-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Epigenetic Repression of by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP
As an inhibitor of cyclin-dependent kinases, p16INK4A is an important tumour suppressor and inducer of cellular senescence that is often inactivated during the development of cancer by promoter DNA methylation. Using newly established lymphoblastoid cell lines (LCLs) expressing a conditional EBNA3C from recombinant EBV, we demonstrate that EBNA3C inactivation initiates chromatin remodelling that resets the epigenetic status of p16INK4A to permit transcriptional activation: the polycomb-associated repressive H3K27me3 histone modification is substantially reduced, while the activation-related mark H3K4me3 is modestly increased. Activation of EBNA3C reverses the distribution of these epigenetic marks, represses p16INK4A transcription and allows proliferation. LCLs lacking EBNA3A express relatively high levels of p16INK4A and have a similar pattern of histone modifications on p16INK4A as produced by the inactivation of EBNA3C. Since binding to the co-repressor of transcription CtBP has been linked to the oncogenic activity of EBNA3A and EBNA3C, we established LCLs with recombinant viruses encoding EBNA3A - and/or EBNA3C-mutants that no longer bind CtBP. These novel LCLs have revealed that the chromatin remodelling and epigenetic repression of p16INK4A requires the interaction of both EBNA3A and EBNA3C with CtBP. The repression of p16INK4A by latent EBV will not only overcome senescence in infected B cells, but may also pave the way for p16INK4A DNA methylation during B cell lymphomagenesis.
Published in the journal: Epigenetic Repression of by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog 6(6): e32767. doi:10.1371/journal.ppat.1000951
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000951Summary
As an inhibitor of cyclin-dependent kinases, p16INK4A is an important tumour suppressor and inducer of cellular senescence that is often inactivated during the development of cancer by promoter DNA methylation. Using newly established lymphoblastoid cell lines (LCLs) expressing a conditional EBNA3C from recombinant EBV, we demonstrate that EBNA3C inactivation initiates chromatin remodelling that resets the epigenetic status of p16INK4A to permit transcriptional activation: the polycomb-associated repressive H3K27me3 histone modification is substantially reduced, while the activation-related mark H3K4me3 is modestly increased. Activation of EBNA3C reverses the distribution of these epigenetic marks, represses p16INK4A transcription and allows proliferation. LCLs lacking EBNA3A express relatively high levels of p16INK4A and have a similar pattern of histone modifications on p16INK4A as produced by the inactivation of EBNA3C. Since binding to the co-repressor of transcription CtBP has been linked to the oncogenic activity of EBNA3A and EBNA3C, we established LCLs with recombinant viruses encoding EBNA3A - and/or EBNA3C-mutants that no longer bind CtBP. These novel LCLs have revealed that the chromatin remodelling and epigenetic repression of p16INK4A requires the interaction of both EBNA3A and EBNA3C with CtBP. The repression of p16INK4A by latent EBV will not only overcome senescence in infected B cells, but may also pave the way for p16INK4A DNA methylation during B cell lymphomagenesis.
Introduction
In vitro, EBV can very efficiently induce the activation and continuous proliferation of resting human B lymphocytes. The resulting lymphoblastoid cell lines (LCLs) carry the viral genome as extra-chromosomal episomes and express only nine ‘latent’ EBV proteins. There are six nuclear antigens (EBNAs 1, 2, 3A, 3B, 3C & LP), three membrane associated proteins (LMP1, LMP2A & 2B) and in addition several untranslated RNA species. Together these factors activate the quiescent B cells and sustain their proliferation while maintaining the viral episome in its extra-chromosomal state (reviewed in [1]). Current data on the persistence of EBV in humans are consistent with the viral genome residing long-term in the resting memory B cell compartment. This occurs in at least of 90% of the world's population. However, to establish persistence EBV generally infects resting (naïve) B cells in Waldeyer's ring of the oropharynx and drives these to proliferate as activated B-blasts. This transient expansion of an infected B-blast population is generally accompanied by migration into germinal centres and differentiation to become centroblasts and centrocytes and finally resting memory B cells. The precise series of events that the EBV-positive B cells undergo to reach the memory compartment is unknown, but it appears to involve the regulated, sequential silencing of EBV genes encoding latency-associated proteins [2], [3].
Although infection is generally asymptomatic, EBV can be the causative agent in the benign self-limiting lymphoproliferation, infectious mononucleosis (IM). Uncontrolled proliferation of latently infected B cells in the immunocompromised may result in a chronic form of IM, a chronic polyclonal B-lymphoproliferative disorder called post-transplant lymphoproliferative disease (PTLD) or rarely the development of malignant lymphoma. Individuals co-infected with malaria (mainly children) or HIV (mainly adults) may be at increased risk of developing EBV-associated B cell lymphomas, including Burkitt's lymphoma (BL) and diffuse large B cell lymphoma [4].
EBNA3A, EBNA3B and EBNA3C are considered to comprise a family which probably arose in primate gamma-herpesvirus evolution by a series of gene duplication events since they have the same gene structure (ie a short 5′ coding exon and a long 3′ coding exon), are arranged in tandem in the EBV genome and share limited but significant amino acid sequence homology. EBNA3 transcripts are alternatively spliced from very long mRNAs generally initiated at the Cp latency promoter and LCLs have only a few copies of these transcripts per cell, suggesting their expression is very tightly regulated and the turnover of the EBNA3s is slow [1], [5]. Although they are related and the three proteins have limited homology, there is nothing to suggest that they have extensively redundant functions. Genetic studies using recombinant viruses originally indicated that EBNA3A and EBNA3C are essential for the efficient in vitro transformation or immortalisation of B cells, whereas EBNA3B is dispensable [6], [7]. However, under the appropriate conditions, with feeder cells present, it has been possible to establish EBNA3A-negative LCLs ([8]; our unpublished data).
Each EBNA3 protein binds to the cellular DNA-binding factor RBP-JK (also known as CBF1). This is the same protein that binds to, and targets to DNA, the EBV transactivator EBNA2 and the NOTCH-IC effector of the NOTCH signalling pathway. EBNA3A, EBNA3B and EBNA3C can repress Cp reporter plasmids and plasmids containing multiple RBP-JK/CBF1 binding sites derived from Cp ([9], [10], [11], [12]; P. Young and MJA, unpublished data). Since Cp is generally the promoter for all EBNA mRNA initiation in LCL cells, the EBNA3 proteins probably contribute to a negative auto-regulatory loop. In addition all three EBNA3s exhibit robust repressor activity when targeted directly to DNA by fusion with the DNA-binding domain of Gal4 and they all interact with one or more cellular factor(s) involved in transcriptional repression or silencing; these include histone deacetylases (HDACs) and CtBP ([11], [12], [13], [14], [15], [16], [17]; P. Young and MJA, unpublished data).
CtBP (C-terminal binding protein) was initially discovered as a cellular factor interacting with the C-terminus of adenovirus E1A oncoprotein and subsequently identified as one of a highly conserved family of co-repressors of transcription (reviewed in [18]). Most of the factors that bind to CtBP and negatively regulate transcription contain at least one conserved Pro-Leu-Asp-Leu-Ser (“PLDLS”) CtBP-interaction domain (or close variant) that is necessary and probably sufficient for the interaction. CtBP-containing complexes can coordinate biochemical and enzymatic events that convert transcriptionally active chromatin directly to a repressive or silent state ([19], [20]). Moreover there is also good evidence that CtBP is involved in the regulation of cell proliferation since it has been shown that CtBP forms a link between human polycomb group (PcG) proteins and pRb [21] and that CtBP and PcG complexes both regulate elements in the CDKN2A locus [22], [23].
EBNA3A and EBNA3C each bind to CtBP in vitro and in vivo but this binding correlates only partially with their ability to repress transcription when targeted to DNA in transient reporter assays. However, the interaction correlates extremely well with their ability to behave as cooperating nuclear oncogenes when introduced into primary rodent fibroblasts with oncogenic Ha-RAS [16], [17], [24]. Since in this type of assay, the oncogene Ha-RAS alone triggers exit from the cell cycle and premature senescence via the induction of the p16INK4A and/or p19ARF proteins encoded by the CDKN2A locus [23], [25], this suggests that EBNA3A and EBNA3C can each rescue primary fibroblasts from growth arrest and senescence.
Further evidence that EBNA3C deregulates the cell cycle came with the demonstration that when over-expressed it overcomes a mitotic metaphase checkpoint and induces polyploidy and multi-nucleation, eventually leading to cell death [26]. However, the molecular details of its action in mitosis have remained elusive. A reported interaction with CHK2 suggests that it could have a role in the transition from G2 to mitosis, but this has not yet been proven [27] and although it has been reported that EBNA3C may repress the transcription of the mitotic regulator BUBR1 in one B cell line, this has not been extended to LCLs [28]. Several recent reports indicate that EBNA3C can also directly associate with multiple other factors involved in the regulation of cell cycle progression and/or the G1/S checkpoint. These include Cyclin A; the ubiquitin ligase SCFSKP2; the tumour suppressor pRb; the oncoprotein MYC; MDM2 in a complex with p53 and p53 alone [29], [30], [31], [32], [33], [34]. It remains to be determined if these interactions occur in infected B cells and whether they are functionally significant.
The most direct and compelling evidence that EBNA3C modulates the cell cycle during EBV-mediated transformation of B cells into LCLs comes from Maruo and colleagues. Using a recombinant Akata EBV made conditional for EBNA3C function by fusing EBNA3C with a modified oestrogen receptor, they revealed that EBNA3C represses expression of the cyclin-dependent kinase inhibitor p16INK4A in LCLs. Removing the inducer of EBNA3C activity (4-hydroxytamoxifen) from the culture medium results in an accumulation of both p16INK4A mRNA and protein and in reduced cell proliferation [35].
EBNA3A also cooperates with Ha-RAS in the transformation and immortalization of REFs and there is again a remarkably good correlation between EBNA3A binding to CtBP and its ability to cooperate with oncogenic ras [17]. Furthermore, a 4-hydroxytamoxifen-dependent LCL conditional for EBNA3A function showed that in the absence of EBNA3A, cell proliferation gradually declines. Although it was not indicated whether this involved regulation of the CDKN2A locus [36], a more recent report of a microarray screen has indicated that repression of p16INK4A transcription in LCLs is associated with EBNA3A expression [8].
We recently demonstrated that EBV represses transcription of the gene encoding the pro-apoptotic BCL-2-related family member BIM [37], [38]. The repression of BIM transcription initially involves a polycomb repressive complex, PRC2, where the histone methyltransferase EZH2, which together with SUZ12 and EED, is responsible for establishing the epigenetic modification H3K27me3 (tri-methylation of lysine 27 on histone H3) (reviewed in [39], [40]). H3K27 methylation of BIM may then be followed by DNA methylation of sites within the CpG-island flanking the BIM transcriptional initiation site ([38] and our unpublished data). Since EBNA3A and EBNA3C are necessary for the chromatin remodelling and epigenetic repression of BIM, here we have examined the p16INK4A locus and the roles EBNA3A and EBNA3C play in regulating its epigenetic status.
Materials and Methods
Generation of recombinant EBV-BACs
An EBNA3C-HT fusion protein (3CHT) in the B95-8 background was constructed with the same linking sequence and 4-hydroxytamoxifen-sensitive murine estrogen receptor that has previously been described in the Akata background [35]. The connection between the 3C and HT is a single proline residue between the last amino acid of EBNA3C and amino acid 281 of the murine estrogen receptor alpha (modified by G525R to make it 4-hydroxytamoxifen-specific. This fusion was recombined into the B95-8 bacterial artificial chromosome (BAC) [41] using previously described methods [37], [42] to produce two independent BACs containing 3CHT (A and C).
A set of CtBP-binding-mutant viruses were generated in which the EBNA3A and/or EBNA3C binding site(s) for CtBP were replaced with previously characterised mutations that lack the ability to bind CtBP [16], [17]. This was achieved by a sequential set of recombinations, initially mutating the pair of CtBP binding sites in EBNA3A (to generate the 3ACtBP mutant). The EBNA3C binding site for CtBP in this was then mutated to create a virus genome lacking all CtBP binding sites among the EBNA3s (E3CtBP). Then the EBNA3A mutant sequence was replaced with wild-type sequence, leaving only the EBNA3C sequence as mutant (3CCtBP) and finally the EBNA3C sequence was reverted to wild-type sequence, generating the CtBP revertant (revCtBP).
