-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Yeasts: How Many Species Infect Humans and Animals?
article has not abstract
Published in the journal: Yeasts: How Many Species Infect Humans and Animals?. PLoS Pathog 10(2): e32767. doi:10.1371/journal.ppat.1003892
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003892Summary
article has not abstract
The Main Significance of Malassezia Yeasts and Their Position in the Tree of Life
Malassezia species are lipophilic yeasts that are members of the normal mycobiota of the skin and mucosal sites of a variety of homeothermic animals. They are also among the few basidiomycetous fungi, such as some Cryptococcus spp., Rhodotorula spp., and Trichosporon spp., that can produce disease in man and animals. However, in contrast with these other species, which are quite often involved in disseminated infections in immunosuppressed patients, Malassezia yeasts are associated mainly with certain skin diseases [1].
This special lipophilic group of yeasts is unique among the fungi. Phylogenetically, they form a well-defined cluster of skin-living yeasts, surrounded by plant pathogens and phylloplane-inhabiting fungi (e.g., Ustilago, Tilletiopsis). However, the taxonomic position of the genus Malassezia in the classes of the phylum Basidiomycota is not yet totally well defined. Moreover, the sexual form of these yeasts is still unknown. Recently, a region corresponding to the mating type locus (MAT) has been identified for these yeasts, and it has been suggested that if there is an extant sexual cycle for some of these yeasts that it is more likely to be bipolar, with just two mating types, rather than tetrapolar, with many mating types [2].
In the last higher-level fungal phylogenetic classification revision [3], the monophyletic genus Malassezia was the only genus included in the order Malasseziales, which has an uncertain taxonomic position in the subphylum Ustilagomycotina (e.g., smut fungi). Very recently, the class Malasseziomycetes has been proposed to accommodate these fungi (2013, provided from an anonymous reviewer; unreferenced). They are taxonomically distant to the orders which include the other commented pathogenic basidiomycetous yeasts of the genera Cryptococcus (Filobasidiales) and Trichosporon in Agaricomycotina (e.g., mushrooms) and of the polyphyletic genus Rhodotorula (Sporidiobolales and Cystobasidiales) in Pucciniomycotina (e.g., rust fungi).
Spectrum of Malassezia Species That Infect Humans and Animals
At present, the genus Malassezia includes 14 species ([4]; Table 1), all of which infect or colonize humans or animals. However, until the late 1980s, this genus remained limited to only to two species; one of these, M. furfur (sensu lato), was considered a heterogeneous group of lipid-dependent yeasts living on human skin and requiring long-chain fatty acids to grow, while the lipophilic but non–lipid-dependent species M. pachydermatis was restricted to animal skin. The latter is the only species in the genus that does not require lipid supplementation for development in culture medium. M. sympodialis, a lipid-dependent species isolated from human skin, was the third species accepted in the genus, a century after the description of M. furfur [5]. Later, the genus Malassezia was revised on the basis of morphological, physiological, and rRNA gene sequencing studies, and four new lipid-dependent species were described [6]. At the same time, different studies [7]–[9] confirmed that the skin of healthy animals could also be colonized by lipid-dependent species, in addition to the non–lipid-dependent species M. pachydermatis. These lipid-dependent species are the major component of the lipophilic mycobiota occurring on the skin of horses and various ruminants [10]. Some of these yeasts isolated from animals were described subsequently as new species, such as M. nana [11], M. equina, or M. caprae [12]. Nowadays, Malassezia yeasts have been isolated from almost all domestic animals, different wild animals held in captivity, and also from wildlife [1]. Despite this, the occurrence of Malassezia yeasts on the skin of most animals remains unknown. The observed host specificity of some of these species made it possible to anticipate an increase in the number of new species in this genus, particularly if other animal species, mainly wild species, were studied.
