mRNA target in the rav2 knockout background (Fig. 4C, lanes 1 and 2). In addition, accumulation of GUS primary siRNAs was reduced in the rav2 compared to the RAV2 background and was similar to that in 306/6b4 plants without HC-Pro (Fig. 4C, lanes 1–4). Accumulation of secondary siRNAs, which are diagnostic of transitive silencing, was suppressed in HC-Pro transgenic plants even in the rav2 knockout background (Fig. 4C, lanes 1–3), suggesting that HC-Pro-suppression of transitive silencing is RAV2-independent. In this experiment, however, we cannot rule out the possibility that the rav2 knockout itself eliminates accumulation of secondary siRNAs. Therefore, our results suggest that RAV2 is required for the HC-Pro-mediated block in primary siRNA activity, but not for HC-Pro suppression of transitive silencing.
RAV2 is required for suppression of hairpin transgene-induced silencing by the carmovirus suppressor of silencing, P38
To determine if RAV2 plays a general role in viral suppression of silencing, we used the 306/6b4 hairpin transgene silencing system to investigate whether Turnip Crinkle Virus (TCV) P38, a viral suppressor of silencing from a different virus family than TuMV HC-Pro [39], requires RAV2 to block silencing. The rav2 knockout line was crossed to a 306/6b4 line that expresses P38, and the resultant F1 plants were allowed to self-fertilize. F2 plants were genotyped, and individuals containing the 306/6b4/P38 loci in the homozygous rav2 mutant background were identified along with control plants containing all three loci in the RAV2 background.
We used northern blot analysis to confirm the expected pattern of expression of RAV2 and P38 in these two sets of plants (Fig. 4D) and to examine suppression of silencing by P38 in the presence and absence of RAV2. Previous experiments showed that P38 behaves much like HC-Pro in the 306/6b4 transgene silencing system, blocking silencing and allowing 6b4 GUS mRNA to accumulate, even though high levels of GUS primary siRNAs also accumulate [7]. Similar to HC-Pro, P38 also blocks transitive silencing in this system as indicated by the absence of GUS secondary siRNAs [7]. In the current work, P38 transgenic 306/6b4 plants with at least one copy of the wild type RAV2 locus replicated those earlier results, showing P38 suppression of silencing, with a concomitant increase in accumulation of GUS primary siRNAs and elimination of GUS secondary siRNAs (Fig. 4E, compare lanes 3 and 4). In contrast, P38 suppression of silencing was strongly diminished in the rav2 knockout background (Fig. 4E, lanes 1 and 2). Similar to our results with HC-Pro, accumulation of primary siRNAs in plants expressing P38 was much reduced in the rav2 compared to the RAV2 background, whereas secondary siRNA accumulation was unaffected by the loss of RAV2 and remained undetectable (Fig. 4E, compare lanes 1 and 2 with lane 3). The variability in accumulation of primary siRNAs observed in the rav2 background (Fig. 4E, lanes 1 and 2) probably reflects the facts that individual plants were tested and accumulation of primary siRNAs is greatly reduced, but not eliminated in the absence of RAV2. Altogether our results indicate that RAV2 plays similar roles in suppression of silencing by P38 and HC-Pro. Interestingly, in both cases, RAV2 function is required for suppression of primary siRNA-directed target degradation, but dispensable for the block to transitive silencing.
RAV2 is required for some of the phenotypic defects induced by HC-Pro, but not for HC-Pro-mediated defects in the miRNA pathway
Arabidopsis plants expressing TuMV HC-Pro display a number of developmental anomalies: the plants are dwarfed with serrated leaves and have abnormal flower morphology associated with severely reduced fertility (Fig. 5A; [14],[33]). The phenotype of homozygous rav2 knockout plants, however, is indistinguishable from that of wild type plants (data not shown). To determine if RAV2 is required for any of the HC-Pro associated developmental anomalies, we compared the phenotype of HC-Pro plants in the wild type RAV2 background to that of plants expressing approximately equal levels of HC-Pro mRNA, but in the rav2 knockout background. The HC-Pro-mediated defects in flower morphology and fertility are completely alleviated in the absence of RAV2 (Fig. 5 and data not shown). In addition, both the dwarfing and serrated leaf phenotypes are mitigated - but not eliminated - in the rav2 knockout background, resulting in an intermediate phenotype that is most visible when the plants are young (Fig. 5A), but becomes less distinguishable from that of wild type after the plants have flowered (Fig. 3A, 5A, and data not shown). These observations indicate that RAV2 is required for HC-Pro-mediated flower and fertility defects and contributes to the defects in plant size and leaf shape.
