-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
“Persisters”: Survival at the Cellular Level
article has not abstract
Published in the journal: “Persisters”: Survival at the Cellular Level. PLoS Pathog 7(7): e32767. doi:10.1371/journal.ppat.1002121
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1002121Summary
article has not abstract
Introduction
Rather than being slowly eroded and destroyed, countless numbers of varied forms of life adapt to the diverse aspects of an ever changing environment. However, the amount of variation is maintained at a practical optimum, as too much variation would make the population ill-adapted in a stable environment, while too little variation would render it unable to adapt to environmental stresses. This principle is perhaps well exemplified by a phenomenon described for microbial cells termed “persistence” where in the face of antibiotics bacterial populations avoid extinction by harboring a subpopulation of drug-insensitive dormant cells. Although this phenomenon poses a major obstacle for the treatment of infectious diseases, persistence has been underappreciated for some time as a mechanism for bacteria to evade antibiotics. But the mechanisms of bacterial persistence are becoming clearer and so are ways to combat them. This article highlights the phenomenon of survival and persistence in cells as diverse as microbial and human and summarizes the recent advances that have taken us one step closer to understanding what persistence is all about.
Microbial “Persister” Cells
In the early 1940s, it was only appropriate for Joseph Bigger to refer to a small subpopulation of bacterial cells that survived killing by penicillin, as “persisters” [1]. These small numbers of cells were then proposed to be dormant and nongrowing phenotypic variants of the general cell population [2], [3]. This theory of “persisters” has since been established in various bacterial populations. However, more recently the existence of a small cell subpopulation that can remain viable at high concentrations of an antifungal agent was described for the fungal pathogen Candida albicans [4]–[6]. Therefore, it has become clear that the ability to avoid killing is a key characteristic common to all microbial persisters that are not mutants, but rather phenotypic variants that can survive antimicrobial treatment. However, unlike drug resistance, drug tolerance appears to be a transient and reversible physiological state in a small subpopulation of genetically identical cells [7], [8]. When the antimicrobial agent is removed, these persisting microbial cells not only resume growth, but their progeny is sensitive to the antimicrobial agent (Figure 1) [7], [8].
Fig. 1. Progression of persister cell development and enhanced drug tolerance. 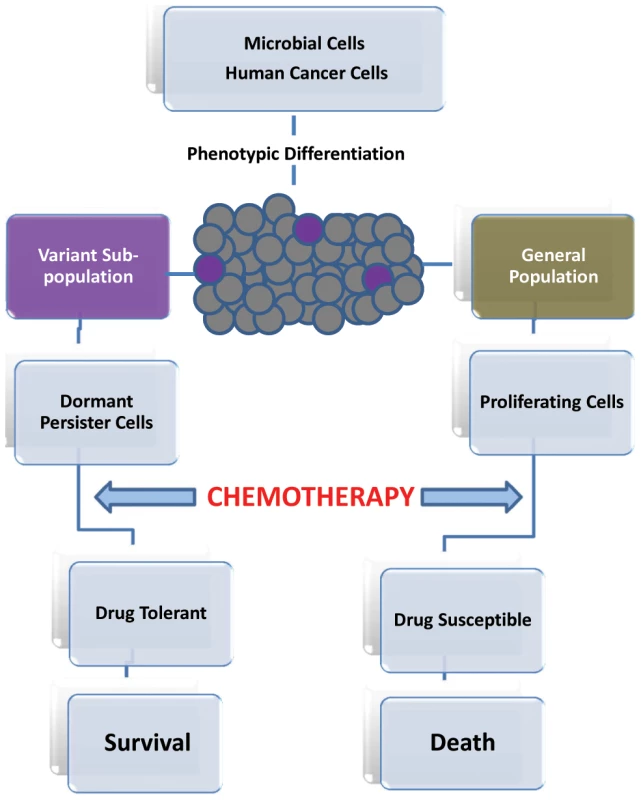
Formation of Drug Tolerant Persisters
Persisters have been described to arise spontaneously on the basis of random stochastic events [2], [3], [9]. Stochasticity can be advantageous in providing flexibility for the cells to adapt to fluctuating environments and sudden stresses and, therefore, stochastic mechanisms are thought to lead to the emergence of phenotypically distinct subgroups within isogenic cell populations [9]. However, defined inducible mechanisms have been recently identified to play a role in persister cell formation [10].
