-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Indifferent, Affectionate, or Deceitful: Lifestyles and Secretomes of Fungi
article has not abstract
Published in the journal: Indifferent, Affectionate, or Deceitful: Lifestyles and Secretomes of Fungi. PLoS Pathog 8(3): e32767. doi:10.1371/journal.ppat.1002515
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1002515Summary
article has not abstract
Introduction
Fungi occupy a myriad of niches. They can be free-living (indifferent) as saprophytes recycling nutrients in the natural environment and/or have a range of relationships (affectionate and deceitful) with insect, animal, or plant hosts. Interactions with plants can be a continuum and range from obligate biotrophy where fungi cannot be cultured outside living hosts to necrotrophy where fungi kill and live on released nutrients. Biotrophic fungi need to avoid or suppress defence responses. They include symbionts, which confer a benefit to the host, and pathogens, which can cause devastating diseases such as stem rust, which threatens production of wheat worldwide [1]. Mycorrhizae colonise roots of >80% of land plants and are symbiotic, increasing nitrogen and phosphorus uptake from the soil, while feeding on sugars from the host photosynthate. Secreted proteins are on the front line of host–fungal interactions, and a particular class, effectors, is a hot topic. Here, we examine a range of fungi and consider their complement of secreted proteins (secretome) and roles of effectors in fungal lifestyles.
For Some Fungi, There Is a Relationship between Lifestyle and Secretome Size
The Fungal Secretome Database (FSD) [2] predicts secreted proteins using SignalP, which identifies secretion signal peptides within proteins. We applied SignalP to several recently completed genomes and examined whether the ratio of secretome size to total gene number reflects the predominant lifestyles (Figure 1). The total gene number ranges from 4,000 to 20,000, and the proportion of secreted proteins from 4% to 14%. Fungi with biphasic lifestyles have a large proportion of secreted proteins. These include the hemibiotrophic rice blast fungus Magnaporthe oryzae, the corn smut fungus Ustilago maydis, and Piriformospora indica, which colonizes dead roots saprophytically and live roots as a biotrophic symbiont [3]. Its biphasic lifestyle is reflected in its transcriptome; many genes induced during growth on living roots are similar to those of the symbiont Laccaria bicolor, whereas genes induced during saprophytic growth are similar to those of the saprophyte Coprinus cinereus. The insect pathogens Metarhizium anisopliae and Metarhizium acridum also have large secretomes [4]. Many saprophytes have similarly sized secretomes as necrotrophs, as noted previously [2], which may reflect the fact that necrotrophs often have an extended saprophytic phase as part of their life cycle. Animal pathogens have fewer genes than saprophytes or plant-interacting fungi do, and a lower proportion of predicted secreted proteins.
Fig. 1. Relationship between predicted secreted protein number and total gene content of fungi. 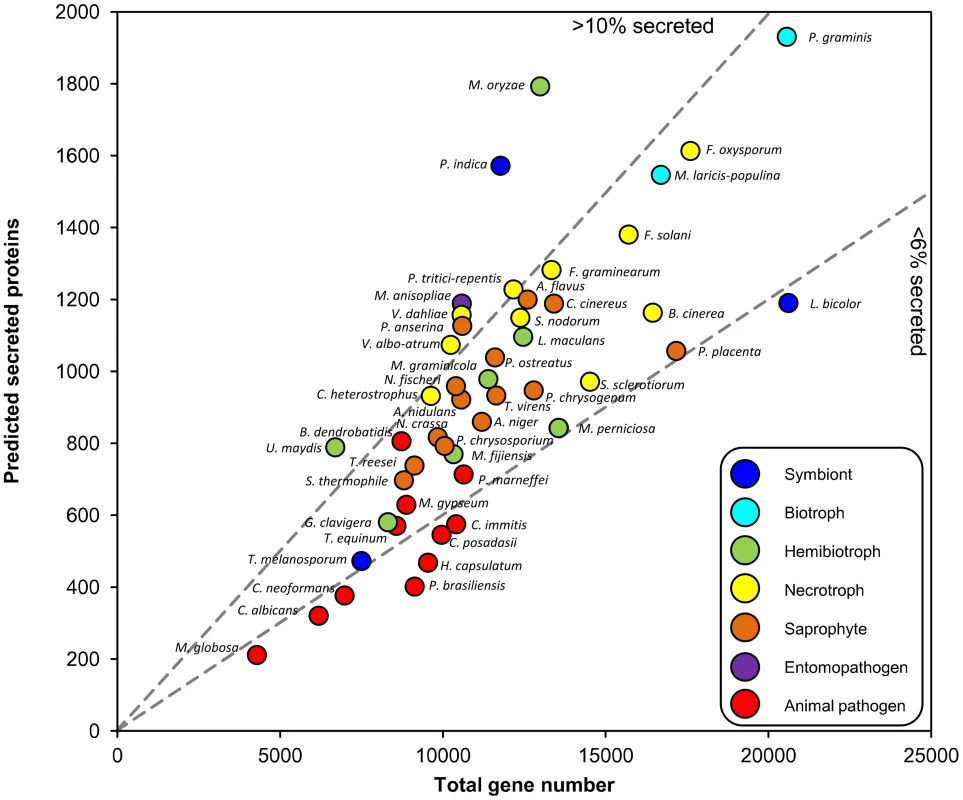
Data are from [3], or by applying SignalP to genome releases (indicated by *). Dashed lines discriminate between fungi with high (>10) or low (<6) % secreted proteins. Animal pathogens: Batrachochytrium dendrobatidis, Candida albicans, Coccidioides immitis, C. posadasii, Cryptococcus neoformans, Histoplasma capsulatum, Malassezia globosa, Microsporum gypseum, Paracoccidioides brasiliensis, Penicillium marneffei, Trichophyton equinum. Hemibiotrophs: Grosmania clavigera*, Leptosphaeria maculans* [14], Magnaporthe oryzae*, Mycosphaerella fijiensis, M. graminicola, Moniliophthora perniciosa, Ustilago maydis. Entomopathogen: Metarhizium anisopliae* [4]. Necrotrophs: Botrytis cinerea, Cochliobolus heterostrophus, Fusarium