Cell culture
Established LCLs were cultured in RPMI-1640 medium (Invitrogen) supplemented with 10% fetal calf serum, penicillin and streptomycin. LCL 3CHT were cultured with addition of 400nM of 4-hydroxytamoxifen (HT, Sigma). After the infection of primary B cells, LCLs were grown to a volume and density suitable for freezing multiple aliquots (typically about 60ml at a density of 3×105 cells/ml or greater). This took 4–8 weeks for WT-EBV, revertant and 3CHT LCLs and 6–12 weeks for the EBNA3A and CtBP mutant LCLs.
Cells recovered from liquid nitrogen were cultured for about 10 days (with HT if necessary) before the start of any experiment. At the end of an experiment the cells were discarded. Twenty-four hours before any experimental treatment, cells were seeded at a density of 2.5×105 cells/ml.
Infection of 1° B cells with recombinant EBV
Virus was produced by transfection of recombinant BACs into HEK293 cells [(ATCC, CRL-1573), a kind gift of Claire Shannon-Lowe, University of Birmingham] and selection of clonal Hygromycin B-resistant cell lines, which were screened for integrity of EBV genome by episome rescue and pulsed-field gel analysis of BAC restriction digests (not shown). Infectious virus was produced by the transfection of EBV-BAC-containing 293 cells with BZLF1 and BALF4 expression constructs [43], and after 4 days, supernatant was filtered through 0.45 µm filters. Virus titre was assessed by infection of Raji cells and counting green cells on an inverted fluorescent microscope after enhancement of GFP expression by overnight treatment with 5 nM TPA and 1.25 mM sodium butyrate. Virus titres were typically in the range of 50 to 250 raji green units (rgu) per microlitre of tissue culture supernatant.
B-cells for generation of LCL-3CHT-A and -C and EBNA3A-knockout LCLs (and for limiting dilution experiments – see below) were isolated from buffy coat residues (UK blood transfusion services) by centrifugation over ficoll. CtBP mutant LCLs and 3CHT-LCL B, D and E (described herein) were generated by infection of PBLs isolated from donated EBV-seronegative blood (a kind gift of Ingo Johannessen, University of Edinburgh). Three donors were used (D1, D2 and D3). 3CHT-LCL B, D and E were made by infection of in blood from donors 1, 3 and 2 respectively with EBV-3CHT-A virus. Essentially, between 50 µl and 1 ml of virus was added to 106 PBLs (typically 2–8% of which are B-cells by FACS for CD20; not shown) in a well of a 24 well plate, and cultured initially in RPMI supplemented with 15% FCS, supplemented with Cyclosporine A (500 ng/ml) for the first 2–3 weeks. Once LCLs had grown out into large culture volumes, the FCS level in the medium was reduced to 10%.
Limiting dilution and cell growth assays
Virus stocks were diluted to 2×104 Raji green units (rgu) per ml in RPMI supplemented with 10% FCS, and ten-fold serial dilutions used to generate virus concentrations down to 0.2 rgu/ml. The virus was added to an equal volume of PBLs at 2×106 cells per ml, and 1 ml was seeded per well in a 24 well plates. Two virus preparations from independent 293 cell producer lines were used for each virus mutant, and 6 wells for each virus concentration. 24 wells with no virus were used to control for spontaneous immortalisation of B-cells. After a week, the culture volume was increased to 2 ml and half the culture volume was replaced weekly thereafter. For the first 2 weeks, the medium was supplemented with cyclosporine A (500 ng/ml). Cell growth was monitored and wells were scored positive or negative after 40 days, based on the presence of clumps of cells characteristic of LCLs. To assess growth rate of CtBP-mutant LCLs, 5×104 cells per ml of cells were seeded in 10 ml in a flask. 0.5 ml was removed every day and live cells (by trypan blue staining to exclude dead cells) were counted on a haemocytometer.
Flow cytometry
To analyse cell cycle distribution, 2×106 LCL cells were fixed in 80% ethanol, incubated in PI solution [PBS containing 18 µg/ml propidium iodide (PI) and 8 µg/ml RNase A (Sigma Aldrich)] at 4°C for 1 h before flow cytometric analysis. To quantify cells in S phase, cells were pulsed with 10 µM 5-bromo-2′-deoxyuridine (BrdU) (Sigma Aldrich) for 1 h at 37°C, harvested immediately after the pulse, fixed in 80% ethanol and co-stained with FITC-conjugated anti-BrdU mAb (Becton Dickinson) for 1 h at room temperature and PI solution for 1 h at 37°C.
Western immunoblotting
Western blotting was performed essentially as described previously [16]. Briefly, proteins extracted using RIPA buffer, or in some cases whole cell lysates were resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to Protran nitrocellulose membranes (Schleicher and Schuell Bioscience, Dassel, Germany). Membranes were blocked with 5% milk powder in PBS/0.05% Tween 20, probed with appropriate primary and HRP-conjugated secondary antibodies. ECL kit (Amersham Biosciences, Chalfont St Giles, UK) was used for visualization. Following primary antibodies were used: mouse monoclonal anti-γ-tubulin (Sigma, T6557), sheep polyclonal anti-EBNA3A (Exalpha, USA), mouse monoclonal anti-EBNA3C (A10, kind gift from Prof. Martin Rowe, University of Birmingham), mouse monoclonal phospho-independent anti-Rb (BD Pharmingen, 554136), rabbit polyclonal anti-phospho-Rb (Ser 807–811) (Cell Signaling, 9308), mouse monoclonal anti-E2F1 (Millipore, mixed clones cKH20 and KH95, 05-379), mouse monoclonal anti-p16INK4A (clone JC8, kind gift from Dr Gordon Peters, Cancer Research UK), rabbit polyclonal anti-p130 (Santa-Cruz, c-20, sc-317), rabbit polyclonal anti-p107 (Santa Cruz, c-18, sc-138), mouse monoclonal anti-EBNA2 (clone PE2, Dako), mouse monoclonal anti-LMP1 (clone CS1-4, Dako), mouse monoclonal anti-EBNA-LP (clone JF186, gift of Paul Farrell), rat monoclonal anti-LMP2A (clone 14B7, AbCam 59026), rabbit polyclonal anti-CtBP [16] and rat monoclonal anti-EBNA3B (clone 6C9, provided by Elizabeth Kremmer, Munich; Popp et al, submitted for publication).
mRNA and Quantitative real time PCR (Q RT-PCR)
For Q RT-PCR, RNA was extracted from approximately 5×106 cells for each cell line using the RNeasy mini kit from Qiagen and following the manufacturer's instructions. One microgram of each RNA sample was reverse-transcribed using SuperScript III First-Strand Synthesis Supermix for qRT-PCR (Invitrogen). Between 0.5–1% of cDNA product (equivalent to 5–10 ng RNA) was used per qPCR reaction. qPCR was performed on an ABI 7900HT real-time PCR machine using Platinum Sybr Green qPCR SuperMix UDG kit (Invitrogen). The cycling conditions were 95°C for 20 sec, followed by 40 cycles of 1 sec at 95°C, 20 sec at 60°C on a fast block. Dissociation curve analysis was performed during each run to confirm absence of non-specific products. Sequences of the assays used are listed in Table S1.
Standard curves, used to standardise amplification efficiency, were produced by six 5-fold serial dilutions of a mixture containing all cDNA samples used. Results were analyzed with qbasePLUS software (Biogazelle, Ghent, Netherlands). Precise normalization was achieved using internal average control calculated from the controls (housekeeping genes – bold in Table S1) with highest stability rating (usually ALAS1 and GNB2L1). The calculated errors in the graphs are the standard errors from three replicate qPCR reactions for each mRNA.
Chromatin immunoprecipitations (ChIP)
Chromatin immunoprecipitations for methylated histone H3 were performed essentially as described previously [38]. Precipitated DNA was cleaned using QIAquick Gel Extraction Kit (Qiagen) and was assayed by qPCR. Input DNA Ct was adjusted from the 5% used in the qPCR to 100% equivalent by subtracting 4.32 (Log2 of 20) cycles. ‘Percent input’ precipitated was then calculated by 100×2∧ (Ct adjusted input – Ct IP). Non-specific background was estimated by precipitation with IgG (data not shown since all values were below 0.03% of input). The error bars represent standard deviations from triplicate pPCR reactions for both input and IP. Sequences of the primers used in these assays are listed in Table S2.
Results
Inactivation of EBNA3C leads to proliferative arrest of LCL 3CHT that can be reversed by re-adding HT
In order to examine in detail the regulation of p16INK4A and cell proliferation by EBV, initially a recombinant EBV encoding a conditional EBNA3C was constructed. We employed the B95.8 EBV-BAC system used previously to generate EBNA3-knockout (KO) viruses and the design of fusion proteins described by Maruo and colleagues for fusing the carboxyl-terminus of EBNA3C to a modified estrogen receptor [35], [37]. EBNA3C fused to this estrogen receptor is dependent on 4-hydroxytamoxifen (HT) in the culture medium for its function and stability. Two independently generated viruses encoding the EBNA3C-HT-fusion (3CHT) were used to establish multiple lymphoblastoid cell lines (called LCL 3CHT).
LCL 3CHT – with one notable exception described below – required HT in the culture medium for their proliferation. When HT was removed, after 3–7 days there was a dramatic reduction in the amount of EBNA3C detected by western blotting (for a representative example see Figure 1). This reduced expression is probably because the inactivated fusion binds to heat shock proteins and is targeted for proteasome-mediated degradation and is consistent with the behaviour of the equivalent fusion in the Akata strain EBV [35]. The LCLs were validated further by western blot analyses probing for each of the EBV latent proteins. As was previously reported in Akata, no consistent differences in steady state levels were seen. In the absence of HT a very slight increase in LMP1 was sometimes observed – this was also reported previously (Figure S1; [35] and data not shown).