Quiescence and Biofilms
Recent findings from studies examining the rate of bacterial persister-cell formation over time showed that the highest frequency (∼1%) occurs in the nongrowing stationary phase [5]. Interestingly, when the culture was kept in early exponential phase by repeated regrowth, persister cells disappeared, indicating that persisters are preformed rather than produced in response to stress [5]. Furthermore, gene expression studies demonstrated the downregulation of transcription of genes involved in energy production and nonessential functions concomitant with upregulation in genes associated with cellular arrest [2]. These findings are consistent with the description of persister cells as being dormant, a transient state of existence that would impede the ability of drugs to corrupt their target molecules in the microbial cell [7]. In that respect, entry into quiescence is advantageous; however, it is more beneficial for a cell to be a dividing cell than a dormant cell. Therefore, it is more likely that the optimal cell strategy is not to enter into persistence, suggesting that the persister state is an altruistic behavior to ensure the continuation of the population.
The simplest strategy to trigger entry into dormancy would be to overproduce proteins or toxins that inhibit cellular processes and growth [3], [11]. One such identified factor is the high persistence gene hipA, which encodes a toxin (HipA) that inhibits translation in Escherichia coli. This toxin was identified to be implicated in forming persisters because its overexpression increased the frequency of persistence by 10,000-fold and resulted in drug tolerance [12]. HipA is normally neutralized by HipB, a transcription repressor that counteracts HipA by attaching to it preventing it from shutting down protein production and, therefore, hipBA has been categorized as a toxin/antitoxin (TA) module [11], [12]. Recently, through extensive studies including structural analyses, Schumacher et al. [12] identified HipA to be a protein kinase that phosphorylates the translation factor EF-Tu. These findings demonstrating that HipA bound the EF-Tu peptide supported the hypothesis that HipA mediates persistence by phosphorylating one or more target proteins. On the basis of these new insights into the mechanisms by which HipA mediates persistence, the authors suggested that inhibitors that specifically target the substrate-binding sites of HipA may prove effective against persistence.
Perhaps, the best defined mechanism by which persister bacterial cells arise comes from the fact that DNA damage induces one or more components of the protective SOS stress response, a signaling pathway that upregulates DNA repair functions [13]. Specifically, in E. coli, exposure to a DNA-damaging antibiotic triggered the gene encoding a small membrane-acting peptide TisB, which decreases proton motive force and ATP levels suggesting that TisB protein may induce dormancy by shutting down cell metabolism [14]. These speculations were substantiated by the findings demonstrating that deletion of the tisB gene resulted in decreased frequency of persisters tolerant to DNA-damaging antibiotic [14]. Interestingly, although overexpression of tisB resulted in cell death, minor overproduction of the peptide induced persister formation suggesting that induction of TisB is involved in the production of multidrug tolerant cells, in turn identifying tisB as a persister gene [14]. Combined, these observations are in accordance with the perception that dormancy and SOS response represent strategies of cell survival.
Persister cells are highly enriched in biofilms, which are complex and highly organized surface-attached communities of microbes embedded in a polymeric matrix [8], [15]. Biofilms form on abiotic surfaces and host tissue and are responsible for infections of indwelling medical devices. It is estimated that over 65% of all infections are biofilm-associated, which tend to be difficult to eradicate because of enhanced resistance to antimicrobials [2], [16]. The biofilm environment is advantageous to the microbial populations, however, when nutrients become limited metabolic dormancy becomes the viable option [8]. In a clinical setting, when most cells in a biofilm are readily killed by low concentrations of antibiotics, the small metabolically dormant phenotypes progress to become tolerant persister cells. By virtue of their dormancy, this subpopulation of cells confer benefits to the general cell population and are in turn responsible for the high tolerance of bacterial biofilms to antimicrobial agents [3], [7].
Persisters are formed by all bacterial species studied and are present at 0.1%–1% in the biofilms of Pseudomonas aeruginosa, E. coli, and Staphylococcus aureus [17]. Recently, the existence of a small cell subpopulation that can remain viable at high concentrations of antifungal agent has been described in fungal biofilms, specifically for the human pathogen C. albicans [4], [6]. Clinically, candidal infections may resolve upon antifungal therapy but often remain recalcitrant to treatment. In a recent study, on the basis of tolerance to high doses of an antifungal agent, invariably all C. albicans isolates recovered from nonresolving infections appeared to be high-persister variants. Similar to bacteria, C. albicans forms adherent biofilms, which are essentially recalcitrant to antifungals [16], [18]. The mechanism of C. albicans biofilm antifungal resistance remains largely unknown; however, biofilms have been described to exhibit a biphasic killing pattern in response to antimicrobial agents, indicating that a subpopulation of highly tolerant cells existed [6], [7]. Interestingly, reinoculation of surviving cells produced a new biofilm with a new subpopulation of persisters. These observations suggest that C. albicans persisters, analogous to their bacterial counterparts, are not mutants but phenotypic variants and that attachment to a surface is what initiates dormancy that leads to the formation of persisters [5], [6].