Fig. 5. RAV2 is Required for Many HC-Pro-associated Morphological Anomalies but not for Defects in MicroRNA Biogenesis. 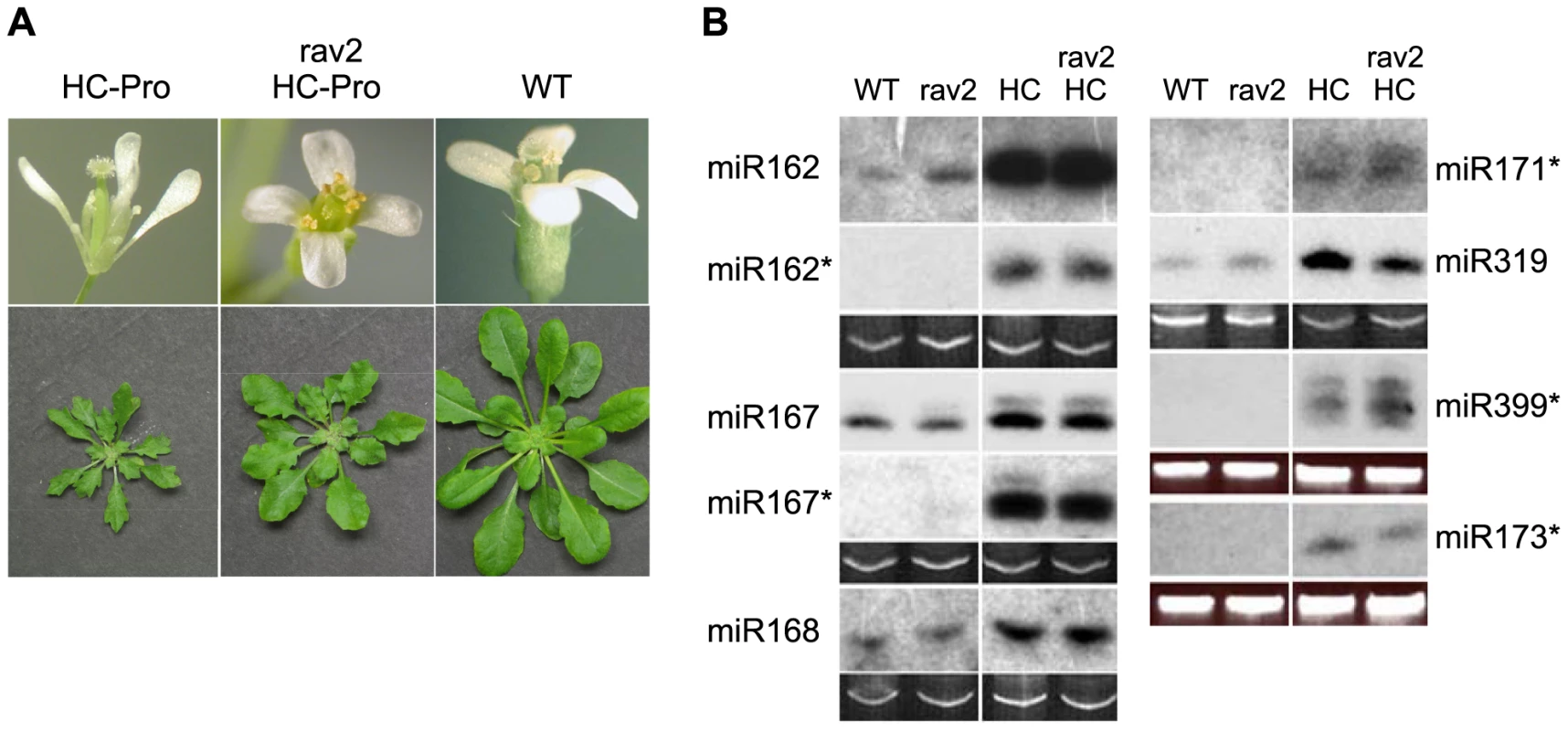
(A) Flower morphological defects in HC-Pro transgenic plants (top left panel) are rescued in the rav2 knockout background (top middle panel) resulting in flower phenotype indistinguishable from wild type (top right panel). Rosette dwarfing and leaf serration in transgenic plants (bottom left panel) are partially rescued in the rav2 knockout background (bottom middle panel) resulting in a phenotype intermediate between wild type (bottom right panel) and Hc-Pro plants. (B) The accumulation of the indicated miRNAs and miRNA*s was determined from RNA gel blot analysis of low molecular weight RNA from wild type (WT), rav2 knockout plants (rav2), HC-Pro plants (HC) and HC-Pro plants in the rav2 knockout background (rav2, HC). Ethidium bromide (EtBr) staining of the predominant RNA species in the low molecular weight fraction is shown as a loading control. In addition to its role in suppression of silencing, HC-Pro also causes defects in the biogenesis and function of certain endogenous small RNAs, including miRNAs, a class of small regulatory RNAs that plays critical roles in development. MiRNAs arise by processing of stem-loop primary transcripts by a Dicer-like enzyme, usually DCL1. The initial product is a 21-nt duplex, composed of the mature miRNA and the imperfectly complementary opposite strand, which is called miRNA*. The two strands separate and the mature miRNA binds to an AGO protein, forming the core of the miRNA effector complex. In HC-Pro transgenic plants, the level of many miRNAs is increased, often dramatically [33],[40]. Despite the increased level of the miRNA in the HC-Pro plants, the miRNA-targeted messenger RNAs also show an increased accumulation, suggesting that the miRNAs have reduced function [33],[41]. In addition, the miRNA* strand, which is unstable and fails to accumulate in wild type plants, characteristically accumulates to high levels in HC-Pro transgenic plants [33]. Together these results have led to the idea that HC-Pro impedes the proper separation of the strands of the miRNA:miRNA* duplex, leading to reduced association of the mature miRNA with AGO and thereby reducing miRNA function.
Because RAV2 is required for HC-Pro effects on the biogenesis and function of primary siRNAs, as well as for many of the HC-Pro-associated developmental anomalies, we hypothesized that RAV2 might also be required for HC-Pro-mediated defects in the miRNA pathway. To address the role of RAV2 in HC-Pro-associated defects in miRNA biogenesis, we compared the levels of a variety of miRNAs and their corresponding miRNA* strands in HC-Pro plants in the presence and absence of RAV2. In all cases, the levels of miRNA and miRNA* were independent of RAV2 (Fig. 5B). These results indicate that RAV2 is not required for the HC-Pro-associated defects in miRNA biogenesis.
To determine if RAV2 is involved in HC-Pro-associated defects in miRNA function, we compared the levels of a set of known miRNA-targeted messenger RNAs in RAV2/HC-Pro plants to those in rav2/HC-Pro plants using whole genome tiling microarray data (see following section for details of the tiling array experiments). Because HC-Pro interferes with the activity of some miRNAs [33],[41], we expected the tiling array data to show increased expression of at least some miRNA-targeted genes in HC-Pro plants. The tiling array data supported this expectation. Specifically, out of 146 verified miRNA targets [42],[43],[44],[45], we found that 39 showed altered expression in the HC-Pro transgenic line compared to the wild type control. Of these, 35 had increased expression, and only one of these was up-regulated in HC-Pro/RAV2 versus HC-Pro/rav2 plants (Table S1), suggesting that RAV2 does not play a general role in HC-Pro inhibition of miRNA activity. Altogether, the results suggest that, although RAV2 is required for many of the morphological anomalies in HC-Pro transgenic plants, it is not required for the HC-Pro-mediated defects in either the biogenesis or function of miRNAs.