Fig. 1. EBNA3C inactivation and reactivation in LCL 3CHT. 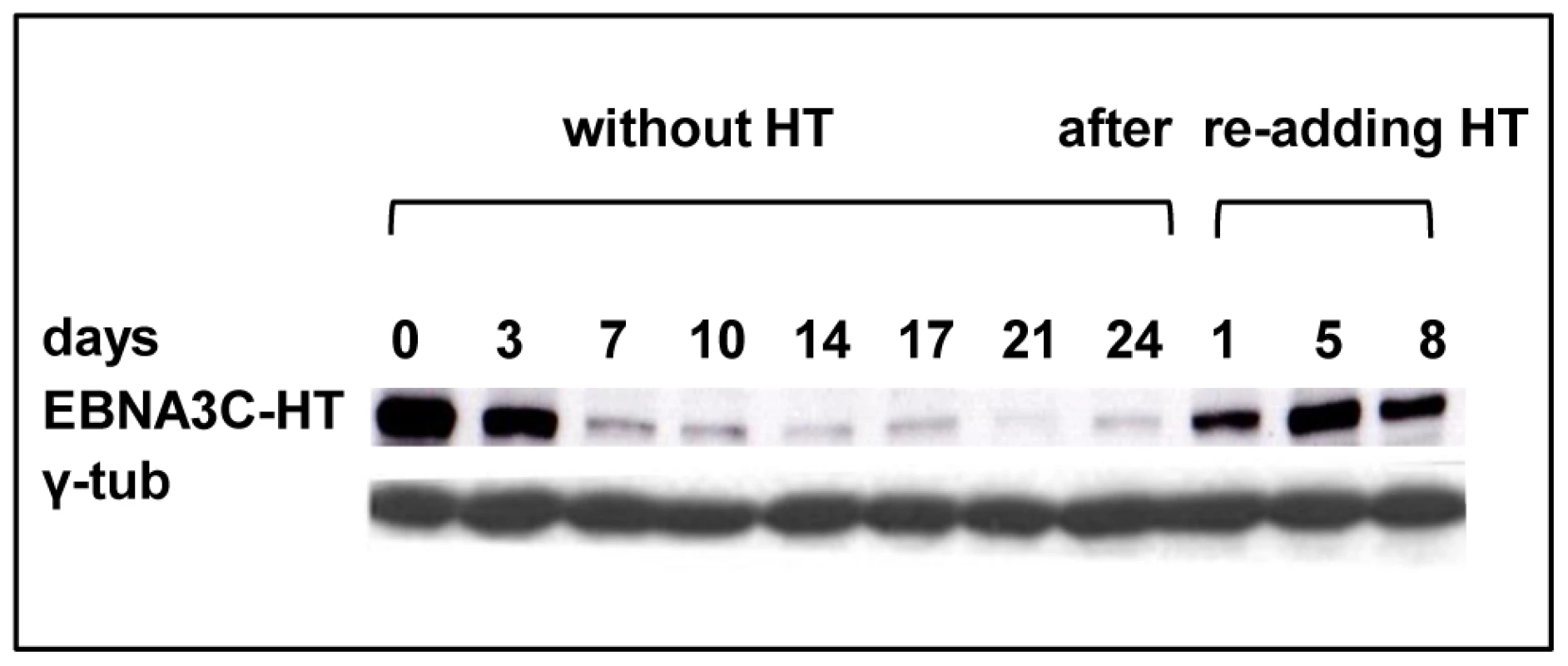
Cells that had been grown in culture medium containing HT were washed and re-suspended in medium with HT omitted. After culturing for the number of days indicated, protein extracts were western blotted and probed with an anti-EBNA3C MAb to reveal EBNA3C-HT. After 17 days some cells were transferred to medium containing HT and after 1, 5 or 8 days samples were again taken for western blotting. The western blot was re-probed with anti-γ-tubulin to ensure equal loading of the proteins. In order to confirm that inactivation of EBNA3C compromises cellular DNA synthesis, BrdU incorporation was assessed. Two LCL 3CHT (-A & -C) cells were cultured with HT, then for 14 or 33 days after HT had been removed from the culture medium. Cells were then pulsed for 1 hour with BrdU, harvested, fixed and stained with anti-BrdU-FITC and propidium iodide. The cells were analysed by flow cytometry (see for example Figure 2A). In both LCL 3CHT, BrdU incorporation after 14 days without HT was reduced to about 40% of that in similar control cells cultured with HT and after 33 days the reduction in the proportion of cells entering S phase was even more profound (Figure 2B).
Fig. 2. Proliferation of LCL 3CHT after inactivation and reactivation of EBNA3C. 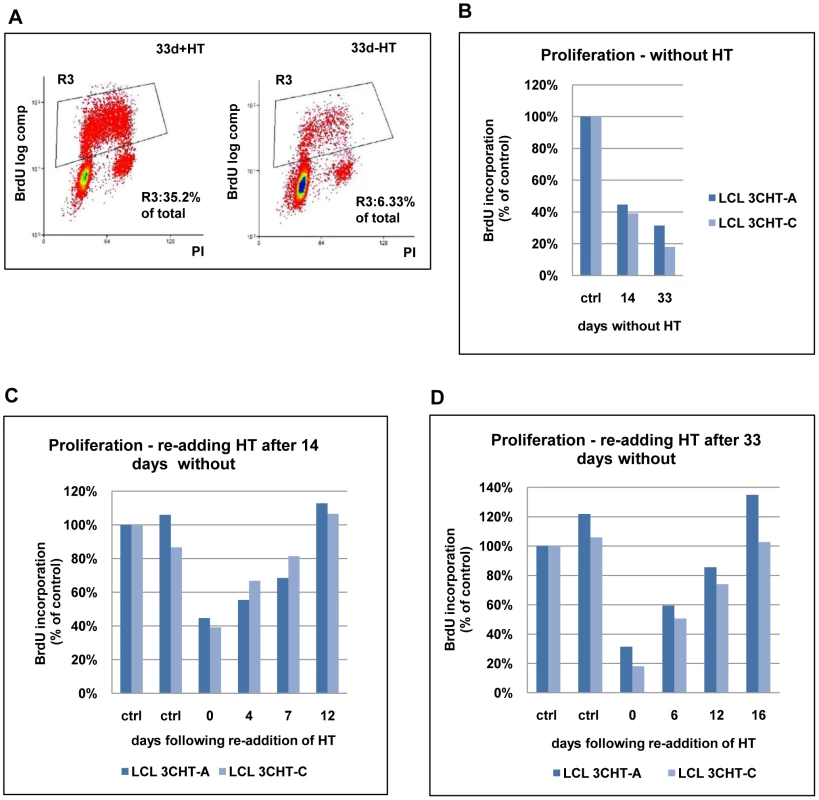
(A) LCL 3CHT cells were cultured either with HT (+HT) or for 33 days after the removal of HT (-HT) from the growth medium. Cells were pulsed for 1 hour with BrdU, harvested, fixed and stained with anti-BrdU-FITC and propidium iodide. The cells were analysed by flow cytometry. The gated BrdU-positive population is reduced in the absence of HT. (B) Two LCL 3CHT (-A and -C) generated from independent 3CHT-BACs using PBL from a single donor were analysed as in (A) after 14 and 33 days without HT. Histograms show BrdU incorporation relative to the control, cycling population. The control (ctrl) was a proliferating population of LCL 3CHT grown in medium with HT. (C) LCLs 3CHT-A and -C were cultured for 14 days in the absence of HT (0) and BrdU-incorporation was assayed as in (A). The incorporation of BrdU was also assayed as above after 4, 7 and 12 days after re-adding HT. (D) A similar experiment was performed on cells cultured without HT for 33 days. To establish whether the growth arrest could be reversed upon reactivation of EBNA3C-HT, HT was re-added after 14 or 33 days growth in its absence. Surprisingly, although the levels of EBNA3C-HT were re-established quite rapidly (within about 24 hours: see Figure 1), there was a significant delay before BrdU incorporation matched that of the controls. These cells clearly do not behave like a synchronised G1/S-arrested population released into S phase. When HT was re-added after 14 days without HT, it took 12 days to return to the level of proliferation seen in control cells (Figure 2C). When the HT was re-added after 33 days without HT, the period required to achieve full proliferation was extended to 16 days (Figure 2D). Since the restoration of cell proliferation after reactivation of EBNA3C is dependent on the time elapsed without functional EBNA3C, it is possible that a subpopulation of cells is being driven into in a state of irreversible arrest similar to senescence. These would be refractory to further pro-proliferative signals from EBNA3C and they would gradually accumulate. Alternatively there may be an intrinsic delay associated with the molecular processes necessary to re-induce cell cycle progression in individual arrested cells. It should be noted that, although viable, these non-proliferating cells did not stain positive for the operational mark of senescence β-galactosidase (data not shown).
Proliferation of LCL 3CHT correlates with the regulation of p16INK4A expression
Maruo and colleagues described how in the absence of functional EBNA3C, the cyclin-dependent kinase inhibitor p16INK4A accumulates [35]. We needed to establish that similar changes in p16INK4A levels occur in the B95.8-derived LCL 3CHT, and determine whether the response to EBNA3C reactivation correlated with (and could account for) the reduction in p16INK4A expression and the changes in proliferation of LCL 3CHT described above.
The DNA sequence of the unique first exon of p16INK4A is GC rich and is predicted to have extensive secondary structure under the conditions used for qPCR (based on DNA sequence analysis using Visual OMP). A quantitative RT-PCR assay specific for p16INK4A transcripts, which detects the amplicon located within the unique first exon of p16INK4A (Table S1), is quantitative only in the presence of large amounts of template. Therefore, a second assay that quantifies an amplicon located within the second and third exon shared by p16INK4A and p14ARF was designed (CDKN2A assay; Table S1), which is quantitative over a 5-log range of template concentration. These assays were used to show that the regulation of p16INK4A in B95.8-BAC LCL 3CHT is reversible (Figure 3). After 14 days in culture in the absence of HT, there was a 2-2.5-fold increase in CDKN2A transcripts relative to the control population. After re-adding HT into the medium, CDKN2A transcripts gradually decreased over the next 12 days (Figure 3A). This result was confirmed with the p16INK4A-specific assay (Figure 3B). Extending the period of culture in the absence of HT to 33 days resulted in a 3 to 5-fold increase in the level of CDKN2A transcripts. After re-adding HT to the medium, CDKN2A transcripts gradually decreased to the levels found in an actively proliferating culture, but it seemed to require at least 16 days (Figure 3C). This was also consistent with the results using the p16INK4A-specific primer set (Figure 3D).
Fig. 3. Repression of p16INK4A following reactivation of EBNA3C. 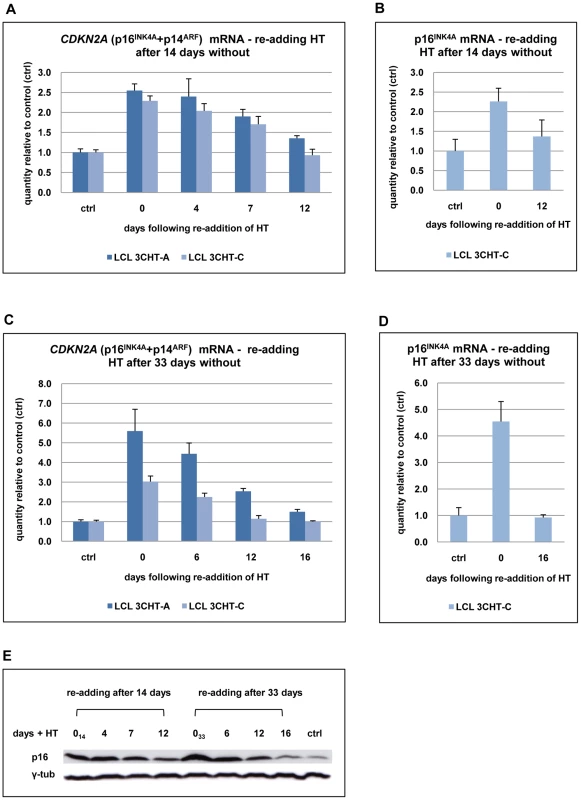
(A) After 14 days without HT (0), total RNA was extracted from aliquots from two LCL 3CHT populations (-A and -C). HT was re-added to the remaining cells and further RNA samples were taken at the times indicated. Real time quantitative RT-PCR (qRT-PCR) was performed to quantify CDKN2A transcripts. The histogram corresponds to CDKN2A mRNA relative to that in control cycling populations of each LCL 3CHT. (B) As in (A) but using a p16INK4A-specific qPCR. (C) & (D) Similar assays to those described in (A) and (B) after 33 days without HT. (E) Western blots – probed with a p16INK4A-specific MAb – of protein extracts from LCL 3CHT-C cells to which HT was re-added after 14 or 33 days in its absence (014 and 033 respectively). The control (ctrl) is LCL 3CHT-C cells continuously cultured in medium with HT. Echoing the transcript data, p16INK4A protein expression was gradually reduced after reactivation of EBNA3C in LCL 3CHT (Figure 3E). We conclude that regulation of p16INK4A expression in LCL 3CHT undergoing changes in proliferation seems to be exclusively or at least predominantly at the level of transcription as has been previously shown in various types of cell, including LCLs [23], [35].
Taken together, the data confirm that p16INK4A accumulates in LCL 3CHT cultured without HT and this inversely correlates with the proportion of cells entering S phase. The data show for the first time that when EBNA3C is reactivated by the re-addition of HT to the culture medium, the reverse occurs and the correlation between EBNA3C activity, p16INK4A transcription and proliferation holds true.