Cancer Persister Cells
Similar to the obstacle in treatment of patients that develop resistance to antimicrobials, acquisition of resistance to anticancer drugs is a major problem in cancer therapy. Most treatments, even ones that work, fail over time because tumor cells become resistant. Different mechanisms of resistance have been described for cancer cells such as modification of drug target and active extrusion of drugs by efflux pumps and, therefore, it was largely assumed that random gene changes confer resistance to drugs [19]. However, this does not explain an increasingly observed phenomenon in cancer chemotherapy; “retreatment response” [20], [21]. In this model, it is proposed that once a small number of cells that survive exposure to drugs that killed the majority of the cells are given a “drug holiday,” they eventually regain their sensitivity to the drug [22]. These observations indicate that acquired resistance to cancer drugs may not necessarily result from stable genetic mutations but may also involve a reversible “drug-tolerant” state [22], [23].
In a recent study by Sharma et al. [22], drug-sensitive cells were treated with antitumor drugs at concentrations exceeding 100 times the established IC50 values. Following three rounds of 72-h treatments, the authors consistently detected a small subpopulation of reversibly “drug-tolerant” cells demonstrating >100-fold reduced drug sensitivity. Further analyses demonstrated that these cells maintained viability via engagement of insulin-like growth factor 1 (IGF1) receptor signaling and an altered chromatin state and treatment with IGF1 receptor inhibitors or chromatin-modifying agents selectively ablated the drug-tolerant subpopulation
Cancer-initiating cells are proposed as a potential resistant subpopulation because of their ability to escape the effect of drug treatment by becoming quiescent [24]. This transient drug-tolerant state could provide a mechanism that allows a small subpopulation of tumor cells to withstand an initial destructive attack of drug to enable their survival, until more permanent resistance mechanisms can be established [22]. Intriguingly, this transient ability to endure anticancer drugs was recently reported to be highly reminiscent of the drug-tolerant microbial “persister” subpopulations [22], [25]. In that sense, it is plausible to regard slow-growing cancerous cells in highly proliferating tumors to be analogous to microbial persister cells in biofilm (Figure 1).
Conclusion and Future Directions
Whether microbial or human in nature, it appears that cells have evolved analogous redundant strategies where the function of survival is assigned to a small dormant subpopulation of cells within a more rapidly proliferating population. With most of the currently available chemotherapeutic agents targeting exponentially growing cells, our therapeutic arsenal is ineffective in eradicating these dormant persister cells. Coupled with the increasing emergence of drug resistance and failure of therapies despite our medical advances, it has become critical to develop novel classes of drugs. The prospect that persisters are responsible for the persistence of chronic infections and, more gravely, recalcitrance of disseminating cancers have identified these culprit cells as viable targets for new therapies. However, such discoveries rely heavily on the depth of our understanding the nature of these intriguing cells, which would provide us with fundamental insights into the mechanisms involved in the development of drug tolerance. Inopportunely, their transient nature and low abundance, has impeded experimental advancements to elucidate the dynamics of the formation of these specialized cells that neither die nor grow. Nevertheless, the recent unearthing of an inherent tactical approach shared by diverse cellular insurgents will undoubtedly herald a new era of research into the new field of “persisters.”