Whole genome tiling analysis links HC-Pro suppression of silencing to the network of host defense pathways
Because RAV2 is a transcription factor, we expected that it might be required for some HC-Pro-mediated changes in gene expression and that identifying these genes could provide insight into the role of RAV2 in HC-Pro suppression of silencing. To address this idea, we employed whole genome tiling microarray experiments to determine if the global pattern of gene expression is altered in HC-Pro transgenic plants and, if so, whether any of the changes are dependent on RAV2 function. Arabidopsis plants with four different genotypes were used in this experiment: 1) a rav2 mutant line, 2) an HC-Pro expressing line, 3) the rav2 mutant line expressing HC-Pro, and 4) the wild type (Columbia ecotype) control. We grew all four genotypes under identical conditions, extracted total RNA from plants just before bolting and used poly-A RNA to generate probes for hybridization to the Arabidopsis tiling arrays as previously described [46],[47]. TileMap [48] was used to identify genes that are significantly up - or down-regulated in each line as compared to wild type plants, as well as to compare the pattern of gene expression in RAV2/HC-Pro plants versus rav2/HC-Pro plants (Tables S2–S9). To check the tiling results, the expression of ten genes in these plant lines was additionally examined using real-time quantitative PCR (RT qPCR). This analysis confirmed the relative levels of expression of these genes determined by the tiling array in 33 of 40 two-way comparisons between the four genotypes (Fig. 6A and B and Fig. S2).
Fig. 6. Tiling Microarray Analysis and RT qPCR Show RAV2-dependent Up-regulation of Silencing-associated Genes by HC-Pro. 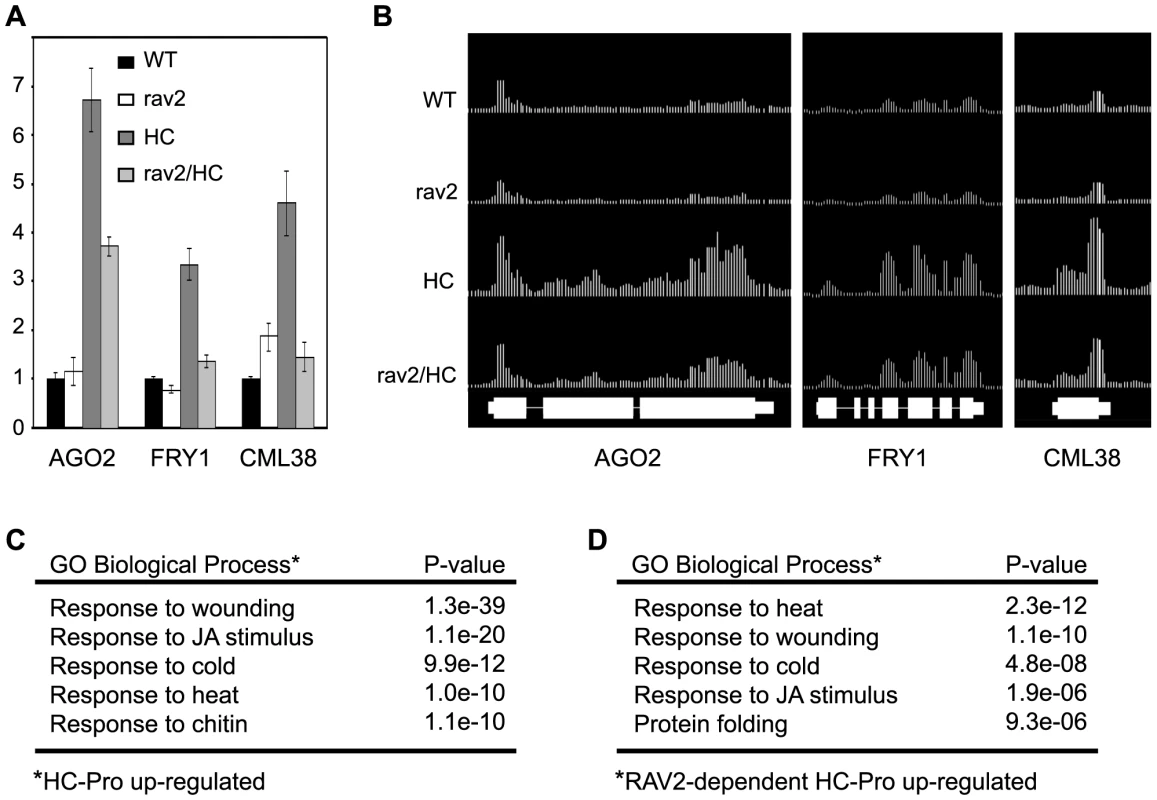
(A) The mRNA levels for AGO2 (At1g31280), FRY1 (At5g63980) and CML38 (At1g76650) in rav2 knockout plants (rav2), HC-Pro transgenic plants (HC), HC plants in the rav2 knockout background (rav2/HC) and wild type control plants (WT) were determined by oligo(dT)-primed RT qPCR analysis. Error bars, ±SD. (B) The mRNA levels for the same genes shown in (A) were determined by Arabidopsis whole-genome tiling microarray expression analysis. The top four tracks show the level of these mRNAs in the genotypes indicated to the left of the track. The bottom track indicates the annotated gene models for the three loci. (C) Gene ontology (GO) analysis results for genes that are up-regulated in HC-Pro transgenic plants as compared to wild type plants. The top five over-represented biological processes categories and the associated hypergeometric distribution P-values are shown. (D) GO analysis results for genes that are up-regulated by HC-Pro in a RAV2-dependent manner. The top five over-represented biological processes categories and the associated hypergeometric distribution P-values are shown. One of the first questions we addressed was whether genes involved in antiviral silencing and other small RNA pathways were affected by HC-Pro and RAV2. Unexpectedly, none of the genes encoding components of the silencing machinery or otherwise known to be required for silencing were down-regulated in the HC-Pro plants. Expression of RAV2 itself was also not altered in HC-Pro plants. However, a number of silencing-associated genes were up-regulated in HC-Pro plants. The up-regulated genes included three of the ten Arabidopsis AGO family members, AGO2, AGO3, and AGO4. AGO4 is required for some kinds of transcriptional silencing. The roles of AGO2 and AGO3 are unknown, but neither has been associated with antiviral silencing [49],[50]. Interestingly, two genes implicated as endogenous suppressors of silencing were also up-regulated in HC-Pro: Arabidopsis FIERY1 (FRY1), which negatively regulates transitive silencing [51], and CML38 (At1g76650), which is a likely Arabidopsis homolog of rgsCaM, an endogenous suppressor of antiviral silencing in tobacco [23]. Like RAV2, rgsCaM was originally identified as an HC-Pro interacting protein [23]; however, it is not yet known whether rgsCaM is required for HC-Pro to suppress silencing. RT qPCR confirmed the relative expression levels of AGO2, FRY1, and CML38 in the HC-Pro expressing line compared to wild type plants (compare Fig. 6A and 6B). The RT qPCR data also showed that increases in both FRY1 and CML38 expression required RAV2, whereas the increase in AGO2 expression was only partially dependent on RAV2 (Fig. 6A). These results argue that the mechanism for HC-Pro suppression of silencing does not involve down-regulation of genes required for silencing, but rather a RAV2-dependent up-regulation of genes that potentially antagonize antiviral silencing.