Inactivation of EBNA3C leads to dephosphorylation of the retinoblastoma protein (Rb), reduced expression of p107 and an increase in p130; activation of EBNA3C reverses these processes
In order to determine the consequences of p16INK4A accumulation on the rest of the Rb-axis in the absence of functional EBNA3C, LCL 3CHT were cultured with HT (controls) and without HT for 14 and 33 days. Subsequently, HT was re-added on day 14 or on day 33. In arrested cells, Rb became hypophosphorylated (as revealed by both pan-specific and phospho-specific anti-Rb antibodies); as the arrest intensified after extended time without HT, reduced phosphorylation was accompanied by a slight reduction in the expression of Rb (Figures 4A and B). Simultaneously the Rb-related p130 protein accumulated and the amount of p107 was reduced (Figure 4B, annotated 014 and 033). This was particularly apparent after 33 days. Consistent with the data showing recovery after re-addition of HT and the reconstitution of functional EBNA3C, Rb was gradually re-phosphorylated and up-regulated (Figure 4B); concomitantly, expression of p130 decreased as cells entered the proliferation cycle, and p107 expression increased.
Fig. 4. Inactivation of EBNA3C and accumulation of p16INK4A leads to de-phosphorylation of Rb, reduced expression of p107 and an increase in p130; activation of EBNA3C reverses these processes. 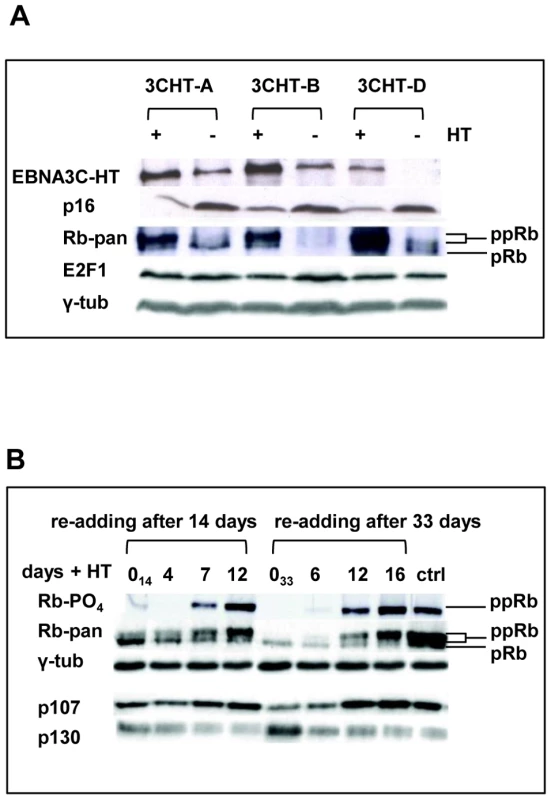
(A) Western blot analyses of three LCL 3CHT lines performed on whole cell lysates from cells cultured with HT (+) without HT for 26 days (−). As the amount of EBNA3C protein reduces (and is inactivated) and p16INK4A accumulates, so hyperphosphorylated Rb (ppRb) protein disappears and only hypophosphorylated Rb (pRb) is detected. The levels of E2F1 and γ-tubulin (γ-tub) remain unchanged irrespective of the culture conditions. (B) Western blot analysis of extracts from cells after re-addition of HT to cultures starved of HT for either 14 or 33 days (014 and 033 respectively). A pan-specific anti-Rb MAb and phospho-specific MAb (Ser 807–811) both show an increase in hyperphosphorylated Rb (ppRb) after HT was added. After 12–16 days the degree of phosphorylation is equivalent to that in the proliferating control LCL 3CHT population (ctrl). As Rb becomes phosphorylated, expression of p107 increases and expression of p130 decreases. The level of γ-tubulin (γ-tub) did not alter throughout the experiments. Regulation of H3K27me3 and H3K4me3 across the p16INK4A locus in LCL 3CHT requires EBNA3C
The repression of p16INK4A by PcG silencing complexes adding H3K27me3 marks to chromatin has been well characterized in primary fibroblasts. In pre-senescent proliferating fibroblasts the H3K27me3 mark forms a broad peak centred on the first exon of p16INK4A; induction of senescence is associated with displacement of PcG silencers and a profound reduction of H3K27me3 on the chromatin associated with p16INK4A exon 1 [40], [44], [45], [46].
We hypothesised that the up-regulation of p16INK4A transcription in the absence of functional EBNA3C might result from loss of PcG-mediated repression. Therefore ChIP analyses were performed to assess the level of H3K27me3 in the p16INK4A locus (see schematic in Figure 5A) in LCL 3CHT cultured with and without HT. Upon EBNA3C inactivation by the removal of HT, H3K27me3 gradually decreased at the p16INK4A exon 1, as transcription increased (Figure 5B and C). This process could be reversed by reconstitution of functional EBNA3C after re-addition of HT (see below). The time taken for H3K27me3 depletion correlated with the length of time in culture without functional EBNA3C and was consistent with the rate of p16INK4A induction. Equivalent results were obtained when similar ChIP experiments were performed on an independent LCL 3CHT and using two different sets of p16INK4A exon 1-specific primers (Figure S2 and data not shown).
Fig. 5. ChIP analysis to quantify the H3K27me3 and H3K4me3 marks on p16INK4A exon 1 when EBNA3C is inactivated and re-activated in LCL 3CHT. 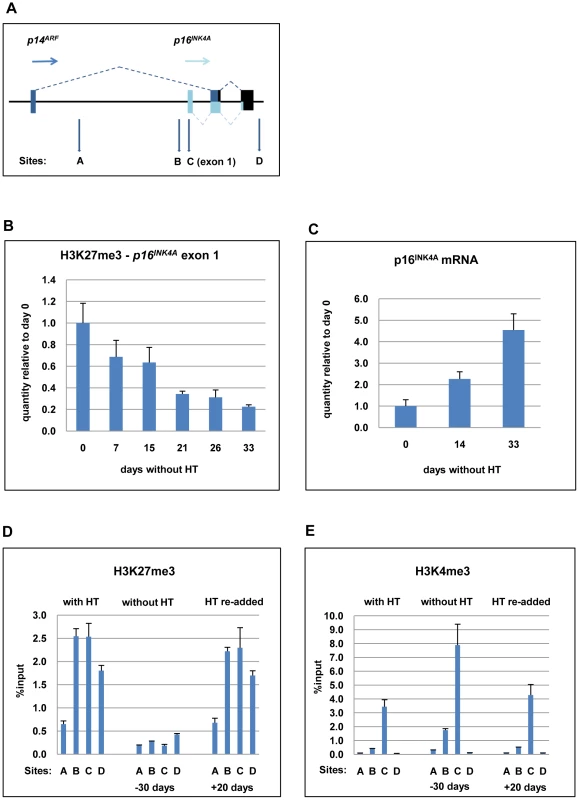
(A) Schematic of the human p16INK4A-ARF locus showing the location of coding exons (boxes) and transcription start sites (horizontal arrows) – not drawn to scale. The vertical (A-D) arrows refer to the approximate locations of primer pairs used for qPCR analysis of precipitated chromatin (as described in Materials and Methods). (B) ChIP analysis of H3K27me3 distribution on exon 1 (site C) of p16INK4A. The histogram shows a decline in H3K27me3 relative to a cycling LCL 3CHT (day 0). (C) Corresponding changes in p16INK4A mRNA quantified by qRT-PCR. (D) ChIP analysis of H3K27me3 distribution across the p16INK4A-ARF locus in LCL 3CHT proliferating in the presence of HT, after 30 days without HT and 20 days after re-adding HT. (E) ChIP analysis of H3K4me3 the p16INK4A-ARF locus in LCL 3CHT proliferating in the presence of HT, after 30 days without HT and 20 days after re-adding HT. Recently it became clear that the transcriptional status of p16INK4A is not determined by H3K27 tri-methylation alone, but rather by the interplay between H3K27me3 and H3K4me3 modifications [44]. Therefore ChIP analyses were performed to assess the quantities of both modifications at the p16INK4A locus in LCL 3CHT cultured with and without HT (Figures 5D and E). EBNA3C inactivation (labelled -30 days) affected epigenetic marks at the p16INK4A locus in a manner consistent with transcriptional activation; the repressive H3K27me3 mark was reduced while the activation-related H3K4me3 increased. After EBNA3C reactivation (labelled +20 days), the epigenetic modifications at p16INK4A locus were apparently reversed.
Further ChIP assays were used to confirm the specificity of p16INK4A regulation. The quantities of H3K27me3 and H3K4me3 at p16INK4A exon 1 (primer set C – indicated by boxes) were compared to the quantities at various other sites in the CDKN2A locus, including a region (site A) located 4.5kb downstream of p14ARF transcription start site (Figures 5A, D and E). As described previously [45] in human primary fibroblasts H3K27me3 marks are broadly distributed across the CDKN2A locus, peaking in exon 1, but extending into the region corresponding to A. Similarly, this region in LCL 3CHT was associated with some degree of H3K27me3 – particularly in cycling cells. However – whether or not EBNA3C is active – H3K4me3 is detected on exon 1 (site C), but at site A it is always completely absent. In parallel, ChIP experiments were performed with an IgG antibody of the same isotype as the anti-H3K27me3 or -H3K4me3 antibodies to assess the level of background and no significant binding was observed at any site (data not shown). The presence of two inversely regulated modifications excludes the possibility that the reduction in histone methylation is due to nucleosome re-positioning away from p16INK4A resulting in a reduction of the total histone H3 at the locus.
Relatively high quantities of H3K4me3 were detected at the p16INK4A exon 1 (site C) compared to site A, even when EBNA3C was active (in LCL 3CHT with HT and with HT re-added) and therefore repressing p16INK4A. This suggests that p16INK4A exon 1 in LCLs might contain a ‘bivalent’ or ‘poised’ chromatin domain [47].
EBNA3C-mediated regulation of p16INK4A does not require Rb
To avoid biases due to the genetic background of a single donor, we decided to confirm our findings using newly established LCL 3CHT lines from several different donors. It was soon noted that one of these cell lines did not arrest after the removal of HT from the medium. Western blotting with a pan-specific anti-Rb antibody failed to detect Rb protein in this cell line (LCL 3CHT-E) cultured with or without HT (Figure 6A). Consistent with this, qPCR showed that LCL 3CHT-E expressed low levels of Rb mRNA in comparison to other LCL 3CHT lines (Figure 6B). It is well established that functional Rb can regulate p16INK4A levels through a negative feedback loop and that Rb-negative tumours (such as carcinoma of cervix) and tumour-derived cell lines can express high levels of p16INK4A [23], [48]. However, even in this LCL in which Rb cannot be detected, p16INK4A was still repressed in the presence of active EBNA3C, and this was relieved after EBNA3C inactivation by the removal of HT (Figure 6A). When ChIP analyses for H3K27Me3 (and H3K4Me3) marks across the CDKN2A locus were performed on the LCL expressing no detectable Rb (LCL 3CHT-E) similar patterns to those seen for LCLs expressing Rb protein were seen (compare Figures 6C and D with Figures 5D and E). That is, high levels of H3K27Me3 occupied exon 1 (site C) in the presence HT, while there were low levels of this repressive mark in its absence. The reverse was true for H3K4me3. This reinforces our view that the epigenetic regulation of p16INK4A by EBNA3C is independent of Rb expression.