graminearum, F. oxysporum, F. solani, Pyrenophora tritici-repentis, Stagonospora nodorum*, Sclerotinia sclerotiorum, Verticillium albo-atrum, V. dahliae. Biotrophs: Melampsora laricis-populina* [15], Puccinia graminis f.sp. tritici. Saprophytes: Aspergillus flavus, A. nidulans, A. niger, Coprinus cinereus, Neurospora crassa, Neosartorya fischeri, Podospora anserina, Penicillium chrysogenum, Phanerochaete chrysosporium, Pleurotus ostreatus, Postia placenta, Sporotrichum thermophile, Trichoderma reesei, T. virens. Symbionts: Laccaria bicolor, Piriformospora indica* [3], Tuber melanosporum* [16]. The Fungal Secretome Includes Carbohydrate-Degrading Enzymes and Effectors
Within the secretome there are different classes of proteins. Below we discuss two of them. Carbohydrate-degrading enzymes encoded by multigene families are secreted copiously by saprophytes to feed from complex molecules in the environment. Insect and plant pathogens that have to breach the host surface to gain entry also have large numbers, as do necrotrophs, which feed from tissue after they kill it. In contrast, biotrophs have few such families, as previously noted for mycorrhizae [5]; consequently, there is minimal release of pathogen-associated molecular patterns (PAMPs) from the plant cell wall. Accordingly, basal innate immunity, a mechanism common to animals and plants, is not triggered. Another class is effectors, which facilitate infection and/or induce defence responses [6]. Effectors are generally <300 amino acids, cysteine-rich, and lack transmembrane domains. They are often species-specific, polymorphic between isolates, and highly transcribed in planta. They can be avirulence proteins, which are complementary to plant resistance proteins in “gene for gene” interactions, host-specific toxins, or interfere with innate immunity by dampening or strengthening defence responses. Many proteins with effector-like properties have unknown functions.
Effectors of Biotrophic Fungi Modulate Plant Responses
There are few genome sequences of biotrophic fungi, and as gene knockouts are difficult to carry out in such fungi, few effectors have been functionally analysed. Three that elicit plant responses have been characterized recently, two from symbionts and one from a pathogen. MISS7 from L. bicolor is the most highly upregulated gene during symbiosis with poplar. The encoded protein, which is crucial for successful symbiosis, moves to the nucleus where it modulates expression of poplar genes, including ones that alter root architecture [7]. A highly expressed effector from the mycorrhiza Glomus intraradices, SP7, moves to the plant nucleus where it interacts with pathogenesis-related transcription factor ERF19 and helps establish symbiosis, probably by dampening host defence [8]. A third effector that modulates plant responses is a chorismate mutase from U. maydis. This enzyme dimerises with a chorismate mutase from corn, and suppresses the production of salicylic acid, a key molecule in plant defence signaling [9].
Effector Genes Can Move within and between Kingdoms
Effector genes are often located within repeat-rich regions, near telomeres, or even on lineage-specific chromosomes; as a result, these genes are readily lost, gained, or mutated [10]. The gene encoding the host-specific toxin ToxA of Stagonospora nodorum is located near a transposase and is present in another wheat pathogen, Pyrenophora tritici-repentis [11]. Transfer of ToxA from S. nodorum to P. tritici-repentis probably occurred by horizontal gene transfer (HGT) on co-infected wheat leaves about 50 years ago. Genes not only move between fungal genera, but also across kingdoms. Phylogenetic analyses have revealed transfer of effector genes from fungi to oomycetes with 8% of the secretome of the sudden oak death oomycete Phytophthora ramorum proposed to be derived by HGT from fungi [12].