The tiling array analysis was used to identify global HC-Pro-mediated changes in gene expression and determine which, if any, depended on RAV2. A significant number of genes were differentially regulated in the HC-Pro plants; 2580 were up-regulated (Table S2) and 2060 were down-regulated (Table S3). Many fewer genes were differentially affected in RAV2/HC-Pro compared to rav2/HC-Pro plants (Tables S4 and S5). Of 265 genes that showed dependence on RAV2 for up-regulation by HC-Pro (Table S10), only a small number showed changed expression in rav2 mutant plants in the absence of HC-Pro as compared to wild type (20 of 265 were up-regulated; 17 of 265 were down-regulated). Similarly, of 433 genes that showed dependence on RAV2 for down-regulation by HC-Pro (Table S11), a relatively small number showed changed expression in the rav2 knockout plants in the absence of HC-Pro as compared to wild type (15 of 433 were up-regulated; 98 of 433 were down-regulated). Together, these results suggest that HC-Pro causes major changes in global gene expression patterns, some of which are mediated by RAV2. Interestingly, based on comparison of the set of genes with altered expression in rav2 mutant plants with the set altered by HC-Pro in a RAV2-dependent manner, it appears that HC-Pro changes the scope and spectrum of genes that are controlled by RAV2.
Gene Ontology (GO) term analysis was used to give a functional characterization of the tiling array results [52]. A key finding of this analysis was that multiple stress and defense responses were induced in HC-Pro expressing plants. The top four biological process categories that were over-represented among genes up-regulated in HC-Pro compared to wild type plants were: response to wounding (67 of 119 genes), response to jasmonic acid (JA) stimulus (48 of 119 genes), cold stress (49 of 197 genes) and heat stress (33 of 109 genes) (Fig. 6C). Strikingly, genes in these same four categories were also over-represented among the genes that are up-regulated by HC-Pro in a RAV2-dependent manner (Fig. 6D). Tables showing the specific genes that are up-regulated by HC-Pro in each of these GO categories, as well as the subsets that require RAV2 for HC-Pro up-regulation are in the Supplementary Tables (Tables S12–15). These results indicate that RAV2 plays a role in altered expression of stress and defense pathways in HC-Pro plants. Interestingly, FRY1 and CML38, both of which have been implicated as suppressors of silencing [23],[51] and are induced by HC-Pro in a RAV2-dependent manner (Fig. 6B), have GO annotations of response to cold and wounding, respectively, suggesting a link between silencing and other stress and defense pathways.
Discussion
It has been over a decade since the first plant viral suppressors of RNA silencing were reported [53],[54],[55], providing an early clue that silencing serves as an anti-viral defense in plants and leading to the identification of many other such silencing suppressors [56]. However, the mechanisms by which these viral proteins manipulate silencing have remained largely elusive. Here we report the identification of a host protein, the transcription factor RAV2, that is required for suppression of silencing mediated by two unrelated viral proteins, potyviral HC-Pro and carmoviral P38. RAV2 is part of a gene family that comprises six members, two of which (RAV1; At1g13260 and RAV2-like; At1g25560) are very closely related to RAV2. Surprisingly, however, neither of these related genes is able to compensate for the loss of RAV2 with respect to suppression of silencing mediated by either HC-Pro or P38. This result indicates that RAV2 provides a unique function in suppression of silencing. The identification of RAV2 as an important element in viral suppression of silencing provides a handle for identifying additional host partners and thereby unraveling the pathway of host involvement in that process.
The discovery that plant viruses from many unrelated families encode suppressors of silencing has underscored the importance of silencing in antiviral defense. Similarly, we expect our finding that viral suppressors from two unrelated viruses have evolved independently to require RAV2 underscores the importance of host proteins in viral counter-defense. In addition, it suggests that RAV2 represents an effective and readily subverted control point – either for suppression of silencing in general or for a subset of suppressors with some mechanistic features in common. It will be interesting to see how general the requirement for RAV2 is in viral suppression of silencing.
How could a transcription factor such as RAV2 be used to suppress silencing? Two reports have identified RAV2 as a repressor of at least some target genes [57],[58]. Therefore, it seemed reasonable to hypothesize that the role of RAV2 in HC-Pro suppression of silencing is to repress transcription of genes that encode components of the silencing machinery for the anti-viral branch of the silencing pathway. However, our global analysis of genome expression indicates that the expression of genes known to be required for RNA silencing is unchanged in HC-Pro transgenic plants as compared to wild type controls. Instead, our data shows that RAV2 is required for HC-Pro-mediated up-regulation of some stress and defense response genes. Earlier work showing that induction of both biotic and abiotic stresses interferes with RNA silencing induced by a viral amplicon in tobacco is consistent with a mechanism in which induction of other defense responses can divert the host from antiviral silencing [59]. The observation that RAV2 is induced by the ethylene defense pathway and is also required for viral suppression of silencing emphasizes the importance of crosstalk among defense pathways and supports the idea that RAV2 constitutes an important control point for the integration of defense responses during virus infection.