Fig. 6. EBNA3C-mediated regulation of p16INK4A does not require Rb. 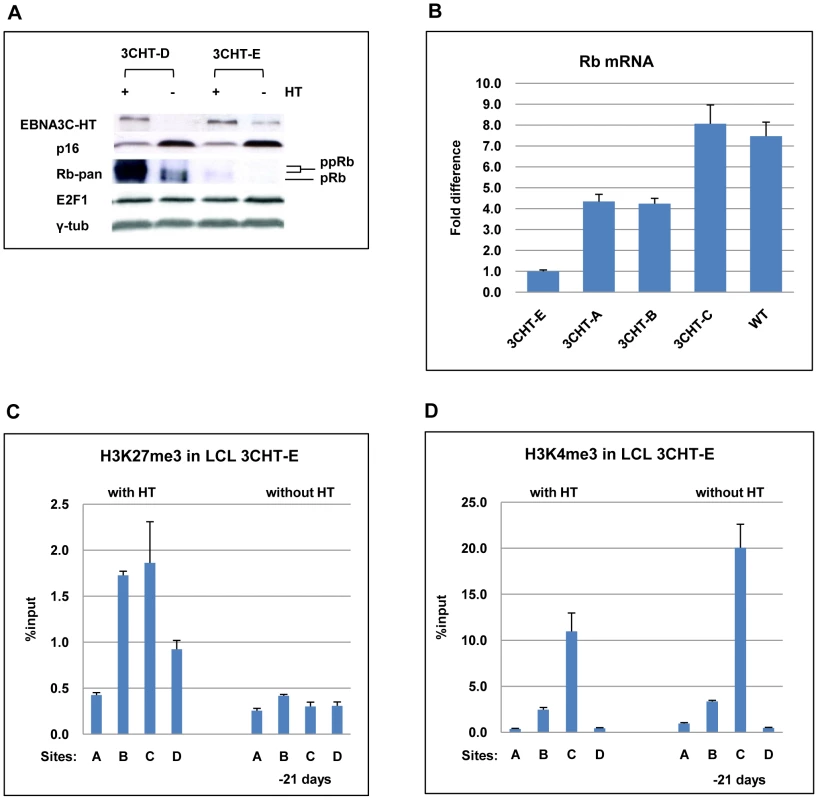
(A) Western blot analysis of whole cell lysates from two LCL 3CHT (-D and -E) cultured with (+) or without (-) HT for 26 days. Although Rb is undetectable in LCL 3CHT-E, when HT is removed from the growth medium, EBNA3C decreases and p16INK4A (p16) increases. E2F1 and γ-tubulin (γ-tub) levels do not alter. (B) Steady-state levels of Rb mRNA in LCL 3CHT-E relative to 3 other LCL 3CHT and a WT-BAC LCL all cultured with HT, quantified by qRT-PCR. (C) ChIP analysis of H3K27Me3 distribution across the p16INK4A-ARF locus in LCL 3CHT-E cells (expressing little or no Rb) with HT or without HT in the growth medium. (D) ChIP analysis of H3K4me3 distribution across the p16INK4-ARF locus essentially as described in (C). EBNA3A also contributes to the regulation of p16INK4A
Recently EBNA3A was shown to regulate p16INK4A in a microarray study using EBNA3A-knockout (KO) LCLs [8]. The efficiency with which LCLs can be established using EBNA3A-KO is lower than with wild type virus, but by infecting peripheral blood leukocytes (PBLs) including macrophages that transiently act as feeder cells, we generated two independent EBNA3A-KO LCLs.
Since EBNA3A and EBNA3C co-operate to epigenetically regulate the cellular gene BIM in BL31 cells, we wanted to ask whether EBNA3A might also cooperate with EBNA3C in modifying chromatin at the p16INK4A locus. The two EBNA3A-KO LCL were validated by probing western blots of protein extracts for the major latent EBV proteins; as previously described for these knockouts in a BL background there were no consistent differences in EBV gene expression ([37]; data not shown). Although only two independent EBNA3A-KO lines were investigated, further characterization showed that – consistent with the report from Hertle and colleagues [8] – p16INK4A protein expression is elevated in both EBNA3A-KO LCLs relative to WT (B95.8)-BAC infected LCL (Figure 7A).
Fig. 7. EBNA3A contributes to the regulation of p16INK4A. 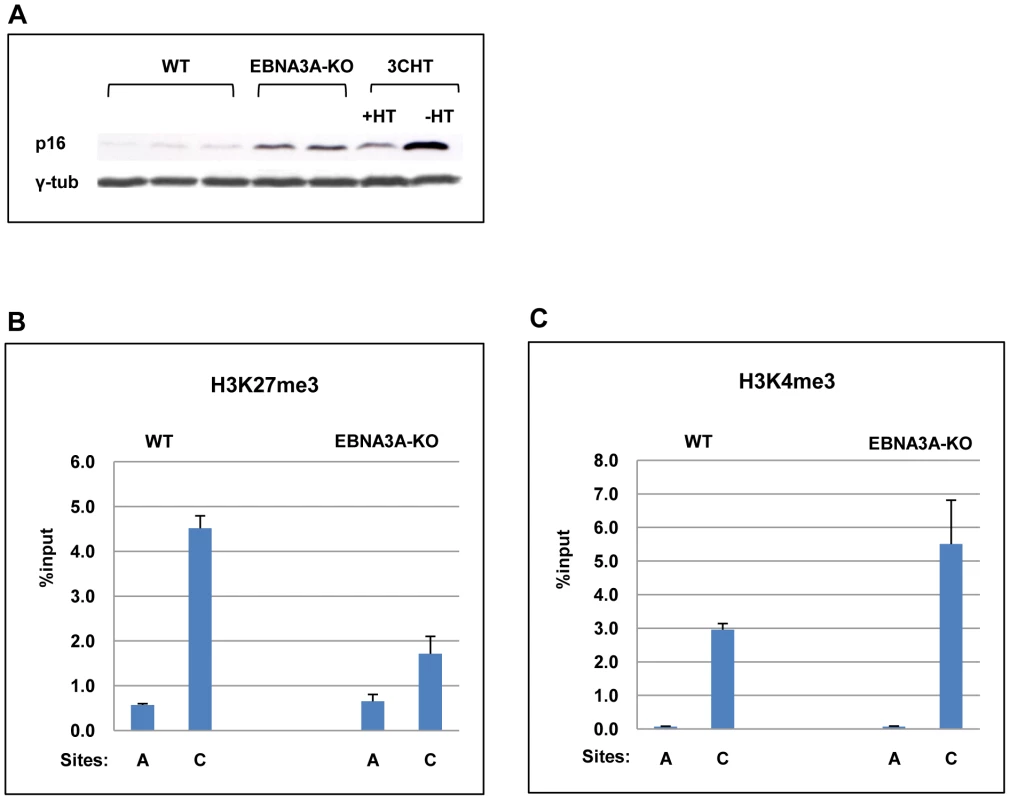
(A) Western blot analysis of three LCLs established with virus derived from B95.8-EBV BAC (WT) and two established with EBNA3A-KO recombinants. The steady state levels of p16INK4A (p16) are elevated in the EBNA3A-KO cells relative to the WT-EBV infected cells. It should be noted that similar results were reported using a larger panel of EBNA3A-KO LCLs [8]. An LCL 3CHT (3CHT) with (+) or without (-) HT is shown for comparison. (B) & (C) ChIP analysis of H3K27me3 and H3K4me3 distribution on exon 1 (site C) and site A in the p16INK4A-ARF locus from an EBNA3A-KO LCL (3AKO) and a WT-EBV LCL (WT). A comparison of steady state levels of H3K27me3 and H3K4me3 at the p16INK4A locus revealed that in the absence of EBNA3A, the ratio of H3K27me3 to H3K4me3 associated with exon 1 was reversed relative to LCLs established with WT EBV BACs (Figures 7B and C). The low level of H3K27me3 and high level of H3K4me3 are consistent with a more transcriptionally active locus and the higher levels of p16INK4A protein detected in the EBNA3A-knockout lines. This suggests that EBNA3A, together with EBNA3C, is involved in the chromatin remodelling of p16INK4A.
In the EBNA3A-KO LCLs used for microarray analysis, the level of Rb transcripts was reported to be lower than in LCLs carrying WT EBV [8]. The two EBNA3A-KO LCLs described here expressed similar levels of Rb to the WT EBV infected cells. However, there was substantially more of the hypophosphorylated form when EBNA3A was not expressed (data not shown).
LCLs expressing CtBP-binding mutants of EBNA3A and EBNA3C grow relatively poorly and express higher than normal levels of p16INK4A
We showed previously that EBNA3A and EBNA3C mutants that are unable to bind CtBP are severely impaired in their ability to transform primary rat embryo fibroblasts in co-operation with activated Ha-RAS [16], [17]. Since this assay, in part, measures the ability of proteins to overcome p16INK4A-mediated premature senescence and because CtBP is involved in the repression of p16INK4A in primary human fibroblasts and keratinocytes [22], it seemed appropriate to ask whether the interaction of CtBP with EBNAs 3A and C is necessary for the modulation of p16INK4A expression in LCLs.
Mutations in EBNA3A and EBNA3C that completely ablate their capacity to bind CtBP have been described ([16], [17]; schematic in Figure 8A). These mutations were serially engineered into the B95.8 EBV-BAC creating CtBP-binding mutants of EBNA3A and EBNA3C, both individually and together, along with revertant virus (Figure 8B). These viruses were used to establish LCLs from PBL and fully validated by mutation-specific PCR, CtBP IP and western blotting for EBV latent proteins (Figure S3). Although latent EBV gene expression generally appeared unaffected by these point mutations it was soon noticeable that population growth was impaired. Limiting dilution analysis of infected PBL was performed in an attempt to quantify this impairment. In terms of the number of wells that contained clumps of cells characteristic of LCL outgrowth after 40 days, there was no substantial difference between the immortalization efficiency of the wild-type and revertant viruses, as compared to the CtBP mutants (data not shown). However, it was clear that the rate of outgrowth of the CtBP mutant LCLs was considerably slower than the wild-type LCLs. This effect is strikingly illustrated by the image of the plate in which the LCLs were grown (Figure 8C). Both the colour of the culture medium and the visible cell clumps in the wells show the much more rapid growth of the revertant LCLs as compared with the mutants. Also notable was the tendency of the mutant EBVs to sometimes grow as a single large clump of cells in the presence of a large number of smaller clumps (Figure 8C, eg wells A4 and C5). This may indicate natural selection driving phenotypic changes in the cells to allow the more robust clones to emerge as LCLs. Even once LCLs are established, CtBP-mutant LCLs continue to show a growth defect, exhibiting reduced rate of population growth relative to wild-type and revertant LCLs (Figure 8D). They also tended to have a much lower maximum cell density, with CtBP-mutant LCLs struggling to grow much beyond 0.7×106 cells/ml.
Fig. 8. LCLs expressing CtBP-binding mutants of EBNA3A and EBNA3C grow relatively poorly. 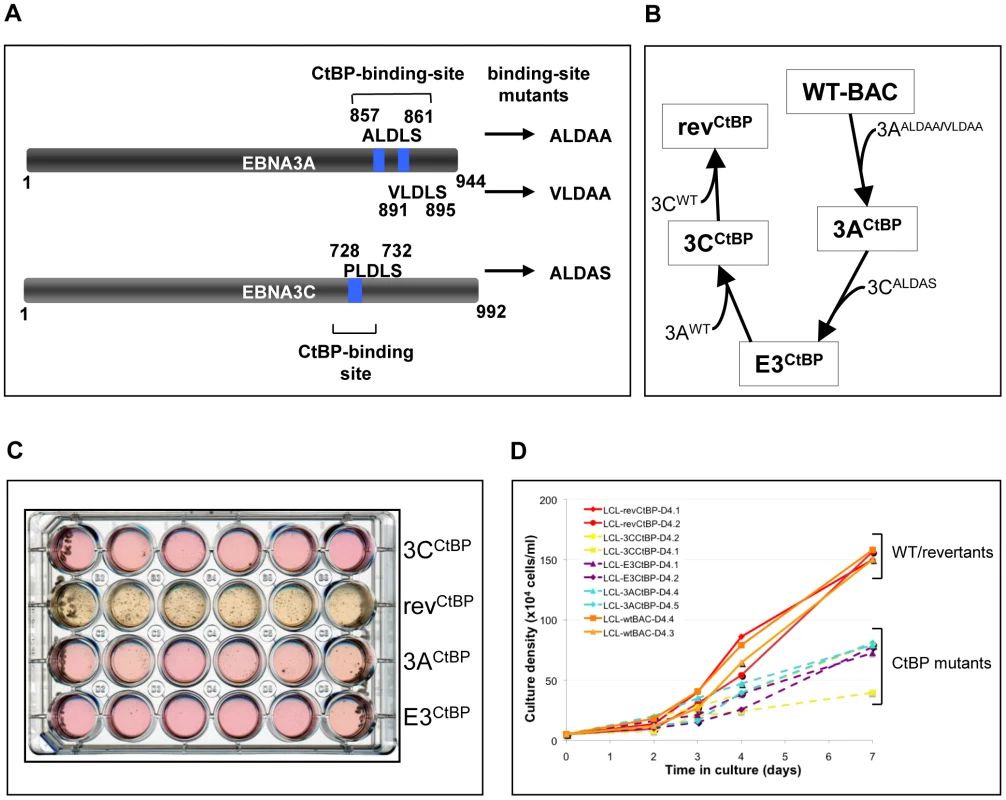
(A) Schematic showing mutations of CtBP-binding sites in EBNA3A and EBNA3C that were introduced into the CtBPM recombinant viruses. EBNA3C includes a single consensus PLDLS site that was mutated to ALDAS [16]. EBNA3A includes two non-canonical CtBP-binding sites that synergise to produce very efficient binding to CtBP; the ALDLS site was mutated to ALDAA and the VLDLS site was mutated to VLDAA [17]. (B) Schematic showing the generation of a series of CtBP-binding mutant EBVs, by serially mutating the EBNA3 CtBP binding sites, and then reverting them, creating both single and double CtBP mutant EBVs, and a revertant. (C) A plate of cells 40 days after infection of PBLs with 1000 rgu of virus per well. Difference in the colour of the medium and the visible density/size of cell clumps in the wells are clear indicators that CtBP-mutant LCLs grow out more slowly than wild-type (not shown) and revertant LCLs. This appears to be more striking in 3CCtBP and E3CtBP LCLs. (D) Established LCLs (approximately 3 months post-infection) were seeded at 5×104 cells/ml and counted each day for a week. CtBP-mutants (dashed lines) grow more slowly, and/or have a lower maximum cell density than wild-type and revertant LCLs (solid lines). Two cell lines were tested for each mutant. When p16INK4A transcripts and protein were quantified by qRT-PCR and western blotting respectively (Figures 9A, B and C) it was apparent that all the mutants express more p16INK4A mRNA and protein than either the WT-BAC or revertant-LCL cells. We assume that this increase in p16INK4A contributes to the impaired outgrowth of the mutant-carrying LCLs.