Tab. 1. Current described Malassezia species, authorities, year of the description, and their main hostsa [24]. ![Current described <i>Malassezia</i> species, authorities, year of the description, and their main hosts<em class="ref">a</em> <em class="ref">[24]</em>.](https://www.prelekara.sk/media/cache/resolve/media_object_image_small/media/image/9c9a2d755777d1119f3dd907b460d522.png)
Cited only those species confirmed by rDNA sequencing analysis. Malassezia Yeasts and Disease
The pathogenic role of Malassezia yeasts in skin diseases has always been a matter of controversy. Commensal Malassezia yeasts are clearly implicated in human skin diseases without the presence of inflammation but with heavy fungal load, such as pityriasis versicolor. They are also associated with other skin disorders with characteristic inflammation, such as seborrheic dermatitis, atopic dermatitis, folliculitis, and psoriasis, where their role in the pathogenesis is less clear and, in some cases, speculative [13]. Emerging evidence demonstrates that the interaction of Malassezia yeasts with the skin is multifaceted and entails constituents of the fungal wall, enzymes, and metabolic products, as well as the cellular components of the epidermis. Some skin disorders can be exacerbated by the interactions between Malassezia yeasts and the host immune system [2].
Although M. globosa was initially reported to be the main species associated with pityriasis versicolor, subsequent studies have shown that the distribution of Malassezia species from healthy and diseased skin is equivalent, thus failing to substantiate the existence of a pathogenic species not only in pityriasis versicolor but also for the other Malassezia-associated diseases. M. globosa and M. restricta are the most commonly found species on healthy and diseased human skin [14]. However, other species such as M. sympodialis or M. furfur have been also associated with various human skin disorders [15].
On the other hand, mainly M. furfur and M. pachydermatis have been reported to be the cause of a low percentage of yeast systemic infections. However, fungemia produced by these yeasts may be underdiagnosed by modern automated blood systems for fungal detection if culture media with lipids are not included in the diagnostic protocol [16]. The majority of published case reports and miniepidemics have involved infants, children, and adults with profound immunosuppression, serious concurrent health problems, and the infusion of total parenteral nutrition with lipid supplementation through central vascular catheters. The main ingredients of this nutrition system (i.e., linoleic, oleic, and palmitic acids) are potent growth stimulants for Malassezia species [15].
Skin colonization by Malassezia species of healthy human neonates does not include M. pachydermatis, whereas the occurrence of other species such as M. sympodialis and M. globosa begins at birth and increases in the first weeks of life. [17]. This fact corroborates the animal origin of M. pachydermatis in human infections. Furthermore, it should be noted that zoonotic transfer of M. pachydermatis has been documented from dogs to neonates by healthcare workers who own dogs [18].
M. pachydermatis, the only species in the genus that does not require lipid supplementation for development in culture medium, is considered to be zoophilic, and is frequently found on wild and domestic carnivores. This species is usually associated with otitis externa and different kinds of dermatitis in domestic animals, especially in dogs (Figure 1). This species is more frequently isolated from dogs than cats and appears to be a relatively infrequent pathogen in other animals. This yeast seems to have an opportunistic nature, and it may become pathogenic with any detected alteration in the skin surface microclimate or in the host defense. In some canine breeds, hypersensitivity conditions such as flea allergy dermatitis, food hypersensitivity or atopy, and antimicrobial or corticosteroid therapy may be factors favoring proliferation of these yeasts. Lipid-dependent species seem to be found more frequently in cats than in dogs, but very little is known about their pathogenic role in animal skin [19].