One puzzle raised by the observation that HC-Pro, which is a cytoplasmic protein [60],[61], interacts with a host transcription factor is: How and where do the two proteins have the opportunity to meet? Although HC-Pro has been shown to accumulate in nuclear inclusions in certain potyviral infections, it is thought that such inclusions represent storage of excess protein [61]. Thus, it seems more likely that HC-Pro and RAV2 interact in the cytoplasm. Sequestering transcription factors in the cytoplasm is a common mechanism used in eukaryotic organisms for controlling the activity of such proteins [62],[63]. The interaction of HC-Pro with RAV2 in the cytoplasm could either reflect a direct involvement of RAV2 itself in suppression of silencing or interference by HC-Pro in the cellular control of RAV2 – either to block activation or promote inappropriate activation – thereby changing host gene expression in such a way that promotes suppression of silencing. Elucidating these issues, as well as examining whether P38 also physically interacts with RAV2, is likely to be a fruitful area of research.
Another particularly interesting aspect of our results is the differential requirement for RAV2 in suppression of different small RNA-mediated processes. Both HC-Pro and P38 suppress transitive silencing in the absence of RAV2; yet, both suppressors require RAV2 for suppression of target degradation via the activity of primary siRNAs. Furthermore, although HC-Pro requires RAV2 to block the activity of primary siRNAs, RAV2 is not required for HC-Pro-mediated defects in miRNA activity. Our present work does not distinguish whether these differential requirements for RAV2 indicate a fundamental difference in the mechanisms responsible for suppression of these processes or simply a difference in the cofactor requirements of a common mechanism.
One current model for viral suppression of small RNA pathways posits a general mechanism in which small RNA duplexes are bound by the suppressor, thereby blocking the incorporation of one strand of the duplex into an active effector complex [17],[64]. Our data showing a role for RAV2 in suppression of silencing does not directly support this proposed mechanism, but is also not inconsistent with it. Indeed, it has been shown that small RNA binding by HC-Pro in vitro is enhanced by unknown cellular factors [17],[64]. Thus, RAV2 might be one such factor, acting either directly or indirectly to enhance small RNA binding.
Expression of HC-Pro in transgenic plants causes a set of morphological anomalies that have been attributed to defects in the biogenesis and function of endogenous miRNAs [33]. However, there is emerging evidence that suggests that the phenotypic changes are largely independent of the miRNA pathway [14],[15],[20],[65]. In support of this notion, the data we have presented here indicate that many of the HC-Pro-mediated morphological anomalies are RAV2-dependent whereas the defects in the miRNA pathway are RAV2-independent, arguing against a causative role for miRNAs in most HC-Pro-associated morphological anomalies
Although the mechanism by which HC-Pro uses RAV2 to suppress silencing is not yet clear, the results of our tiling array analysis suggest two interesting, though speculative, possibilities. The first of these relates to the induction of AGO2 and a subset of other AGO genes in HC-Pro transgenic plants, an effect that is only partially dependent on RAV2. The AGO genes that are up-regulated by HC-Pro are not required for post-transcriptional gene silencing (PTGS). These results suggest that an alteration of the mix of AGO proteins in the cell might tip the balance away from PTGS towards other small RNA pathways that are not directly involved in anti-viral defense. The recent demonstration that changing the 5′ nucleotide of a miRNA so as to favor binding to AGO2 instead of AGO1 inactivates that miRNA [66] supports the idea that an overabundance of the wrong AGO proteins could contribute to suppression of silencing. The second interesting possibility suggested by our tiling data concerns the result that HC-Pro requires RAV2 to induce expression of FRY1 and CML38, both of which have been implicated as endogenous suppressors of silencing and both of which are associated with stress or defense responses. Induction of endogenous suppressors of silencing may be more widespread than we know because most have probably not yet been identified [51]. It is tempting to speculate that the induction of stress and defense pathways by HC-Pro might have the counter-productive result - from the plant's perspective - of inducing a set of endogenous suppressors of antiviral silencing.
Materials and Methods
Plant Material and Transgenic Lines
The tobacco 6b5 [32] and Arabidopsis TuMV HC-Pro [CT25 [14]], TCV-P38 [39], 306 and 6b4 [35] lines have been previously described. The Arabidopsis rav2/edf2 (At1g68840) T-DNA insertion line (SALK_070847) was used and did not express detectable levels of RAV2 mRNA as assayed by northern analysis. See Text S1 for the procedures used to generate the 35S:ntRAV tobacco transgenic line and genotyping of the SALK_070847 T-DNA insertion line. All Arabidopsis plants were of the Columbia ecotype.
GUS Histochemical Staining
Histochemical staining for GUS activity was carried out as described [53].
VIGS Silencing Assays
The silencing of endogenous CH42 expression using the geminivirus CaLCV vector was performed exactly as described previously [14].
RNA Isolation and Northern Analysis
RNA isolation and RNA gel blot analysis of high and low molecular weight RNA were performed exactly as previously described [14],[40],[67]. Probes for detection of TuMV HC-Pro, TCV-P38 and 6b4 mRNAs, miRNA as well as those for primary and secondary siRNAs from the 6b4/306 transgene silencing system were previously described [7]. The RAV2 probe was generated using the primer set (5′ primer-TTGGAAAGTTCGGTCTGGTC and 3′ primer-TAATACGACTCACTATAGGGACCGCAAACATATCATCAACATCTC), which generate a 152 bp fragment from the 3′ end of the gene. The 3′ RAV2 primer contains T7 promoter sequences and a 4 nucleotide spacer at its 5′ end to facilitate synthesis of the probe using T7 polymerase.