Fig. 9. Expression of p16INK4A is increased in CtBPM LCLs relative to revertant and WT LCLs. 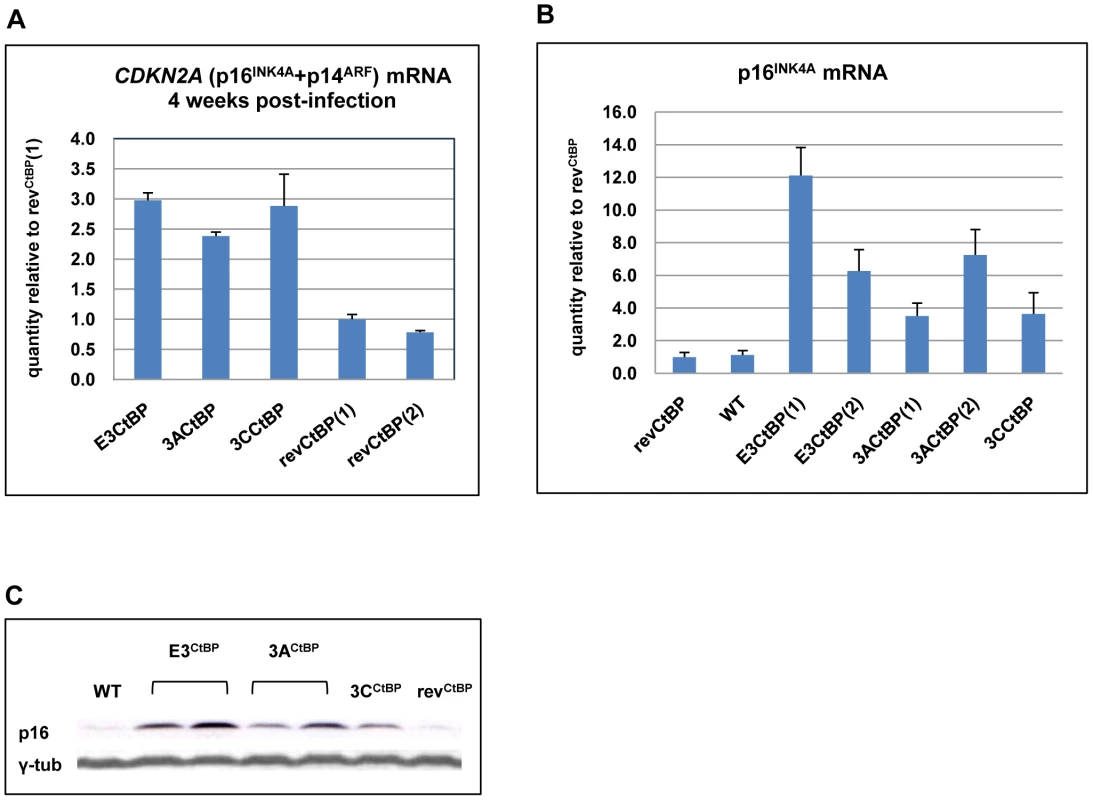
(A) E3CtBP, 3ACtBP, 3CCtBP and two revCtBP LCLs were harvested 28 days after the infection of primary B cells with the recombinant EBVs. RNA was extracted and the relative levels of CDKN2A transcripts were quantified by qRT-PCR. (B) RNA was extracted from two established E3CtBP LCLs, two 3ACtBP LCLs, 3CCtBP LCL, a revertant (revCtBP) and a WT-EBV LCL. The relative levels of p16INK4A transcripts were quantified by qRT-PCR. (C) Western blot analysis of protein extracts from two E3CtBP LCLs, two 3ACtBP LCLs and a 3CCtBP LCL all established from a single donor. A revertant (revCtBP) and a WT-EBV LCL are shown for comparison. Levels of p16INK4A (p16) are shown relative to γ-tubulin (γ-tub). Interaction of EBNA3A and EBNA3C with CtBP is necessary for the chromatin remodelling associated with the repression of p16INK4A
CtBP has been implicated in PcG-mediated repression [19], [49], [50], [51] and may be directly involved in the chromatin remodelling of p16INK4A [22]. ChIP analysis within p16INK4A exon 1 showed that all the CtBP-mutant LCLs exhibited a significant reduction of the H3K27Me3 mark relative to a WT-EBV LCL. There was generally a corresponding increase in the level of the activation mark H3K4me3 relative to WT (Figure 10A). A detailed comparison of a WT-BAC LCL and a revertant LCL (revCtBP) with a double CtBP-mutant LCL (E3CtBP) across the p16INK4A locus was also performed. The double CtBPM mutant profile closely resembled those of LCL 3CHT cells grown without HT and therefore lacking a functional EBNA3C. In contrast the WT and revertant profiles – as would be expected – resembled LCL 3CHT cultured in the presence of HT (compare Figures 10B and C with Figures 5D and E). We conclude that the interaction of both EBNA3A and EBNA3C with CtBP is important for EBV-mediated chromatin remodelling and repression of p16INK4A during B cell transformation. This may explain why these protein:protein interactions are particularly important for the efficient outgrowth and establishment of LCLs.
Fig. 10. The interaction of EBNA3A and EBNA3C with CtBP is necessary for the chromatin remodelling associated with the repression of p16INK4A. 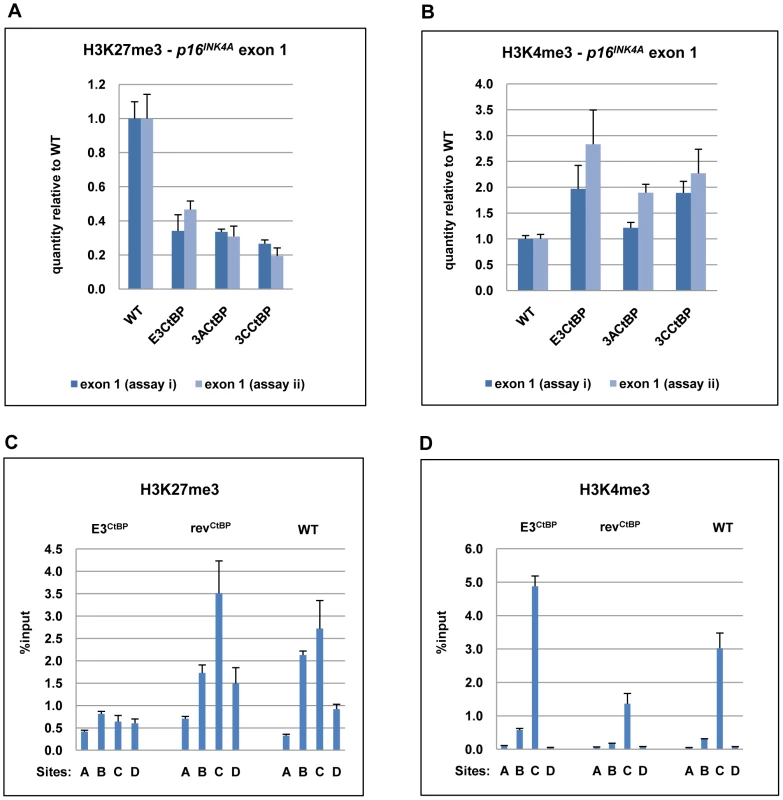
(A) & (B) ChIP analysis of H3K27me3 and H3K4me3 in p16INK4A exon 1 (site C) performed in duplicate (assays i & ii) on an E3CtBP LCL, a 3ACtBP and a 3CCtBP. Similar analysis of a WT-EBV LCL is shown for comparison. (C) & (D) ChIP anaysis of H3K27me3 and H4Kme3 distribution across the p16INK4A-ARF locus in an E3CtBP LCL. Similar assays performed on revertant (revCtBP) and WT-EBV LCLs are shown for comparison. The sites correspond to those shown in Figure 5A. Although the E3CtBP LCL examined here appears to have an active p16INK4A locus and relatively high levels of p16INK4A protein, the cells are clearly capable of cell division – leading us to suspect that some other element of the Rb-axis might be compromised. An examination of these cells, and other CtBP-mutant cells, showed that proliferation was probably not completely inhibited because in both E3CtBP and 3ACtBP LCLs expression of Rb is often reduced (see for example Figure S4). Since there will be considerable selection pressure during the establishment of an LCL, this is probably the result of unidentified compensatory lesions occurring in the Rb-axis early after infection and the outgrowth of the more robustly proliferating clones.
Discussion
Here we show that EBNA3A, EBNA3C and CtBP are all involved in the epigenetic repression of p16INK4A expression that is necessary for the proliferation of EBV-transformed B cells.
The CDKN2A locus in the human genome encodes two tumour suppressor proteins – p16INK4A and p14ARF – that are encoded by transcripts consisting of a unique first exon and shared second and third exons (Figure 5A). However, the proteins are structurally unrelated since p14ARF utilizes an alternative reading frame to that used for p16INK4A [23]. The p16INK4A promoter region is epigenetically regulated by the interplay of repressive modifications, eg H3K27me3, and activating trimethylation of lysine 4 at histone H3 (H3K4me3) in human stem cells, primary fibroblasts and cancer cells [44], [45], [52]. In stem cells H3K27me3 and H3K4me3 modifications co-localize at the p16INK4A promoter and form what is known as a ‘bivalent’ chromatin domain. Such domains are thought to repress or silence genes while keeping them poised for activation [47], [53]. This epigenetic repression of the CDKN2A locus is mediated by polycomb group proteins (PcG) which are thought to silence or repress target genes through the formation of higher order chromatin structures (eg chromatin looping) and by ubiquitinylating H2A at K119 – a histone modification that interferes with the binding of RNA Polymerase II and transcriptional elongation [39], [54]. The H3K27me3 demethylase JMJD3 contributes to plasticity of the p16INK4A locus, and functions in a switch from the repressive chromatin in response to oncogenic stress [45], [46]
The data presented here have shown that compromising EBNA3C function initiates chromatin remodelling that resets the epigenetic status of p16INK4A locus. In the presence of active EBNA3C, the p16INK4A locus displays repressive chromatin and the locus is closed for transcription, but when EBNA3C is inactivated the chromatin is remodelled into an active state that supports transcription. These changes in epigenetic modifications of the p16INK4A locus might be a consequence of EBNA3C acting on the p16INK4A locus either directly or indirectly. Results of experiments performed with EBNA3A-KO LCLs and CtBP-binding mutants of both EBNA3A and 3C are all consistent with EBNA3A cooperating with EBNA3C, and both interacting with CtBP in the regulation of these epigenetic chromatin modifications of the p16INK4A locus.