Fig. 1. Gram stain of a smear (A) and culture (B) from an otic swab of a dog with otitis externa, showing numerous M. pachydermatis cells (A) and colonies (B). 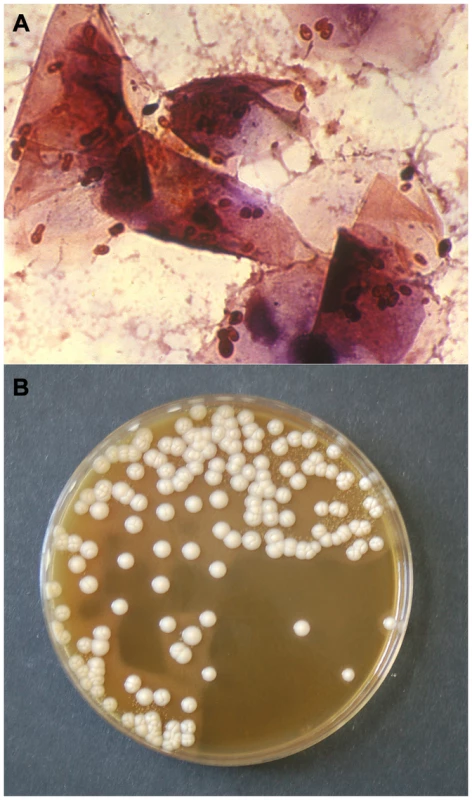
This species is a controversial pathogen that is now recognized as an important cause of dermatitis and otitis externa in dogs. Author: F. Javier Cabañes. Difficulties in Determining the Biodiversity and Significance of Malassezia Yeasts
The study of some Malassezia yeasts continues to be difficult, due mainly to their low viability and lack of suitable methods for their isolation and preservation. The majority of yeasts can be stored at temperatures between 4 and 12°C and subcultured at intervals of 6 to 8 months. However, Malassezia spp. do not fit this pattern. Freezing at −80°C is the only successful method to maintain viable all Malassezia spp., particularly M. globosa, M. restricta, and M. obtusa, which have been reported as difficult species to maintain in vitro [20].
In most surveys, these yeasts have been identified only on the basis of phenotypic characteristics without confirmation through molecular analysis. Difficulties remain in obtaining a high level of certainty in the identification of some lipid-dependent strains by means of physiological tests (e.g., Tween physiological tests) without molecular characterization. Although some Malassezia yeasts may be distinguished using these tests, sequencing of some genes (e.g., ITS-5.8S and D1D2 26S rRNA, β-tubulin) [4] or the use of new tools such as MALDI-TOF mass spectrometry [21] are necessary for a proper identification.
Recently, the spectrum of fungal species in the human skin has been explored using culture and culture-independent methods [22]. In this study, Malassezia yeasts predominated on most of the sampled body sites. Moreover, 11 of the 14 species (all of them, with the exception of M. caprae, M. cuniculi, and M. equina) (2013 letter from K. Findley to me; unreferenced) were directly identified by rRNA gene sequencing from different clinical samples of ten healthy volunteers. Other DNA sequences that may represent unidentified Malassezia spp. were also detected. Some species predominated in certain body sites (e.g., M. globosa on the back). Using culturing methods, apparently the most abundant Malassezia species on human skin (e.g., M. globosa, M. restricta, and M. sympodialis) were isolated in this study. These authors [22] used only Sabouraud glucose agar (SGA) with olive oil containing chloramphenicol and cycloheximide for recovering Malassezia species from different body sites.
The use of SGA overlaid with olive oil has been used frequently in the past, but only some Malassezia species grow well on this medium [23], [24]. For an exhaustive survey, the samples must be inoculated onto more complex culture media, such as modified Dixon agar (mDA) or Leeming and Notman agar (LNA), which facilitate the recovery of the more fastidious Malassezia species from the skin. These culture media include, among other ingredients, a mixture of fatty acids such as oleic acid, whole-fat cow milk, and some polyoxyethylene sorbitanesters (e.g., Tween 40, Tween 60). However, the exact nutritional requirements of Malassezia species in culture are yet to be fully determined, and this hinders the study of these yeasts. Moreover, it is difficult and expensive to obtain fatty acids of sufficient purity to fully establish the fatty-acid requirements of each Malassezia species [25].
A recent example of the difficulties inherent to the recovery of these fastidious yeasts from the skin is M. cuniculi. In the description of this species [26], only a few lipid - dependent Malassezia yeasts were recovered from two of the 11 rabbits investigated. They grew scarcely on LNA and no growth was obtained either on mDA or SGA. They were also not able to grow on glucose peptone agar supplemented with Tweens (20, 40, 60, and 80) and Cremophor EL as sole sources of lipids, which are used to phenotypically characterize these species [6], [27]. This inhibition of growth may be related to the toxic effects of these mixtures of fatty acids at higher concentrations. LNA contains, among other components, Tween 60 at a 10-fold lower concentration (0.05%) than that used in the Tween physiological tests [6].