GST Pulldown Assays
The production of the HC-Pro-GST fusion protein and 35S-methionine labeled ntRAV is described in Text S1. To determine if HC-Pro-GST and ntRAV interact, approximately equimolar amounts of GST or HC-Pro-GST fusion protein were added to 20 µl of glutathione sepharose 4B beads (GE Healthcare) in GLB buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 1 mM PMSF) supplemented to contain 100 µg/ml BSA and 0.1% NP-40 (Roche) and shaken gently for 1 hour at 4°C. After rinsing with supplemented GLB, an equal amount of 35S-methionine labeled ntRAV was added to each sample, shaken gently at 4°C for 2 hours and rinsed again with supplemented GLB. Bound protein was eluted from the beads with Laemmli sample buffer, resolved by SDS-PAGE, and transferred to PVDF membrane. 35S-methionine labeled ntRAV was visualized by autoradiography.
Co-immunoprecipitation of RAV2 and HC-Pro
Protein was extracted from 0.5 g of Arabidopsis rosette leaf tissue by the following procedure. Tissue was frozen in liquid nitrogen, ground into powder with a mortar and pestle, homogenized in 4 ml of protein extraction buffer (40 mM Tris-Cl, pH 8.0, 200 mM NaCl, 2.5 mM EDTA, 1% Triton X-100, 0.1% NP-40) containing protease inhibitor cocktail (Roche), and centrifuged (12,000 g at 4°C). The supernatant was incubated with 100 µl pre-washed anti-FLAG M2 agarose beads (Sigma F2426) at 4°C for two hours. Agarose beads containing protein complexes were washed three times with extraction buffer, boiled in SDS sample buffer, resolved on a 10% SDS polyacrylamide gel, and subjected to western blotting. The presence of RAV2 protein was detected using a rabbit anti-RAV2 peptide antibody generated from the peptide GGKRSRDVDDMFALRC, and a rabbit anti-HC-Pro peptide antibody generated from the peptide KEFTKVVRDKLVGE was used to detect HC-Pro. Both RAV2 and HC-Pro peptide antibodies were produced by Sigma-Genosys.
Tiling Microarray Analysis
Total RNA was isolated as described above from the above ground portions of six week old plants that had not yet bolted. Generation of probes to poly-A RNA and hybridization to the tiling arrays were performed as described previously [46],[47]. The data was analyzed using the program TileMap with a posterior probability of 0.8 [48]. The TileMap program identifies sequences that have significant changes in expression compared to controls, but does not provide fold-differences in expression levels. GO analysis was performed using ProfCom [68].
Supporting Information
Zdroje
1. FujitaM
FujitaY
NoutoshiY
TakahashiF
NarusakaY
2006
Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks.
Curr Opin Plant Biol
9
436
442
2. KunkelBN
BrooksDM
2002
Cross talk between signaling pathways in pathogen defense.
Current Opinion in Plant Biology
5
325
331
3. SunkarR
ZhuJK
2004
Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis.
Plant Cell
16
2001
2019
Epub 2004 Jul 2016
4. HamiltonAJ
BaulcombeDC
1999
A species of small antisense RNA in posttranscriptional gene silencing in plants.
Science
286
950
952
5. RatcliffF
HarrisonBD
BaulcombeDC
1997
A Similarity Between Viral Defense and Gene Silencing in Plants.
Science
276
1558
1560
6. DingSW
VoinnetO
2007
Antiviral immunity directed by small RNAs.
Cell
130
413
426
7. MlotshwaS
PrussGJ
PeragineA
EndresMW
LiJ
2008
DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis.
PLoS ONE
3
e1755
doi:10.1371/journal.pone.0001755
8. FagardM
BoutetS
MorelJB
BelliniC
VaucheretH
2000
AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals.
Proc Natl Acad Sci U S A
97
11650
11654
9. MourrainP
BeclinC
ElmayanT
FeuerbachF
GodonC
2000
Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance.
Cell
101
533
542
10. ChuckG
CandelaH
HakeS
2009
Big impacts by small RNAs in plant development.
Curr Opin Plant Biol
12
81
86
11. MalloryAC
VaucheretH
2006
Functions of microRNAs and related small RNAs in plants.
Nat Genet
38
S31
36
12. SunkarR
ChinnusamyV
ZhuJ
ZhuJK
2007
Small RNAs as big players in plant abiotic stress responses and nutrient deprivation.
Trends Plant Sci
12
301
309
13. QuF
YeX
MorrisTJ
2008
Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1.
Proc Natl Acad Sci U S A
105
14732
14737
14. MlotshwaS
SchauerSE
SmithTH
MalloryAC
HerrJMJr
2005
Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways.
Plant Cell
17
2873
2885
15. Diaz-PendonJA
DingSW
2008
Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis.
Annu Rev Phytopathol
46
303
326
16. BurgyanJ
2008
Role of silencing suppressor proteins.
Methods Mol Biol
451
69
79
17. LakatosL
CsorbaT
PantaleoV
ChapmanEJ
CarringtonJC
2006
Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors.
Embo J
25
2768
2780
18. MeraiZ
KerenyiZ
KerteszS
MagnaM
LakatosL
2006
Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing.
J Virol
80
5747
5756
19. BaumbergerN
TsaiCH
LieM
HaveckerE
BaulcombeDC
2007
The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation.
Curr Biol
17
1609
1614
20. DelerisA
Gallego-BartolomeJ
BaoJ
KasschauKD
CarringtonJC
2006
Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense.
Science
313
68
71
21. GlickE
ZrachyaA
LevyY
MettA
GidoniD
2008
Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein.
Proc Natl Acad Sci U S A
105
157
161
22. ZhangX
YuanYR
PeiY
LinSS
TuschlT
2006
Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense.
Genes Dev
20
3255
3268
23. AnandalakshmiR
MaratheR
GeX
HerrJMJr
MauC
2000
A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants.