Animal Pathogens May Not Need Many Effectors
Generally, there are few barriers for a fungus to overcome when infecting animals. In many cases a fungus needs to be small enough to enter the host, survive at 37°C (in the case of mammalian pathogens), and evade immune responses [13]. Animal pathogens are often soil saprophytes that infect opportunistically, but, unlike most plant pathogens, some mammalian pathogens are not highly adapted to their hosts. Perhaps because of this, many animal fungal pathogens, in contrast to most plant fungal pathogens, do not display host specificity. An obvious exception to this is the insect pathogenic genus Metarhizium, which displays species specificity [4]. Furthermore, there is generally no intimate cellular relationship between fungus and animal as exists for obligate biotrophs or symbionts, which are enveloped in fungal and plant plasmalemmas. Thus, effectors may not be necessary to mediate deceit in all fungal–animal interactions, but are likely to be crucial for interactions that deceitful and affectionate fungi have with plants.
Zdroje
1. SinghRPHodsonDPHuerta-EspinoJJinYBhavaniS 2011 The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Ann Rev Phytopathol 49 465 481
2. ChoiJParkJKimDJungKKangS 2010 Fungal Secretome Database: integrated platform for annotation of fungal secretomes. BMC Genomics 11 105
3. ZuccaroALahrmannUGüldenerULangenGPfiffiS 2011 Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog 7 e1002290 doi:10.1371/journal.ppat.1002290
4. GaoQJinKYingS-HZhangYXiaoG 2011 Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet 7 e1001264 doi:10.1371/journal.pgen.1001264
5. PlettJMMartinF 2011 Blurred boundaries: lifestyle lessons from ectomycorrhizal fungal genomes. Trends Genet 27 14 22
6. De WitPJMehrabiRVan den BurgHAStergiopoulosI 2009 Fungal effector proteins: past, present and future. Mol Plant Pathol 10 735 747
7. PlettJMKemppainenMKaleSDKohlerALegueV 2011 A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol 21 1197 1203
8. KloppholzSKuhnHRequenaN 2011 A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21 1204 1209
9. DjameiASchipperKRabeFGhoshAVinconV 2011 Metabolic priming by a secreted fungal effector. Nature 478 395 398
10. RepMKistlerHC 2010 The genomic organization of plant pathogenicity in Fusarium species. Curr Opin Plant Biol 13 420 426
11. FriesenTLStukenbrockEHLiuZMeinhardtSLingH 2006 Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet 38 953 956
12. RichardsTASoanesDMJonesMDMVasievaOLeonardG 2011 Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Nat Acad Sci U S A 108 15258 15263
13. HowlettBJ 2011 Fungal pathogenesis in plants and animals; similarities and differences. ChoffnesERRelmanDA Fungal diseases: an emerging challenge to human, animal and plant health Washington (D.C.) National Academies Press 264 273
14. RouxelTGrandaubertJHaneJKHoedeCvan de WouwAP 2011 Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nature Commun 2 202
15. DuplessisSCuomoCALinY-CAertsATisserantE 2011 Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A 108 9166 9171
16. MartinFKohlerAMuratCBalestriniRCoutinhoPM 2010 Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464 1033 1038
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2012 Číslo 3- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- An Entomopathogenic Nematode by Any Other Name
- Induced Release of a Plant-Defense Volatile ‘Deceptively’ Attracts Insect Vectors to Plants Infected with a Bacterial Pathogen
- A Foot in the Door for Dermatophyte Research
- New Insights into spp.: A Potential Link with Irritable Bowel Syndrome
- Indifferent, Affectionate, or Deceitful: Lifestyles and Secretomes of Fungi
- Mutation and Selection of Prions
- Sleeping with the Enemy: How Intracellular Pathogens Cope with a Macrophage Lifestyle
- Taste for Blood: Hemoglobin as a Nutrient Source for Pathogens
- Direct Recognition of by the NK Cell Natural Cytotoxicity Receptor NKp46 Aggravates Periodontal Disease
- A20 () Deficiency in Myeloid Cells Protects against Influenza A Virus Infection
- Anthrax Lethal Factor Cleavage of Nlrp1 Is Required for Activation of the Inflammasome
- Differential Function of Lip Residues in the Mechanism and Biology of an Anthrax Hemophore
- PK-sensitive PrP Is Infectious and Shares Basic Structural Features with PK-resistant PrP
- A Peptidoglycan Fragment Triggers β-lactam Resistance in
- Capsule Type of Determines Growth Phenotype
- Additive Function of MARTX and VvhA Cytolysins Promotes Rapid Growth and Epithelial Tissue Necrosis During Intestinal Infection
- A Novel Mouse Model of Egg-Induced Immunopathology
- Redundant Notch1 and Notch2 Signaling Is Necessary for IFNγ Secretion by T Helper 1 Cells During Infection with
- The Novel Transporter Dur31 Is a Multi-Stage Pathogenicity Factor
- Short ORF-Dependent Ribosome Shunting Operates in an RNA Picorna-Like Virus and a DNA Pararetrovirus that Cause Rice Tungro Disease
- Structural Insights into a Unique Effector LidA Recognizing Both GDP and GTP Bound Rab1 in Their Active State
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- A Foot in the Door for Dermatophyte Research
- Structural Insights into a Unique Effector LidA Recognizing Both GDP and GTP Bound Rab1 in Their Active State
- An Entomopathogenic Nematode by Any Other Name
- New Insights into spp.: A Potential Link with Irritable Bowel Syndrome
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