In other recent surveys performed in soil nematodes [28], marine sponges [29], and coral colonies [30], Malassezia yeasts have been tentatively identified exclusively on the basis of some genotypic characteristics by culture-independent methods. However, although other habitats for Malassezia yeasts may exist, their significance and the real identity of these yeasts still remain unknown.
Zdroje
1. Sugita J, Boekhout T, Velegraki A, Guillot J, Hadina S, et al.. (2010) Epidemiology of Malassezia-related skin diseases. In: Boekhout T, Guého E, Mayser P, Velegraki A, editors. Malassezia and the skin. Berlin Heidelberg: Springer-Verlag. pp. 65–119.
2. SaundersCW, ScheyniusA, HeitmanJ (2012) Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLOS Pathog 8: e1002701 doi:10.1371/journal.ppat.1002701
3. HibbettDS, BinderM, BischoffJF, BlackwellM, CannonPF, et al. (2007) A higher level phylogenetic classification of the Fungi. Mycol Res 111 : 509–547.
4. CastelláG, CoutinhoSD, CabañesFJ (2013) Phylogenetic relationships of Malassezia species based on multilocus sequence analysis. Med Mycol. E-pub ahead of print doi:10.3109/13693786.2013.815372
5. SimmonsRB, GuéhoE (1990) A new species of Malassezia. Mycol Res 94 : 1146–1149.
6. GuéhoE, MidgleyG, GuillotJ (1996) The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek 69 : 337–355.
7. GuillotJ, ChermetteR, GuéhoE (1994) Prévalence du genre Malassezia chez les mammifères. J Mycol Méd 4 : 72–79.
8. BondR, HowellSA, HaywoodPJ, LloydDH (1997) Isolation of Malassezia sympodialis and Malassezia globosa from healthy pet cats. Vet Rec 141 : 200–201.
9. CrespoMJ, AbarcaML, CabañesFJ (1999) Isolation of Malassezia furfur from a cat. J Clin Microbiol 37 : 1573–1574.
10. CrespoMJ, AbarcaML, CabañesFJ (2002) Occurrence of Malassezia spp. in horses and domestic ruminants. Mycoses 45 : 333–337.
11. HiraiA, KanoR, MakimuraK, DuarteER, HamdanJS, et al. (2004) Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals. Int J Syst Evol Microbiol 54 : 623–627.
12. CabañesFJ, TheelenB, CastelláC, BoekhoutT (2007) Two new lipid dependent Malassezia species from domestic animals. FEMS Yeast Res 7 : 1064–1076.
13. Crespo Erchiga V, Hay RJ (2010) Pityriasis versicolor and other Malasssezia skin diseases. In: Boekhout T, Guého E, Mayser P, Velegraki A, editors. Malassezia and the skin. Berlin Heidelberg: Springer-Verlag. pp. 175–199.
14. GaitanisG, VelegrakiA, MayserP, BassukasID (2013) Skin diseases associated with Malassezia yeasts: facts and controversies. Clin Dermatol 31 : 455–463.
15. GaitanisG, MagiatisP, HantschkeM, BassukasID, VelegrakiA (2012) The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25 : 106–141.
16. ArendrupMC, BoekhoutT, AkovaM, MeisJF, CornelyOA, et al. (2013) ESCMID/ECMM joint clinical guideline for the diagnosis and management of rare invasive yeast infections. Clin Microb Rev 18 E-pub ahead of print. doi:10.1111/1469-0691.12360
17. BernierV, WeillFX, HirigoyenV, ElleauC, FeylerA, et al. (2002) Skin colonization by Malassezia species in neonates: a prospective study and relationship with neonatal cephalic pustulosis. Arch Dermatol 138 : 215–218.
18. ChangH, MillerH, WatkinsN, ArduinoM, AshfordD, et al. (1998) An epidemic of Malassezia pachydermatis in an intensive care nursery associated with colonization of health care workers' pet dogs. N Engl J Med 338 : 706–711.
19. Bond R, Guillot J, Cabañes FJ (2010) Malassezia yeasts in animal diseases. In: Boekhout T, Guého E, Mayser P, Velegraki A, editors. Malassezia and the skin. Berlin Heidelberg: Springer-Verlag. pp. 271–299.