Potential mechanisms
Although at least one virus – Paramecium bursaria Chlorella virus 1 – encodes its own histone methyltransferase that catalyses the methylation of histone H3K27 and represses a multitude of host genes [55], this is unlikely to be the case of EBNA3C or EBNA3A. Even though the crystal structure of neither is available and their secondary structure is difficult to predict [5], sequence homology studies fail to identify a potential methyltransferase domain in either EBNA3A or EBNA3C.
The consensus of opinion is that the regulation of p16INK4A is primarily under the control of members of the polycomb group of proteins (PcG). As already indicated, these multi-component repressor complexes generate histone modifications – including H3K27me3 – that are characteristic of silent chromatin. These marks are heritable and may affect the whole CDKN2A locus [23]. In Drosophila, repression by PcG complexes spreads from polycomb response elements (PREs) located within a genomic regulatory locus, but the equivalent of PREs have not been identified in mammalian cells. The mechanism of targeting PcGs specifically to the p16INK4A locus is unknown and, more generally, how the DNA-binding specificity of PcG complexes is achieved remains a key question in mammalian biology. The most likely candidates for PcG recruiting factors are sequence-specific transcription factors or long non-coding RNAs (nc-RNAs) [40], so a major challenge for the immediate future is to establish whether EBNAs 3A and 3C together deregulate or interact with sequence specific transcription factors or nc-RNAs that normally regulate p16INK4A and/or BIM.
No consistent changes in the expression of PcG proteins or the H3K27me3-specific demethylase JMJD3 have been identified in LCLs expressing inactivated or mutated EBNAs 3A or 3C ([8]; our unpublished observations). Furthermore, although it has been shown that LMP1 is a negative regulator of p16INK4A in fibroblasts and epithelial cells [56], [57], LMP1 expression was not reduced in any of the mutant LCLs studied here or reported elsewhere (Figures S1 and S3; [8], [35]. We therefore do not think LMP1 signalling plays a significant role in the repression of p16INK4A in B cells.
Consequences of epigenetic repression in lymphomagenesis
Epigenetic changes are by definition heritable, but not always irreversible. Certain loci exhibit a high degree of plasticity and are poised for rapid activation. Examples include families of developmental genes such as those in the HOX locus and tumour suppressor genes that become active when aberrant pro-proliferative signals are detected [23], [40]. Our data suggest that the repression of p16INK4A locus by EBNA3C (presumably cooperating with EBNA3A which is constitutively expressed) in the LCL 3CHT system is reversible. H3K27me3 is known to cause the local formation of heterochromatin that is labile or readily reversible, but this type of histone methylation can also facilitate methylation of DNA of the same region, particularly in the development of cancer. DNA methylation represents a more stable modification and can ‘fix’ the repression of the locus. It has been shown that promoter regions bound by PcG which remain largely unmethylated on CpG dinucleotides in normal tissues, serve during tumorigenesis as a map to direct DNA methylation. CpG islands methylated de novo in cancer have often been previously marked by the presence of PcG proteins and H3K27me3 [58], [59], [60], [61], [62], [63]. We have shown previously that this is the case for BIM in EBV-positive lymphomas [38]. It is therefore also likely that during EBV-mediated lymphomagenesis p16INK4A is repressed as a result of EBNA3A/C-induced histone modifications that then act as a focus for DNA methyltransferases to initiate the more stable cancer-associated CpG methylation. This is certainly consistent with the limited data on EBV-positive BL, that suggest the p16INK4A locus is nearly always silenced by DNA methylation of exon 1 in tumour-derived cell lines and in many BL biopsies [64], [65].
Limitations of the conditional EBNA3C-HT system
EBNA3C is absolutely essential for the transformation of primary B cells by EBV, therefore the successful establishment of LCL 3CHT reassures us that EBNA3C modified by a C-terminal fusion retains most of the functions required for transformation. However, there appear to be at least two limitations of the EBNA3C-HT fusion system. Firstly, even in the presence of HT the repression of p16INK4A is not complete. The amount of p16INK4A is higher than in WT-EBV LCL and is almost equivalent to the accumulation seen in EBNA3A-KO and CtBP-mutant LCLs (Figures 7A and 9C). The elevated levels of p16INK4A in LCL 3CHT, even in the presence of HT might indicate that EBNA3C activation is compromised in a subpopulation of cells and these cells exit from the cell cycle. Alternatively, and perhaps more likely, fusion of EBNA3C with the large estrogen receptor domain might partially inhibit EBNA3C function in all cells in the population, even in the presence of HT.
A second limitation to the utility of these cells for studying EBNA3C function is the considerable delay between removing or re-adding HT from/to the culture medium and the change in levels of p16INK4A. The first signs of p16INK4A increase are seen about 10 days after removing HT from the medium and the first signs of p16INK4A repression are apparent no sooner than two days after re-addition of HT. This may be caused by the EBNA3C-HT fusion not being quite equivalent to WT EBNA3C (as discussed above) or could indicate that p16INK4A is not a direct target of EBNA3C. A similar phenomenon occurs during the induction of p16INK4A by activated RAS. It takes approximately 5 days to fully activate p16INK4A by RAS. Since such an extended timeframe is not compatible with the signalling dynamics of the RAF-MEK-ERK cascade, it has been suggested that mutant RAS leads to the accumulation of intracellular stresses and subsequent activation of p38 [23]. It is possible that EBV latent gene expression minus EBNA3C or EBNA3A sets up some sort of dis-equilibrium that results in intracellular stress, but preliminary data suggest that this is not mediated by p38 activation (data not shown). Alternatively, it has been suggested that the slow kinetics of p16INK4A activation might reflect the need to displace either repressive histone or repressive PcG-mediated DNA methylation [23], [60], [66] and this could be the case in B cells latently infected with EBV.
The role of CtBP
The role of CtBP in EBNA3A/3C-mediated repression of p16INK4A is intriguing but not yet understood. CtBP was discovered because of its binding to adenoviral oncoprotein E1A. Binding of E1A to CtBP antagonizes the function of CtBP, and E1A mutants unable to bind CtBP show enhanced efficiency of transformation [67]. In contrast, EBNA3A and EBNA3C mutants unable to bind CtBP were less effective in transforming and immortalizing primary rat embryo fibroblasts in cooperation with Ha-RAS [16], [17]. Furthermore we showed that Marek's disease virus, a herpesvirus that induces T cell lymphoma in chickens, requires its nuclear oncoprotein MEQ to bind CtBP for tumorigenesis [68]. Consistent with the role of CtBP in rodent cell transformation by EBNA3A and EBNA3C, LCLs produced by immortalization with CtBP-binding mutant viruses grow out much more slowly than WT-EBV LCLs and fail to effectively repress p16INK4A. Taken together the data suggest that binding of EBNA3A and EBNA3C to CtBP augments transformation efficiency and LCL outgrowth by aiding the establishment or maintenance of the H3K27me3 mark on p16INK4A exon 1. CtBP-containing complexes have been previously linked to chromatin remodelling including demethylation of H3K4, and CtBP has been shown to recruit PcG silencers to certain genes in mammalian cells [19], [50]. Although both EBNA3A and EBNA3C can be immunoprecipitated from LCLs with CtBP [16], [17] and under standard conditions EBNA3A and EBNA3C can be reciprocally co-precipitated (our unpublished data) we have been unable to ChIP EBNA3A/3C-CtBP complexes on either the p16INK4A or BIM promoters (data not shown). We do not know whether this is because of technical limitations of the reagents that are available or whether these promoters are not actually direct targets of the complexes. It may be necessary to develop new regents to address this issue. Although we do not yet understand this requirement for CtBP-binding, the data are consistent with recent reports that the C-terminus of EBNA3C – and specifically the PLDLS CtBP-binding site – is necessary to completely rescue proliferation in Akata-derived E3C-HT LCLs cultured without HT [69], [70].
Selection for the loss of Rb
Serendipitously an LCL 3CHT cell line with no detectable Rb protein was produced. In this cell line, p16INK4A was repressed in the presence of functional EBNA3C and de-repressed after EBNA3C inactivation. We therefore assume that regulation of p16INK4A by EBNA3C in LCL 3CHT is an Rb-independent phenomenon. Repression of p16INK4A in the Rb-negative LCL 3CHT line is unlikely to be functionally relevant, because the main target of p16INK4A is absent. Since EBNA3C repressed p16INK4A even when no additional proliferation advantage was to be gained, this implies that repression of p16INK4A is a specific consequence of EBNA3C working in collaboration with EBNA3A and CtBP. Decreased expression of Rb protein was also seen in E3CtBP and 3ACtBP LCLs, (Figure S4) and decreased Rb mRNA has been reported in EBNA3A-KO LCLs [8]. Since the absence of Rb will confer a common proliferative advantage, it is probable that elevated p16INK4A expression in these various cell lines creates a strong selection pressure for the loss, or reduced expression of Rb during transformation.
In summary, we have described a novel mechanism used by EBV to overcome stress-induced growth arrest by preventing the induction of p16INK4A and, by a similar mechanism, enhance cell survival by preventing the induction of pro-apoptotic BIM [37], [38]. To our knowledge this ability to epigenetically inactivate a crucial cell cycle inhibitor and a potent inducer of cell death makes EBV unique among the known DNA ‘tumour viruses’ [71]. Understanding the precise molecular details of EBV-mediated chromatin remodelling of p16INK4A and the role that CtBP plays in this process should cast new light on the nature of viral oncogenesis and the mysteries of polycomb-mediated gene repression.