Zdroje
1. BiggerJ 1944 Treatment of staphylococcal infections with penicillin. Lancet 244 497 500
2. LewisK 2007 Persister cells, dormancy and infectious disease. Nat Rev Micro 5 48 56
3. LewisK 2010 Persister cells. Ann Rev Microbiol 64 357 372
4. Al-DhaheriRSDouglasJ 2008 Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob Agents Chemother 52 1884 1887
5. KerenIKaldaluNSpoeringAWangYLewisK 2004 Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230 13 18
6. LaFleurMDKumamotoCALewisK 2006 Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother 50 3839 3846
7. LewisK 2008 Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322 107 31
8. RobertsMEStewartPS 2005 Modeling protection from antimicrobial agents in biofilms through the formation of persister cells. Microbiol 151 75 80
9. KaernM 2005 Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6 451 464
10. JayaramanAWoodTK 2008 Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Ann Rev Biomed Eng 10 145 167
11. Vazquez-LaslopNLeeHNeyfakhAA 2006 Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol 188 3494 3497
12. SchumacherMAPiroKMXuWHansenSLewisK 2009 Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323 396 401
13. MillerCThomsenLEGaggeroCMosseriRIngmerH 2004 SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305 1629 1631
14. DörrTVulićMLewisK 2010 Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8 e1000317 doi:10.1371/journal.pbio.1000317
15. LewisK 2001 Riddle of biofilm resistance. Antimicrob Agents Chemother 45 999 1007
16. KumamotoCA 2002 Candida biofilms. Curr Opin Microbiol 5 608 611
17. KerenIShahDSpoeringAKaldaluNLewisK 2004 Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186 872 880
18. Jabra-RizkMAFalklerWAJrMeillerTF 2004 Fungal biofilms and drug resistance. Emerg Infect Dis 10 14 19
19. RedmondKMWilsonTRJohnstonPGLongleyDB 2008 Resistance mechanisms to cancer chemotherapy. Front Biosci 13 5138 5154
20. KurataTTamuraKKanedaHNogamiTUejimaH 2004 Effect of re-treatment with gefitinib after acquisition of resistance. Ann Oncol 15 173 174
21. YanoSNakatakiEOhtsukaSInayamaMTomimotoH 2005 Retreatment of lung adenocarcinoma patients with gefitinib who had experienced favorable results from their initial treatment with this selective epidermal growth factor receptor inhibitor: a report of three cases. Oncol Res 15 107 111
22. SharmaSVLeeDYLiBQuinlanMPTakahashiF 2010 A Chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141 69 80
23. GlasspoolRMTeodoridisJBrownR 2006 Epigenetics as a mechanism driving polygenic clinical drug resistance. Br J Cancer 94 1087 1092
24. FrankNSchattonTFrankMH 2010 The therapeutic promise of the cancer stem cell concept. J Clin Invest 20 41 50
25. BalabanNQMerrinJChaitRKowalikLLeiblerS 2004 Bacterial persistence as a phenotypic switch. Science 305 1622 1625
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek A Typhimurium-Typhi Genomic Chimera: A Model to Study Vi Polysaccharide Capsule Function In VivoČlánek SUMO Pathway Dependent Recruitment of Cellular Repressors to Herpes Simplex Virus Type 1 GenomesČlánek A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate ReceptorsČlánek Dynamic Evolution of Pathogenicity Revealed by Sequencing and Comparative Genomics of 19 IsolatesČlánek Widespread Endogenization of Genome Sequences of Non-Retroviral RNA Viruses into Plant GenomesČlánek The Cost of Virulence: Retarded Growth of Typhimurium Cells Expressing Type III Secretion System 1Článek A Role for the Chemokine RANTES in Regulating CD8 T Cell Responses during Chronic Viral Infection
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 7- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- What Do We Really Know about How CD4 T Cells Control ?