20. CrespoMJ, AbarcaML, CabañesFJ (2000) Evaluation of different preservation and storage methods for Malassezia spp. J Clin Microbiol 38 : 3872–3875.
21. KoleckaA, KhayhanK, ArabatzisM, VelegrakiA, KostrzewaM, et al. (2013) Efficient identification of Malassezia yeasts by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Br J Dermatol E-pub ahead of print. doi:10.1111/bjd.12680
22. FindleyK, OhJ, YangJ, ConlanS, DemingC, et al. (2013) Topographic diversity of fungal and bacterial communities in human skin. Nature 498 : 367–370.
23. MidgleyG (2000) The lipophilic yeasts: state of the art and prospects. Med Mycol 38(Suppl. I): 9–16.
24. Guého E, Boekhout T, Begerow D (2010) Biodiversity, phylogeny and ultrastructure In: Boekhout T, Guého E, Mayser P, Velegraki A, editors. Malassezia and the skin. Berlin Heidelberg: Springer-Verlag. pp. 17–63.
25. BatraR, BoekhoutT, GuéhoE, CabañesFJ, DawsonTL, GuptaAK (2005) Malassezia Baillon, emerging clinical yeasts. FEMS Yeast Res 5 : 1101–1113.
26. CabañesFJ, VegaS, CastelláG (2011) Malassezia cuniculi sp. nov., a novel yeast species isolated from rabbit skin. Med Mycol 49 : 40–48.
27. MayserP, HazeP, PapavassilisC, PickelM, GruenderK, GuéhoE (1997) Differentiation of Malassezia species: selectivity of cremophor EL, castor oil and ricinoleic acid for M. furfur. Br J Dermatol 137 : 208–213.
28. RenkerC, AlpheiJ, BuscotF (2003) Soil nematodes associated with the mammal pathogenic fungal genus Malassezia (Basidiomycota: Ustilaginomycetes) in Central European forests. Biol Fertil Soils 37 : 70–72.
29. GaoZ, LiB, ZhengC, WangG (2008) Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl Environ Microbiol 74 : 6091–6101.
30. AmendAS, BarshisDJ, OliverTA (2012) Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME J 6 : 1291–1301.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic HelixČlánek Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron MobilizationČlánek AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for ImmunityČlánek Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden HamstersČlánek Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 2- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Viral Enhancer Mimicry of Host Innate-Immune Promoters
- The Epstein-Barr Virus-Encoded MicroRNA MiR-BART9 Promotes Tumor Metastasis by Targeting E-Cadherin in Nasopharyngeal Carcinoma
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- The Consequences of Reconfiguring the Ambisense S Genome Segment of Rift Valley Fever Virus on Viral Replication in Mammalian and Mosquito Cells and for Genome Packaging
- Substrate-Induced Unfolding of Protein Disulfide Isomerase Displaces the Cholera Toxin A1 Subunit from Its Holotoxin
- Male-Killing Induces Sex-Specific Cell Death via Host Apoptotic Pathway
- Highly Active Antiretroviral Therapies Are Effective against HIV-1 Cell-to-Cell Transmission
- The microRNAs in an Ancient Protist Repress the Variant-Specific Surface Protein Expression by Targeting the Entire Coding Sequence
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Type III Secretion Protein MxiI Is Recognized by Naip2 to Induce Nlrc4 Inflammasome Activation Independently of Pkcδ
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
- Induction of Type I Interferon Signaling Determines the Relative Pathogenicity of Strains
- Structure of the Membrane Anchor of Pestivirus Glycoprotein E, a Long Tilted Amphipathic Helix
- Foxp3 Regulatory T Cells Delay Expulsion of Intestinal Nematodes by Suppression of IL-9-Driven Mast Cell Activation in BALB/c but Not in C57BL/6 Mice
- Iron Acquisition in : The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization
- MicroRNA Editing Facilitates Immune Elimination of HCMV Infected Cells
- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Identification of Host-Targeted Small Molecules That Restrict Intracellular Growth
- A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection
- Engagement of NKG2D on Bystander Memory CD8 T Cells Promotes Increased Immunopathology following Infection
- Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress Expression