Science
290
142
144
24. KagayaY
OhmiyaK
HattoriT
1999
RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants.
Nucleic Acids Res
27
470
478
25. SohnKH
LeeSC
JungHW
HongJK
HwangBK
2006
Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance.
Plant Mol Biol
61
897
915
26. KimSY
KimYC
LeeJH
OhSK
ChungE
2005
Identification of a CaRAV1 possessing an AP2/ERF and B3 DNA-binding domain from pepper leaves infected with Xanthomonas axonopodis pv. glycines 8ra by differential display.
Biochim Biophys Acta
1729
141
146
27. KagayaY
HattoriT
2009
Arabidopsis transcription factors, RAV1 and RAV2, are regulated by touch-related stimuli in a dose-dependent and biphasic manner.
Genes Genet Syst
84
95
99
28. FowlerS
ThomashowMF
2002
Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway.
Plant Cell
14
1675
1690
29. AlonsoJM
StepanovaAN
LeisseTJ
KimCJ
ChenH
2003
Genome-wide insertional mutagenesis of Arabidopsis thaliana.
Science
301
653
657
30. BroekaertWF
DelaureSL
De BolleMF
CammueBP
2006
The role of ethylene in host-pathogen interactions.
Annu Rev Phytopathol
44
393
416
31. GuoH
EckerJR
2004
The ethylene signaling pathway: new insights.
Curr Opin Plant Biol
7
40
49
32. ElmayanT
VaucheretH
1996
Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally.
Plant Journal
9
787
797
33. KasschauKD
XieZ
AllenE
LlaveC
ChapmanEJ
2003
P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function.
Dev Cell
4
205
217
34. TurnageMA
MuangsanN
PeeleCG
RobertsonD
2002
Geminivirus-based vectors for gene silencing in Arabidopsis.
Plant J
30
107
114
35. BeclinC
BoutetS
WaterhouseP
VaucheretH
2002
A branched pathway for transgene-induced RNA silencing in plants.
Curr Biol
12
684
688
36. DaxingerL
HunterB
SheikhM
JauvionV
GasciolliV
2008
Unexpected silencing effects from T-DNA tags in Arabidopsis.
Trends Plant Sci
13
4
6
37. MaratheR
SmithTH
AnandalakshmiR
BowmanLH
FagardM
2000
Plant viral suppressors of post-transcriptional silencing do not suppress transcriptional silencing.
Plant J
22
51
59
38. MetteMF
MatzkeAJ
MatzkeMA
2001
Resistance of RNA-mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing.
Curr Biol
11
1119
1123
39. ChapmanEJ
ProkhnevskyAI
GopinathK
DoljaVV
CarringtonJC
2004
Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step.
Genes Dev
18
1179
1186
40. MalloryAC
ReinhartBJ
BartelD
VanceVB
BowmanLH
2002
From the Cover: A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco.
Proc Natl Acad Sci U S A
99
15228
15233
41. DunoyerP
LecellierCH
ParizottoEA
HimberC
VoinnetO
2004
Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing.
Plant Cell
16
1235
1250
42. AllenE
XieZ
GustafsonAM
CarringtonJC
2005
microRNA-directed phasing during trans-acting siRNA biogenesis in plants.
Cell
121
207
221
43. GermanMA
PillayM
JeongDH
HetawalA
LuoS
2008
Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends.
Nat Biotechnol
26
941
946
44. FahlgrenN
HowellMD
KasschauKD
ChapmanEJ
SullivanCM
2007
High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes.
PLoS ONE
2
e219
doi:10.1371/journal.pone.0000219
45. HowellMD
FahlgrenN
ChapmanEJ
CumbieJS
SullivanCM
2007
Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA - and tasiRNA-directed targeting.
Plant Cell
19
926
942
46. GregoryBD
O'MalleyRC
ListerR
UrichMA
Tonti-FilippiniJ
2008
A link between RNA metabolism and silencing affecting Arabidopsis development.
Dev Cell
14
854
866
47. ChekanovaJA
GregoryBD
ReverdattoSV
ChenH
KumarR
2007
Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome.
Cell
131
1340
1353
48. JiH
WongWH
2005
TileMap: create chromosomal map of tiling array hybridizations.
Bioinformatics
21
3629
3636
49. MalloryAC
ElmayanT
VaucheretH
2008
MicroRNA maturation and action–the expanding roles of ARGONAUTEs.
Curr Opin Plant Biol
11
560
566
50. VaucheretH
2008
Plant ARGONAUTES.
Trends Plant Sci
13
350
358
51. GyI
GasciolliV
LauresserguesD
MorelJB
GombertJ
2007
Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors.
Plant Cell
19
3451
3461
52. ThomasPD
MiH
LewisS
2007
Ontology annotation: mapping genomic regions to biological function.
Curr Opin Chem Biol
11
4
11
53. AnandalakshmiR
PrussGJ
GeX
MaratheR
MalloryAC
1998
A viral suppressor of gene silencing in plants.
Proc Natl Acad Sci U S A
95
13079
13084
54. BrignetiG
VoinnetO
LiWX
JiLH
DingSW
1998
Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana.
Embo J
17
6739
6746
55. KasschauKD
CarringtonJC
1998
A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing.
Cell
95
461
470
56. RothBM
PrussGJ
VanceVB
2004
Plant viral suppressors of RNA silencing.
Virus Res
102
97
108
57. CastillejoC
PelazS
2008
The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering.
Curr Biol
18
1338
1343
58. IkedaM
Ohme-TakagiM
2009
A novel group of transcriptional repressors in Arabidopsis.
Plant Cell Physiol
50
970
975
59. TalianskyM
KimSH
MayoMA
KalininaNO
FraserG
2004
Escape of a plant virus from amplicon-mediated RNA silencing is associated with biotic or abiotic stress.
Plant J
39
194
205
60. MlotshwaS
VerverJ
Sithole-NiangI
GopinathK
CaretteJ
2002
Subcellular location of the helper component-proteinase of Cowpea aphid-borne mosaic virus.