Supporting Information
Zdroje
1. BornkammGW
HammerschmidtW
2001 Molecular virology of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci 356 437 459
2. Thorley-LawsonDA
GrossA
2004 Persistence of the Epstein-Barr Virus and the Origins of Associated Lymphomas. The New England journal of medicine 350 1328 1337
3. RoughanJE
Thorley-LawsonDA
2009 The intersection of Epstein-Barr virus with the germinal center. J Virol 83 3968 3976
4. CrawfordDH
2001 Biology and disease associations of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci 356 461 473
5. TouitouR
O'NionsJ
HeaneyJ
AlldayMJ
2005 Epstein-Barr virus EBNA3 proteins bind to the C8/alpha7 subunit of the 20S proteasome and are degraded by 20S proteasomes in vitro, but are very stable in latently infected B cells. J Gen Virol 86 1269 1277
6. TomkinsonB
RobertsonE
KieffE
1993 Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol 67 2014 2025
7. TomkinsonB
KieffE
1992 Use of second-site homologous recombination to demonstrate that Epstein-Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J Virol 66 2893 2903
8. HertleML
PoppC
PetermannS
MaierS
KremmerE
2009 Differential gene expression patterns of EBV infected EBNA-3A positive and negative human B lymphocytes. PLoS Pathogens 5 e1000506
9. Le RouxA
KerdilesB
WallsD
DedieuJF
PerricaudetM
1994 The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology 205 596 602
10. WaltzerL
PerricaudetM
SergeantA
ManetE
1996 Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J kappa-EBNA2-activated transcription by inhibiting the binding of RBP-J kappa to DNA. J Virol 70 5909 5915
11. RadkovSA
TouitouR
BrehmA
RoweM
WestM
1999 Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol 73 5688 5697
12. CludtsI
FarrellPJ
1998 Multiple functions within the Epstein-Barr virus EBNA-3A protein. J Virol 72 1862 1869
13. BainM
WatsonRJ
FarrellPJ
AlldayMJ
1996 Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J Virol 70 2481 2489
14. BourillotPY
WaltzerL
SergeantA
ManetE
1998 Transcriptional repression by the Epstein-Barr virus EBNA3A protein tethered to DNA does not require RBP-Jkappa. J Gen Virol 79 (Pt2) 363 370
15. KnightJS
LanK
SubramanianC
RobertsonES
2003 Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J Virol 77 4261 4272
16. TouitouR
HickabottomM
ParkerG
CrookT
AlldayMJ
2001 Physical and Functional Interactions between the Corepressor CtBP and the Epstein-Barr Virus Nuclear Antigen EBNA3C. J Virol 75 7749 7755
17. HickabottomM
ParkerGA
FreemontP
CrookT
AlldayMJ
2002 Two nonconsensus sites in the Epstein-Barr virus oncoprotein EBNA3A cooperate to bind the co-repressor carboxyl-terminal-binding protein (CtBP). J Biol Chem 277 47197 47204
18. ChinnaduraiG
2007 Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol 39 1593 1607
19. ShiY
SawadaJ
SuiG
Affar elB
WhetstineJR
2003 Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422 735 738
20. ShiY
LanF
MatsonC
MulliganP
WhetstineJR
2004 Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119 941 953
21. DahiyaA
WongS
GonzaloS
GavinM
DeanDC
2001 Linking the Rb and polycomb pathways. Mol Cell 8 557 569
22. MrozEA
BairdAH
MichaudWA
RoccoJW
2008 COOH-terminal binding protein regulates expression of the p16INK4A tumor suppressor and senescence in primary human cells. Cancer Res 68 6049 6053
23. GilJ
PetersG
2006 Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7 667 677
24. ParkerGA
CrookT
BainM
SaraEA
FarrellPJ
1996 Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 13 2541 2549
25. SerranoM
LinAW
McCurrachME
BeachD
LoweSW
1997 Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88 593 602
26. ParkerGA
TouitouR
AlldayMJ
2000 Epstein-Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene 19 700 709
27. ChoudhuriT
VermaSC
LanK
MurakamiM
RobertsonES
2007 The ATM/ATR signaling effector Chk2 is targeted by Epstein-Barr virus nuclear antigen 3C to release the G2/M cell cycle block. J Virol 81 6718 6730
28. GruhneB
SompallaeR
MasucciMG
2009 Three Epstein-Barr virus latency proteins independently promote genomic instability by inducing DNA damage, inhibiting DNA repair and inactivating cell cycle checkpoints. Oncogene 28 3997 4008
29. KnightJS
RobertsonES
2004 Epstein-Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J Virol 78 1981 1991
30. KnightJS
SharmaN
RobertsonES
2005 Epstein–Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci U S A 102 18562 18566
31. KnightJS
SharmaN
RobertsonES
2005 SCFSkp2 complex targeted by Epstein-Barr virus essential nuclear antigen. Mol Cell Biol 25 1749 1763
32. BajajBG
MurakamiM
CaiQ
VermaSC
LanK
2008 Epstein-Barr virus nuclear antigen 3C interacts with and enhances the stability of the c-Myc oncoprotein. J Virol 82 4082 4090
33. SahaA
MurakamiM
KumarP
BajajB
SimsK
2009 Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J Virol 83 4652 4669
34. YiF
SahaA
MurakamiM
KumarP
KnightJS
2009 Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology 388 236 247
35. MaruoS
WuY
IshikawaS
KandaT
IwakiriD
2006 Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci U S A 103 19500 19505
36. MaruoS
JohannsenE
IllanesD
CooperA
KieffE
2003 Epstein-Barr Virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J Virol 77 10437 10447
37. AndertonE
YeeJ
SmithP
CrookT
WhiteRE
2008 Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene 27 421 433
38. PaschosK
SmithP
AndertonE
MiddeldorpJM
WhiteRE
2009 Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathogens 5 e1000492
39. SimonJA
KingstonRE
2009 Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10 697
40. BrackenAP
HelinK
2009 Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9 773 784
41. DelecluseHJ
HilsendegenT
PichD
ZeidlerR
HammerschmidtW
1998 Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95 8245 8250
42. WhiteRE
CalderwoodMA
WhitehouseA
2003 Generation and precise modification of a herpesvirus saimiri bacterial artificial chromosome demonstrates that the terminal repeats are required for both virus production and episomal persistence. J Gen Virol 84 3393 3403
43. NeuhierlB
FeederleR
HammerschmidtW
DelecluseHJ
2002 Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc Natl Acad Sci U S A 99 15036 15041
44. KiaSK
GorskiMM
GiannakopoulosS
VerrijzerCP
2008 SWI/SNF Mediates Polycomb Eviction and Epigenetic Reprogramming of the INK4b-ARF-INK4a Locus. Mol Cell Biol 28 3457 3464
45. BarradasM
AndertonE
AcostaJC
LiS
BanitoA
2009 Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev 23 1177 1182
46. AggerK
CloosPA
RudkjaerL
WilliamsK
AndersenG
2009 The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene - and stress-induced senescence. Genes Dev 23 1171 1176
47. BernsteinBE
MikkelsenTS
XieX
KamalM
HuebertDJ
2006 A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125 315 326
48. LukasJ
ParryD
AagaardL
MannDJ
BartkovaJ
1995 Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 375 503 506
49. SewaltRG
GunsterMJ
van der VlagJ
SatijnDPE
OtteAP
1999 C-Terminal Binding Protein Is a Transcriptional Repressor That Interacts with a Specific Class of Vertebrate Polycomb Proteins. Mol Cell Biol 19 777 787
50. AtchisonL
GhiasA
WilkinsonF
BoniniN
AtchisonML
2003 Transcription factor YY1 functions as a PcG protein in vivo. EMBO Journal 22 1347 1358
51. SrinivasanL
AtchisonML
2004 YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev 18 2596 2601
52. LiH
ColladoM
VillasanteA
StratiK
OrtegaS
2009 The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460 1136 1139
53. KrivtsovAV
ArmstrongSA
2007 MLL translocations, histone modifications and leukaemia stem-cell development. Nature Rev Cancer 7 823 833
54. TiwariVK
McGarveyKM
LicchesiJD
OhmJE
HermanJG
2008 PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biology 6 2911 2927
55. MujtabaS
ManzurKL
GurnonJR
KangM
Van EttenJL
2008 Epigenetic transcriptional repression of cellular genes by a viral SET protein. Nat Cell Biol 10 1114 1122
56. YangX
HeZ
XinB
CaoL
2000 LMP1 of Epstein-Barr virus suppresses cellular senescence associated with the inhibition of p16INK4a expression. Oncogene 19 2002 2013
57. OhtaniN
BrennanP
GaubatzS
SanijE
HertzogP
2003 Epstein-Barr virus LMP1 blocks p16INK4a-RB pathway by promoting nuclear export of E2F4/5. J Cell Biol 162 173 183
58. KeshetI
SchlesingerY
FarkashS
RandE
HechtM
2006 Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 38 149 153
59. SchlesingerY
StraussmanR
KeshetI
FarkashS
HechtM
2007 Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39 232 236
60. VireE
BrennerC
DeplusR
BlanchonL
FragaM
2006 The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439 871 874
61. OhmJE
McGarveyKM
YuX
ChengL
SchuebelKE
2007 A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39 237 242
62. WidschwendterM
FieglH
EgleD
Mueller-HolznerE
SpizzoG
2007 Epigenetic stem cell signature in cancer. Nat Genet 39 157 158
63. CedarH
BergmanY
2009 Linking DNA methylation and histone modification: patterns and paradigms. Nature Rev Genet 10 295 304
64. KlangbyU
OkanI
MagnussonKP
WendlandM
LindP
1998 p16/INK4a and p15/INK4b gene methylation and absence of p16/INK4a mRNA and protein expression in Burkitt's lymphoma. Blood 91 1680 1687
65. LindstromMS
WimanKG
2002 Role of genetic and epigenetic changes in Burkitt lymphoma. Semin Cancer Biol 12 381 387
66. Hernandez-MunozI
TaghaviP
KuijlC
NeefjesJ
van LohuizenM
2005 Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol 25 11047 11058
67. SchaeperU
BoydJM
VermaS
UhlmannE
SubramanianT
1995 Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci U S A 92 10467 10471
68. BrownAC
BaigentSJ
SmithLP
ChattooJP
PetherbridgeLJ
2006 Interaction of MEQ protein and C-terminal-binding protein is critical for induction of lymphomas by Marek's disease virus. Proc Natl Acad Sci U S A 103 1687 1692
69. LeeS
SakakibaraS
MaruoS
ZhaoB
CalderwoodMA
2009 Epstein-Barr virus nuclear protein 3C domains necessary for lymphoblastoid cell growth: interaction with RBP-Jkappa regulates TCL1. J Virol 83 12368 12377
70. MaruoS
WuY
ItoT
KandaT
KieffED
2009 Epstein-Barr virus nuclear protein EBNA3C residues critical for maintaining lymphoblastoid cell growth. Proc Natl Acad Sci U S A 106 4419 4424
71. HowleyPM
LivingstonDM
2009 Small DNA tumor viruses: large contributors to biomedical sciences. Virology 384 256 259
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin NeutralizationČlánek Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane KinasesČlánek Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 6- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Cognitive Dysfunction Is Sustained after Rescue Therapy in Experimental Cerebral Malaria, and Is Reduced by Additive Antioxidant Therapy
- DNA Watermarking of Infectious Agents: Progress and Prospects
- Self-Protection against Gliotoxin—A Component of the Gliotoxin Biosynthetic Cluster, GliT, Completely Protects Against Exogenous Gliotoxin
- Modifies the Tsetse Salivary Composition, Altering the Fly Feeding Behavior That Favors Parasite Transmission
- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- The Enteropathogenic Effector EspF Targets and Disrupts the Nucleolus by a Process Regulated by Mitochondrial Dysfunction
- Epithelial p38α Controls Immune Cell Recruitment in the Colonic Mucosa
- Coexpression of PD-1, 2B4, CD160 and KLRG1 on Exhausted HCV-Specific CD8+ T Cells Is Linked to Antigen Recognition and T Cell Differentiation
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- EspA Acts as a Critical Mediator of ESX1-Dependent Virulence in by Affecting Bacterial Cell Wall Integrity
- An RNA Element at the 5′-End of the Poliovirus Genome Functions as a General Promoter for RNA Synthesis
- Tetherin Restricts Productive HIV-1 Cell-to-Cell Transmission
- Epstein-Barr Virus-Encoded LMP2A Induces an Epithelial–Mesenchymal Transition and Increases the Number of Side Population Stem-like Cancer Cells in Nasopharyngeal Carcinoma
- Protein Kinase A Dependent Phosphorylation of Apical Membrane Antigen 1 Plays an Important Role in Erythrocyte Invasion by the Malaria Parasite
- Requirement for Ergosterol in V-ATPase Function Underlies Antifungal Activity of Azole Drugs
- The SOCS-Box of HIV-1 Vif Interacts with ElonginBC by Induced-Folding to Recruit Its Cul5-Containing Ubiquitin Ligase Complex
- A Crucial Role for Infected-Cell/Antibody Immune Complexes in the Enhancement of Endogenous Antiviral Immunity by Short Passive Immunotherapy
- Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator
- Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step
- Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane Kinases
- Epigenetic Repression of by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP
- Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
- Role of PKR and Type I IFNs in Viral Control during Primary and Secondary Infection
- A Kinome RNAi Screen Identified AMPK as Promoting Poxvirus Entry through the Control of Actin Dynamics
- Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity
- NleG Type 3 Effectors from Enterohaemorrhagic Are U-Box E3 Ubiquitin Ligases
- Human Cytomegalovirus UL29/28 Protein Interacts with Components of the NuRD Complex Which Promote Accumulation of Immediate-Early RNA
- Complement Receptor 1 Is a Sialic Acid-Independent Erythrocyte Receptor of
- A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs
- Immunotoxin Complementation of HAART to Deplete Persisting HIV-Infected Cell Reservoirs
- Protein Expression Redirects Vesicular Stomatitis Virus RNA Synthesis to Cytoplasmic Inclusions
- Entry and Fusion of Emerging Paramyxoviruses
- Paramyxovirus Entry and Targeted Vectors for Cancer Therapy
- Suppressing Glucose Transporter Gene Expression in Schistosomes Impairs Parasite Feeding and Decreases Survival in the Mammalian Host
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Fungal Cell Gigantism during Mammalian Infection
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
- Rotavirus Structural Proteins and dsRNA Are Required for the Human Primary Plasmacytoid Dendritic Cell IFNα Response
- The Epigenetic Landscape of Latent Kaposi Sarcoma-Associated Herpesvirus Genomes
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