- CIB1 Synergizes with EphrinA2 to Regulate Kaposi's Sarcoma-Associated Herpesvirus Macropinocytic Entry in Human Microvascular Dermal Endothelial Cells
- A Gammaherpesvirus Bcl-2 Ortholog Blocks B Cell Receptor-Mediated Apoptosis and Promotes the Survival of Developing B Cells
- Metabolic Reprogramming during Purine Stress in the Protozoan Pathogen
- The Post-transcriptional Regulator / Activates T3SS by Stabilizing the 5′ UTR of , the Master Regulator of Genes, in
- Tailored Immune Responses: Novel Effector Helper T Cell Subsets in Protective Immunity
- AvrBsT Acetylates ACIP1, a Protein that Associates with Microtubules and Is Required for Immunity
- Epstein-Barr Virus Large Tegument Protein BPLF1 Contributes to Innate Immune Evasion through Interference with Toll-Like Receptor Signaling
- The Major Cellular Sterol Regulatory Pathway Is Required for Andes Virus Infection
- Insights into the Initiation of JC Virus DNA Replication Derived from the Crystal Structure of the T-Antigen Origin Binding Domain
- Domain Shuffling in a Sensor Protein Contributed to the Evolution of Insect Pathogenicity in Plant-Beneficial
- Lectin-Like Bacteriocins from spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor
- A Compositional Look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects
- Exploits Asparagine to Assimilate Nitrogen and Resist Acid Stress during Infection
- Interleukin-33 Increases Antibacterial Defense by Activation of Inducible Nitric Oxide Synthase in Skin
- Protective Vaccination against Papillomavirus-Induced Skin Tumors under Immunocompetent and Immunosuppressive Conditions: A Preclinical Study Using a Natural Outbred Animal Model
- Gem-Induced Cytoskeleton Remodeling Increases Cellular Migration of HTLV-1-Infected Cells, Formation of Infected-to-Target T-Cell Conjugates and Viral Transmission
- Viral MicroRNA Effects on Pathogenesis of Polyomavirus SV40 Infections in Syrian Golden Hamsters
- Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection
- Inflammatory Monocytes Orchestrate Innate Antifungal Immunity in the Lung
- Quantitative and Qualitative Deficits in Neonatal Lung-Migratory Dendritic Cells Impact the Generation of the CD8+ T Cell Response
- Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response
- The Master Regulator of the Cellular Stress Response (HSF1) Is Critical for Orthopoxvirus Infection
- Code-Assisted Discovery of TAL Effector Targets in Bacterial Leaf Streak of Rice Reveals Contrast with Bacterial Blight and a Novel Susceptibility Gene
- Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri ORF57 to the Mammalian Export Adaptor ALYREF
- The Type III Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including TepP, a Tyrosine-phosphorylated Protein Required for the Recruitment of CrkI-II to Nascent Inclusions and Innate Immune Signaling
- Yeasts: How Many Species Infect Humans and Animals?
- Clustering of Pattern Recognition Receptors for Fungal Detection
- Distinct Antiviral Responses in Pluripotent versus Differentiated Cells
- Igniting the Fire: Virulence Factors in the Pathogenesis of Sepsis
- Inactivation of the Host Lipin Gene Accelerates RNA Virus Replication through Viral Exploitation of the Expanded Endoplasmic Reticulum Membrane
- Inducible Deletion of CD28 Prior to Secondary Infection Impairs Worm Expulsion and Recall of Protective Memory CD4 T Cell Responses
- Clonal Expansion during Infection Dynamics Reveals the Effect of Antibiotic Intervention
- The Secreted Triose Phosphate Isomerase of Is Required to Sustain Microfilaria Production
- Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2
- ‘Death and Axes’: Unexpected Ca Entry Phenologs Predict New Anti-schistosomal Agents
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Reversible Silencing of Cytomegalovirus Genomes by Type I Interferon Governs Virus Latency
- Implication of PMLIV in Both Intrinsic and Innate Immunity
- Transmission-Blocking Antibodies against Mosquito C-Type Lectins for Dengue Prevention
- Lundep, a Sand Fly Salivary Endonuclease Increases Parasite Survival in Neutrophils and Inhibits XIIa Contact Activation in Human Plasma
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