- “Persisters”: Survival at the Cellular Level
- E6 and E7 from Beta Hpv38 Cooperate with Ultraviolet Light in the Development of Actinic Keratosis-Like Lesions and Squamous Cell Carcinoma in Mice
- Selection of Resistant Bacteria at Very Low Antibiotic Concentrations
- The Extracytoplasmic Domain of the Ser/Thr Kinase PknB Binds Specific Muropeptides and Is Required for PknB Localization
- CD39/Adenosine Pathway Is Involved in AIDS Progression
- Hypoxia and a Fungal Alcohol Dehydrogenase Influence the Pathogenesis of Invasive Pulmonary Aspergillosis
- One Is Enough: Effective Population Size Is Dose-Dependent for a Plant RNA Virus
- Effects of Interferon-α/β on HBV Replication Determined by Viral Load
- A Typhimurium-Typhi Genomic Chimera: A Model to Study Vi Polysaccharide Capsule Function In Vivo
- Dual Chaperone Role of the C-Terminal Propeptide in Folding and Oligomerization of the Pore-Forming Toxin Aerolysin
- Rotavirus Stimulates Release of Serotonin (5-HT) from Human Enterochromaffin Cells and Activates Brain Structures Involved in Nausea and Vomiting
- Dissociation of Infectivity from Seeding Ability in Prions with Alternate Docking Mechanism
- The Impact of Recombination on dN/dS within Recently Emerged Bacterial Clones
- The Regulation of Sulfur Metabolism in
- Illumination of Parainfluenza Virus Infection and Transmission in Living Animals Reveals a Tissue-Specific Dichotomy
- A Permeable Cuticle Is Associated with the Release of Reactive Oxygen Species and Induction of Innate Immunity
- A Concerted Action of Hepatitis C Virus P7 and Nonstructural Protein 2 Regulates Core Localization at the Endoplasmic Reticulum and Virus Assembly
- SUMO Pathway Dependent Recruitment of Cellular Repressors to Herpes Simplex Virus Type 1 Genomes
- Re-localization of Cellular Protein SRp20 during Poliovirus Infection: Bridging a Viral IRES to the Host Cell Translation Apparatus
- Divergent Effects of Human Cytomegalovirus and Herpes Simplex Virus-1 on Cellular Metabolism
- A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate Receptors
- Transformation of Natural Genetic Variation into Genomes
- EBV Latency Types Adopt Alternative Chromatin Conformations
- Global mRNA Degradation during Lytic Gammaherpesvirus Infection Contributes to Establishment of Viral Latency
- Dynamic Evolution of Pathogenicity Revealed by Sequencing and Comparative Genomics of 19 Isolates
- Microbial Virulence as an Emergent Property: Consequences and Opportunities
- Widespread Endogenization of Genome Sequences of Non-Retroviral RNA Viruses into Plant Genomes
- Structural Basis of Chemokine Sequestration by CrmD, a Poxvirus-Encoded Tumor Necrosis Factor Receptor
- Cross-Species Transmission of a Novel Adenovirus Associated with a Fulminant Pneumonia Outbreak in a New World Monkey Colony
- An Interaction between KSHV ORF57 and UIF Provides mRNA-Adaptor Redundancy in Herpesvirus Intronless mRNA Export
- Elevated 17β-Estradiol Protects Females from Influenza A Virus Pathogenesis by Suppressing Inflammatory Responses
- The Role of IL-15 Deficiency in the Pathogenesis of Virus-Induced Asthma Exacerbations
- Fluorescence Lifetime Imaging Unravels Metabolism and Its Crosstalk with the Host Cell
- Programmed Death (PD)-1-Deficient Mice Are Extremely Sensitive to Murine Hepatitis Virus Strain-3 (MHV-3) Infection
- Hemoglobin Promotes Nasal Colonization
- Crystallography of a Lewis-Binding Norovirus, Elucidation of Strain-Specificity to the Polymorphic Human Histo-Blood Group Antigens
- The Cost of Virulence: Retarded Growth of Typhimurium Cells Expressing Type III Secretion System 1
- A Genome-Wide Approach to Discovery of Small RNAs Involved in Regulation of Virulence in
- Requires Glycerol for Maximum Fitness During The Tick Phase of the Enzootic Cycle
- C Metabolic Flux Analysis Identifies an Unusual Route for Pyruvate Dissimilation in Mycobacteria which Requires Isocitrate Lyase and Carbon Dioxide Fixation
- A Role for the Chemokine RANTES in Regulating CD8 T Cell Responses during Chronic Viral Infection
- Glycosaminoglycans and Sialylated Glycans Sequentially Facilitate Merkel Cell Polyomavirus Infectious Entry
- Regulation of Stomatal Tropism and Infection by Light in : Evidence for Coordinated Host/Pathogen Responses to Photoperiod?
- Multiple Translocation of the Effector Gene among Chromosomes of the Rice Blast Fungus and Related Species
- Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens
- Unique Cell Adhesion and Invasion Properties of O:3, the Most Frequent Cause of Human Yersiniosis
- C-Terminal Region of EBNA-2 Determines the Superior Transforming Ability of Type 1 Epstein-Barr Virus by Enhanced Gene Regulation of LMP-1 and CXCR7
- Novel Chikungunya Vaccine Candidate with an IRES-Based Attenuation and Host Range Alteration Mechanism
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Requires Glycerol for Maximum Fitness During The Tick Phase of the Enzootic Cycle
- Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens
- The Role of IL-15 Deficiency in the Pathogenesis of Virus-Induced Asthma Exacerbations
- “Persisters”: Survival at the Cellular Level
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