Virus Genes
25
207
216
61. RiedelD
LesemannDE
MaissE
1998
Ultrastructural localization of nonstructural and coat proteins of 19 potyviruses using antisera to bacterially expressed proteins of plum pox potyvirus.
Arch Virol
143
2133
2158
62. GarciaAV
ParkerJE
2009
Heaven's Gate: nuclear accessibility and activities of plant immune regulators.
Trends Plant Sci
14
479
487
63. LeeY
LeeHS
LeeJS
KimSK
KimSH
2008
Hormone - and light-regulated nucleocytoplasmic transport in plants: current status.
J Exp Bot
59
3229
3245
64. SilhavyD
BurgyanJ
2004
Effects and side-effects of viral RNA silencing suppressors on short RNAs.
Trends Plant Sci
9
76
83
65. Diaz-PendonJA
LiF
LiWX
DingSW
2007
Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs.
Plant Cell
19
2053
2063
66. MiS
CaiT
HuY
ChenY
HodgesE
2008
Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide.
Cell
133
116
127
67. MlotshwaS
YangZ
KimY
ChenX
2006
Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana.
Plant Mol Biol
61
781
793
68. AntonovAV
SchmidtT
WangY
MewesHW
2008
ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data.
Nucleic Acids Res
36
W347
351
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Polyoma Virus-Induced Osteosarcomas in Inbred Strains of Mice: Host Determinants of MetastasisČlánek Nutrient Availability as a Mechanism for Selection of Antibiotic Tolerant within the CF AirwayČlánek Type I Interferon Induction Is Detrimental during Infection with the Whipple's Disease Bacterium,
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 1- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- CD8+ T Cell Control of HIV—A Known Unknown
- The Deadly Chytrid Fungus: A Story of an Emerging Pathogen
- Characterization of the Oral Fungal Microbiome (Mycobiome) in Healthy Individuals
- Polyoma Virus-Induced Osteosarcomas in Inbred Strains of Mice: Host Determinants of Metastasis
- Within-Host Evolution of in Four Cases of Acute Melioidosis
- The Type III Secretion Effector NleE Inhibits NF-κB Activation
- Protease-Sensitive Synthetic Prions
- Histone Deacetylases Play a Major Role in the Transcriptional Regulation of the Life Cycle
- Parasite-Derived Plasma Microparticles Contribute Significantly to Malaria Infection-Induced Inflammation through Potent Macrophage Stimulation
- β-Neurexin Is a Ligand for the MSCRAMM SdrC
- Structure of the HCMV UL16-MICB Complex Elucidates Select Binding of a Viral Immunoevasin to Diverse NKG2D Ligands
- Nutrient Availability as a Mechanism for Selection of Antibiotic Tolerant within the CF Airway
- Like Will to Like: Abundances of Closely Related Species Can Predict Susceptibility to Intestinal Colonization by Pathogenic and Commensal Bacteria
- Importance of the Collagen Adhesin Ace in Pathogenesis and Protection against Experimental Endocarditis
- N-glycan Core β-galactoside Confers Sensitivity towards Nematotoxic Fungal Galectin CGL2
- Two Plant Viral Suppressors of Silencing Require the Ethylene-Inducible Host Transcription Factor RAV2 to Block RNA Silencing
- A Small-Molecule Inhibitor of Motility Induces the Posttranslational Modification of Myosin Light Chain-1 and Inhibits Myosin Motor Activity
- Temporal Proteome and Lipidome Profiles Reveal Hepatitis C Virus-Associated Reprogramming of Hepatocellular Metabolism and Bioenergetics
- Marburg Virus Evades Interferon Responses by a Mechanism Distinct from Ebola Virus
- B Cell Activation by Outer Membrane Vesicles—A Novel Virulence Mechanism
- Killing a Killer: What Next for Smallpox?
- PPARγ Controls Dectin-1 Expression Required for Host Antifungal Defense against
- TRIM5α Modulates Immunodeficiency Virus Control in Rhesus Monkeys
- Immature Dengue Virus: A Veiled Pathogen?
- Panton-Valentine Leukocidin Is a Very Potent Cytotoxic Factor for Human Neutrophils
- In Vivo CD8+ T-Cell Suppression of SIV Viremia Is Not Mediated by CTL Clearance of Productively Infected Cells
- Placental Syncytiotrophoblast Constitutes a Major Barrier to Vertical Transmission of
- Type I Interferon Induction Is Detrimental during Infection with the Whipple's Disease Bacterium,
- The M/GP Glycoprotein Complex of Porcine Reproductive and Respiratory Syndrome Virus Binds the Sialoadhesin Receptor in a Sialic Acid-Dependent Manner
- Social Motility in African Trypanosomes
- Melanoma Differentiation-Associated Gene 5 (MDA5) Is Involved in the Innate Immune Response to Infection In Vivo
- Protection of Mice against Lethal Challenge with 2009 H1N1 Influenza A Virus by 1918-Like and Classical Swine H1N1 Based Vaccines
- Upregulation of xCT by KSHV-Encoded microRNAs Facilitates KSHV Dissemination and Persistence in an Environment of Oxidative Stress
- Persistent ER Stress Induces the Spliced Leader RNA Silencing Pathway (SLS), Leading to Programmed Cell Death in
- Evolutionary Trajectories of Beta-Lactamase CTX-M-1 Cluster Enzymes: Predicting Antibiotic Resistance
- Nucleoporin 153 Arrests the Nuclear Import of Hepatitis B Virus Capsids in the Nuclear Basket
- CD8+ Lymphocytes Control Viral Replication in SIVmac239-Infected Rhesus Macaques without Decreasing the Lifespan of Productively Infected Cells
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Panton-Valentine Leukocidin Is a Very Potent Cytotoxic Factor for Human Neutrophils
- CD8+ T Cell Control of HIV—A Known Unknown
- Polyoma Virus-Induced Osteosarcomas in Inbred Strains of Mice: Host Determinants of Metastasis
- The Deadly Chytrid Fungus: A Story of an Emerging Pathogen
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



